Abstract
The current diagnostic work‐up of inborn errors of metabolism (IEM) is rapidly moving toward integrative analytical approaches. We aimed to develop an innovative, targeted urine metabolomics (TUM) screening procedure to accelerate the diagnosis of patients with IEM. Urinary samples, spiked with three stable isotope‐labeled internal standards, were analyzed for 258 diagnostic metabolites with an ultra‐high performance liquid chromatography‐quadrupole time‐of‐flight mass spectrometry (UHPLC‐QTOF‐MS) configuration run in positive and negative ESI modes. The software automatically annotated peaks, corrected for peak overloading, and reported peak quality and shifting. Robustness and reproducibility were satisfactory for most metabolites. Z‐scores were calculated against four age‐group‐matched control cohorts. Disease phenotypes were scored based on database metabolite matching. Graphical reports comprised a needle plot, annotating abnormal metabolites, and a heatmap showing the prioritized disease phenotypes. In the clinical validation, we analyzed samples of 289 patients covering 78 OMIM phenotypes from 12 of the 15 society for the study of inborn errors of metabolism (SSIEM) disease groups. The disease groups include disorders in the metabolism of amino acids, fatty acids, ketones, purines and pyrimidines, carbohydrates, porphyrias, neurotransmitters, vitamins, cofactors, and creatine. The reporting tool easily and correctly diagnosed most samples. Even subtle aberrant metabolite patterns as seen in mild multiple acyl‐CoA dehydrogenase deficiency (GAII) and maple syrup urine disease (MSUD) were correctly called without difficulty. Others, like creatine transporter deficiency, are illustrative of IEM that remain difficult to diagnose. We present TUM as a powerful diagnostic screening tool that merges most urinary diagnostic assays expediting the diagnostics for patients suspected of an IEM.
Keywords: diagnostics, inborn error of metabolism, mass spectrometry, urine metabolomics, targeted
1. INTRODUCTION
First‐line diagnostic screening for inborn errors of metabolism (IEM) comprises several dedicated assays aimed at a specific selection of biomarkers. 1 New analytical approaches providing broad coverage of metabolism with a short turn‐around‐time would improve the biochemical diagnostic workflow and facilitate functional elucidation of the many variants of unknown significance (VUS) that are found in molecular medicine. Since its introduction, researchers have embraced high‐resolution mass spectrometry to create comprehensive analytical approaches providing an all‐inclusive view of metabolism including known and “orphan” metabolites. What we now call ([un]targeted) metabolomics is a powerful tool to gather insight into disease mechanism, but also for diagnostics. 2 , 3
Application of single‐platform untargeted metabolomics in diagnostics decreases the need for different methods, operated on different analytical platforms, and would speed up the diagnostic process. However, bringing untargeted single‐platform metabolomics to diagnostics is a daunting challenge.
Current metabolomics methods have many time‐consuming steps, such as feature identification, and require large numbers of samples to obtain proper statistical power. 2 In reality, diagnostic results are needed on short notice to enable a rapid diagnosis and treatment of IEM patients. Often, they deal with emergency situations and have to work with a single sample of a single patient. This underlines the need for suitable, targeted single‐platform metabolomics since methods based on within‐run comparison are unfit for emergency diagnostics in an N = 1 setting. Unfortunately, the chemical properties of IEM‐associated metabolites are extremely diverse and for now prohibit an all‐in‐one test. Therefore, the combination of the diagnostic question addressed, together with the body fluid analysed, directs the choice of analytical platform and subsequent data analysis.
Previously, we have implemented an ultra‐high performance liquid chromatography‐quadrupole time‐of‐flight mass spectrometry (UHPLC‐QTOF‐MS) method for fast and accurate quantitative detection of 71 metabolites, replacing GC‐MS for the analysis of organic acids. 4 This was already promising in the light of metabolomics literature 5 , 6 since we obtained explicit biochemical signatures for 16 of the 18 organic acidurias included in the clinical validation.
This method provided the basis for the targeted semi‐quantitative single‐platform metabolomics method presented here. We expanded the number of metabolites to 258, covering 153 phenotypes described in IEMbase (http://iembase.org/). We call this method targeted urine metabolomics (TUM).
Our primary goal was the consolidation of several commonly applied dedicated assays to speed up diagnostics of (treatable) IEM based on known detectable metabolites in urine. The focus was on intermediary metabolism (amino acids, acylcarnitines, purines and pyrimidines, and sugars) and we excluded metabolites known as biomarkers for lysosomal storage disorders and congenital defects in glycosylation. Generalized, but not single enzyme peroxisomopathies were identified through increased excretion of pipecolic acid.
An automated pipeline extracted a panel of metabolite‐associated features in a targeted manner and processed them resulting in age‐corrected z‐scores. Next, the interpretation and reporting tool analysed the results to aid interpretation by a laboratory specialist. We applied TUM to 289 urine samples, covering 78 IEM. The results we present here prove the suitability of this novel screening method for routine and emergency metabolic screening.
2. MATERIALS AND METHODS
2.1. Chemicals and (internal) standards
All organic solvents and water used in sample and mobile phase preparation were UPLC‐MS‐grade and purchased from BIOSOLVE Chemicals (Valkenswaard, The Netherlands). Reference standards for 250 metabolites were analysed to annotate retention time (Rt) and spectral information (Table S1a lists the chemicals used and Table S1b lists analytical information per compound). For the remaining eight, we used patient urine samples. The three stable isotope‐labeled internal standards (IS) were 13C6‐galactitol (positive and negative electron spray ionization modes [ESI±], for all metabolites with a Rt < 1 minute), D4‐sebacic acid (ESI−, Rt > 1 minute) and D3‐hexanoylglycine (ESI+, Rt > 1 minute).
2.2. Control cohort and clinical validation samples
The control cohort was composed of 261 random urine samples from patients without an IEM or any abnormal biochemical findings. The clinical validation included 289 urine samples supplied by multiple laboratories, covering 78 IEM. Most samples were from the archive of the Maastricht Laboratory of Clinical Genetics or were from the ERNDIM Diagnostic Proficiency Testing (DPT) schemes. Colleagues kindly provided other samples as mentioned in the acknowledgement. We re‐analysed samples that were collected over 2 years ago with current dedicated assays to ensure suitability and proper comparison between methods.
2.3. UHPLC‐QTOF‐MS method
A 25 μL of the sample diluted with water to ±1 mM creatinine was mixed with 275 μL 0.1% formic acid in UPLC‐MS water and 25 μL of the internal standard (IS) mixture containing 187 μM 13C6 galactitol, 8.9 μM D3‐hexanoylglycine and 11.6 μM D4‐sebacic acid. Samples were analysed in ESI+ and ESI‐− modes with LC‐conditions as described before. 4 The injection volume that was 1 μL for ESI+ and 0.5 μL for ESI−. Our approach is nonenantioselective.
2.4. Data processing and statistics
The QTOF‐MS acquired untargeted data were imported into Agilent MassHunter Quant software, extracted for the relative concentration of 540 targeted analytes of 258 metabolites. The results were compared and aberrant results presenting as (a) a divergent retention time (cut‐off 0.05 minutes), (b) a too low peak height (cut‐off 2500), or (c) a too low mass match score (cut‐off 80) were marked. For peaks with an overloading problem (cut‐off fold difference = 1.3 and cut‐off height = 300 000), the signal at 50 ppm was used for further data processing. For the remaining peaks signals at 10 ppm were used.
The corrected relative response was normalized to creatinine. Z‐scores were calculated against an age‐matched control cohort: 0 to 18 months (N = 58); 18 months to 10 years (N = 73); 10 to 20 years (N = 48) and > 20 years (N = 82) (see Figure S1 for the calculations). The age groups were based on IEMbase (2018) and Blau (2014). Outliers were removed from the control cohort.
A sample's z‐score profile was compared to our urine metabolite database (172 phenotypes, 232 OMIM diseases) and for each phenotype a match score was calculated (scale 0‐100, z‐score cut‐off 2 and 3). Listed are the top 10 of potential phenotypes for either z‐score cut‐off 2 or 3.
Data were processed in R environment (version R 3.5) and results were presented in excel sheets.
2.5. Data visualization
The sample's z‐score profile was visualized in needle plots labeled with metabolite names for analytes with absolute z‐score values above the cut‐off.
The prioritized phenotypes with match scores were plotted in a heatmap showing their theoretical z‐score profiles. Plots were made using ggplot2 package (v3.1.1) and pheatmap packages (v1.012) in R.
2.6. Interpretation of data
Four laboratory specialists performed blinded interpretation of the needle‐plots, the heatmaps and the excel sheets with results. Consensus determined if a diagnosis could be made, or if the results were indicative and warranted further analysis.
3. RESULTS
3.1. Analytical characteristics of the UPLC‐QTOF/MS method
The QTOF‐MS method separated and identified 258 metabolites represented by 540 analytes expressed as relative response ratios (RRR) to the respective internal standards.
We already reported on the extensive analytical validation of 68 metabolites (organic acids) regarding linearity, recovery and within‐ and between‐run variation. 4 In this study, we introduced new metabolite classes, including amino acids, acylcarnitines, and purines and pyrimidines. The first two metabolite classes gave better responses in ESI+ than ESI− which we took into account in the final protocol. We validated this semi‐quantitative method for within‐ and between‐run variation.
3.2. Within‐ and between‐run variation
We determined within and between‐run variation using three quality control (QC) samples: (a) a mixture of urine samples from patients with an IEM, (b) a mixture of urine samples from controls without biochemical abnormalities, and (c) urine samples spiked with specific metabolites.
The average within‐run variation was 4%, and under 10% for 270 of 285 analytes (95%).
Between‐run variation was determined for 271 analytes with a peak signal >1*e4. The between run variation was 16 ± 9% for 234 of the 271 analytes. The variation was >25% for 37 of the 271 analytes, probably because of in source fragmentation, suboptimal peak shape and concentrations near detection limits. Among the metabolites with high variation were several amino acids (Table 2) and glycolic acid. These few metabolites with a higher variation pose little problem since the diagnostic match‐score incorporates information from several metabolites. Diagnosis of hyperoxaluria type 1 (OMIM 259900) might be problematic because its biomarkers are glycolic acid and oxalic acid, of which the latter has low sensitivity and solubility.
3.3. Clinical validation
We applied TUM to 289 patient urines representing 78 OMIM phenotypes belonging to 12/15 disease groups according to the 2011 society for the study of inborn errors of metabolism (SSIEM) classification of IEM (Figures 1 and 2; http://www.ssiem.org/images/centralstore/resources/SSIEMClassificationIEM2011.pdf). Most of the 289 urine samples represented disorders of amino acid and peptide metabolism (n = 162). Because 36 patients were on diagnosis‐specific treatment, we expected biochemical abnormalities to be less pronounced compared to those detected in samples of untreated patients.
FIGURE 1.
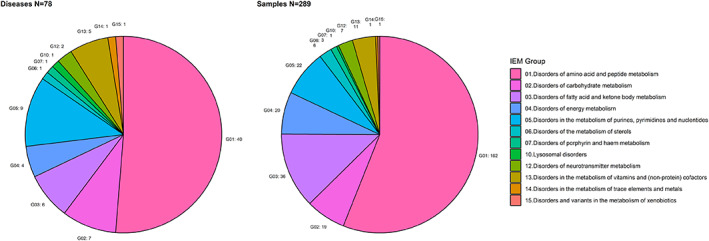
Summary of phenotypes and samples in clinical validation. Diseases are grouped according to the SSIEM classification of Inborn Errors 2011 (http://www.ssiem.org/images/centralstore/resources/SSIEMClassificationIEM2011.pdf). Left: the distribution of the 78 different IEM in the clinical validation. Right: the disease distribution of 289 urine samples in the clinical validation. The number of disease/samples in each group are indicated next to the pie plots
FIGURE 2.
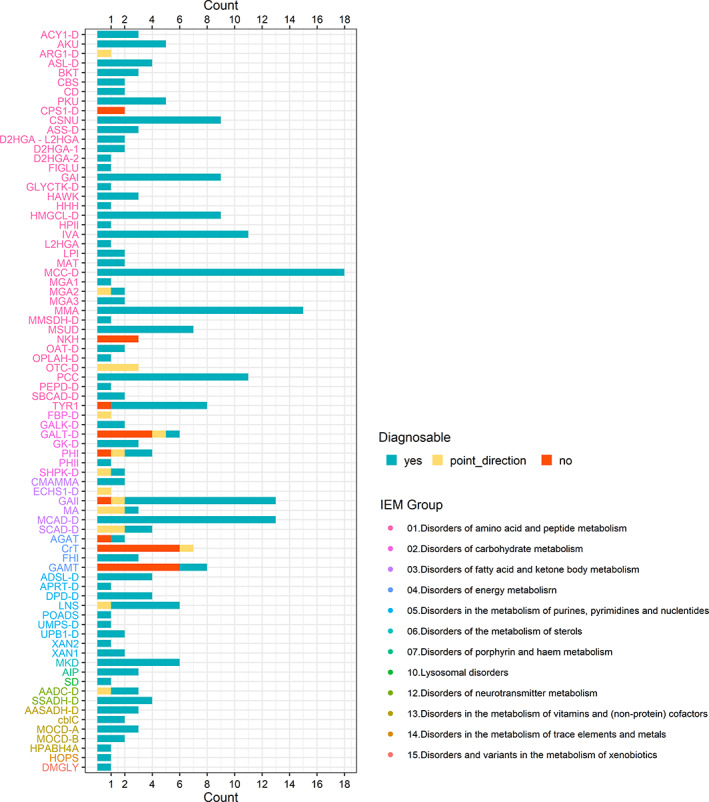
Clinical validation summary of IEM disease groups included in clinical validation. IEM are shown on the y‐axis and grouped according to the SSIEM classification of Inborn Errors 2011 (http://www.ssiem.org/images/centralstore/resources/SSIEMClassificationIEM2011.pdf). Disease groups are color coded per SSIEM category (legend on the right). The number of patient samples per IEM is indicated on the x‐axis. Turquoise = diagnosable with this screening, yellow = result points in the right direction, red = not diagnosable with this screening. A list of abbreviations can be found in Table S3
3.3.1. SSIEM disease group 1—disorders of amino acid and peptide metabolism
TUM detected most of the 40 IEM represented by 162 samples, or it pointed toward an IEM (see Figure 2 for details). In three instances, we faced challenges diagnosing urea cycle disorders (UCD) inherent to treatment, sample condition, or biochemistry as explained by the following examples. Argininemia (OMIM 207800) and ornithine transcarbamylase deficiency (OMIM 311250) were not unequivocally diagnosed; however, the phenotype prediction score only pointed in the direction of an urea cycle disorder. This was due to degradation of the sample and proper clinical treatment, respectively. Hyperammonemia in the absence of increased orotic acid and other pyrimidines could indicate a carbamoylphosphate synthetase I deficiency (OMIM 237300). When biochemically stable, these patients are indistinguishable from controls, as was the case in the sample we obtained.
TUM diagnosed all 18 organic acidurias (Figure 3A, isovaleric aciduria as an example). One patient with Barth syndrome had subtle but detectable biomarker changes discernible in TUM and in the dedicated organic acid analysis. 4 In disorders of phenylalanine and tyrosine metabolism, one tyrosinemia type I patient was hard to identify because of NTBC treatment. Patients with defects in sulfur‐amino acid metabolism and patients with glutamate formiminotransferase deficiency (OMIM 229100) and d‐glyceric aciduria (OMIM 220120) were found by TUM.
FIGURE 3.
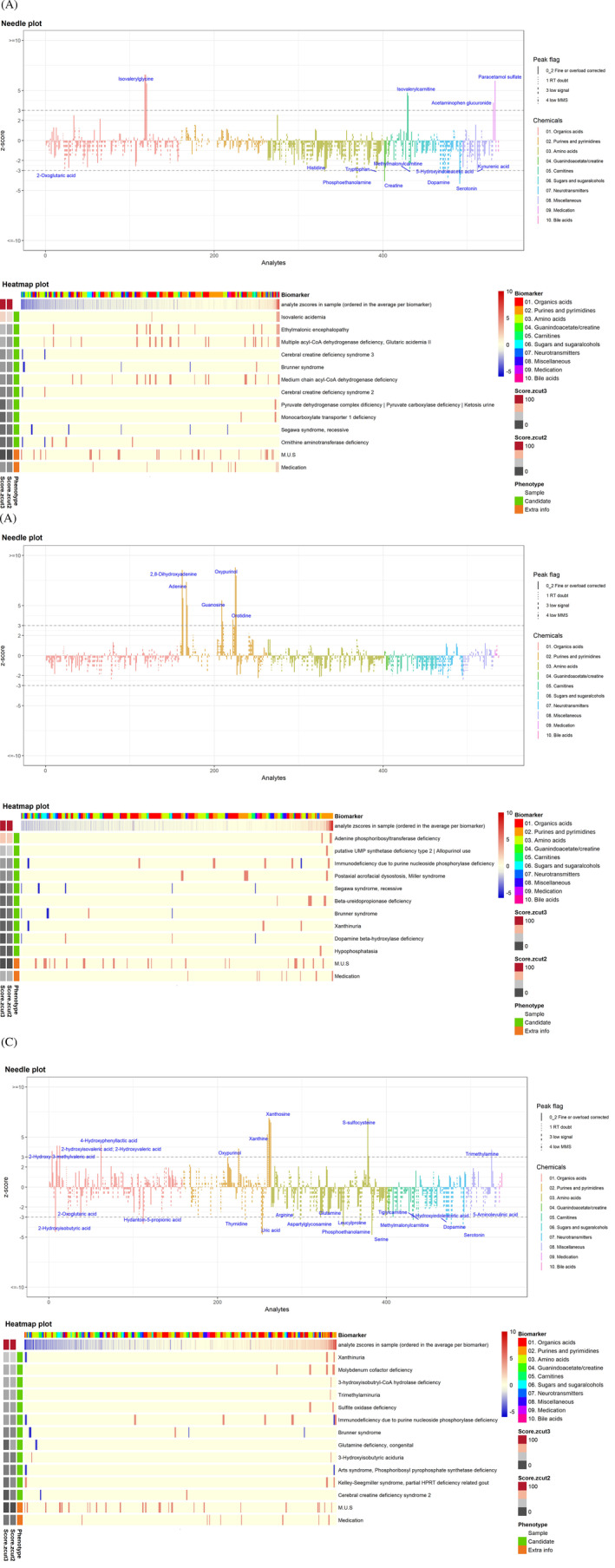
Representative needle plots and heatmaps. A, Isovaleric aciduria with isovaleryl carnitine and isovaleryl glycine as primary metabolites, B, APRT deficiency with a prominent 2,8‐dihydroxyadenine peak, C, molybdenum cofactor deficiency with S‐sulphocysteine, xanthosine, xanthine and oxypurinol. Needle plot—The z‐scores of all unique metabolite/analyte combinations are plotted. The chemical category of the metabolite is color coded. The peak score is visualized by different line styles. Solid line = analytes with a reliable peak integration. Three types of dashed lines = score 1, 3 or 4, implying a doubtful analytical peak. The metabolites/analytes are sorted by metabolite category and name. The z‐score cut‐off = 3 is shown here and indicated as a dotted line in the plot. Metabolites whose analyte (ion) reaches a |z‐score| above the cut‐off is labeled. Heatmap plot—The first line of the heatmap shows the distribution of the metabolites in the sample. The metabolites are colour coded by chemical category. The second line shows the z‐score profile of the sample (increased metabolites are shown in red and decreased metabolites in blue). The theoretical z‐score profiles for the candidate phenotypes are plotted underneath. The left two columns show the phenotype match scores (colour scale: more reddish indicates a higher matching score) at cut‐off = 2 and cut‐off = 3. The corresponding phenotype names can be read on the right
Regarding mild forms of amino acid metabolism deficiencies, we correctly identified the MSUD patient and two GAII patients in this clinical validation.
3.3.2. SSIEM disease group 2—disorders of carbohydrate metabolism
We included 7 of the 44 phenotypes suitable for TUM. Galactitol is the prime biomarker for galactokinase deficiency (OMIM 230200) and galactose‐1‐phosphate:uridyltransferase deficiency (OMIM 230400). Samples from patients on galactose restriction showed nondiscriminant z‐scores.
While glycerol and sedoheptulose are not quantified in dedicated assays, we can analyze them in TUM. Even in samples with many abnormal biomarkers, these specific metabolites resulted in high‐ranking prediction scores for untreated patients with fructose‐1,6‐biphosphate deficiency (OMIM 229700) or sedoheptulose kinase deficiency (OMIM 617213).
Untreated patients with disorders in glyoxylate metabolism can be diagnosed based on glycolic and glyceric acid.
3.3.3. SSIEMdisease group 3—disorders of fatty acid and ketone body metabolism
TUM gave comparable results to urine organic acid analysis for the disorders included here, including six disorders of ketone body metabolism, malonyl‐CoA decarboxylase deficiency (OMIM 248360), and combined malonic and methylmalonic aciduria (OMIM 614265). We excluded defects of long‐chain fatty acid oxidation (LCHADD, VLCADD), as they have no (specific) urinary biomarkers.
3.3.4. SSIEM disease group 4—disorders of energy metabolism
Our validation included fumarase deficiency (OMIM 136850) and disorders of creatine metabolism. In case of fumarase deficiency (Figure 4), TUM even resulted in the identification of previously unknown, but theoretically logical, disease‐associated metabolites.
FIGURE 4.
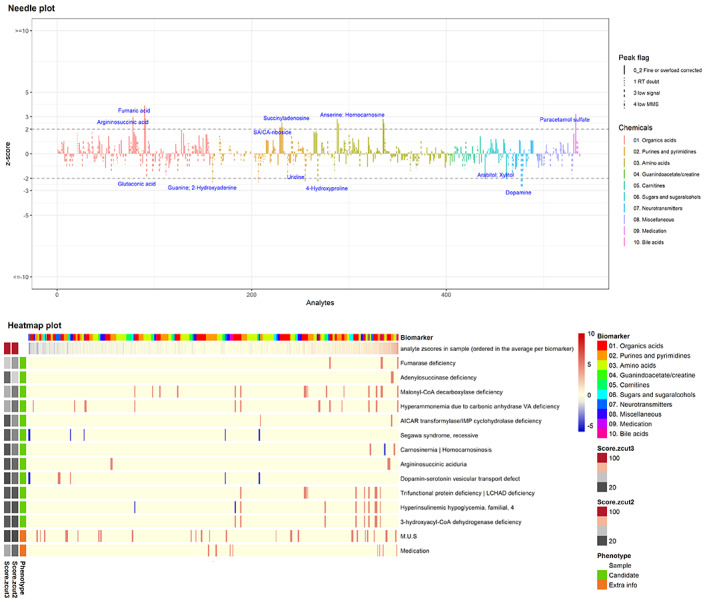
Needle plot and heatmap from a patient with fumarase deficiency displaying new metabolites due to a secondary adenylosuccinate lyase deficiency
Disorders of creatine biosynthesis and transport lead to subtle metabolite abnormalities and proved more difficult to diagnose. In addition, creatine treatment disturbs the phenotype prediction. In case of arginine:glycine amidinotransferase (AGAT, n = 2) deficiency (OMIM 612718) the diagnosis was obvious in one sample, whereas in other sample, we observed multiple abnormalities.
TUM correctly called two of the eight samples of patients with cerebral creatine deficiency syndrome 2 (guanidinoacetate methyltransferase [GAMT]; OMIM 612736). In two cases, treatment with creatine disturbed the prediction algorithm. In the remaining four samples, z‐scores for guanidinoacetate and creatine were within reference. Creatine Transporter (CrT) deficiency was difficult to diagnose with our pipeline.
3.3.5. SSIEM disease group 5—disorders in the metabolism of purines, pyrimidines, and nucleotides
TUM detected all disorders of purine and pyrimidine metabolism included in the clinical validation (Figure 2). All needle plots and heatmaps were obvious (Figure 3B needle plot and heatmap of APRT deficiency as an example).
3.3.6. SSIEM disease group 6—disorders of the metabolism of sterols
Mevalonate kinase deficiency (OMIM 610377) is the sole disorder in this group with a known urinary biomarker, and we successfully identified it.
3.3.7. SSIEM disease group 7—disorders of porphyrin and heme metabolism
We included metabolites for acute intermittent porphyria (AIP, OMIM 176000) and diagnosed it in the four samples analysed.
3.3.8. SSIEM disease group 10—lysosomal disorders
We included one urine sample from a patient with Salla disease (OMIM 269920) and found the correct diagnosis.
3.3.9. SSIEM disease group 12—disorders of neurotransmitter metabolism
TUM easily called both aromatic l‐amino decarboxylase (AADC) deficiency (OMIM 608643) and succinic semi‐aldehyde dehydrogenase (SSADH) deficiency (OMIM 271980).
3.3.10. SSIEM disease group 13—disorders in the metabolism of vitamins and cofactors
The diagnoses of cobalamin C deficiency (OMIM 277400), alpha‐amino adipic semialdehyde (α‐AASA) dehydrogenase deficiency (PDE‐ALDH7A1, OMIM 266100) and two disorders in molybdenum cofactor metabolism (MOCS1 and MOCS2 deficiency, respectively; OMIM 603707 and 603 708) were straightforward. Molybdenum cofactor deficiency type A (OMIM 252150) (Figure 3C) was easily detected by the combination of metabolites of purine metabolism and S‐sulphocysteine. The prominent α‐AASA excretion in the α‐AASA‐DH deficiency resulted in an obvious pattern.
3.3.11. SSIEM disease group 14—disorders in the metabolism of trace elements and metals
Hypophosphatasia (OMIM 241500) which was easily detected by the prominent phosphoethanolamine peak.
3.3.12. SSIEM disease group 15—disorders and variants in the metabolism of xenobiotics
The group of disorders and variants in the metabolism of xenobiotics only includes dimethylglycinuria (OMIM 605850), which was correctly diagnosed.
4. DISCUSSION
We describe a single platform high‐resolution UHPLC‐QTOF‐MS metabolomics approach for analysis of urine using targeted ion extraction, automated data processing, age‐group‐matched z‐scores and a phenotype prediction score. We tested 78 of the total 232 OMIM diseases in our urine disease‐metabolite database and found the correct diagnosis in 68 samples. In 7 out of the remaining 10 samples a normalized metabolite pattern, due to treatment, hampered detection of the correct diagnosis.
As TUM is intended as a screening method, we therefore foresee no problems using TUM to diagnose yet untreated patients with IEM within context. Our primary goal was the consolidation of dedicated assays in urine, for example, organic acids, amino acids, purines and pyrimidines, and acylcarnitines. We aimed to include screening for acute intermittent porphyria, which is often performed in an emergency setting requiring short turn‐around time, and screening for creatine biosynthesis and transport disorders. Combining these assays in TUM streamlines the diagnostic process, reduces the time to results to 24 hours in case of emergency, and gives an unbiased overview of metabolites potentially leading to discovery of new disease‐associated metabolite profiles. 7 , 8
We successfully combined the mentioned dedicated assays into TUM, and disorders in amino acid, fatty acid, ketone body, purines and pyrimidines, neurotransmitters, and vitamin metabolism could be identified. Disorders in carbohydrate, sterol, and xenobiotics metabolism and porphyrias were detected (Figure 2).
Main issues affecting sensitivity were (a) suboptimal separation of metabolites, (b) metabolites with great analytical variation, and (c) high biological variability for a few metabolites in the youngest age‐group. Regarding the separation, polar metabolites are not optimally separated as they have limited retention on a reversed‐phase column, which holds true for amino acids and sugars. Theoretically, parallel runs with, for example, a hydrophilic interaction liquid chromatography column could be performed to reach improved coverage of these polar metabolites. However, this would decrease time efficiency, and therefore we opt for performing a dedicated assay as a second tier test in case of inconclusive results or high suspicion of a certain IEM based on TUM results. Regarding analytical variability, certain metabolites (methylglutaconic acid and creatine) show a high within‐run variation seldom resulting in z‐score values above |3| in patients. Unfortunately, these metabolites are equally troublesome in dedicated assays. Thus, biochemical diagnosis of creatine transporter deficiency can be challenging, regardless of the chosen analytical method. Regarding the youngest age group, the large variation in intensity of some metabolites could potentially result in false‐normal z‐scores. We require more control samples in this group allowing us to further specify the current age group of 0‐18 months to several age groups in order to obtain more reliable z‐scores. Until that moment we advise to apply TUM for patients aged >18 months.
The goal of diagnostic metabolomics in whatever matrix, be it urine, plasma or CSF, is a comprehensive screening using a stringently limited number of runs to detect diagnostically relevant metabolites for as many IEM as possible, with emphasis on the treatable conditions. 9 , 10 As mentioned in the introduction, a single run analysis is not feasible because of the chemical spectrum of the relevant biomarkers. However, to be as complete as possible we can strive to develop logistics for the parallel runs needed and make concomitant analysis pipelines as fast as possible. Our TUM workflow takes 24 hours and could complement data from additional analysis of macromolecules or lipidomics, having different sample work‐up and separation.
Our partner laboratory has recently developed (un)targeted plasma metabolomics. 3 We envision the consolidation of our two workflows allowing for ultrafast analysis of plasma samples parallel to TUM to broaden our metabolite profiling with apolar metabolites. Additionally, the raw data is still available for different processing to probe the untargeted data for discovery of potential novel biomarkers. In a pilot study, we performed untargeted metabolomics on urine samples using the pipeline described by Coene et al., 3 and the data look promising (data not published).
This integrative approach of TUM with untargeted data‐extraction also led to the expansion of biomarker profiles for some well‐known IEM. For instance, in two out of three urine samples of fumarase deficiency (OMIM 606812) SAICA‐riboside (SAICAr), succinyladenosine (SAdo) and argininosuccinate were increased next to the primary metabolite fumarate. The former two metabolites are associated with adenylosuccinate lyase (ADSL) deficiency (OMIM 103050) and the latter with argininosuccinate lyase (ASL) deficiency (OMIM 207900). The accumulating fumarate in fumarase deficiency inhibits the fumarate‐releasing reactions of ADSL and ASL, 11 , 12 thereby causing secondary deficiencies of these enzymes and leading to the corresponding metabolic profile. 13 , 14 This sheds a new light on fumarase deficiency, and we await further confirmation of these findings in additional samples.
In conclusion, we have developed an innovative, targeted urine metabolomics procedure, TUM, that includes a prediction software tool to assist the diagnosis of IEM. We present TUM as a powerful diagnostic screening tool that merges most urinary diagnostic assays, thereby accelerating the diagnostic work‐up for patients suspected of IEM.
CONFLICT OF INTEREST
Laura K.M. Steinbusch, Ping Wang, Huub W.A.H. Waterval, Fons A.P.M. Stassen, Karlien L.M. Coene, Udo F.H. Engelke, Daphna D.J. Habets, Jörgen Bierau, Irene M.L.W. Körver‐Keularts declare that they have no conflict of interest.
INFORMED CONSENT
All patients have agreed to the anonymous use of their left‐over material from routine diagnostics for laboratory development and validation purposes.
AUTHOR CONTRIBUTIONS
Laura K.M. Steinbusch collected samples, analysed data, performed the validation and wrote the manuscript. Ping Wang developed the prediction and report tool, analysed data and participated in writing the manuscript. Huub W.A.H. Waterval did all QTOF analyses, performed the analytical validation, analysed data, participated in the clinical validation and writing of the manuscript. Fons A.P.M. Stassen developed the software program for automated peak checking and analysed data. Karlien LM Coene gave input for study design, clinical validation and revised the manuscript. Udo F.H. Engelke gave input for study design, clinical validation and revised the manuscript. Daphna D.J. Habets performed the validation and edited the manuscript. Jörgen Bierau performed the validation and edited the manuscript. Irene MLW Körver‐Keularts conceptualized the project, designed the clinical validation and wrote the manuscript; The name of the corresponding author: Irene M.L.W. Körver‐Keularts.
Supporting information
Supplemental Figure 1 Flow chart of z‐score calculation in control cohort and for individual samples.
Left: Control cohort derived z‐score parameters describing decision points and calculations used. Right: Sample z‐score calculation describing decision points and calculations used.
Supplemental Table 1 Biomarkers in TUM.
Reference standards for metabolites in TUM mentioning biomarker name, CAS‐number, supplier and CAT number.
Supplemental Table 1a. List of standards that were used in the targeted urine metabolomics analysis
Supplemental Table 2 Metabolites with high between‐run variation.
All metabolites with high between‐run variation are listed in this table.
Supplemental Table 3 List of IEM disease abbreviations
Table 3 includes the disease abbreviations used in TUM, OMIM disease name, SSIEM category, OMIM number and gene name.
ACKNOWLEDGMENTS
We thank Dr. G.J.G. Ruijter and Dr. E.H. Jacobs from the Metabolic Laboratory, Department of Clinical Genetics, Erasmus University Medical Center in Rotterdam, the Netherlands for kindly providing samples for the clinical validation of 24 IEMs. In addition, we thank Dr. François Boemer and Prof. Dr. François‐Guillaume Debray from, the Biochemical Genetics Laboratory Liège (Human Genetics, CHU Sart‐Tilman, Liege, Belgium) and Marie‐Françoise Vincent and Sandrine Marie from UC Louvain 5 (Bruxelles, Belgium) for kindly proving samples for the clinical validation.
[Correction added on 15 June 2021, after first online publication: The Acknowledgements section has been updated in this version.]
Steinbusch LKM, Wang P, Waterval HWAH, et al. Targeted urine metabolomics with a graphical reporting tool for rapid diagnosis of inborn errors of metabolism. J Inherit Metab Dis. 2021;44:1113–1123. 10.1002/jimd.12385
Communicating Editor: Sander M Houten
REFERENCES
- 1. Bonte R, Bongaerts M, Demirdas S, et al. Untargeted metabolomics‐based screening method for inborn errors of metabolism using semi‐automatic sample preparation with an UHPLC‐ Orbitrap‐MS platform. Metabolites. 2019;9(12):1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ivanisevic J, Want EJ. From samples to insights into metabolism: uncovering biologically relevant information in LC‐HRMS metabolomics data. Metabolites. 2019;9(12):1‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coene KLM, Kluijtmans LAJ, van der Heeft E, et al. Next‐generation metabolic screening: targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J Inherit Metab Dis. 2018;41(3):337‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Korver‐Keularts I, Wang P, Waterval H, et al. Fast and accurate quantitative organic acid analysis with LC‐QTOF/MS facilitates screening of patients for inborn errors of metabolism. J Inherit Metab Dis. 2018;41(3):415‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haijes HA, van der Ham M, Prinsen H, et al. Untargeted metabolomics for metabolic diagnostic screening with automated data interpretation using a knowledge‐based algorithm. Int J Mol Sci. 2020;21(3):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khamis MM, Adamko DJ, El‐Aneed A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom Rev. 2017;36(2):115‐134. [DOI] [PubMed] [Google Scholar]
- 7. Gao Y. Urine‐an untapped goldmine for biomarker discovery? Sci China Life Sci. 2013;56(12):1145‐1146. [DOI] [PubMed] [Google Scholar]
- 8. Kennedy AD, Miller MJ, Beebe K, et al. Metabolomic profiling of human urine as a screen for multiple inborn errors of metabolism. Genet Test Mol Biomarkers. 2016;20(9):485‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sirrs SM, Lehman A, Stockler S, van Karnebeek CD. Treatable inborn errors of metabolism causing neurological symptoms in adults. Mol Genet Metab. 2013;110(4):431‐438. [DOI] [PubMed] [Google Scholar]
- 10. van Karnebeek CD, Stockler S. Treatable inborn errors of metabolism causing intellectual disability: a systematic literature review. Mol Genet Metab. 2012;105(3):368‐381. [DOI] [PubMed] [Google Scholar]
- 11. Toth EA, Yeates TO. The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway. Structure. 2000;8(2):163‐174. [DOI] [PubMed] [Google Scholar]
- 12. Nagamani SCS, Erez A, Lee B. Argininosuccinate Lyase deficiency. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews([R]). Seattle, WA: University of Washington; 2011:1‐21. [PubMed] [Google Scholar]
- 13. Zeman J, Krijt J, Stratilova L, et al. Abnormalities in succinylpurines in fumarase deficiency: possible role in pathogenesis of CNS impairment. J Inherit Metab Dis. 2000;23(4):371‐374. [DOI] [PubMed] [Google Scholar]
- 14. Tregoning S, Salter W, Thorburn DR, et al. Fumarase deficiency in dichorionic diamniotic twins. Twin Res Hum Genet. 2013;16(6):1117‐1120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Flow chart of z‐score calculation in control cohort and for individual samples.
Left: Control cohort derived z‐score parameters describing decision points and calculations used. Right: Sample z‐score calculation describing decision points and calculations used.
Supplemental Table 1 Biomarkers in TUM.
Reference standards for metabolites in TUM mentioning biomarker name, CAS‐number, supplier and CAT number.
Supplemental Table 1a. List of standards that were used in the targeted urine metabolomics analysis
Supplemental Table 2 Metabolites with high between‐run variation.
All metabolites with high between‐run variation are listed in this table.
Supplemental Table 3 List of IEM disease abbreviations
Table 3 includes the disease abbreviations used in TUM, OMIM disease name, SSIEM category, OMIM number and gene name.


