Abstract
Given their intrinsic pleiotropism, microRNAs (miR) play complex biological roles, in both normal and pathological conditions. Often the same miR can act as oncogene or oncosuppressor, depending on the biological process dysregulated in each specific tissue. miR‐223 does not represent an exception to this rule and its functions greatly differ in different contexts. miR‐223 has been widely studied in the hematopoietic compartment, where it plays a central role in innate immune response, regulating myeloid differentiation and granulocytes function. Accordingly, dysregulated expression of miR‐223 has been associated to different inflammatory disorders and tumors arising from the immune compartment. Most carcinomas, breast cancer being the most studied, display loss of miR‐223. However, in gastro‐esophageal cancers miR‐223 is frequently overexpressed and correlates with worse prognosis. A link between miR‐223 and response to CDK4/6‐inhibitors has been recently proposed, suggesting a role as biomarker of therapeutic response. The notion that one of the most commonly mutated protein in cancer, mutant p53, binds the promoter of miR‐223 and suppresses its transcription, adds a further level of complexity to the full understanding of miR‐223 in cancer. In this review, we will summarize the current knowledge on the molecular networks that alter or are altered by miR‐223, in different cancer types. We will discuss if the times are ready for the exploitation of miR‐223 as predictive biomarker of treatment response or, even, as therapeutic target, in specific settings. Finally, we will suggest which could be the next steps to be taken for a realistic clinical application of miR‐223.
This article is categorized under:
RNA in Disease and Development > RNA in Disease
Keywords: biomarker, breast cancer, cancer, microRNA, miR‐223
miR‐223–centered view of the Hallmarks of Cancer.
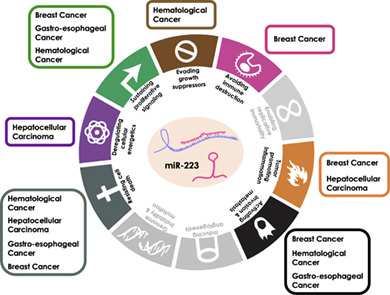
Many of the hallmarks of cancer can be affected by miR‐223 up‐regulation or down‐regulation, via different modulation of its targets and depending on the tumor type. These aspects of miR‐223 biology support the possibility that it could serve as prognostic/predictive biomarker and therapeutic target, in certain tumoral contexts.
1. INTRODUCTION
microRNAs, also known as miRNAs or miRs, are small non‐coding RNA sequences of approximately 22 nucleotides that can regulate gene expression at post‐transcriptional level (Gebert & MacRae, 2019; O'Brien et al., 2018; Treiber et al., 2019).
The generation of miRNAs starts with the transcription of long primary miRNAs (pri‐miRNAs) in the cell nucleus, their processing into a 60–110 nucleotides‐long pre‐miRNA and their export to the cytoplasm, where they are further processed to eventually form the mature miRNA. This is initially a duplex, made by a 3p strand, originating from the 3′ end, and a 5p strand deriving from the 5′ end of the pre‐miRNA hairpin, that is then unwound forming the so‐called guide strand and the passenger strand. The guide strand is the one that will be actually loaded onto the miRNA‐induced silencing complex (miRISC) to functionally silence its mRNA targets.
The miRNA specificity derives from the “seed” sequence of the miRNA, a 2–8 nucleotides‐long sequence that binds complementary sequences in different mRNA regions. miRNA preferentially binds the 3′‐UTR sequence of target mRNA; however, the binding to 5′‐UTR and to the coding sequence of the mRNA are also contemplated, in particular situations. As a consequence of the binding, gene silencing through translation repression and mRNA decay is accomplished, with mRNA decay responsible for most of the silencing effect (Gebert & MacRae, 2019).
It is estimated that approximately 60% of human genes contain multiple seed binding sites, thus highlighting how complex and critical the role of miRNA is in regulating gene expression (Gebert & MacRae, 2019; O'Brien et al., 2018; Treiber et al., 2019). Taking in consideration the action of miRNAs together with the process of messenger RNA decay, we can easily appreciate that as much as 50% of the changes in gene expression are at the level of mRNA stability (Garneau et al., 2007; Heck & Wilusz, 2018).
In accord with the widespread distribution of seed binding sites among mRNAs, miRNAs are involved in almost every cellular process, from development to differentiation and homeostasis, and their dysregulation is associated with a wide number of pathologies, including cancer (Peng & Croce, 2016).
In the context of cancer, depending from the cellular or tissue context, a single miRNA can act both as tumor promoter or suppressor, pinpointing the fact that the use of miRNA as prognostic marker or potential target for cancer therapies has to be deeply contextualized (Peng & Croce, 2016).
The complex roles played by miRNA in normal and pathological conditions have been extensively and exhaustively reviewed elsewhere (Haneklaus et al., 2013; O'Connell et al., 2010; Paul et al., 2018; Peng & Croce, 2016; Tan et al., 2018). In this review we will focus on the role(s) of one specific miRNA, miRNA‐223 (hereafter miR‐223), in normal and pathological conditions.
miR‐223 expression has been predominantly studied in the hematopoietic compartment, where it is abundant especially in neutrophils, macrophages, and myeloid progenitors (Fazi et al., 2005). However, besides its central role in the innate immune response where it regulates myeloid differentiation and granulocytes function, abnormal expression of miR‐223 has been associated to many inflammatory disorders and cancer (Yuan et al., 2018).
Here, we will describe the experimental and clinical settings in which miR‐223 activity has been implicated and the different roles, even opposing, that it can play, depending on the tissue context or the cancer type. Most of studies have focused on miR‐223‐3p and only a few on miR‐223‐5p; however, in most cases, the strand is not specified.
We will also discuss how the collected preclinical observations on miR‐223 may turn into promising future clinical applications, both as a predictive biomarker and as a potential therapeutic target.
Finally, we will highlight the caveats and the controversies that may currently prevent the use of miR‐223 in human cancer.
2. MIR‐223 IN INFLAMMATION AND INNATE IMMUNITY
Inflammation is the response of immune system against harmful stimuli and involves a huge variety of cellular and molecular mediators, including microRNAs (C.‐Z. Chen et al., 2004). This complex biological reaction leads to the production and release of several inflammatory cytokines that activate immune cells, like neutrophils, macrophages and, later on, adaptive immune cells (S.‐C. Sun, 2017).
miR‐223 has been initially identified bioinformatically and then found to be expressed in the hematopoietic system, where it regulates the differentiation of myeloid progenitors (C.‐Z. Chen et al., 2004).
Great contribution to the understanding of the functional roles of miR‐223 in normal and pathological conditions came from the generation and study of the miR‐223 knock‐out (KO) mouse (Johnnidis et al., 2008). These mice were born at normal Mendelian ratios, were fertile and displayed no gross abnormalities. At a deeper level of investigation, it was found that they displayed an increased number of circulating neutrophils probably resulting from enhanced differentiation and proliferation of the granulocyte progenitor pool. Neutrophils are an essential part of the innate immune response as they are critical for the first line of defense against bacteria and fungi. While no significant difference was found in the ability of mutant neutrophils to extravasate, migrate or to phagocytose, the absence of miR‐223 led to increased neutrophil activation and killing. It was hypothesized that loss of miR‐223 could lead to neutrophil‐mediated disease and, accordingly, it was observed that the lungs of old miR‐223 KO animals (>1.2 year) displayed inflammatory lung pathology characterized by areas of atelectasis, increased cellularity within the parenchyma, and inflammatory infiltration into the interstitial tissue (Johnnidis et al., 2008). In partial contrast, others reported that miR‐223 negatively regulated activation and chemotaxis of neutrophils. Accordingly, miR‐223 KO mice displayed an increased number of neutrophils in bone marrow, blood and lungs, causing mild inflammatory phenotypes under normal conditions. These neutrophils were hypersensitive to TLR4 stimulation and therefore, when challenged, they produced an excessive amount of pro‐inflammatory cytokines (Dorhoi et al., 2013; Jeffries et al., 2019; Neudecker et al., 2017). In line with these notions, it was shown that neutrophils are able to transfer miR‐223 to pulmonary epithelial cells, thus dampening acute lung injury through repression of PARP1, in a model of ventilator‐induced acute lung injury in mice (Dorhoi et al., 2013; Neudecker et al., 2017). The active transfer of miR‐223 has been shown also in other settings. miR‐223‐3p originating from neutrophils and macrophages can be transported in the circulation by high‐density lipoprotein (HDL) and delivered to recipient endothelial cells where it inhibits inflammation blocking neutrophils adhesion to the vascular endothelium and preventing infiltration into the sub‐intimal space (Cuesta Torres et al., 2019; J. Li, Tan, et al., 2017; Vickers et al., 2011).
Further, both in neutrophils and macrophages, miR‐223 inhibits the activity of the NLRP3 inflammasome by binding the 3′‐UTR of NLRP3 mRNA, thereby decreasing NLRP3 (NOD‐, LRR‐, and pyrin domain‐containing protein 3) availability. Accordingly, miR‐223 deficiency can lead to the sustained activation of NLRP3‐IL‐1β leading to acute lung injury/acute respiratory distress syndrome in patients with pulmonary inflammatory injury. Elimination of peripheral Ly6G+ neutrophils and/or pharmacological blockade of the miR‐223‐NLRP3‐IL‐1β signaling axis could alleviate acute lung injury (Bauernfeind et al., 2012; Feng et al., 2017).
With regards to macrophages, it was shown that miR‐223 KO mice display enhanced Toll‐like receptor 4 (TLR4)‐activated M1 (pro‐inflammatory) and reduced IL‐4‐stimulated M2 (anti‐inflammatory) responses. At molecular level, absence of miR‐223 released STAT3 from miR‐223‐driven inhibition, leading to the production of inflammatory IL‐1β and IL‐6 (Q. Chen et al., 2012). Consistently, when TLR4 was activated it led to downregulation of miR‐223, thereby inducing the same phenotypes.
Notably, in other settings the same molecules have been reported to act in completely different manners. In E6‐driven cervical squamous cell carcinoma (CSCC) transcriptional activity of STAT3 has been reported to mediate miR‐223‐3p expression, which, in turn, induced IL‐6 secretion by cocultured monocyte/macrophage, forming a positive activation feedback loop (J. Zhang et al., 2020).
The promoter sequence of miR‐223 contains three PPARδ regulatory elements, which enhance miR‐223 expression in bone marrow‐derived macrophages, regulating their polarization (Ying et al., 2015). The PPARδ/miR‐223 regulatory axis also targets the alternative pathway of macrophage activation, acting on RASA1 and NFAT5, altogether highlighting the central role of miR‐223 in controlling macrophage polarization (Ying et al., 2015).
It was also shown that miR‐223 KO mice display low platelet counts and prolonged time to occlusive thrombosis in comparison to their wild‐type counterpart, after carotid injury. The mechanism responsible for this phenotype involved, at least in part, the downregulation of IGFR1 by platelet miR‐223, and suggests inhibition of platelet miR‐223 or activation of its downstream target IGFR1 as potential therapeutic strategies to prevent arterial thrombosis (H. Wang et al., 2017).
Furthermore, deficiency of platelet miR‐223 in KO mice leads to enhanced vascular smooth muscle cell (VSMC) differentiation, thereby impairing the process of vessel injury repair induced by arterial wire injury experiments in diabetes mellitus mouse model. This phenotype is caused by the action of miR‐223 which targets platelet‐derived growth factor receptor β (PDGFR β). In the absence of miR‐223, PDGFR β is upregulated thus contributing to VSMC proliferation and hyperplasia (Zeng et al., 2019).
Finally, in natural killer (NK) cells, innate lymphocytes important for early host defense against pathogens and for surveillance against malignant transformation, miR‐223 was found to be significantly downregulated following cytokine activation. In this context, while miR‐223 specifically targeted the 3′‐UTR of granzyme B in resting NK cells, miR‐223 was promptly downregulated upon NK cell activation, for example, via IL‐15 (Fehniger et al., 2010).
Altogether, the regulation of miR‐223 expression in different types of immune cells is crucial to induce their activation and/or mediate the inflammatory response properly. Increased plasma levels of myeloid cells, such as polymorphonucleated neutrophils and macrophages/monocytes, are associated with unstable coronary artery disease and cardiovascular events and mis‐regulation of neutrophil activation is associated with several inflammatory and autoimmune diseases, strongly supporting that pharmacological manipulation of miR‐223 may hold great promise in many clinical settings.
3. MIR‐223 IN CANCER ONSET AND PROGRESSION
3.1. Hematological tumors
Given the important role played in controlling myeloid progenitor differentiation and immune response, it was largely expected that miR‐223 was frequently dysregulated in tumors arising from the hematological/immune compartment. Indeed, the expression of miR‐223 in these pathologies can be either up‐regulated or down‐regulated, depending on the tumor type.
In T‐cell acute lymphoblastic leukemia (T‐ALL), a very aggressive cancer of the bone marrow, the expression of miR‐223 is increased compared with their normal counterpart, due to the action of the Notch pathway, which is subjected to activating mutations in most T‐ALL cases, and the NF‐κB pathway (Chiaretti et al., 2010; Mavrakis et al., 2011; Shu et al., 2020). In vitro experiments demonstrate that miR‐223‐3p targets the 3′‐UTR of ARRB1, a member of the β‐arrestin family which facilitates Notch1 ubiquitination and degradation. As a consequence, ARRB1 is expressed at low levels in T‐ALL patients. Conversely, when ARRB1 is exogenously expressed, T‐ALL proliferation is impaired and survival of T‐ALL xenograft animals improved. Together, these data show how miR‐223‐3p impairs the tumor suppressive role of ARRB1 in T‐ALL, thus upregulating Notch and creating a positive feedback loop that fosters T‐ALL progression (Shu et al., 2020). miR‐223 is also involved in another important pathway which is commonly dysregulated in T‐ALL, the TAL1/FBXW7 axis. TAL1 complex, a well‐known oncogenic transcription factor in T‐ALL, binds miR‐223 promoter enhancing its transcription (Sanda & Leong, 2017). Mansour et al. demonstrated that aberrant upregulation of miR‐223 by TAL1 complex is important for the growth of TAL1‐positive T‐ALL cells, and sustained expression of miR‐223 partially rescues T‐ALL cells after TAL1 knockdown (Mansour et al., 2013). Furthermore, TAL1 regulation of miR‐223 also impacts on FBXW7, an E3 ubiquitin ligase that plays an important tumor suppressor role in T‐ALL by priming a pool of oncogenes like Myc, Myb, and Notch1 for degradation via proteasome and that is targeted by miR‐223. Accordingly, loss‐of‐function of FBXW7 is present in 20% of T‐ALL patients (Welcker & Clurman, 2008) and Fbxw7 knockout mice spontaneously develop T‐ALL (Onoyama et al., 2007).
Treatment with γ‐secretase inhibitors (GSI) to inhibit Notch signaling in T‐ALL cells can have differing effects on miR‐223. Kumar et al. report that treatment with GSI in vitro increased endogenous levels of miR‐223 in GSI‐resistant T‐ALL cells, whereas it lowered them in GSI‐sensitive cells (Kumar et al., 2014). On one side, the GSI inhibited Notch activation by blocking Notch cleavage, on the other it increased the expression of miR‐223 transcriptional activator C/EBPα, but only in GSI‐resistant cells. Ectopic modulation of miR‐223 expression subverted the GSI sensitivity of GSI‐resistant T‐ALL cells. Moreover, GSI treatment resulted in decreased expression of FBXW7 in GSI‐resistant T‐ALL cells, whereas it did not affect FBXW7 levels in GSI‐sensitive ones (Kumar et al., 2014). Altogether, these data point to a key role of miR‐223 in regulating the response to GSI and suggest that blockade of miR‐223 may be exploited in T‐ALL target therapy to revert GSI resistance.
In acute myeloid leukemia (AML), a cancer of the myeloid line of blood cells, miR‐223 show the opposite trend, with low levels of miR‐223 correlating with poor prognosis of the patients (Gentner et al., 2015; Guopan Yu et al., 2020). Pulikkan et al. discovered an autoregulatory negative feedback loop linking C/EBPα, miR‐223 and E2F1 normally regulating granulopoiesis and dysregulated in AML (Pulikkan et al., 2010). C/EBPα function is frequently altered by various mechanisms in AML and C/EBPα mutations are reported in approximately 10% of patients. When this happens, transcription of miR‐223 by C/EBPα is decreased, resulting in accumulation of E2F1, that would be targeted by miR‐223 under normal conditions. Overexpressed E2F1 then binds to miR‐223 promoter acting as repressor, further inhibiting miR‐223 transcription through a negative feedback loop, eventually resulting in myeloid cell differentiation blockage, proliferation and AML progression. This hypothesis was confirmed both in vivo, since protein levels of E2F1 were increased in bone marrow neutrophils derived from miR‐223 null mice, and in vitro, since overexpression of miR‐223 led to downregulation of E2F1 protein levels, overall confirming the ability of miR‐223 to modulate E2F1 expression (Armenia et al., 2014; Pulikkan et al., 2010). Low levels of miR‐223 correlate with poor prognosis also in AML patients carrying the AML1/ETO fusion protein. AML1/ETO, the most common chromosomal alteration in AML, exerts its oncogenic activity increasing the affinity for histone deacetylase (HDAC) and DNA methyltransferase, thus acting as a potent transcriptional repressor (Fazi et al., 2007). Fazi et al. demonstrated that AML1/ETO chromatin remodeling complex binds the pre‐miR‐223 sequence, leading to transcriptional silencing of miR‐223. Moreover, ectopic expression of miR‐223, RNAi against AML1/ETO, or demethylating treatment, enhances miR‐223 levels and restore cell differentiation, thus highlighting a new important function for this leukemia fusion protein (Fazi et al., 2007).
Also in chronic lymphocytic leukemia (CLL) patients low miR‐223 expression levels decrease with disease progression and significantly predict treatment‐free survival and overall survival (Stamatopoulos et al., 2009). An explanation of the mechanism that might explain the correlation between low levels of miR‐223 and poor prognosis was provided by the group of Rodríguez‐Vicente et al. (2015). In CLL, Heat shock proteins (HSPs) are upregulated and they might play a role in cancer cell survival (Castro et al., 2005). The work of Rodríguez‐Vicente shows that a common polymorphism present in 24% of CLL patients (rs2307842) disrupts the binding site for miR‐223 in HSP90B1, leading to its overexpression in B lymphocytes (Rodríguez‐Vicente et al., 2015).
Little is known about miR‐223 role in chronic myeloid leukemia (CML). However, it is known that the BCR‐ABL oncoproteins, the leukemia‐specific gene products of the Philadelphia chromosome translocation, inhibits the translation of C/EBPα, the principal regulator of granulocytic differentiation and a well‐known transcriptional regulator of miR‐223 (Perrotti et al., 2002). Other transcription factors involved in miR‐223 expression are also dysregulated in CML, together with the presence of a chronic granulocytic maturation arrest in which miR‐223 might be involved. MEF2C, a transcription factor required for cell survival and proliferation which plays also a role in lymphopoiesis and myelopoiesis (Stehling‐Sun et al., 2009), has been described as a miR‐223 target in CML. Also PTBP2, an RNA‐binding neuronal‐specific protein involved in the splicing of nuclear transcript and in the stabilization of certain miRNAs in the cytoplasm, is a target of miR‐223 in CML (Agatheeswaran et al., 2013). Together, these data strongly suggest that miR‐223 plays a tumor‐suppressive role in CML.
In mantle cell lymphoma (MCL), a very aggressive lymphoid malignancy, purified CD19+ lymphocytes from patients display downregulated miR‐223 levels in comparison with those from healthy donors (K. Zhou et al., 2018). miR‐223 overexpressing MCL cells showed decreased cell proliferation, increased cell‐cycle arrest and a twofold increase of apoptotic cells, compared with controls. SOX11, a highly specific marker for both cyclin D1‐positive and D1‐negative MCL, was identified as target of miR‐223 in MCL. Accordingly, miR‐223 and SOX11 expression levels inversely correlated to each other and high miR‐223 predicted better prognosis in MCL patients (Mozos et al., 2009; K. Zhou et al., 2018).
In pediatric lymphoblastic T‐cell lymphoma (T‐LBL), a pathology that shares many clinical features with T‐ALL, miR‐223 was frequently overexpressed and was an integral part of a six‐genes T‐LBL specific miR‐signature (Mussolin et al., 2014). In line with what observed in T‐ALL, increased Notch1 activity, via direct targeting of FBXW7, a negative modulator of Notch1, was identified as a main mechanism of miR‐223 pro‐oncogenic activity in these T cells. The work of Pomari et al. confirmed the observed high level of miR‐223 in T‐LBL patients, associated with worse prognosis and decreased PFS (Shaw, 2009). They also discovered that SIK1, a key regulator of anoikis associated with metastatic spread and survival of disseminated cells in solid tissues, was a miR‐223 target in T‐LBL. Protein levels of SIK1 were significantly reduced in patients expressing high levels of miR‐223, while they were almost undetectable in relapsed patients, in which very high level of miR‐223 were present, altogether pointing to a possible role as onco‐miR for miR‐223.
Together, the literature on hematological cancers shows that miR‐223 is extensively involved in the pathogenesis and progression of these diseases, being either down‐regulated or up‐regulated.
3.2. Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) represents the fourth most common cause of cancer‐related death worldwide and it is well reported that altered miRs expression is associated with both physiological and pathological liver conditions, impacting on liver metabolism, injury, fibrosis and, eventually, also on tumor development (X. Wang et al., 2021; J. D. Yang et al., 2019).
In alcohol‐associated liver disease (ALD) and in non‐alcoholic fatty liver disease (NAFLD) patients or mouse models miR‐223 levels are elevated in serum and liver. However, ablation of miR‐223 expression exacerbated ALD (M. Li, He, et al., 2017) and NAFLD (He et al., 2019) in mice. Accordingly, the murine model of miR‐223 KO fed with a high‐fat diet (HFD) displayed increased NAFLD that could evolve into non‐alcoholic steatohepatitis (NASH), liver fibrosis, higher levels of hepatic neutrophils and macrophages, inflammatory cytokines, proliferation markers and HCC markers. After 1 year of HFD, this pre‐tumoral condition evolved in HCC in a much higher rate in miR‐223 KO mice compared with wild‐type ones (He et al., 2019). This could be due, at mechanistic level, to the fact that miR‐223 inhibits IL‐6 expression and, subsequently, attenuates the IL‐6 pathway in neutrophils, thereby protecting against ALD (M. Li, He, et al., 2017). Reduction of miR‐223 led to subsequent upregulation of the miR‐223 target gene transcriptional activator with PDZ‐binding motif (TAZ), a well‐known factor to promote NASH fibrosis (Hou et al., 2020). Mice with a deletion of the granulocyte‐specific miR‐223 gene showed an impairment of the phenotypic switch of proinflammatory macrophages into a restorative profile that could be reversed by treating animals with miR‐223‐3p mimic or by infusion of neutrophils from wild‐type mice (Calvente et al., 2019; He et al., 2021). Just recently, He et al. demonstrated that neutrophils were able to deliver miR‐223 in hepatocytes by secreting miR‐223‐enriched extracellular vesicles (EV) and, once internalized by hepatocytes, the EV‐derived miR‐223 acted to inhibit hepatic inflammatory. However, if this uptake was for any reason impaired, the progression of steatosis to NASH was accelerated (He et al., 2021).
miR‐223 is commonly repressed in HCC and cell lines and it is known to modulate cell viability interacting with many different targets (Y. W. Dong et al., 2014; X. Wang et al., 2021; Wong et al., 2008; Guifang Yu et al., 2016). In vitro overexpression of miR‐223 significantly suppressed cell proliferation and induced apoptosis in HCC cell lines (Z. Dong et al., 2017). Overexpression of miR‐223 induced downregulation of mTOR and p70S6K signaling axis activation and decreased levels of the antiapoptotic protein Bcl‐2. This modulation of the mTOR pathway and Bcl‐2 was mediated by Rab1, a member of the Rab small GTPase family, that was a confirmed target of miR‐223 in HCC (Z. Dong et al., 2017).
Another well‐established target of miR‐223 in HCC is stathmin 1, a cytosolic phosphoprotein acting as key regulator of microtubule dynamics and whose expression in tumors and cell lines inversely correlated with the one of miR‐223 (Wong et al., 2008). Other studies pinpointed that miR‐223 levels were particularly low in highly metastatic HCC cell lines and its overexpression hampered their migration in vitro (Y. W. Dong et al., 2014). The modulation of αv integrin by miR‐223 partially accounted for this behavior. Accordingly, both cell lines and patient samples displayed an inverse correlation between miR‐223 and αv integrin levels, with enhanced miR‐223 expression that impaired the αv integrin‐mediated cell migration (Y. W. Dong et al., 2014). Previous studies reported that sulfatide, a class of glycolipids that contains a sulfate group, induced the expression of αv integrin (Zhong Wu et al., 2004) and was also able to downregulate miR‐223 expression through decreased recruitment of acetylated histone H3 and C/EBPα to its promoter, unveiling a novel regulator of miR‐223 expression (Y. W. Dong et al., 2014). Moreover, miR‐223 has been implicated in the modulation of ABCB1, also known as multidrug resistance protein 1, reducing the accumulation of drugs and inducing multidrug resistance (T. Yang et al., 2013). Yang et al. demonstrated that ABCB1 is a target of miR‐223 in HCC and overexpression of miR‐223 in vitro increased sensitivity to doxorubicin and paclitaxel by suppressing ABCB1 expression, while its inhibition showed the opposite effect (T. Yang et al., 2013). Finally, a downmodulation of miR‐223 expression was observed as part of a six‐miRNA signature that was reported as independent predictor of overall survival and recurrence‐free survival in HCC patients (Han et al., 2012).
3.3. Gastro‐esophageal cancer
Gastric cancer (GC) is the third cause of cancer‐related death worldwide and its incidence, as well as its clinicopathologic features, differ among geographical regions, highlighting how GC is a heterogeneous group of diseases whose development is caused by a wide number of predisposing and etiologic factors (Russo & Strong, 2019). It is known that GC cells and tissues display increased levels of miR‐223‐3p in comparison to healthy controls and these levels correlate with tumor invasion, lymph node metastasis and recurrence risk (X. Li et al., 2011; X. Zhou et al., 2015; Zhu et al., 2020). Accordingly, miR‐223‐3p has been found inside exosomes deriving from peritoneal fluids of metastatic GC patients, correlating with peritoneal cancer index (Ohzawa et al., 2020). miR‐223 can exert its oncogenic role in gastric cancer through different pathways, some of which are not fully clarified. In vitro experiments demonstrated that miR‐223 was upregulated upon Helicobacter pylori infection and this upregulation was NF‐κB‐dependent (Link et al., 2015; F. Yang, Xu, et al., 2018). Interestingly, miR‐223 overexpression significantly dropped following the bacterium eradication, suggesting that miR‐223 upregulation could result, at least in part, from the inflammatory infiltrate triggered by the H. pylori infection. NF‐κB bound and activated miR‐223 promoter and pre‐treatment with NF‐κB inhibitor or its silencing abrogated miR‐223 upregulation. High levels of miR‐223‐3p enhanced GC cell proliferation and migration, via direct modulation of ARID1A (AT‐Rich Interaction Domain 1A), a regulator of transcription, member of the of the SWI/SNF family (F. Yang, Xu, et al., 2018). Twist is a pro‐migratory factor dysregulated in GC, whose high expression levels correlate with metastasis in gastric cancer (Yan‐Qi et al., 2007). In vitro experiments demonstrated that Twist bound and activated the putative promoter of miR‐223, which, in turn, targeted EPB41L3 (Erythrocyte Membrane Protein Band 4.1 Like 3), a tumor suppressor gene that modulates the activity of protein arginine N‐methyltransferases. The Twist/miR‐223/EPB41L3 axis may, at least in part, account for the role of miR‐223 in GC metastasis (X. Li et al., 2011).
The potential role of miR‐223 in GC drug resistance has been also investigated. FBXW7, a target of miR‐223 in GC, has been involved in both cisplatin and trastuzumab miR‐223‐mediated drug resistance (Eto et al., 2015; X. Zhou et al., 2015). Accordingly, miR‐223 and FBXW7 expression displayed an inverse correlation both in cells and tissue samples from GC patients. Inhibition of miR‐223 or FBXW7 overexpression partially restored the sensitivity to cisplatin in resistant cell lines, increasing cell‐cycle arrest and apoptosis, pointing to the miR‐223/FBXW7 axis as a predictive marker of cisplatin response and a novel target for GC patients (X. Zhou et al., 2015). This regulatory axis also modulated trastuzumab response. Overexpression of miR‐223 reduced FBXW7 levels and the sensitivity of HER2‐positive GC cells to trastuzumab, while suppression of miR‐223 led to the opposite effects (Eto et al., 2015). Collectively, these results highlight a key role of miR‐223/FBXW7 axis in regulating drug resistance in GC cells.
In contrast with all GC studies cited above, Kang et al. report a low expression of miR‐223 in GC and, as observed in hepatocellular carcinoma, an inverse correlation with the microtubule destabilizing protein, stathmin 1, upregulated in both GC cell lines and primary gastric adenocarcinomas (Kang et al., 2012).
In line with what has been reported in most GC studies, esophageal cancer also displays an up‐regulation of miR‐223. An early and progressive miR‐223 upregulation during the different steps of gastric mucosa transformation and during Barrett esophageal adenocarcinoma (BAc) evolution was reported, with a significant overexpression in Barrett mucosa in comparison to naïve esophageal mucosa (Fassan et al., 2017; Streppel et al., 2013). Accordingly, also the circulating levels of miR‐223 were higher in GC and BAc patients, suggesting the measurement of circulating miR‐223 as potential early biomarker for these pathologies (Fassan et al., 2017). Notably, the authors were able to demonstrate that the epithelial compartment of BAc expressed miR‐223, displaying higher migratory and invasive properties in cell autonomous manner (Fassan et al., 2017; Streppel et al., 2013). Although the progressive upregulation of miR‐223 during BAc pathogenesis seemed to contribute to the aggressive phenotype, the authors propose that, due to miR‐223 regulation of PARP1 in this context, these patients may benefit from DNA‐damaging chemotherapeutic treatments (Streppel et al., 2013). The fact that longstanding inflammation is often the leitmotiv in GC and BAc development and that miR‐223 plays a driving role as “oncomiR” in these two cancer types, suggests that inflammation‐driven miR‐223 may modulate targets and trigger effects in a different way from what it does in other contexts.
Finally, inadequate dietary zinc intake is implicated in the etiology of esophageal squamous cell carcinoma (ESCC) in many populations, including people with heavy alcohol consumption. Notably, prolonged zinc deficiency also leads to a cancer‐associated inflammatory program, that first results in esophageal hyperplasia and then, upon subsequent exposure to environmental carcinogens, evolves to ESCC (Fong et al., 2016). Strong positive staining for miR‐223, accompanied by downregulation of its target FBXW7, was detected in moderately to poorly differentiated ESCC samples, whereas the staining was almost absent in the normal mucosa adjacent to the tumor site (Fong et al., 2016). Together, these results identified a miRNA signature that may underlie the molecular pathogenesis of ESCC in zinc‐deficient population (Fong et al., 2016) and further support the different role played by miR‐223 in inflammatory contexts.
3.4. Breast cancer
Dysregulated miR‐223 expression and function have been observed in many other cancer types, such as ovarian, cervical, head and neck carcinomas and sarcomas, often sharing common mechanisms of regulation. However, its precise role has been often less investigated and it is therefore not completely clear.
Here, we will now focus on the reported role(s) of miR‐223 in breast cancer, in which miR‐223 plays mainly oncosuppressive functions.
Breast cancer (BC) represents the first most common cause of cancer‐related death in women worldwide. As it is a highly heterogeneous disease, treatment strategies depend on the molecular subtype (Harbeck et al., 2019). Indeed, expression profile of miRNAs has also been proposed to distinguish between cancer and healthy samples, but also to classify specific molecular subtypes of breast cancer including HER2, Luminal A, Luminal B, and TNBC. As expected, many miRNAs have been deeply involved in the regulation of different pathway in BC, such as those involved in proliferation, apoptosis, metastasis, recurrence, and drug resistance (Loh et al., 2019). At this regard, it has been reported that downregulation of miR‐223‐3p, as a part of a 4‐miR‐signature, is associated with HER2 status in the TCGA cohort (Kunc et al., 2020).
It has been reported that miR‐223 can target the epidermal growth factor (EGF) thereby dampening the activation of the EGF pathway, a key pathway in normal mammary gland development and breast cancer (Fabris et al., 2016). In BC patients, in the setting of surgery and intra‐operative radiotherapy (IORT), an up‐regulation of miR‐223 expression was observed in the peri‐tumoral normal mammary gland following IORT (Fabris et al., 2016). This peak of miR‐223 was able to attenuate EGF local release and the autocrine/paracrine EGF/EGFR signaling that is strongly induced by the surgical act (Fabris et al., 2016). Dampening of the EGF pathway, thanks to this miR‐223 local upregulation, eventually resulted in decreased survival of residual isolated cancer cells, left behind after surgery, which, in turn, translated into the improved recurrence‐free survival observable in IORT‐treated patients (Fabris et al., 2016). Accordingly, circulating miR‐223 levels were found to be diminished post‐surgically in matched samples harvested pre‐operatively and post‐operatively from BC patients (Kodahl et al., 2014; Kudela et al., 2020).
In recent work, the analysis of miR‐223 expression in a large cohort of different subtypes of BC patients demonstrated that miR‐223 is downregulated in BC, mainly in the luminal and HER2 positive subtypes (Citron et al., 2020). Furthermore, it was observed that miR‐223 levels decreased in the transition from healthy mammary tissue to ductal carcinoma in situ (DCIS) and, even further, to invasive ductal carcinoma (IDC), suggesting miR‐223 as a marker for identifying pre‐cancerous lesions that will likely progress to cancer (Citron et al., 2020). However, it is to note that Yoshikawa et al., who looked at circulating exosomal miR‐223‐3p levels, found these levels to be significantly higher in patients with IDC compared with DCIS (Yoshikawa et al., 2018), indicating that the use of miR‐223 as biomarker in this setting is still quite controversial.
At mechanistic level, it has been reported that miR‐223 impinges on BC cell proliferation in different ways. In vitro, oncogene activation mediated early miR‐223 suppression through the promotion of E2F1 activity; then, low miR‐223 levels further allowed for increased E2F1 expression, since E2F1 is also a target of miR‐223 (Armenia et al., 2014; Citron et al., 2020). Others recently reported that chaperonin‐containing TCP1 complex 3 (CCT‐3) may promote breast cancer cell proliferation by directly binding to miR‐223, thus weakening the regulation of miR‐223 on the Wnt/b‐catenin pathway (Qu et al., 2020).
miR‐223 together with other regulatory miRNAs inhibits proliferation and migration of BC cells in vitro, impairing the activity of STIM1 (Kulkarni et al., 2019). STIM1, a protein involved in controlling the entry of calcium ions into cells, displays high levels of expression in BC tissues and inversely correlate with miR‐223 ones (Y. Yang, Jiang, et al., 2018). Similarly, caprin1, a protein involved in the proliferation of human breast cancer cells, may be targeted by miR‐223 and the expression levels of caprin1 and miR‐223 inversely correlated in breast cancer cell lines (Gong et al., 2013).
The interaction between miR‐223 and NLRP3, critical in the control of the inflammation process, has been also demonstrated in BC. The inhibition of the NLRP3 inflammasome, driven by miR‐223‐3p, blocks the growth and the immunosuppressive ability of BC, both in vitro and in vivo, representing a new potential therapeutic strategy in BC management (L. Zhang et al., 2019).
Little is known about miR‐223 in triple‐negative breast cancer (TNBC). However, downregulation of miR‐223 in TNBC stem cells has been demonstrated and miR‐223 overexpression re‐sensitized TNBC stem cells to TRAIL‐induced apoptosis, by targeting HAX1 (HCLS1 associated protein X‐1), a protein known to associate with hematopoietic cell‐specific Lyn substrate 1, a substrate of Src family tyrosine kinases, and whose mutation results in autosomal recessive severe congenital neutropenia (X. Sun et al., 2016).
Finally, particularly in BC, an interesting role of miR‐223 has been unveiled in extracellular vesicles (EV), especially exosomes. EV can be secreted by the primary tumor, as well as by stromal and immune cells, and are now widely recognized as carriers of key messages that can affect sites both near and far from the primary tumor. This horizontal signaling may be responsible for formation of the pre‐metastatic niches, for promotion of quiescence and, also, for drug resistance (Lakshmi et al., 2021).
It has been reported that miR‐223 can be transmitted from the bone marrow stroma to the BC cells to induce dormancy of bone marrow metastases, via dampening of CXCL12 signaling (Lim et al., 2011). Furthermore, mesenchymal stem cells, primed by BC cells to release exosomes containing miR‐223, promoted quiescence in a subset of cancer cells and conferred drug resistance. The possibility to target these dormant BC cells, by systemic administration of MSC loaded with antagomiRs, has been proposed (Bliss et al., 2016).
Given the fact that these noncycling cells are substantially invisible to the immune system and to traditional therapies and that BC can recur even decades after initial diagnosis, implying that they display a very long‐term survival, the development of treatments targeting these dormant cells represent a very promising approach to eradicate BC.
3.5. miR‐223 and p53
TP53, the gene coding for the p53 tumor suppressor gene, is the most mutated gene in human cancer. Presence of TP53 mutations correlates with poor prognosis in many cancer types (Kandoth et al., 2013). p53 pathway is activated during normal cellular responses, such as senescence, apoptosis and stress responses, such as DNA damage (Lakin & Jackson, 1999). Most of TP53 mutations generate p53 variants that are no more capable to bind the classical p53 targets on DNA (loss‐of‐function). In addition, some mutants gain the ability to repress the activity of the wild‐type counterpart, either through interaction with other transcription factors or by direct binding to secondary DNA structures (gain‐of‐function) (Göhler et al., 2005; Kim & Lozano, 2018; Mantovani et al., 2019).
p53 can also bind and regulate miRNA expression and activity. Masciarelli et al. demonstrated that mutant p53 binds the miR‐223 promoter in breast cancer and colon cancer cell lines, suppressing its transcription (Masciarelli et al., 2014). However, this interaction only occurred in the presence of the transcriptional repressor ZEB‐1. Moreover, in vitro overexpression of miR‐223 in cell lines harboring mutant p53 sensitizes them to DNA‐damaging agents and decreased S phase accumulation or the percent of cells in S phase, promoted by the gain‐of‐function mutation of p53. Mutant p53‐mediated repression of miR‐223 eventually impacted on the expression of its target stathmin 1, a well‐recognized oncoprotein that eventually mediates cellular resistance to chemotherapeutic drugs (Masciarelli et al., 2014).
Also, in lung squamous cell carcinoma, mutant p53 binds to miR‐223 promoter and represses its transcription, thus increasing cell proliferation and migration. Furthermore, miR‐223‐3p was able to bind mutant p53, thereby creating a fine‐tuned feedback loop (Luo et al., 2019).
Together, all these data highlight the important role of miR‐223 in p53 pathway, suggesting a possible role of miR‐223 as therapeutic target and/or prognostic factor.
4. MIR‐223 AS POTENTIAL BIOMARKER
As summarized in Table 1, miR‐223 expression levels in tissues are actively regulated by several biological processes. Thus, it is conceivable that miR‐223 profile might be considered as diagnostic biomarker in many types of cancer. However, there are several obstacles limiting this approach, especially in terms of accessibility to tissue specimens. Interestingly, an increasing number of papers report that also the circulating levels of miR‐223 are associated with different steps of tumorigenesis, and they could be easily evaluated with molecular biology assays. Since microRNA molecules can also be secreted from cells and are remarkably stable in plasma, they have been frequently proposed as a new class of disease biomarkers. Although several technical and biological issues need to be overcome before a practical clinical application may be reached, they have most of the features that an excellent biomarker must possess. For this reason, the research in this field in general, and in the field of miR‐223 in particular, is very active during these days.
TABLE 1.
miR‐223 in cancer
| Cancer type | Expression | Pathway | Target(s) | Hallmark/process | References | |
|---|---|---|---|---|---|---|
| Up | Down | |||||
| T‐cell acute lymphoblastic leukemia (T‐ALL) | ↑ | Notch; NF‐kB | ARRB1 | Proliferation | Chiaretti et al. (2010); Mavrakis et al. (2011); Shu et al. (2020) | |
| ↑ | – | TAL1/FBXW7 | Proliferation | Sanda and Leong (2017); Mansour et al. (2013); Welcker and Clurman (2008); Onoyama et al. (2007); Kumar et al. (2014) | ||
| Acute myeloid leukemia (AML) | ↓ | C/EBPα | E2F1 | Differentiation and proliferation | Pulikkan et al. (2010) | |
| ↓ | AML1/ETO | – | Differentiation | Fazi et al. (2007) | ||
| Chronic lymphocytic leukemia (CLL) | ↓ | Heat shock proteins (HSPs) | HSP90B1 | Survival | Rodríguez‐Vicente et al. (2015); Castro et al. (2005) | |
| Chronic myeloid leukemia (CML) | ↓ | C/EBPα | MEF2C/PTBP2 | Granulocytic maturation | Perrotti et al. (2002); Stehling‐Sun et al. (2009); Agatheeswaran et al. (2013) | |
| Mantle cell lymphoma (MCL) | ↓ | – | SOX11 | Survival | Mozos et al. (2009); K. Zhou et al. (2018) | |
| T‐cell lymphoblastic lymphoma (T‐LBL) | ↑ | Notch 1 | FBXW7/SIK1 | Survival, proliferation, and metastasis | Mussolin et al. (2014); Shaw (2009) | |
| Hepatocellular carcinoma (HCC) | ↓ | mTOR/p70S6K; BCL‐2 | Rab1 | Survival and proliferation | Dong et al. (2017) | |
| ↓ | p53 | Stathmin 1 | Survival, proliferation, and metastasis | Wong et al. (2008); Han et al. (2012) | ||
| ↓ | – | ABCB1 | Drug response | Yang et al. (2013) | ||
| ↓ | C/EBPα | αv integrin | Drug response, migration, and metastasis | Dong et al. (2014); Zhong Wu et al. (2004) | ||
| Hepatocellular carcinoma (HCC), alcohol‐associated liver disease (ALD), non‐alcoholic fatty liver disease (NAFLD), non‐alcoholic steatohepatitis (NASH) | ↓ | IL‐6 | TAZ | Inflammation and proliferation | Li, He, et al. (2017); Li, Tan, et al. (2017); He et al. (2019); Hou et al. (2020); Calvente et al. (2019); He et al. (2021) | |
| Gastric cancer (GC) | ↑ | NF‐kB | ARID1A | Proliferation and migration | Link et al. (2015); F. Yang, Xu, et al. (2018) | |
| ↑ | Twist | EPB41L3 | Metastasis | Li et al. (2011) | ||
| ↑ | – | FBXW7 | Drug response | Zhou et al. (2015); Eto et al. (2015) | ||
| ↓ | p53 | Stathmin 1 | Survival, proliferation, and metastasis | Kang et al. (2012) | ||
| Esophageal squamous cell carcinoma (ESCC) | ↑ | DNA damage response | PARP1 | Transformation and drug response | Streppel et al. (2013) | |
| ↑ | Zn‐deficiency | FBXW7 | Transformation and progression | Fong et al. (2016) | ||
| Breast cancer (BC) | ↓ | EGFR | EGF/E2F1 | Survival and proliferation | Armenia et al. (2014); Fabris et al. (2016) | |
| ↓ | E2F1 | – | Drug response | Citron et al. (2020) | ||
| ↓ | Wnt/β‐Catenin | CCT‐3/β‐Catenin | Proliferation | Qu et al. (2020) | ||
| ↓ | Ca2+ entry | STIM1 | Migration | Kulkarni et al. (2019); Y. Yang, Jiang, et al. (2018) | ||
| ↓ | – | Caprin 1 | Proliferation and migration | Gong et al. (2013) | ||
| ↓ | NLRP3 | – | Proliferation and immunosuppression | Zhang et al. (2019) | ||
| ↓ | TRAIL | HAX1 | Survival | Sun et al. (2016) | ||
| ↓ | Quiescence, dormancy | CXCL12 | MSC migration and drug response | Lim et al. (2011); Bliss et al. (2016) | ||
| Breast cancer (BC) and melanoma | ↓ | p53/ZEB‐1 | Stathmin 1 | Proliferation and drug response | Masciarelli et al. (2014) | |
Note: Table reports the current literature describing the role of miR‐223 in different cancer types. Arrows indicate whether the miR‐223 is upregulated or downregulated in that tumor setting.
Regarding GC, in which miR‐223 levels are increased compared with normal tissue, it was first hypothesized that the plasma levels might show some alterations. In fact, many studies demonstrated that miR‐223 plasma levels are significantly higher in GC patients, but they are independent of the metastasis status (B. Li et al., 2012; Sierzega et al., 2017). Further analysis demonstrated that also Barrett esophageal adenocarcinoma patients display higher level of circulating miR‐223 in comparison to healthy control (Fassan et al., 2017). Moreover, circulating levels of miR‐223 seem to be regulated during the infection process, in fact they increase upon Helicobacter pylori infection both in GC patients and in healthy donors (B. Li et al., 2012). Other studies also showed that the peritoneal expression profile of miR‐223 reflected the tumor burden in GC patients with peritoneal metastasis, thus suggesting miR‐223 as both a biomarker and a target in the treatment of peritoneal metastasis (Tokuhisa et al., 2015).
On the other side, a wide number of studies focusing on tumors in which miR‐223 levels are decreased have reported the possibility to look at diminished circulating levels of miR‐223 as a biomarker of disease. For instance, in HCC patients miR‐223‐3p serum levels are low and could be exploited to discriminate between HCC patients, chronic liver injury patients and healthy donors (Bao et al., 2017; Bhattacharya et al., 2016; Giray et al., 2014; Khairy et al., 2016).
Interestingly, serum levels of miR‐223‐3p have been validated as reproducible and reliable biomarker of early‐stage non‐small cell lung cancer (NSCLC), independent of age and smoking habit (D'Antona et al., 2019). The evaluation of circulating miR‐223 levels was sufficient to discriminate with stages I–II NSCLC from tumor‐free controls. Many signatures have been proposed for detection of early‐stage NSCLC, most using by qRT‐PCR and predominantly comprising several miRs (from three to 32). However, for clinical transferability, a simple and consistently reproducible signature, including very few miRNAs, would be desirable. So, this study reached two mandatory objectives for the implementation of a valid biomarker. First, precision and reproducibility of measurement was guaranteed by the use of droplet digital PCR (ddPCR) in place of traditional qRT‐PCR. Moreover, the use of a training set of samples followed by validation in external cohorts reassured on the robustness of the findings and, thus, on translatability to clinical application. Second, they were able to identify an extremely simple and cost‐effective miR‐based signature, actually composed by the only miR‐223, which can be realistically transferable to a population‐screening setting.
Reliability, simplicity and feasibility are in fact the main features to be looked for, if the use of a new biomarker is to be implemented.
In AML patients, circulating levels of miR‐223 were found under‐expressed, especially in the intermediate and unfavorable cytogenetic risk groups. Moreover, low serum levels of miR‐223 have been correlated with aggressive clinical variables and shorter survival rates. Interestingly, AML patients display a significant increase in miR‐223 levels after treatment, suggesting a strong reliability of miR‐223 serum levels as diagnostic and prognostic biomarker in acute myeloid leukemia patients (Guopan Yu et al., 2020).
The properties of miR‐223 as biomarker have been investigated also in breast cancer. First, it has been shown that miR‐223 expression is progressively lost when analyzing healthy mammary tissue DCIS lesions and IDC (Citron et al., 2020). Since not all DCIS lesions evolve in IDC it would be extremely interesting to evaluate the possibility that loss of miR‐223 expression might correlate with clinical outcome, thus investigating a possible role of miR‐223 as predictive biomarker in the progression from DCIS to IDC.
With the advent of targeted therapies there is urgent need to find reliable biomarkers. On one side they are strongly needed to identify and select patients that may respond from those who may not; on the other, they can allow to follow‐up the response to the therapy. Recent data have shown that miR‐223 levels might represent a reliable marker of treatment efficacy for CDK4/6 inhibitor (Palbociclib). Upon treatment with Palbociclib, a rapid increase in miR‐223 expression was observed both in vitro, in BC cells, and in vivo, in Δ16HER2 mice, an aggressive mouse model of HER2+ BC (Citron et al., 2020). These preclinical results strongly stimulate the interest for the evaluation of circulating levels of miR‐223 in BC patients, in which it might be very useful to predict the clinical response to Palbociclib treatment.
However, if we think toward to the development of miR‐223 as early biomarker of mammary cell transformation or of response to anti‐tumoral treatment, such as to Palbociclib, many still unanswered questions will need to be answered. First of all, validation in larger cohorts of patients; then, specific identification of the cells that lose the expression of miR‐223 in the transition from in situ to invasive carcinoma; finally, evaluation of whether the circulating levels of miR‐223, free or loaded into exosomes, in breast cancer patients treated with Palbociclib robustly predicts response to this therapy; only after all these criteria are met, the robustness of miR‐223 as biomarker may be established.
It is to note that the feasibility of using miR‐223 serum levels as biomarker is not only being exploited in cancer patients, but also in a wide range of other pathologies, ranging from neurodegenerative diseases like Parkinson, Alzheimer and dementia (Jia & Liu, 2016; Mancuso et al., 2019; Wei et al., 2018) to arthritis (Dunaeva et al., 2018), diabetes (Parrizas et al., 2020) and bacterial peritonitis (Brook et al., 2019). Given the deep involvement of miR‐223 in inflammation and immune response, particularly in the respiratory tract (Bauernfeind et al., 2012; Dorhoi et al., 2013; Feng et al., 2017; Neudecker et al., 2017), it will not be surprising to discover that miR‐223 plays a central role in SARS‐CoV 2 pathogenesis and may be exploitable as either biomarker or therapeutic target.
In conclusion, the emerging role of miR‐223 as potential diagnostic, prognostic and/or treatment response biomarker confirms its pivotal function(s) in different pathologies. These data highlight that the assessment of miR‐223 serum level variations may be relevant and soon ready for a transfer to the clinical practice.
5. THERAPEUTIC POTENTIAL OF MIR‐223
The potential of miRs to selectively silence any gene of interest and their capacity to target multiple mRNAs makes them interesting candidates as both cancer therapeutics and therapeutic targets. Even more intriguingly, manipulation of miR expression may potentially enable the targeting of so far “undruggable” targets, making them potentially unique tools, in many cancer types. Therefore, the therapeutic potential of miRs is under investigation since many years now and the manipulation of their expression in cancer seems a tangible possibility in the near future. Many exosomal miRNAs, and among them miR‐223, have been reported in preclinical studies to have a potential therapeutic application. However, the translation of these studies into clinical trials and, ultimately, into real therapeutic utility, is still a long way to go (Lakshmi et al., 2021).
So far, the most advanced miR trial involves use of anti‐miR‐122 (Miravirsen) for hepatitis C therapy (Janssen et al., 2013). In cancer, most clinical trials that involve miRs are of the observational type, and miRs are used as biomarkers for patient stratification, prognosis, and drug efficacy, with breast cancer being the most advanced in the field (ClinicalTrials.gov; Hanna et al., 2019). Moreover, in most cases these trials are not specific for one miR, but rather involve a global expression‐profiling platform for the mining of most promising biomarkers in each setting.
However, in addition to these biomarker studies, miRNA and anti‐miRNA constructs are extensively under investigation as potential therapeutic agents for cancer. Both miRNA mimics and miRNA antagonists are synthetic double stranded small RNA molecules that can either restore or suppress miRNA expression and function, to re‐establish the normal balance. Currently, the main obstacle for the use of miRs in clinical practice is the availability of an effective system that could guarantee the delivery to the specific tissue and cell type, without being degraded by circulating nucleases abundantly present in the serum.
Given the diversity of roles played by miR‐223 in cancer, miRNA‐based therapeutics may need be applied either to restore miR‐223 expression (and function) or to suppress its abundance. Restoration may be achieved by the use of miRNA mimics and would be pursued in those contexts in which miR‐223 acts as tumor suppressor and its expression is lost, such as in breast cancer. On the other hand, suppression of its expression may be achieved by the use of miRNA antagonists and would be pursued in those contexts in which miR‐223 is overexpressed, such as in gastric cancer.
Many studies, both in vitro and in vivo, support the effectiveness of miR‐223‐based treatments. In 3D‐mammary acini formation assays, depletion of miR‐223 led to the loss of normal acini structure by normal mammary epithelial cells. On the other hand, miR‐223 overexpression in fully transformed BC cells led to the acquisition of smaller 3D‐structures with partially restored apico‐basal polarity, and to the growth of smaller HER2+ tumors, in vivo (Citron et al., 2020; Fabris et al., 2016). These data, together with the observation that DCIS express higher levels of miR‐223 than IDC (Citron et al., 2020), support the possibility that restoring miR‐223 expression in patients with DCIS may prevent the evolution of these lesions into invasive carcinomas.
Further findings also support the possibility that restoration of miR‐223 levels may be used not only to target tumor cells, but also to enhance the efficacy of other therapies, as reported for Palbociclib in breast cancer mouse models (Citron et al., 2020). Similarly, it was reported that ectopic modulation of miR‐223 expression subverted the sensitivity to γ‐secretase inhibitors (GSI) of GSI‐resistant T‐ALL cells (Kumar et al., 2014). On the contrary, ablation of miR‐223 could be an effective approach to re‐sensitize gastric cancer cells to cisplatin and trastuzumab (Eto et al., 2015, p. 7; X. Zhou et al., 2015, p. 7).
Altogether, the study of microRNA‐based therapies is still in progress but the knowledge has expanded at a remarkable rate. Once that nanoparticles for specific delivery are optimized, it is quite clear that miR clinical utility will become a reality in a very near future.
6. CONTROVERSIES AND CURRENT RESEARCH GAPS
Since miRNA regulation is highly tissue specific, it is possible (and quite likely) that the same miR may play different, or even opposite roles in different organs. Since dysregulation of miR expression may be caused by several mechanisms, including miR genes amplification or deletion, dysregulated transcriptional control, epigenetic changes, and defects in the miR biogenesis machinery, it is evident that they can function as either tumor suppressor or oncogene depending from the context and the circumstances (Peng & Croce, 2016).
miR‐223 is not an exception to this rule and while it clearly acts as a gatekeeper of cell proliferation in certain tumor types (e.g., breast), it promotes transformation in others (e.g., liver, gastric). Since the expression of miR‐223 is very high in the hematopoietic compartment and it actively participates to the innate immune response, the final action of miR‐223 in the context of cancer is even more complicated by the different extent by which the immune compartment is involved in the different cancer types. Inflammation may induce miR‐223 local upregulation even if the neoplastic cells per se are negative or low for miR‐223 expression. It is intriguing to note that when miR‐223 acts as “oncomiR” (see T‐ALL and gastric cancer) FBXW7 represents a frequent target, while p53/stathmin1 are more common targets in context where miR‐223 is downregulated (see hepatocellular carcinoma and breast cancer). Whether there is any significance of this in relation to the different types of cancer is not clear yet. However, it is conceivable that different triggering signals, such as pro‐inflammatory or pro‐proliferative or pro‐differentiation signals, not only act differently on miR‐223 expression but also depend on the cell‐specificity and tissue‐specificity, making the action of the same miR quite specific in each context.
The fact that miR‐223 can also be secreted by some cells (Bliss et al., 2016; Ohzawa et al., 2020; Tokuhisa et al., 2015; Yoshikawa et al., 2018) and then up‐taken by neighboring or distant ones, makes the context even more difficult to disentangle, if a use of miR‐223 as biomarker is envisaged.
These aspects, which regard all miRs in general and miR‐223 in particular, are something to keep well in mind when evaluating the possibility of using circulating miR for diagnostic/predictive purposes.
Finally, as mentioned above, the main barrier for the therapeutic usage of miRs is currently the lack of an effective delivery system. Nanoparticles efficiently protecting miRs from degradation and designed to effectively deliver high concentrations of active miRs to the target organs are strongly needed to take this last step toward clinical application.
Last, we have seen that miR‐223 can be exported from cells in extracellular vesicles. This mechanism is currently under deep investigation for its huge potential in diagnostic and therapeutic fields. However, once loaded onto exosomes, the miR can potentially reach any cell and organ of the body, in which, as we have described, the same miR can lead to even opposing consequences. Thus, the study of the potential systemic effects deriving from this transport will need to be taken into great account, since they may become apparent only when studied in patients.
7. CONCLUSION AND PERSPECTIVES
As summarized in Figure 1, miR‐223 can play many roles in different cancer types. With few exception, miR‐223 expression is often downmodulated during cellular transformation and cancer progression, supporting the possibility that miR‐223 mainly plays a tumor suppressive function in cancer control. However, the fact that gastro‐esophageal and some hematological cancers display the opposite behavior highlights that miR‐223 works in a highly context‐dependent manner and that different cell types may regulate and exploit miR‐223 function in different ways (Figure 1).
FIGURE 1.
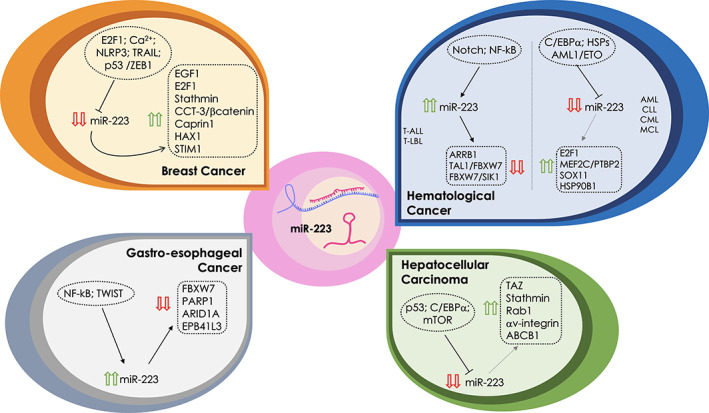
Roles of miR‐223 in cancer. The alternative molecular functions played by miR‐223 in different cancer types are depicted. Four main cancer types have been represented, that is, breast cancer (orange panel, upper left), hematological cancers (blue panel, upper right), hepatocellular carcinoma (green panel, lower right), and gastro‐esophageal cancer (gray panel, lower left). The molecular players that alter or are altered by miR‐223 are summarized in each panel. The expression levels of miR‐223 in each context are depicted by red (down) or green (up) arrows
The strong predictive and prognostic value of miRNA has been widely recognized in cancer and we expect that its role in precision medicine will soon be established.
In view of a therapeutic exploitation of miR‐223, as well as of other miRs, a challenge that will need to be faced is the development of efficient and specific delivery systems. Up to now, efficacy of miR‐based treatments is limited by poor delivery, targeting ability and high instability, due to miR degradation by nucleases in circulation. Several approaches have been studied to obtain an efficient delivery system, from chemical modification to vehicle complexation. Viral vectors or nonviral carriers have been tested but both of them show some disadvantages. Recent evidence suggests the possibility to use cell‐derived membrane vesicles, such as exosomes or microvesicles and, thanks to these advancements, some miR drug candidates are currently in clinical development or in phase 1 and 2 clinical trials (Y. Chen et al., 2015; Hanna et al., 2019).
Finally, an alternative way to overcome the limitation of inefficient miR delivery could certainly be the identification of the miR critical target(s), in each cancer setting. Since miR‐223, as well as every miR, has multiple targets, assessing their effective contribution to malignant transformation and progression, together with their druggability, could be of primary importance for the clinical exploitation of miR‐223.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Andrea Favero: Conceptualization; data curation; visualization; writing‐original draft. Ilenia Segatto: Data curation; visualization; writing‐original draft. Tiziana Perin: Data curation; writing‐review & editing. Barbara Belletti: Conceptualization; data curation; funding acquisition; supervision; visualization; writing‐review & editing.
RELATED WIREs ARTICLES
Artificial miRNAs as therapeutic tools: Challenges and opportunities
ACKNOWLEDGMENTS
We thank present and past members of the SCICC lab for their valuable contributions. We apologize to all colleagues whose work, for space limitations, was not cited. This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) to B. Belletti (IG 20061) and I. Segatto (#18171); by CRO Intramural Research Grant (5X1000_2016_MdS) to I. Segatto; by CRO Ricerca Corrente core grant of Ministero della Salute to B. Belletti (linea 1) and T. Perin (linea 4); by L.R. 17/2014‐Regione FVG (TNBCneo) to B. Belletti. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish these results.
Favero A, Segatto I, Perin T, Belletti B. The many facets of miR‐223 in cancer: Oncosuppressor, oncogenic driver, therapeutic target, and biomarker of response. WIREs RNA. 2021;12:e1659. 10.1002/wrna.1659
Edited by: Jeff Wilusz, Editor‐in‐Chief
Tiziana Perin and Barbara Belletti contributed equally to this study.
Funding information Associazione Italiana per la Ricerca sul Cancro, Grant/Award Numbers: #18171, IG 20061; CRO Intramural Research Grant, Grant/Award Number: 5X1000_2016_MdS; Ministero della Salute, Ricerca Corrente core grant; Regione Autonoma Friuli Venezia Giulia, Grant/Award Number: L.R. 17/2014‐Regione FVG (TNBCneo)
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Agatheeswaran, S. , Singh, S. , Biswas, S. , Biswas, G. , Chandra Pattnayak, N. , & Chakraborty, S. (2013). BCR‐ABL mediated repression of miR‐223 results in the activation of MEF2C and PTBP2 in chronic myeloid leukemia. Leukemia, 27(7), 1578–1580. 10.1038/leu.2012.339 [DOI] [PubMed] [Google Scholar]
- Armenia, J. , Fabris, L. , Lovat, F. , Berton, S. , Segatto, I. , D'Andrea, S. , Ivan, C. , Cascione, L. , Calin, G. A. , Croce, C. M. , Colombatti, A. , Vecchione, A. , Belletti, B. , & Baldassarre, G. (2014). Contact inhibition modulates intracellular levels of miR‐223 in a p27kip1‐dependent manner. Oncotarget, 5(5), 1185–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, S. , Zheng, J. , Li, N. , Huang, C. , Chen, M. , Cheng, Q. , Yu, K. , Chen, S. , Zhu, M. , & Shi, G. (2017). Serum microRNA levels as a noninvasive diagnostic biomarker for the early diagnosis of hepatitis B virus‐related liver fibrosis. Gut and Liver, 11(6), 860–869. 10.5009/gnl16560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind, F. , Rieger, A. , Schildberg, F. A. , Knolle, P. A. , Schmid‐Burgk, J. L. , & Hornung, V. (2012). NLRP3 Inflammasome activity is negatively controlled by miR‐223. The Journal of Immunology, 189(8), 4175–4181. 10.4049/jimmunol.1201516 [DOI] [PubMed] [Google Scholar]
- Bhattacharya, S. , Steele, R. , Shrivastava, S. , Chakraborty, S. , Di Bisceglie, A. M. , & Ray, R. B. (2016). Serum miR‐30e and miR‐223 as novel noninvasive biomarkers for hepatocellular carcinoma. The American Journal of Pathology, 186(2), 242–247. 10.1016/j.ajpath.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss, S. A. , Sinha, G. , Sandiford, O. A. , Williams, L. M. , Engelberth, D. J. , Guiro, K. , Isenalumhe, L. L. , Greco, S. J. , Ayer, S. , Bryan, M. , Kumar, R. , Ponzio, N. M. , & Rameshwar, P. (2016). Mesenchymal stem cell‐derived Exosomes stimulate cycling quiescence and early breast cancer dormancy in bone marrow. Cancer Research, 76(19), 5832–5844. 10.1158/0008-5472.CAN-16-1092 [DOI] [PubMed] [Google Scholar]
- Brook, A. C. , Jenkins, R. H. , Clayton, A. , Kift‐Morgan, A. , Raby, A.‐C. , Shephard, A. P. , Mariotti, B. , Cuff, S. M. , Bazzoni, F. , Bowen, T. , Fraser, D. J. , & Eberl, M. (2019). Neutrophil‐derived miR‐223 as local biomarker of bacterial peritonitis. Scientific Reports, 9(1), 10136. 10.1038/s41598-019-46585-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvente, C. J. , Tameda, M. , Johnson, C. D. , Del Pilar, H. , Lin, Y. C. , Adronikou, N. , De Mollerat Du Jeu, X. , Llorente, C. , Boyer, J. , & Feldstein, A. E. (2019). Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA‐223. The Journal of Clinical Investigation, 129(10), 4091–4109. 10.1172/JCI122258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, J. E. , Prada, C. E. , Loria, O. , Kamal, A. , Chen, L. , Burrows, F. J. , & Kipps, T. J. (2005). ZAP‐70 is a novel conditional heat shock protein 90 (Hsp90) client: Inhibition of Hsp90 leads to ZAP‐70 degradation, apoptosis, and impaired signaling in chronic lymphocytic leukemia. Blood, 106(7), 2506–2512. 10.1182/blood-2005-03-1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.‐Z. , Li, L. , Lodish, H. F. , & Bartel, D. P. (2004). MicroRNAs modulate hematopoietic lineage differentiation. Science (New York, N.Y.), 303(5654), 83–86. 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- Chen, Q. , Wang, H. , Liu, Y. , Song, Y. , Lai, L. , Han, Q. , Cao, X. , & Wang, Q. (2012). Inducible microRNA‐223 down‐regulation promotes TLR‐triggered IL‐6 and IL‐1β production in macrophages by targeting STAT3. PLoS One, 7(8), e42971. 10.1371/journal.pone.0042971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Gao, D.‐Y. , & Huang, L. (2015). In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Advanced Drug Delivery Reviews, 81, 128–141. 10.1016/j.addr.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaretti, S. , Messina, M. , Tavolaro, S. , Zardo, G. , Elia, L. , Vitale, A. , Fatica, A. , Gorello, P. , Piciocchi, A. , Scappucci, G. , Bozzoni, I. , Fozza, C. , Candoni, A. , Guarini, A. , & Foà, R. (2010). Gene expression profiling identifies a subset of adult T‐cell acute lymphoblastic leukemia with myeloid‐like gene features and over‐expression of miR‐223. Haematologica, 95(7), 1114–1121. 10.3324/haematol.2009.015099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron, F. , Segatto, I. , Vinciguerra, G. L. R. , Musco, L. , Russo, F. , Mungo, G. , D'Andrea, S. , Mattevi, M. C. , Perin, T. , Schiappacassi, M. , Massarut, S. , Marchini, C. , Amici, A. , Vecchione, A. , Baldassarre, G. , & Belletti, B. (2020). Downregulation of miR‐223 expression is an early event during mammary transformation and confers resistance to CDK4/6 inhibitors in luminal breast cancer. Cancer Research, 80(5), 1064–1077. 10.1158/0008-5472.CAN-19-1793 [DOI] [PubMed] [Google Scholar]
- Cuesta Torres, L. F. , Zhu, W. , Öhrling, G. , Larsson, R. , Patel, M. , Wiese, C. B. , Rye, K.‐A. , Vickers, K. C. , & Tabet, F. (2019). High‐density lipoproteins induce miR‐223‐3p biogenesis and export from myeloid cells: Role of scavenger receptor BI‐mediated lipid transfer. Atherosclerosis, 286, 20–29. 10.1016/j.atherosclerosis.2019.04.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antona, P. , Cattoni, M. , Dominioni, L. , Poli, A. , Moretti, F. , Cinquetti, R. , Gini, E. , Daffrè, E. , Noonan, D. M. , Imperatori, A. , Rotolo, N. , & Campomenosi, P. (2019). Serum miR‐223: A validated biomarker for detection of early‐stage non‐small cell Lung cancer. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 28(11), 1926–1933. 10.1158/1055-9965.EPI-19-0626 [DOI] [PubMed] [Google Scholar]
- Dong, Y. W. , Wang, R. , Cai, Q. Q. , Qi, B. , Wu, W. , Zhang, Y. H. , & Wu, X. Z. (2014). Sulfatide epigenetically regulates miR‐223 and promotes the migration of human hepatocellular carcinoma cells. Journal of Hepatology, 60(4), 792–801. 10.1016/j.jhep.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Dong, Z. , Qi, R. , Guo, X. , Zhao, X. , Li, Y. , Zeng, Z. , Bai, W. , Chang, X. , Hao, L. , Chen, Y. , Lou, M. , Li, Z. , & Lu, Y. (2017). MiR‐223 modulates hepatocellular carcinoma cell proliferation through promoting apoptosis via the Rab1‐mediated mTOR activation. Biochemical and Biophysical Research Communications, 483(1), 630–637. 10.1016/j.bbrc.2016.12.091 [DOI] [PubMed] [Google Scholar]
- Dorhoi, A. , Iannaccone, M. , Farinacci, M. , Faé, K. C. , Schreiber, J. , Moura‐Alves, P. , Nouailles, G. , Mollenkopf, H.‐J. , Oberbeck‐Müller, D. , Jörg, S. , Heinemann, E. , Hahnke, K. , Löwe, D. , Del Nonno, F. , Goletti, D. , Capparelli, R. , & Kaufmann, S. H. E. (2013). MicroRNA‐223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. The Journal of Clinical Investigation, 123(11), 4836–4848. 10.1172/JCI67604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva, M. , Blom, J. , Thurlings, R. , & Pruijn, G. J. M. (2018). Circulating serum miR‐223‐3p and miR‐16‐5p as possible biomarkers of early rheumatoid arthritis. Clinical and Experimental Immunology, 193(3), 376–385. 10.1111/cei.13156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto, K. , Iwatsuki, M. , Watanabe, M. , Ishimoto, T. , Ida, S. , Imamura, Y. , Iwagami, S. , Baba, Y. , Sakamoto, Y. , Miyamoto, Y. , Yoshida, N. , & Baba, H. (2015). The sensitivity of gastric cancer to trastuzumab is regulated by the miR‐223/FBXW7 pathway. International Journal of Cancer, 136(7), 1537–1545. 10.1002/ijc.29168 [DOI] [PubMed] [Google Scholar]
- Fabris, L. , Berton, S. , Citron, F. , D'Andrea, S. , Segatto, I. , Nicoloso, M. S. , Massarut, S. , Armenia, J. , Zafarana, G. , Rossi, S. , Ivan, C. , Perin, T. , Vaidya, J. S. , Avanzo, M. , Roncadin, M. , Schiappacassi, M. , Bristow, R. G. , Calin, G. , Baldassarre, G. , & Belletti, B. (2016). Radiotherapy‐induced miR‐223 prevents relapse of breast cancer by targeting the EGF pathway. Oncogene, 35(37), 4914–4926. 10.1038/onc.2016.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassan, M. , Saraggi, D. , Balsamo, L. , Realdon, S. , Scarpa, M. , Castoro, C. , Coati, I. , Salmaso, R. , Farinati, F. , Guzzardo, V. , Arcidiacono, D. , Munari, G. , Gasparini, P. , Veronese, N. , Luchini, C. , Valeri, N. , & Rugge, M. (2017). Early miR‐223 upregulation in gastroesophageal carcinogenesis. American Journal of Clinical Pathology, 147(3), 301–308. 10.1093/ajcp/aqx004 [DOI] [PubMed] [Google Scholar]
- Fazi, F. , Racanicchi, S. , Zardo, G. , Starnes, L. M. , Mancini, M. , Travaglini, L. , Diverio, D. , Ammatuna, E. , Cimino, G. , Lo‐Coco, F. , Grignani, F. , & Nervi, C. (2007). Epigenetic silencing of the myelopoiesis regulator microRNA‐223 by the AML1/ETO oncoprotein. Cancer Cell, 12(5), 457–466. 10.1016/j.ccr.2007.09.020 [DOI] [PubMed] [Google Scholar]
- Fazi, F. , Rosa, A. , Fatica, A. , Gelmetti, V. , De Marchis, M. L. , Nervi, C. , & Bozzoni, I. (2005). A minicircuitry comprised of microRNA‐223 and transcription factors NFI‐A and C/EBPalpha regulates human granulopoiesis. Cell, 123(5), 819–831. 10.1016/j.cell.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Fehniger, T. A. , Wylie, T. , Germino, E. , Leong, J. W. , Magrini, V. J. , Koul, S. , Keppel, C. R. , Schneider, S. E. , Koboldt, D. C. , Sullivan, R. P. , Heinz, M. E. , Crosby, S. D. , Nagarajan, R. , Ramsingh, G. , Link, D. C. , Ley, T. J. , & Mardis, E. R. (2010). Next‐generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Research, 20(11), 1590–1604. 10.1101/gr.107995.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Qi, S. , Zhang, Y. , Qi, Z. , Yan, L. , Zhou, J. , He, F. , Li, Q. , Yang, Y. , Chen, Q. , Xiao, S. , Li, Q. , Chen, Y. , & Zhang, Y. (2017). Ly6G+ neutrophil‐derived miR‐223 inhibits the NLRP3 inflammasome in mitochondrial DAMP‐induced acute lung injury. Cell Death & Disease, 8(11), e3170–e3170. 10.1038/cddis.2017.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, L. Y. , Taccioli, C. , Jing, R. , Smalley, K. J. , Alder, H. , Jiang, Y. , Fadda, P. , Farber, J. L. , & Croce, C. M. (2016). MicroRNA dysregulation and esophageal cancer development depend on the extent of zinc dietary deficiency. Oncotarget, 7(10), 10723–10738. 10.18632/oncotarget.7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau, N. L. , Wilusz, J. , & Wilusz, C. J. (2007). The highways and byways of mRNA decay. Nature Reviews Molecular Cell Biology, 8(2), 113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Gebert, L. F. R. , & MacRae, I. J. (2019). Regulation of microRNA function in animals. Nature Reviews Molecular Cell Biology, 20(1), 21–37. 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner, B. , Pochert, N. , Rouhi, A. , Boccalatte, F. , Plati, T. , Berg, T. , Sun, S. M. , Mah, S. M. , Mirkovic‐Hösle, M. , Ruschmann, J. , Muranyi, A. , Leierseder, S. , Argiropoulos, B. , Starczynowski, D. T. , Karsan, A. , Heuser, M. , Hogge, D. , Camargo, F. D. , Engelhardt, S. , … Kuchenbauer, F. (2015). MicroRNA‐223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Experimental Hematology, 43(10), 858–868.e7. 10.1016/j.exphem.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giray, B. G. , Emekdas, G. , Tezcan, S. , Ulger, M. , Serin, M. S. , Sezgin, O. , Altintas, E. , & Tiftik, E. N. (2014). Profiles of serum microRNAs; miR‐125b‐5p and miR223‐3p serve as novel biomarkers for HBV‐positive hepatocellular carcinoma. Molecular Biology Reports, 41(7), 4513–4519. 10.1007/s11033-014-3322-3 [DOI] [PubMed] [Google Scholar]
- Göhler, T. , Jäger, S. , Warnecke, G. , Yasuda, H. , Kim, E. , & Deppert, W. (2005). Mutant p53 proteins bind DNA in a DNA structure‐selective mode. Nucleic Acids Research, 33(3), 1087–1100. 10.1093/nar/gki252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, B. , Hu, H. , Chen, J. , Cao, S. , Yu, J. , Xue, J. , Chen, F. , Cai, Y. , He, H. , & Zhang, L. (2013). Caprin‐1 is a novel microRNA‐223 target for regulating the proliferation and invasion of human breast cancer cells. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 67(7), 629–636. 10.1016/j.biopha.2013.06.006 [DOI] [PubMed] [Google Scholar]
- Han, Z.‐B. , Zhong, L. , Teng, M.‐J. , Fan, J.‐W. , Tang, H.‐M. , Wu, J.‐Y. , Chen, H.‐Y. , Wang, Z.‐W. , Qiu, G.‐Q. , & Peng, Z.‐H. (2012). Identification of recurrence‐related microRNAs in hepatocellular carcinoma following liver transplantation. Molecular Oncology, 6(4), 445–457. 10.1016/j.molonc.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneklaus, M. , Gerlic, M. , O'Neill, L. A. J. , & Masters, S. L. (2013). miR‐223: Infection, inflammation and cancer. Journal of Internal Medicine, 274(3), 215–226. 10.1111/joim.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, J. , Hossain, G. S. , & Kocerha, J. (2019). The potential for microRNA therapeutics and clinical research. Frontiers in Genetics, 10, 478. 10.3389/fgene.2019.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbeck, N. , Penault‐Llorca, F. , Cortes, J. , Gnant, M. , Houssami, N. , Poortmans, P. , Ruddy, K. , Tsang, J. , & Cardoso, F. (2019). Breast cancer. Nature Reviews Disease Primers, 5(1), 66. 10.1038/s41572-019-0111-2 [DOI] [PubMed] [Google Scholar]
- He, Y. , Hwang, S. , Cai, Y. , Kim, S.‐J. , Xu, M. , Yang, D. , Guillot, A. , Feng, D. , Seo, W. , Hou, X. , & Gao, B. (2019). MicroRNA‐223 ameliorates nonalcoholic Steatohepatitis and cancer by targeting multiple inflammatory and oncogenic genes in hepatocytes. Hepatology (Baltimore, MD), 70(4), 1150–1167. 10.1002/hep.30645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Rodrigues, R. M. , Wang, X. , Seo, W. , Ma, J. , Hwang, S. , Fu, Y. , Trojnár, E. , Mátyás, C. , Zhao, S. , Ren, R. , Feng, D. , Pacher, P. , Kunos, G. , & Gao, B. (2021). Neutrophil‐to‐hepatocyte communication via LDLR‐dependent miR‐223–enriched extracellular vesicle transfer ameliorates nonalcoholic steatohepatitis. Journal of Clinical Investigation, 131(3), e141513. 10.1172/JCI141513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, A. M. , & Wilusz, J. (2018). The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harbor Perspectives in Biology, 10(5), a032839. 10.1101/cshperspect.a032839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X. , Yin, S. , Ren, R. , Liu, S. , Yong, L. , Liu, Y. , Li, Y. , Zheng, M.‐H. , Kunos, G. , Gao, B. , & Wang, H. (2020). Myeloid cell‐specific IL‐6 signaling promotes miR‐223‐enriched exosome production to attenuate NAFLD‐associated fibrosis. Hepatology (Baltimore, MD). 10.1002/hep.31658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, H. L. A. , Reesink, H. W. , Lawitz, E. J. , Zeuzem, S. , Rodriguez‐Torres, M. , Patel, K. , van der Meer, A. J. , Patick, A. K. , Chen, A. , Zhou, Y. , Persson, R. , King, B. D. , Kauppinen, S. , Levin, A. A. , & Hodges, M. R. (2013). Treatment of HCV infection by targeting MicroRNA. New England Journal of Medicine, 368(18), 1685–1694. 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- Jeffries, J. , Zhou, W. , Hsu, A. Y. , & Deng, Q. (2019). MiRNA‐223 at the crossroads of inflammation and cancer. Cancer Letters, 451, 136–141. 10.1016/j.canlet.2019.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, L.‐H. , & Liu, Y.‐N. (2016). Downregulated serum miR‐223 servers as biomarker in Alzheimer's disease. Cell Biochemistry and Function, 34(4), 233–237. 10.1002/cbf.3184 [DOI] [PubMed] [Google Scholar]
- Johnnidis, J. B. , Harris, M. H. , Wheeler, R. T. , Stehling‐Sun, S. , Lam, M. H. , Kirak, O. , Brummelkamp, T. R. , Fleming, M. D. , & Camargo, F. D. (2008). Regulation of progenitor cell proliferation and granulocyte function by microRNA‐223. Nature, 451(7182), 1125–1129. 10.1038/nature06607 [DOI] [PubMed] [Google Scholar]
- Kandoth, C. , McLellan, M. D. , Vandin, F. , Ye, K. , Niu, B. , Lu, C. , Xie, M. , Zhang, Q. , McMichael, J. F. , Wyczalkowski, M. A. , Leiserson, M. D. M. , Miller, C. A. , Welch, J. S. , Walter, M. J. , Wendl, M. C. , Ley, T. J. , Wilson, R. K. , Raphael, B. J. , & Ding, L. (2013). Mutational landscape and significance across 12 major cancer types. Nature, 502(7471), 333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, W. , Tong, J. H. M. , Chan, A. W. H. , Lung, R. W. M. , Chau, S. L. , Wong, Q. W. L. , Wong, N. , Yu, J. , Cheng, A. S. L. , & To, K. F. (2012). Stathmin1 plays oncogenic role and is a target of microRNA‐223 in gastric cancer. PLoS One, 7(3), e33919. 10.1371/journal.pone.0033919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairy, A. , Hamza, I. , Shaker, O. , & Yosry, A. (2016). Serum miRNA panel in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. Asian Pacific Journal of Cancer Prevention: APJCP, 17(5), 2699–2703. [PubMed] [Google Scholar]
- Kim, M. P. , & Lozano, G. (2018). Mutant p53 partners in crime. Cell Death and Differentiation, 25(1), 161–168. 10.1038/cdd.2017.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodahl, A. R. , Zeuthen, P. , Binder, H. , Knoop, A. S. , & Ditzel, H. J. (2014). Alterations in circulating miRNA levels following early‐stage estrogen receptor‐positive breast cancer resection in post‐menopausal women. PLoS One, 9(7), e101950. 10.1371/journal.pone.0101950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudela, E. , Samec, M. , Koklesova, L. , Liskova, A. , Kubatka, P. , Kozubik, E. , Rokos, T. , Pribulova, T. , Gabonova, E. , Smolar, M. , & Biringer, K. (2020). MiRNA expression profiles in luminal a breast cancer‐implications in biology, prognosis, and prediction of response to hormonal treatment. International Journal of Molecular Sciences, 21(20), 7691. 10.3390/ijms21207691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, R. P. , Elmi, A. , Alcantara‐Adap, E. , Hubrack, S. , Nader, N. , Yu, F. , Dib, M. , Ramachandran, V. , Najafi Shoushtari, H. , & Machaca, K. (2019). MiRNA‐dependent regulation of STIM1 expression in breast cancer. Scientific Reports, 9(1), 13076. 10.1038/s41598-019-49629-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V. , Palermo, R. , Talora, C. , Campese, A. F. , Checquolo, S. , Bellavia, D. , Tottone, L. , Testa, G. , Miele, E. , Indraccolo, S. , Amadori, A. , Ferretti, E. , Gulino, A. , Vacca, A. , & Screpanti, I. (2014). Notch and NF‐kB signaling pathways regulate miR‐223/FBXW7 axis in T‐cell acute lymphoblastic leukemia. Leukemia, 28(12), 2324–2335. 10.1038/leu.2014.133 [DOI] [PubMed] [Google Scholar]
- Kunc, M. , Popęda, M. , Niemira, M. , Szałkowska, A. , Bieńkowski, M. , Pęksa, R. , Łacko, A. , Radecka, B. S. , Braun, M. , Pikiel, J. , Litwiniuk, M. , Pogoda, K. , Iżycka‐Świeszewska, E. , Krętowski, A. , Żaczek, A. J. , Biernat, W. , & Senkus‐Konefka, E. (2020). MicroRNA expression profile in single hormone receptor‐positive breast cancers is mainly dependent on HER2 status‐a pilot study. Diagnostics (Basel, Switzerland), 10(9), 617. 10.3390/diagnostics10090617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin, N. D. , & Jackson, S. P. (1999). Regulation of p53 in response to DNA damage. Oncogene, 18(53), 7644–7655. 10.1038/sj.onc.1203015 [DOI] [PubMed] [Google Scholar]
- Lakshmi, S. , Hughes, T. A. , & Priya, S. (2021). Exosomes and exosomal RNAs in breast cancer: A status update. European Journal of Cancer (Oxford, England: 1990), 144, 252–268. 10.1016/j.ejca.2020.11.033 [DOI] [PubMed] [Google Scholar]
- Li, B. , Zhao, Y. , Guo, G. , Li, W. , Zhu, E. , Luo, X. , Mao, X. , Zou, Q. , Yu, P. , Zuo, Q. , Li, N. , Tang, B. , Liu, K. , & Xiao, B. (2012). Plasma microRNAs, miR‐223, miR‐21 and miR‐218, as novel potential biomarkers for gastric cancer detection. PLoS One, 7(7), e41629. 10.1371/journal.pone.0041629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Tan, M. , Xiang, Q. , Zhou, Z. , & Yan, H. (2017). Thrombin‐activated platelet‐derived exosomes regulate endothelial cell expression of ICAM‐1 via microRNA‐223 during the thrombosis‐inflammation response. Thrombosis Research, 154, 96–105. 10.1016/j.thromres.2017.04.016 [DOI] [PubMed] [Google Scholar]
- Li, M. , He, Y. , Zhou, Z. , Ramirez, T. , Gao, Y. , Gao, Y. , Ross, R. A. , Cao, H. , Cai, Y. , Xu, M. , Feng, D. , Zhang, P. , Liangpunsakul, S. , & Gao, B. (2017). MicroRNA‐223 ameliorates alcoholic liver injury by inhibiting the IL‐6‐p47phox‐oxidative stress pathway in neutrophils. Gut, 66(4), 705–715. 10.1136/gutjnl-2016-311861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zhang, Y. , Zhang, H. , Liu, X. , Gong, T. , Li, M. , Sun, L. , Ji, G. , Shi, Y. , Han, Z. , Han, S. , Nie, Y. , Chen, X. , Zhao, Q. , Ding, J. , Wu, K. , & Daiming, F. (2011). MiRNA‐223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Molecular Cancer Research: MCR, 9(7), 824–833. 10.1158/1541-7786.MCR-10-0529 [DOI] [PubMed] [Google Scholar]
- Lim, P. K. , Bliss, S. A. , Patel, S. A. , Taborga, M. , Dave, M. A. , Gregory, L. A. , Greco, S. J. , Bryan, M. , Patel, P. S. , & Rameshwar, P. (2011). Gap junction‐mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Research, 71(5), 1550–1560. 10.1158/0008-5472.CAN-10-2372 [DOI] [PubMed] [Google Scholar]
- Link, A. , Schirrmeister, W. , Langner, C. , Varbanova, M. , Bornschein, J. , Wex, T. , & Malfertheiner, P. (2015). Differential expression of microRNAs in preneoplastic gastric mucosa. Scientific Reports, 5(1), 8270. 10.1038/srep08270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, H.‐Y. , Norman, B. P. , Lai, K.‐S. , Rahman, N. M. A. N. A. , Alitheen, N. B. M. , & Osman, M. A. (2019). The regulatory role of MicroRNAs in breast cancer. International Journal of Molecular Sciences, 20(19), 4940. 10.3390/ijms20194940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, P. , Wang, Q. , Ye, Y. , Zhang, J. , Lu, D. , Cheng, L. , Zhou, H. , Xie, M. , & Wang, B. (2019). MiR‐223‐3p functions as a tumor suppressor in lung squamous cell carcinoma by miR‐223‐3p‐mutant p53 regulatory feedback loop. Journal of Experimental & Clinical Cancer Research: CR, 38(1), 74. 10.1186/s13046-019-1079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso, R. , Agostini, S. , Hernis, A. , Zanzottera, M. , Bianchi, A. , & Clerici, M. (2019). Circulatory miR‐223‐3p discriminates between Parkinson's and Alzheimer's patients. Scientific Reports, 9(1), 9393. 10.1038/s41598-019-45687-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour, M. R. , Sanda, T. , Lawton, L. N. , Li, X. , Kreslavsky, T. , Novina, C. D. , Brand, M. , Gutierrez, A. , Kelliher, M. A. , Jamieson, C. H. M. , von Boehmer, H. , Young, R. A. , & Look, A. T. (2013). The TAL1 complex targets the FBXW7 tumor suppressor by activating miR‐223 in human T cell acute lymphoblastic leukemia. The Journal of Experimental Medicine, 210(8), 1545–1557. 10.1084/jem.20122516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, F. , Collavin, L. , & Del Sal, G. (2019). Mutant p53 as a guardian of the cancer cell. Cell Death and Differentiation, 26(2), 199–212. 10.1038/s41418-018-0246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciarelli, S. , Fontemaggi, G. , Di Agostino, S. , Donzelli, S. , Carcarino, E. , Strano, S. , & Blandino, G. (2014). Gain‐of‐function mutant p53 downregulates miR‐223 contributing to chemoresistance of cultured tumor cells. Oncogene, 33(12), 1601–1608. 10.1038/onc.2013.106 [DOI] [PubMed] [Google Scholar]
- Mavrakis, K. J. , Van Der Meulen, J. , Wolfe, A. L. , Liu, X. , Mets, E. , Taghon, T. , Khan, A. A. , Setty, M. , Setti, M. , Rondou, P. , Vandenberghe, P. , Delabesse, E. , Benoit, Y. , Socci, N. B. , Leslie, C. S. , Van Vlierberghe, P. , Speleman, F. , & Wendel, H.‐G. (2011). A cooperative microRNA‐tumor suppressor gene network in acute T‐cell lymphoblastic leukemia (T‐ALL). Nature Genetics, 43(7), 673–678. 10.1038/ng.858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozos, A. , Royo, C. , Hartmann, E. , De Jong, D. , Baró, C. , Valera, A. , Fu, K. , Weisenburger, D. D. , Delabie, J. , Chuang, S.‐S. , Jaffe, E. S. , Ruiz‐Marcellan, C. , Dave, S. , Rimsza, L. , Braziel, R. , Gascoyne, R. D. , Solé, F. , López‐Guillermo, A. , Colomer, D. , … Campo, E. (2009). SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1‐negative subtype. Haematologica, 94(11), 1555–1562. 10.3324/haematol.2009.010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolin, L. , Holmes, A. B. , Romualdi, C. , Sales, G. , D'Amore, E. S. G. , Ghisi, M. , Pillon, M. , Rosolen, A. , & Basso, K. (2014). An aberrant microRNA signature in childhood T‐cell lymphoblastic lymphoma affecting CDKN1B expression, NOTCH1 and growth factor signaling pathways. Leukemia, 28(9), 1909–1912. 10.1038/leu.2014.134 [DOI] [PubMed] [Google Scholar]
- Neudecker, V. , Brodsky, K. S. , Clambey, E. T. , Schmidt, E. P. , Packard, T. A. , Davenport, B. , Standiford, T. J. , Weng, T. , Fletcher, A. A. , Barthel, L. , Masterson, J. C. , Furuta, G. T. , Cai, C. , Blackburn, M. R. , Ginde, A. A. , Graner, M. W. , Janssen, W. J. , Zemans, R. L. , Evans, C. M. , … Eltzschig, H. K. (2017). Neutrophil transfer of miR‐223 to lung epithelial cells dampens acute lung injury in mice. Science Translational Medicine, 9(408), eaah5360. 10.1126/scitranslmed.aah5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, J. , Hayder, H. , Zayed, Y. , & Peng, C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology, 9, 402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, R. M. , Rao, D. S. , Chaudhuri, A. A. , & Baltimore, D. (2010). Physiological and pathological roles for microRNAs in the immune system. Nature Reviews Immunology, 10(2), 111–122. 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- Ohzawa, H. , Kumagai, Y. , Yamaguchi, H. , Miyato, H. , Sakuma, Y. , Horie, H. , Hosoya, Y. , Kawarai Lefor, A. , Sata, N. , & Kitayama, J. (2020). Exosomal microRNA in peritoneal fluid as a biomarker of peritoneal metastases from gastric cancer. Annals of Gastroenterological Surgery, 4(1), 84–93. 10.1002/ags3.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoyama, I. , Tsunematsu, R. , Matsumoto, A. , Kimura, T. , de Alborán, I. M. , Nakayama, K. , & Nakayama, K. I. (2007). Conditional inactivation of Fbxw7 impairs cell‐cycle exit during T cell differentiation and results in lymphomatogenesis. The Journal of Experimental Medicine, 204(12), 2875–2888. 10.1084/jem.20062299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrizas, M. , Mundet, X. , Castaño, C. , Canivell, S. , Cos, X. , Brugnara, L. , Giráldez‐García, C. , Regidor, E. , Mata‐Cases, M. , Franch‐Nadal, J. , & Novials, A. (2020). MiR‐10b and miR‐223‐3p in serum microvesicles signal progression from prediabetes to type 2 diabetes. Journal of Endocrinological Investigation, 43(4), 451–459. 10.1007/s40618-019-01129-z [DOI] [PubMed] [Google Scholar]
- Paul, P. , Chakraborty, A. , Sarkar, D. , Langthasa, M. , Rahman, M. , Bari, M. , Singha, R. S. , Malakar, A. K. , & Chakraborty, S. (2018). Interplay between miRNAs and human diseases. Journal of Cellular Physiology, 233(3), 2007–2018. 10.1002/jcp.25854 [DOI] [PubMed] [Google Scholar]
- Peng, Y. , & Croce, C. M. (2016). The role of MicroRNAs in human cancer. Signal Transduction and Targeted Therapy, 1, 15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti, D. , Cesi, V. , Trotta, R. , Guerzoni, C. , Santilli, G. , Campbell, K. , Iervolino, A. , Condorelli, F. , Gambacorti‐Passerini, C. , Caligiuri, M. A. , & Calabretta, B. (2002). BCR‐ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nature Genetics, 30(1), 48–58. 10.1038/ng791 [DOI] [PubMed] [Google Scholar]
- Pulikkan, J. A. , Dengler, V. , Peramangalam, P. S. , Peer Zada, A. A. , Müller‐Tidow, C. , Bohlander, S. K. , Tenen, D. G. , & Behre, G. (2010). Cell‐cycle regulator E2F1 and microRNA‐223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood, 115(9), 1768–1778. 10.1182/blood-2009-08-240101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, H. , Zhu, F. , Dong, H. , Hu, X. , & Han, M. (2020). Upregulation of CCT‐3 induces breast cancer cell proliferation through miR‐223 competition and Wnt/β‐catenin signaling pathway activation. Frontiers in Oncology, 10, 533176. 10.3389/fonc.2020.533176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Vicente, A. E. , Quwaider, D. , Benito, R. , Misiewicz‐Krzeminska, I. , Hernández‐Sánchez, M. , De Coca, A. G. , Fisac, R. , Alonso, J.‐M. , Zato, C. , De Paz, J. F. , García, J. L. , Sarasquete, M. E. , Hernández, J. Á. , Corchado, J. M. , González, M. , Gutiérrez, N. C. , & Hernández‐Rivas, J.‐M. (2015). MicroRNA‐223 is a novel negative regulator of HSP90B1 in CLL. BMC Cancer, 15, 238. 10.1186/s12885-015-1212-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, A. E. , & Strong, V. E. (2019). Gastric cancer etiology and Management in Asia and the west. Annual Review of Medicine, 70, 353–367. 10.1146/annurev-med-081117-043436 [DOI] [PubMed] [Google Scholar]
- Sanda, T. , & Leong, W. Z. (2017). TAL1 as a master oncogenic transcription factor in T‐cell acute lymphoblastic leukemia. Experimental Hematology, 53, 7–15. 10.1016/j.exphem.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Shaw, R. J. (2009). Tumor suppression by LKB1: SIK‐ness prevents metastasis. Science Signaling, 2(86), pe55. 10.1126/scisignal.286pe55 [DOI] [PubMed] [Google Scholar]
- Shu, Y. , Wang, Y. , Lv, W.‐Q. , Peng, D.‐Y. , Li, J. , Zhang, H. , Jiang, G.‐J. , Yang, B.‐J. , Liu, S. , Zhang, J. , Chen, Y.‐H. , Tang, S. , Wan, K.‐X. , Yuan, J.‐T. , Guo, W. , Fu, G. , Qi, X.‐K. , Liu, Z.‐D. , Liu, H.‐Y. , … Zou, L. (2020). ARRB1‐promoted NOTCH1 degradation is suppressed by OncomiR miR‐223 in T‐cell acute lymphoblastic leukemia. Cancer Research, 80(5), 988–998. 10.1158/0008-5472.CAN-19-1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierzega, M. , Kaczor, M. , Kolodziejczyk, P. , Kulig, J. , Sanak, M. , & Richter, P. (2017). Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of miR‐21 and miR‐331. British Journal of Cancer, 117(2), 266–273. 10.1038/bjc.2017.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatopoulos, B. , Meuleman, N. , Haibe‐Kains, B. , Saussoy, P. , Van Den Neste, E. , Michaux, L. , Heimann, P. , Martiat, P. , Bron, D. , & Lagneaux, L. (2009). MicroRNA‐29c and microRNA‐223 down‐regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood, 113(21), 5237–5245. 10.1182/blood-2008-11-189407 [DOI] [PubMed] [Google Scholar]
- Stehling‐Sun, S. , Dade, J. , Nutt, S. L. , DeKoter, R. P. , & Camargo, F. D. (2009). Regulation of lymphoid versus myeloid fate “choice” by the transcription factor Mef2c. Nature Immunology, 10(3), 289–296. 10.1038/ni.1694 [DOI] [PubMed] [Google Scholar]
- Streppel, M. M. , Pai, S. , Campbell, N. R. , Hu, C. , Yabuuchi, S. , Canto, M. I. , Wang, J. S. , Montgomery, E. A. , & Maitra, A. (2013). MicroRNA 223 is upregulated in the multistep progression of Barrett9s esophagus and modulates sensitivity to chemotherapy by targeting PARP1 . Clinical Cancer Research, 19(15), 4067–4078. 10.1158/1078-0432.CCR-13-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S.‐C. (2017). The non‐canonical NF‐κB pathway in immunity and inflammation. Nature Reviews Immunology, 17(9), 545–558. 10.1038/nri.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Li, Y. , Zheng, M. , Zuo, W. , & Zheng, W. (2016). MicroRNA‐223 increases the sensitivity of triple‐negative breast cancer stem cells to TRAIL‐induced apoptosis by targeting HAX‐1. PLoS One, 11(9), e0162754. 10.1371/journal.pone.0162754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, W. , Liu, B. , Qu, S. , Liang, G. , Luo, W. , & Gong, C. (2018). MicroRNAs and cancer: Key paradigms in molecular therapy. Oncology Letters, 15(3), 2735–2742. 10.3892/ol.2017.7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhisa, M. , Ichikawa, Y. , Kosaka, N. , Ochiya, T. , Yashiro, M. , Hirakawa, K. , Kosaka, T. , Makino, H. , Akiyama, H. , Kunisaki, C. , & Endo, I. (2015). Exosomal miRNAs from peritoneum lavage fluid as potential prognostic biomarkers of peritoneal metastasis in gastric cancer. PLoS One, 10(7), e0130472. 10.1371/journal.pone.0130472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber, T. , Treiber, N. , & Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nature Reviews Molecular Cell Biology, 20(1), 5–20. 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- Vickers, K. C. , Palmisano, B. T. , Shoucri, B. M. , Shamburek, R. D. , & Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nature Cell Biology, 13(4), 423–433. 10.1038/ncb2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wang, Q. , Kleiman, K. , Guo, C. , & Eitzman, D. T. (2017). Hematopoietic deficiency of miR‐223 attenuates thrombosis in response to photochemical injury in mice. Scientific Reports, 7(1), 1606. 10.1038/s41598-017-01887-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , He, Y. , Mackowiak, B. , & Gao, B. (2021). MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut, 70(4), 784–795. 10.1136/gutjnl-2020-322526 [DOI] [PubMed] [Google Scholar]
- Wei, H. , Xu, Y. , Xu, W. , Zhou, Q. , Chen, Q. , Yang, M. , Feng, F. , Liu, Y. , Zhu, X. , Yu, M. , & Li, Y. (2018). Serum Exosomal miR‐223 serves as a potential diagnostic and prognostic biomarker for dementia. Neuroscience, 379, 167–176. 10.1016/j.neuroscience.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Welcker, M. , & Clurman, B. E. (2008). FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Reviews Cancer, 8(2), 83–93. 10.1038/nrc2290 [DOI] [PubMed] [Google Scholar]
- Wong, Q. W.‐L. , Lung, R. W.‐M. , Law, P. T.‐Y. , Lai, P. B.‐S. , Chan, K. Y.‐Y. , To, K. , & Wong, N. (2008). MicroRNA‐223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology, 135(1), 257–269. 10.1053/j.gastro.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Yang, F. , Xu, Y. , Liu, C. , Ma, C. , Zou, S. , Xu, X. , Jia, J. , & Liu, Z. (2018). NF‐κB/miR‐223‐3p/ARID1A axis is involved in helicobacter pylori CagA‐induced gastric carcinogenesis and progression. Cell Death & Disease, 9(1), 12. 10.1038/s41419-017-0020-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. D. , Hainaut, P. , Gores, G. J. , Amadou, A. , Plymoth, A. , & Roberts, L. R. (2019). A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nature Reviews Gastroenterology & Hepatology, 16(10), 589–604. 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T. , Zheng, Z.‐M. , Li, X.‐N. , Li, Z.‐F. , Wang, Y. , Geng, Y.‐F. , Bai, L. , & Zhang, X.‐B. (2013). MiR‐223 modulates multidrug resistance via downregulation of ABCB1 in hepatocellular carcinoma cells. Experimental Biology and Medicine (Maywood, N.J.), 238(9), 1024–1032. 10.1177/1535370213497321 [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Jiang, Z. , Ma, N. , Wang, B. , Liu, J. , Zhang, L. , & Gu, L. (2018). MicroRNA‐223 targeting STIM1 inhibits the biological behavior of breast cancer. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology, 45(2), 856–866. 10.1159/000487180 [DOI] [PubMed] [Google Scholar]
- Yan‐Qi, Z. , Xue‐Yan, G. , Shuang, H. , Yu, C. , Fu‐Lin, G. , Fei‐Hu, B. , Shi‐Ren, S. , Xu‐Feng, W. , Jie, D. , & Dai‐Ming, F. (2007). Expression and significance of TWIST basic helix‐loop‐helix protein over‐expression in gastric cancer. Pathology, 39(5), 470–475. 10.1080/00313020701570053 [DOI] [PubMed] [Google Scholar]
- Ying, W. , Tseng, A. , Chang, R. C.‐A. , Morin, A. , Brehm, T. , Triff, K. , Nair, V. , Zhuang, G. , Song, H. , Kanameni, S. , Wang, H. , Golding, M. C. , Bazer, F. W. , Chapkin, R. S. , Safe, S. , & Zhou, B. (2015). MicroRNA‐223 is a crucial mediator of PPARγ‐regulated alternative macrophage activation. The Journal of Clinical Investigation, 125(11), 4149–4159. 10.1172/JCI81656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M. , Iinuma, H. , Umemoto, Y. , Yanagisawa, T. , Matsumoto, A. , & Jinno, H. (2018). Exosome‐encapsulated microRNA‐223‐3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncology Letters, 15(6), 9584–9592. 10.3892/ol.2018.8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Chen, X. , Chen, S. , Ye, W. , Hou, K. , & Liang, M. (2016). MiR‐19a, miR‐122 and miR‐223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. Journal of Translational Medicine, 14(1), 122. 10.1186/s12967-016-0888-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Yin, Z. , He, H. , Zheng, Z. , Chai, Y. , Xuan, L. , Lin, R. , Wang, Q. , Li, J. , & Xu, D. (2020). Low serum miR‐223 expression predicts poor outcome in patients with acute myeloid leukemia. Journal of Clinical Laboratory Analysis, 34(3), e23096. 10.1002/jcla.23096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, X. , Berg, N. , Lee, J. W. , Le, T.‐T. , Neudecker, V. , Jing, N. , & Eltzschig, H. (2018). MicroRNA miR‐223 as regulator of innate immunity. Journal of Leukocyte Biology, 104(3), 515–524. 10.1002/JLB.3MR0218-079R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. , Xia, L. , Fan, X. , Ostriker, A. C. , Yarovinsky, T. , Su, M. , Zhang, Y. , Peng, X. , Xie, Y. , Pi, L. , Gu, X. , Chung, S. K. , Martin, K. A. , Liu, R. , Hwa, J. , & Tang, W. H. (2019). Platelet‐derived miR‐223 promotes a phenotypic switch in arterial injury repair. Journal of Clinical Investigation, 129(3), 1372–1386. 10.1172/JCI124508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Jiang, M. , Qian, L. , Lin, X. , Song, W. , Gao, Y. , & Zhou, Y. (2020). The STAT3‐miR‐223‐TGFBR3/HMGCS1 axis modulates the progression of cervical carcinoma. Molecular Oncology, 14(9), 2313–2331. 10.1002/1878-0261.12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Li, H. , Zang, Y. , & Wang, F. (2019). NLRP3 inflammasome inactivation driven by miR‐223‐3p reduces tumor growth and increases anticancer immunity in breast cancer. Molecular Medicine Reports, 19(3), 2180–2188. 10.3892/mmr.2019.9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Wu, X. , Honke, K. , Long Zhang, Y. , Liang Zha, X. , & Taniguchi, N. (2004). Lactosylsulfatide expression in hepatocellular carcinoma cells enhances cell adhesion to vitronectin and intrahepatic metastasis in nude mice. International Journal of Cancer, 110(4), 504–510. 10.1002/ijc.20127 [DOI] [PubMed] [Google Scholar]
- Zhou, K. , Feng, X. , Wang, Y. , Liu, Y. , Tian, L. , Zuo, W. , Yi, S. , Wei, X. , Song, Y. , & Qiu, L. (2018). MiR‐223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Experimental Hematology, 58, 27–34. 10.1016/j.exphem.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Jin, W. , Jia, H. , Yan, J. , & Zhang, G. (2015). MiR‐223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. Journal of Experimental & Clinical Cancer Research: CR, 34, 28. 10.1186/s13046-015-0145-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Li, K. , Yan, L. , He, Y. , Wang, L. , & Sheng, L. (2020). MiR‐223‐3p promotes cell proliferation and invasion by targeting Arid1a in gastric cancer. Acta Biochimica et Biophysica Sinica, 52(2), 150–159. 10.1093/abbs/gmz151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


