Abstract
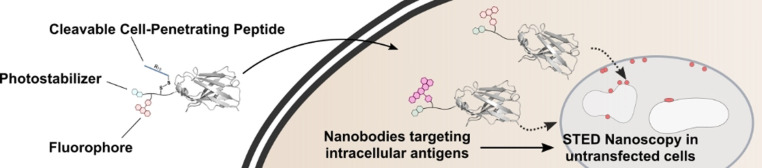
Super‐resolution microscopy in living cells can be restricted by the availability of small molecule probes, which only exist against few targets and genetically encoded tags. Here, we expand the applicability of live‐cell STED by engineering cell‐permeable and highly fluorescent nanobodies as intracellular targeting agents. To ensure bright fluorescent signals at low concentrations we used the concept of intramolecular photostabilization by ligating a fluorophore along with the photostabilizer trolox to the nanobody using expressed protein ligation (EPL). Furthermore, these semi‐synthetic nanobodies are equipped with a cleavable cell‐penetrating peptide for efficient cellular entry, which enables super‐resolution imaging of GFP and mCherry, as well as two endogenous targets, nuclear lamins and the DNA replication and repair protein PCNA. We monitored cell division and DNA replication via confocal and STED microscopy thus demonstrating the utility of these new intracellular tools for functional analysis.
Keywords: cell-penetrating peptides, nanobody, semi-synthesis, STED, super-resolution microscopy
Cell‐permeable nanobodies equipped with a fluorophore and a photostabilizer are introduced as powerful tools for live‐cell super‐resolution microscopy. By using intramolecular photostabilization dynamic processes can be monitored in untransfected living cells.
Introduction
Still a rather recent discovery, super‐resolution microscopy techniques have already established themselves as powerful methods in resolving biological structures at the nanoscale. [1] However, applying these techniques to living cells, especially for monitoring dynamic processes, poses unique challenges. [2] Out of all super‐resolution techniques, stimulated emission–depletion (STED) microscopy is particularly useful in living systems due to its flexible acquisition speed and infrared depletion lasers. [3] However, STED can be phototoxic when employing fluorescent proteins, which often have blue excitation and orange STED depletion wavelengths. This has led to the development of fluorescent proteins with near‐infrared excitation wavelengths. [4]
Small‐molecule organic fluorophores can have highly advantageous photophysical properties for STED microscopy. [5] However, labeling an intracellular protein of interest with a small molecule fluorophore can also be a challenge in itself, since the target has to be efficiently modified inside cells ideally with minimal perturbance. [6] Typically, target labeling is accomplished using immunocytochemistry, which requires cell fixation and permeabilization. However, such experiments can significantly alter the ultrastructure compared to living cells [7] and cell permeabilization is generally not compatible with living systems. Some STED‐ and live‐cell‐compatible fluorescent probes exist, [8] but they are only available against few targets. A powerful alternative for living cells are self‐labeling enzymes such as the Halotag [9] or small‐molecule ligands against genetically encodable tags, [10] which are very effective but require genetic modification of the protein of interest as well as cell‐permeable, ideally fluorogenic, labels. [11] Dual‐color imaging then also requires cotransfection of two enzymes as well as two orthogonal, cell‐permeable labels. [12]
Recently, attempts to improve the photophysical properties of labels have been made by generating “self‐healing” fluorophores via attaching a triplet state quencher or photostabilizer to the fluorophore, a concept referred to as “intramolecular photostabilization”. [13] As the dyes are then less prone to bleaching, this approach may be highly useful for the acquisition of time‐lapse experiments. Photostabilizers and fluorophores have been successfully conjugated to proteins, [14] and have been used in imaging of the endosomal compartment in living cells. [15] However, such a system has not yet been proven effective in combination with intracellular targeting in living cells, since the modified targeting agent has to be delivered into the cytosol effectively.
Herein, we present a semi‐synthetic strategy in which a fluorophore and a photostabilizer are ligated to a versatile targeting platform. As intracellular targeting moieties we employ single domain antibodies (camelid‐derived nanobodies). [16] Nanobodies are excellent tools for super‐resolution microscopy due to their small size, leading to a small linkage error, that is, a small distance between target and label. [17] In order to deliver the fluorescently labeled nanobodies into cells, we take advantage of a cell‐penetrating peptide additive strategy to efficiently deliver proteins into cells at low μm concentrations. [18] Briefly, by mixing a cysteine‐containing protein with an excess of a thiol‐reactive cell‐penetrating peptide (CPP), the CPP will reversibly modify the protein and the remaining CPP will be covalently anchored to the cell surface to facilitate cellular uptake.
Results and Discussion
Semi‐Synthesis and Evaluation of Nanobody‐Fluorophore‐Photostabilizer Conjugates
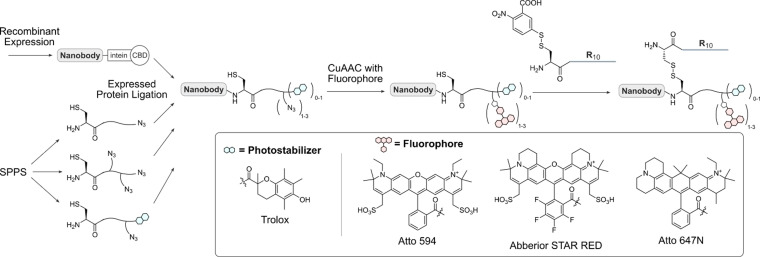
The ideal nanobody conjugate for live‐cell microscopy should be highly fluorescent and stable inside cells. To that end, we first evaluated different strategies for the fluorescent labeling of an anti‐GFP nanobody GBP1. To access fluorescent nanobodies, we took advantage of a previously developed modular semi‐synthetic approach using expressed protein ligation (EPL) for the C‐terminal modification of nanobodies (Scheme 1), [19] in which the nanobodies are expressed as intein fusions. This strategy is advantageous, as the antigen‐binding region of nanobodies is typically facing away from the C‐terminus of the protein, and C‐terminal modifications may therefore be less likely to disrupt binding.
Scheme 1.
Semi‐synthetic strategy to obtain fluorescent, cell‐permeable nanobodies. Nanobodies are recombinantly expressed as intein fusion proteins. The intein is cleaved and replaced with a synthetic azide containing peptide, generating attachment sites for a fluorophore and a cell‐penetrating peptide via click chemistry and a disulfide linkage, respectively. The peptides contain either one or three azides and/or the photostabilizer trolox and can be modified with different fluorophores depending on the experimental setup.
The nanobodies were then ligated to peptides synthesized via solid‐phase peptide synthesis (SPPS) carrying one or three azides for the attachment of fluorophores using classical Cu‐catalyzed azide–alkyne cycloaddition (CuAAC), along with the photostabilizers nitrophenylalanine and trolox ((±)‐6‐hydroxy‐2,5,7,8‐tetramethylchromane‐2‐carboxylic acid, peptide analytical data in SI Figure 1).
Whereas all peptides were successfully synthesized, we observed that ligation of the nitrophenylalanine peptide to the nanobody was not successful, as the protein spontaneously decomposed (noted by the appearance of small molecular weight bands on SDS‐PAGE gels, data not shown). This problem did not occur when using the photostabilizer trolox, which allowed us to obtain three nanobody‐Atto594‐fluorophore conjugates incorporating one fluorophore, three fluorophores in a row, or one fluorophore and trolox (Figure 1 a–d, analytical data in SI Figure 2).
Figure 1.
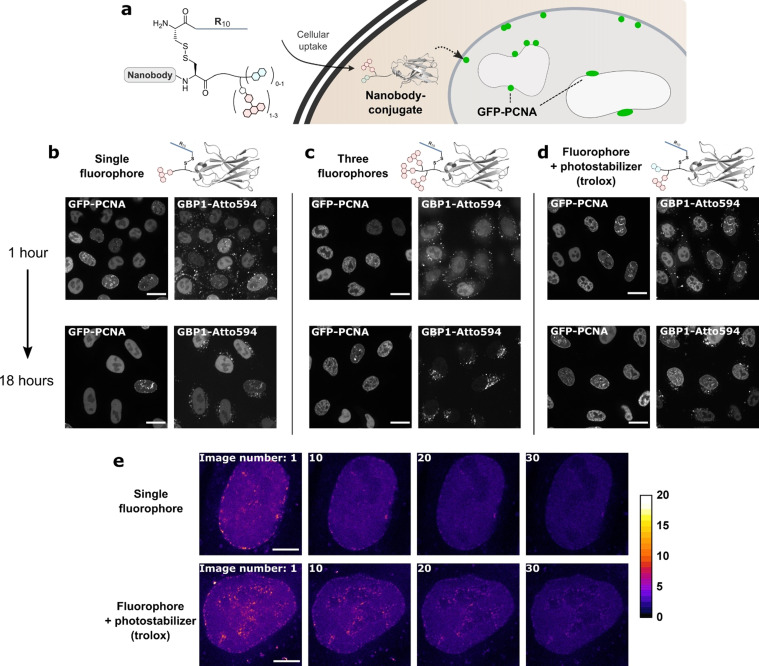
Evaluation of nanobody–fluorophore conjugates for super‐resolution microscopy. a) The fluorescent, R10‐functionalized GFP‐binding nanobody GBP1 enters cells after which the R10 peptide is cleaved off. b–d) HeLa kyoto cells expressing GFP–PCNA were treated with 2 μm of the GPB1 nanobody variants. Colocalization with GFP and intracellular stability were assessed by confocal microscopy after 1 and 18 hours post‐treatment of the cells. Scale bars 20 μm. e) STED images from the time series of the nanobody–fluorophore conjugates in living cells. Scale bars 5 μm.
Next, we tested the three nanobody–fluorophore conjugates in their performance in confocal and super‐resolution microscopy on a HeLa Kyoto cell line stably expressing GFP–PCNA (proliferating cell nuclear antigen) in the nucleus. [20] PCNA is generally broadly distributed throughout the nucleus but forms discrete “replication foci” where DNA replication occurs during S‐Phase. [21] The cellular uptake of the proteins was easily achieved by mixing the nanobodies (which contain a free cysteine at the ligation site [19] ) with the thiol‐reactive TNB‐R10 peptide and then applying the mixture to cells for 1 hour. [18] During our confocal microscopy measurements, we observed that the nanobody carrying three fluorophores showed a more diffuse intracellular localization compared to the other variants (Figure 1 b–d, upper row, Pearson's correlation coefficients (PCC) with GFP, from left to right: 0.64, 0.61, 0.85). The difference became even more apparent after 18 hours in the cells, at which point the construct showed no more colocalization with GFP but rather endosomal localization, pointing towards possible instability of the nanobody (Figure 1 c, lower row, PCC, from left to right: 0.81, 0.09, 0.59). The other two constructs showed good colocalization with GFP and were stable over at least 18 hours inside cells.
Subsequently, we investigated whether the trolox moiety increases the stability of the Atto594 fluorophore attached next to it. After internalization of the nanobody into the same GFP–PCNA expressing cell line, we performed a bleaching test using STED super‐resolution microscopy. We took 30 STED images in the same position and quantified the total intranuclear fluorescence. Indeed, the trolox‐modified nanobody showed an increased resistance to bleaching compared to the single fluorophore–nanobody conjugate and still allowed the resolution of single replication foci even after 30 consecutive STED images (Figure 1 e, bleaching curves in SI Figure 3). Compared to previous work,[ 13b , 13c ] the effect of photostabilization we observed was relatively modest. To investigate this further, we generated a new construct of the GFP‐binding nanobody GBP1 with the fluorophore Cy5, which is more prone to bleaching (SI Figure 4). We then compared photostabilization in live and fixed cells and found that the effect is more noticeable in the fixed sample (SI Figure 5), which may be due to diffusion of molecules in and out of plane in living, dynamic systems. Nevertheless, we concluded that the incorporation of a suitable fluorophore together with the trolox moiety is the most promising strategy for subsequent super‐resolution microscopy measurements.
A Cell‐Permeable mCherry Nanobody Allows STED‐Microscopy of mCherry Fusion Proteins in Living Cells
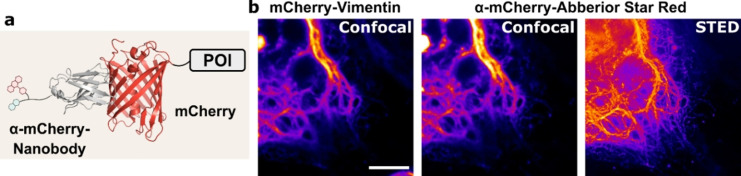
As the trolox conjugate proved to be most effective for the GBP1 nanobody, we decided to use the same strategy in the semi‐synthesis of the nanobody LaM4 that binds to DsRed and mCherry (Figure 2 a). [22] We initially found poor conversion of the nanobody in the EPL reaction and introduced a short helical linker sequence at the C‐terminus of the protein ((EAAAK)3). [23] We could then successfully generate a conjugate of the nanobody with the commercially available sulfated near‐infrared fluorophore Abberior STAR RED and trolox (SI Figure 7). As before, we conjugated the fluorophore to the nanobody using CuAAC followed by disulfide attachment of the R10 peptide (analytical data in SI Figure 5). The resulting nanobody conjugate showed good uptake into a cell line transfected with mCherry–vimentin, and good colocalization in confocal microscopy with its antigen in live cells (Figure 2 b). Afterwards we performed STED microscopy with the mCherry fusion protein, demonstrating the successful staining of commonly used fluorescent tags with a fluorescent nanobody in living cells (Figure 2 b).
Figure 2.
The cell‐permeable mCherry nanobody in STED microscopy. a) Schematic of the Abberior STAR RED‐labeled anti‐mCherry nanobody bound to an mCherry fusion protein. b) STED microscopy of HeLa Kyoto cells transfected with mCherry–Vimentin and treated with 2 μm of the cell‐permeable nanobody. Scale bar 5 μm.
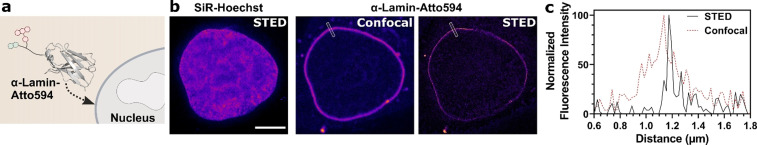
Super‐Resolution Microscopy of Endogenous Nuclear Lamins with a Fluorescent Cell‐Permeable Lamin Nanobody
Protein delivery using cell‐penetrating peptides can be used in experiments in which transfection is difficult. To demonstrate this aspect, we applied our cell‐permeable highly fluorescent nanobodies to target endogenous proteins. We started our investigations by using a nanobody binding nuclear lamins. [24] The nuclear lamina can be misshapen in cells overexpressing fusions of lamins with fluorescent proteins; [25] consequently, a cell‐permeable nanobody presents a promising strategy for live‐cell microscopy. As the nuclear lamina is 30–100 nm thick, i.e., below the diffraction limit for visible light, STED measurements should allow sub‐diffraction resolution with our approach.
We again used the EPL strategy with the helical linker ((EAAAK)3) on the C‐terminus to furnish a cell‐permeable fluorescent nanobody for nuclear lamina staining in living cells (Figure 3 a, analytical data in SI Figure 8).
Figure 3.
The cell‐permeable lamin nanobody in STED microscopy. a) Schematic of the Atto594‐labeled anti‐lamin nanobody binding to the nuclear lamina. b) STED microscopy of HeLa Kyoto cells treated with 2 μm of the cell‐permeable nanobody with 500 nm SiR‐Hoechst. Scale bar 5 μm. c) Histogram of the normalized fluorescence intensity over a line ROI (see white box in (b)).
We performed STED microscopy with the lamin nanobody in combination with the DNA‐stain SiR‐Hoechst [8b] (Figure 3 b), which allowed resolution of the nuclear lamina down to 40 nm thickness (half‐maximal width, Figure 3 c). We could also use the nanobody to monitor cell division using confocal microscopy (SI Figure 9).
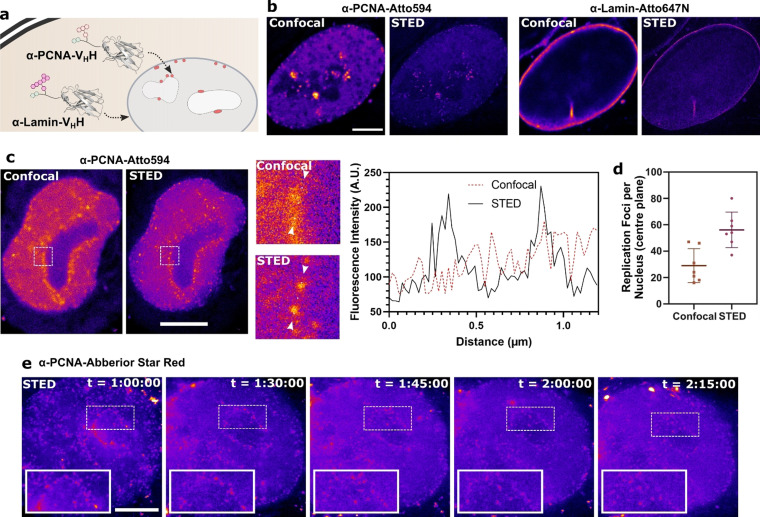
The Cell‐Permeable PCNA‐Nanobody Allows Super‐Resolution Time‐Lapse Imaging of Replication Foci
Next, we applied our semi‐synthesis protocol to a nanobody targeting endogenous PCNA. PCNA belongs to the family of DNA sliding clamps and tethers DNA polymerases to the nascent DNA strand during replication. [21] As such, it is a marker of the discrete foci where DNA replication occurs during S‐phase, so‐called “replication foci”. [26] We were able to express and label a PCNA nanobody analogously to the lamin nanobody (analytical data in SI Figure 10).
Delivery of the nanobody into HeLa Kyoto cells expressing GFP‐labeled PCNA showed good colocalization of the PCNA nanobody and GFP (SI Figure 11 a), confirming that the nanobody binds its antigen in living cells. We could also confirm through conventional immunofluorescence on fixed cells that the PCNA nanobody localizes to the same replication foci as the commonly used monoclonal PCNA antibody PC10 (SI Figure 11 b).
Next, we questioned whether we could co‐deliver two different nanobodies at the same time. For this, we conjugated the cell‐permeable lamin nanobody with Atto647N (as a far‐red fluorophore with little overlap with Atto594) and the PCNA nanobody with Atto594. Adding both nanobodies to cells simultaneously allowed co‐staining in STED microscopy of both the nuclear lamina and of replication sites within the same living cell (Figure 4 a,b).
Figure 4.
The cell‐permeable PCNA nanobody in STED microscopy. a) Schematic of the double labeling experiment using the Atto594‐labeled PCNA nanobody together with the Atto647N‐labeled lamin nanobody binding their nuclear antigens. b) STED microscopy of HeLa Kyoto cells treated with 2 μm of each of the cell‐permeable nanobodies. c) STED and confocal images of a nucleus stained with 2 μm of the cell‐permeable PCNA nanobody. The white squares are shown enlarged. A histogram of the fluorescence intensity was plotted between the white arrows. d) Replication foci were in a single plane of nuclei in confocal and STED microscopy. e) Time‐lapse STED microscopy of a cell in S‐phase stained with the cell‐permeable PCNA nanobody. “t” indicates the timepoints after addition of the nanobody to the cells. Scale bars 5 μm.
It became immediately visible from our STED microscopy imaging of PCNA that the super‐resolution technique allowed resolution of more individual replication foci when compared to the confocal microscopy image (Figure 4 c). We counted replication foci in a single plane of several nuclei and could distinguish approximately two times more replication foci in the super‐resolution images compared to the confocal image (Figure 4 d).
Finally, we synthesized a conjugate of the PCNA nanobody with the far‐red STED fluorophore Abberior STAR RED (for minimal cytotoxicity) along with the photostabilizer trolox. With this conjugate in hand, we were able to record a super‐resolution time‐lapse of cells in S‐phase (Figure 4 e), monitoring replication foci throughout.
Conclusion
We present here the to our knowledge first application of intramolecular photostabilization in live‐cell imaging. Through conjugation of a photostabilizer and fluorophore to cell‐permeable nanobodies, we can demonstrate that the photostabilizer variant outperforms a nanobody with multiple fluorophores. Furthermore, we demonstrated the simultaneous dual‐color staining of intracellular targets by delivering two different nanobodies. With these findings applied to the engineering of a cell‐permeable PCNA‐nanobody we got an unprecedented look at replication foci within the cell. Future work will include the application of this nanobody construct to monitor individual replication foci and analyze their formation, dissolution, and motility, which was previously done only with photoactivatable fluorophores. [27] Along with the application in difficult‐to‐transfect cell lines, we aim to improve our understanding of DNA replication and cell division in more complex systems. The performance of our probes is mainly limited by two factors: intracellular degradation and background signal (e.g. from endosomes), and finding solutions to these limitations will also be key in developing the system further. We believe that our design concept can be applied to many other intracellular antigens, and that intracellular intramolecular photostabilization has tremendous potential for live‐cell nanoscopy.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
The authors thank Kristin Kemnitz‐Hassanin and Ines Kretzschmar for technical support, and Profs. M. Cristina Cardoso and Heinrich Leonhardt for helpful discussions and plasmids. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SPP1623, RTG2473, Project number: 392923329) to C.P.R.H. (HA 4468/9‐1), the Einstein Foundation Berlin (Leibniz–Humboldt Professorship), the GIF, the German–Israeli Foundation for Scientific Research and Development and the Boehringer–Ingelheim Foundation (Plus 3 award) to C.P.R.H., the Fonds der Chemischen Industrie (FCI) to C.P.R.H. and A.F.L.S. (Chemiefonds fellowship). Open access funding enabled and organized by Projekt DEAL.
A. F. L. Schneider, L. S. Benz, M. Lehmann, C. P. R. Hackenberger, Angew. Chem. Int. Ed. 2021, 60, 22075.
References
- 1. Sahl S. J., Hell S. W., Jakobs S., Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. [DOI] [PubMed] [Google Scholar]
- 2. Godin A. G., Lounis B., Cognet L., Biophys. J. 2014, 107, 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Westphal V., Rizzoli S. O., Lauterbach M. A., Kamin D., Jahn R., Hell S. W., Science 2008, 320, 246–249; [DOI] [PubMed] [Google Scholar]
- 3b. Urban N. T., Willig K. I., Hell S. W., Nagerl U. V., Biophys. J. 2011, 101, 1277–1284; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3c. Jahr W., Velicky P., Danzl J. G., Methods 2020, 174, 27–41; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3d. Kilian N., Goryaynov A., Lessard M. D., Hooker G., Toomre D., Rothman J. E., Bewersdorf J., Nat. Methods 2018, 15, 755–756; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3e. Nagerl U. V., Willig K. I., Hein B., Hell S. W., Bonhoeffer T., Proc. Natl. Acad. Sci. USA 2008, 105, 18982–18987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Wegner W., Ilgen P., Gregor C., van Dort J., Mott A. C., Steffens H., Willig K. I., Sci. Rep. 2017, 7, 11781; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b. Matlashov M. E., Shcherbakova D. M., Alvelid J., Baloban M., Pennacchietti F., Shemetov A. A., Testa I., Verkhusha V. V., Nat. Commun. 2020, 11, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang L., Frei M. S., Salim A., Johnsson K., J. Am. Chem. Soc. 2019, 141, 2770–2781. [DOI] [PubMed] [Google Scholar]
- 6. Schneider A. F. L., Hackenberger C. P. R., Curr. Opin. Biotechnol. 2017, 48, 61–68. [DOI] [PubMed] [Google Scholar]
- 7. Schnell U., Dijk F., Sjollema K. A., Giepmans B. N., Nat. Methods 2012, 9, 152–158. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Belov V. N., Stoldt S., Ruttger F., John M., Seikowski J., Schimpfhauser J., Hell S. W., J. Org. Chem. 2020, 85, 7267–7275; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Lukinavicius G., Blaukopf C., Pershagen E., Schena A., Reymond L., Derivery E., Gonzalez-Gaitan M., D'Este E., Hell S. W., Gerlich D. W., Johnsson K., Nat. Commun. 2015, 6, 8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erdmann R. S., Baguley S. W., Richens J. H., Wissner R. F., Xi Z., Allgeyer E. S., Zhong S., Thompson A. D., Lowe N., Butler R., Bewersdorf J., Rothman J. E., St Johnston D., Schepartz A., Toomre D., Cell Chem. Biol. 2019, 26, 584–592 e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wombacher R., Heidbreder M., van de Linde S., Sheetz M. P., Heilemann M., Cornish V. W., Sauer M., Nat. Methods 2010, 7, 717–719. [DOI] [PubMed] [Google Scholar]
- 11. Grimm J. B., Muthusamy A. K., Liang Y., Brown T. A., Lemon W. C., Patel R., Lu R., Macklin J. J., Keller P. J., Ji N., Lavis L. D., Nat. Methods 2017, 14, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bottanelli F., Kromann E. B., Allgeyer E. S., Erdmann R. S., Wood Baguley S., Sirinakis G., Schepartz A., Baddeley D., Toomre D. K., Rothman J. E., Bewersdorf J., Nat. Commun. 2016, 7, 10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. van der Velde J. H., Uusitalo J. J., Ugen L. J., Warszawik E. M., Herrmann A., Marrink S. J., Cordes T., Faraday Discuss. 2015, 184, 221–235; [DOI] [PubMed] [Google Scholar]
- 13b. van der Velde J. H., Oelerich J., Huang J., Smit J. H., Aminian Jazi A., Galiani S., Kolmakov K., Guoridis G., Eggeling C., Herrmann A., Roelfes G., Cordes T., Nat. Commun. 2016, 7, 10144; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13c. Altman R. B., Terry D. S., Zhou Z., Zheng Q., Geggier P., Kolster R. A., Zhao Y., Javitch J. A., Warren J. D., Blanchard S. C., Nat. Methods 2012, 9, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tessier R., Ceballos J., Guidotti N., Simonet-Davin R., Fierz B., Waser J., Chem 2019, 5, 2243–2263. [Google Scholar]
- 15. Zheng Q., Jockusch S., Zhou Z., Altman R. B., Zhao H., Asher W., Holsey M., Mathiasen S., Geggier P., Javitch J. A., Blanchard S. C., Chem. Sci. 2017, 8, 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schumacher D., Helma J., Schneider A. F. L., Leonhardt H., Hackenberger C. P. R., Angew. Chem. Int. Ed. 2018, 57, 2314–2333; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 2336–2357. [Google Scholar]
- 17.
- 17a. Ries J., Kaplan C., Platonova E., Eghlidi H., Ewers H., Nat. Methods 2012, 9, 582–584; [DOI] [PubMed] [Google Scholar]
- 17b. Klein A., Hank S., Raulf A., Joest E. F., Tissen F., Heilemann M., Wieneke R., Tampe R., Chem. Sci. 2018, 9, 7835–7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schneider A. F. L., Kithil M., Cardoso M. C., Lehmann M., Hackenberger C. P. R., Nat. Chem. 2021, 13, 530–539. [DOI] [PubMed] [Google Scholar]
- 19. Herce H. D., Schumacher D., Schneider A. F. L., Ludwig A. K., Mann F. A., Fillies M., Kasper M. A., Reinke S., Krause E., Leonhardt H., Cardoso M. C., Hackenberger C. P. R., Nat. Chem. 2017, 9, 762–771. [DOI] [PubMed] [Google Scholar]
- 20. Chagin V. O., Casas-Delucchi C. S., Reinhart M., Schermelleh L., Markaki Y., Maiser A., Bolius J. J., Bensimon A., Fillies M., Domaing P., Rozanov Y. M., Leonhardt H., Cardoso M. C., Nat. Commun. 2016, 7, 11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moldovan G. L., Pfander B., Jentsch S., Cell 2007, 129, 665–679. [DOI] [PubMed] [Google Scholar]
- 22. Fridy P. C., Li Y., Keegan S., Thompson M. K., Nudelman I., Scheid J. F., Oeffinger M., Nussenzweig M. C., Fenyo D., Chait B. T., Rout M. P., Nat. Methods 2014, 11, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T., Protein Eng. 2001, 14, 529–532. [DOI] [PubMed] [Google Scholar]
- 24. Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M. C., Leonhardt H., Nat. Methods 2006, 3, 887–889. [DOI] [PubMed] [Google Scholar]
- 25. Broers J. L., Machiels B. M., van Eys G. J., Kuijpers H. J., Manders E. M., van Driel R., Ramaekers F. C., J. Cell Sci. 1999, 112, 3463–3475. [DOI] [PubMed] [Google Scholar]
- 26. Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I., Science 2005, 310, 1821–1824. [DOI] [PubMed] [Google Scholar]
- 27. Zessin P. J., Sporbert A., Heilemann M., Sci. Rep. 2016, 6, 18779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information