Abstract
Eukaryotic gene expression is closely regulated by translation and turnover of mRNAs. Recent advances highlight the importance of translation in the control of mRNA degradation, both for aberrant and apparently normal mRNAs. During translation, the information contained in mRNAs is decoded by ribosomes, one codon at a time, and tRNAs, by specifically recognizing codons, translate the nucleotide code into amino acids. Such a decoding step does not process regularly, with various obstacles that can hinder ribosome progression, then leading to ribosome stalling or collisions. The progression of ribosomes is constantly monitored by the cell which has evolved several translation‐dependent mRNA surveillance pathways, including nonsense‐mediated decay (NMD), no‐go decay (NGD), and non‐stop decay (NSD), to degrade certain problematic mRNAs and the incomplete protein products. Recent progress in sequencing and ribosome profiling has made it possible to discover new mechanisms controlling ribosome dynamics, with numerous crosstalks between translation and mRNA decay. We discuss here various translation features critical for mRNA decay, with particular focus on current insights from the complexity of the genetic code and also the emerging role for the ribosome as a regulatory hub orchestrating mRNA decay, quality control, and stress signaling. Even if the interplay between mRNA translation and degradation is no longer to be demonstrated, a better understanding of their precise coordination is worthy of further investigation.
This article is categorized under:
RNA Turnover and Surveillance > Regulation of RNA Stability
Translation > Translation Regulation
RNA Interactions with Proteins and Other Molecules > RNA‐Protein Complexes
Keywords: decay, degradation, mRNA, ribosome, translation
The process of mRNA translation is tightly monitored by eukaryotic cells to ensure the correct progression of ribosomes. A multitude of different pathways have evolved to mediate co‐translational mRNA degradation in order to restrict expression of aberrant mRNAs but also that of functional mRNAs.

1. INTRODUCTION
Messenger RNAs (mRNAs) are ephemeral intermediates between DNA, the carrier of genetic information, and proteins that display a functional role. Their primary sequence contains not only the information ribosomes need to synthesize proteins but also additional sequence and structural elements that regulate the translation process as well as other steps including transcription, splicing, polyadenylation, nuclear export, localization, and degradation. These elements can be located within untranslated regions (UTR) or in the coding region, owing to the degeneracy of the genetic code that allows some sequence flexibility. An intricate set of regulatory elements is therefore embedded within genes, under selective pressure, to play a specific role during the expression, function, or turnover of mRNAs.
mRNA translation is a sophisticated process that involves a large number of protein and RNA actors assisting ribosomal subunits in binding and assembling onto the mRNA. Thereafter, the ribosome translocates over the coding region and dissociates once the stop codon is reached, consequently releasing the newly synthesized protein. As such, translation is tightly regulated through a multitude of pathways involving cis‐acting features and trans‐acting factors that can affect the process in a transcript‐specific or global manner. Although the initiation of translation is generally considered as the rate‐limiting step, it has long been known that elongation is not a uniform process and termination is also tightly regulated, although these studies were technically challenging. The advent of high‐throughput sequencing and the ribosome profiling protocol (Ingolia et al., 2009), together with structural biology approaches, have greatly contributed to the characterization of ribosome dynamics from the recruitment of the 40S to the elongation of 80S ribosomes across the open reading frame and the recycling of post‐termination ribosomes. Collectively, these works have led to the discovery of new mechanisms involved in the regulation of translation and highlighted the numerous crosstalks between translation and other processes such as co‐translational protein folding and mRNA turnover. This review summarizes recent findings that contribute to improve our understanding of the intricate relationship between mRNA decay and the translation process, with particular emphasis on how the translating ribosome has emerged as a central regulatory hub not only for the quality control pathways that degrade faulty mRNAs but also for canonical mRNA degradation.
2. TRANSLATION‐DEPENDENT mRNA SURVEILLANCE PATHWAYS
To ensure high fidelity of gene expression, eukaryotic cells have evolved three major cytosolic mRNA surveillance pathways that rely on translation to recognize and trigger degradation of aberrant transcripts. These pathways, known as nonsense‐mediated decay, non‐stop decay, and no‐go decay, respectively, signal RNAs with a premature termination codon (PTC), RNAs lacking a termination codon and RNAs containing diverse ribosome‐stalling sequences (Powers et al., 2020; Shoemaker & Green, 2012). Despite their several mechanistic differences, the three mRNA surveillance pathways survey mRNA quality through careful inspection of the translation process, alerting when ribosomes are not progressing normally, in order to degrade aberrant mRNAs and their nascent polypeptides and to recycle ribosomes.
2.1. Nonsense‐mediated decay
This surveillance pathway causes decay of transcripts harboring a PTC that can be brought in by genetic mutations or introduced into an mRNA through stochastic errors in transcription or splicing. In addition to its role in eliminating aberrant mRNAs with a PTC, nonsense‐mediated decay (NMD) also degrades a large number of normal transcripts that contain an upstream open reading frame (uORF) or a long 3′‐untranslated region (UTR; Kurosaki et al., 2019; Nasif et al., 2018). In all cases, it is assumed that RNA features downstream of a termination codon must interfere with translating ribosomes and, depending on the NMD trigger, the termination codon is defined as premature or genuine. The core NMD factors comprise a set of up‐frameshift (UPF) proteins (UPF1, 2, and 3B) and suppressors with morphogenetic effects on genitalia (SMG) proteins (SMG1, 5, 6, and 7). The NMD machinery is thought to be recruited to target mRNAs because translation termination at a PTC is aberrant. In normal translation, ribosomes decode transcripts until they encounter a stop codon, usually present in the most 3′‐exon in close proximity to the poly(A)‐binding protein (PABPC1 in mammals, Pab1 in yeast) bound to the poly(A) tail. PABPC1 promotes translation termination by stimulating the recruitment of eukaryotic release factors eRF1 and eRF3a to the ribosomal A‐site, which releases the nascent peptidic chain (Ivanov et al., 2016). Afterwards, ribosomes are dissociated into 40S and 60S ribosomal subunits that are recycled by the factor ABCE1 (Rli1 in yeast) with help of several initiation factors (Figure 1(a)).
FIGURE 1.
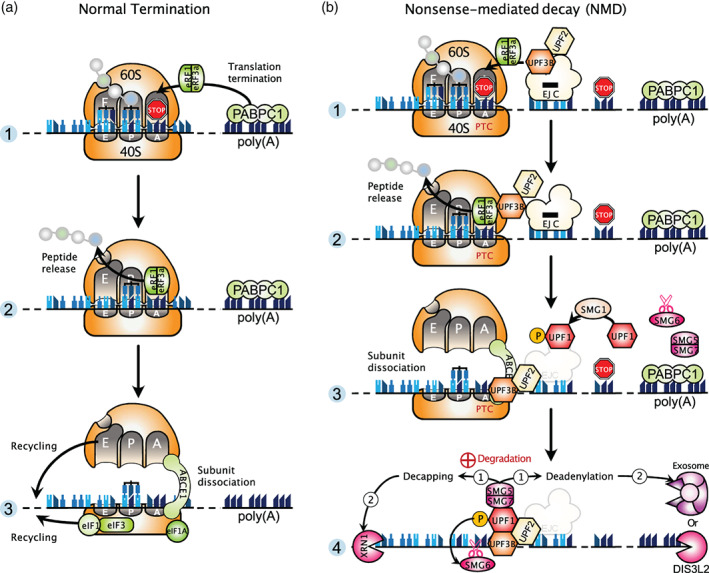
Translation termination at a normal stop codon and at a premature termination codon. (a) Normal translation termination. Panel 1: The translating 80S ribosome, composed of 60S and 40S subunits, encounters the STOP codon (red STOP sign) located near the poly(a) tail bound by PABPC1 in mammals. Ribosomal A‐site, P‐site, and E‐site are indicated. tRNAs are bound in the P‐site and E‐site. PABPC1 promotes the recruitment of the eRF1 and eRF3a release factors to the ribosomal A‐site. Panel 2: After GTP hydrolysis by eRF3a, eRF1 triggers hydrolysis of the peptidyl‐tRNA, releasing the completed nascent protein. This leaves a ribosome still bound to the mRNA, which has to be disassembled and recycled. Panel 3: The ribosome recycling factor ABCE1 is recruited and it supports, with help of other initiation factors, the dissociation of the 80S ribosome into 40S and 60S subunits, which can then be recycled for extra rounds of translation. (b) Translation termination at a premature termination codon (PTC) and induction of nonsense‐mediated mRNA decay (NMD). Panel 1: The ribosome stops at a PTC located upstream of an EJC and distant from the normal stop codon. The presence of the NMD factor UPF3B at the EJC allows the association of eRF1 and eRF3a in the ribosomal A‐site. Panel 2: A truncated protein product is released through the action of eRF3a and eRF1. Panel 3: Ribosome dissociation is mediated by ABCE1 together with UPF3B. Subsequently, UPF2 and UPF3B are thought to activate the SMG1 kinase for UPF1 phosphorylation. Panel 4: Phosphorylated UPF1 mainly associates with the SMG5‐SMG7 complex which recruits decapping and deadenylation activities (1), followed by the exoribonucleases with 5′‐to‐3′ (XRN1) and 3′‐to‐5′ (exosome or DIS3L2) activities (2). In addition, phospho‐UPF1 can recruit the endonuclease SMG6 that cleaves RNA in the vicinity of the PTC
In the case of a PTC, two predominant models of NMD activation have been put forward. The first, referred to as the exon junction complex (EJC) model, postulates that, during the pioneer round of translation, when a ribosome encounters a stop codon located sufficiently upstream of an EJC, a warning signal is sent to the NMD pathway to degrade the mRNA carrying a PTC (Nagy & Maquat, 1998; Thermann et al., 1998; Zhang et al., 1998). In this scenario, PABPC1 cannot interact with eRF3a due to their distance and, instead, the EJC allows the recruitment of NMD factors which, in turn, associate with eRF3a. Importantly, it has been reported that eRF3a interacts directly with UPF3B, but not UPF1 as previously thought, and UPF3B acts to dissociate post‐termination ribosomes (Neu‐Yilik et al., 2017). In addition, the ribosome recycling factor ABCE1, required for ribosome dissociation at a normal termination codon, is also essential for termination at a PTC, contrary to what was previously believed (Zhu et al., 2020). Indeed, loss of ABCE1 was found to induce an accumulation of unrecycled ribosomes in the 3′‐UTR of NMD targets, leading to displacement of downstream EJC, followed by an impaired recruitment of the central NMD factor UPF1. Following proper ribosome dissociation, UPF1, activated by SMG1‐mediated phosphorylation and aided by its partners UPF2 and UPF3B, can remodel the 3′‐UTR and recruit the enzymes involved in mRNA decay (Figure 1(b)). The second model for NMD, called the faux UTR model, has been proposed in yeast. This model postulates that the inherently aberrant nature of premature translation termination allows the binding of UPF factors to mRNAs, thus triggering NMD. Supporting this model, early results showed that translation termination at a PTC is inefficient, likely because the PTC is distant from the poly(A)‐binding protein Pab1 located at the 3′‐end of the ORF (Amrani et al., 2004). In vivo experiments in yeast now substantiate these previous findings by demonstrating that the efficiency of translation termination increases as PTCs are closer to ORF 3′‐ends, and furthermore this positional effect depends on Pab1 (W. Chan et al., 2020). The authors proposed from their results that NMD is activated as soon as the efficiency of termination is below a certain threshold.
By developing a method that allows real‐time imaging of both translation and NMD of single mRNA molecules in live human cells, a recent work provided a unifying model for NMD activation by recapitulating most aspects of NMD, but this study also unveils that the mechanism of NMD activation is more complicated than previously believed (Hoek et al., 2019). This powerful method to study NMD kinetics and heterogeneity reveals that each ribosome (the first or one of the following ribosomes) that terminates translation at the PTC has an equal probability of triggering NMD. In addition, NMD occurs with the same probability during the pioneer round of translation (CBC‐bound mRNAs) or during a later round (eIF4E‐bound mRNAs). Surprisingly, for a given NMD substrate, a fraction ranging from 5 to 30% of mRNA molecules is resistant to NMD. This could be explained by some variability in splicing or EJC deposition. Contrariwise, NMD probability can vary substantially depending on several key parameters such as the distance between the PTC and the downstream EJC, the number of EJCs, or the mRNA sequence downstream of the PTC. On the other hand, changing the 3′‐UTR length of the mRNA reporters did not have a major effect on NMD efficiency. This finding agrees with a recent study reporting that NMD in human cells occurs independently of stable ribosome stalling at PTCs (Karousis et al., 2020). To monitor ribosome density at stop codons, the authors developed a toeprinting assay based on in vitro translation of NMD reporter mRNAs with human cell lysates. Their results demonstrated a comparable ribosomal density at stop codons of NMD‐sensitive and NMD‐insensitive reporters. In line with this, ribosome profiling analyses also did not show any increase in ribosome occupancy at termination codon of endogenous NMD‐sensitive transcripts, compared with NMD‐insensitive mRNAs. Together, both studies do support that NMD is induced by a mechanism more complicated than just ribosome stalling at stop codons.
Another work was designed to define NMD targets and decay intermediates on a transcriptome‐wide level (Kurosaki et al., 2018). Because NMD substrates are degraded by nucleases common to other RNA decay pathways and the UPF1 factor is involved in NMD but also in other decay mechanisms, the authors took advantage of the fact that, in human embryonic kidney (HEK) 293T cells, UPF1 is activated by phosphorylation exclusively during NMD and not during other decay pathways. The use of an antibody specific of phosphorylated UPF1 combined with high‐throughput sequencing allowed them to selectively identify direct NMD targets and their decay intermediates. Importantly, their findings provided evidence that NMD initiates on mRNAs that are bound by one or more translating ribosomes. This strongly suggests that NMD does not occur in P‐bodies (see Box 1), in agreement with previous studies (Eulalio, Behm‐Ansmant, Schweizer, & Izaurralde, 2007; Stalder & Mühlemann, 2009) but in disagreement with others (Durand et al., 2007; Sheth & Parker, 2006). Their data also support that phospho‐UPF1 predominantly associates with the SMG5‐SMG7 complex which recruits decapping and deadenylation activities, followed by the action of exoribonucleases with 5′‐to‐3′ (XRN1) and 3′‐to‐5′ (exosome or DIS3L2) activities. Alternatively, phospho‐UPF1 can interact with the endonuclease SMG6, which cleaves in close proximity to the PTC (Figure 1(b)). Finally, the authors found that NMD decay intermediates were modified by uridylation at their 3′‐ends by the poly(U) polymerases TUT4 and TUT7, which further increases 3′‐to‐5′ decay and also XRN1‐mediated 5′‐to‐3′ decay.
BOX 1. Processing‐bodies.
Processing‐bodies (P‐bodies) are cytosolic membraneless organelles that form through a process known as liquid–liquid phase separation (for a comprehensive review see Luo et al., 2018; Standart & Weil, 2018). P‐bodies are composed of aggregates of untranslating mRNAs and proteins related to RNA metabolism. Among these proteins are factors involved in mRNA degradation (including decapping factors, the 5′‐3′ exonuclease XRN1, deadenylation factors, miRNA‐mediated silencing factors, and nonsense‐mediated decay factors), as well as proteins involved in translational repression (for an exhaustive list of proteins see Eulalio, Behm‐Ansmant, & Izaurralde, 2007; Parker & Sheth, 2007). The enrichment of such factors together with that of mRNAs targeted by mRNA degradation pathways such as miRNAs or ARE‐elements, led to the hypothesis that P‐bodies were sites of active mRNA degradation. However, this model has been recently challenged by studies showing that mRNAs found within P‐bodies are intact (no specific detection of degradation intermediates), thus suggesting that P‐bodies are sites of regulated mRNA storage instead of degradation (Courel et al., 2019; Standart & Weil, 2018). Further supporting this hypothesis, mRNAs stored in P‐bodies have been shown under specific conditions to exit P‐bodies and reengage in translation (Bhattacharyya et al., 2006; Brengues, 2005). Nevertheless, mRNAs enriched in P‐bodies appear to follow a distinct pathway for their degradation that involves the action of the PAT1B protein (Courel et al., 2019).
2.2. No‐go decay
The no‐go decay (NGD) pathway acts to recognize and degrade transcripts containing sequence features that induce ribosome stalling during translation elongation (Doma & Parker, 2006; Tsuboi et al., 2012). The most prevalent stalling features into mRNAs comprise stable secondary structures as stem‐loop motifs or GC‐rich sequences, tracts of rare codons that leave the ribosomal A‐site empty, or damaged RNA bases. Such obstacles that hinder the movement of ribosomes along mRNAs lead to ribosome stalling and potentially ribosome collision, which implies a collision between a stalled ribosome and the elongating ribosome behind it. Depending on the context, two different mechanisms evolved to cope with the event of ribosomal arrest. In the case of a stalled ribosome at the end of a truncated mRNA, which leaves empty the ribosomal A‐site, it was found that the yeast complex Dom34‐Hbs1 (or Pelota‐Hbs1 in mammals) binds to the A‐site and is thought to recruit an endonuclease for mRNA cleavage in the vicinity of the stalled ribosome (Doma & Parker, 2006). The ensuing cleavage products are degraded by either the Xrn1 exonuclease or the Ski‐exosome complex. Some mechanistic details were obtained using high resolution ribosome profiling and biochemistry approaches in yeast (D'Orazio et al., 2019). The authors identified Cue2 as the primary endonuclease that cleaves mRNA precisely within the A‐site of the stalled ribosome and they found that exonucleolytic decay of RNA intermediates proceeds primarily with Xrn1 rather than the exosome. Furthermore, the NGD process closely couples decay of faulty mRNAs with ribosome rescue and degradation of the nascent polypeptide. Dom34/Pelota and Hbs1, which are structural homologs of the canonical termination factors eRF1 and eRF3, recruit the ribosome‐recycling factor Rli1/ABCE1 to promote dissociation of the stalled ribosome (Tsuboi et al., 2012). Because Dom34/Pelota is unable to manage peptidyl‐tRNA hydrolysis, the nascent peptide remains attached to the 60S ribosomal subunit and is then handled by the ribosome‐associated quality control (RQC) pathway for subsequent ubiquitination, extraction, and degradation by the proteasome (Shao et al., 2013; Verma et al., 2013; Figure 2(a)).
FIGURE 2.
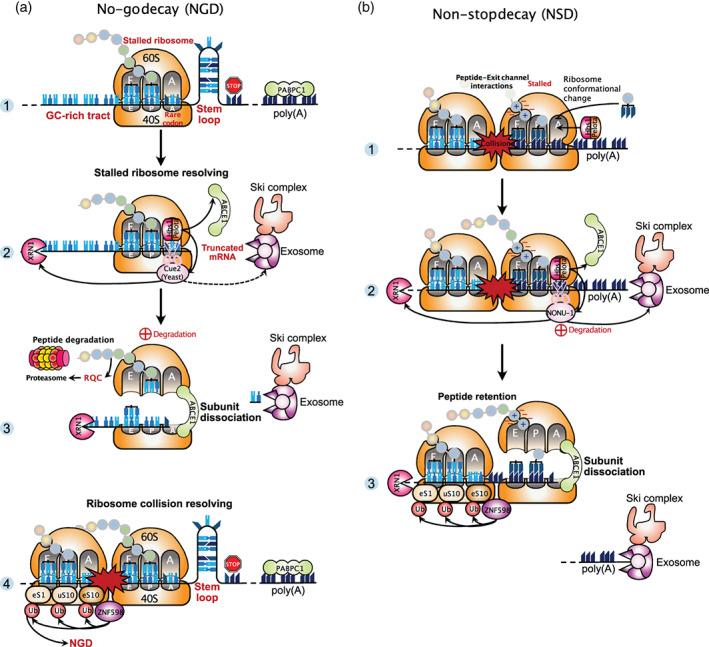
No‐go decay and non‐stop decay. (a) No‐go decay (NGD). Panel 1: The ribosome stops before reaching the stop codon because it encounters a stalling feature such as a stem‐loop motif, a GC‐rich tract, or rare codons. These obstacles lead to ribosome stall and potentially ribosome collision. Two different mechanisms evolved to cope with this particular event of ribosomal arrest. The first mechanism, aimed to resolve a stalled ribosome, is illustrated on panels 2 and 3. The mammalian complex Pelota‐Hbs1 (Dom34‐Hbs1 in yeast) binds to the empty ribosomal A‐site and recruits the ribosome recycling factor ABCE1 (Rli1 in yeast) and also the Cue2 endonuclease (in yeast), which cleaves the mRNA in the A‐site of the stalled ribosome, further allowing exonucleolytic decay mediated primarily by the XRN1 exonuclease rather than by the exosome assisted with the Ski complex. Panel 3: NGD closely couples decay of faulty mRNA with ribosome recycling and degradation of the nascent polypeptide. Because Pelota‐Hbs1 cannot hydrolyze the peptidyl‐tRNA bound to the P‐site, peptide release does not occur. The nascent peptide still attached to the 60S ribosomal subunit is handled by the ribosome‐associated quality control (RQC) pathway and degraded via the proteasome. Panel 4 illustrates the second mechanism aimed to resolve collided ribosomes. This event induces conformational changes of the ribosome that are recognized by the ubiquitin ligase ZNF598 (in mammals). ZNF598 ubiquitinates several ribosomal proteins and these modifications are required for robust induction of NGD. (b) Non‐stop decay (NSD). NSD relies on the same factors as NGD, but differs in its mRNA targets. Panel 1: Due to the lack of a stop codon, the ribosome translates a poly(A) sequence and translation elongation progressively slows down and eventually stops. Activation of NSD results from interactions between the positively charged lysine residues of the nascent polypeptide and the negatively charged exit channel of the ribosome. Panel 2: The complex Pelota‐Hbs1 in the A‐site recruits the endonuclease NONU‐1 (in C. elegans, homolog of Cue2 in S. cerevisiae). The interactions between positively‐charged residues with the exit channel result in the retention of the nascent peptide when the ribosome subunits are dissociated. Panel 3: Ribosome stalling caused by translation of poly(A) sequences is recognized by ZNF598. Ubiquitination of ribosomal proteins by ZNF598 are important modifications for ribosome rescue during NSD
A second major mechanism is employed during NGD to resolve ribosome collision events. This mechanism does not involve external factors interfacing with ribosomes, but it relies on direct modifications of ribosomes. When the trailing ribosome collides with the leading stalled ribosome, the complex formed by the two is called a disome. The structure of the disome was solved by cryo‐electron microscopy (cryo‐EM) and revealed a unique conformation suitable for recognition and modification of some ribosomal proteins by the ubiquitin ligase ZNF598 (Hel2 in yeast; Ikeuchi et al., 2019; Juszkiewicz et al., 2018). In particular, ubiquitination of the ribosomal protein uS10 (RPS20) was found to be required for elimination of the disome unit by the NGD pathway in yeast (Ikeuchi et al., 2019). Consistent with these findings, it was demonstrated that ribosome collisions are crucial for robust induction of the NGD process (Simms et al., 2017; Figure 2(a), Panel 4).
2.3. Non‐stop decay
Despite its name, the non‐stop decay (NSD) pathway is not dedicated to the decay of all transcripts lacking a stop codon but primarily dedicated to the decay of transcripts inappropriately polyadenylated in their coding region, resulting in formation of nonstop mRNAs (Frischmeyer et al., 2002). Therefore, although they differ in their substrate specificity and ribosome stalling mechanism, NSD and NGD both degrade problematic RNAs with stalled ribosomes and collided ribosomes if severe ribosome slowdown. In line with this, NSD relies on the same factors as NGD to induce mRNA decay and ribosome rescue (Figure 2(b)). These include Dom34/Pelota‐Hbs1 and Rli1/ABCE1 (Tsuboi et al., 2012). Recently, NONU‐1 was identified in C. elegans as a novel endonuclease that induces RNA cleavage in the vicinity of stalled ribosomes during both NSD and NGD (Glover et al., 2020). Homology searches revealed that one homolog of NONU‐1 in S. cerevisiae is Cue2, which was previously identified as the primary endonuclease functioning in NGD (D'Orazio et al., 2019). Mechanistically, earlier studies suggested that ribosome stalling during NSD was caused by the interaction of the nascent polypeptide, positively charged because of consecutive lysine residues encoded by the poly(A) sequence, with the ribosomal exit channel that is charged negatively (Koutmou et al., 2015; Lu & Deutsch, 2008). Further biochemical and structural studies in mammalian systems have elucidated the mechanism by which ribosomes stall during translation of a poly(A) tail (Chandrasekaran et al., 2019). By cryo‐EM, the authors visualized a suboptimal peptidyl‐tRNA conformation for peptide bond formation and a reconfiguration of the decoding center by the poly(A) mRNA that conflict with incoming aminoacyl‐tRNA, thus impeding further elongation. Accordingly, they proposed that NSD is selectively activated upon coincident detection by the ribosome of poly‐lysine chains in the exit tunnel and poly(A) tracts in the decoding center. Additionally, it was found that ribosome stalling caused by translation of poly(A) sequences is recognized by ZNF598. This E3 ligase, also involved in NGD, ubiquitinates the 40S ribosomal proteins eS10 (RPS10), uS10 (RPS20), and eS1 (RPS3A) in mammals (Figure 2(b), Panel 3), and these modifications are crucial to detect and resolve ribosomes that have stalled during decoding of poly(A) sequences (Garzia et al., 2017; Juszkiewicz & Hegde, 2017; Sundaramoorthy et al., 2017). In yeast, similar ubiquitinations catalyzed by Hel2 were also described, yet targeting distinct ribosomal proteins (Matsuo et al., 2017).
3. TRANSLATION INITIATION AND mRNA DECAY
In eukaryotic cells, coupling between mRNA translation and decay seems to be widespread and extends beyond the aforementioned mRNA surveillance mechanisms. While individual steps of translation and decay processes are rather well elucidated, less is known about the upstream determinants that control mRNA fate. Translation initiation is generally viewed as the most influential step in translation and numerous studies have attempted to address its impact on mRNA decay. Early observations in S. cerevisiae have shown that active translation initiation protects certain mRNAs from degradation (Schwartz & Parker, 1999). For those transcripts, inhibition of translation initiation by mutation of initiation factors led to higher rates of mRNA decay, associated with an increase in both decapping and deadenylation activities. A transcriptome‐wide study in S. cerevisiae revealed that this relationship between mRNA translation initiation and decay is broadly generalizable (P. P. Chan & Lowe, 2016). By combining a minimally invasive metabolic labeling‐based assay to measure mRNA decay rates with selective perturbations of the translation process, these authors found that slowing translation initiation caused a higher rate of transcript degradation on a transcriptome‐wide level. Consistent with this global destabilization of mRNAs upon inhibition of translation initiation, they observed an enhanced formation of P‐bodies, which are sites of mRNA storage and decay. By contrast, they found that slowing translation elongation led to an overall stabilization of transcripts and no change on P‐body formation. Collectively, their results provide further evidence that translation initiation is an important determinant of mRNA stability in yeast. Two mutually non‐exclusive models have been proposed to explain how translation may affect mRNA stability. The first model, referred to as the stalled ribosome‐triggered decay model, predicts that a slow rate of translation elongation at suboptimal codons is perceived as a signal to trigger mRNA decay. The second model, referred to as the translation factor‐protection model, predicts that the translation initiation machinery protects mRNAs from decay by a competition mechanism for mRNA binding. The data obtained by (P. P. Chan & Lowe, 2016) are consistent with mRNA decay being regulated by translation initiation but not elongation.
In higher eukaryotes, a study now provides a broad and comprehensive view regarding the impact of the 5′‐UTR on mRNA translatability and stability (L. Jia et al., 2020). By designing a mRNA library of over one million 5′‐UTR variants with a randomized sequence preceding a small uORF and downstream GFP, the authors identified several elements in the 5′‐UTR that contribute to reporter mRNA outcomes in mammalian cells. While translation of the GFP‐encoding main ORF preserves mRNAs from degradation, uORF translation was found to inhibit translation of downstream GFP, by triggering mRNA decay in a way reminiscent of NMD since this uORF‐mediated decay depends on both UPF1 and translation. Additionally, enrichment of GGC motifs in the 5′‐UTR inhibits translation and confers a shorter half‐life to mRNAs. These sequence elements form RNA G‐quadruplex (RG4) structures that impair the scanning process and relocate the RG4 reporter mRNAs into P‐bodies for DCP2‐dependent or XRN1‐dependent decay. Finally, a 5′‐UTR unstructured A‐rich motif was uncovered as an internal ribosome‐entry site to mediate cap‐independent translation. This poly(A) tract element, while stabilizing mRNAs engaged in translation, destabilizes ribosome‐free mRNAs likely by recruiting nucleases. Together, these findings reveal that the 5′‐UTR contributes to a tight coupling between mRNA translation initiation and degradation. Furthermore, this coupling can occur through various mechanisms depending on the 5′‐UTR sequence features.
4. THE RIBOSOME AS A HUB FOR mRNA DECAY, QUALITY CONTROL, AND STRESS SIGNALING
To ensure protein homeostasis, eukaryotic cells have evolved co‐translational quality control mechanisms that respond to abnormal translation by targeting the associated mRNA and nascent polypeptide chain for degradation and by recycling ribosomes (Joazeiro, 2019). Emerging evidence highlights crucial roles for the ribosome in quality control mechanisms, acting as a hub for recruiting factors that determine the fate of mRNAs and polypeptides. In addition, ubiquitin modifications at the ribosome emerge as part of the quality control pathways. Furthermore, recent work has revealed that the ribosome is a new central player to trigger various stress responses.
4.1. The ribosome and mRNA decay
It was long believed that mRNA degradation required the removal of ribosomes first until a pioneering study in yeast uncovered that mRNA decay could occur while transcripts were still bound by translating ribosomes (Hu et al., 2009). The prevailing model of cytoplasmic mRNA decay is that it starts with shortening of the poly(A) tail by the Ccr4‐Not deadenylase complex and removal of the 5′‐cap structure by the Dcp1–Dcp2 decapping complex. Then, mRNA decay proceeds through 5′‐to‐3′ degradation by the exoribonuclease Xrn1 and/or 3′‐to‐5′ exonucleolytic degradation by the exosome assisted with the Ski complex (Figure 3(a)). The study by Hu et al. showed that decapping and 5′‐to‐3′ exonucleolytic degradation take place on certain mRNAs still associated with ribosomes. Their findings indicate that removal of ribosomes is not a prerequisite for initiation of mRNA decay and, furthermore, suggest that Xrn1‐mediated mRNA degradation from the 5′‐end follows the last elongating ribosome. By using high‐throughput sequencing of 5′‐phosphorylated mRNA degradation intermediates, another work revealed that up to approximately 35% of transcripts in S. cerevisiae undergo co‐translational decay, which is inferred from the characteristic 3‐nucleotide periodicity pattern observed in the mRNA coding region (Pelechano et al., 2015). Additionally, the use of novel sequencing technologies to identify RNA decay intermediates has shown that co‐translational mRNA degradation is much more widespread than previously thought (Ibrahim et al., 2018). Ibrahim et al. have developed a new transcriptome‐wide method coined “Akron‐seq” to simultaneously capture and sequence the 3′‐ends of capped mRNAs (Akron3) or the 5′‐ends of polyadenylated transcripts (Akron5) in human cells (Ibrahim et al., 2018). Their results showed that most 3′ and 5′ RNA ends are mapped within the mRNA body. Specifically, up to 63% of the 3′‐ends of capped mRNAs are mapped within coding sequences and only 11% of capped transcripts contain a poly(A) sequence at their 3′‐end, thus indicating that fragmented mRNAs are much more represented in cells than full‐length RNA molecules. Importantly, relative positioning analyses of Akron reads with ribosome profiling data revealed that 5′‐end and 3′‐end reads exhibit a striking 3‐nucleotide periodicity for all transcripts, suggesting that canonical mRNA degradation may be coupled with translating ribosomes. Furthermore, their positioning analyses stipulated that RNA intermediates are not generated by exonuclease activities but by a repeated ribosome‐associated endonucleolytic activity, which cleaves translated RNAs at the exit site of mRNA ribosome channel. This cleavage generates a 5′‐capped mRNA fragment with potentially active elongating ribosomes. When the leader ribosome on this 5′‐capped fragment reaches the 3′‐cleavage site, it undergoes a hard stall (the 3′‐end of the mRNA being in the A‐site) that induces a new endonucleolytic cleavage at the exit site of mRNA ribosome channel, generating a final degradation product of 16 nt in length. Ibrahim et al called “ribothrypsis” this novel process of ribosome‐phased endonucleolysis, for which the endonuclease remains to be identified (Figure 3(b)). Beyond that, their study provides evidence that translation‐dependent mRNA degradation is much more widespread than previously thought, as it is very widely used in canonical mRNA decay. Final ribothrypsis products are similar to the small ribosome footprints (15–18 nt) observed at known sites of mRNA truncation in yeast lacking the auxiliary exosome factor Ski2 (Guydosh & Green, 2014). However, final ribothrypsis products are naturally generated by a ribosome‐associated endonuclease while the 5′‐end of the small ribosome footprints observed in yeast are generated by the RNaseI treatment following cell lysis. It is, therefore, not known yet whether a mechanism similar to ribothrypsis is conserved in lower eukaryotes.
FIGURE 3.
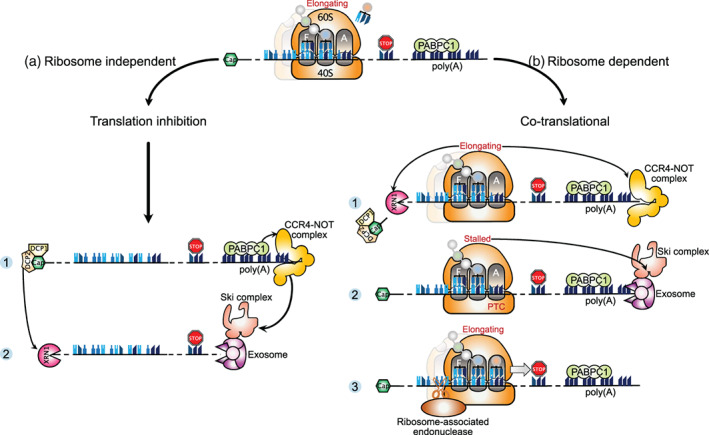
Conventional mRNA decay pathways in yeast and mammals. Cytoplasmic mRNA decay occurs independently of the translation process for the most part (a), but it can also proceed co‐translationally (b). (a) The translation‐independent mRNA decay starts with deadenylation by the CCR4–Not complex, followed by removal of the 5′‐cap by the DCP1–DCP2 complex (Panel 1). Thereafter, mRNA decay proceeds through 5′‐to‐3′ degradation by the exoribonuclease XRN1 and/or 3′‐to‐5′ degradation by the exosome assisted with the Ski complex (Panel 2). (b) Co‐translational mRNA decay is initiated when, for various reasons, a ribosome is not loaded by a new tRNA in its A‐site. As above, mRNA decay starts with deadenylation by the CCR4–Not complex, then decapping by the DCP1–DCP2 complex, leading to exonucleolytic cleavage by XRN1 (5′‐to‐3′) and the ski‐exosome complex (3′‐to‐5′). In mammals, the mRNA decay pathways are not redundant, but instead perform specialized functions. XRN1 predominantly mediates bulk mRNA decay with the aid of normal translation (Panel 1). The Ski‐exosome complex functions in global mRNA surveillance and triggers mRNA decay in the case of aberrant translation events as stalled ribosomes at a premature stop codon (Panel 2). Alternatively, mRNA decay can occur through a repeated ribosome‐associated endonucleolytic activity, for which the endonuclease remains to be identified (Panel 3)
Recently, Tuck et al. wanted to find out whether decay factors could interact with ribosomes in mammals, and to what extent mRNA decay was coupled to translation in higher eukaryotes (Tuck et al., 2020). Using mouse embryonic stem cells, they first performed a global comparison of transcripts bound by XRN1 or by the Ski complex helicase SKIV2L (homolog of yeast Ski2), and found a different specificity for a large number of transcripts, suggesting that the two cytoplasmic decay pathways could have specialized functions. Their results showed that XRN1 and SKIV2L are both recruited on translating ribosomes but their distribution is different, with XRN1 associating uniformly throughout the mRNA length and SKIV2L accumulating specifically on RNAs occupied with stalled ribosomes. This work, as a whole, sustains the idea that XRN1 mediates bulk mRNA decay with the aid of normal translation, whereas the Ski complex functions in global mRNA surveillance and assists the exosome in triggering mRNA decay in the case of aberrant translation events as stalled ribosomes (Figure 3(b)).
Consistent with mRNA decay occurring co‐translationally, a cryo‐EM structure of yeast Xrn1 bound to a ribosome was solved and the overall architecture of the Xrn1–ribosome complex is compatible with Xrn1 binding at the mRNA exit site of the ribosome (Tesina et al., 2019). Also, Schmidt et al. determined the structure of a yeast ribosome associated with the Ski2–Ski3–Ski8 complex and found that the Ski complex is positioned near the ribosomal mRNA entry tunnel (Schmidt et al., 2016). Additionally, a ribosome tagging method developed in mouse embryonic stem cells revealed that the mammalian “ribo‐interactome” is composed of a repertoire of approximately 400 proteins including the expected ribosomal proteins and translation factors but also many unexpected proteins controlling diverse cellular processes (Simsek et al., 2017). Moreover, this dataset contains several RNA helicases including SKIV2L and UPF1, as well as certain subunits of the Ccr4‐Not deadenylase complex and m6A‐binding proteins, all of which are known to regulate the efficiency of mRNA translation and decay.
4.2. The ribosome and selective co‐translational degradation of functional mRNAs
Besides the role of ribosomes in orchestrating bulk mRNA degradation, other pathways involving co‐translational degradation of selected functional transcripts exist. These involve ribosome interacting proteins that can either contact nascent peptides or specific RNA motifs within translated mRNAs to trigger their degradation.
Staufen‐mediated mRNA decay (SMD) is a translation‐dependent degradation pathway mediated by the mammalian double‐stranded RNA (dsRNA) binding proteins Staufen1 and Staufen2 (Y. K. Kim et al., 2005; E. Park & Maquat, 2013). Staufen proteins bind dsRNA structures located in the 3′‐UTR of specific mRNAs or resulting from intermolecular interactions between two different RNA species (de Lucas et al., 2014; Gong et al., 2013; Gong & Maquat, 2011; Ricci et al., 2014; Sugimoto et al., 2015). Staufen proteins interact directly with ribosomes as well as with UPF1, inducing phosphorylation of the latter and stimulating its helicase activity (Y. K. Kim et al., 2005; E. Park & Maquat, 2013). Similarly to NMD, SMD requires active translation of the target mRNA to trigger its decay. Interestingly, Staufen proteins and UPF2 were shown to bind to UPF1 at overlapping regions thus suggesting that their association to UPF1 is mutually exclusive (Gong et al., 2009). As a consequence, NMD and SMD were shown to be in competition, with the down‐regulation of Staufen proteins activating NMD while UPF2 down‐regulation stimulates SMD (Gong et al., 2009). However, biochemical reconstitution of UPF1 recruitment to Stau1 in vitro indicates a crucial role for UPF2 in facilitating UPF1 binding to Stau1 and its activation, indicating that the observed competition between NMD and SMD does not result from a competition of UPF2 and Stau1 to bind UPF1 (Gowravaram et al., 2019). Regarding its biological roles, SMD has been implicated in the regulation of myogenesis, adipogenesis, and in the local degradation of neuronal mRNAs upon their transport in axons, among others (Cho et al., 2012; Gong et al., 2009; J. Y. Kim et al., 2017).
Similarly to Staufen proteins, Regnase‐1 is an RNA‐binding protein with RNase activity that mediates translation‐dependent mRNA degradation of specific transcripts through UPF1 (Mino et al., 2015). Regnase‐1 binds to specific RNA stem‐loops in the 3′‐UTRs of inflammation‐related mRNAs and plays an essential role in modulating the immune response (Cui et al., 2017; Kidoya et al., 2019; Mino et al., 2015; Tanaka et al., 2019; Uehata et al., 2013). Regnase‐1 can bind to target mRNAs before they are translated but, in the absence of translation, this interaction does not result in mRNA degradation (Mino et al., 2019). Following the pioneer round of translation, UPF1 interacts with Regnase‐1 and this association induces UPF1 phosphorylation and stimulates its RNA‐helicase activity. As a consequence, UPF1 unwinds the RNA stem‐loop structure bound to Regnase‐1 and this conformational switch licenses Regnase‐1 to cleave the target mRNA (Mino et al., 2019). Together, SMD‐mediated and Regnase‐1‐mediated mRNA decay depend on specific RNA structures to recognize their target RNAs and exploit UPF1 to mediate translation‐dependent mRNA decay, although through different mechanisms.
Translation‐dependent mRNA degradation can occur independently of a sequence or a structural motif within the target mRNA but instead rely on the co‐translational recognition of nascent peptides. Such a mechanism has been described for the degradation of mRNAs coding for secretory proteins whenever they fail to be correctly addressed to the endoplasmic reticulum (ER). Secretory proteins contain an N‐terminal signal peptide that mediates co‐translational recruitment of the signal recognition particle (SRP; Walter et al., 1981). Following its recruitment to the nascent peptide, the SRP induces a transient arrest of elongation and interacts with its receptor located in the rough ER to mediate the correct translocation of the nascent protein into the ER lumen before translation is resumed (Wolin & Walter, 1988, 1989). Mutations of the signal peptide that impair SRP recruitment lead to a decrease in protein expression and degradation of the corresponding mRNA (Karamyshev et al., 2014). Similarly, when SRP levels are down‐regulated, the mRNAs coding for secretory proteins are destabilized within the cell. The absence of the SRP at the ribosome exit tunnel was shown to allow the Argonaute2 (Ago2) protein to interact with the nascent peptide and the ribosome to trigger mRNA degradation through a still uncharacterized mechanism that is independent of its slicing activity (Karamyshev et al., 2014). However, Ago2 is not required for the degradation of all mRNAs encoding secretory proteins that fail to recruit SRP, thus suggesting that other proteins could be involved in this degradation pathway (Pinarbasi et al., 2018). Furthermore, it is not clear how the cell discriminates between mRNAs coding for secretory proteins that fail to recruit an SRP from mRNAs coding for cytosolic proteins.
Another example of co‐translational mRNA degradation through recognition of nascent peptides occurs for the regulation of tubulin expression. Indeed, the levels of alpha and beta tubulins are tightly controlled within cells, through an autoregulation mechanism that induces the degradation of tubulin mRNAs when the concentration of soluble tubulin proteins (i.e., non‐polymerized) is above a certain threshold (Cleveland, 1988; Cleveland et al., 1981; Gasic et al., 2019). This regulation occurs co‐translationally and depends on the recognition of the first four residues of the alpha and beta tubulin proteins (Met–Arg–Glu–Ile for β‐tubulin and Met–Arg–Glu–Cys for α‐tubulin) by the tetratricopeptide protein 5 (TTC5) (Bachurski & Cleveland, 1994; Gay et al., 1989; Lin et al., 2020; Pachter, 1987; Theodorakis & Cleveland, 1992; Yen et al., 1988). TTC5 associates with nascent tubulin peptides and contacts the ribosome near the exit tunnel through interactions with 28S rRNA and ribosomal protein uL24 (Z. Lin et al., 2020). Mutation of TTC5 residues that interact with ribosomes leads to loss of the regulation. The authors also found that, when soluble tubulin levels are low, the binding of TTC5 to nascent tubulin peptides is prevented by a still unknown factor. In contrast, the activity of this factor is inhibited whenever cells detect an excess of soluble tubulin, thus allowing TTC5 to bind to nascent tubulin peptides and ribosomes and to induce mRNA degradation. The precise mechanism by which TTC5 mediates mRNA degradation is yet to be characterized. It could rely on the recruitment of mRNA decay factors to TTC5‐mediated stalled ribosomes or on the recognition of a specific ribosomal conformational change induced by TTC5.
Altogether, these studies highlight the crucial role of ribosomes in integrating multiple signals originating from the nascent peptide or from cis‐acting elements within mRNAs to mediate co‐translational degradation of functional mRNAs.
4.3. The ribosome and associated quality control mechanisms
In addition to the emerging role of the ribosome as a platform for a large number of interactors, ribosome ubiquitination appears to be crucial, particularly in the context of ribosome collisions. Obviously, collided ribosomes are detrimental to protein homeostasis and thus require mechanisms to cope with them. The vast majority of insights about ribosome collisions has been obtained from model substrates consisting of aberrant mRNAs with stalling sequences which, when translated, engender the leading ribosome to stall, causing collisions with trailing ribosomes (see the Sections 2.2 and 2.3 about the NGD and NSD mRNA surveillance pathways). A few studies have now sought to systematically identify the proteins associated with collided ribosomes. Widespread ribosomal collisions can be induced using low‐doses of the elongation inhibitor emetine. Using quantitative proteomics on polysome fractions from cells treated with low‐dose emetine, two groups identified the protein Endothelial differentiation‐related factor 1 (EDF1) as being the new first‐acting ribosome collision sensor (Juszkiewicz et al., 2020; Sinha et al., 2020). Cryo‐EM structure reveals that EDF1 and its highly conserved yeast homolog Mbf1 bind at the disome interface close to the mRNA entry channel (Sinha et al., 2020). If collisions persist, EDF1 is needed for stabilization of both the E3 ubiquitin ligase ZNF598 and the protein Grb10‐interacting GYF (glycine‐tyrosine‐phenylalanine) domain protein 2 (GIGYF2) at collided ribosomes (Juszkiewicz et al., 2020; Sinha et al., 2020). GIGYF2 and ZNF598 were previously found to interact with the eIF4E‐homologous protein (4EHP) (Morita et al., 2012), which inhibits translation by competing with eIF4E for binding to the 5′‐cap structure (Rom et al., 1998). Several findings suggest that both GIGYF2‐4EHP and ZNF598 aim to minimize ribosome collisions: the translational repressor GIGYF2‐4EHP blocks new rounds of translation onto problematic mRNAs and marks these transcripts for decay whereas ZNF598 catalyzes 40S ribosomal subunit ubiquitinations required for ribosome rescue by the ribosome‐associated quality control (RQC) pathway (Hickey et al., 2020; Juszkiewicz et al., 2020; Sinha et al., 2020; Weber et al., 2020; Figure 4). In particular, ZNF598 is able to discriminate between transient or long‐lasting collisions and is essential to resolve ribosome queuing that can form on mRNAs containing ribosome stalling sequences (Goldman et al., 2021). In addition to ZNF598, a second E3 ubiquitin ligase was reported to function in RQC. The makorin ring finger protein 1 (MKRN1) binds to prematurely polyadenylated transcripts and ubiquitinates the ribosomal protein eS10 and PABPC1 (Hildebrandt et al., 2019). While it is clear that ubiquitinations are key for proper rescue of stalled ribosomes, their precise role and temporal sequence are just beginning to emerge. It has been found that the ZNF598‐catalyzed ubiquitinations are reversed by the deubiquitinating enzymes USP21 and OTUD3 (Garshott et al., 2020). Furthermore, the authors reported that ubiquitination events are hierarchically organized, suggesting the existence of an ubiquitin code on ribosomes. Alternatively, ubiquitination marks may serve as a scaffold for recruiting downstream quality control factors involved in resolution of ribosome stalls.
FIGURE 4.
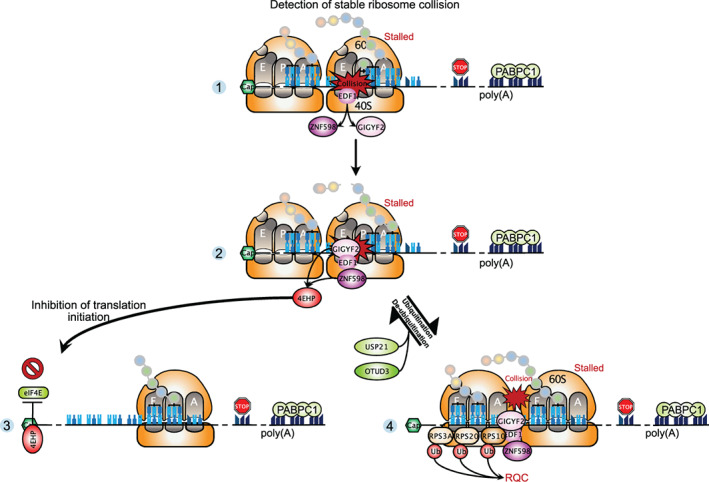
Ribosome‐associated quality control mechanisms. Upon ribosome collision induced by problematic mRNAs, EDF1 is recruited first at the disome interface (Panel 1), and then it stabilizes the interaction of ZNF598, GIGYF2, and 4EHP with collided ribosomes (Panel 2). These proteins act in two distinct ways to minimize ribosome collision: (1) GIGYF2‐4EHP represses initiation of new rounds of translation by competing with eIF4E for binding to the cap structure and marks problematic mRNAs for decay (Panel 3). (2) ZNF598 ubiquitinates several 40S ribosomal proteins and these ubiquitinations are required for the ribosome‐associated quality control involved in degradation of the nascent peptide and rescue of collided ribosomes. The ubiquitinations can be reversed by the deubiquitinases USP21 and OTUD3 (Panel 4)
4.4. The ribosome and associated stress responses
A novel function was ascribed to ribosome collisions, apart from their role in quality control. Two studies have now shown that general cellular stress conditions induce high levels of ribosome collisions and that the resulting colliding ribosomes are a key signaling node to coordinately regulate stress responses (C. Wu et al., 2020; Yan & Zaher, 2020). Specifically, a first study in human cells revealed that cellular stress caused by amino acid starvation or UV irradiation induce a large number of colliding ribosomes that serve as a platform to recruit the kinase ZAKα (C. Wu et al., 2020). This protein is a common activator of the p38/JNK‐mediated ribotoxic stress response and the integrated stress response (ISR), the core event of which is the phosphorylation of the initiation factor eIF2α by one of four kinases, including GCN2. The second study in yeast showed that alkylation or oxidation stresses promote ribosome collisions and trigger both the Gcn2‐mediated ISR pathway and the RQC pathway activated by the Hel2 ubiquitin ligase (ZNF598 in mammals; Yan & Zaher, 2020). Notably, an apparent competition was observed between these two pathways, suggesting that ISR and RQC must be coordinated somehow. What seems to emerge from these two studies is that the extent of ribosome collisions is the major determinant of which signaling pathway should be activated. In unstressed cells, basal levels of colliding ribosomes, which reflect occasional problematic mRNAs, are suggested to induce the RQC pathway in order to degrade the aberrant transcript and nascent peptide. In stressed cells, intermediate levels of ribosome collisions could activate the GCN2‐mediated ISR pathway that blocks global translation initiation but induces translation of specific mRNAs to promote cell survival. In contrast, high levels of colliding ribosomes are thought to trigger the p38/JNK‐mediated stress response in order to induce apoptotic cell death. From this perspective, ZAKα would have a role in monitoring the extent of ribosome collisions.
5. CODON AND AMINO ACID USAGE MODULATE mRNA STABILITY
Because the genetic code is degenerate, most amino acids, except methionine and tryptophan, are encoded by multiple codons known as synonymous codons. In eukaryotes, the number of tRNA genes in the genome can vary from 275 in yeast (S. cerevisiae) to 416 in humans, and more than 8000 in zebrafish (P. P. Chan & Lowe, 2016). However, in all known organisms there are fewer tRNA anticodons than the 61 codons encoding the 20 canonical amino acids. The limited number of available anticodons is compensated by the possibility of non‐Watson–Crick base pairings between the first position of the anticodon and the third position of the codon, known as the wobble decoding (Crick, 1966).
Usage of synonymous codons is not random and differs between species and within genes for a given organism in what is described as “codon usage bias” (Grantham et al., 1980). Codon usage bias is thought to depend on global genomic GC content, resulting from mutational bias, and on selective pressure acting on coding sequences (Chen et al., 2004). Codon usage has been shown to affect ribosome decoding speed and co‐translational folding of nascent proteins (Drummond & Wilke, 2008; Yu et al., 2015; Zhao et al., 2017; T. Zhou et al., 2009). In some bacteria, lower eukaryotes (such as S. cerevisiae, C. elegans, and D. melanogaster) and plants (A. thaliana), codon usage has been shown to be biased towards the most abundant tRNAs (Duret & Mouchiroud, 1999; Grantham et al., 1980; Gutman & Hatfield, 1989; Ikemura, 1985). Furthermore, codon usage bias is positively correlated with transcript abundance in these species, leading to the most abundant transcripts being further enriched for codons corresponding to abundant tRNAs (Duret & Mouchiroud, 1999; Gouy & Gautier, 1982; Ikemura, 1985; Sharp & Li, 1986, 1987). These features support a “translational selection” model, where codons decoded by the most abundant tRNAs are positively selected among highly expressed genes, to optimize translational efficiency and accuracy (Akashi, 1994; Moriyama, 1998). Consistent with this hypothesis, ribosome profiling assays in S. cerevisiae and N. crassa in the absence of translation inhibitors show a negative correlation between tRNA abundance and ribosome elongation dwell time (Dana & Tuller, 2014; Gardin et al., 2014; Hanson et al., 2018; Hussmann et al., 2015; Weinberg et al., 2016). Nevertheless, tRNA levels do not exclusively explain codon decoding rates and other factors including wobble base‐pairing, the nature of the amino acids at the P‐site and A‐site (defining peptidyl transfer time) and at proximity of the exit tunnel of the ribosome have also been shown to modulate the dwell time on a given codon (Gardin et al., 2014; Stadler & Fire, 2011).
To quantify the degree of synonymous codon usage bias within an organism and between species, different metrics have been developed, each with their own pros and cons. For example, the Codon Adaptation Index (CAI) calculates the relative synonymous codon usage (RSCU) value from a restricted set of highly expressed transcripts in a given organism (Sharp & Li, 1987). The obtained values are then used to calculate a score for each gene within that same organism. It is a simple and powerful metric to monitor RSCU, but it is based on the assumption that translational selection occurs within the studied organism. To quantify translational selection, the tRNA adaptation index (tAI) was created (Reis et al., 2004) that takes into account tRNA abundance and the binding strength of the codon–anticodon coupling to measure how well a given mRNA is adapted to the available tRNA pool (using tRNA gene number as a proxy for tRNA abundance). Using the tAI, authors were able to study the degree of co‐adaptation between codon usage and tRNA levels in a wide range of organisms and quantify the extent of translational selection acting on codon usage (Reis et al., 2004). Interestingly, their results indicate that genome size and tRNA gene number act in combination to determine the extent of translational selection on codon usage bias. Above a certain genome size, translational selection should no longer be observed. This raises many questions regarding the impact of codon usage and translational selection in lower eukaryotes with small genomes, and clear evidence of translational selection, compared with higher eukaryotes with larger genomes and a relatively restricted number of tRNA gene copies.
5.1. Codon usage is a major determinant of mRNA decay rates in yeast
Evidence for codon usage affecting mRNA stability was first proposed in S. cerevisiae, by manipulating the codon content of the highly expressed gene PGK1, which is enriched for synonymous codons corresponding to the most abundant tRNA species (Hoekema et al., 1987). Replacing “optimal” codons (i.e., those corresponding to the most abundant tRNAs and over‐represented in highly expressed genes) within the PGK1 coding region by “non‐optimal” synonymous codons resulted in a 10‐fold reduction of protein output and a 3‐fold reduction of steady‐state mRNA levels, which was attributed to decreased mRNA stability (Hoekema et al., 1987). Further evidence linking codon usage to mRNA stability was obtained by direct measurement of mRNA decay rates for 20 different yeast endogenous mRNAs, where authors found that mRNA half‐life was correlated to the percentage of rare codons within the ORF (Herrick et al., 1990). Proof of a direct role for translation elongation and codon usage was obtained by expressing chimeric mRNAs corresponding to in‐frame fusions of the stable PGK1 or ACT1 transcripts with that of the unstable MATα1 transcript (Caponigro et al., 1993; Parker & Jacobson, 1990). This approach led to the identification of a stretch of rare codons in the coding region of MATα1 that was required, but not sufficient, to induce decay of chimeric mRNAs. Both insertion of a stop codon upstream of the stretch of rare codons from MATα1 or translation inhibition with cycloheximide led to a stabilization of the chimeric mRNAs, implying that ribosomes elongating across the stretch of rare codons are required to induce mRNA degradation (Caponigro et al., 1993; Parker & Jacobson, 1990). This observation was generalized in a transcriptome‐wide study monitoring mRNA decay in S. cerevisiae (Presnyak et al., 2015). Using a new metric described as the codon occurrence to mRNA stability correlation coefficient (CSC), that corresponds to the Pearson correlation between the frequency of each codon within the coding region in mRNAs and their corresponding half‐lives, Presnyak et al were able to identify codons associated with mRNA stability or instability. Interestingly, CSC values in S. cerevisiae were found to be positively correlated with codon optimality as defined by the tAI metric (Reis et al., 2004), and linked to the speed of ribosome decoding (Figure 5). More precisely, mRNA stability correlates with the codon‐specific elongation rate at the A‐site of the ribosome but not with amino acid identity or codon usage at the P‐site and E‐site (Hanson et al., 2018). This indicates that among all steps that can influence the dynamics of translation elongation, the kinetics of tRNA recruitment at the A‐site is the only one that is probed by the cell to induce mRNA decay in yeast. Further dissection of the molecular determinants linking codon usage and mRNA decay in S. cerevisiae identified the DEAD‐box protein Dhh1p (DDX6 in mammals) as a key factor implicated in monitoring the decoding speed of ribosomes and in inducing translational repression and degradation of transcripts with non‐optimal codons (Radhakrishnan et al., 2016; Sweet et al., 2012).
FIGURE 5.
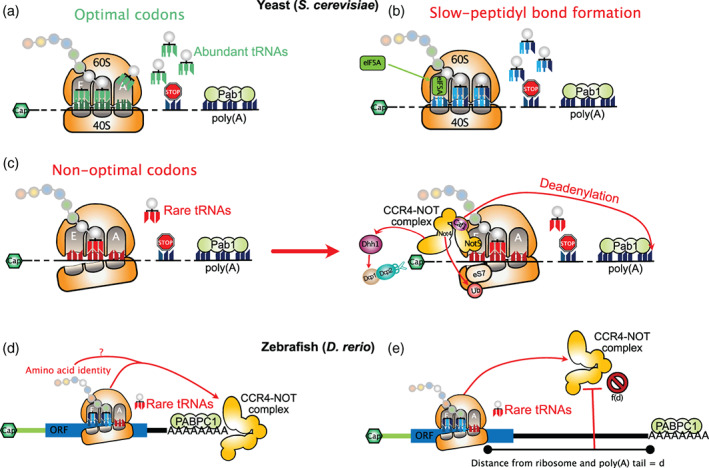
Codon‐dependent mRNA decay in yeast and zebrafish. In yeast, the kinetics of tRNA recruitment at the A‐site of elongating ribosomes is a major determinant of mRNA stability. (a) mRNAs with optimal codons (corresponding to abundant tRNAs) mediate efficient recruitment of tRNAs in the A‐site of elongating ribosomes and display long half‐lives. (b) If peptidyl transfer is slow but an optimal codon resides at the ribosome A‐site, the ribosome will bear an empty E‐site and an accommodated A‐site. This conformation allows recruitment of eIF5A to the E‐site to promote peptidyl transfer activity and rescue the stalled ribosome. (c) If a non‐optimal codon is present at the ribosome A‐site, tRNA accommodation will take longer than for an optimal codon and this increases the probability of the ribosome to bear empty E‐site and A‐site. This specific post‐translocation conformation favors the recruitment of the Ccr4–Not complex in the E‐site, which induces eS7 ubiquitination, mRNA deadenylation through the Caf1 subunit and stimulates Dhh1p recruitment to induce mRNA degradation. (d) In zebrafish, a similar Ccr4–Not‐dependent mechanism probes codon optimality to induce mRNA degradation in a translation‐dependent manner. However, in addition to codon usage, amino‐acid content can also mediate mRNA destabilization through a still uncharacterized mechanism. (e) In zebrafish, the distance between the non‐optimal codon and the poly(A) tail acts as a buffer against the deadenylase activity of the Ccr4–Not complex
In addition to Dhh1p, the Ccr4–Not complex is involved in monitoring codon optimality. The yeast Ccr4–Not core complex, composed of two catalytic subunits (Ccr4 and Caf1) and five Not proteins (Not1 to 5) (H.‐Y. Liu et al., 1998), catalyzes mRNA deadenylation. Furthermore, the Ccr4–Not complex interacts with 80S ribosomes and represses translation of mRNAs with stalled ribosomes (Preissler et al., 2015). Moreover, the Not4 and Not5 core subunits have been shown to mediate ubiquitination of the ribosomal protein eS7 and translational control (Panasenko & Collart, 2012; Villanyi et al., 2014). In a study aimed at understanding the role of Pab1 in regulating the activity of the Ccr4–Not complex, Webster et al (Webster et al., 2018) showed a direct interaction between Pab1 and the Ccr4–Not complex, which is required for efficient bulk poly(A) shortening mostly catalyzed by Ccr4. In contrast, Caf1 is not necessary for bulk mRNA deadenylation but is required for deadenylation of mRNAs bearing non‐optimal codons (Figure 5(c)). In this case, Caf1 acts in a translation‐dependent manner, as introduction of a stem–loop structure in the 5′‐UTR to block ribosome loading relieves Caf1‐dependent deadenylation of such transcripts. Interestingly, the authors observed a positive correlation between Pab1 binding and codon optimality, arguing that translation elongation rates could have an impact on the relative abundance of Pab1 associated with transcripts. In this model, mRNAs with optimal codons and high Pab1 loading are deadenylated by the Ccr4–Not complex through the action of the Ccr4 nuclease alone, while deadenylation of mRNAs with non‐optimal codons and low Pab1 binding is enhanced by the combined action of Ccr4 and Caf1. This step acts upstream of the activity of Dhh1p, since yeast lacking the DHH1 gene still show Caf1‐dependent deadenylation of mRNAs with non‐optimal codons. Further refining the role of Ccr4–Not in linking codon optimality to mRNA deadenylation and decay in yeast, a second study by Buschauer et al. purified ribosomes bound to Ccr4–Not to perform cryo‐EM analyses (Buschauer et al., 2020). Their results indicate an association of the entire Ccr4–Not complex with monosomes and polysomes and, more specifically, an interaction of the N‐terminal domain (NTD) of Not5 with the E‐site of post‐translocation ribosomes carrying a peptidyl‐tRNA in the P‐site and a vacant A‐site (Figure 5(c)). Incubation of cells with cycloheximide, that favors the pre‐translocation state with an accommodated tRNA in the A‐site, decreases Ccr4–Not binding to ribosomes and increases eIF5A binding to the E‐site, indicating specific recruitment of the Ccr4–Not complex to ribosomes with a vacant A‐site (Figure 5(c)). Furthermore, ribosome profiling of Ccr4–Not‐bound ribosomes revealed an enrichment for non‐optimal codons at the A‐site of ribosome footprints, as defined by their CSC or tAI values. Deletion of Not5–NTD impairs Dhh1p recruitment on ribosomes with empty A‐sites and reduces their decapping, therefore linking the role of Ccr4–Not in monitoring translation elongation rates with the downstream activity of Dhh1p in mediating mRNA decapping leading to decay. Binding of Not5–NTD to the ribosome E‐site was finally shown by authors to depend on the prior ubiquitination of eS7 by Not4 (Figure 5(c)). Such a mechanism allows cells to monitor translation elongation speed and to discriminate between ribosomes displaying slow decoding and those showing slow peptidyl transfer: slow‐decoding ribosomes, characterized by a peptidyl‐tRNA loaded in the P‐site and a vacant A‐site, induce Ccr4–Not deadenylation and Dhh1p decapping followed by 5′‐to‐3′ degradation (Figure 5(c)) whereas ribosomes displaying slow peptidyl transfer, characterized by a peptidyl‐tRNA loaded in the P‐site and a tRNA accommodated in the A‐site, are rescued by eIF5A (Figure 5(b)), which promotes peptidyl transferase center activity (Schuller et al., 2017).
5.2. Codon and amino acid usage coupled to GC content regulate mRNA decay in metazoans
In addition to yeast, several pieces of evidence support that the role of codon usage in mediating mRNA decay may be conserved across most eukaryotes, including metazoans. However, the constraints imposed by multicellular organisms with large genomes suggest a more complex relationship between codon usage, tRNA expression, and mRNA decay than the one observed in yeast. In particular, additional factors such as amino acid identity, GC content, UTR length, and RNA binding proteins appear to modulate the impact of codon usage on mRNA stability or even drive translation‐dependent mRNA decay independently from it.
Two studies performed in zebrafish uncovered a role for codon usage in mediating translation‐dependent degradation of maternal mRNAs during the maternal to zygotic transition (MZT), similar to yeast (Bazzini et al., 2016; Mishima & Tomari, 2016). During the late stages of oogenesis, the maternal genome becomes transcriptionally silent and relies on stored mRNAs and proteins to undergo the meiotic maturation, fertilization and first steps of embryo development (Tadros & Lipshitz, 2009). Degradation of maternal mRNAs during MZT is an essential step during embryogenesis that allows reprogramming of gene expression from the maternal genome to the zygotic genome. In order to identify cis‐acting factors involved in maternal mRNA degradation, Bazzini et al. used an elegant approach relying on the use of synthetic reporter mRNAs with invariable 5′‐UTR and 3′‐UTR sequence, but with variable codon composition, to demonstrate that mRNA stability during the MZT was modulated by codon usage (Bazzini et al., 2016). Similar results were obtained with endogenous mRNAs upon transcriptional inhibition, which allowed them to identify optimal and non‐optimal codons, using the CSC metric described by Presnyak et al. (2015). Consistent with findings obtained in S. cerevisiae, the authors found a mild but significant correlation between tRNA levels and codon optimality as well as a clear correlation between codon usage in the transcriptome and codon optimality (Figure 5(d)). Moreover, mRNAs with non‐optimal codons display shorter poly(A) tails and a lower translational efficiency. Finally, the authors uncovered a translation‐dependent effect of specific amino‐acids in regulating mRNA stability (Figure 5(d)), thus suggesting that metazoans, in contrast to S. cerevisiae, have evolved additional mechanisms to probe for translation elongation dynamics. Mishima and Tomari (2016) also found that codon usage is a major determinant of maternal mRNA stability during the MZT in zebrafish and that the exonuclease activity of the Ccr4–Not complex is responsible for co‐translational poly(A) tail shortening of mRNAs bearing uncommon codons (Figure 5(e)). Surprisingly, the authors also found that long 3′‐UTRs could protect mRNAs with uncommon codons from degradation. This protective role was sequence‐independent but, instead, relied on the distance between the poly(A) tail and the uncommon codons (Figure 5(e)). Altogether, these results argue that Ccr4–Not acts as a sensor of ribosomes with vacant A‐sites, both in yeast and metazoans. Further supporting this conclusion, Not5 (CNOT3 in metazoans), which is involved in anchoring Ccr4–Not to the ribosome is highly conserved across eukaryotes (Buschauer et al., 2020).
Interestingly, although zebrafish has a large genome (1.5 billion base pairs), it does possess an unusual large number of tRNA genes (>8000) (P. P. Chan & Lowe, 2016), which could be compatible with translational selection acting on codon usage (Reis et al., 2004), as observed in S. cerevisiae. However, organisms with large genomes and a limited number of tRNA genes (as it is the case for most mammals) should be less prone to translational selection on codon usage, thus raising the question of whether codon usage could have an impact on mRNA translation and stability in those species. Numerous studies have addressed this question in different mammalian species (human, mouse, and Chinese hamster) using different technical approaches to monitor mRNA decay rates and the impact of codon usage (Forrest et al., 2020; Narula et al., 2019; Shu et al., 2020; Q. Wu et al., 2019). Q. Wu et al. (2019) and Narula et al. (2019) both used a library of reporter genes with fixed UTRs and variable composition of the coding sequence, combined with non‐invasive metabolic labeling of nascent transcripts or transcription inhibitors, to monitor the impact of codon usage on mRNA stability in HEK 293T, HeLa, RPE, and K562 cells. Forrest et al. (2020) and Shu et al. (2020) relied on metabolic labeling of nascent transcripts to monitor mRNA stability of endogenous transcripts in HeLa, CHO, and primary mouse neurons. Interestingly, despite the methodological differences, most studies have yielded similar CSC values, which validates a role for codon usage in regulating mRNA stability in mammals and indicates a conservation of optimal and non‐optimal codons between these organisms, independent of the cell type (Shu et al., 2020). Although the results are not as robust as those observed in yeast, the authors showed that, in human cells, the effects of codon usage on mRNA decay rates are explained, to some extent, by the tRNA abundance and speed of codon decoding (Forrest et al., 2020; Narula et al., 2019; Shu et al., 2020; Q. Wu et al., 2019; Figure 6(a)). In agreement with this, overexpression of specific tRNAs in metastatic cells was shown to increase translation and stability of mRNAs enriched with cognate codons (Goodarzi et al., 2016). Similarly to findings in yeast and zebrafish, transcripts with non‐optimal codons in mammals display a shorter poly(A) tail. However, the presence of a poly(A) tail is not required for codon‐dependent mRNA degradation, since transcripts with non‐optimal codons bearing a histone tail instead of a poly(A) tail are still prone to degradation (Q. Wu et al., 2019). Finally, in addition to a role of synonymous codons in inducing mRNA decay, Forrest et al also identified several amino‐acids whose incorporation is associated with mRNA stabilization or destabilization (Forrest et al., 2020) (Figure 6(a)). Therefore, both synonymous codons and specific amino acids are able to modulate mRNA stability in mammals, similarly to zebrafish but contrary to S. cerevisiae.
FIGURE 6.
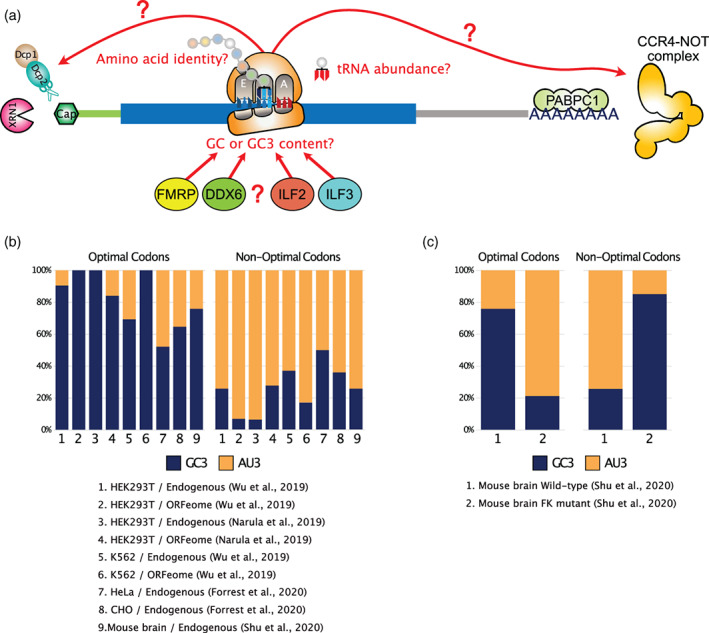
GC3 content, amino acid identity, and RBPs modulate translation‐dependent mRNA degradation in mammals. (a) Possible factors implicated in codon‐dependent mRNA destabilization in mammals. (b) Barplot representing the percentage of GC3 and AU3 codons among optimal and non‐optimal codons as defined for endogenous mRNAs or ORFeome reporters with fixed UTRs and variable codons in the CDS from different cell types as obtained from the literature. In most studies, optimal codons are mainly GC3, while non‐optimal codons are enriched in AU3. (c) FMRP deficiency in mouse primary neurons leads to an inversion in the GC3 and AU3 content of optimal and non‐optimal codons
Surprisingly, in all but one of the aforementioned studies, a clear bias in the GC content at the wobble position can be observed among optimal and non‐optimal codons (Figure 6(b)), although this specific finding was not highlighted by authors. Independently of whether endogenous mRNAs or ORFeome reporters were studied, codons enriched for cytidine and guanine at the wobble position (GC3) are typically associated with positive CSCs and classified as optimal (Figure 6(b), see “Optimal Codons”), while codons enriched for adenine and uridine at the wobble position (AU3) are typically associated with negative CSCs and classified as non‐optimal (Figure 6(b), see “Non‐optimal Codons”). GC3 bias is particularly strong in HEK 293T and K562 cells, where up to 100% of optimal codons are GC3 and up to 93% of non‐optimal codons are AU3 (Figure 6(b), Conditions 1–6). Further supporting a role of GC3 content in modulating translation‐dependent mRNA degradation in mammals, a study by Hia et al. performed an analysis of the frequency of codons across all human coding sequences and found that 22% of the total variance is explained by GC3 content, correlating with CSC values obtained from HEK 293 cells (Hia et al., 2019). They also found that GC3‐rich transcripts tend to be more stable than AU3‐rich transcripts, both on a transcriptome‐wide scale and when using reporter constructs with different GC3 content, which agrees with the GC3 bias observed between optimal and non‐optimal codons in other studies. Authors also found that genes with optimal GC3‐rich codons display higher translation efficiencies than transcripts enriched with AU3 codons, as measured by ribosome profiling (Hia et al., 2019).
In agreement with these observations, it has been shown that 70% of synonymous codon usage within human genes is explained by local GC3 content, which is caused by the process of GC‐biased gene conversion (gBGC) that occurs during meiotic recombination in mammals, favoring transmission of GC‐alleles over AT‐alleles (Pouyet et al., 2017). Accordingly, there should be limited room for other selective processes to act on codon usage at a global scale in mammalian genomes. This does not exclude an impact of synonymous codon usage and GC3 content on the regulation of mRNA translation and decay, as clearly shown by the above‐mentioned studies. However, it raises many questions regarding the mechanisms evolved by mammalian cells to cope with the gBGC‐driven content bias.
One possible mechanism could be through the regulated expression or aminoacylation of tRNAs. Indeed, it has been proposed that genes related to cellular proliferation have different codon usage than those involved in differentiation. Particularly, proliferation related genes tend to have an A or T at the third codon position, while differentiation related genes tend to be enriched in C or G (Gingold et al., 2014). In the same study, authors also found evidence of coordination in the gene expression pattern of tRNAs and mRNAs during cell differentiation and cancer where tRNA corresponding to the codons enriched in proliferation or differentiation are differentially expressed to match the codon demand associated with differential transcript expression. A GC content bias in the codon composition of differentially expressed genes in differentiating and proliferating cells has also been described in other studies although no‐associated changes in tRNA expression were found (Bornelöv et al., 2019; Guimaraes et al., 2020; Rudolph et al., 2016). Nevertheless, changes in tRNA charging or editing of specific anticodon loops were found to occur and mediate changes in ribosome density that could potentially regulate mRNA stability (Bornelöv et al., 2019; Guimaraes et al., 2020).
A second possible mechanism to explain the impact of GC3 content in regulating mRNA stability could be through the action of RNA‐binding proteins (RBPs) with biased binding specificities. Hia et al identified several RBPs, preferentially bound to either GC3‐rich or AU3‐rich transcripts (Hia et al., 2019). Among these, ILF2 and ILF3 were shown to interact with AU3‐rich transcripts and induce their decay. The RBP Fragile X mental retardation protein (FMRP) also functions in regulating mRNA decay through codon usage in mouse neurons (Shu et al., 2020) and CSC values in neurons show a clear bias in favor of GC3 among optimal codons and AU3 among non‐optimal codons (Figure 6(c)). Loss of FMRP induces a complete reshuffling of CSC values in neurons, leading to optimal codons showing a clear bias in favor or AU3 and non‐optimal codons enriched for GC3 (Figure 6(c)). FMRP was shown by authors to bind preferentially to mRNAs with optimal codons, independently of the overall transcript GC content (but not GC3 content), thus suggesting a requirement for active translation of GC3‐rich codons to mediate FMRP‐dependent effects (Shu et al., 2020). Finally, GC and GC3 content have been shown to directly regulate mRNA storage within P‐bodies and dictate the decay pathway of AU‐rich and GC‐rich transcripts in human cells (Courel et al., 2019). AU‐rich transcripts are typically enriched in P‐bodies, contain rare codons and are poorly translated, while GC‐rich transcripts contain frequent codons and are efficiently translated. Interestingly, in this study, DDX6 (the mammalian homolog of Dhh1p) was shown to bind to GC‐rich transcripts enriched with high usage codons and to regulate their degradation via the XRN1 5′‐to‐3′ exonuclease. On the contrary, DDX6 regulates the translation of AU‐rich transcripts that are mainly degraded by the PAT1B 3′‐to‐5′ exonuclease (Courel et al., 2019). Together, these results indicate an important role for several RBPs in affecting stability of mammalian transcripts in a codon and GC specific manner. However, their precise mechanism of action remains to be fully characterized. In particular, the role of DDX6 could apparently differ between mammals and yeast. Additionally, it is currently unclear to what extent the differences in translation and stability observed between GC‐rich and AU‐rich transcripts are driven by the primary sequence or RNA secondary structures (GC content and secondary structures being highly correlated). In an attempt to disentangle the two factors, a study reported that both secondary structure and codon usage appear to modulate translation efficiency and the functional half‐life of transfected synthetic transcripts in a coherent but independent manner (Mauger et al., 2019). Transcripts with extensive secondary structures display higher ribosome loading and longer functional half‐lives. Additional work will be necessary to better understand the precise role of codon usage, GC content and RNA secondary structure in regulating translation and decay of mammalian mRNAs.
Altogether, the role of codon usage in modulating mRNA degradation appears much more complex in mammals than in unicellular organisms. This is likely due to additional layers of gene regulation and differences in the selective pressures that shape the nucleotide composition at the genomic scale. Nevertheless, in spite of this complexity, there is evidence that, tRNA availability plays an important role in addition to RNA‐binding proteins. In humans, expression of tRNAs is not perfectly correlated to tRNA copy number (Behrens et al., 2021), thus indicating a potential level of regulation. Accordingly, the expression of tRNA isodecoders has been shown to differ significantly between tissues (Pinkard et al., 2020; Schmitt et al., 2014). However, in spite of the observed differences in isodecoder expression, the global anticodon pool (integrating all isodecoders together) remains very similar across tissues (Pinkard et al., 2020; Schmitt et al., 2014). More work is necessary to understand how tRNA expression and post‐transcriptional modification are dynamically regulated during cell differentiation and proliferation and how these changes affect translation and mRNA stability. Furthermore, numerous changes in tRNA expression or sequence are linked to pathological conditions such as cancer and there is evidence that these changes can affect both translation and stability of mRNAs (Goodarzi et al., 2016; Lant et al., 2019; Schaffer et al., 2019).
5.3. Codon pair identity regulates mRNA translation and stability in yeast and mammals
Similarly to synonymous codon usage bias, synonymous codon pairs display a non‐random frequency distribution that is independent from codon usage bias in many organisms (Buchan et al., 2006; Gutman & Hatfield, 1989). Codon pair usage has been shown to regulate translation elongation and translation efficiency in yeast (Gamble et al., 2016). In this process, inhibitory codon pairs were shown to involve wobble decoding to slow‐down ribosome elongation as the expression of non‐native tRNAs with exact match to anticodons was able to suppress the inhibitory effect of codon pairs. In addition to the observed effect on translation elongation, inhibitory codon pairs in yeast are associated with shorter mRNA half‐lives, even when taking into account the GC content, individual codon information and the nature of dipeptides (Harigaya & Parker, 2017). Therefore, individual codons and codon‐pairs are able to modulate mRNA stability independently in yeast, although the molecular mechanism of the latter has not been characterized yet.
In mammals, modifying codon pair usage to favor rare codon pairs is a common strategy to attenuate virus replication (Coleman et al., 2008; Shen et al., 2015). Moreover, specific synonymous codon pairs at the P‐site and A‐site of elongating ribosomes have been shown to be more frequently associated with ribosome collisions (Arpat et al., 2020). However, it is unclear from these works whether codon pair bias (CPB) itself or bias in dinucleotide composition (CpG in particular since they are underrepresented in vertebrates genomes) are responsible for the observed effects. Using a set of recoded Influenza A virus mutants with variable codon pair scores and CpG content, Groenke et al. were able to disentangle the contribution of both features on virus attenuation (Groenke et al., 2020). They found that codon pair deoptimization, but not changes in CpG frequency, are significantly associated with lower expression of viral proteins and virus attenuation both in cultured cells and in vivo. Codon pair deoptimization is associated with a significant increase in the degradation rates of viral RNAs, with a concomitant decrease in translation efficiency as measured by ribosome profiling and metabolic labeling of nascent transcripts. Interestingly, a premature arrest of ribosomes at a discrete position upstream of the stop codon was observed for one of the deoptimized transcripts, suggesting that codon pair identity can effectively affect ribosome progression and lead to transcript degradation, probably through no‐go decay. Therefore, in addition to codon usage, CPB can also represent an important determinant of mRNA stability in eukaryotic cells.
6. IMPACT OF THE M6A EPITRANSCRIPTOMIC MODIFICATION ON TRANSLATION AND STABILITY OF mRNAs
It was long thought that the fate of mRNAs was strongly dependent on the nucleotide sequence, which has direct effects on mRNA structure. In recent years, there is mounting evidence that nucleotides can be modified in ways that confer additional regulation to mRNAs, leading to an emerging field termed “epitranscriptomics.” Recent advances in RNA sequencing technologies have greatly helped to define the transcriptome‐wide distribution and function of these dynamic modifications that affect many steps of mRNA metabolism (X. Li et al., 2017). In this section, we aim to highlight recent progress made in the elucidation of the molecular mechanisms by which the most prevalent mRNA modification in eukaryotes, N6‐adenosine methylation (m6A), regulate mRNA translation and/or stability.
The m6A mark, estimated to occur in half of human transcripts, is modulated by the coordinated action of multiple proteins referred to as “writers,” “erasers,” or “readers,” which, respectively, add, remove, or bind a methyl group at the 6‐amino position of adenosine to generate N6‐methyladenosine (for a recent review see Shi et al., 2019). In brief, m6A is co‐transcriptionally catalyzed by the methyltransferase‐like (METTL)3–14 heterodimer, in association with several auxiliary proteins (J. Liu et al., 2014). In addition, for a small number of transcripts, methylation is installed by the METTL16 enzyme (Pendleton et al., 2017). The m6A modification can be reversed by the alkylated DNA repair protein alkB homolog 5 (ALKBH5) or the demethylase fat mass and obesity‐associated (FTO) protein (G. Jia et al., 2011; Zheng et al., 2013). A variety of m6A‐associated molecular events are mediated through multiple m6A readers that can be roughly divided into three groups depending on their binding mechanism (Shi et al., 2019). One group comprises the family of YTH domain‐containing proteins (YTHDF1/2/3, YTHDC1/2) that directly and robustly bind to m6A marks on mRNAs. A second category of m6A readers includes proteins that bind around or nearby m6A (HNRNPC, HNRNPG, and HNRNPA2B1). Binding of these proteins to mRNAs is enhanced because the epitranscriptomic modification induces a local remodeling of the RNA structure. A third class corresponds to RNA binding proteins that exhibit higher affinity for m6A‐containing transcripts (IGF2BP1/2/3 and FMR1). The exact underlying mechanism is unknown to date. Additionally, the eIF3 translation initiation factor can bind m6A sites within the 5′‐UTR of specific transcripts (Meyer et al., 2015). Lastly, the METTL3 methylase, which acts as a m6A writer in the nucleus, was also shown to function as a m6A reader in the cytoplasm by recognizing the mark present on 3′‐UTRs (S. Lin et al., 2016).
A number of studies have now provided valuable information on how m6A methylation can impact the fate of mRNA. Several molecular machineries with different functions are recruited to m6A‐modified transcripts via distinct reader proteins. The emerging view is that m6A marks can promote translation and/or affect stability of certain transcripts, depending on which reader protein is present, the mRNA region enriched in m6A, and the specific cellular context. Regarding the m6A readers belonging to the YTH domain family (YTHDF1/2/3), the three proteins share a set of common target mRNAs, the fate of which appears to be regulated in a cooperative and integrated manner (Shi et al., 2017). Specifically, YTHDF1 binds preferentially m6A within the 3′‐UTR and increases translation efficiency by interacting with the eIF3 initiation factor (Wang et al., 2015). YTHDF3 also recognizes m6A sites in 3′‐UTRs and exerts a translation‐promoting effect by functioning together with YTHDF1 (Shi et al., 2017). Additionally, YTHDF3 promotes mRNA decay, either by recruiting the PAN2‐PAN3 deadenylase complex (J. Liu et al., 2020) or by binding to YTHDF2 (Shi et al., 2017), which emerges as the main factor involved in decay of m6A‐modified transcripts. Recent reports have shown that YTHDF2 can promote mRNA degradation in two distinct ways. Firstly, YTHDF2 recruits the CCR4–Not deadenylase complex to m6A‐modified mRNAs, via its direct interaction with the CNOT1 component, thereby triggering deadenylation and subsequent 3′‐to‐5′ decay of m6A‐containing transcripts (Du et al., 2016; J. Liu et al., 2020). Secondly, degradation of m6A transcripts bound by YTHDF2 occurs via their association with the RNase P and its close relative RNase MRP, two ribonucleoprotein complexes that function as endoribonucleases (O. H. Park et al., 2019). The authors uncovered that the association of YTHDF2 with RNase P/MRP complex is bridged by the adaptor protein HRSP12. Consequently, the YTHDF2–HRSP12–RNase P/MRP complex triggers the cleavage of m6A transcripts and the resulting RNA fragments are subsequently processed by 5′‐to‐3′ and 3′‐to‐5′ exoribonucleases. Currently, it is not known which particular cellular contexts preferentially lead to the activation of one or the other of these two YTHDF2‐mediated decay pathways.
In addition to the aforementioned combinatorial control between YTHDF1, YTHDF2, and YTHDF3 on stability of certain m6A mRNAs, METTL3 binds to m6A marks in 3′‐UTRs and promotes translation by interacting with the eIF3 initiation factor (Choe et al., 2018; S. S. Lin et al., 2016). Alternatively, eIF3 can directly bind to m6A marks in 5′‐UTRs, particularly under various stress conditions associated with severe attenuation of cap‐dependent translation, and with a large redistribution of m6A bases from 3′‐UTRs to 5′‐UTRs (Meyer et al., 2015). In line with this, 5′‐UTR m6A methylation is strongly increased in response to heat shock stress (J. Zhou et al., 2015). Furthermore, in response to amino acid starvation, translation of the ATF4 transcript containing uORFs is up‐regulated, in part, by the dynamic removal of m6A in the 5′‐UTR (J. Zhou et al., 2018). m6A in the 5′‐UTR of ATF4 acts by controlling ribosome scanning and alternative start codon selection. Together, these studies highlight the functional importance of m6A modification in the regulation of translation initiation. Because the initiation step is a primary determinant of mRNA stability in eukaryotes (see the Section 3 about translation initiation and mRNA decay), it is reasonable to think that these m6A‐related molecular mechanisms which have an impact on the initiation of translation are also very likely to influence the stability of target mRNAs. Conversely, a m6A‐linked stabilizing effect on transcripts also seems to confer a higher translation efficiency. The m6A readers IGF2BP1/2/3, which bind to m6A sites predominantly in the 3′‐UTR of thousands of transcripts, increase the stability of their target RNAs and this stabilizing effect was found to enhance their translatability (Huang et al., 2018).
In addition to the m6A‐related effects mediated by the reader proteins, it has been suggested that m6A modifications within the mRNA coding region could directly and locally influence the dynamics of translation elongation by affecting the mRNA decoding process, meaning that the ribosome itself could be a m6A “reader‐like.” Using a single molecule‐based in vitro translation system reconstituted from E. coli, Choi et al. found that codons with m6A engenders non‐optimal interactions with cognate anticodons in the ribosomal A‐site, thus leading to a slower translation elongation dynamics (Choi et al., 2016). Another study in mammalian cells also found that the presence of m6A sites within the coding region of endogenous transcripts delayed translation elongation but, unexpectedly, if the formation of the m6A marks was prevented, a supplemental decrease of translation was observed (Mao et al., 2019). In exploring this counter‐intuitive finding further, the authors found that m6A methylation within the coding region facilitates translation elongation by resolving mRNA secondary structures. Moreover, YTHDC2, which is the only m6A reader with an RNA helicase activity, is essential for destabilization of these mRNA structures. Based on their results, Mao et al proposed that m6A residues located in coding regions reduce translation efficiency of unstructured mRNAs but promote that of structured mRNAs, by resolving stable secondary structures. Consistent with this, it was previously reported that YTHDC2 increases the efficiency of translation and subsequent degradation of its m6A target transcripts (Hsu et al., 2017). Mechanistically, the effects of YTHDC2 on translation and decay of structured transcripts could be explained by the interaction of YTHDC2 with the two helicases MOV10 and UPF1, and also the exoribonuclease XRN1 (Hsu et al., 2017; Kretschmer et al., 2018). Collectively, all the results presented in this section and illustrated on Figure 7 emphasize that the m6A modification increases the level of sophistication in the tight interplay between mRNA translation and decay.
FIGURE 7.
Impact of the m6A methylation on mRNA fate. The m6A modification can regulate the fate of modified mRNAs in different ways, that is, enhanced translation, mRNA decay, and mRNA stabilization, depending on which reader protein is present, the mRNA region enriched in m6A, and the specific cellular context. The m6A readers that promote translation are YTHDF1, YTHDF3, METTL3, YTHDC2, and eIF3. The m6A readers that promote mRNA decay are YTHDF2, YTHDF3, and YTHDC2. YTHDF2 induces mRNA decay by recruiting either the CCR4–Not complex or the endonuclease RNase P/MRP via HRSP12. YTHDF3 promotes mRNA degradation by recruiting the PAN2–PAN3 deadenylase complex. YTHDC2 exerts a dual function of stimulating both translation and decay of its targeted transcripts. Also, YTHDF1/2/3 share common target mRNAs and their fate seems to be regulated in a coordinated manner. The ribosome can serve as a scaffold for the m6A reader YTHDC2. Lastly, IGF2BP1/2/3 stabilize their target mRNAs
7. MICRORNAS, RIBOSOME DYNAMICS, AND mRNA DEGRADATION
The microRNAs (miRNAs) are a conserved class of small RNAs (~22 nt) present in metazoans that associate with Argonaute proteins to mediate post‐transcriptional regulation of mRNA targets (for a recent review see Bartel, 2018). miRNAs recognize and bind their targets through Watson–Crick base pairing, either in a full complementarity manner that induces target mRNA cleavage, or in a partial complementary manner allowing the recruitment of effector proteins that mediate translational repression and mRNA degradation (Bartel, 2018).
The precise mechanism by which miRNAs mediate gene silencing is still a source of debate (for a comprehensive review see Wilczynska & Bushell, 2015). Some studies suggest a two‐step process where miRNAs first induce translational repression and then mRNA decay (Bazzini et al., 2012; Béthune et al., 2012; Djuranovic et al., 2012; Fabian et al., 2009; Huntzinger et al., 2013; Larsson & Nadon, 2013; Meijer et al., 2013; Y. Mishima et al., 2012; Selbach et al., 2008), while others suggest that miRNAs predominantly act through mRNA destabilization (Bagga et al., 2005; Eisen et al., 2020; Eulalio et al., 2008; Guo et al., 2010). It is therefore possible that miRNAs could repress gene expression through a variety of mechanisms depending on the RNA target and miRNA‐associated partners. Nevertheless, at least for some miRNA‐targeted mRNAs, there is strong evidence for a co‐translational degradation, in a process involving deadenylation followed by decapping and 5′‐to‐3′ degradation by XRN1 (Antic et al., 2015; Tat et al., 2016). Furthermore, the miRNA‐induced silencing complex (miRISC) has been found to be associated with ribosomes (Jannot et al., 2011; Maroney et al., 2006; Nottrott et al., 2006), thus suggesting a role for ribosomes in the degradation of miRNA‐targeted mRNAs. Interestingly, a recent report by Biasini et al. (2020) has shown that translation is required for degradation of miRNA‐targeted transcripts in mammalian cells. Indeed, the authors showed that cytoplasmic long non‐coding RNAs, although they are bound by Argonaute proteins, are not efficiently degraded by miRNAs contrary to protein‐coding mRNAs. However, when placed downstream of a protein‐coding sequence, the long non‐coding RNAs are able to destabilize the mRNA in a miRNA‐dependent manner. Finally, in zebrafish, codon usage was recently shown to modulate the extent of miRNA‐mediated mRNA degradation (Medina‐Muñoz et al., 2021), thus suggesting a cross‐talk between these two co‐translational regulatory pathways. Nevertheless, more work is needed to understand the dynamics of these processes.
Besides the active role of miRNAs in inducing mRNA translational repression and degradation, translation can also have an impact in modulating miRNA/siRNA target accessibility. Using single‐molecule tracking of translating ribosomes and that of a reporter mRNA bearing siRNA‐targets in its coding sequence, Ruijtenberg et al. have recently shown that ribosome progression across the coding sequence can transiently dissolve RNA secondary structures, which facilitates siRNA binding and subsequent RNA cleavage (Ruijtenberg et al., 2020). They also show that translation can alleviate RNA secondary structures immediately downstream of the stop codon and improve siRNA accessibility and cleavage of targets located in the 3′ UTR. This is consistent with a previous report indicating that miRNAs are more effective in inducing target mRNA repression when bound shortly downstream the stop codon (Grimson et al., 2007).
A similar mechanism involving an interplay between translation and targeting of coding regions by small RNAs has been shown to mediate clearance of maternal mRNAs during the MZT in C. elegans (Quarato et al., 2021). In C. elegans, the Argonaute protein CSR‐1 and its associated guide RNAs (22G‐RNAs) are maternally inherited in embryos and are essential for embryonic development. CSR‐1 is implicated in the clearance of maternally inherited mRNAs through its slicing activity. Interestingly, a large fraction of CSR‐1‐associated 22G‐RNAs are directed against the coding sequence of maternally inherited mRNAs and the efficiency of mRNA slicing by CSR‐1 is negatively correlated with the translational efficiency of maternal mRNAs. Elongating ribosomes are therefore responsible for delaying the degradation of maternal mRNAs by CSR‐1 and could then act as a molecular clock to define the precise timing of clearance through a mechanism that could be coordinated with 3′‐UTR‐dependent regulation of translation.
Taken together, these results open new perspectives regarding the role of translation in regulating the binding of small RNAs and proteins within the coding sequence of mRNAs.
8. RIBOSOME‐GUIDED ENDONUCLEOLYTIC CLEAVAGE OF piRNA PRECURSOR TRANSCRIPTS
As described throughout this review, the dynamics of ribosome progression is under constant scrutiny by the cell in order to trigger rapid clearance of aberrant mRNAs and also to fine tune the half‐life of functional transcripts, which depends on their sequence as well as structural and epitranscriptomic features. All the aforementioned mechanisms participate in the catabolism of protein‐coding mRNAs. However, a recent study has highlighted an unconventional mechanism of ribosome‐guided endonucleolytic cleavage that is involved in small RNA biogenesis (Sun et al., 2020).
PIWI‐interacting RNAs (piRNAs) are a class of small non‐coding RNAs (21–35 nucleotides) that are generally expressed in germ cells of animals and participate in transposon silencing through their association with PIWI proteins of the Argonaute family (for a recent review see Ozata et al., 2019). Mouse piRNA genes are transcribed by RNA polymerase II to generate capped, spliced, and polyadenylated precursor transcripts that were thought to be translationally silent (X. Z. Li et al., 2013). Instead, Sun et al. showed that a specific class of piRNA precursors expressed at the pachytene stage of meiosis during spermatogenesis in mammals and birds (namely pachytene piRNAs) co‐sediment with actively translating ribosomes (Sun et al., 2020). Through ribosome profiling experiments, the authors showed that piRNA precursors display ribosome occupancy with a clear 3‐nucleotide periodicity on predicted ORFs near the 5′‐end (Figure 8(a)). Surprisingly, they could also detect ribosome occupancy downstream of annotated ORFs but without any clear periodicity (Figure 8(b)). Instead, they noticed that the 5′‐extremity of ribosome‐protected fragments originating downstream from ORFs coincided with the 5′‐ends of mature piRNAs bound to PIWI proteins, thus suggesting that the PLD6 endonuclease (an essential endonuclease in the piRNA processing pathway) uses ribosome protection to generate the 5′‐end of mature piRNAs (Figure 8(c,d)). Further understanding of the mechanism of ribosome‐mediated piRNA biogenesis identified the RNA helicase MOV10L1 as being essential for ribosome translocation downstream of 5′‐proximal ORFs upon their translation (Figure 8(c)). Consistent with this, incubation with puromycin or inactivation of MOV10L1 expression within testis leads to a decrease in ribosome occupancy downstream of ORFs and impairs mature piRNA expression. Upon ribosome‐templated RNA cleavage by PLD6, 5′‐cleaved RNA fragments are loaded onto PIWI proteins and their 3′ extremity further trimmed by PNLDC1 to produce a fully mature PIWI‐bound piRNA (Figure 8(e)).
FIGURE 8.
Ribosome‐dependent piRNA processing during mouse spermatogenesis. (a) piRNA precursors are capped and polyadenylated RNAs that reach the cytoplasm and associate with the translational machinery. Ribosomes translate the 5′ proximal short ORFs to produce short peptides of unknown function. (b) Upon reaching the stop codon of short ORFs, ribosomes fail to dissociate from the piRNA precursor and instead translocate downstream in a MOV10L1‐dependent non‐canonical manner. (c) The endonuclease PLD6 cleaves the 5′ end of the ribosome‐protected piRNA precursor, followed by the non‐canonical translocation of the ribosome to the next downstream PLD6 cleavage site. (d) A PIWI protein is recruited at the 5′‐extremity of the PLD6‐generated fragment while a second ribosome‐protected PLD6‐dependent cleavage event occurs downstream. (e) The 5′ PIWI‐bound RNA fragment is trimmed at its 3′ end by PNLDC1 to produce the mature piRNA sequence that is further chemically modified. The 3′ ribosome‐bound fragment associates with a new PIWI protein while the ribosome translocates downstream to mark the next PLD6 cleavage site
The molecular details responsible for ribosome translocation downstream of ORFs and piRNA loading from ribosomes to PIWI‐proteins are not yet understood, nor what distinguishes piRNA precursors from canonical protein‐coding mRNAs in order to trigger ribosome‐guided endonucleolytic cleavage. Nevertheless, these results highlight the multiple roles played by ribosomes beyond translation and open up exciting new questions about the regulation of these processes within cells.
9. CONCLUSION
For many years, it was believed that translation‐dependent mRNA decay was restricted to aberrant mRNAs, and that the bulk of mRNA turnover occurred mainly through a two‐step process, first with repression of translation coupled with ribosome dissociation, followed by decapping and exonucleolytic degradation of RNAs within P‐bodies (Franks & Lykke‐Andersen, 2008; Parker & Sheth, 2007). However, since the discovery of co‐translational decapping and exonucleolytic degradation, first in yeast and later in mammals, it has become very clear that translation‐dependent mRNA degradation is a major mechanism of regulation of mRNA half‐lives at the transcriptome level. From unicellular eukaryotic organisms to metazoans, the translation process appears to be constantly scrutinized by the cell, in order to trigger mRNA degradation through a myriad of pathways capable of monitoring different aspects of ribosome progression. It is now known that pathways that were initially thought to participate uniquely in mRNA quality control are also implicated in the fine tuning of a large spectrum of functional transcripts. These widespread ribosome‐dependent RNA decay mechanisms have even been harnessed by cells in order to couple translation‐dependent RNA decay with the biogenesis of functional small RNAs.
Not surprisingly, ribosomes themselves are emerging as central players in this process, acting as hubs for the recruitment of mRNA decay factors or as signaling substrates to orchestrate the various steps required for conflict resolution or mRNA degradation. However, although the molecular mechanisms that trigger translation‐dependent mRNA decay are now well characterized, more work is required to fully understand their regulations. Also, how the cell discriminates between benign situations that can be resolved from those requiring mRNA degradation awaits further investigations. Furthermore, since many of these pathways converge on similar effectors, including XRN1 or Ccr4–Not complex, it will be important to characterize their interplay, as recently done by the Bazzini laboratory for the use of codons, m6A marks, and microRNAs (Medina‐Muñoz et al., 2021). Finally, although co‐translational mRNA degradation is emerging as a major pathway to regulate the levels of functional transcripts, mRNA degradation does not occur exclusively in association with ribosomes. How cells coordinate mRNA degradation in a translation‐dependent or translational‐independent manner remains an open question.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Christelle Morris: Writing‐original draft; writing‐review & editing. David Cluet: Writing‐review & editing. Emiliano Ricci: Conceptualization; funding acquisition; writing‐original draft; writing‐review & editing.
FURTHER READING
Coller, J. , & Parker, R. (2005). General translational repression by activators of mRNA decapping. Cell, 122(6), 875–886. 10.1016/j.cell.2005.07.012
Leon, Y. C. , Mugler, C. F. , Heinrich, S. , Vallotton, P. , & Weis, K. (2018). Non‐invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. eLife, 7, e32536. 10.7554/eLife.32536
RELATED WIREs ARTICLES
Interrelations between translation and general mRNA degradation in yeast
Controlling translation via modulation of tRNA levels
Ribosome‐based quality control of mRNA and nascent peptides
Regulation of cytoplasmic RNA stability: Lessons from Drosophila
ACKNOWLEDGMENTS
Work in our laboratory is supported by the European Research Council (ERC‐StG‐LS6‐805500) under the European Union's Horizon 2020 research and innovation programs; ATIP‐Avenir program; Fondation FINOVI; Agence Nationale des Recherches sur le SIDA et les Hépatites Virales (ANRS‐ECTZ3306) and Agence Nationale de la Recherche (ANR‐20‐CE15‐0025).
Morris C, Cluet D, Ricci EP. Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny. WIREs RNA. 2021;12:e1658. 10.1002/wrna.1658
Edited by: Jeff Wilusz, Editor‐in‐Chief
Funding information Agence Nationale de la Recherche, Grant/Award Number: ANR‐20‐CE15‐0025; Agence Nationale de Recherches sur le Sida et les Hepatites Virales, Grant/Award Number: ANRS‐ECTZ3306; Fondation Innovations en Infectiologie; H2020 European Research Council, Grant/Award Number: ERC‐StG‐LS6‐805500
Contributor Information
Christelle Morris, Email: christelle.desbois@ens-lyon.fr.
Emiliano P. Ricci, Email: emiliano.ricci@ens-lyon.org.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Akashi, H. (1994). Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics, 136(3), 927–935. 10.1093/genetics/136.3.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani, N. , Ganesan, R. , Kervestin, S. , Mangus, D. A. , Ghosh, S. , & Jacobson, A. (2004). A faux 3′‐UTR promotes aberrant termination and triggers nonsense‐mediated mRNA decay. Nature, 432(7013), 112–118. 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- Antic, S. , Wolfinger, M. T. , Skucha, A. , Hosiner, S. , & Dorner, S. (2015). General and MicroRNA‐mediated mRNA degradation occurs on ribosome complexes in Drosophila cells. Molecular and Cellular Biology, 35(13), 2309–2320. 10.1128/MCB.01346-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat, A. B. , Liechti, A. , Matos, M. D. , Dreos, R. , Janich, P. , & Gatfield, D. (2020). Transcriptome‐wide sites of collided ribosomes reveal principles of translational pausing. Genome Research, 30, 985–999. 10.1101/gr.257741.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, C. J. , & Cleveland, D. W. (1994). An amino‐terminal tetrapeptide specifies cotranslational degradation of beta‐tubulin but not alpha‐tubulin mRNAs. Molecular and Cellular Biology, 14, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga, S. , Bracht, J. , Hunter, S. , Massirer, K. , Holtz, J. , Eachus, R. , & Pasquinelli, A. E. (2005). Regulation by let‐7 and lin‐4 miRNAs results in target mRNA degradation. Cell, 122(4), 553–563. 10.1016/j.cell.2005.07.031 [DOI] [PubMed] [Google Scholar]
- Bartel, D. P. (2018). Metazoan microRNAs. Cell, 173(1), 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini, A. A. , Lee, M. T. , & Giraldez, A. J. (2012). Ribosome profiling shows that miR‐430 reduces translation before causing mRNA decay in zebrafish. Science (New York, N.Y.), 336(6078), 233–237. 10.1126/science.1215704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini, A. A. , del Viso, F. , Moreno‐Mateos, M. A. , Johnstone, T. G. , Vejnar, C. E. , Qin, Y. , Yao, J. , Khokha, M. K. , & Giraldez, A. J. (2016). Codon identity regulates mRNA stability and translation efficiency during the maternal‐to‐zygotic transition. The EMBO Journal, 35(19), 2087–2103. 10.15252/embj.201694699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, A. , Rodschinka, G. , & Nedialkova, D. D. (2021). High‐resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim‐tRNAseq. Molecular Cell, 81, 1–14. 10.1016/j.molcel.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béthune, J. , Artus‐Revel, C. G. , & Filipowicz, W. (2012). Kinetic analysis reveals successive steps leading to miRNA‐mediated silencing in mammalian cells. EMBO Reports, 13(8), 716–723. 10.1038/embor.2012.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, S. N. , Habermacher, R. , Martine, U. , Closs, E. I. , & Filipowicz, W. (2006). Relief of microRNA‐mediated translational repression in human cells subjected to stress. Cell, 125(6), 1111–1124. 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- Biasini, A. , Abdulkarim, B. , Pretis, S. , Tan, J. Y. , Arora, R. , Wischnewski, H. , Dreos, R. , Pelizzola, M. , Ciaudo, C. , & Marques, A. C. (2020). Translation is required for miRNA‐dependent decay of endogenous transcripts. The EMBO Journal, 40, e104569. 10.15252/embj.2020104569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelöv, S. , Selmi, T. , Flad, S. , Dietmann, S. , & Frye, M. (2019). Codon usage optimization in pluripotent embryonic stem cells. Genome Biology, 20(1), 119. 10.1186/s13059-019-1726-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues, M. (2005). Movement of eukaryotic mRNAs between Polysomes and cytoplasmic processing bodies. Science, 310(5747), 486–489. 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, R. J. , Aucott, L. S. , & Stansfield, I. (2006). TRNA properties help shape codon pair preferences in open reading frames. Nucleic Acids Research, 34(3), 1015–1027. 10.1093/nar/gkj488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschauer, R. , Matsuo, Y. , Sugiyama, T. , Chen, Y.‐H. , Alhusaini, N. , Sweet, T. , Ikeuchi, K. , Cheng, J. , Matsuki, Y. , Nobuta, R. , Gilmozzi, A. , Berninghausen, O. , Tesina, P. , Becker, T. , Coller, J. , Inada, T. , & Beckmann, R. (2020). The Ccr4‐not complex monitors the translating ribosome for codon optimality. Science, 368(6488), eaay6912. 10.1126/science.aay6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G. , Muhlrad, D. , & Parker, R. (1993). A small segment of the MATalpha1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Molecular and Cellular Biology, 13(9), 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, P. P. , & Lowe, T. M. (2016). GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Research, 44(D1), D184–D189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, W. , Roy, B. , He, F. , Yan, K. , & Jacobson, A. (2020). Poly(a)‐binding protein regulates the efficiency of translation termination. Cell Reports, 33(7), 108399. 10.1016/j.celrep.2020.108399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran, V. , Juszkiewicz, S. , Choi, J. , Puglisi, J. D. , Brown, A. , Shao, S. , Ramakrishnan, V. , & Hegde, R. S. (2019). Mechanism of ribosome stalling during translation of a poly(a) tail. Nature Structural & Molecular Biology, 26, 1132–1140. 10.1038/s41594-019-0331-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. L. , Lee, W. , Hottes, A. K. , Shapiro, L. , & McAdams, H. H. (2004). Codon usage between genomes is constrained by genome‐wide mutational processes. Proceedings of the National Academy of Sciences, 101(10), 3480–3485. 10.1073/pnas.0307827100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. , Kim, K. M. , Han, S. , Choe, J. , Park, S. G. , Choi, S. S. , & Kim, Y. K. (2012). Staufen1‐mediated mRNA decay functions in Adipogenesis. Molecular Cell, 46, 495–506. 10.1016/j.molcel.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Choe, J. , Lin, S. , Zhang, W. , Liu, Q. , Wang, L. , Ramirez‐Moya, J. , Du, P. , Kim, W. , Tang, S. , Sliz, P. , Santisteban, P. , George, R. E. , Richards, W. G. , Wong, K.‐K. , Locker, N. , Slack, F. J. , & Gregory, R. I. (2018). MRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature, 561(7724), 556–560. 10.1038/s41586-018-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Ieong, K.‐W. , Demirci, H. , Chen, J. , Petrov, A. , Prabhakar, A. , O'Leary, S. E. , Dominissini, D. , Rechavi, G. , Soltis, S. M. , Ehrenberg, M. , & Puglisi, J. D. (2016). N 6 ‐methyladenosine in mRNA disrupts tRNA selection and translation‐elongation dynamics. Nature Structural & Molecular Biology, 23(2), 110–115. 10.1038/nsmb.3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D. W. (1988). Autoregulated instability of tubulin mRNAs, 5. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W. , Lopata, M. A. , Sherline, P. , & Kirschner, M. W. (1981). Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell, 25(2), 537–546. 10.1016/0092-8674(81)90072-6 [DOI] [PubMed] [Google Scholar]
- Coleman, J. R. , Papamichail, D. , Skiena, S. , Futcher, B. , Wimmer, E. , & Mueller, S. (2008). Virus attenuation by genome‐scale changes in codon pair bias. Science, 320(5884), 1784–1787. 10.1126/science.1155761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courel, M. , Clément, Y. , Bossevain, C. , Foretek, D. , Vidal Cruchez, O. , Yi, Z. , Bénard, M. , Benassy, M. , Kress, M. , Vindry, C. , Ernoult‐Lange, M. , Antoniewski, C. , Morillon, A. , Brest, P. , Hubstenberger, A. , Roest Crollius, H. , Standart, N. , & Weil, D. (2019). GC content shapes mRNA storage and decay in human cells. eLife, 8, 1–32. 10.7554/eLife.49708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick, F. H. C. (1966). Codon‐anticodon pairing. Journal of Molecular Biology, 19(2), 548–555. 10.1016/S0022-2836(66)80022-0 [DOI] [PubMed] [Google Scholar]
- Cui, X. , Mino, T. , Yoshinaga, M. , Nakatsuka, Y. , Hia, F. , Yamasoba, D. , Tsujimura, T. , Tomonaga, K. , Suzuki, Y. , Uehata, T. , & Takeuchi, O. (2017). Regnase‐1 and Roquin nonredundantly regulate Th1 differentiation causing cardiac inflammation and fibrosis. The Journal of Immunology, 199(12), 4066–4077. 10.4049/jimmunol.1701211 [DOI] [PubMed] [Google Scholar]
- Dana, A. , & Tuller, T. (2014). The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Research, 42(14), 9171–9181. 10.1093/nar/gku646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas, S. , Oliveros, J. C. , Chagoyen, M. , & Ortín, J. (2014). Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Research, 42(7), 4516–4526. 10.1093/nar/gku073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic, S. , Nahvi, A. , & Green, R. (2012). MiRNA‐mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science, 336(6078), 237–240. 10.1126/science.1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma, M. K. , & Parker, R. (2006). Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature, 440(7083), 561–564. 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazio, K. N. , Wu, C. C.‐C. , Sinha, N. , Loll‐Krippleber, R. , Brown, G. W. , & Green, R. (2019). The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. eLife, 8, 1–27. 10.7554/eLife.49117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, D. A. , & Wilke, C. O. (2008). Mistranslation‐induced protein misfolding as a dominant constraint on coding‐sequence evolution. Cell, 134(2), 341–352. 10.1016/j.cell.2008.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Zhao, Y. , He, J. , Zhang, Y. , Xi, H. , Liu, M. , Ma, J. , & Wu, L. (2016). YTHDF2 destabilizes m 6 A‐containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nature Communications, 7(1), 12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, S. , Cougot, N. , Mahuteau‐Betzer, F. , Nguyen, C.‐H. , Grierson, D. S. , Bertrand, E. , Tazi, J. , & Lejeune, F. (2007). Inhibition of nonsense‐mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P‐bodies. Journal of Cell Biology, 178(7), 1145–1160. 10.1083/jcb.200611086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L. , & Mouchiroud, D. (1999). Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 96(8), 4482–4487. 10.1073/pnas.96.8.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, T. J. , Eichhorn, S. W. , Subtelny, A. O. , & Bartel, D. P. (2020). MicroRNAs cause accelerated decay of short‐tailed target mRNAs. Molecular Cell, 77, 775–785. 10.1016/j.molcel.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A. , Huntzinger, E. , Nishihara, T. , Rehwinkel, J. , Fauser, M. , & Izaurralde, E. (2008). Deadenylation is a widespread effect of miRNA regulation. RNA, 15(1), 21–32. 10.1261/rna.1399509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A. , Behm‐Ansmant, I. , & Izaurralde, E. (2007). P bodies: At the crossroads of post‐transcriptional pathways. Nature Reviews Molecular Cell Biology, 8(1), 9–22. 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Eulalio, A. , Behm‐Ansmant, I. , Schweizer, D. , & Izaurralde, E. (2007). P‐body formation is a consequence, not the cause, of RNA‐mediated gene silencing. Molecular and Cellular Biology, 27(11), 3970–3981. 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, M. R. , Mathonnet, G. , Sundermeier, T. , Mathys, H. , Zipprich, J. T. , Svitkin, Y. V. , Rivas, F. , Jinek, M. , Wohlschlegel, J. , Doudna, J. A. , Chen, C.‐Y. A. , Shyu, A.‐B. , Yates, J. R. , Hannon, G. J. , Filipowicz, W. , Duchaine, T. F. , & Sonenberg, N. (2009). Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP‐dependent deadenylation. Molecular Cell, 35(6), 868–880. 10.1016/j.molcel.2009.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, M. E. , Pinkard, O. , Martin, S. , Sweet, T. J. , Hanson, G. , & Coller, J. (2020). Codon and amino acid content are associated with mRNA stability in mammalian cells. PLoS One, 15(2), e0228730. 10.1371/journal.pone.0228730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, T. M. , & Lykke‐Andersen, J. (2008). The control of mRNA decapping and P‐body formation. Molecular Cell, 32(5), 605–615. 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer, P. A. , Van Hoof, A. , O'Donnell, K. , Guerrerio, A. L. , Parker, R. , & Dietz, H. C. (2002). An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science, 295(5563), 2258–2261. 10.1126/science.1067338 [DOI] [PubMed] [Google Scholar]
- Gamble, C. E. , Brule, C. E. , Dean, K. M. , Fields, S. , & Grayhack, E. J. (2016). Adjacent codons act in concert to modulate translation efficiency in yeast. Cell, 166(3), 679–690. 10.1016/j.cell.2016.05.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin, J. , Yeasmin, R. , Yurovsky, A. , Cai, Y. , Skiena, S. , & Futcher, B. (2014). Measurement of average decoding rates of the 61 sense codons in vivo. eLife, 3, 1–20. 10.7554/eLife.03735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshott, D. M. , Sundaramoorthy, E. , Leonard, M. , & Bennett, E. J. (2020). Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. eLife, 9, 1–22. 10.7554/eLife.54023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia, A. , Jafarnejad, S. M. , Meyer, C. , Chapat, C. , Gogakos, T. , Morozov, P. , Amiri, M. , Shapiro, M. , Molina, H. , Tuschl, T. , & Sonenberg, N. (2017). The E3 ubiquitin ligase and RNA‐binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nature Communications, 8(1), 1–10. 10.1038/ncomms16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic, I. , Boswell, S. A. , & Mitchison, T. J. (2019). Tubulin mRNA stability is sensitive to change in microtubule dynamics caused by multiple physiological and toxic cues. PLoS Biology, 17(4), e3000225. 10.1371/journal.pbio.3000225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay, D. A. , Sisodia, S. S. , & Cleveland, D. W. (1989). Autoregulatory control of f3‐tubulin mRNA stability is linked to translation elongation. Proceedings of the National Academy of Sciences of the United States of America, 86, 5763–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold, H. , Tehler, D. , Christoffersen, N. R. , Nielsen, M. M. , Asmar, F. , Kooistra, S. M. , Christophersen, N. S. , Christensen, L. L. , Borre, M. , Sørensen, K. D. , Andersen, L. D. , Andersen, C. L. , Hulleman, E. , Wurdinger, T. , Ralfkiær, E. , Helin, K. , Grønbæk, K. , Ørntoft, T. , Waszak, S. M. , … Pilpel, Y. (2014). A dual program for translation regulation in cellular proliferation and differentiation. Cell, 158(6), 1281–1292. 10.1016/j.cell.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Glover, M. L. , Burroughs, A. M. , Monem, P. C. , Egelhofer, T. A. , Pule, M. N. , Aravind, L. , & Arribere, J. A. (2020). NONU‐1 encodes a conserved endonuclease required for mRNA translation surveillance. Cell Reports, 30(13), 4321–4331. 10.1016/j.celrep.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, D. H. , Livingston, N. M. , Movsik, J. , Wu, B. , & Green, R. (2021). Live‐cell imaging reveals kinetic determinants of quality control triggered by ribosome stalling. Molecular Cell, 81, 1–11. 10.1016/j.molcel.2021.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, C. , Kim, Y. K. , Woeller, C. F. , Tang, Y. , & Maquat, L. E. (2009). SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes & Development, 23(1), 54–66. 10.1101/gad.1717309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, C. , & Maquat, L. E. (2011). LncRNAs transactivate STAU1‐mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature, 470(7333), 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, C. , Tang, Y. , & Maquat, L. E. (2013). MRNA–mRNA duplexes that autoelicit Staufen1‐mediated mRNA decay. Nature Structural & Molecular Biology, 20(10), 1214–1220. 10.1038/nsmb.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi, H. , Nguyen, H. C. B. , Zhang, S. , Dill, B. D. , Molina, H. , & Tavazoie, S. F. (2016). Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell, 165(6), 1416–1427. 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy, M. , & Gautier, C. (1982). Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Research, 10(22), 7055–7074. 10.1093/nar/10.22.7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowravaram, M. , Schwarz, J. , Khilji, S. K. , Urlaub, H. , & Chakrabarti, S. (2019). Insights into the assembly and architecture of a Staufen‐mediated mRNA decay (SMD)‐competent mRNP. Nature Communications, 10(1), 5054. 10.1038/s41467-019-13080-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham, R. , Gautier, C. , & Gouy, M. (1980). Codon frequencies in 19 individual genes confrm consistent choices of degenerate bases according to genome type. Nucleic Acids Research, 8(9), 1893–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson, A. , Farh, K. K.‐H. , Johnston, W. K. , Garrett‐Engele, P. , Lim, L. P. , & Bartel, D. P. (2007). MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Molecular Cell, 27(1), 91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenke, N. , Trimpert, J. , Merz, S. , Conradie, A. M. , Wyler, E. , Zhang, H. , Hazapis, O.‐G. , Rausch, S. , Landthaler, M. , Osterrieder, N. , & Kunec, D. (2020). Mechanism of virus attenuation by codon pair Deoptimization. Cell Reports, 31(4), 107586. 10.1016/j.celrep.2020.107586 [DOI] [PubMed] [Google Scholar]
- Guimaraes, J. C. , Mittal, N. , Gnann, A. , Jedlinski, D. , Riba, A. , Buczak, K. , Schmidt, A. , & Zavolan, M. (2020). A rare codon‐based translational program of cell proliferation. Genome Biology, 21(1), 44. 10.1186/s13059-020-1943-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Ingolia, N. T. , Weissman, J. S. , & Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature, 466(7308), 835–840. 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman, G. A. , & Hatfield, G. W. (1989). Nonrandom utilization of codon pairs in Escherichia coli . Proceedings of the National Academy of Sciences, 86(10), 3699–3703. 10.1073/pnas.86.10.3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh, N. R. , & Green, R. (2014). Dom34 rescues ribosomes in 3′ untranslated regions. Cell, 156(5), 950–962. 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, G. , Alhusaini, N. , Morris, N. , Sweet, T. , & Coller, J. (2018). Translation elongation and mRNA stability are coupled through the ribosomal A‐site. RNA (New York, NY), 24, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya, Y. , & Parker, R. (2017). The link between adjacent codon pairs and mRNA stability. BMC Genomics, 18(1), 364. 10.1186/s12864-017-3749-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick, D. , Parker, R. , & Jacobson, A. (1990). Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae . Molecular and Cellular Biology, 10(5), 2269–2284. 10.1128/MCB.10.5.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hia, F. , Yang, S. F. , Shichino, Y. , Yoshinaga, M. , Murakawa, Y. , Vandenbon, A. , Fukao, A. , Fujiwara, T. , Landthaler, M. , Natsume, T. , Adachi, S. , Iwasaki, S. , & Takeuchi, O. (2019). Codon bias confers stability to human mrna s. EMBO Reports, 20(11), 1–19. 10.15252/embr.201948220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, K. L. , Dickson, K. , Cogan, J. Z. , Replogle, J. M. , Schoof, M. , D'Orazio, K. N. , Sinha, N. K. , Hussmann, J. A. , Jost, M. , Frost, A. , Green, R. , Weissman, J. S. , & Kostova, K. K. (2020). GIGYF2 and 4EHP inhibit translation initiation of defective messenger RNAs to assist ribosome‐associated quality control. Molecular Cell, 79(6), 950–962. 10.1016/j.molcel.2020.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt, A. , Brüggemann, M. , Rücklé, C. , Boerner, S. , Heidelberger, J. B. , Busch, A. , Hänel, H. , Voigt, A. , Möckel, M. M. , Ebersberger, S. , Scholz, A. , Dold, A. , Schmid, T. , Ebersberger, I. , Roignant, J.‐Y. , Zarnack, K. , König, J. , & Beli, P. (2019). The RNA‐binding ubiquitin ligase MKRN1 functions in ribosome‐associated quality control of poly(a) translation. Genome Biology, 20(1), 1–20. 10.1186/s13059-019-1814-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek, T. A. , Khuperkar, D. , Lindeboom, R. G. H. , Sonneveld, S. , Verhagen, B. M. P. , Boersma, S. , Vermeulen, M. , & Tanenbaum, M. E. (2019). Single‐molecule imaging uncovers rules governing nonsense‐mediated mRNA decay. Molecular Cell, 75(2), 324–339. 10.1016/j.molcel.2019.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema, A. , Kastelein, R. A. , Vasser, M. , & De Boer, H. A. (1987). Codon replacement in the PGKI gene of Saccharomyces cerevisiae: Experimental approach to study the role of biased codon usage in gene expression. Molecular and Cellular Biology, 7(8), 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P. J. , Zhu, Y. , Ma, H. , Guo, Y. , Shi, X. , Liu, Y. , Qi, M. , Lu, Z. , Shi, H. , Wang, J. , Cheng, Y. , Luo, G. , Dai, Q. , Liu, M. , Guo, X. , Sha, J. , Shen, B. , & He, C. (2017). Ythdc2 is an N 6‐methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Research, 27(9), 1115–1127. 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W. , Sweet, T. J. , Chamnongpol, S. , Baker, K. E. , & Coller, J. (2009). Co‐translational mRNA decay in Saccharomyces cerevisiae . Nature, 461(7261), 225–229. 10.1038/nature08265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Weng, H. , Sun, W. , Qin, X. , Shi, H. , Wu, H. , Zhao, B. S. , Mesquita, A. , Liu, C. , Yuan, C. L. , Hu, Y.‐C. , Hüttelmaier, S. , Skibbe, J. R. , Su, R. , Deng, X. , Dong, L. , Sun, M. , Li, C. , Nachtergaele, S. , … Chen, J. (2018). Recognition of RNA N 6 ‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature Cell Biology, 20(3), 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger, E. , Kuzuoğlu‐Öztürk, D. , Braun, J. E. , Eulalio, A. , Wohlbold, L. , & Izaurralde, E. (2013). The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Research, 41(2), 978–994. 10.1093/nar/gks1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann, J. A. , Patchett, S. , Johnson, A. , Sawyer, S. , & Press, W. H. (2015). Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genetics, 11(12), e1005732. 10.1371/journal.pgen.1005732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, F. , Maragkakis, M. , Alexiou, P. , & Mourelatos, Z. (2018). Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome‐phased mRNA endonucleolysis. Nature Structural & Molecular Biology, 25(4), 302–310. 10.1038/s41594-018-0042-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura, T. (1985). Codon usage and tRNA content in unicellular and multicellular organisms. Molecular Biology and Evolution, 2(1), 13–34. 10.1093/oxfordjournals.molbev.a040335 [DOI] [PubMed] [Google Scholar]
- Ikeuchi, K. , Tesina, P. , Matsuo, Y. , Sugiyama, T. , Cheng, J. , Saeki, Y. , Tanaka, K. , Becker, T. , Beckmann, R. , & Inada, T. (2019). Collided ribosomes form a unique structural interface to induce Hel2‐driven quality control pathways. The EMBO Journal, 38(5), 1–21. 10.15252/embj.2018100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia, N. T. , Ghaemmaghami, S. , Newman, J. R. S. , & Weissman, J. S. (2009). Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science (New York, NY), 324(5924), 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, A. , Mikhailova, T. , Eliseev, B. , Yeramala, L. , Sokolova, E. , Susorov, D. , Shuvalov, A. , Schaffitzel, C. , & Alkalaeva, E. (2016). PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Research, 44(16), 7766–7776. 10.1093/nar/gkw635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannot, G. , Bajan, S. , Giguère, N. J. , Bouasker, S. , Banville, I. H. , Piquet, S. , Hutvagner, G. , & Simard, M. J. (2011). The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Reports, 12(6), 581–586. 10.1038/embor.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, G. , Fu, Y. , Zhao, X. , Dai, Q. , Zheng, G. , Yang, Y. , Yi, C. , Lindahl, T. , Pan, T. , Yang, Y.‐G. , & He, C. (2011). N‐6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nature Chemical Biology, 7(12), 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, L. , Mao, Y. , Ji, Q. , Dersh, D. , Yewdell, J. W. , & Qian, S.‐B. (2020). Decoding mRNA translatability and stability from the 5′ UTR. Nature Structural & Molecular Biology, 27(9), 814–821. 10.1038/s41594-020-0465-x [DOI] [PubMed] [Google Scholar]
- Joazeiro, C. A. P. (2019). Mechanisms and functions of ribosome‐associated protein quality control. Nature Reviews Molecular Cell Biology, 20(6), 368–383. 10.1038/s41580-019-0118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz, S. , Chandrasekaran, V. , Lin, Z. , Kraatz, S. , Ramakrishnan, V. , & Hegde, R. S. (2018). ZNF598 is a quality control sensor of collided ribosomes. Molecular Cell, 72(3), 469–481. 10.1016/j.molcel.2018.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz, S. , & Hegde, R. S. (2017). Initiation of quality control during poly(a) translation requires site‐specific ribosome ubiquitination. Molecular Cell, 65(4), 743–750. 10.1016/j.molcel.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz, S. , Slodkowicz, G. , Lin, Z. , Freire‐Pritchett, P. , Peak‐Chew, S.‐Y. , & Hegde, R. S. (2020). Ribosome collisions trigger cis‐acting feedback inhibition of translation initiation. eLife, 9, e60038. 10.7554/eLife.60038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev, A. L. , Patrick, A. E. , Karamysheva, Z. N. , Griesemer, D. S. , Hudson, H. , Tjon‐Kon‐Sang, S. , Nilsson, I. , Otto, H. , Liu, Q. , Rospert, S. , von Heijne, G. , Johnson, A. E. , & Thomas, P. J. (2014). Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell, 156(1–2), 146–157. 10.1016/j.cell.2013.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karousis, E. D. , Gurzeler, L.‐A. , Annibaldis, G. , Dreos, R. , & Mühlemann, O. (2020). Human NMD ensues independently of stable ribosome stalling. Nature Communications, 11(1), 4134. 10.1038/s41467-020-17974-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidoya, H. , Muramatsu, F. , Shimamura, T. , Jia, W. , Satoh, T. , Hayashi, Y. , Naito, H. , Kunisaki, Y. , Arai, F. , Seki, M. , Suzuki, Y. , Osawa, T. , Akira, S. , & Takakura, N. (2019). Regnase‐1‐mediated post‐transcriptional regulation is essential for hematopoietic stem and progenitor cell homeostasis. Nature Communications, 10(1), 1072. 10.1038/s41467-019-09028-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. Y. , Deglincerti, A. , & Jaffrey, S. R. (2017). A Staufen1‐mediated decay pathway influences the local transcriptome in axons. Translation, 5(2), e1414016. 10.1080/21690731.2017.1414016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. K. , Furic, L. , Desgroseillers, L. , & Maquat, L. E. (2005). Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell, 120(2), 195–208. 10.1016/j.cell.2004.11.050 [DOI] [PubMed] [Google Scholar]
- Koutmou, K. S. , Schuller, A. P. , Brunelle, J. L. , Radhakrishnan, A. , Djuranovic, S. , & Green, R. (2015). Ribosomes slide on lysine‐encoding homopolymeric a stretches. eLife, 4, 1–18. 10.7554/eLife.05534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer, J. , Rao, H. , Hackert, P. , Sloan, K. E. , Höbartner, C. , & Bohnsack, M. T. (2018). The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA, 24(10), 1339–1350. 10.1261/rna.064238.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki, T. , Miyoshi, K. , Myers, J. R. , & Maquat, L. E. (2018). NMD‐degradome sequencing reveals ribosome‐bound intermediates with 3′‐end non‐templated nucleotides. Nature Structural & Molecular Biology, 25(10), 940–950. 10.1038/s41594-018-0132-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki, T. , Popp, M. W. , & Maquat, L. E. (2019). Quality and quantity control of gene expression by nonsense‐mediated mRNA decay. Nature Reviews Molecular Cell Biology, 20(7), 406–420. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lant, J. T. , Berg, M. D. , Heinemann, I. U. , Brandl, C. J. , & O'Donoghue, P. (2019). Pathways to disease from natural variations in human cytoplasmic tRNAs. Journal of Biological Chemistry, 294(14), 5294–5308. 10.1074/jbc.REV118.002982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, O. , & Nadon, R. (2013). Re‐analysis of genome wide data on mammalian microRNA‐mediated suppression of gene expression. Translation, 1(1), e24557. 10.4161/trla.24557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Xiong, X. , & Yi, C. (2017). Epitranscriptome sequencing technologies: Decoding RNA modifications. Nature Methods, 14(1), 23–31. 10.1038/nmeth.4110 [DOI] [PubMed] [Google Scholar]
- Li, X. Z. , Roy, C. K. , Dong, X. , Bolcun‐Filas, E. , Wang, J. , Han, B. W. , Xu, J. , Moore, M. J. , Schimenti, J. C. , Weng, Z. , & Zamore, P. D. (2013). An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Molecular Cell, 50(1), 67–81. 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Choe, J. , Du, P. , Triboulet, R. , & Gregory, R. I. (2016). The m6A methyltransferase METTL3 promotes translation in human cancer cells. Molecular Cell, 62(3), 335–345. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z. , Gasic, I. , Chandrasekaran, V. , Peters, N. , Shao, S. , Mitchison, T. J. , & Hegde, R. S. (2020). TTC5 mediates autoregulation of tubulin via mRNA degradation. Science, 367(6473), 100–104. 10.1126/science.aaz4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.‐Y. , Badarinarayana, V. , Audino, C. D. , Rappsilber, J. , Mann, M. , & Denis, L. C. (1998). The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. The EMBO Journal, 17(4), 1096–1106. 10.1093/emboj/17.4.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Gao, M. , Xu, S. , Chen, Y. , Wu, K. , Liu, H. , Wang, J. , Yang, X. , Wang, J. , Liu, W. , Bao, X. , & Chen, J. (2020). YTHDF2/3 are required for somatic reprogramming through different RNA deadenylation pathways. Cell Reports, 32(10), 108120. 10.1016/j.celrep.2020.108120 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Yue, Y. , Han, D. , Wang, X. , Fu, Y. , Zhang, L. , Jia, G. , Yu, M. , Lu, Z. , Deng, X. , Dai, Q. , Chen, W. , & He, C. (2014). A METTL3–METTL14 complex mediates mammalian nuclear RNA N 6 ‐adenosine methylation. Nature Chemical Biology, 10(2), 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , & Deutsch, C. (2008). Electrostatics in the ribosomal tunnel modulate chain elongation rates. Journal of Molecular Biology, 384(1), 73–86. 10.1016/j.jmb.2008.08.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Na, Z. , & Slavoff, S. A. (2018). P‐bodies: Composition, properties, and functions. Biochemistry, 57(17), 2424–2431. 10.1021/acs.biochem.7b01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y. , Dong, L. , Liu, X.‐M. , Guo, J. , Ma, H. , Shen, B. , & Qian, S.‐B. (2019). M6A in mRNA coding regions promotes translation via the RNA helicase‐containing YTHDC2. Nature Communications, 10(1), 5332. 10.1038/s41467-019-13317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P. A. , Yu, Y. , Fisher, J. , & Nilsen, T. W. (2006). Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nature Structural & Molecular Biology, 13(12), 1102–1107. 10.1038/nsmb1174 [DOI] [PubMed] [Google Scholar]
- Matsuo, Y. , Ikeuchi, K. , Saeki, Y. , Iwasaki, S. , Schmidt, C. , Udagawa, T. , Sato, F. , Tsuchiya, H. , Becker, T. , Tanaka, K. , Ingolia, N. T. , Beckmann, R. , & Inada, T. (2017). Ubiquitination of stalled ribosome triggers ribosome‐associated quality control. Nature Communications, 8(1), 159. 10.1038/s41467-017-00188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger, D. M. , Cabral, B. J. , Presnyak, V. , Su, S. V. , Reid, D. W. , Goodman, B. , Link, K. , Khatwani, N. , Reynders, J. , Moore, M. J. , & McFadyen, I. J. (2019). MRNA structure regulates protein expression through changes in functional half‐life. Proceedings of the National Academy of Sciences, 116(48), 24075–24083. 10.1073/pnas.1908052116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina‐Muñoz, S. G. , Kushawah, G. , Castellano, L. A. , Diez, M. , DeVore, M. L. , Salazar, M. J. B. , & Bazzini, A. A. (2021). Crosstalk between codon optimality and cis‐regulatory elements dictates mRNA stability. Genome Biology, 22(1), 14. 10.1186/s13059-020-02251-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, H. A. , Kong, Y. W. , Lu, W. T. , Wilczynska, A. , Spriggs, R. V. , Robinson, S. W. , Godfrey, J. D. , Willis, A. E. , & Bushell, M. (2013). Translational repression and eIF4A2 activity are critical for microRNA‐mediated gene regulation. Science, 340(6128), 82–85. 10.1126/science.1231197 [DOI] [PubMed] [Google Scholar]
- Meyer, K. D. , Patil, D. P. , Zhou, J. , Zinoviev, A. , Skabkin, M. A. , Elemento, O. , Pestova, T. V. , Qian, S.‐B. , & Jaffrey, S. R. (2015). 5′ UTR m6A promotes cap‐independent translation. Cell, 163(4), 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino, T. , Iwai, N. , Endo, M. , Inoue, K. , Akaki, K. , Hia, F. , Uehata, T. , Emura, T. , Hidaka, K. , Suzuki, Y. , Standley, D. M. , Okada‐Hatakeyama, M. , Ohno, S. , Sugiyama, H. , Yamashita, A. , & Takeuchi, O. (2019). Translation‐dependent unwinding of stem–loops by UPF1 licenses Regnase‐1 to degrade inflammatory mRNAs. Nucleic Acids Research, 47(16), 8838–8859. 10.1093/nar/gkz628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino, T. , Murakawa, Y. , Fukao, A. , Vandenbon, A. , Wessels, H.‐H. , Ori, D. , Uehata, T. , Tartey, S. , Akira, S. , Suzuki, Y. , Vinuesa, C. G. , Ohler, U. , Standley, D. M. , Landthaler, M. , Fujiwara, T. , & Takeuchi, O. (2015). Regnase‐1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell, 161(5), 1058–1073. 10.1016/j.cell.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Mishima, Y. , Fukao, A. , Kishimoto, T. , Sakamoto, H. , Fujiwara, T. , & Inoue, K. (2012). Translational inhibition by deadenylation‐independent mechanisms is central to microRNA‐mediated silencing in zebrafish. Proceedings of the National Academy of Sciences, 109(4), 1104–1109. 10.1073/pnas.1113350109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima, Y. , & Tomari, Y. (2016). Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Molecular Cell, 61(6), 874–885. 10.1016/j.molcel.2016.02.027 [DOI] [PubMed] [Google Scholar]
- Morita, M. , Ler, L. W. , Fabian, M. R. , Siddiqui, N. , Mullin, M. , Henderson, V. C. , Alain, T. , Fonseca, B. D. , Karashchuk, G. , Bennett, C. F. , Kabuta, T. , Higashi, S. , Larsson, O. , Topisirovic, I. , Smith, R. J. , Gingras, A.‐C. , & Sonenberg, N. (2012). A novel 4EHP‐GIGYF2 translational repressor complex is essential for mammalian development. Molecular and Cellular Biology, 32(17), 3585–3593. 10.1128/MCB.00455-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. (1998). Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli . Nucleic Acids Research, 26(13), 3188–3193. 10.1093/nar/26.13.3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, E. , & Maquat, L. E. (1998). A rule for termination‐codon position within intron‐containing genes: When nonsense affects RNA abundance. Trends in Biochemical Sciences, 23(6), 198–199. 10.1016/S0968-0004(98)01208-0 [DOI] [PubMed] [Google Scholar]
- Narula, A. , Ellis, J. , Taliaferro, J. M. , & Rissland, O. S. (2019). Coding regions affect mRNA stability in human cells. RNA, 25(12), 1751–1764. 10.1261/rna.073239.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasif, S. , Contu, L. , & Mühlemann, O. (2018). Beyond quality control: The role of nonsense‐mediated mRNA decay (NMD) in regulating gene expression. Seminars in Cell & Developmental Biology, 75, 78–87. 10.1016/j.semcdb.2017.08.053 [DOI] [PubMed] [Google Scholar]
- Neu‐Yilik, G. , Raimondeau, E. , Eliseev, B. , Yeramala, L. , Amthor, B. , Deniaud, A. , Huard, K. , Kerschgens, K. , Hentze, M. W. , Schaffitzel, C. , & Kulozik, A. E. (2017). Dual function of UPF3B in early and late translation termination. The EMBO Journal, 36(20), 2968–2986. 10.15252/embj.201797079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott, S. , Simard, M. J. , & Richter, J. D. (2006). Human let‐7a miRNA blocks protein production on actively translating polyribosomes. Nature Structural & Molecular Biology, 13(12), 1108–1114. 10.1038/nsmb1173 [DOI] [PubMed] [Google Scholar]
- Ozata, D. M. , Gainetdinov, I. , Zoch, A. , O'Carroll, D. , & Zamore, P. D. (2019). PIWI‐interacting RNAs: Small RNAs with big functions. Nature Reviews Genetics, 20(2), 89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Pachter, J. (1987). Autoregulation of tubulin expression is achieved through specific degradation of polysomal tubulin mRNAs. Cell, 51(2), 283–292. 10.1016/0092-8674(87)90155-3 [DOI] [PubMed] [Google Scholar]
- Panasenko, O. O. , & Collart, M. A. (2012). Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase: Not4 influence ribosome ubiquitination and assembly. Molecular Microbiology, 83(3), 640–653. 10.1111/j.1365-2958.2011.07957.x [DOI] [PubMed] [Google Scholar]
- Park, E. , & Maquat, L. E. (2013). Staufen‐mediated mRNA decay: Staufen‐mediated mRNA decay. Wiley Interdisciplinary Reviews: RNA, 4(4), 423–435. 10.1002/wrna.1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, O. H. , Ha, H. , Lee, Y. , Boo, S. H. , Kwon, D. H. , Song, H. K. , & Kim, Y. K. (2019). Endoribonucleolytic cleavage of m6A‐containing RNAs by RNase P/MRP complex. Molecular Cell, 74(3), 494–507. 10.1016/j.molcel.2019.02.034 [DOI] [PubMed] [Google Scholar]
- Parker, R. , & Jacobson, A. (1990). Translation and a 42‐nucleotide segment within the coding region of the mRNA encoded by the MATal gene are involved in promoting rapid mRNA decay in yeast. Proceedings of the National Academy of Sciences, 87, 2780–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R. , & Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Molecular Cell, 25(5), 635–646. 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Pelechano, V. , Wei, W. , & Steinmetz, L. M. (2015). Widespread co‐translational RNA decay reveals ribosome dynamics. Cell, 161(6), 1400–1412. 10.1016/j.cell.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton, K. E. , Chen, B. , Liu, K. , Hunter, O. V. , Xie, Y. , Tu, B. P. , & Conrad, N. K. (2017). The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell, 169(5), 824–835. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinarbasi, E. S. , Karamyshev, A. L. , Tikhonova, E. B. , Wu, I.‐H. , Hudson, H. , & Thomas, P. J. (2018). Pathogenic signal sequence mutations in progranulin disrupt SRP interactions required for mRNA stability. Cell Reports, 23(10), 2844–2851. 10.1016/j.celrep.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkard, O. , McFarland, S. , Sweet, T. , & Coller, J. (2020). Quantitative tRNA‐sequencing uncovers metazoan tissue‐specific tRNA regulation. Nature Communications, 11(1), 4104. 10.1038/s41467-020-17879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyet, F. , Mouchiroud, D. , Duret, L. , & Sémon, M. (2017). Recombination, meiotic expression and human codon usage. eLife, 6, 1–19. 10.7554/eLife.27344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, K. T. , Szeto, J.‐Y. A. , & Schaffitzel, C. (2020). New insights into no‐go, non‐stop and nonsense‐mediated mRNA decay complexes. Current Opinion in Structural Biology, 65, 110–118. 10.1016/j.sbi.2020.06.011 [DOI] [PubMed] [Google Scholar]
- Preissler, S. , Reuther, J. , Koch, M. , Scior, A. , Bruderek, M. , Frickey, T. , & Deuerling, E. (2015). Not4‐dependent translational repression is important for cellular protein homeostasis in yeast. The EMBO Journal, 34(14), 1905–1924. 10.15252/embj.201490194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak, V. , Alhusaini, N. , Chen, Y.‐H. , Martin, S. , Morris, N. , Kline, N. , Olson, S. , Weinberg, D. , Baker, K. E. , Graveley, B. R. , & Coller, J. (2015). Codon optimality is a major determinant of mRNA stability. Cell, 160(6), 1111–1124. 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarato, P. , Singh, M. , Cornes, E. , Li, B. , Bourdon, L. , Mueller, F. , Didier, C. , & Cecere, G. (2021). Germline inherited small RNAs facilitate the clearance of untranslated maternal mRNAs in C. elegans embryos. Nature Communications, 12(1), 1441. 10.1038/s41467-021-21691-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan, A. , Chen, Y.‐H. , Martin, S. , Alhusaini, N. , Green, R. , & Coller, J. (2016). The DEAD‐box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell, 167(1), 122–132. 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, M. D. , Savva, R. , & Wernisch, L. (2004). Solving the riddle of codon usage preferences: A test for translational selection. Nucleic Acids Research, 32(17), 5036–5044. 10.1093/nar/gkh834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci, E. P. , Kucukural, A. , Cenik, C. , Mercier, B. C. , Singh, G. , Heyer, E. E. , Ashar‐Patel, A. , Peng, L. , & Moore, M. J. (2014). Staufen1 senses overall transcript secondary structure to regulate translation. Nature Structural & Molecular Biology, 21(1), 26–35. 10.1038/nsmb.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom, E. , Kim, H. C. , Gingras, A.‐C. , Marcotrigiano, J. , Favre, D. , Olsen, H. , Burley, S. K. , & Sonenberg, N. (1998). Cloning and characterization of 4EHP, a novel mammalian eIF4E‐related cap‐binding protein. Journal of Biological Chemistry, 273(21), 13104–13109. 10.1074/jbc.273.21.13104 [DOI] [PubMed] [Google Scholar]
- Rudolph, K. L. M. , Schmitt, B. M. , Villar, D. , White, R. J. , Marioni, J. C. , Kutter, C. , & Odom, D. T. (2016). Codon‐driven translational efficiency is stable across diverse mammalian cell states. PLoS Genetics, 12(5), e1006024. 10.1371/journal.pgen.1006024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg, S. , Sonneveld, S. , Cui, T. J. , Logister, I. , de Steenwinkel, D. , Xiao, Y. , MacRae, I. J. , Joo, C. , & Tanenbaum, M. E. (2020). MRNA structural dynamics shape Argonaute‐target interactions. Nature Structural & Molecular Biology, 27(9), 790–801. 10.1038/s41594-020-0461-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, A. E. , Pinkard, O. , & Coller, J. M. (2019). TRNA metabolism and neurodevelopmental disorders. Annual Review of Genomics and Human Genetics, 20(1), 359–387. 10.1146/annurev-genom-083118-015334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, C. , Kowalinski, E. , Shanmuganathan, V. , Defenouillère, Q. , Braunger, K. , Heuer, A. , Pech, M. , Namane, A. , Berninghausen, O. , Fromont‐Racine, M. , Jacquier, A. , Conti, E. , Becker, T. , & Beckmann, R. (2016). The cryo‐EM structure of a ribosome–Ski2‐Ski3‐Ski8 helicase complex. Science, 354(6318), 1431–1433. 10.1126/science.aaf7520 [DOI] [PubMed] [Google Scholar]
- Schmitt, B. M. , Rudolph, K. L. M. , Karagianni, P. , Fonseca, N. A. , White, R. J. , Talianidis, I. , Odom, D. T. , Marioni, J. C. , & Kutter, C. (2014). High‐resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA–tRNA interface. Genome Research, 24(11), 1797–1807. 10.1101/gr.176784.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller, A. P. , Wu, C. C.‐C. , Dever, T. E. , Buskirk, A. R. , & Green, R. (2017). EIF5A functions globally in translation elongation and termination. Molecular Cell, 66(2), 194–205. 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D. C. , & Parker, R. (1999). Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae . Molecular and Cellular Biology, 19(8), 5247–5256. 10.1128/MCB.19.8.5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach, M. , Schwanhäusser, B. , Thierfelder, N. , Fang, Z. , Khanin, R. , & Rajewsky, N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature, 455(7209), 58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Shao, S. , von der Malsburg, K. , & Hegde, R. S. (2013). Listerin‐dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Molecular Cell, 50(5), 637–648. 10.1016/j.molcel.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P. M. , & Li, W. H. (1987). The codon adaptation index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research, 15(3), 1281–1295. 10.1093/nar/15.3.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P. M. , & Li, W.‐H. (1986). An evolutionary perspective on synonymous codon usage in unicellular organisms. Journal of Molecular Evolution, 24(1–2), 28–38. 10.1007/BF02099948 [DOI] [PubMed] [Google Scholar]
- Shen, S. H. , Stauft, C. B. , Gorbatsevych, O. , Song, Y. , Ward, C. B. , Yurovsky, A. , Mueller, S. , Futcher, B. , & Wimmer, E. (2015). Large‐scale recoding of an arbovirus genome to rebalance its insect versus mammalian preference. Proceedings of the National Academy of Sciences, 112(15), 4749–4754. 10.1073/pnas.1502864112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U. , & Parker, R. (2006). Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell, 125(6), 1095–1109. 10.1016/j.cell.2006.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Wang, X. , Lu, Z. , Zhao, B. S. , Ma, H. , Hsu, P. J. , Liu, C. , & He, C. (2017). YTHDF3 facilitates translation and decay of N 6‐methyladenosine‐modified RNA. Cell Research, 27(3), 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Wei, J. , & He, C. (2019). Where, when, and how: Context‐dependent functions of RNA methylation writers, readers, and erasers. Molecular Cell, 74(4), 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, C. J. , & Green, R. (2012). Translation drives mRNA quality control. Nature Structural & Molecular Biology, 19(6), 594–601. 10.1038/nsmb.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, H. , Donnard, E. , Liu, B. , Jung, S. , Wang, R. , & Richter, J. D. (2020). FMRP links optimal codons to mRNA stability in neurons. Proceedings of the National Academy of Sciences, 117, 30400–30411. 10.1073/pnas.2009161117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms, C. L. , Yan, L. L. , & Zaher, H. S. (2017). Ribosome collision is critical for quality control during no‐go decay. Molecular Cell, 68(2), 361–373. 10.1016/j.molcel.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek, D. , Tiu, G. C. , Flynn, R. A. , Byeon, G. W. , Leppek, K. , Xu, A. F. , Chang, H. Y. , & Barna, M. (2017). The mammalian Ribo‐interactome reveals ribosome functional diversity and heterogeneity. Cell, 169(6), 1051–1065. 10.1016/j.cell.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N. K. , Ordureau, A. , Best, K. , Saba, J. A. , Zinshteyn, B. , Sundaramoorthy, E. , Fulzele, A. , Garshott, D. M. , Denk, T. , Thoms, M. , Paulo, J. A. , Harper, J. W. , Bennett, E. J. , Beckmann, R. , & Green, R. (2020). EDF1 coordinates cellular responses to ribosome collisions. eLife, 9, 1–44. 10.7554/eLife.58828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, M. , & Fire, A. (2011). Wobble base‐pairing slows in vivo translation elongation in metazoans. RNA, 17(12), 2063–2073. 10.1261/rna.02890211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder, L. , & Mühlemann, O. (2009). Processing bodies are not required for mammalian nonsense‐mediated mRNA decay. RNA, 15(7), 1265–1273. 10.1261/rna.1672509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standart, N. , & Weil, D. (2018). P‐bodies: Cytosolic droplets for coordinated mRNA storage. Trends in Genetics, 34(8), 612–626. 10.1016/j.tig.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Sugimoto, Y. , Vigilante, A. , Darbo, E. , Zirra, A. , Militti, C. , D'Ambrogio, A. , Luscombe, N. M. , & Ule, J. (2015). HiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature, 519(7544), 491–494. 10.1038/nature14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. H. , Zhu, J. , Xie, L. H. , Li, Z. , Meduri, R. , Zhu, X. , Song, C. , Chen, C. , Ricci, E. P. , Weng, Z. , & Li, X. Z. (2020). Ribosomes guide pachytene piRNA formation on long intergenic piRNA precursors. Nature Cell Biology, 22(2), 200–212. 10.1038/s41556-019-0457-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy, E. , Leonard, M. , Mak, R. , Liao, J. , Fulzele, A. , & Bennett, E. J. (2017). ZNF598 and RACK1 regulate mammalian ribosome‐associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Molecular Cell, 65(4), 751–760. 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, T. , Kovalak, C. , & Coller, J. (2012). The DEAD‐box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biology, 10(6), e1001342. 10.1371/journal.pbio.1001342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros, W. , & Lipshitz, H. D. (2009). The maternal‐to‐zygotic transition: A play in two acts. Development, 136(18), 3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tanaka, H. , Arima, Y. , Kamimura, D. , Tanaka, Y. , Takahashi, N. , Uehata, T. , Maeda, K. , Satoh, T. , Murakami, M. , & Akira, S. (2019). Phosphorylation‐dependent Regnase‐1 release from endoplasmic reticulum is critical in IL‐17 response. Journal of Experimental Medicine, 216(6), 1431–1449. 10.1084/jem.20181078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tat, T. T. , Maroney, P. A. , Chamnongpol, S. , Coller, J. , & Nilsen, T. W. (2016). Cotranslational microRNA mediated messenger RNA destabilization. eLife, 5, e12880. 10.7554/eLife.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesina, P. , Heckel, E. , Cheng, J. , Fromont‐Racine, M. , Buschauer, R. , Kater, L. , Beatrix, B. , Berninghausen, O. , Jacquier, A. , Becker, T. , & Beckmann, R. (2019). Structure of the 80S ribosome–Xrn1 nuclease complex. Nature Structural & Molecular Biology, 26(4), 275–280. 10.1038/s41594-019-0202-5 [DOI] [PubMed] [Google Scholar]
- Theodorakis, N. G. , & Cleveland, D. W. (1992). Physical evidence for cotranslational regulation of 3‐tubulin mRNA degradation. Molecular and Cellular Biology, 12, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann, R. , Neu‐Yilik, G. , Deters, A. , Frede, U. , Wehr, K. , Hagemeier, C. , Hentze, M. W. , & Kulozik, A. E. (1998). Binary specification of nonsense codons by splicing and cytoplasmic translation. The EMBO Journal, 17(12), 3484–3494. 10.1093/emboj/17.12.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi, T. , Kuroha, K. , Kudo, K. , Makino, S. , Inoue, E. , Kashima, I. , & Inada, T. (2012). Dom34:Hbs1 plays a general role in quality‐control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Molecular Cell, 46(4), 518–529. 10.1016/j.molcel.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Tuck, A. C. , Rankova, A. , Arpat, A. B. , Liechti, L. A. , Hess, D. , Iesmantavicius, V. , Castelo‐Szekely, V. , Gatfield, D. , & Bühler, M. (2020). Mammalian RNA decay pathways are highly specialized and widely linked to translation. Molecular Cell, 77(6), 1222–1236. 10.1016/j.molcel.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehata, T. , Iwasaki, H. , Vandenbon, A. , Matsushita, K. , Hernandez‐Cuellar, E. , Kuniyoshi, K. , Satoh, T. , Mino, T. , Suzuki, Y. , Standley, D. M. , Tsujimura, T. , Rakugi, H. , Isaka, Y. , Takeuchi, O. , & Akira, S. (2013). Malt1‐induced cleavage of Regnase‐1 in CD4+ helper T cells regulates immune activation. Cell, 153(5), 1036–1049. 10.1016/j.cell.2013.04.034 [DOI] [PubMed] [Google Scholar]
- Verma, R. , Oania, R. S. , Kolawa, N. J. , & Deshaies, R. J. (2013). Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife, 2, e00308. 10.7554/eLife.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanyi, Z. , Ribaud, V. , Kassem, S. , Panasenko, O. O. , Pahi, Z. , Gupta, I. , Steinmetz, L. , Boros, I. , & Collart, M. A. (2014). The Not5 subunit of the Ccr4‐not complex connects transcription and translation. PLoS Genetics, 10(10), e1004569. 10.1371/journal.pgen.1004569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P. , Ibrahimi, I. , & Blobel, G. (1981). Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in‐vitro‐assembled polysomes synthesizing secretory protein. Journal of Cell Biology, 91(2), 545–550. 10.1083/jcb.91.2.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Zhao, B. S. , Roundtree, I. A. , Lu, Z. , Han, D. , Ma, H. , Weng, X. , Chen, K. , Shi, H. , & He, C. (2015). N6‐methyladenosine modulates messenger RNA translation efficiency. Cell, 161(6), 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, R. , Chung, M.‐Y. , Keskeny, C. , Zinnall, U. , Landthaler, M. , Valkov, E. , Izaurralde, E. , & Igreja, C. (2020). 4EHP and GIGYF1/2 mediate translation‐coupled messenger RNA decay. Cell Reports, 33(2), 108262. 10.1016/j.celrep.2020.108262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, M. W. , Chen, Y.‐H. , Stowell, J. A. W. , Alhusaini, N. , Sweet, T. , Graveley, B. R. , Coller, J. , & Passmore, L. A. (2018). MRNA deadenylation is coupled to translation rates by the differential activities of Ccr4‐not nucleases. Molecular Cell, 70(6), 1089–1100. 10.1016/j.molcel.2018.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, D. E. , Shah, P. , Eichhorn, S. W. , Hussmann, J. A. , Plotkin, J. B. , & Bartel, D. P. (2016). Improved ribosome‐footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Reports, 14(7), 1787–1799. 10.1016/j.celrep.2016.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska, A. , & Bushell, M. (2015). The complexity of miRNA‐mediated repression. Cell Death & Differentiation, 22(1), 22–33. 10.1038/cdd.2014.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S. L. , & Walter, P. (1988). Ribosome pausing and stacking during translation of a eukaryotic mRNA. The EMBO Journal, 7(11), 3559–3569. 10.1002/j.1460-2075.1988.tb03233.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin, S. L. , & Walter, P. (1989). Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. Journal of Cell Biology, 109(6), 2617–2622. 10.1083/jcb.109.6.2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Peterson, A. , Zinshteyn, B. , Regot, S. , & Green, R. (2020). Ribosome collisions trigger general stress responses to regulate cell fate. Cell, 182(2), 404–416. 10.1016/j.cell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. , Medina, S. G. , Kushawah, G. , DeVore, M. L. , Castellano, L. A. , Hand, J. M. , Wright, M. , & Bazzini, A. A. (2019). Translation affects mRNA stability in a codon dependent manner in human cells. eLife, 8, e45396. 10.7554/eLife.45396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L. L. , & Zaher, H. S. (2020). Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Molecular Cell, 81, 614–628. 10.1016/j.molcel.2020.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, T. J. , Machlin, P. S. , & Cleveland, D. W. (1988). Autoregulated instability of fl‐tubulin mRNAs by recognition of the nascent amino terminus of fl‐tubulin. Nature, 334(6183), 580–585. [DOI] [PubMed] [Google Scholar]
- Yu, C.‐H. , Dang, Y. , Zhou, Z. , Wu, C. , Zhao, F. , Sachs, M. S. , & Liu, Y. (2015). Codon usage influences the local rate of translation elongation to regulate co‐translational protein folding. Molecular Cell, 59(5), 744–754. 10.1016/j.molcel.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Sun, X. , Qian, Y. , LaDuca, J. P. , & Maquat, L. E. (1998). At least one intron is required for the nonsense‐mediated decay of triosephosphate isomerase mRNA: A possible link between nuclear splicing and cytoplasmic translation. Molecular and Cellular Biology, 18(9), 5272–5283. 10.1128/MCB.18.9.5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F. , Yu, C. , & Liu, Y. (2017). Codon usage regulates protein structure and function by affecting translation elongation speed in Drosophila cells. Nucleic Acids Research, 45(14), 8484–8492. 10.1093/nar/gkx501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, G. , Dahl, J. A. , Niu, Y. , Fedorcsak, P. , Huang, C.‐M. , Li, C. J. , Vågbø, C. B. , Shi, Y. , Wang, W.‐L. , Song, S.‐H. , Lu, Z. , Bosmans, R. P. G. , Dai, Q. , Hao, Y.‐J. , Yang, X. , Zhao, W.‐M. , Tong, W.‐M. , Wang, X.‐J. , Bogdan, F. , … He, C. (2013). ALKBH5 is a mammalian RNA Demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell, 49(1), 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wan, J. , Gao, X. , Zhang, X. , Jaffrey, S. R. , & Qian, S.‐B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature, 526(7574), 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wan, J. , Shu, X. E. , Mao, Y. , Liu, X.‐M. , Yuan, X. , Zhang, X. , Hess, M. E. , Brüning, J. C. , & Qian, S.‐B. (2018). N6‐Methyladenosine guides mRNA alternative translation during integrated stress response. Molecular Cell, 69(4), 636–647. 10.1016/j.molcel.2018.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T. , Weems, M. , & Wilke, C. O. (2009). Translationally optimal codons associate with structurally sensitive sites in proteins. Molecular Biology and Evolution, 26(7), 1571–1580. 10.1093/molbev/msp070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Zhang, H. , & Mendell, J. T. (2020). Ribosome recycling by ABCE1 links lysosomal function and Iron homeostasis to 3′ UTR‐directed regulation and nonsense‐mediated decay. Cell Reports, 32(2), 107895. 10.1016/j.celrep.2020.107895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


