Abstract
Introduction
Positron emission tomography (PET) amyloid quantification methods require magnetic resonance imaging (MRI) for spatial registration and a priori reference region to scale the images. Furthermore, different tracers have distinct thresholds for positivity. We propose the AMYQ index, a new measure of amyloid burden, to overcome these limitations.
Methods
We selected 18F‐amyloid scans from ADNI and Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL) with the corresponding T1‐MRI. A subset also had neuropathological data. PET images were normalized, and the AMYQ was calculated based on an adaptive template. We compared AMYQ with the Centiloid scale on clinical and neuropathological diagnostic performance.
Results
AMYQ was related with amyloid neuropathological burden and had excellent diagnostic performance to discriminate controls from patients with Alzheimer's disease (AD) (area under the curve [AUC] = 0.86). AMYQ had a high agreement with the Centiloid scale (intraclass correlation coefficient [ICC] = 0.88) and AUC between 0.94 and 0.99 to discriminate PET positivity when using different Centiloid cutoffs.
Discussion
AMYQ is a new MRI‐independent index for standardizing and quantifying amyloid load across tracers.
Keywords: Alzheimer's disease, amyloid burden standardization, amyloid pet, amyloid pet quantification, centiloid, neuropathology
1. INTRODUCTION
Amyloid beta (Aβ) is one of the pathophysiological markers of Alzheimer's disease (AD). 1 , 2 Amyloid imaging with positron emission tomography (PET) enables the in vivo evaluation of Aβ deposition in the brain. 3 , 4 , 5 Several radiotracers have been developed for this purpose. 11C‐Pittsburgh compound B (PiB), the first tracer, has been used widely since its inception. 5 However, due to the short half‐life of carbon 11, several fluorine‐18 (18F)–labeled radioligands were developed: 18F‐AV45, 18F‐Florbetaben, or 18F‐Flutemetamol. These PET tracers have proved to be a reliable alternative with wider application, and have therefore been accepted by the US Food and Drug Administration (FDA) and the European Medicines agency (EMA). 4 , 6 , 7
Quantification of amyloid deposition in the brain is generally performed using a standardized uptake value ratio (SUVr). 8 This methodology is used widely in both cross‐sectional and longitudinal amyloid PET studies. 9 , 10 , 11 , 12 However, it has some requirements and presents several caveats. First, this procedure depends on two regions of interest, one target cortical region in which to quantify amyloid and one reference region that should not be susceptible to amyloid pathology, used to intensity scale the PET images. 13 The choice of the reference region is a source of variability, yielding different SUVr depending on which is used. 13 , 14 Second, due to the low anatomical information of the amyloid PET images, the SUVr methodology typically requires an individual 3D magnetic resonance image (MRI) for spatial normalization and segmentation. 15 Finally, the different amyloid tracers are not directly comparable and have different thresholds for amyloid positivity. 15 , 16
To overcome these limitations, the Centiloid Working Group was created with the aim of scaling all non‐standard methodologies of amyloid PET quantification into a standard Centiloid scale. 17 This scale has shown good results across the different amyloid tracers. 16 , 18 , 19 , 20 , 21 , 22 The Centiloid scale has been tested against the neuropathological results to determine amyloid positivity thresholds. 19 , 23 , 24 However, the transformation of the SUVr values into Centiloid requires a structural MRI, a reference region, and a set of linear transformations.
Given that structural MRI is not systematically available, and that MRI motion artifacts could influence the accuracy of region of interest (ROI) definition, 25 MRI‐independent approaches have been developed recently to quantify amyloid deposition based on a spatial normalization process that does not require MRI. 26 , 27 Some authors have proposed automated adaptive template methods, where a combination of different images is used to create a single template to mitigate the bias when a mean‐single template is used. 28 , 29 , 30 , 31 , 32 , 33 However, the methodologies developed thus far do not obtain standard amyloid measures across different tracers.
Our objectives were to develop a methodology that would be able: (1) to normalize amyloid PET data without the use of MRI, and (2) to quantify amyloid deposition in a reproducible manner across tracers (AMYQ), that does not require the use of a priori reference regions, or transformations between tracers. We assessed the relationship of AMYQ with standard neuropathological scales, and we compared it to the commonly used Centiloid scale.
2. METHODS
2.1. Participants
We selected a total of 751 subjects with available 18F‐AV45 PET scans and 225 subjects with available 18F‐Florbetaben PET scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort and 198 subjects from the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL) data set with available 18F‐Flutemetamol PET scans. Participants were cognitive normal, had mild cognitive impairment (MCI), or an AD dementia diagnosis. All participants had a 3T T1‐weighted MRI obtained <1 year apart from the PET acquisition. More details about acquisition and pre‐processing steps on PET and MRI data can be found at adni.loni.usc.edu and aibl.csiro.au.
RESEARCH IN CONTEXT
Systematic review: The literature was reviewed using PubMed to identify publications on methodologies of quantification of amyloid positron emission tomography (PET) imaging.
Interpretation: This work proposes a new index that overcomes some of the limitations and practical difficulties that have limited the implementation of amyloid PET quantification methods in clinical practice. AMYQ is consistent across tracers and does not require MRI or the definition of a priori reference and cortical regions of interest. These features would facilitate its use in the clinical practice.
Future directions: Standardization of the quantification of amyloid burden across PET tracers is essential in Alzheimer's disease (AD) studies. The proposed methodology should be replicated in other amyloid PET samples and longitudinal sensitivity should be assessed in order to establish the validity of this measure.
2.2. Neuropathology assessment
Twenty‐six subjects with 18F‐AV45 PET had also a neuropathological assessment in ADNI (the mean interval between PET acquisition and death was 0.93 + ‐0.26 years). Pathological data included several amyloid quantification scales: the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) score, 34 the Thal phase pathologic criteria, 35 and the Alzheimer's disease neuropathic change (ADNC). 36 The CERAD score reflects the amyloid plaque density, particularly neuritic plaques in selected cortical areas, whereas the Thal phases reflect the topography of the amyloid plaques. This last assessment does not distinguish between compact and diffuse amyloid deposits. The ADNC is a composite score that uses the CERAD score and the Thal and Braak neurofibrillary tangle stages to classify the ADNCs. More information can be found at adni.loni.usc.edu.
2.3. Image processing with MRI
All PET images were co‐registered to their corresponding MRI using a rigid body registration and warped into the Montreal Neurological Institute (MNI) space using an MRI‐based affine transformation followed by a non‐linear registration using the Advanced Normalization Tools (ANTs) software. 37 SUVrs were calculated from the spatially normalized images using the defined cortical volume of interest as a composite and the whole cerebellum plus brainstem as the reference region. 14 , 17 Finally, SUVrs for the different tracers were transformed into the Centiloid scale (Supplementary Material). Of note, we also tested other volumes of interest provided by the Centiloid working group (http://www.gaain.org/Centiloid‐project; Supplementary Material).
2.4. Template creation
To normalize the amyloid PET scans to the MNI regardless of MRI, we created a PET template in the MNI space. For each tracer, we iteratively and randomly selected 50 subsamples of MNI‐normalized and scaled PET images of 15 controls, 10 MCI, and 15 AD subjects. For each subsample, we applied a principal component analysis (PCA) to obtain the main components. The final PCA template for each tracer was created as the mean of the 50 permutations of the previously obtained components. For all three tracers, the first two components of the PCA explained >90% of the variance of the data (Figure 1A). The visual inspection of each component identified the first component (PC0) as the non‐specific binding of the amyloid tracer (ie, mainly white matter binding), whereas the second one (PC1) represented the specific cortical binding. More details about template creation are provided in the Supplementary Material.
FIGURE 1.
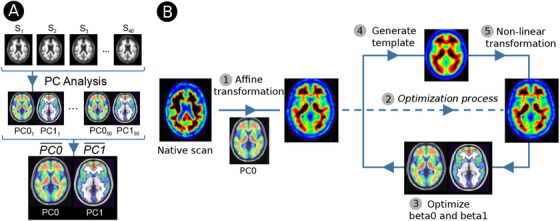
(A) Flow‐chart of the template creation. (B) Adaptive template pipeline to normalize PET scans to the MNI space. (1) Affine transformation from native space to the MNI with the first component of the template. (2) Entering in the normalization optimization process. (3) and (4) Optimize the β0 and β1 to create an adaptive template that minimize the error with the individual PET. (5) Non‐linear transformation to refine the normalization with the template. Steps 3, 4, and 5 are repeated until betas do not change
2.5. Image processing with adaptive template and AMYQ index
All 1174 individual amyloid PET scans were normalized to the MNI space using a linear combination of the two components of the PCA template. Figure 1B shows the complete process. The native PET image was initially normalized with an affine registration to the PC0 using mutual information as similarity measure. To refine the normalization to MNI space, we performed an optimization process. First, we generated a subject‐specific adaptive template optimizing the two betas of the two principal components of the PCA template (β0 for the non‐specific component, and β1 for the cortical component) by maximizing the global correlation between the voxel intensities of the spatially normalized amyloid PET image and the adaptive template. Therefore,
We then repeated the normalization step using the generated adaptive template using non‐linear transformation from ANTs. Then we recomputed the betas to generate a new adaptive template with which to again normalize the images. This process was iteratively conducted until the betas were optimized and did not change from the previous normalization step. The algorithm finally created a unique amyloid‐PET template for each subject using the optimal estimated betas for each of the two components of the template.
We assessed the normalization procedure of each tracer with the corresponding tracer‐specific template (ie, data‐driven specific and unspecific binding), but we also used the different tracer‐specific templates in the other tracer subsamples. We compared the global Pearson's correlation between templates, and they showed high correspondence between their specific and unspecific binding components (Supplementary Material). The 18F‐Flutemetamol template outperformed the two other templates (ie, higher agreement with the Centiloid scale) in all subsamples. Therefore, we present the results using the PCA template created using 18F‐Flutemetamol for all the analyses. For completeness, the results using the two other two templates are shown in the Supplementary Material.
The AMYQ index was determined as the β1/β0 ratio. We used the first component (or PC0) as a data‐driven reference region to scale the cortical component and the second (or PC1) as the data‐driven region in which to measure amyloid deposition. The ratio was then transformed into a 100‐point scale using the same approach as that used by Klunk et al. 17 Taking advantage of the 18F amyloid data set from Global Alzheimer's Association Interactive Network (GAAIN), 20 , 21 , 22 we calculated the mean β1/β0 ratio in the young controls and in the AD patients. The β1/β0 ratio of the young controls was mean centered to 0 AMYQ and to 100 AMYQ in AD patients. Thus the AMYQ for each individual was defined as:
As with the Centiloid scale, higher values of AMYQ represent high amyloid burden (ie, AD subjects mean scaled to 100 AMYQ), whereas values around zero represent low amyloid burden (ie, young controls mean scaled to zero). AMYQ, as the Centiloid scale, can exceed these limits.
2.6. Statistical analysis
Analyses were performed with the R statistical software (v 3.6.3; https://www.r‐project.org). To assess the quality of the spatial normalization of the adaptive template methodology, we computed the similarity structural image similarity (SSIM) 38 index between the two normalized images.
To assess the relationship of AMYQ with post‐mortem amyloid deposition, we computed the Spearman correlation between the four neuropathologic scales and both the AMYQ and the Centiloid scores. Kruskal‐Wallis and Dunn tests were performed to test differences with the neuropathologic assessments for both measures. In addition, paired Wilcoxon signed‐rank tests were performed within each score of the neuropathological scales to test differences between AMYQ and Centiloid measures.
To derive AMYQ thresholds, receiver‐operating characteristic (ROC) curves were calculated to assess the ability of AMYQ and Centiloid to distinguish between controls and AD dementia patients in the three subsamples separately, and in the combined amyloid sample. Youden index was used to establish the optimal thresholds. In addition, the DeLong test was used to compare the performance of both amyloid measures to distinguish controls from AD. Then, between‐clinical group effect sizes (Hedges g) were calculated for both AMYQ and Centiloid measures.
To assess the agreement between AMYQ and Centiloid, we computed the ICC between both metrics for each tracer separately and in the combined amyloid sample. In addition, Bland‐Altman plots were used to compare both metrics. Finally, ROC curves were conducted to assess the power of AMYQ to discriminate between positive and negative amyloid‐PET scans. To dichotomize the data into amyloid‐positive and amyloid‐negative individuals, we used a Centiloid cutoff of 12.2, which has been reported to identify Aβ‐detectable Thal phases and to identify “moderate/frequent” CERAD amyloid burden. 24 Other thresholds were also used and are reported in the Supplementary Material.
3. RESULTS
3.1. Participants
PET images were successfully normalized with the adaptive template in 1148 subjects (of 1174) in contrast to the 1106 scans (of 1174), which where correctly warped to the MNI with the MRI‐based normalization. Of note, 12 subjects failed with both methodologies; 56 failed only with the MRI‐based normalization, whereas only 14 failed particularly with the adaptive template approach. Details on the normalization failure are provided in the Supplementary Material. Therefore, the normalization process was successful with both methodologies in 1092 subjects. Table 1 summarizes the demographics of these 1092 participants included in the subsequent analyses: 434 (39.7%) were cognitive normal, 505 (46.3%) had MCI, and 153 (14.0%) had an AD dementia diagnosis.
TABLE 1.
Demographics for the overall sample and the three subsamples divided by diagnosis (Control, MCI, mild cognitive impairment, and AD, Alzheimer's disease dementia). Median and interquartile range (IQR) are shown for each continuous variable
| Overall | AV45 | Florbetaben | Flutemetamol | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | MCI | AD | Control | MCI | AD | Control | MCI | AD | Control | MCI | AD | |
| N | 434 | 505 | 153 | 150 | 405 | 121 | 144 | 61 | 15 | 140 | 39 | 17 |
| Age (years) (median [IQR]) | 71.60 [67.50, 76.00] | 72.00 [66.20, 77.00] | 75.00 [71.00, 79.70] | 72.80 [68.53, 77.35] | 71.50 [66.00, 76.20] | 75.30 [71.00, 79.50] | 69.70 [67.00, 74.53] | 74.00 [66.90, 78.30] | 73.60 [70.80, 81.55] | 72.00 [68.00, 76.00] | 74.00 [69.00, 78.50] | 75.00 [71.00, 79.00] |
| Gender = M (%) | 186 ( 42.9) | 283 ( 56.0) | 86 ( 56.2) | 73 ( 48.7) | 225 ( 55.6) | 68 ( 56.2) | 56 ( 38.9) | 38 ( 62.3) | 9 ( 60.0) | 57 ( 40.7) | 20 ( 51.3) | 9 ( 52.9) |
| Centiloid (median [IQR]) |
6.68 [−3.98, 26.77] |
26.53 [−0.35, 83.20] |
94.67 [65.37, 122.18] |
6.70 [−7.51, 32.25] |
28.28 [0.27, 83.20] | 94.28 [66.35, 121.74] |
0.99 [−6.63, 15.74] |
6.43 [−6.86, 65.29] |
87.39 [54.71, 109.35] | 12.80 [4.74, 30.60] | 65.50 [11.65, 102.72] | 104.30 [66.09, 143.94] |
| AMYQ (median [IQR]) |
1.06 [−20.78, 27.30] |
22.38 [−10.17, 79.04] |
90.29 [61.86, 111.16] |
−6.40 [−28.43, 27.83] |
22.13 [−10.12, 79.04] |
88.29 [60.55, 107.77] |
−3.79 [−25.83, 17.85] |
12.76 [−26.09, 55.36] |
103.29 [65.47, 122.91] |
7.23 [−5.39, 36.30] |
65.45 [11.01, 94.25] | 90.99 [74.71, 115.85] |
3.2. Assessing quality control of normalization procedure
The estimated similarity between the spatially normalized PET images warped using MRI and the PCA template was accurate across tracers (SSIMAV45 = 0.85 + −0.06, SSIMFlorbetaben = 0.85 + −0.05, and SSIMFlutemetamol = 0.91 + −0.04.
3.3. AMYQ correlates with the different neuropathological scales
Figure 2 shows the distribution of AMYQ and Centiloid scores across the different post‐mortem amyloid quantifications. Both measures increased significantly with higher scores in the four neuropathologic scales (ρAMYQ = 0.474, ρCentiloid = 0.562; ρAMYQ = 0.443, ρCentiloid = 0.551; ρAMYQ = 0.392, ρCentiloid = 0.546; and ρAMYQ = 0.610, ρCentiloid = 0.660 for Thal phases, ADNC, and CERAD neuritic and diffuse, respectively).
FIGURE 2.
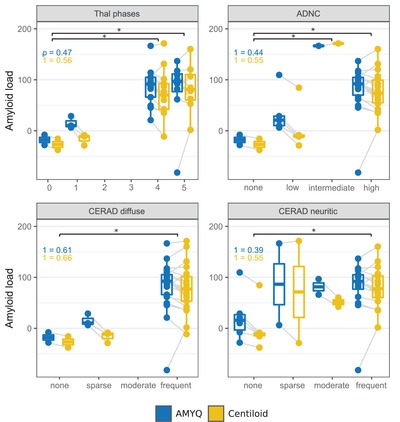
Boxplots showing the distribution of the AMYQ and Centiloid measures through the different neuropathological scales Thal phases, AD neuropathological change (ADNC), CERAD neuritic plaques, and CERAD diffuse plaques. *Indicates significant differences between scores of the different scales with both measures (AMYQ and Centiloid) at P < .05
AMYQ and the Centiloid scale showed similar results in all the amyloid neuropathological stages. There were no statistically significant differences between the Thal phase 0 and phase 1, “none” versus “low” ADNC, or “none” versus “sparse” amyloid pathology using CERAD. Both AMYQ and Centiloid showed differences with the other Thal phases, when compared with Thal phase 0 or “none” pathology (P = .025 and P = .022 vs phase 4; P = .025 and P = .013 vs phase 5); with the ADNC scores (P = .012 and P = .010 vs “intermediate”; P = .024 and P = .015 vs “high”); and for both CERAD neuritic “frequent” (P = .029 and P = .005), and diffuse “frequent” plaques (P = .020 and P = .014 for AMYQ and Centiloid, respectively). The Wilcoxon ranked test did not reveal significant differences between AMYQ and the Centilod scales in any of the scores of the four neuropathological scales.
3.4. AMYQ distinguishes between clinical groups
The ROC curve using AMYQ for the discrimination between clinical groups did not show significant differences between AMYQ and the Centiloid scales. Both showed a good performance across the three tracers for the discrimination between controls and AD dementia patients. The effect sizes for between‐group comparisons were similar across methods and tracers (Table 2). Both AMYQ and the Centiloid scale had high accuracy in the discrimination between controls and AD dementia patients in the combined amyloid sample (AUCAMYQ = 0.84, AUCCentiloid = 0.86, P > .05). The threshold for positivity derived from the Youden index were 48.9 for AMYQ and 41.9 in the Centiloid scale.
TABLE 2.
Effect sizes (Hedges g) and AUC between diagnoses for both Centiloid and AMYQ across tracers
| AV45 | Florbetaben | Flutemetamol | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Centiloid | AMYQ | Centiloid | AMYQ | Centiloid | AMYQ | |||||||
| Hedges g | AUC | Hedges g | AUC | Hedges g | AUC | Hedges g | AUC | Hedges g | AUC | Hedges g | AUC | |
| CN—MCI | 0.52 | 0.65 | 0.60 | 0.67 | 0.41 | 0.56 | 0.30 | 0.58 | 1.10 | 0.71 | 1.06 | 0.75 |
| MCI—AD | 0.87 | 0.73 | 0.86 | 0.74 | 1.04 | 0.75 | 1.27 | 0.82 | 0.77 | 0.71 | 0.74 | 0.71 |
| CN—AD | 1.51 | 0.84 | 1.58 | 0.86 | 1.96 | 0.82 | 1.92 | 0.87 | 2.33 | 0.89 | 2.00 | 0.89 |
3.5. Good agreement between the AMYQ and Centiloid scales
The AMYQ index had a high agreement with the Centiloid for the different PET tracers separately (Figure 3; ICCAV45 = 0.88, ICCFlorbetaben = 0.88, and ICCFlutemetamol = 0.89), and in the combined sample (ICC = 0.88). The Bland‐Altman plot showed a bias of variances across the measurement range (P < .001). When stratifying by amyloid positivity (as determined by a 12.2 Centiloid), the negative subsample showed a wider heterogeneity within the range of values (r = −0.71; P < .001), whereas no bias was found in the positive subsample (r = 0.08; P > .05).
FIGURE 3.
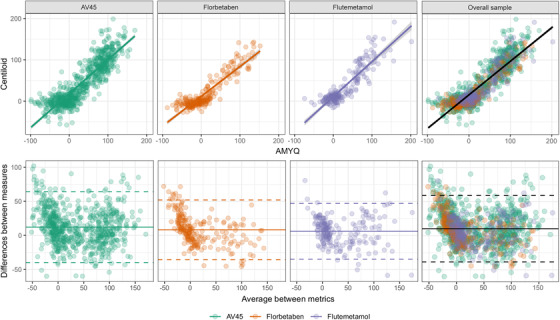
In the upper row, scatterplots showing the agreement between the AMYQ and Centiloid scales in the three tracer subsamples (from left to right: AV45, Florbetaben, and Flutemetamol) and the overall sample (on the right). In the lower row, Bland‐Altman plots showing the difference between Centiloid and AMYQ by tracer and in the overall sample
The discriminatory power of AMYQ to detect amyloid positivity was very high in the combined sample and in the three samples separately (AUCCombined = 0.94, AUCAV45 = 0.94, AUCFlorbetaben = 0.97, and AUCFlutemetamol = 0.89). An AMYQ threshold of 15.6 discriminated between amyloid positivity groups, with 92% specificity and 84% sensitivity in the combined sample. The aforementioned PET‐positivity threshold of 15.6 AMYQ had a sensitivity of 86% and a specificity of 68% in the discrimination between controls and AD dementia patients in the overall sample. Of note, other established Centiloid thresholds to determine PET positivity 23 , 39 yielded AUCs of AMYQ between 0.97 and 0.99 (Supplementary Material).
4. DISCUSSION
The AMYQ index is a new metric to quantify brain amyloid load that does not require an MRI. AMYQ can be immediately computed in the nuclear medicine departments after the amyloid PET acquisition, providing a quantitative assessment that could help in clinical practice, which currently usually relies only on a visual read. Furthermore, AMYQ is interchangeable across tracers, and thus provides a standardized measure for amyloid quantification that could facilitate the comparison of studies in which different tracers are used and in clinical practice.
AMYQ is based on a synthetic amyloid template generated using PCA. The topographical pattern of this template is in agreement with previous tracer‐specific works where one component represents the unspecific binding, mostly comprising white matter regions, and a second component that shows the neocortical uptake. 32 , 33 Applying a linear combination of the specific and non‐specific components by iteratively optimizing the weights of the two components of the PCA template, we created a unique adaptive template for each amyloid scan that will work as standard space to warp the individual image. By computing the ratio between these two weights, we obtained a measure of amyloid burden that accounts for the differences of intensity scale between PET acquisitions. One of the main advantages of this methodology is that it does not depend upon predefined flat (non‐weighted) regions of cortical load or reference region to scale the PET. It is important to note that the weighted data‐driven regions derived from the PCA analysis allow the assigning of more weight to key spatial regions related with the amyloid uptake pattern (eg, for the precuneus or medial frontal regions). Therefore, this approach removes the variability associated with the selection of distinct regions (both the reference region and the cortical region), and facilitates the harmonization between methods. To ensure the robustness of our results, we created a data‐driven template for each tracer and compared the performance of each template onto all subsamples. All had good performances when applied to the other tracers, which is not surprising given that all tracers have been related with Aβ neuropathology and share both the specific and unspecific binding (as shown in Supplementary Material). The 18F‐Flutemetamol template had a higher non‐specific binding in the white matter than the other two tracers, as described previously. 11 , 40 , 41 , 42 In this respect, a recent study using fluorodeoxyglucose (FDG)‐PET, reported a remarkable bias when intensity normalization methods were used (instead of data‐driven methods), and recommended the use of reference regions with large volume and good stability. 43 Accordingly, the data‐driven reference regions derived from the 18F‐Flutemetamol subsample proved to be superior to those derived from the other two subsamples. It is important to emphasize that AMYQ, which can be assimilated to a data‐driven SUVr, is based in larger and more stable cortical and reference regions than those commonly used in a priori reference regions. 11 , 12 , 13 , 15
AMYQ was related to post‐mortem amyloid pathology, both with CERAD the Thal phases, thus indicating that AMYQ is related with not only the abundance of neuritic plaques but also with the spatial‐temporal distribution of amyloid deposition. The correlation between post‐mortem amyloid pathology and the AMYQ and Centiloid scales was similar, and in agreement with previous Centiloid reports. 21 , 23 , 24 These data highlight the association between the amyloid PET quantitative measure and the severity of the neuropathology staging, which supports the concept of monitoring AD disease progression through amyloid PET.
The discriminatory power of AMYQ when differentiating between AD dementia patients and controls showed excellent accuracy, equivalent to that of the Centiloid scale. For all three tracers, the AMYQ and the Centiloid scale index showed similar effect sizes and AUC to discriminate between the different diagnoses. In previous studies, Whittington et al. 33 have shown a very good diagnostic performance of their amyloid load measure in 18F‐AV45 samples. Their measure showed a higher sensitivity than the commonly used SUVr in all comparisons. 33 , 44 Of note, in the 18F‐AV45 subsample of our work, AMYQ showed effect sizes similar to those reported by the Whittington measure. The agreement between the AMYQ and Centiloid values was excellent, both when assessed for the three tracers separately and when assessed in the combined sample. This yielded similar thresholds for AMYQ and the Centiloid scale when comparing controls and AD dementia patients. Of note, the thresholds were similar to those reported in other studies using the Centiloid scale. 45 Of note, these thresholds were also similar to those reported when assessing agreement with CSF p‐Tau/Aβ42 and t‐Tau/Aβ42 ratios in the ADNI cohort. 39 Despite the high correspondence between measures, we observed dissociation of values between the AMYQ and Centiloid in the subsample with low amyloid burden, but not in the amyloid‐positive range.
The ability of AMYQ to detect amyloid positivity using different Centiloid cutoffs was also very high, with AUCs systematically above 0.94 in the overall sample and in the three subsamples separately. Using the Centiloid cutoff of 12.2 as the gold standard (this cutoff has been proposed to differentiate none/sparse from moderate/high CERAD scores and to identify Aβ‐detectable Thal phase), 24 our analyses identified a very similar cutoff of 15.6 for AMYQ. When this threshold was used to differentiate patients with clinical diagnosis of AD dementia from controls, it proved very sensitive but with lower specificity for both scales; this low specificity is probably due to the high percentage of controls with amyloid pathology (40.8%). Higher Centiloid thresholds, between 23.5 and 26.0, might be better suited to identify intermediate/higher ADNC 24 and, importantly, to correlate with visual reads. 23 , 39
Automated adaptive template methods have been used typically to normalize amyloid PET images, but not to replace the use of reference region to scale the intensity of the image. One of the most widely used methodologies to normalize amyloid PET without MRI is the CapAIBL, 18 , 19 , 28 which reported excellent normalization results. Others have also reported an improved normalization using their respective adaptive templates in 18F‐Flutemetamol scans 32 , 46 or 11C‐PiB. 26 The PCA‐template normalization showed a good similarity with the MRI normalization, and showed very good agreement between SUVr in the three tracer subsamples and similar performance to those methods reported in the literature.
The advantage of AMYQ over the aforementioned automated adaptive template methods is that it provides a quantitative assessment of the amyloid load that does not require MRI and is interchangeable across tracers. Thus, AMYQ can be immediately computed in the nuclear medicine departments after the amyloid PET acquisition, and be used to complement the visual read. Visual reads have been classically implemented as a binary assessment. However, the number of borderline cases is non‐negligible and, considering that the subthreshold Aβ positivity could be indicative of faster AD pathology accumulation, the dichotomization may overlook those at‐risk individuals. 47 Thus, AMYQ could aid in clinical practice as an immediate tool for PET quantification readily available at the nuclear medicine department. By using AMYQ, the practical difficulties imposed by the need of MRI usually acquired in a different visit in a different department or center, which have contributed to the limited implementation of quantification methods in clinical practice, 48 are avoided.
Some limitations should be considered. First, this work proves that the AMYQ index is consistent using different templates randomly generated from a wide heterogeneous sample, but does not discard bias on homogenous samples. To determine whether the use of the “universal” Flutemetamol template is preferable over a single tracer template, we would require calculating AMYQ indices in a sample submitted with different radiotracers and PET scanners. Such samples were not available in this study. In addition, this work does not definitely solve whether the “universal” template should always be used instead of a specific template created for a particular study. The later approach might prove to be more accurate within a specific cohort, but it could compromise the comparability of the indices calculated. Second, the relationship between AMYQ and Centiloid in the amyloid‐negative subjects should be further explored against a pathological gold standard. Moreover, this index has not been assessed in cohorts with other amyloid tracers such as 18F‐NAV‐469 or 11C‐PiB and it should be tested in other replication cohorts to validate the reported thresholds, and evaluated longitudinally to assess its sensitivity to change in time as this is a cross‐sectional study. Despite the big sample size in some of the analysis, most of the sample had absence of neuropathological confirmation, which was in addition restricted to the 18F‐AV45 subsample. Further analyses with larger sample size with neuropathological data should be assessed. Finally, Centiloid is widely used in research settings. While AMYQ could be quickly implemented in nuclear medicine departments, its use in research settings in which MRIs are usually acquired simultaneously will depend on its validation and proof of potential additional advantages, such as to measure longitudinal amyloid load or to capture early amyloid deposition.
In summary, our study provides a new index of global amyloid load that does not require a structural MRI and is independent from a priori reference regions. AMYQ could be used directly in clinical practice to quantify amyloid load consistently across amyloid tracers.
COMPETING INTERESTS
AL has served at the scientific advisory boards of Biogen, Fujirebio Europe, Eli Lilly, Novartis, Roche Diagnostics, and Nutricia. JF has served at the scientific advisory boards of AC Inmune and has provided consultancies to Novartis and Merck as part of the Medavante adjudication team in the Merck MK‐8931 and Novartis CAPI015A2201J and CCNP520A2202J trials. DA participated in advisory boards from Fujirebio‐Europe and Roche Diagnostics and received speaker honoraria from Fujirebio‐Europe, Nutricia, and from Krka Farmacéutica S.L. DA, JF, and AL declare a filed patent application (WO2019175379 A1 Markers of synaptopathy in neurodegenerative disease). All other authors report no competing interests.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health grant U01 AG024904) and DOD ADNI (Department of Defense award W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Core funding for the AIBL study was provided in part by the study partners (Australian Commonwealth Scientific and Industrial Research Organization [CSIRO], Edith Cowan University [ECU], Mental Health Research Institute [MHRI], Alzheimer's Australia [AA], National Ageing Research Institute [NARI], Austin Health, CogState Ltd., Hollywood Private Hospital, Sir Charles Gardner Hospital). The study also received support from the National Health and Medical Research Council (NHMRC) and the Dementia Collaborative Research Centres program (DCRC2), as well as ongoing funding from the Science and Industry Endowment Fund (SIEF).
In addition, this study was supported by the Fondo de Investigaciones Sanitario (FIS), Instituto de Salud Carlos III (PI14/01126 and PI17/01019 to JF, PI13/01532 and PI16/01825 to RB, PI18/00435 and INT19/00016 to DA, and PI14/1561 and PI17/01896 to AL) and the CIBERNED program (Program 1, Alzheimer Disease to Alberto Lleó and SIGNAL study, www.signalstudy.es), partly jointly funded by Fondo Europeo de Desarrollo Regional, Unión Europea, Una manera de hacer Europa. This work was also supported by the National Institutes of Health (NIA grants 1R01AG056850 ‐ 01A1; R21AG056974 and R01AG061566 to JF), Departament de Salut de la Generalitat de Catalunya, Pla Estratègic de Recerca i Innovació en Salut (SLT002/16/00408 to A.L), Fundació La Marató de TV3 (20141210 to JF and 044412 to RB); VM is supported by Fondo de Investigaciones Sanitario (FI18/00275). This work was also supported by Generalitat de Catalunya (SLT006/17/00119 to JF, SLT006/17/95 to EV, and SLT006/17/00125 to DA) and a grant from the Fundació Bancaria La Caixa to RB.
Pegueroles J, Montal V, Bejanin A, et al. AMYQ: An index to standardize quantitative amyloid load across PET tracers. Alzheimer's Dement. 2021;17:1499–1508. 10.1002/alz.12317
REFERENCES
- 1. Fortea J, Vilaplana E, Alcolea D, et al. Cerebrospinal fluid β‐amyloid and phospho‐tau biomarker interactions affecting brain structure in preclinical Alzheimer disease. Ann Neurol. 2014;76(2). [DOI] [PubMed] [Google Scholar]
- 2. Mattsson N, Insel PS, Nosheny R, et al. Emerging β‐Amyloid pathology and accelerated cortical atrophy. JAMA Neurol. 2014;71(6):725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jagust W. Is amyloid‐β harmful to the brain? Insights from human imaging studies. Brain. 2016;139(1):23–30. 10.1093/brain/awv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong DF, Rosenberg PB, Zhou Y, et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F‐AV‐45 (flobetapir F 18). J Nucl Med. 2010;51(6):913‐920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klunk WE, Engler H, Nordberg A, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound‐B. Ann Neurol. 2004;55(3):306‐319. [DOI] [PubMed] [Google Scholar]
- 6. Nelissen N, Van Laere K, Thurfjell L, et al. Phase 1 Study of the Pittsburgh compound B derivative 18F‐Flutemetamol in healthy volunteers and patients with probable Alzheimer's disease. J Nucl Med. 2009;50(8):1251‐1259. [DOI] [PubMed] [Google Scholar]
- 7. Rowe CC, Ackerman U, Browne W, et al. Imaging of amyloid β in Alzheimer's disease with 18F‐BAY94‐9172, a novel PET tracer: proof of mechanism. Lancet Neurol. 2008;7(2):129‐135. [DOI] [PubMed] [Google Scholar]
- 8. Cselényi Z, Farde L. Quantification of blood flow‐dependent component in estimates of beta‐amyloid load obtained using quasi‐steady‐state standardized uptake value ratio. J Cereb Blood Flow Metab. 2015;35(9):1485–1493. 10.1038/jcbfm.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aghakhanyan G, Vergallo A, Gennaro M, et al. The precuneus‐a witness for excessive aβ gathering in alzheimer's disease pathology. Neurodegener Dis. 2019:302‐309.Published online. [DOI] [PubMed] [Google Scholar]
- 10. Bullich S, Villemagne VL, Catafau AM, et al. Optimal reference region to measure longituDinal amyloid‐b change with 18F‐Florbetaben PET. J Nucl Med. 2017;58(8):1300‐1306. [DOI] [PubMed] [Google Scholar]
- 11. Mountz JM, Laymon CM, Cohen AD, et al. Comparison of qualitative and quantitative imaging characteristics of [11C]PiB and [18F]flutemetamol in normal control and Alzheimer's subjects. NeuroImage Clin. 2015;9:592‐598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shokouhi S, Mckay JW, Baker SL, et al. Reference tissue normalization in longitudinal 18F‐florbetapir positron emission tomography of late mild cognitive impairment. Alzheimers Res Ther. 2016;8(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiao P, Bedell BJ, Avants B, et al. Impact of reference and target region selection on amyloid PET SUV ratios in the phase 1B PRIME study of aducanumab. J Nucl Med. 2019;60(1):100‐106. [DOI] [PubMed] [Google Scholar]
- 14. Su Y, Flores S, Hornbeck RC, et al. Utilizing the Centiloid scale in cross‐sectional and longitudinal PiB PET studies. NeuroImage Clin. 2018;19(April):406‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Landau SM, Thomas BA, Thurfjell L, et al. Amyloid PET imaging in Alzheimer's disease: a comparison of three radiotracers. Eur J Nucl Med Mol Imaging. 2014;41(7):1398‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su Y, Flores S, Wang G, et al. Comparison of Pittsburgh compound B and florbetapir in cross‐sectional and longitudinal studies. Alzheimer's Dement Diagnosis . Assess Dis Monit. 2019;11:180‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimer's Dement. 2015;11(1):1‐15.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bourgeat P, Doré V, Fripp J, et al. Implementing the centiloid transformation for11C‐PiB and β‐amyloid18F‐PET tracers using CapAIBL. Neuroimage. 2018;183:387‐393. [DOI] [PubMed] [Google Scholar]
- 19. Doré V, Bullich S, Rowe CC, et al. Comparison of 18 F‐florbetaben quantification results using the standard Centiloid, MR‐based, and MR‐less CapAIBL ® approaches: validation against histopathology. Alzheimer's Dement. 2019:1‐10.Published online. [DOI] [PubMed] [Google Scholar]
- 20. Battle MR, Pillay LC, Lowe VJ, et al. Centiloid scaling for quantification of brain amyloid with [18F]flutemetamol using multiple processing methods. EJNMMI Res. 2018;8(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Navitsky M, Joshi AD, Kennedy I, et al. Standardization of amyloid quantitation with florbetapir standardized uptake value ratios to the Centiloid scale. Alzheimer's Dement. 2018;14(12):1565‐1571. [DOI] [PubMed] [Google Scholar]
- 22. Rowe CC, Doré V, Jones G, et al. 18F‐Florbetaben PET beta‐amyloid binding expressed in Centiloids. Eur J Nucl Med Mol Imaging. 2017:2053‐2059.Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amadoru S, Doré V, McLean CA, et al. Comparison of amyloid PET measured in Centiloid units with neuropathological findings in Alzheimer's disease. Alzheimer's Res Ther. 2020;12(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. La Joie R, Ayakta N, Seeley WW, et al. Multisite study of the relationships between antemortem [11 C]PIB‐PET Centiloid values and postmortem measures of Alzheimer's disease neuropathology. Alzheimer's Dement. 2019;15(2):205‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imabayashi E, Matsuda H, Tabira T, et al. Comparison between brain CT and MRI for voxel‐based morphometry of Alzheimer's disease. Brain Behav. 2013;3(4):487‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akamatsu G, Ikari Y, Ohnishi A, et al. Automated PET‐only quantification of amyloid deposition with adaptive template and empirically pre‐defined ROI. Phys Med Biol. 2016;61(15):5768‐5780. [DOI] [PubMed] [Google Scholar]
- 27. Edison P, Carter SF, Rinne JO, et al. Comparison of MRI based and PET template based approaches in the quantitative analysis of amyloid imaging with PIB‐PET. Neuroimage. 2013;70:423‐433. [DOI] [PubMed] [Google Scholar]
- 28. Bourgeat P, Dore V, Fripp J, Villemagne VL, Rowe CC, Salvado O, Computational analysis of PET by AIBL CapAIBL: a cloud‐based processing pipeline for the quantification of PET images [abstract]. J Nucl Med. 2015;56(suppl 3):149. [Google Scholar]
- 29. Bourgeat P, Villemagne VL, Dore V, et al. Comparison of MR‐less PiB SUVR quantification methods. Neurobiol Aging. 2015;36:S159‐S166. [DOI] [PubMed] [Google Scholar]
- 30. Kang SK, Seo S, Shin SA, et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 2018;39(9):3769‐3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lilja J, Leuzy A, Chiotis K, Savitcheva I, Sörensen J, Nordberg A. Spatial normalization of [18 F]flutemetamol PET images utilizing an adaptive principal components template. J Nucl Med. 2018:jnumed.118.207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lundqvist R, Lilja J, Thomas BA, et al. Implementation and validation of an adaptive template registration method for 18F‐Flutemetamol Imaging Data. J Nucl Med. 2013;54(8):1472‐1478. [DOI] [PubMed] [Google Scholar]
- 33. Whittington A, Gunn RN. Amyloid load: a more sensitive biomarker for amyloid imaging. J Nucl Med. 2019;60(4):536‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mirra SS, Heyman A, McKeel D, et al. The consortium to establish a registry for Alzheimer's disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41(4):479‐486. [DOI] [PubMed] [Google Scholar]
- 35. Thal DR, Rüb U, Orantes M, Braak H. Phases of Aβ‐deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58(12):1791‐1800. [DOI] [PubMed] [Google Scholar]
- 36. Hyman BT, Phelps CH, Beach TG, et al. National Institute on Aging‐Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease. Alzheimer's Dement. 2012;8(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross‐correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13(4):600‐612. [DOI] [PubMed] [Google Scholar]
- 39. Salvadó G, Molinuevo JL, Brugulat‐Serrat A, et al. Centiloid cut‐off values for optimal agreement between PET and CSF core AD biomarkers. Alzheimers Res Ther. 2019;11(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lowe VJ, Lundt E, Knopman D, et al. Comparison of [18F]Flutemetamol and [11C]Pittsburgh Compound‐B in cognitively normal young, cognitively normal elderly, and Alzheimer's disease dementia individuals. NeuroImage Clin. 2017;16(July):295‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adamczuk K, Schaeverbeke J, Nelissen N, et al. Amyloid imaging in cognitively normal older adults: comparison between 18F‐flutemetamol and 11C‐Pittsburgh compound B. Eur J Nucl Med Mol Imaging. 2016;43(1):142‐151. [DOI] [PubMed] [Google Scholar]
- 42. Cho SH, Choe YS, Kim YJ, et al. Head‐to‐Head comparison of 18f‐florbetaben and 18f‐flutemetamol in the cortical and striatal regions. J Alzheimer's Dis. 2020:1‐10.Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. López‐González FJ, Silva‐Rodríguez J, Paredes‐Pacheco J, et al. Intensity normalization methods in brain FDG‐PET quantification. Neuroimage. 2020;222(July). [DOI] [PubMed] [Google Scholar]
- 44. Tanaka T, Stephenson MC, Nai YH, et al. Improved quantification of amyloid burden and associated biomarker cut‐off points: results from the first amyloid Singaporean cohort with overlapping cerebrovascular disease. Eur J Nucl Med Mol Imaging. 2020;47(2):319‐331. [DOI] [PubMed] [Google Scholar]
- 45. Leuzy A, Chiotis K, Hasselbalch SG, et al. Pittsburgh compound B imaging and cerebrospinal fluid amyloid‐β in a multicentre European memory clinic study. Brain. 2016;139(9):2540‐2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lilja J, Leuzy A, Chiotis K, Savitcheva I, Sörensen J, Nordberg A. Spatial normalization of 18 F‐flutemetamol PET images using an adaptive principal‐component template. J Nucl Med. 2019;60(2):285‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bischof GN, Jacobs HIL. Subthreshold amyloid and its biological and clinical meaning: long way ahead. Neurology. 2019;93(2):72‐79. [DOI] [PubMed] [Google Scholar]
- 48. Mainta IC, Vargas MI, Trombella S, Frisoni GB, Unschuld PG, Garibotto V. Hybrid PET‐MRI in Alzheimer's disease research. In: methods in Molecular Biology. Humana Press Inc. 2018;1750:185‐200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


