Abstract
Expression of the cyclin-dependent kinase inhibitor p21 is highly induced by many stresses, including exposure to short-wavelength UV light (UVC), which increases p21 mRNA stability. Investigation into the mechanisms underlying this stabilization process revealed that proteins present in cytoplasmic lysates of human RKO colorectal carcinoma cells formed complexes with p21 mRNA that were inducible by treatment with UVC and other stress agents. The ubiquitous Elav-type RNA-binding protein HuR was identified within the p21 mRNA-protein complexes, as antibodies recognizing HuR supershifted these complexes and revealed HuR-immunoreactive proteins complexing with p21 mRNA on Western blots. Lowering of endogenous HuR levels through expression of antisense HuR decreased p21 RNA-protein complexes, greatly reduced the UVC inducibility and half-life of p21 mRNA, and prevented UVC-mediated induction of luciferase activity in p21 3′ untranslated region-containing reporter constructs. Our findings indicate that HuR plays a major role in regulating stress-induced p21 expression by enhancing p21 mRNA stability and that these effects are coupled to HuR's elevated presence in the cytoplasm.
Expression of p21, a universal inhibitor of cyclin-dependent kinases, has been found to increase following exposure to a wide variety of stress agents, including genotoxins, oxidants, and metabolic perturbations. Indeed, increased p21 expression is believed to participate in mediating the growth arrest that follows exposure to such insults (15, 24, 62) and affects profoundly the outcome of the stressed cell, frequently favoring cell survival (23, 25, 26, 51, 62). Not unexpectedly, therefore, p21 expression is tightly regulated at multiple levels. The best-understood transcriptional activator of the p21 gene is the tumor suppressor p53, particularly following exposure to several DNA-damaging agents (16, 44). Numerous other transcription factors, including Sp1, p300, CEBPβ, and STATs, are also known to participate in the transcriptional activation of the p21 gene (5, 8, 11, 45). Another mode of regulation of p21 levels is through alterations in the stability of the p21 protein; an example of control at this level is the association of p21 with the transcription factor CEBPα, which results in considerable extension in the half-life of the p21 protein (60).
Regulation of the stability of the p21 mRNA also constitutes a critical rate-limiting step in p21 expression (56). Recently, we reported that p21 induction by short-wavelength UV light (UVC) was mediated through stabilization of the p21 mRNA (27) and have observed various other inducers of p21 which do not significantly elevate the transcription of the p21 gene but instead seem to exert their influence by altering the half-life of the p21 transcripts. Changes in mRNA stability contribute to regulating the expression of many eukaryotic genes (53, 54, 64). Many robustly transcribed genes are not expressed because of the intrinsic instability of their mRNA transcripts. Although the mechanisms determining mRNA turnover are poorly understood, they are generally believed to involve RNA-binding proteins recognizing specific RNA sequences (33). There has been growing interest in a particular pathway which regulates mRNA stability and is mediated by AU-rich elements (AREs), usually found in the 3′ untranslated region (UTR) of short-lived mRNAs (10, 37). AREs may act as repressors of gene expression, since their insertion into stable mRNAs renders them unstable (27, 57, 61). There is relatively little sequence similarity among AREs, but most contain multiple copies of the sequence AUUUA (66).
The stability of ARE-containing mRNAs can change dramatically in response to a variety of extracellular signals (9, 42, 65). Recently it has become clear that this response can be effected by the Elav-like RNA-binding proteins. These proteins, first identified as tumor antigens, are the homologues of Elav, a Drosophila protein necessary for neuronal differentiation (52, 59). Four highly conserved Elav-like proteins have been identified: HuD, HuC, and Hel-N1, expressed in terminally differentiated neurons and neuroendocrine tumors (4, 13, 14, 43, 49, 59), and HuR, which is expressed ubiquitously (21, 40, 59). The protein products of all four genes bind with high affinity and specificity to AREs in a variety of mRNAs, among which are those encoding vascular endothelial growth factor (VEGF), GLUT-1, and c-Fos (1, 13, 17, 20, 29, 30, 35, 38, 41, 46), and are believed to increase their stability.
In a recent report, recombinant HuD was found to bind in vitro to the p21 mRNA (30). This observation and the fact that p21 displayed a ubiquitous pattern of expression, not restricted to neuronal tissues, prompted us to examine whether the HuD-related protein HuR might also bind to the p21 mRNA and contribute to its regulation during UVC stress. Here, we have examined the mechanisms contributing to p21 mRNA stabilization after exposure to UVC and other stresses. We provide direct evidence that endogenous HuR binds to the p21 mRNA in a stress-inducible manner and that HuR mediates the UVC-induced stabilization of p21 mRNA. Interestingly, UVC and other stresses enhance the cytoplasmic localization of HuR, possibly through transport processes such as those serving to regulate the intracellular location of various RNA-binding proteins (47, 48) and HuR specifically (2, 18, 50), suggesting that the subcellular localization of HuR may be critically linked to its function as a regulator of mRNA stability.
MATERIALS AND METHODS
Cell culture, treatments, transfection, and DAPI staining.
Human colorectal carcinoma RKO cells (neo [with wild-type p53 function] and E6 [p53 deficient]) (34) were cultured in minimum essential medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah) and antibiotics. Hydrogen peroxide, lithium acetate, prostaglandin A2 (PGA2), methyl methanesulfonate (MMS), actinomycin D, and ionomycin were from Sigma (St. Louis, Mo.). Unless otherwise specified, cells were irradiated with UVC (20 J/m2) and collected 6 h later. To establish lines expressing antisense HuR mRNA constitutively, RKO cells were transfected with pZeoSV2(−) HuR by calcium phosphate precipitation and selected in zeocin (600 μg/ml; Invitrogen, Carlsbad, Calif.). Clonal transfectants were stored as frozen aliquots and used within 3 weeks of thawing. Transient transfection of RKO cultures with either pGL3, pGL3-FL, or pGL3-ΔB2 was carried out by the calcium phosphate precipitation method. Cotransfection of pSV-βgal served as an internal control. Luciferase and β-galactosidase activities were measured with a luciferase assay system (Promega, Madison, Wis.) and Galacto-light Plus (Tropix, Bedford, Mass.), respectively, following the manufacturers' instructions. All luciferase measurements were normalized to β-galactosidase measurements from the same sample. Transient transfection of RKO cultures with pEGFP-HuR were carried out with the DMRIE-C reagent (Gibco BRL) as recommended by the manufacturer. Cells were incubated with the transfection medium for 10 h and were then trypsinized and replated on chamber slides; transfection efficiency was about 20%.
Construction of pEGFP-HuR, pGL3-FL, and pGL3-ΔB2.
For construction of the vector expressing the fusion protein green fluorescent protein (GFP)-HuR, the open reading frame of HuR was amplified by PCR using HuR cDNA as the template with primers 5′-ATGTCTAATGGTTATGAAGACCAC-3′ and 5′-TTATTTGTGGGACTTGTTGGTTTTG-3′ and cloned into pEGFP-C1 (Clontech, Palo Alto, Calif.). For visualizing nuclei with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma), cells were washed three times with phosphate-buffered saline, fixed with 4% paraformaldehyde, and incubated with DAPI for 30 min. GFP and DAPI signals were examined by fluorescence microscopy. For the construction of pGL3-FL and pGL3-ΔB2, PCR products were prepared with 5′ primers 5′-GGACTAGTCCGCCCACAGGAAGCCTGC-3′ and 5′-GGACTAGTTCTCCTTTTCCTCTCTCCC-3′, respectively, and the 3′ primer 5′-GGACTAGTAAGTCACTAAGAATCATTTATTGAGCACC-3′ and cloned into the XbaI site of plasmid pGL3 (Promega).
Northern blot analysis.
Total RNA was isolated and Northern blot analysis was carried out as described elsewhere (27). For detection of p21 and β-actin mRNAs, random primer-labeled cDNA inserts excised from pCEP-Waf1 (16) and pBS-β-Actin (27) were used. Normalization of Northern signals was performed with an oligomer recognizing the 18S rRNA (5′-ACGGTATCTGATCGTCTTCGAACC-3′; Integrated DNA Technologies, Coralville, Iowa) that was end labeled as previously described (27). Northern signals were visualized and quantitated with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Subcellular fractionation.
To obtain cytoplasmic fractions, cells were trypsinized, rinsed with phosphate-buffered saline, incubated in 200 μl of hypotonic buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2) supplemented with inhibitors (leupeptin [1 μg/ml], aprotinin [1 μg/ml], and 0.5 mM phenylmethylsulfonyl fluoride) on ice, and lysed by addition of 25 μl of buffer A containing 2.5% Nonidet P-40 plus inhibitors. Nuclei were pelleted (3,500 rpm, 4 min, 4°C), and supernatants were saved, freeze-thawed five times, and centrifuged (10 min, 3,500 rpm, 4°C). Cytosolic fractions were prepared by subjecting cytoplasmic lysates to an additional step of high-speed centrifugation (14,000 rpm for 60 min at 4°C) and discarding any pelleted material. For preparing nuclear fractions, nuclear pellets were incubated in extraction buffer C (20 mM HEPES [pH 7.9], 0.45 M NaCl, 1 mM EDTA) plus inhibitors and centrifuged (10 min, 14,000 rpm, 4°C), and supernatants were saved. The efficiency and quality of nuclei preparation were monitored with a hemacytometer at the end of the nucleus isolation procedure. Whole-cell lysates were prepared in 20 mM HEPES (pH 7.4)–50 mM β-glycerophosphate–1% Triton X-100–10% glycerol–2 mM EGTA–1 mM dithiothreitol–protease inhibitors.
Western blot analysis.
Whole-cell (20 μg), cytoplasmic (40 μg), cytosolic (40 μg), or nuclear (10 μg) lysates were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes. Equal loading and transferring of samples were confirmed by staining membranes with Ponceau red (Sigma) before hybridization. HuR was detected with monoclonal antibody 19F12 (M. Orlian, H. Cheng, B. Joseph, and H. Furneaux, submitted for publication), p21 was detected with a monoclonal antibody (Calbiochem, San Diego, Calif.), actin was detected with a monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), c-Jun was detected with a polyclonal antibody (Santa Cruz Biotechnology), BAF57c was detected with a polyclonal antibody (63), AUF1 was detected with polyclonal antiserum (67), and hnRNP C was detected with a monoclonal antibody (12). Following incubation with the appropriate secondary antibody, signals were detected with an enhanced chemiluminescence system (Amersham, Arlington Heights, Ill.). Where indicated, samples were incubated with 2 to 10 fmol of radiolabeled B2 or B5, digested with RNase T1, and cross-linked before Western blot analysis. Visualization of radioactive samples on membranes, which required a 24-h period of exposure to autoradiographic film, was carried out before Western blotting, which required only a 30-s exposure to autoradiographic film, thus dismissing all concerns that radioactive signals might have been mistakenly detected by Western blot analysis.
Preparation of transcripts.
pCEP4-Waf1 was used as a template for PCR amplification of different p21 cDNA regions. All 5′ oligonucleotides contained the T7 promoter sequence 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′ (T7). For generating the A1 template, oligonucleotides A (5′-T7GCCGAAGTCAGTTCCTTGTG-3′) and 1 (5′-TTCCAGGACTGCAGGCTTC-3′), corresponding to positions 1 to 20 and 600 to 582 of the p21 mRNA, respectively, were used. For B2 and B5 templates, oligonucleotide B (5′-T7CCAAGAGGAAGCCCTAATCC-3′), corresponding to positions 554 to 573, was used along with either 2 (5′-GAAAAGGAGAACACGGGATG-3′) or 5 (5′-AAAGTCACTAAGAATCATTTATTG-3′) at positions 851 to 832 and 2102 to 2079, respectively. For fragment C5, oligonucleotides C (5′-T7CATCCCGTGTTCTCCTTTTC-3′, at positions 832 to 851) and 5 were used. RNA fragments A1, B2, B5, and C5 (radiolabeled unless otherwise indicated) were synthesized from PCR-generated DNA fragments as described previously (22), purified through spin columns, and used at 100,000 cpm/μl (2 to 10 fmol/μl). Unlabeled RNA was synthesized with 2 mM UTP instead of [32P]UTP.
RNA-protein binding reactions and supershift assays.
Reaction mixtures (10 μl) containing 1 μg of tRNA, 2 to 10 fmol of RNA, and 10 μg of protein in reaction buffer (15 mM HEPES [pH 7.9], 10 mM KCl, 10% glycerol, 0.2 mM dithiothreitol, 5 mM MgCl2) were incubated for 30 min at 25°C and digested with RNase T1 (100 U/reaction) for 15 min at 37°C. Complexes were resolved by electrophoresis, either through native gels (7% acrylamide in 0.25× Tris-borate-EDTA buffer) without loading buffer (160 V, 2 h, 4°C) or through SDS–15% acrylamide gels, after cross-linking of complexes through delivery of 1,800 J/m2 with a Stratalinker (Stratagene, La Jolla, Calif.) and denaturation with Laemmli buffer. Cross-linking before or after RNase T1 digestion yielded identical results. Native and SDS-gels were dried, and radioactivity was visualized with a PhosphorImager. For supershifts, 4 μg of antibody were incubated with lysates for 1 h on ice before addition of radiolabeled RNA; all subsequent steps were as described for native gels. All antibodies used in supershift assays were from Pharmingen (San Diego, Calif.) except those recognizing HuR.
RNase T1 selection and gel retardation assays.
RNase T1 selection assays were carried out by incubating 20 fmol of RNA and recombinant glutathione S-transferase (GST) or GST-HuR, as described elsewhere (30). Reaction mixtures were digested with RNase T1, analyzed by electrophoresis on 12% polyacrylamide–50% urea gels, fixed, and exposed to X-ray film. The same recombinant proteins were used in gel retardation assays, carried out as described previously (30).
RESULTS
Identification of UVC-inducible p21 mRNA-protein complexes.
UVC irradiation induces p21 mRNA levels by increasing p21 mRNA half-life (27). Since stabilization of short-lived mRNAs is known to involve binding of proteins that recognize certain mRNA sequences, we examined whether proteins present in lysates from human colorectal carcinoma RKO cells bound to p21 mRNA. Depicted in Fig. 1 are the p21 mRNA and the RNA molecules used for analysis, transcripts A1, B2, B5, and C5, prepared after PCR amplification of the corresponding cDNA fragments (see Materials and Methods). In this initial characterization, binding to p21 RNA was also tested in lysates from RKO cells lacking p53 function (via expression of the E6 viral oncoprotein). This was of interest since we previously observed that UVC-triggered p21 mRNA stabilization required functional p53 (27). As shown in Fig. 1A, incubation with B5 (radiolabeled, unless otherwise stated) revealed RNA-protein complexes in all lysates tested. Excess B5 RNA was degraded with RNase T1. Both cytoplasmic and nuclear proteins bound to B5, but only complexes forming with cytoplasmic lysates exhibited UVC inducibility, despite the abundance of nuclear complexes. However, no differences in complex formation were detected when we compared RKO cells with wild-type (neo) versus deficient (E6) p53 function.
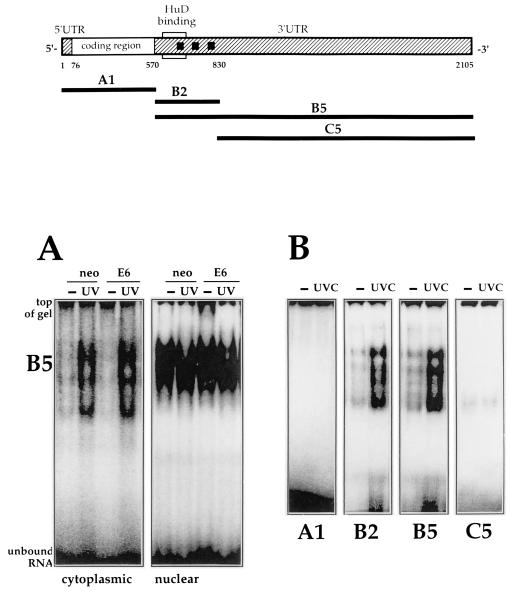
FIG. 1.
Structure of the p21 mRNA and fragment analysis. Top, structure of the full-length p21 mRNA, indicating the coding and untranslated regions (5′ and 3′ UTRs). Squares represent AUUUA sequences; the region previously shown to bind HuD (30) is indicated. Heavy lines indicate the RNAs used in this study. Corresponding PCR-amplified regions served as templates for in vitro transcription of these RNA molecules (see Materials and Methods). (A) B5 was incubated with cytoplasmic and nuclear lysates of RKO cells (neo and E6) 6 h following treatment with UVC (20 J/m2; UV) or no treatment (−); unbound transcript was digested with RNase T1. Reaction products were resolved by electrophoresis through native 7% polyacrylamide gels. (B) Cytoplasmic lysates from either untreated or UVC-treated cells were tested for binding to the various transcripts shown in panel A. Binding assays and electrophoresis were carried out as described in Materials and Methods.
To better characterize this binding activity, various RNAs corresponding to different regions of the p21 mRNA were incubated with lysates from RKO cells. As shown, only B2 and B5 fragments formed complexes that increased after UVC treatment (Fig. 1B). Incubation of A1, B2, and C5 with nuclear lysates showed binding patterns similar to those seen with B5 (not shown). The time dependency of complex formation is shown in Fig. 2A. The panel below depicts Northern blot analysis of p21 mRNA expression in cells treated similarly, illustrating the correlation between p21 mRNA induction and binding of proteins to the p21 mRNA. The specificity of binding to the B5 RNA was evidenced by the fact that addition of unlabeled B2 effectively competed for binding of B5, while A1 failed to do so, even at a 50-fold molar excess (Fig. 2B). To further assess the specificity of binding and to aid in identifying the protein(s) involved, we performed UV cross-linking experiments to covalently bind the radiolabeled RNA to the protein(s) in the complex and resolved them by SDS-PAGE. Cytoplasmic complexes forming with B2 (and B5 [not shown]) revealed a major UVC-inducible band of about 37 to 40 kDa (Fig. 2C). However, cross-linked bands were absent from nuclear preparations. It is possible that the RNA-binding protein(s) recognizing B2 in the nucleus differ from those in the cytoplasm. Alternatively, the same protein(s) may interact in ways that cannot be cross-linked. The kinetics of formation of cross-linked complexes after UVC treatment (Fig. 2D) were similar to those observed with complexes resolved on native gels (Fig. 2A).
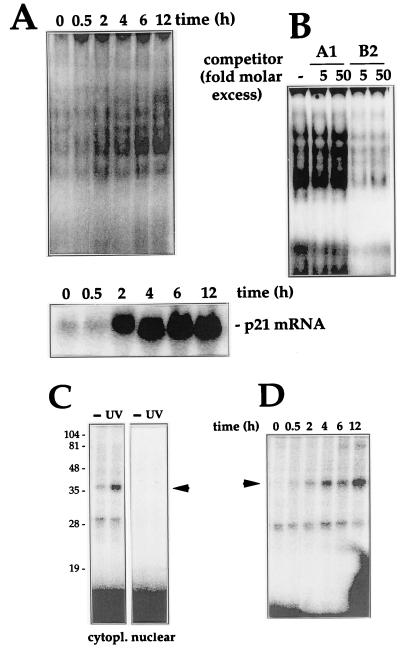
FIG. 2.
Characterization of binding activity: time course, specificity, and cross-linking assays. (A) After UVC irradiation of RKO cells, cytoplasmic lysates were prepared at the times indicated and assayed for binding to B2 (top), or total mRNA, prepared and p21 mRNA expression assayed by Northern blotting (bottom); RNA was loaded evenly, as assessed after stripping of membranes and hybridization using a probe recognizing 18S (not shown). (B) Binding specificity was tested in cytoplasmic lysates from UVC-treated cells and B5, in the absence (−) or presence of the indicated molar excesses of unlabeled B2 or A1 RNAs. (C) Subcellular fractions were incubated with B2, digested with RNase T1, cross-linked, and resolved by SDS-PAGE (15% gel). Gels were dried and exposed to X-ray film to visualize radiolabeled complexes. Numbers denote sizes (in kilodaltons) of molecular weight markers. (D) At the times indicated after UVC irradiation, cytoplasmic lysates were prepared and binding to B2 was assayed. Processing of samples was carried out as described for panel A. Arrowheads indicate inducible complexes.
HuR is a specific RNA-binding component of the p21 mRNA-protein complex.
Based on the apparent size of the complexes, their cytoplasmic localization, the specific region of the p21 3′ UTR recognized, and its presence in nonneuronal cells, we postulated that the Elav-like protein HuR may form part of these p21 RNA-protein complexes. To test this possibility, we assayed the ability of antibodies recognizing HuR to supershift the p21 RNA-protein complexes on native gels. As shown (Fig. 3A), HuR indeed forms part of the complexes, as a prominent band of slower electrophoretic mobility was detected when the anti-HuR antibody was added to B2-protein complexes. Supershifted bands were readily observed in cytoplasmic lysates; of the approximately four bands seen routinely, addition of anti-HuR antibody depleted most clearly the second slower-mobility band, indicating that HuR formed part of at least this shifted complex. Despite their abundance, nuclear complexes could not be supershifted, even when assayed under a variety of binding conditions. Binding of the anti-HuR antibody to cytoplasmic proteins was specific, as antibodies recognizing unrelated proteins failed to supershift the complex (Fig. 3B).
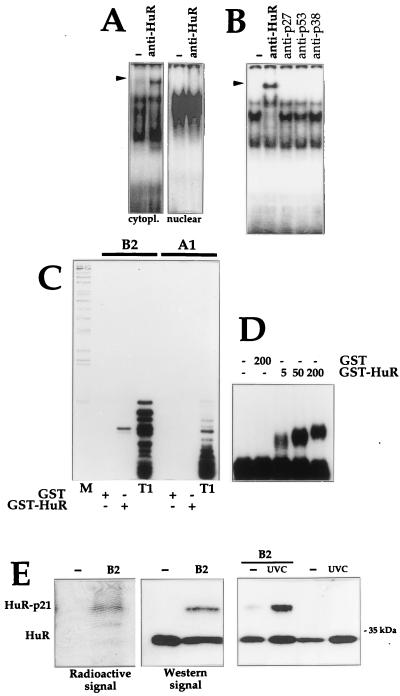
FIG. 3.
HuR binds the p21 mRNA in vivo and in vitro. (A) Monoclonal antibody 19F12 (4 μg) was incubated with cytoplasmic or nuclear lysates of UVC-treated cells and B2. Complexes were resolved in native 7% polyacrylamide gels. (B) The indicated antibodies were tested for their ability to supershift complexes forming between cytoplasmic proteins and B2. Arrowheads indicate positions of specific supershifted complexes. (C) RNase T1 selection assay was carried out with B2 and A1, incubated with 10 nM GST or GST-HuR (see Materials and Methods). T1, digestions with RNase T1 alone; M, molecular weight markers. (D) Gel retardation assays using B2 and the indicated concentrations of either GST or GST-HuR. (E) Left, cytoplasmic fractions were either incubated with B2 or not, cross-linked, digested with RNase T1, resolved by SDS-PAGE (15% gel), and transferred onto polyvinylidene difluoride membranes, which were sequentially exposed to X-ray film for 24 h (Radioactive signal) and subjected to Western blot analysis to detect HuR (Western signal); exposure time, 30 s. Right, Lysates from UVC-treated or untreated cells were incubated with B2 and then subjected to Western blot analysis. Estimated size of the HuR-p21 complexes, 37 to 40 kDa.
Further indication that HuR bound the p21 mRNA was obtained in assays using recombinant GST-HuR. In RNase T1 selection assays, two fragments, 15 and 16 nucleotides long, were selected after incubation of transcript B2 with GST-HuR; as anticipated, such fragments were not seen in incubations with either GST or the A1 transcript (Fig. 3C). The pattern seen after incubation with B2 resembles that previously seen with HuD (30). As determined in gel retardation assays (Fig. 3D), B2 and GST-HuR exhibited high-affinity interaction, with an apparent Kd of 4 nM.
Additional evidence that HuR was present in complexes with p21 RNA came from experiments where cross-linked complexes were fractionated by SDS-PAGE and transferred onto membranes that were first exposed to X-ray film for 24 h to visualize the radioactive p21 RNA-protein complexes and subsequently subjected to Western blot analysis to detect HuR (both free and p21 mRNA bound) after a short exposure period of 30 s. The difference in exposure time required, as well as the order of signal detection (radioactive first, ECL-Western second) helped ensure that radioactive signals were not detected by Western blotting. As shown (Fig. 3E), the electrophoretic mobility of the major radioactive band was identical to that of the signal detected on Western blots. Indeed, the HuR-p21 radioactive and Western signals overlapped precisely. The complex migrated with an apparent molecular mass of 37 to 40 kDa, somewhat larger than that of unbound HuR protein, which has a size of approximately 34 kDa. As anticipated, complexes formed with either B2 or B5 increased following UVC treatment (Fig. 3E).
HuR is required for the UVC-induced stabilization of p21 mRNA.
To assess the significance of HuR binding to the p21 mRNA, we developed RKO lines expressing reduced levels of HuR protein through transfection with a plasmid constitutively expressing an antisense HuR (AS HuR) transcript. HuR levels in selected clonal isolates (Fig. 4A) were between four- and sixfold lower than those seen in untransfected cells and in cells transfected with an empty vector (not shown). Expression of other RNA-binding proteins such as hnRNP C (Fig. 4A) or AUF1 (not shown) was not affected by transfection with AS HuR. As anticipated, lysates from these cells exhibited substantially reduced binding to the p21 RNA (B2 or B5), and this decrease was apparent in lysates from both control and UVC-treated cells (Fig. 4B). Interestingly, we observed a reduction in all shifted bands but are uncertain why we observe this global decrease in binding activity (see Discussion). Using these transfectants, we investigated whether HuR influenced UVC-induced p21 mRNA expression by Northern blot analysis. As shown in Fig. 4C, clonal isolates of cells expressing low levels of HuR showed significantly lower induction of p21 mRNA relative to parental lines (four- to fivefold reduction); induction of p21 protein by UVC irradiation was also reduced two- to threefold in AS HuR-expressing cells (Fig. 4D). To assess whether these reductions were due to a secondary effect of AS HuR transfection, resulting in an impairment in the cell's responsiveness to UVC, we examined the expression of the UVC-responsive protein c-Jun. As shown, c-Jun expression was unchanged among untreated populations with different HuR levels and underwent a comparable activation (phosphorylation) in each of the three lines examined following UVC irradiation. This observation supports the notion that the responsiveness to UVC is intact in cells with lower HuR. Finally, to ascertain if the reduced p21 mRNA expression was due to decreased p21 mRNA stability, mRNA half-life was measured in cells expressing either normal or reduced HuR levels after addition of actinomycin D (Fig. 4E). The p21 mRNA half-life in RKO cells with normal HuR levels was about 6 h (higher than seen in other cell lines), and treatment with UVC increased it markedly, with an estimated half-life of greater than 10 h. By contrast, the p21 mRNA half-life was much lower in AS.1 cells (2.8 h in untreated cells, 3 h after UVC irradiation) and in AS.7 cells (2.5 h in untreated cells, 2.8 h in UVC-treated cells). Importantly, the half-life of p21 mRNA did not increase substantially after UVC irradiation in AS.1 or AS.7 cells. The change in stability of the p21 mRNA was specific for this transcript, as the rate of elimination of a control mRNA (encoding β-actin) was essentially the same among the cell lines tested (Fig. 4E). Together, these observations strongly support a role for HuR in the stabilization of p21 mRNA, resulting in increased p21 mRNA levels.
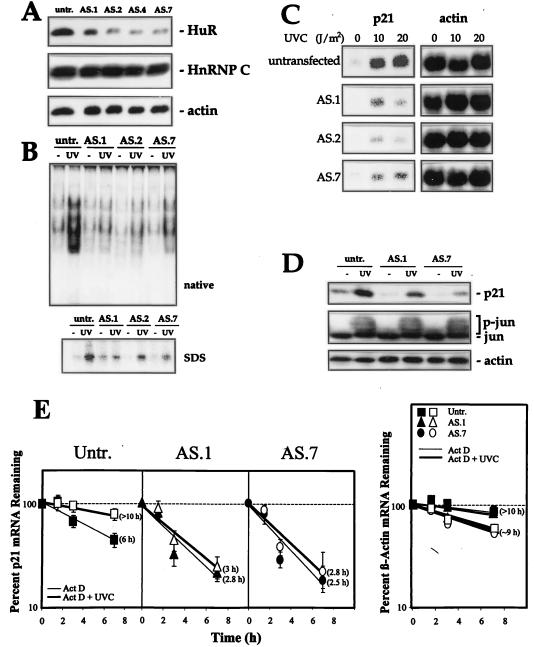
FIG. 4.
Decreased HuR expression lowers binding to the p21 3′ UTR and reduces p21 mRNA stability and p21 induction by UVC. (A) Western blot analysis of HuR expression in RKO cells, either untransfected (untr.) or transfected with pZeoSV2(−)HuR, expressing AS HuR. Chosen clonal isolates are shown. Blots were sequentially stripped and rehybridized with an antibody recognizing actin (43 kDa), to visualize differences in loading and transfer, and with an antibody recognizing hnRNP C (43 kDa). (B) B5 binding activity in lysates from untransfected and AS HuR-expressing cells 6 h after UVC irradiation. (C) Northern blot analysis of p21 mRNA expression in untransfected and AS HuR-expressing RKO cells 8 h after either no treatment (−) or exposure to the indicated UVC doses. Evenness in loading and transfer among samples was assessed after stripping the membrane and rehybridizing it with an oligomer probe recognizing 18S rRNA. (D) Western blot analysis to assess the expression of p21, c-Jun (39 kDa), and actin in untransfected and AS HuR-expressing RKO cells 10 h after either no treatment or exposure to 20 J/m2 UVC. p-jun, phosphorylated Jun. (E) Graphs depict the rate of loss of p21 and β-actin mRNAs in cells with different HuR levels after actinomycin D (2 μg/ml) addition with or without UVC irradiation. At the times indicated, total RNA was extracted and p21 and β-actin mRNAs were monitored by Northern blotting; signals were quantitated with a PhosphorImager, normalized against 18S (not shown), and plotted on a logarithmic scale. The mRNA half-life in each treatment group is indicated in parentheses. Values represent means ± standard errors of the means of three independent experiments.
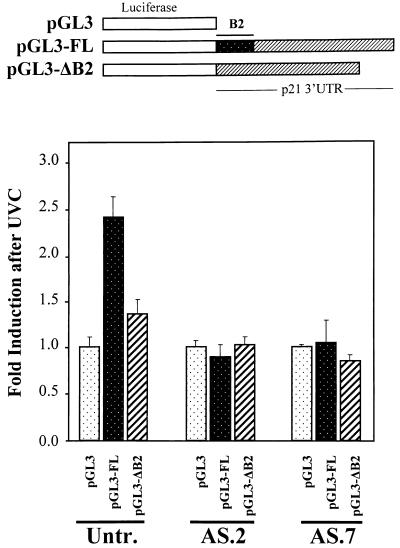
Independent assessment of the stabilizing and inducing effect of HuR on p21 expression was sought through the creation and analysis of luciferase reporter constructs linked to either the full-length p21 3′ UTR or a mutation thereof lacking the B2 HuR-binding region (Fig. 5). Following transient transfection with the parent vector pGL3, luciferase activity in UVC-irradiated RKO cells was no different than that measured in untreated populations. By contrast, pGL3-FL, expressing a chimeric mRNA encoding luciferase and the full-length p21 3′ UTR, exhibited a readily inducible luciferase activity that was 2.4-fold higher after UVC irradiation than that in untreated cells of the same transfection group (Fig. 5). This induction was abolished when pGL3-ΔB2, a mutant construct lacking the HuR-binding region B2, was used: UVC irradiation of cells transfected with this construct yielded luciferase activities that were only 1.35-fold higher than those seen in untreated cells (Fig. 5). Similar transfection of AS HuR clones AS.2 and AS.7 failed to display such UVC inducibility of pGL3 or pGL3-ΔB2 constructs over that of pGL3, indicating that HuR was required for the UVC-triggered induction of pGL3-FL. Of note, however, when we assayed AS.7 cells, all transfection groups, pGL3, pGL3-FL and pGL3-ΔB2, showed a twofold higher level of luciferase activity following UVC irradiation. In summary, the p21 3′ UTR fragment B2, recognized by HuR, confers UVC inducibility of a heterologous reporter gene while deletion of B2 greatly diminishes this induction. HuR is required for the UVC inducibility of the chimeric construct containing B2.
FIG. 5.
Effect of the full-length and mutant p21 3′ UTR on expression of a luciferase reporter construct. (Top) Expression vectors pGL3, pGL3-FL, and pGL3-ΔB2 (see Materials and Methods) were transiently cotransfected into RKO parental (untransfected [Untr.]), AS.2, or AS.7 cells along with pSV-βgal (used to normalize for transfection efficiency); cells were irradiated with UVC (20 J/m2) or left untreated, and luciferase and β-galactosidase activities were examined 24 h later. (Bottom) Relative fold increase in luciferase activity after UVC exposure, seen with either pGL3-FL or pGL3-ΔB2 compared with that seen with the control vector pGL3. Values represent means ± standard errors of the means of five independent experiments.
HuR is primarily nuclear but becomes cytoplasmic after exposure to UVC.
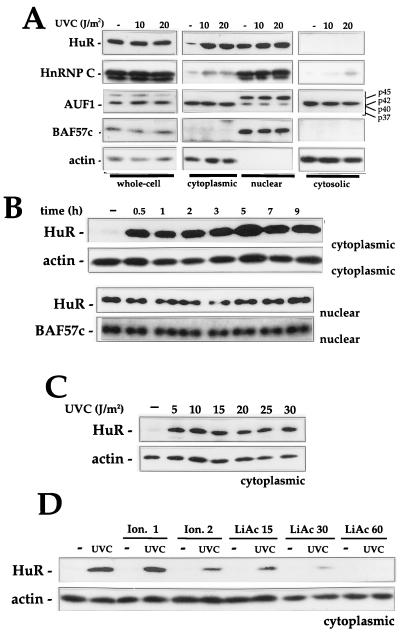
The evidence presented thus far indicates that UVC irradiation enhances the formation of cytoplasmic HuR-p21 RNA complexes. Therefore, we sought to directly examine the subcellular location of HuR following exposure to UVC and other stresses. Shown in Fig. 6 are Western blots examining the expression of HuR and other RNA-binding proteins in whole-cell lysates and various cellular fractions. UVC irradiation had no significant effect on total cellular HuR, but it greatly increased cytoplasmic HuR levels (Fig. 6A); while mechanisms such as stabilization of the HuR protein in the cytoplasm might be invoked to explain this increase, we favor the view that HuR is being transported to the cytoplasm, possibly through transport systems such as those described for other RNA-binding proteins (47, 48) or for HuR specifically (2, 18, 50). The lack of appreciable differences in nuclear HuR can be explained by the fact that HuR is very abundant in the nucleus (approximately 30-fold higher in resting cells) and therefore only a small fraction of nuclear HuR is mobilized to the cytoplasm. In addition, cytosolic fractions were devoid of HuR, as determined by Western blot analysis. This loss is presumably due to the pelleting of HuR bound to ribosome-associated mRNP complexes and is in agreement with our findings that cytosolic fractions exhibit no RNA-binding activity (data not shown). It was of interest to examine if other RNA-binding proteins also redistributed among intracellular compartments following UVC irradiation; therefore, we examined the expression levels and relative distribution of hnRNP C and AUF1. hnRNP C was more abundant in the nucleus, but cytoplasmic hnRNP C increased two- to threefold in the cytoplasmic and cytosolic fractions following UVC irradiation. AUF1, on the other hand, underwent no enhancement in cytoplasmic presence following UVC irradiation; however, the relative abundance of the p45, p42/p40, and p37 isoforms did vary: p45 was markedly more abundant in the nucleus, while the p42 and p40 isoforms were more abundant in the cytoplasm. Levels of the AUF1 p37 isoform were very low in all cell compartments examined.
FIG. 6.
Western blot analysis of HuR expression and subcellular localization. (A) Six hours after irradiation with the indicated doses of UVC, whole-cell (20 μg), cytoplasmic (40 μg), nuclear (10 μg), and cytosolic (40 μg) lysates were prepared and subjected to Western blot analysis to monitor the expression of HuR, hnRNP C, AUF1, BAF57c (57 kDa), and actin. Cell lysates were collected at the times indicated after irradiation with UVC (20 J/m2) (B) or 6 h after irradiation with the indicated doses of UVC (C), and Western blot analysis of HuR expression performed on cytoplasmic (40 μg) and nuclear (10 μg) fractions. (D) Indicated doses of ionomycin (Ion.; micromolar) or lithium acetate (LiAc; millimolar) were added to cells 1 h before UVC irradiation with 20 J/m2 and Western blot analysis of cytoplasmic HuR. Hybridization using antibodies against actin and BAF57c was carried out to assess uniformity in loading and transfer among cytoplasmic and nuclear samples, respectively.
Verification that nuclear proteins did not leak into the cytoplasmic fractions during the fractionation process was obtained through subsequent hybridizations of Western membranes to detect BAF57c (BRG1-associated protein [63]), a protein that localizes exclusively in the nucleus; BAF57c was detected only in nuclear fractions (Fig. 6A). Conversely, actin was detected only in the cytoplasmic fractions. These observations indicate that cytoplasmic HuR increases specifically and dramatically following exposure UVC. Such cytoplasmic HuR increases specifically and dramatically following exposure UVC. Such cytoplasmic enrichment was found to be rapid (Fig. 6B) and occurred even after exposure to very low doses of UVC (Fig. 6C).
To further explore the process of HuR transport to the cytoplasm, we tested the involvement of potential regulatory pathways. First, we tested the effect of inhibitors of UVC-triggered signaling cascades such as those involving mitogen-activated protein kinases, protein kinase C, phosphatidylinositol-3 kinase, S6 kinase, and other kinases by using pharmacological inhibitors, dominant-negative regulators, etc., but none of them influenced the cytoplasmic localization of HuR after UVC irradiation (data not shown). From the panel of treatments tested, only two agents that modulate intracellular signalling were found to prevent the cytoplasmic accumulation of HuR: ionomycin, a Ca2+ ionophore (3, 28, 58), and lithium acetate, which blocks inositol phosphate-mediated signalling, inhibits glycogen synthase kinase 3β, and perturbs intracellular Ca2+ (6, 7, 36). Among the wide range of cellular effects elicited by ionomycin and lithium acetate are alterations in nucleocytoplasmic transport (55, 58) and specifically promotion of nuclear import of proteins (32). As shown, ionomycin pretreatment prior to UVC irradiation moderately prevented UVC-mediated increase in cytoplasmic HuR, while lithium acetate pretreatment had a more profound influence, effectively preventing increases in cytoplasmic HuR following UVC irradiation (Fig. 6D). It is possible that these two treatments inhibit HuR's transport out of the nucleus, as reported for other proteins (32), but further experimentation is required to ascertain if this hypothesis is correct. Interestingly, though, pretreatment with either ionomycin or lithium acetate also prevented p21 mRNA induction by UVC on Northern blots (not shown), supporting the hypothesis that cytoplasmic HuR mediates stabilization of, and hence induces, the p21 mRNA.
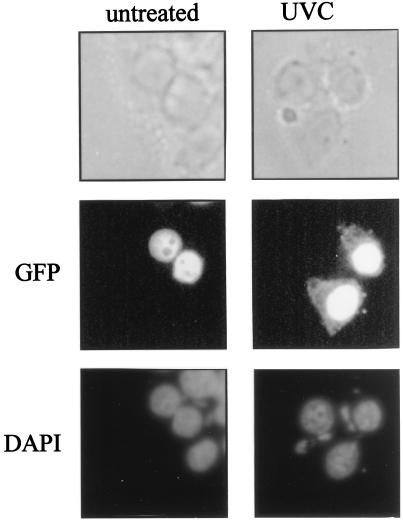
Additional evidence that HuR's presence increases in the cytoplasm after exposure to UVC was obtained with a GFP-HuR fusion construct. By fluorescence microscopy, signal of the chimeric protein was undetectable in the cytoplasm of untreated cells and was seen only in the nucleus. However, GFP-HuR signal in the cytoplasm increased greatly following UVC irradiation (Fig. 7). Not unexpectedly, however, abundant GFP-HuR signal was still seen in the nucleus after UVC irradiation, an observation in keeping with our Western data results showing that HuR remains very abundant in the nucleus even after UVC irradiation (Fig. 6A).
FIG. 7.
Subcellular localization of HuR. GFP-HuR was visualized by fluorescence microscopy in transiently transfected RKO cells that were either left untreated or treated with 20 J of UVC/m2 (4 h earlier). DAPI staining served to visualize the nucleus. Note the distinct overlap of DAPI and GFP-HuR signals in untreated cells; while UVC-irradiated cells also exhibit abundant nuclear GFP-HuR, the treatment causes a substantial increase in the cytoplasmic GFP-HuR signal, not seen in untreated cells.
Other stresses elevate cytoplasmic HuR and HuR binding to the p21 mRNA.
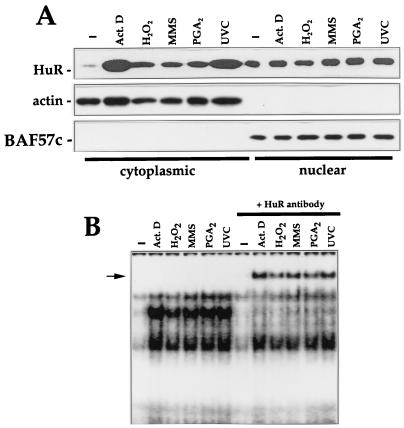
To determine if HuR participated, in a more general sense, in binding to p21 mRNA after exposure to other stresses, we treated cells with other damaging agents, including actinomycin D, hydrogen peroxide, the alkylating agent MMS, and the cyclopentenone PGA2. Each of these treatments have been found to induce p21 expression (reference 24 and unpublished results). Interestingly, all treatments elevated cytoplasmic HuR and binding to the p21 mRNA (Fig. 8); the presence of HuR in such stress-inducible complexes was assessed by supershift analysis (Fig. 8B).
FIG. 8.
Increased cytoplasmic HuR and p21 RNA binding after exposure to stresses. (A) Western blot analysis to monitor HuR expression in cytoplasmic and nuclear fractions after treatment with the indicated agents. Samples were collected 2 h after addition of actinomycin (Act.) D (1 μg/ml) or 4 h after exposure to 100 μM H2O2, MMS (100 μg/ml), 48 μM PGA2, or UVC (20 J/m2). Hybridizations using antibodies against actin and BAF57c were carried out to assess uniformity in loading and transfer among cytoplasmic and nuclear samples, respectively. (B) B2 binding activity in cytoplasmic lysates of cells treated as for panel and supershift analysis of complexes forming after exposure to such stresses.
DISCUSSION
In the present study, we have provided evidence that during conditions of stress, the RNA-binding protein HuR binds to the p21 mRNA and plays an important role in regulating its stability. Our conclusion that endogenous HuR binds to the p21 mRNA is based on four major observations. First, the binding activity mapped to a region of the 3′ UTR previously found to be bound by purified HuD (30) and, as shown here, also by purified HuR. Second, cross-linking experiments to permanently bind complexes of radiolabeled RNAs and proteins revealed radioactive bands that overlapped those detected on HuR Western blots. Third, p21 RNA-protein complexes were supershifted, at least partly, by an antibody that recognizes HuR (Fig. 3). Finally, a reduction in HuR levels through expression of AS HuR led to a dramatic decrease in p21 mRNA-protein binding. In fact, all of the bands were reduced, suggesting that HuR may be a part of each one of the complexes. HuR may associate with additional proteins, thus explaining the different complex sizes, or it may be posttranslationally modified. Alternatively, other UVC-inducible RNA-binding proteins may participate in these complexes, and their expression and/or binding to the p21 RNA may somehow be altered in the AS HuR lines.
Our findings are in keeping with the general view that HuR exerts a stabilizing influence on labile mRNAs. We arrive at this conclusion from data indicating that UVC-stimulated binding of HuR to the p21 mRNA was associated with enhanced p21 mRNA levels (Fig. 2) and also from direct analysis of AS HuR-expressing cells, where lowered HuR levels markedly reduced the degree of p21 induction seen with UVC, the p21 mRNA half-life, and the induction of the p21 protein (Fig. 4). Independent evidence that this stabilization effect led to gene induction was obtained through analysis of reporter constructs expressing chimeric mRNAs where the luciferase coding region was linked to either the full-length p21 3′ UTR or a mutant lacking B2. Chimeric constructs with the full-length p21 3′ UTR were readily induced by UVC, while either deletion of B2 or assaying in cells with reduced HuR levels (Fig. 5) abolished this induction. Precedence for a stabilizing effect of HuR and other Elav-like proteins comes from studies on the VEGF mRNA (39), ARE-bearing mRNAs (17), in vitro mRNA deadenylation/degradation systems (19), and GLUT-1 mRNA (29). HuR action may thus fit a broadly accepted model to explain the function of ARE-binding proteins in mRNA stabilization: binding prevents the association of other destabilizing factor(s) to those mRNAs, perhaps through competition for binding to similar mRNA sequences.
It will be of great interest to elucidate the mechanisms regulating the cytoplasmic localization of HuR, particularly based on our findings that binding to the p21 mRNA appears to be optimal when HuR is in the cytoplasm. The majority of HuR is nuclear in untreated cells and its increased presence in the cytoplasm is likely to result from export of HUR out of the nucleus. This process may occur universally in mammalian cells, as similar stress-triggered changes in subcellular localization of HuR and in binding activity were also seen in mouse embryo fibroblasts, in human diploid fibroblasts, and in human renal carcinoma cells (not shown). In support of a recent model (2, 17–19, 31, 50) whereby HuR can be actively transported to the cytoplasm (or perhaps shuttles between the nucleus and the cytoplasm) are three observations reported here: (i) the rapidity of HuR increase in cytoplasm, which was almost maximal by 30 min; (ii) the inhibition of UVC-triggered cytoplasmic localization of HuR by ionomycin and lithium acetate, two agents that have been shown to prevent cytoplasmic export and induce nuclear import of proteins (32); and (iii) the demonstration that the nucleus-localized GFP-HuR fusion protein is mobilized to the cytoplasm following UVC irradiation. The nature of the transport system(s) and additional posttranslational regulatory events that may be critical for HuR's binding activity deserve close attention, since our observations lend support to the exciting model that HuR cytoplasmic localization is coupled to mRNA stabilization (18, 50).
While we previously demonstrated that the p21 mRNA was preferentially stabilized by UVC in the presence of functional p53 (27), HuR is unlikely to mediate these differences, as p53 status did not influence HuR expression, subcellular location, or binding to the p21 mRNA (not shown).
In conclusion, UVC-triggered elevation in cytoplasmic HuR leads to HuR's increased binding to the p21 mRNA, resulting in the stabilization and enhanced expression of p21 mRNA. That exposure to additional stresses of a different nature (genotoxins, oxidants, etc.) also enhanced cytoplasmic HuR and binding to p21 mRNA suggests that changes in p21 mRNA stability are not limited to UVC treatment but also occur with other stresses and act in concert with transcriptional mechanisms to regulate p21 expression. The relative contribution of the transcriptional and mRNA stabilization processes is likely to vary among stresses, but our findings support a central role for HuR in regulating p21 expression during the stress response. In addition, given the large number of stress-regulated genes with AU-rich UTRs that could potentially bind HuR, many of which are known to be induced by stresses (c-Fos, VEGF, GLUT-1, etc.), it is likely that HuR plays a broad role in regulating gene expression during stress, permitting the cell to adapt to environmental changes.
ACKNOWLEDGMENTS
We thank M. B. Kastan for the RKO cells, B. Vogelstein for pCEP4Wafl, W. Wang for the BAF57c antibody, G. Dreyfuss for the hnRNP C antibody, and G. Brewer for the AUF1 antibody. We are also grateful to our colleagues S. Lin, S. Shack, and D. L. Longo for critical reading of the manuscript.
REFERENCES
- 1.Antic D, Lu N, Keene J D. ELAV tumor antigen, hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atasoy U, Watson J, Patel D, Keene J D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 3.Badminton M N, Kendall J M, Rembold C M, Campbell A K. Current evidence suggests independent regulation of nuclear calcium. Cell Calcium. 1998;23:79–86. doi: 10.1016/s0143-4160(98)90105-1. [DOI] [PubMed] [Google Scholar]
- 4.Barami K, Iversen K, Furneaux H, Goldman S. Hu protein as an early marker of neuronal phenotypic differentiation by subependymal zone cells of the adult songbird forebrain. J Neurobiol. 1995;28:82–101. doi: 10.1002/neu.480280108. [DOI] [PubMed] [Google Scholar]
- 5.Bellido T, O'Brien C A, Roberson P K, Manolagas S C. Transcriptional activation of the p21WAF1, CIP1, SDI1 gene by interleukin-6 type cytokines. A prerequisite for their pro-differentiating and anti-apoptotic effects on human osteoblastic cells. J Biol Chem. 1998;273:21137–21144. doi: 10.1074/jbc.273.33.21137. [DOI] [PubMed] [Google Scholar]
- 6.Berridge M J, Downes C P, Hanley M R. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 7.Berridge M J. The Albert Lasker Medical Awards. Inositol trisphosphate, calcium, lithium, and cell signaling. JAMA. 1989;262:1834–1841. [PubMed] [Google Scholar]
- 8.Biggs J R, Kudlow J E, Kraft A S. The role of the transcription factor Sp1 in regulating the expression of the WAF1/CIP1 gene in U937 leukemic cells. J Biol Chem. 1996;271:901–906. doi: 10.1074/jbc.271.2.901. [DOI] [PubMed] [Google Scholar]
- 9.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 10.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 11.Chinery R, Brockman J A, Peeler M O, Shyr Y, Beauchamp R D, Coffey R J. Antioxidants enhance the cytotoxicity of chemotherapeutic agents in colorectal cancer: a p53-independent induction of p21WAF1/CIP1 via C/EBPβ. Nat Med. 1997;3:1233–1241. doi: 10.1038/nm1197-1233. [DOI] [PubMed] [Google Scholar]
- 12.Choi Y D, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984;99:1997–1204. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 14.Dalmau J, Furneaux H, Cordon-Cardo C, Posner J B. The expression of the Hu (paraneoplastic encephalomyelitis/sensory neuronopathy) antigen in human normal and tumor tissues. Am J Pathol. 1992;141:1–6. [PMC free article] [PubMed] [Google Scholar]
- 15.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry W, Tokino T, Velculescu V, Levy D, Parsons R, Trent J, Lin D, Mercer W, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan X C, Steitz J A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford L P, Watson J, Keene J D, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao F B, Carson C C, Levine C T, Keene J. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good P J. A conserved family of Elav-like genes in vertebrates. Proc Natl Acad Sci USA. 1995;89:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorospe M, Baglioni C. Degradation of unstable interleukin-1α mRNA in a rabbit reticulocyte cell-free system. Localization of an instability determinant to a cluster of AUUUA motifs. J Biol Chem. 1994;269:11845–11851. [PubMed] [Google Scholar]
- 23.Gorospe M, Holbrook N J. Role of p21 in prostaglandin A2-mediated cellular arrest and death. Cancer Res. 1996;56:475–479. [PubMed] [Google Scholar]
- 24.Gorospe M, Martindale J M, Sheikh M S, Fornace A J, Jr, Holbrook N J. Regulation of p21CIP1/WAF1 expression by cellular stress: p53-dependent and -independent mechanisms. Mol Cell Differ. 1996;4:47–65. [Google Scholar]
- 25.Gorospe M, Wang X, Guyton K, Holbrook N J. Protective role of p21Waf1/Cip1 against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol Cell Biol. 1996;16:6654–6660. doi: 10.1128/mcb.16.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi M, Holbrook N J. p21Waf1/Cip1 protects against p53-mediated apoptosis of human melanoma cells. Oncogene. 1997;14:929–935. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- 27.Gorospe M, Wang X, Holbrook N J. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartigan J A, Johnson G V. Transient increases in intracellular calcium result in prolonged site-selective increases in Tau phosphorylation through a glycogen synthase kinase 3 beta-dependent pathway. J Biol Chem. 1999;274:21395–21401. doi: 10.1074/jbc.274.30.21395. [DOI] [PubMed] [Google Scholar]
- 29.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph B, Orlian M, Furneaux H. p21waf1 mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 31.Keene J D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehlenbach R H, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 34.Kessis T D, Slebos R J, Nelson W D, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Human papilloma virus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King P H, Levine T D, Fremeau R T, Jr, Keene J D. Mammalian homologs of Drosophila ELAV localized to a neuronal subset can bind in vitro to the 3′ UTR of mRNA encoding the Id transcriptional repressor. J Neurosci. 1994;14:1943–1952. doi: 10.1523/JNEUROSCI.14-04-01943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein P S, Melton D A. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 40.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 41.Ma W J, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malter J S, Hong Y. A redox switch and phosphorylation are involved in the post-translational up-regulation of the adenosine-uridine binding factor by phorbol ester and ionophore. J Biol Chem. 1991;266:3167–3171. [PubMed] [Google Scholar]
- 43.Marusich H M, Furneaux H, Henion P, Weston J A. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 44.Michieli P, Chedid D, Lin D, Pierce J, Mercer W, Givol D. Inhibition of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 45.Mutoh H, Naya F J, Tsai M J, Leiter A B. The basic helix-loop-helix protein BETA2 interacts with p300 to coordinate differentiation of secretin-expressing enteroendocrine cells. Genes Dev. 1998;12:820–830. doi: 10.1101/gad.12.6.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 48.Nakielny S, Fischer U, Michael W M, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 49.Okano H J, Darnell R B. A hierarchy in Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poluha W, Poluha D K, Chang B, Crosbie N E, Schonhoff C M, Kilpatrick D L, Ross A H. The cyclin-dependent kinase inhibitor p21WAF1 is required for survival of differentiating neuroblastoma cells. Mol Cell Biol. 1996;16:1335–1341. doi: 10.1128/mcb.16.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinow S, Campos A R, Yao K M, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- 53.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 54.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 55.Shibasaki F, Price E R, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 56.Schwaller J, Koeffler H P, Niklaus G, Loetscher P, Nagel S, Fey M F, Tobler A. Posttranscriptional stabilization underlies p53-independent induction of p21WAF1/CIP1/SDI1 in differentiating human leukemic cells. J Clin Investig. 1995;95:973–979. doi: 10.1172/JCI117806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shyu A B, Greenberg M E, Belasco J G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 58.Strubing C, Chapman D E. Active nuclear import and export is independent of lumenal Ca2+ stores in intact mammalian cells. J Gen Physiol. 1999;113:239–248. doi: 10.1085/jgp.113.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szabo A, Dalmau J, Manley G, Rosenfeld M R, Wong E, Henson J, Posner J B, Furneaux H. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 60.Timchenko N A, Wilde M, Nakanishi M, Smith J R, Darlington G J. CCAAT/enhancer-binding protein α (C/EBP α) inhibits cell proliferation through the p21(WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 61.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 62.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;831:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 63.Wang W, Chi T, Xue Y, Kuo A, Zhou S, Crabtree G. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wellington C L, Greenberg M E, Belasco J G. The destabilizing elements in the coding region of c-fos mRNA are recognized as RNA. Mol Cell Biol. 1993;13:5034–5042. doi: 10.1128/mcb.13.8.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wodnar-Filipowicz A, Moroni C. Regulation of interleukin 3 mRNA expression in mast cells occurs at the posttranscriptional level and is mediated by calcium ions. Proc Natl Acad Sci USA. 1990;87:777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu N, Chen C-Y A, Shyu A-B. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]