ABSTRACT
Nutritional immunity involves cellular and physiological responses to invading pathogens, such as limiting iron, increasing exposure to bactericidal copper, and altering zinc to restrict the growth of pathogens. Here, we examine infection of bone marrow-derived macrophages from 129S6/SvEvTac mice by Salmonella enterica serovar Typhimurium. The 129S6/SvEvTac mice possess a functional Slc11a1 (Nramp-1), a phagosomal transporter of divalent cations that plays an important role in modulating metal availability to the pathogen. We carried out global RNA sequencing upon treatment with live or heat-killed Salmonella at 2 h and 18 h postinfection and observed widespread changes in metal transport, metal-dependent genes, and metal homeostasis genes, suggesting significant remodeling of iron, copper, and zinc availability by host cells. Changes in host cell gene expression suggest infection increases cytosolic zinc while simultaneously limiting zinc within the phagosome. Using a genetically encoded sensor, we demonstrate that cytosolic labile zinc increases 45-fold at 12 h postinfection. Further, manipulation of zinc in the medium alters bacterial clearance and replication, with zinc depletion inhibiting both processes. Comparing the transcriptomic changes to published data on infection of C57BL/6 macrophages revealed notable differences in metal regulation and the global immune response. Our results reveal that 129S6 macrophages represent a distinct model system compared to C57BL/6 macrophages. Further, our results indicate that manipulation of zinc at the host-pathogen interface is more nuanced than that of iron or copper. The 129S6 macrophages leverage intricate means of manipulating zinc availability and distribution to limit the pathogen’s access to zinc, while simultaneously ensuring sufficient zinc to support the immune response.
KEYWORDS: metal, zinc, macrophages, Salmonella, Slc11a1, RNA-seq, nutritional immunity, infection
INTRODUCTION
Pathogenic organisms must acquire essential micronutrients such as iron, zinc, and manganese to sustain growth and maintain pathogenicity. To protect itself, a host organism in turn attempts to sequester these nutrients, as unfettered access can allow for the exponential growth of an invasive microbe population (1). Metal ion sequestration occurs on a systemic level, evidenced by the sharp decrease of free metal ions in serum upon infection (2). Additionally, phagocytic immune cells manipulate the intracellular localization of metal ions to limit the viability of engulfed pathogens. Within macrophages, iron (Fe) ions are bound by cytosolic storage proteins and transported out of phagosomal compartments (3), while these same compartments are flooded with bactericidal copper (Cu) ions (4, 5). The role of zinc (Zn) in nutritional immunity in macrophages is more nuanced than that of Fe or Cu, as it can be used either to poison or starve a pathogen (1). Some microbes have developed strategies to counter these tactics, including siderophores to scavenge Fe, oxidases to detoxify Cu, exporters to expunge excess metals, and high-affinity importers to overcome metal limitation (1, 2, 6). There is a growing body of work suggesting that both the pathogen and the host have mechanisms to regulate Zn accessibility (1, 7); however, the overall picture of Zn regulation during infection is not well understood.
Zinc is an essential micronutrient required for growth, proliferation, and several fundamental biological processes. In mammals, Zn is a requisite cofactor for approximately 10% of the proteome (8), including many transcription factors and enzymes. As with other metal ions, excess Zn can be toxic, and cells tightly regulate its availability. Mammalian cells contain hundreds of micromolar total Zn, but most of this is bound to proteins, enzymes, and other ligands, such that labile Zn, as measured by fluorescent sensors, is typically in the hundreds of picomolar range in the cytosol (9–11). There are fewer tools for, and hence less consensus on, the concentration of labile Zn in organelles. However, multiple different fluorescent sensor platforms have suggested that labile Zn distribution in organelles is heterogeneous (11–15). Zn is regulated by membrane-specific Zn transporters, including Slc30a1-10 (also referred to as ZnT1-10), which transports zinc out of the cytosol, and Slc39a1-14 (also referred to as Zip1-14), which transports Zn [as well as Fe and manganese (Mn)] into the cytosol (16). Cells also contain metallothioneins (Mt), which serve to bind and buffer Zn, and a metal-dependent transcription factor (Mtf1) that translocates to the nucleus upon binding excess Zn to regulate the expression of genes that lower cytosolic Zn levels. One major role of Mtf1 is regulating the expression of Mt genes in response to increased cytosolic Zn (17). Consequently, increased Mt expression is often used as a proxy for increased cytosolic Zn.
How macrophages use Zn to fight infection seems to depend on the pathogen it encounters, as well as the nature of the host cell. Macrophages poison Mycobacterium tuberculosis, Salmonella pneumoniae, Escherichia coli, and Helicobacter pylori by transporting toxic levels of Zn into pathogen-containing phagosomes (18–21). Conversely, challenge with Histoplasma capsulatum induces macrophages to sequester Zn and increase ROS production to counter the infection (22). There does not appear to be consensus regarding Zn manipulation upon infection with Salmonella Typhimurium. Salmonella possess genes that aid in surviving Zn toxicity and starvation (23–26), and studies done in different types of macrophages have suggested that Zn can both facilitate and impair infection. In a study of macrophages derived from primary human monocytes and the human monocyte THP-1 cell line, infection induced Zn to accumulate in punctate vesicular compartments, though Salmonella managed to avoid these toxic Zn compartments via a SPI-1-dependent mechanism (27). In contrast, Salmonella infection of the mouse RAW264.7 macrophage cell line induced Zn mobilization, and increased Zn correlated with impaired bacterial clearance (26). On the other hand, multiple studies have revealed that the high-affinity ZnuABC zinc uptake system and associated accessory proteins are critical virulence factors in Salmonella, suggesting that Salmonella experience Zn starvation inside the host (23, 24, 28). Critical comparison of Zn at the host-pathogen interface may be confounded by differences in the macrophage models used across studies. For example, both BALB/c and C57BL/6 mice lack functional Slc11a1 (Nramp-1), a phagosomal transporter for magnesium (Mg), Fe, Mn, and Zn (29). Nramp-1 (natural resistance-associated macrophage protein) was initially recognized as instrumental in phagosomal maturation, and macrophages from mice lacking this protein are more susceptible to infection and have a reduced capacity to clear intracellular pathogens (30). RAW264.7 cells, a popular mouse macrophage cell line used to study intracellular pathogens, are derived from C57BL/6 mice and therefore equally impaired. Additionally, functional Slc11a1 has recently been identified as the key factor in metal ion deprivation of phagocytosed Salmonella (31), underscoring its importance in nutritional immunity and the relevance of characterizing a model system expressing this gene.
In this study, we carried out global RNA sequencing upon infection of macrophages from 129S6 mice with Salmonella Typhimurium and examined the changes in gene expression via clustering and gene set enrichment analysis (GSEA). We deliberately used the 129S6/SvEvTac (hereafter referred to as 129S6) mouse because it contains a functional Slc11a1 protein, has been used as a model system for chronic systemic infection (30), and the transcriptional changes upon infection have not been reported. We examined changes in metal-regulatory and metal-dependent genes and compared our results to published data on infection of C57BL/6 mice. While some changes in metal-regulatory genes were similar between the two different model systems, suggesting common mechanisms of altering metals at the host-pathogen interface, there were also notable differences, indicating that macrophages from 129S6 represent a distinct model system with an altered nutritional immunity landscape within the host. We also found that cytosolic Zn levels increased over the course of infection and that Zn availability alters infection outcome. Specifically, we found that Zn supports increased replication of intracellular bacteria, but also facilitates clearance of bacteria from macrophages. Our results indicate that changes in gene expression, including the immune response, metal regulation in response to infection, and infection outcomes are significantly different in macrophages from different mouse models.
RESULTS
Infection induces widespread changes in gene expression.
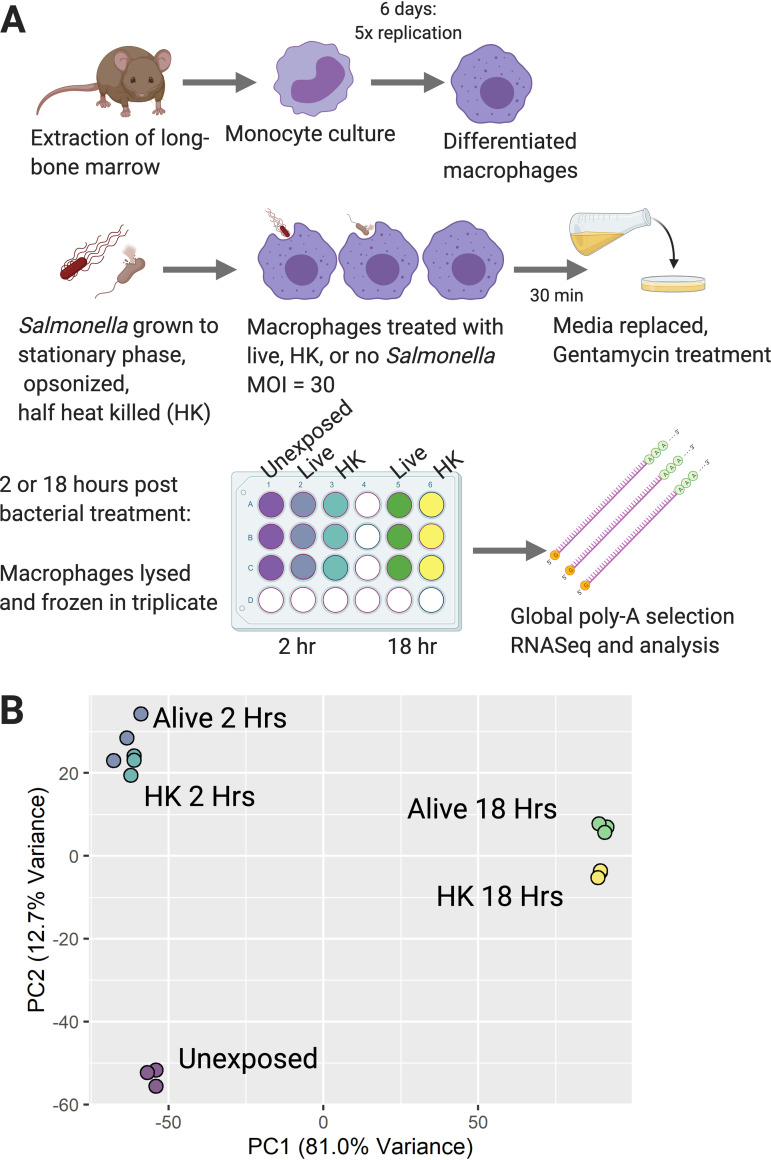
To identify changes in 129S6 macrophage gene expression in response to infection, macrophages were treated with live or heat-killed Salmonella enterica serovar Typhimurium (hereafter referred to as Salmonella Typhimurium). To examine both early and late immune responses, macrophages were lysed at 2 or 18 h post treatment and subjected to global RNA sequencing (Fig. 1A). Macrophages that were subjected to medium changes but not exposed to bacteria were used as a control. Principal-component analysis (PCA) shows that samples clustered predominantly by the time postinfection and status of bacteria (alive versus heat killed, Fig. 1B).
FIG 1.
Salmonella treatment of macrophages induces global RNA expression changes across two primary axes: time post treatment and Salmonella status (alive versus heat killed). (A) Schematic of infection experimental design for 129S6 BMDMs exposed to either live Salmonella Typhimurium SL1344 (live), heat killed Salmonella Typhimurium SL1344 (HK), or no bacteria (unexposed). (B) Global principal-component analysis (PCA) with the x axis showing differential expression correlated with time post treatment and the y axis showing differential expression correlated with Salmonella status (live, heat killed [HK] or unexposed [UE]). Figure created with BioRender.com.
We identified 7,766 genes that were differentially expressed and performed hierarchical clustering to examine the patterns of gene expression changes. We limited analysis to genes with significant differential expression (adjusted P value [padj] < 0.01), as determined by a DESeq2 log ratio test. An expression level cutoff was also applied (see Materials and Methods). An unsupervised algorithm (degPatterns function in DEGreport R package) was used to cluster genes based on similar expression profiles across all conditions. Twenty groups, each with at least 15 genes, emerged. Some groups showed time-dependent expression, with little change between bacterial conditions, while others showed significant differences in expression upon treatment with alive versus heat-killed bacteria. Additionally, some groups were different based primarily on expression levels in control cells.
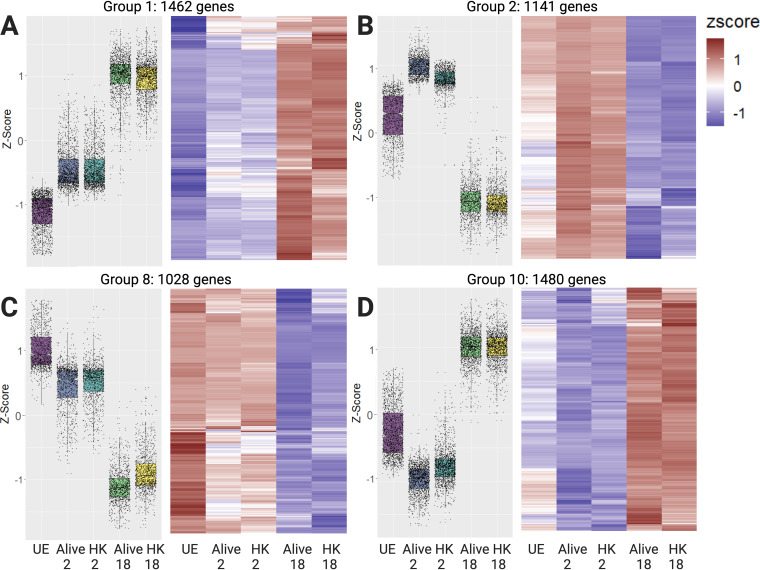
In Fig. 2, we highlight the four different expression patterns that encompass the largest gene groups. The remaining groups are presented in Fig. S1, S2, S3, and S4 in the supplemental material and the genes associated with each group are presented in Data Set S1. We used DAVID Bioinformatics Resources 6.8 to annotate molecular functions and biological processes associated with the different gene expression clusters. Figure 2A presents group 1, which consists of genes with time-dependent increases in expression. These genes respond to the presence of Salmonella Typhimurium regardless of its live or heat-killed (HK) status, and are enriched for mitochondrial function (e.g., oxidative phosphorylation or ATP production), ER-Golgi transport and protein folding, immune response, sugar metabolism, and protein degradation (including proteasome and unfolded protein response genes). Specific genes that are characteristic of the M2 anti-inflammatory immune response (32), such as il4rα, arg1, timp1, and fcgr2b, are present in this group, along with master regulators of the lipopolysaccharide (LPS) immune response, ifit1, stat2, and irf7. Genes involved in pH and redox homeostasis, such as sod1, nos2, and hif1α, carbonic anhydrases car2/4/13, as well as metal-regulatory slc11a1, iron-sequestering lcn2, and ferritin iron storage fth1, are found in this group. Finally, s100a8, a metal-binding protein that is a component of calprotectin and plays a prominent role in nutritional immunity, exhibited a similar expression pattern, with low expression at 2 h and a significant increase at 18 h (Fig. S1D). These results indicate that the M2 anti-inflammatory response, along with many metal-associated genes, are upregulated over time in response to infection, regardless of whether the bacteria are alive or HK.
FIG 2.
Clustered data expose multiple distinct expression trends in global data. Four of the 20 groups that emerged from clustering analysis are presented as a box and whisker plot of gene expression levels as Z-scores (normalized across all DE genes) and a hierarchically clustered heat map of gene expression as rlog-scaled DESeq2 normalized counts. (A) Group 1 contains 1,462 genes that increase in expression at 2 h and increase further at 18 h, without significant differences between the alive and HK conditions. (B) Group 2 contains 1,141 genes that increase in expression at 2 h and then decrease below baseline at 18 h. (C) Group 8 contains 1,028 genes that are expressed in unexposed cells, but then decrease as a function of time postinfection. (D) Group 10 contains 1,480 genes that decrease in expression at 2 h and increase at 18 h. UE, unexposed control cells; HK, stimulation with heat-killed bacteria. Figure created with BioRender.com.
The genes in group 2 (Fig. 2B) are upregulated at 2 h, with higher expression in alive compared to HK conditions, and are subsequently suppressed below baseline at 18 h. This group is enriched in proinflammatory M1 immune genes (32), including tnf, the inflammasome component nlrp3, and irf1, which is responsible for activating cytokine production. This group also contains multiple genes involved in membrane transport of zinc (slc30a4 and slc39a6/a8/a10). Another hallmark proinflammatory M1 marker, tlr2, and metal-regulatory genes (the zinc-dependent transcription factor mtf1 and iron storage ferritin light chain ftl1) are present in group 3 (Fig. S1A), which is similar to group 2 across time but differs in alive versus HK expression at 2 h. Combined, these two groups indicate that in macrophages with functional slc11a1, much of the proinflammatory response is activated at early time points and then suppressed later in infection.
Figure 2C (group 8) shows genes that are distinctly downregulated upon infection over time. This group is notable for its relatively small number of Gene Ontology (GO) annotations, which are focused on cell cycle, cell motility, zinc binding and transport, and transcription. Finally, Fig. 2D demonstrates group 10, in which genes are suppressed at 2 h postinfection and upregulated above untreated cells at 18 h. This group is enriched for metabolism, protein export, and transcription. Specifically, these genes are involved in ER and Golgi trafficking, respiration, lysosomal activity, zinc transport (slc39a3/a7), iron-dependent functions (cytochrome bs, glutaredoxin2, NADH dehydrogenase), mRNA and phospholipid transport, and autophagy. Approximately 10% of these genes are involved in transcription, and half of these transcription factors bind zinc. Also in this cluster are key inflammation-related genes, including tlr4, which binds lipopolysaccharide (LPS), and irf2/3/10, which mediate the type I interferon (IFN) response. Combined, these gene expression patterns reveal remodeling of metal homeostasis, metabolism, and transcription during infection.
GSEA reveals pathways differentially affected by live versus heat-killed bacteria.
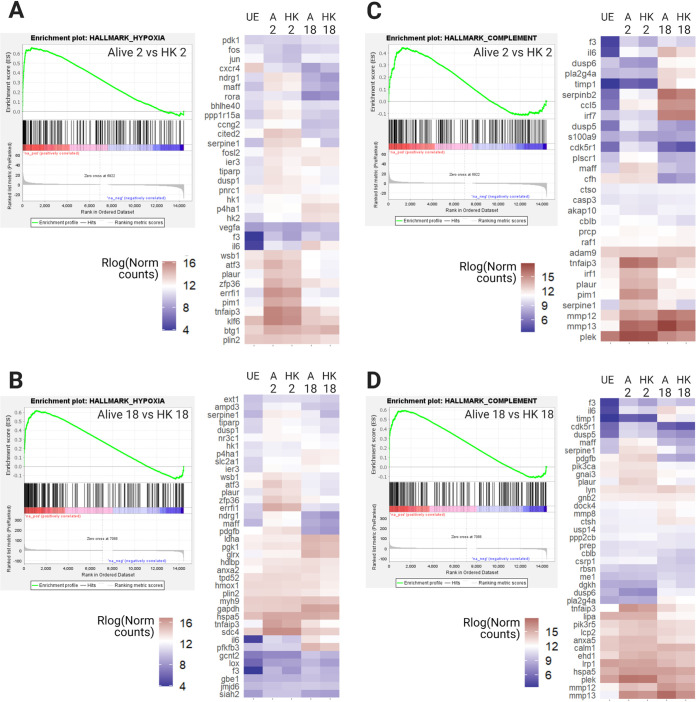
In analyzing global transcription patterns, we found that some groups contain differences in gene expression between live and heat-killed bacterial treatments (see groups 4, 5, 7, and 16 as examples), revealing genes that respond differently to a generalized LPS trigger (HK bacteria) versus Salmonella Typhimurium with active virulence mechanisms. To identify biological processes or signaling pathways underlying these differences, GSEA (33) was performed on the differentially expressed ranked gene list from live versus HK at 2 h, and live versus HK at 18 h. When using the hallmark gene set from the Molecular Signatures Database (34), we found 47 gene sets enriched at 2 h and 14 gene sets enriched at 18 h for live versus HK treatment (false discovery rate of q < 0.05, Data set S2). One gene set that was significantly enriched in live versus HK exposure at both time points was hypoxia (Fig. 3A and B). Hypoxia is an important component of the in vivo immune response to infection, as hypoxia accompanies inflammation and a hypoxic environment may affect macrophage M1/M2 polarization (32, 35). Hypoxia is also accompanied by decreased glucose, increased lactate, and decreased pH and, as discussed above, we observed an increase in hif1α, carbonic anhydrases, and nos2. Multiple studies have shown activation of hypoxia-induced factors upon infection, and hypoxia is generally associated with macrophage stress as well as defense against bacterial replication (35–37).
FIG 3.
GSEA reveals enrichment of the hypoxia and complement hallmark gene sets in BMDMs infected with live Salmonella compared to treatment with heat-killed bacteria. Each panel shows a GSEA enrichment plot for a hallmark gene set and a hierarchically clustered expression heat map of the leading edge genes from that GSEA plot. In GSEA plots, each gene is represented by a vertical black line and leading edge genes are on the positive end of the spectrum, up to the point of maximum enrichment score. (A) GSEA and heat map of leading edge genes for the hallmark hypoxia gene set enriched in the 2 h alive condition compared to 2 h HK (q = 0.000, NES = 2.12). (B) GSEA and heat map of leading edge genes for the hallmark hypoxia gene set enriched in the 18 h alive condition compared to 18 h HK. At 18 h, analysis of the hypoxia gene set shows 50 leading edge genes versus 36 at 2 h. Only 20 of these genes are in the leading edge at both time points (q = 0.024, NES = 1.65). (C) GSEA and heat map of leading edge genes for the hallmark complement gene set enriched in the 2 h alive condition versus 2 h HK (q = 0.062, NES = 1.44). (D) GSEA and heat map of leading edge genes for the hallmark complement gene set enriched in the 18 h alive condition versus 18 h HK (q = 0.042, NES = 1.57). A total of 42 complement genes appear in the leading edge, compared to 36 at 2 h. While there is a core set of 16 genes upregulated at both time points, the remaining genes are unique. UE, unexposed control; HK, heat killed; q, false discovery rate; NES, normalized enrichment score from GSEA. Figure created with BioRender.com.
The complement gene set is activated more strongly in macrophages infected with live Salmonella Typhimurium. The complement system is part of the innate immune response and mediates host responses such as opsonization, inflammation, and direct bacterial lysis (38). Complement systems respond to bacterial coat components (39) and, therefore, we were surprised that many of these genes showed upregulation in live versus HK conditions (Fig. 3C and D). At 2 h postinfection, the complement leading edge is enriched for zymogens, enzymes, and secreted proteins, while at 18 h there are more chemokine- and Fc-signaling related genes. At both time points, live conditions show upregulation of matrix metalloproteinases mmp12 and mmp13 and their inhibitor timp1. Additionally, this gene set includes members of the coagulation pathway (plaur and serpin, f3), which overlaps and interacts with the complement system to trap and kill microbes.
Tumor necrosis factor α (TNF-α) signaling via NF-κB is also more strongly activated by live Salmonella Typhimurium. During infection, multiple signaling cascades activate the transcription factor NF-κB, including the proinflammatory cytokine TNF-α (TNF in mice). NF-κB responds to TNF activation with proinflammatory and antiapoptotic signaling. At both 2 h and 18 h, live infected samples were enriched for proinflammatory genes, while each time point was enriched in a different set of antiapoptotic genes (Fig. S5). At 2 h, btg2, egr3, jun, vegfa, klf4, and nr4a were enriched in alive conditions compared to HK, while at 18 h bcl6, smad3, stat5a, plaur, and socs3 were upregulated in alive versus HK. Most genes present in the leading edge for both time points were more highly expressed at 2 h, reinforcing the trend of macrophages moving toward an M2-like state later in infection.
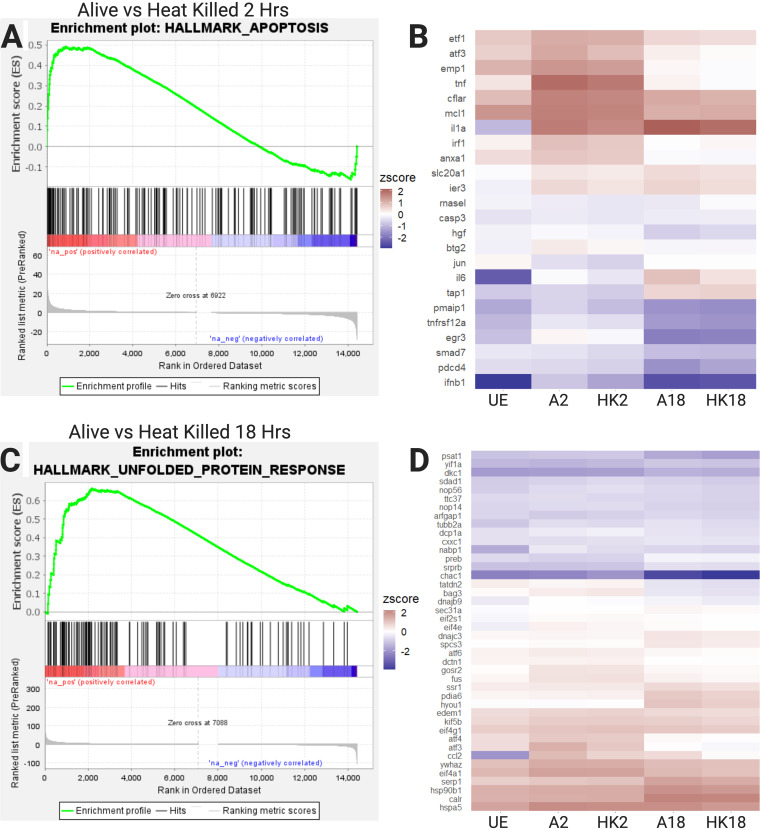
We also observed gene sets that were enriched only at 2 h or 18 h. For example, the apoptosis gene set was enriched at 2 h, and the unfolded protein response (UPR) set was enriched at 18 h (Fig. 4). Late-log-phase growth induces maximal expression of Salmonella Typhimurium pathogenicity island 1 (SPI1) genes, which allow Salmonella Typhimurium to be highly invasive and often induce apoptosis in macrophages (40). Several proapoptosis genes are upregulated by infection with live bacteria at 2 h, including the cAMP-dependent transcription factors atf3, jun, and pmaip1. The fact that the apoptotic pathway is not enriched at 18 h suggests that the macrophages manage this response. While UPR and apoptosis are both possible responses to metabolic and ER stress, the upregulation of UPR at 18 h could represent an attempt to restore cellular homeostasis. The transition from apoptosis genes at 2 h to UPR genes at 18 h correlates with a shift in macrophage polarization and may imply decreased ER stress over time.
FIG 4.
GSEA reveals enrichment of the apoptosis hallmark gene set at 2 h and the UPR gene set at 18 hin BMDMs treated with live versus heat-killed Salmonella. (A) GSEA curve of the hallmark apoptosis gene set, showing enrichment in the 2 h alive condition versus the 2 h HK condition (q = 0.023, NES = 1.58). (B) Heatmap of 25 leading edge apoptosis genes. (C) GSEA curve of the hallmark UPR gene set, showing enrichment in the 18 h alive condition versus the 18 h HK condition (q = 0.021, NES = 1.69). (D) Heatmap of the 47 leading edge UPR genes. UE, unexposed control; HK, heat killed; q, false discovery rate; NES, normalized enrichment score from GSEA. Figure created with BioRender.com.
Significant remodeling of metal homeostasis in response to infection.
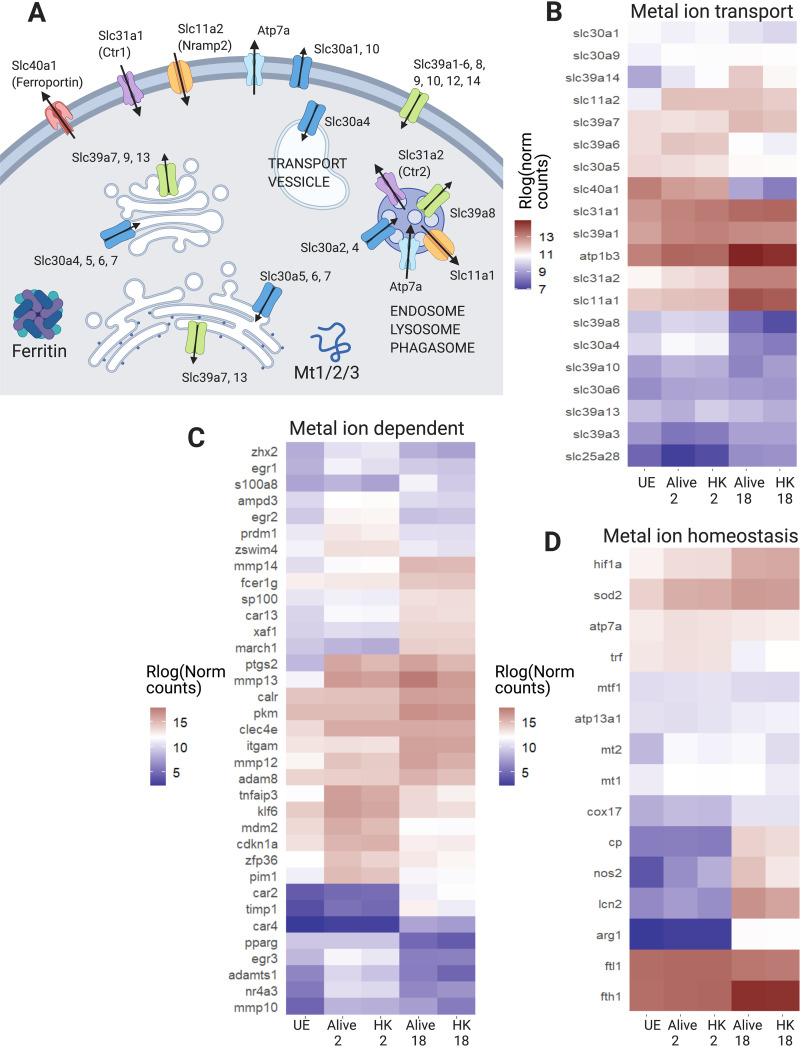
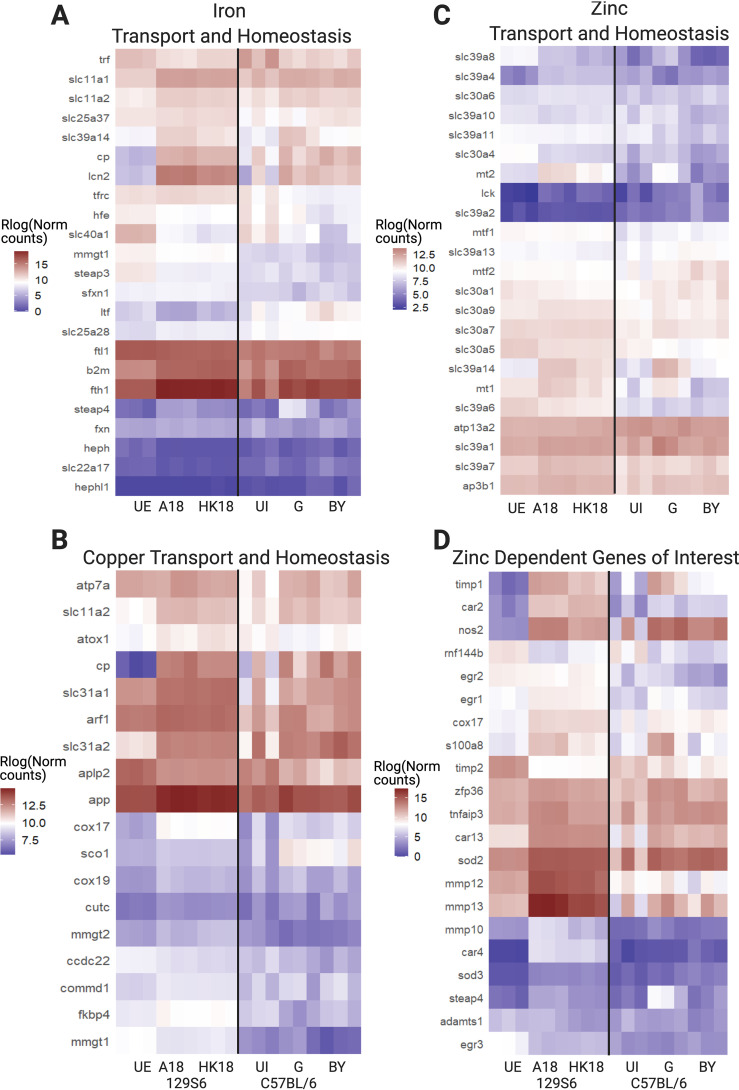
One of our primary goals in carrying out transcriptome sequencing (RNA-seq) upon infection of 129S6 bone marrow-derived macrophages (BMDMs) was to define infection-related changes in transition metal-regulatory and metal-dependent genes in a model system with functional Slc11a1 (Nramp-1). This protein plays a critical role in regulating metal homeostasis and its expression allows us to inform our understanding of nutritional immune responses within 129S6 cells (29, 30). Figure 5A shows several major metal-regulatory proteins in macrophages. This includes Slc39a1-14 (Zip) transporters that facilitate entry of Zn, as well as Mn and Fe for some Slc39a members, into the cytosol, Slc30a1-10 (Znt) transporters that remove Zn from the cytosol, the Fe exporter ferroportin (Slc40a1), and copper (Cu) transporters Atp7a and Ctr1/2 (Slc31a1/2). Also shown are metallothioneins that store and buffer cytosolic Zn, and the Fe storage protein ferritin (Fth1 [ferritin heavy chain] and Ftl1[ferritin light chain]), which is protective against Fe-induced oxidative stress in macrophages.
FIG 5.
There are widespread changes in metal transport, metal-dependent, and metal homeostasis genes. (A) Schematic of major metal ion transporters and buffering proteins in macrophages. Slc30a1 to 10 are also referred to as ZnT1 to 10. Slc39a1 to 14 are also referred to as Zip1 to 14. (B to D) Heatmaps of subsets of metal transport, metal-dependent, and metal homeostasis genes that are highly expressed and differentially expressed in our data set (log ratio test q < 0.01, mean normalized counts > 100). These gene sets are curated to focus on nutritional immunity metals, including Fe, Zn, Cu, and Mn. Genes that fit in multiple categories are only listed once. (B) Metal ion transporters. (C) Metal-dependent and metal-responsive genes. (D) Metal ion homeostasis genes. UE, unexposed control cells; HK, heat killed bacteria exposure. Figure created with BioRender.com.
Infection of 129S6 BMDMs leads to broad changes in genes that control metal transport (Fig. 5B, Data Set S3). There is a significant increase in slc11a2 at 2 h and slc11a1 at 18 h in live versus HK Salmonella Typhimurium. Slc11a1 localizes to the phagosomal membrane, where it exports divalent metal cations (Fe, Zn, and Mn) while simultaneously acidifying the compartment. There are also significant changes in transporters dedicated to import, export, and distribution of Fe, Cu, and Zn (Fig. 5B). There is a downregulation of the transferrin (trf) Fe import pathway and the Fe exporter slc40a1. The Cu transporters slc31a1 (plasma membrane) and slc31a2 (endosome) are significantly increased in expression at 18 h, while endosomal atp7a is upregulated at 2 h. Changes in Zn transporters include upregulation of slc30a4, slc39a14, slc39a1, and slc39a7 and downregulation of slc30a5. The increase in slc39a14 is especially strong at 18 h. Decreased expression of Golgi importer slc30a5 and increased expression of ER exporter slc39a7 suggests diminished Zn in the secretory pathway. Slc30a4, which transports Zn into the phagosome and secretory vesicles, is upregulated at 2 h and downregulated below control at 18 h. Combined, there are widespread changes in metal transport that indicate limitation of Fe, accumulation of Cu, and redistribution of Zn.
In addition to metal transport, there are significant changes in genes that are metal dependent (Fig. 5C) and genes related to metal homeostasis (Fig. 5D). There is a change in ferritin genes that suggests increased Fe storage capacity; fth1 increases while ftl1 drops in expression, reflecting the fact that, during infection, heavy chains polymerize together to the exclusion of light chains (41). It should also be noted that ferritin chains are further regulated at the translational level (42). Similarly, Zn buffering capacity is increased due to the increase in metallothioneins (mt1/2). There is also a significant increase in expression of Zn-dependent carbonic anhydrases that regulate pH, Zn-dependent proteases (mmp10, 12, 13, 14, timp1, adamts1, and adam8), and the ferroxidase ceruloplasmin (Fig. 5C). Additionally, there is an increase in expression of lipocalin, a protein involved in Fe homeostasis, inflammation, and hypoxic response to ischemia-reperfusion (43). Finally, there are significant increases in genes involved in oxidative stress, particularly at late stages of infection, including hif1a, Cu/Zn-dependent sod2 and sod3, Mn-dependent arg1, and Zn-dependent nos2. These changes indicate that remodeling metal transport is accompanied by significant changes in the expression of metal-dependent enzymes and metal storage.
Cytosolic labile Zn increases at late stages of infection.
The widespread changes in Zn transporter expression upon infection suggest remodeling of Zn homeostasis, and would be expected to lead to an increase in cytosolic Zn and decreased Zn in the secretory pathway. Previous studies in different model systems have suggested different changes in intracellular labile Zn pools in response to infection (27, 44). However, all previous studies have used small molecule indicators such as FluoZin3 or Zinpyr-1, which often exhibit variable and unpredictable localization within a cell and can confound measurement of Zn in a defined location. For example, FluoZin3-AM has been shown to localize to the cytosol, Golgi, lysosomes, and vesicles, and localizations differ among cell types (10, 45, 46). Previous studies in Raw264.7 cells used FluoZin3 to determine that there was an increase in labile Zn upon infection via fluorescence-activated cell sorting (FACS). Unfortunately, the absence of fluorescence images precluded evaluation of where the Zn increase occurred. In human monocyte-derived macrophages, FluoZin3 localizes to vesicles or early endosomes (27) and a study in these cells indicated that vesicular Zn increases upon infection. Of the studies that have attempted to measure Zn inside activated or infected macrophages, none have unambiguously addressed whether there are changes in cytosolic Zn.
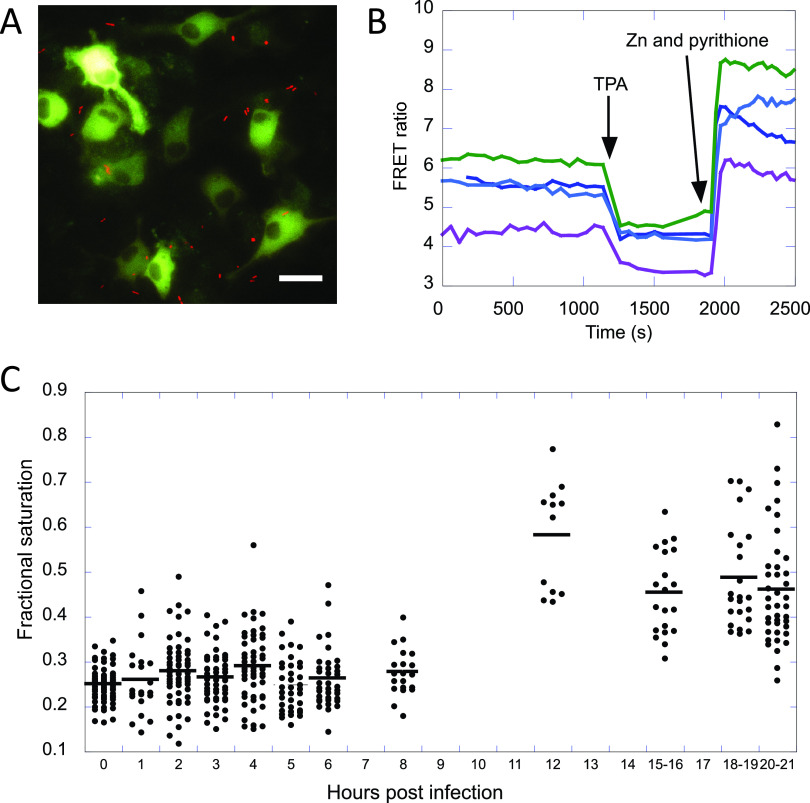
To directly measure cytosolic Zn in 129S6 BMDMs at various time points postinfection, we used the genetically encoded zinc sensor NES-ZapCV2 (47, 48). Cells were subsequently subjected to an in situ calibration to determine the minimum and maximum Förster resonance energy transfer (FRET) ratio in each cell (Fig. 6A and B) and the calibration parameters were used to calculate the fractional saturation of each cell under resting conditions and the concentration of labile cytosolic Zn (see Materials and Methods) (Fig. 6C). Uninfected cells have a mean fractional saturation of 0.30, which corresponds to a Zn concentration of 490 pM. At 12 h and beyond, the fractional saturation increased to ∼ 0.51, which corresponds to ∼ 22 nM Zn. Our data show that labile Zn in the cytosol does indeed increase later in the infection (45-fold), consistent with the observed increase in expression of mt1 and mt2.
FIG 6.
Changes in labile cytosolic Zn in 129S6 BMDMs upon infection with Salmonella. (A) Representative image of BMDMs expressing the NES-ZapCV2 fluorescent Zn sensor (green) treated with Salmonella constitutively expressing mCherry (red). Scale bar = 20 μm. (B) Representative in situ calibration of the NES-ZapCV2 sensor showing the FRET ratio (R, background-corrected FRET channel divided by the background-corrected CFP channel) over time. The Rmin is obtained upon addition of 50 μM TPA and the Rmax is obtained upon addition of 23.8 nM buffered Zn, 0.001% saponin, and 0.75 μM pyrithione. Each trace represents a single cell. (C) Fractional saturation of the NES-ZapCV2 sensor at different time points postinfection. Each dot represents an individual cell. Data represent at least two independent experiments per time point.
Zn promotes bacterial clearance as well as bacterial replication.
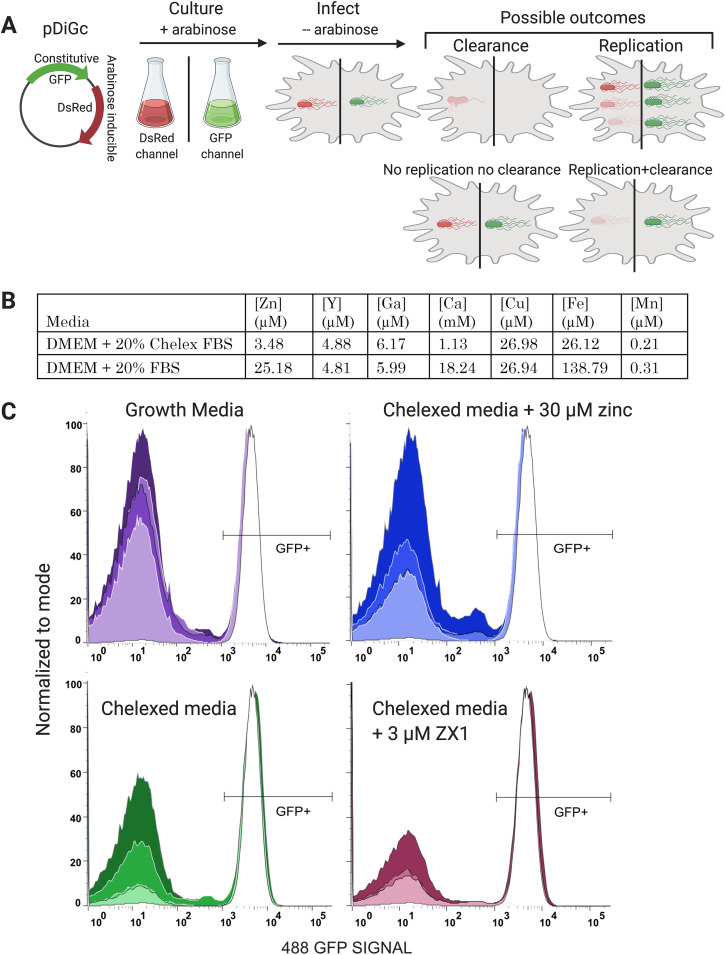
Having established that Zn regulation and Zn availability in the cytosol are altered upon infection, we evaluated the influence of Zn availability on infection outcome. While a previous study examined increases and decreases in Zn on Salmonella Typhimurium infection of Raw264.7 cells (26), we explored the impact of Zn on infection outcome in macrophages with functional Slc11a1. To examine infection outcomes, we incorporated the pDiGc plasmid (Fig. 7A) into Salmonella Typhimurium. On this plasmid, DsRed is expressed under an arabinose-inducible promoter, while green fluorescent protein (GFP) is constitutively expressed (49), allowing infected cells to be identified by GFP fluorescence and bacterial replication to be tracked by dilution of DsRed signal upon cell division. Comparison of green and red signals across time yields a picture of bacterial division versus macrophage clearance of bacteria.
FIG 7.
Bacterial clearance as a function of time and Zn availability. (A) Schematic of the experimental protocol. Salmonella expressing the pDiGc plasmid expresses GFP constitutively and DSRed under arabinose induction. Bacteria are cultured with arabinose and aeration to the stationary phase, at which point they express both fluorescent proteins. Arabinose is removed at infection, and infected macrophages are fluorescent in both red and green channels by flow cytometry. As infection proceeds, the green to red fluorescence ratio “tracks” the bacteria. (B) Metal content of normal growth medium and medium containing Chelex-treated FBS, as determined by inductively coupled plasma mass spectrometry (ICP-MS). (C) Histograms of the GFP signal in macrophages expressing pDiGc in 4 different media and at 4 time points. Each experimental condition was done in triplicate, and 10,000 cells were measured per sample. Replicates were essentially identical, so data for the replicates were concatenated. Increasing color saturation indicates time post Salmonella exposure (2, 10, 18, or 24 h). The right peak in each graph corresponds to the GFP+ macrophages, indicating infection. As Salmonella are cleared, the macrophages become GFP− and shift to the left peak on each graph. At the initial inoculation (white curve in each panel), macrophages show almost 100% infection rate. Normal macrophage growth medium and Chelexed medium supplemented with 30 μM Zn (replete-Zn conditions) show almost 100% bacterial clearance at 24 h. Chelexed medium (low Zn) shows decreased bacterial clearance, while Chelexed medium with 3 μM extracellular zinc chelator ZX1, (essentially no Zn) shows poor bacterial clearance. Figure created with BioRender.com.
To alter Zn availability, we manipulated Zn levels in the macrophage medium after Salmonella Typhimurium had been internalized. Four media conditions were used in this study to span the range from Zn replete to Zn deficient. The four conditions were (i) standard macrophage growth medium (Dulbecco’s modified Eagle medium [DMEM] + 20% fetal bovine serum [FBS]) containing 25 μM Zn (Fig. 7B); (ii) medium prepared with Chelex-100-treated FBS to deplete metal ions (4 μM Zn, Fig. 7B); (iii) medium further depleted of Zn (Chelex medium + 3 μM ZX1, ∼ 0 μM Zn); or (iv) medium reconstituted with replete Zn (Chelex medium + 30 μM Zn, 34 μM Zn total). The Zn-replete medium has slightly higher Zn than standard macrophage growth medium, but is in line with mammalian culture media, which have Zn levels ranging from 1 to 40 μM (50).
Macrophages were infected with Salmonella Typhimurium expressing pDiGc in macrophage growth medium and infection proceeded for 45 min before experimental media conditions were introduced. This avoided the confounding variable of Zn’s effect on the phagocytosis of bacteria. A control sample fixed immediately after the 45 min infection revealed that 91% of macrophages contained GFP-positive bacteria at the beginning of the experiment. Figure 7C demonstrates the loss of GFP signal in macrophages in each growth medium as a function of time. The increase in the fraction of cells with no GFP is indicative of bacterial clearance. In each panel, the white curve is from the initial infection and increased color saturation indicates time postinfection. In normal growth medium, clearance of bacteria is strong at 2 h, with 70% of initially infected cells having lost all GFP signal. Clearance further increases over time, with 76%, 74%, and 85% clearance at 10, 18, and 24 h, respectively. Control and Zn-replete media contained similar Zn levels but different amounts of calcium (Ca) and Fe due to Chelex treatment. At early time points (2, 10, and 18 h), there is 56%, 64%, and 59% bacterial clearance, respectively, in the Zn-replete media compared to the control, likely due to either depleted Ca or Fe. By 24 h, Zn-replete samples reached a similar level of infection clearance, with 87% clearance. In contrast, macrophages in Chelexed medium with low Zn and ZX1-treated medium with effectively no Zn showed significantly impaired bacterial clearance. At 24 h, Chelex and Zx1 conditions had achieved 73% and 53% clearance, respectively. Direct comparison of the different media conditions at each time point are presented in Fig. S6. These results confirm that 129S6 macrophages are competent at clearing Salmonella Typhimurium throughout infection and that Zn availability positively correlates with the effectiveness of bacterial clearance.
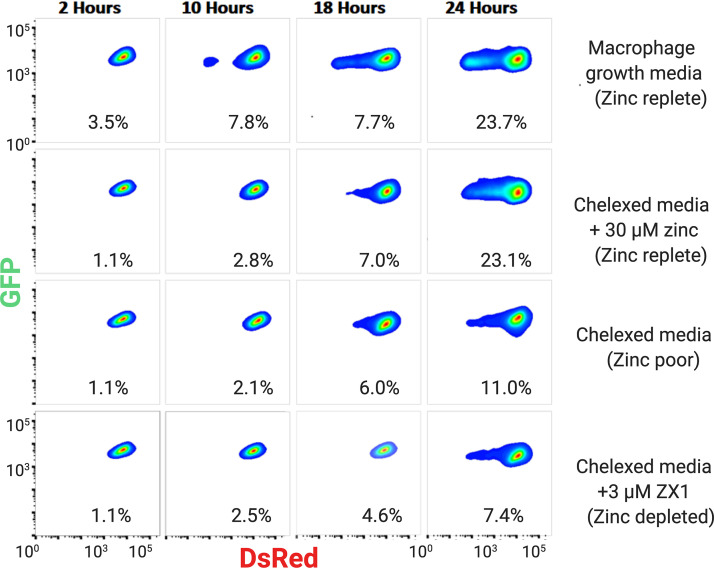
In addition to measuring bacterial clearance, the pDiGc plasmid can also be used to measure bacterial replication, as the DsRed signal is diluted with every cell division. We compared the DsRed signal versus GFP signal as a function of time and different media conditions (Fig. 8). In standard macrophage growth medium (top row), the DsRed signal decreases over time in 23% of the GFP+ cells, indicative of bacterial replication. It is important to note that although 129S6 macrophage are efficient at clearing the Salmonella Typhimurium infection, here we are looking at the small portion of the macrophage population that remains infected (GFP+). The Zn-replete medium (2nd row) shows bacterial replication comparable to standard growth medium, with 23% of GFP+ macrophages showing diluted DSRed signal at 24 h. In contrast, in Chelexed medium with depleted Zn and Chelexed medium containing ZX1 (no Zn), there is weaker bacterial replication (11% and 7%, respectively), as indicated by the lack of a fluorescent “tail” at lower DsRed intensity. Interestingly, this result suggests that while Zn facilitates host clearance of bacteria, it also helps bacteria to replicate in cells that do not clear the infection. This may suggest that bacterial clearance occurs more easily in dividing bacteria, as persistent infections are mainly nonreplicating.
FIG 8.
Bacterial replication as a function of time and Zn availability. The 129S6 BMDMs were infected with Salmonella Typhimurium expressing pDiGc. The GFP and DsRed signals in BMDMs were measured by flow cytometry and plotted against one another. These density graphs show only macrophages containing GFP+ Salmonella Typhimurium, which indicates active infection. Percentages on each plot indicate the number of macrophages with diluted DSRed signal, an indicator of Salmonella Typhimurium division. Dilution of DsRed is a function of time and zinc availability, with all media conditions showing an increase in bacterial division over time, but control and zinc-replete conditions (first and second rows) show much more robust division. Figure created with BioRender.com.
Macrophages from 129S6 and C57BL/6 strains differ in nutritional immune response to infection.
Because the Slc11a1 transporter specifically aids in depriving pathogens of metal ions (31), we hypothesized that 129S6 macrophages and macrophages without functional Slc11a1 would handle metals differently during Salmonella Typhimurium infection, and that this would be reflected in differential gene expression. To assess this, we searched the literature for an RNA-seq data set with conditions comparable to our study. We performed a comprehensive analysis of changes in expression in 129S6 macrophages (this study) and macrophages derived from C57BL/6 mice (RNA-seq study by Stapels et al. [51]) at 18 h postinfection. Specifically, we compared our unexposed control (UE), 18 h live bacteria, and 18 h HK conditions against uninfected macrophages (UI), those containing growing Salmonella Typhimurium (G), and bystander macrophages not containing Salmonella Typhimurium at 18 h (BY) from Stapels et al. All data were analyzed on the same pipeline and normalized together within DESeq2 to make the data as comparable as possible (Data set S4).
To evaluate potential differences in genes involved in nutritional immunity between the two data sets, we examined all genes with UNIPROT keywords for iron, zinc, or copper transport or homeostasis. Figure 9 presents heat maps for Fe, Cu, and Zn transport and Zn-dependent genes. There are a number of similarities in regulation of metal transport, metal homeostasis, and metal-dependent genes, indicating that there are common mechanisms for managing metal ions at the host-pathogen interface, even when there are differences in the host. In particular, there is a significant decrease in Fe import (trf), export (slc40a1), and an increase in ferritin storage (fth1) in both studies (Fig. 9A). Similarly, both studies show upregulation of the copper importer slc31a1 (Fig. 9B). With respect to Zn regulation, both studies show upregulation of the Zn importer slc39a14, downregulation of the Golgi importer slc30a5, and an increase in expression of Zn buffers mt1 and mt2 (Fig. 9B and C). Combined, these results suggest a general pattern of limiting the availability of Fe, increasing exposure to Cu, and increasing cytosolic Zn.
FIG 9.
Changes in metal transport and homeostasis genes reveal differences in metal regulation in BMDMs derived from 129S6 mice compared to C57BL/6 mice. RNA-seq data from this study are compared to data from Stapels et al., GEO Accession GSE104785. Data from both studies were processed through the same pipeline and normalized together. Gene lists were compiled using Uniprot keywords and the mouse genome, and filtered for mean normalized count > 5 in at least one data set. (A) Genes involved in Fe transport and homeostasis. (B) Genes involved in Cu transport and homeostasis. (C) Genes involved in Zn transport and homeostasis. UE, 129S6 BMDMs unexposed to Salmonella; HK, heat-killed Salmonella treatment; UI, uninfected C57BL/6 BMDMs; G, C57BL/6 BMDMs containing growing Salmonella; BY, bystander C57BL/6 BMDMs. Figure created with BioRender.com.
Despite these similarities, there are also notable differences that suggest changes in metal regulation at the host-pathogen interface. With respect to Fe, lactoferrin (ltf), a major Fe-binding factor with bactericidal properties, steap3, which encodes a ferrireductase that reduces ferric Fe released from transferrin in the endosome, and mmgt1, which encodes a Mg transporter that ferries multiple metal ions across membranes, are all downregulated in infected 129S6 cells but do not respond to infection in C57BL/6 cells. With respect to Cu, the Cu importer slc31a2 is upregulated in 129S6, while there is no change in C57BL/6. The copper chaperone atox1 is upregulated in 129S6, but downregulated in C57BL/6; cox17 is upregulated in 129S6, but not C57BL/6 (q > 0.06). Finally, sco1 is upregulated in C57BL/6 but not 129S6. There are also differences in the magnitude of gene expression changes, with more marked changes in 129S6, including for ferroportin (Slc40a1), multicopper enzyme ceruloplasmin (cp), and lcn2, which reduces bacterial replication by sequestering Fe bound to microbial siderophores. Lcn2 is also intimately involved in regulating the expression of proapoptotic bcl2l11, which is strongly downregulated in 129S6 but remains unaffected in C57BL/6 cells, suggesting these two cell types have disparate apoptotic tendencies at 18 h postinfection.
There are also notable differences in infection-induced expression changes in genes encoding Zn transporters and Zn-dependent proteins between the two systems (Fig. 9C and D). Specifically, upregulation of slc39a7 and downregulation of slc30a4 in 129S6 but not C57BL/6 cells suggest limitation of Zn in the secretory pathway and phago-lysosomal compartment in response to infection in 129S6 BMDMs. Additionally, slc39a8 is downregulated in 129S6 but upregulated in C57BL/6. In primary human lung macrophages, the Zn importer slc39a8 is upregulated in response to LPS and negatively regulates proinflammatory responses (52). Zn-dependent proteins, such as proteases (mmp9/10/12/13) and carbonic anhydrases (car 4/11), are upregulated in 129S6 but not C57BL/6. Finally, there are differences in the magnitude of gene expression changes, including car2, car4, and nos2. These differences suggest changes in Zn distribution between the two model systems and altered regulation of Zn-dependent enzymes. A comparison of the some of the notable changes in metal-dependent genes and how they are similar or different between the two model systems is presented in Fig. 10.
FIG 10.
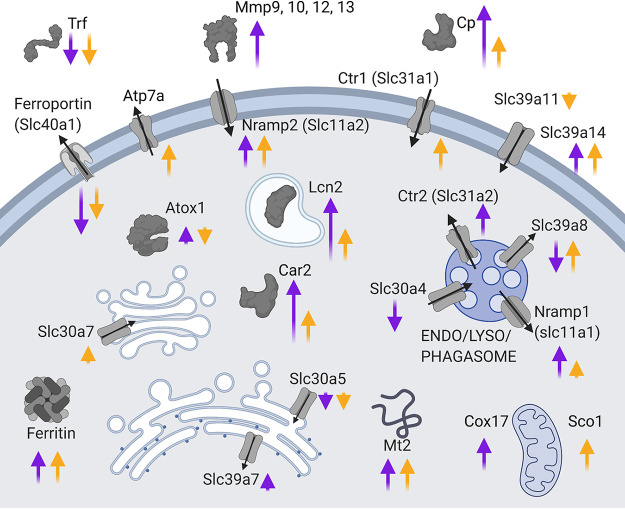
Summary of major metal-related gene expression changes in 129S6 and C57BL/6 macrophages. Purple arrows represent the log2 fold change between 129S6 unexposed (UE) and 18 h alive conditions, yellow arrows represent the log2 fold change between C57BL/6 uninfected (UI) and growing (G) conditions. A full list of metal-related gene expression data can be found in Data Set S4 in the supplemental material. Figure created with BioRender.com.
A comparison between A18 versus UE and G versus UI revealed global expression differences as well. A total of 1,991 genes are differentially expressed (padj < 0.01) between G and UI that were not differential (padj > 0.03) between A18 and UE. Further, 2,723 genes were significantly different between A18 and UE, but not in G versus UI. When these gene lists were scrutinized using DAVID annotation clustering, the genes differential in 129S6 were enriched for cell cycle functions, and the majority were downregulated. Interestingly, the genes differentially expressed in C57BL/6 were enriched for apoptosis, and almost all of these genes were upregulated. These data suggest that these two cell types are moving toward different fates by 18 h post Salmonella Typhimurium infection, with 129S6 cells entering quiescence but C57BL/6 cells experiencing apoptosis.
DISCUSSION
This work presents a systematic analysis of changes in global gene expression, as well as a more targeted analysis of changes in genes related to transition metal ions in 129S6 macrophages infected with Salmonella Typhimurium. The 129S6 model system was chosen because these mice contain functional Slc11a1 (Nramp-1), a divalent metal ion transporter that localizes to phagosomes and regulates metal availability to the intracellular pathogen. Slc11a1 confers resistance to intracellular pathogens and the 129S6 mouse has been used to model long-term chronic infection by Salmonella Typhimurium, as the mice do not succumb to initial acute infection as do C57BL/6 or BALB/c mice (30, 31, 53). A recent study examined the Salmonella Typhimurium proteome upon infection of mice homozygous or heterozygous for functional versus nonfunctional Scl11a1 and found an abundance of Mg, Zn, Fe, and Mn uptake systems expressed in Salmonella Typhimurium in resistant mice but not Salmonella Typhimurium that colonized susceptible BALB/c mice (31). This study indicates that Slc11a1 plays a major role in affecting metal ion availability for uptake by Salmonella Typhimurium. However, how metal regulation in the host is altered in a model system with functional Slc11a1 has not been explored. Given that metal manipulation is a key component of nutritional immunity, we sought to define how genes encoding metal transporters, metal-dependent proteins, and metal-regulatory proteins were affected by treatment with live or heat-killed Salmonella Typhimurium at early (2 h postinfection) and late (18 h postinfection) time points.
We observed widespread changes in metal transport, metal-dependent, and metal homeostasis genes, suggesting remodeling of Fe, Cu, and Zn availability by host cells. There is significant upregulation of slc11a1, especially at late stages of infection, consistent with the host cell limiting Fe, Mg, Mn, and Zn in the phagosome. Further changes in gene expression are consistent with decreased Fe uptake (trf and steap3), export (slc40a1), and increased storage (fth1); increased Cu uptake (slc31a1), import into the phagosome (atp7a), and altered distribution (Cu chaperones atox1, sco1, cox17); and increased transport of Zn from medium into the cytosol (slc39a1 and slc39a14) coupled with decreased transport into the secretory pathway and endo-lysosomal system (slc39a7, slc30a4, and slc30a5). Increased expression of mt1 and mt2 genes further suggests an increase in cytosolic Zn. We also observed numerous changes in proteins and enzymes associated with metal ions, many of which play an important role in nutritional immunity (lcn2, ltf, cp, steap3, sod2, arg1, nos2, mmp, car, and s100a8). Some of these changes have been seen in a variety of mouse model systems in response to diverse intracellular pathogens, suggesting widespread or universal strategies for the host to combat infection. In particular, the changes in gene expression to limit Fe (6, 54) and accumulate Cu appear to be common strategies for nutritional immunity (4). Indeed, the changes in expression of trf, slc40a1, fth1, slc31a1, lcn2, and cp were also observed in analysis of RNA-seq data from infection of macrophages from C57BL/6 mice at 18 h postinfection. However, it is notable that many of the changes are significantly more pronounced (larger fold changes) in 129S6 cells, indicating a more robust nutritional immunity response in the 129S6 model system. This observation is consistent with the findings of Cunrath and Bumann, which found that only in infection of mice with functional Slc11a1 were bacterial metal uptake systems induced (31), suggesting that functional Slc11a1 is necessary to activate nutritional immunity mechanisms. Interestingly, the changes we observed in expression in slc11a1, as well as key Fe and Cu regulatory genes, were comparable in macrophages infected with live bacteria or treated with HK bacteria. This suggests that the host response is driven not only by active pathogen virulence factors, but also pathogen-associated cues, such as LPS. It is important to note that the expression of many metal transporters can be due to posttranscriptional regulation of mRNA stability in response to changes in the concentration of metal ions.
As noted previously, there does not appear to be a universal strategy for host manipulation of Zn at the host-pathogen interface, with examples of host mechanisms to both starve and poison an intracellular pathogen (2, 5, 22). Cellular regulation of Zn is more complex than Fe or Cu, with 24 Zn transporters that control transport into and out of the cytosol, as well as intracellular organelles. Indeed, pathogens and immune system modulators have been shown to have different effects on Zn transporter expression (7), including upregulation of slc39a8 (52) in response to LPS and TNF-α, upregulation of slc30a4 and slc30a7 upon stimulation with granulocyte-macrophage colony-stimulating factor (GM-CSF) (55), and upregulation of slc39a14 in response to LPS (56). Previous studies have suggested that Salmonella Typhimurium experiences Zn starvation inside the host, as the high-affinity bacterial Zn uptake system ZnuABC (23) and associated accessory protein ZinT (24) and low-affinity uptake system ZupT (57) contribute to virulence in mice. But the concept of Zn starvation is hard to reconcile with reports of Zn accumulation in puncta (27) and the suggestion that Salmonella Typhimurium induces Zn elevation to evade bacterial clearance (26). To provide a more complete picture of Zn modulation within the host, we analyzed changes in the expression of Zn regulatory genes, measured labile Zn levels in the cytosol at different times postinfection, and examined the effect of manipulation of Zn levels in media on bacterial clearance and bacterial replication. We were particularly interested in comparing how the host cell remodels Zn availability in macrophages with functional Slc11a1, and hence a robust nutritional immunity response.
The changes in Zn transporters and homeostasis genes predict an increase in cytosolic Zn and a decrease in Zn in the ER/Golgi/phagosome. Specifically, we observed significant upregulation of slc39a1, slc39a14, slc11a1, mt1, and mt2, with more pronounced changes at late stages of infection (18 h). Using a genetically encoded Zn sensor explicitly targeted to the cytosol, we measured an increase in Zn from ∼ 490 pM to ∼ 22 nM at 12 h postinfection that remained elevated through the last measurement time (21 h postinfection). Intriguingly, mt1, mt2, and slc39a14 were more highly upregulated upon infection with live Salmonella Typhimurium compared with heat-killed bacteria, suggesting an active recognition of and response to live bacteria. We speculate that the host cell increases cytosolic Zn to support metabolic and transcriptional proteins that fight infection. This is consistent with our observation that a large fraction of differentially expressed genes has DAVID annotations that include “Zn binding”. Our study does not resolve the source of the increase in cytosolic Zn. RNA-seq suggests it could result from increased import of Zn across the plasma membrane or increased transport out of the phagosome and into the cytosol. While inductively coupled plasma mass spectrometry (ICP-MS) can be used to measure total Zn, the increase in cytosolic Zn is 5 orders of magnitude smaller (nM) than the total Zn in mammalian cells (hundreds of μM).
The increase in slc39a7 and slc11a1 coupled with the decrease in slc30a5 and slc30a4 suggest decreased Zn in the ER/Golgi/phagosome. The changes in slc39a7, a5, and a4 were comparable in macrophages treated with live Salmonella Typhimurium versus HK bacteria, suggesting a general host response to the pathogen and pathogen-associated cues (for example LPS). Limitation of Zn in the secretory and phago-lysosomal pathway would lead to Zn restriction for the pathogen. This finding is consistent with previous observations that Salmonella Typhimurium upregulates bacterial Zn uptake mechanisms, especially in the presence of functional Slc11a1. Combined, our results indicate that manipulation of Zn at the host-pathogen interface is more nuanced than Fe or Cu, where the host leverages its intricate means of manipulating Zn availability and distribution to limit the ability of the pathogen to access Zn, while simultaneously ensuring sufficient Zn to support the immune response.
In addition to representing a distinct model system for studying nutritional immunity, we anticipated that the 129S6 model system would have a different global immune response as well. To critically compare infection-induced changes in the transcriptome, we identified an RNA-seq study that was carried out in macrophages from C57BL/6 mice (51), with strong parallels to our conditions, and reanalyzed the data along with ours in an identical pipeline. We found that 4,500 genes were differentially expressed in one study but not the other, with differences in expression at baseline as well as in response to infection. This supports our hypothesis that macrophages from these mice represent distinct model systems, with differences that extend beyond Slc11a1 expression. Indeed, mouse lines possessing one or two functional slc11a1 alleles on a C57BL/6 background (30, 31) experienced lower infection loads but still ultimately succumbed to Salmonella Typhimurium infection, indicating that Slc11a1 alone does not recapitulate the 129S6 phenotype. Annotation analysis of the differential gene list indicates that these cell types tend toward different fates in response to the stress of Salmonella Typhimurium infection, with 129S6 cells showing suppression of cell cycle genes but C57BL/6 cells showing upregulation of apoptotic genes.
In assessing the GSEA and GO term enrichment present in our data, we find recurring themes of inflammation, hypoxia, apoptosis, and the unfolded protein response (UPR), which are inextricably linked (37, 58). Both intracellular hypoxia and Salmonella-induced inflammation can result in apoptosis or the UPR, which is an attempt to regain cellular homeostasis under stress. When comparing live versus HK Salmonella Typhimurium conditions across time, we see enrichment of hypoxia-related genes along with enrichment of apoptosis at 2 h, which transitions to enrichment of UPR at 18 h. This apparent transition in the cellular stress response over time demonstrates the ability of 129S6 macrophages to withstand Salmonella Typhimurium infection. In contrast, infection of C57BL/6 macrophages showed marked enrichment of apoptosis genes at 18 h. This finding is supported by multiple previous studies which found that Salmonella Typhimurium induces apoptosis in macrophages from a number of mouse models (40, 59).
Macrophages experience a range of phenotypes, from inflammatory and cytotoxic (termed M1 polarization) to anti-inflammatory and wound healing (M2 polarization), depending on their milieu. These states are fluid, differing in gene expression and metabolism, and are heterogeneous across in vivo cell populations (37). The bactericidal mechanisms available to M1 macrophages enable them to kill internalized pathogens efficiently. The lack of these mechanisms in M2-like macrophages makes them a more permissive niche for facultative intracellular bacteria like Salmonella Typhimurium. Although a number of previous studies have tried to identify individual genes that distinguish M1 and M2, for example, arg1 versus nos2 (60) or egr2 versus CD38 (61), the increasing availability of whole-transcriptome sequencing makes it possible to define a more comprehensive genetic signature of M1 and M2. Single cell RNA-seq by Saliba et al. (32) enabled identification of gene clusters related to M1 and M2 and how these gene clusters correlated with infection (naive macrophages, bystanders, macrophages with growing bacteria, and macrophages with nongrowing bacteria). Dual RNA-seq of both the pathogen and host by Stapels et al. (51) further enables correlation of the M1 and M2 gene clusters within the host with expression of Salmonella Typhimurium virulence genes. Indeed, this analysis revealed that in C57BL/6 macrophages, the M2 polarization correlates with increased SPI2 expression, suggesting that Salmonella Typhimurium activates virulence mechanisms in this niche.
Examination of M1 and M2 gene clusters further reinforces that 129S6 macrophages represent a distinct intracellular niche with respect to immune response to infection. As shown in Fig. 2, polarization genes follow a temporal pattern of expression, with the majority of M1-related genes strongly upregulated by 2 h postinfection but sharply downregulated by 18 h. Alternatively, we see slower time-dependent activation of M2-related genes. This observation is supported at 2 h by enrichment in glycolysis, the metabolic state of inflammatory or hypoxic macrophages, and at 18 h by enrichment in oxidative phosphorylation and fatty acid metabolism, which are the metabolic markers of M2 macrophages (37, 58). Importantly, at each time point we see little difference in M1/M2 associated genes between live and HK Salmonella Typhimurium conditions, especially when normalized to C57BL/6 data (Fig. S7). This is in contrast to findings in macrophages from C57BL/6 mice, which indicate that dividing Salmonella Typhimurium may actively induce M2 polarization to enhance bacterial survival, and that macrophages exposed to but not infected by Salmonella Typhimurium maintain an M1 phenotype late in infection (32, 51). While our work is at a population level and therefore contains macrophage and Salmonella Typhimurium heterogeneity, exposure to HK Salmonella Typhimurium does not lead to retention of an M1 phenotype at 18 h, nor does it seem that 129S6 cells are induced to M2 by live Salmonella Typhimurium.
Our study recapitulates the ability of 129S6 macrophages to fight infection at all time points, which occurs regardless of the expression profile of M1 or M2 genes. At 2 h, 70% of initially infected macrophages had cleared the infection, which correlated with the transcriptional induction of M1 polarization. However, despite the shift toward an M2-like transcriptional profile at 18 h, the rate of bacterial clearance increased slightly over time to 74%. Concurrently, the proportion of infected macrophages showing evidence of bacterial replication also increased over time, from 8% at 10 and 18 h to 24% by 24 h. This high clearance rate and relatively low replication rate resulted in only 3% of initially infected macrophages harboring dividing bacteria at 24 h (Data Set S5). This is in contrast to studies of Nramp-1+/+ 129X1 mice (62) and Slc11a1 nonfunctional macrophages, which report that replicating Salmonella Typhimurium is preferentially found in M2 cells (32, 36, 51). It is also notable that 129S6 macrophages surviving to 24 h while harboring dividing Salmonella Typhimurium are not allowing high division rates, as indicated by only modest decreases in the inducible fluorescent signal (Fig. 9), whereas macrophages from other model systems experience high levels of Salmonella Typhimurium division at 24 h (36, 49). When we manipulated Zn availability in the medium, we found that Zn-replete conditions led to robust clearance of infected bacteria, suggesting that elevated Zn may help the host fight infection. On the other hand, Zn repletion in the medium also enables bacterial replication, albeit at a very low rate. While these results clearly indicate Zn availability can influence infection outcomes, they do not resolve the mechanism. Future studies are needed to determine how changes in Zn in the media affect total Zn, cytosolic Zn, and phagosomal Zn, and how the different media conditions affect antimicrobial immune pathways within the host, as well as virulence pathways in the pathogen. Finally, it is important to note that Chelex treatment reduced Fe and Ca as well as Zn. Figure 7 suggests reduced Fe and Ca in the medium slows bacterial clearance; however, by 24 h, clearance (87%) matches that of regular growth medium (85%).
This study reveals that 129S6 macrophages differ significantly from other model systems of Salmonella Typhimurium infection. First, 129S6 macrophages show a robust remodeling of metal homeostasis and expression of metal-dependent enzymes, indicating a distinct nutritional immunity response. Second, remodeling of Zn regulatory proteins leads to an increase in cytosolic Zn and a likely restriction of Zn for the pathogen. We speculate that this ensures sufficient cytosolic Zn to fight infection and promote bacterial clearance, while still limiting Zn availability to fight against bacterial replication. Third, transcription of M2-related genes doesn’t correlate exclusively with live Salmonella Typhimurium or with a lack of bactericidal capacity, suggesting that in this model system, the M2 polarization does not promote bacterial survival. Rather, activation of M1-related genes early and M2-related genes at later time points correlates with the switch from expression of apoptosis genes to UPR genes, suggesting a choreographed response to fight infection without killing the host cell. Together, these results imply that 129S6 macrophages do not conform well to the M1/M2 expression dichotomy derived largely from studies of Nramp-1 nonfunctional macrophages and mice. Instead, they appear to present a mixed phenotype that effectively uses zinc to quell Salmonella Typhimurium infection.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the Institutional Animal Care and Use Committee at the University of Colorado Boulder. The animal subjects plan protocol number is 2547.
Monocyte extraction from bone marrow.
Animal work followed protocol 2547, approved by the University of Colorado IACUC. The 8- to12-week-old 129S6 female mice (Taconic Laboratories) were euthanized by CO2 inhalation according to IACUC guidelines, followed by cervical dislocation. Femur, tibia, and humerus bones were extracted, then scraped and flushed with ice cold phosphate-buffered saline (PBS). Approximately 1 ml of PBS was used per bone. PBS with marrow cells was passed through a 70-micron nylon mesh Falcon Cell Strainer (Corning 352350), then overlaid onto an equal volume of Histopaque-1083 (Sigma-Aldrich) in centrifuge tubes. Tubes were centrifuged at 500 × g for 30 min and allowed to slow with no brake. Monocytes appear as a fuzzy layer at the fluid interface. These were removed to clean tubes and washed twice with 14 ml PBS. After the final wash, monocytes were resuspended in a small amount of macrophage growth medium and counted with a hemocytometer. Growth medium consisted of Dulbecco’s modified Eagle medium (DMEM) (Sigma-Aldrich) supplemented with 20% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin/streptomycin (50 IU/ml penicillin and 50 μg/ml streptomycin), and 10 pg/μl recombinant murine macrophage colony-stimulating factor (PeproTech, Inc.).
Primary cell culture and infection.
Monocytes were plated into 12-well plates at a density of 200,000 cells per well. Then, 2 ml of macrophage growth medium was added to each well and plates were incubated at 37°C and 5% CO2 for 6 days. Medium was refreshed 3 days after plating. Wild-type Salmonella Typhimurium SL1344 was cultured to stationary phase in LB with ampicillin (100 μM) and streptomycin (50 μM) at 37°C with aeration. Bacteria were washed with PBS and opsonized for 30 min at room temperature in a 1:1 solution of mouse serum (Sigma) and cell culture medium (Gibco). Bacteria were then pelleted at 13,000 × g for 1 min and resuspended in PBS. Half the bacteria were heat killed at 60° C for 3 min, then placed on ice. One well of macrophages was scraped and counted, and bacteria were diluted in macrophage medium without antibiotics to a multiplicity of infection (MOI) of 30. An MOI of 30 was chosen to ensure a high infection rate and to enable us to compare this work to other studies that have used MOIs ranging from 5 (51) to 50 (32). Plated macrophages were washed 3 times with PBS, then treated with antibiotic-free macrophage medium containing live bacteria, heat-killed bacteria, or no bacteria. Medium was removed 30 min later, after which cells were washed 3× with PBS and macrophage medium containing 100 g/ml gentamicin was added. After 90 min, medium was again removed. RNEasy lysis buffer was added directly to wells and these were designated as 2-h samples. Macrophage medium with 10 μg/ml gentamicin was added to wells designated as 18-h samples. Lysed samples were collected and frozen at −20°C. At 18 h post Salmonella Typhimurium infection, medium was removed from the remaining wells, lysis buffer was added, and cell lysates were frozen at –20°C. To examine both early and late immune responses, macrophages were lysed at 2 h or 18 h. Macrophages that were subjected to medium changes but not exposed to Salmonella Typhimurium erved as a control. Control samples were lysed at the 2-h time point.
RNA extraction and sequencing.
RNA extraction was done on all samples at once, after one freeze-thaw cycle. An RNEasy kit (Qiagen) was used, and DNase I treatment was done on the column. RNA integrity was checked via Tape Station and library prep was done with an Illumina TruSeq LT kit, which included polyA selection. Paired-end 75 base sequencing was done on a NextSeq 2.1.0 Illumina machine. Tape Station run, library prep, and sequencing were performed by the JSCBB sequencing core.
Data analysis pipeline.
R 3.3.0 on the JSCBB computing core was used for analysis of raw data. Data quality was assessed with fastqc (0.11.2) and the first 10 bases were trimmed with Trimmomatic (0.36). Reads were mapped with TopHat (2.0.6)/Samtools (0.1.18)/bowtie2 (2.0.2) using -b2-very-sensitive, fr-firststrand settings. Read counting was completed with EdgeR Subread featureCounts (1.6.0) using the mm10 gtf file from UCSC (July 2015 version). MetaFeature mapping was paired end, with both ends mapped, and no MultiOverlap allowed. R 4.0.2 was used for all remaining analysis. A principal-component analysis (PCA) of global RNA expression was performed using EdgeR. Analysis of differential expression was done with DESeq2 (1.18.1), but the nature of this data set violates the DESeq2 assumption that most genes do not change expression between conditions. Therefore, all genes with differential expression (padj < 0.05) in binary comparisons were compiled and excluded from the final data set matrix (dds) normalization using the controlGenes parameter. A log ratio test was run on the correctly normalized DESeq data set matrix (dds) with the following function: DESeq (dds, test = “LRT,” reduced ≈ 1), resulting in a list of globally differentially expressed genes. This data set (P < 0.01) was further filtered with two transcripts per million (TPM) requirements. Inclusion in the final analysis indicates that a gene has an overall mean TPM > 5, and at least one condition with a mean TPM > 10. The 7,766 genes meeting these criteria were fed into the DEGReport R package (1.24.1) for rlog transformation, which produced normalized counts, and expression clustering, which produced Z-scores. Expression clustering was done with the following function: degPatterns (cluster_rlog, metadata = metadata, time = “Treatment”). Heatmap gene dendrograms were constructed using the hierarchical clustering packages hclust and ggdendro (0.1.22), and this gene order was used to construct global and cluster heatmaps with ggplot (Tidyverse R package 1.3.0). The median value for each plot was set as white. DAVID annotation enrichment categories were used to assess the makeup of DEGReport clusters. This analysis included functional categories, Gene Ontology, Biocarta, and Kegg pathways, and protein domains.
In brief, DESeq2 binary comparisons between conditions were fed into GSEA (63) to validate GO term enrichment and assess transcription targets. More specifically, adjusted P value ranked lists were created from DESeq2 differential expression data sets comparing 2A versus 2HK or 18A versus 18HK. For each gene in a given data set, a rank was determined from the padj value (false discovery rate) and the direction of fold change value (up or downregulation). Rank = −log(padj) × sign(fold change). All genes with a valid padj value were included. GSEA was used to compare these ranked lists to validated gene sets. These were: Canonical Pathways, Hallmark gene sets, GO_Biological Processes, GO_Molecular Functions, and Transcription Factor Targets. Because GSEA’s default is human gene sets, the mouse genome was collapsed and remapped using the Mouse Gene Symbol Remapping Human Orthologs MsigDB.v7.2.chip. Gene set permutations were used in this analysis, which is a less stringent assessment of significance, and produces more false-positive results. Therefore, a cutoff of q < 0.05 was applied to gene set results.
To assess changes in metal-related gene expression, comprehensive gene lists were compiled from the Uniprot database. All 3,306 reviewed mouse entries with “metal” annotation were downloaded and compared with our RNA sequencing data. Genes meeting the expression cutoff of mean normalized counts > 5 and the differential expression cutoff of log ratio test padj < 0.01 were included in further analysis. These genes were then cross referenced with the keyword annotations of “metal-binding,” “metal transport,” or “metal homeostasis,” and were further curated to establish lists of genes primarily associated with Zn, Cu, or Fe.
Comparison of current study with Stapels et al study.
The Stapels et al. study (51) examined changes in mRNA expression at 18 h upon infection of C57BL6 macrophages with Salmonella Typhimurium strain SL1344 for 25 min at an MOI of 5. The Salmonella strain and time point are identical to our study. The MOI is lower compared to our MOI of 30, and the infection time is comparable (25 min versus 30 min for our study).
FASTQ files (GEO accession GSE104785) were downloaded and run through the above pipeline with the following alterations: Trimmomatic was used to remove Illumina adaptors and alterations were made to mapping and counting based on the unstranded nature of the data. This differs from the pipeline of Stapels et al. in that their data was mapped to the Salmonella Typhimurium genome prior to the mouse mm10 genome. Additionally, their read counts were performed with HTSeq (version 0.6.1) instead of FeatureCounts. To normalize our data with this previously published data set, DESeq2 was run on the combined data with the experimental design ∼Treatment, and normalized counts were calculated. A PCA and dendrogram of the Stapels data set were created and compared with their published heatmap to generally validate our sequencing results (Fig. S8).
To assess differences in metal homeostasis genes as comprehensively as possible, mouse-specific lists of genes involved in the transport and homeostasis of metal ions were compiled from the Uniprot database. Separate lists were compiled for Zn, Fe, and Cu, and each list was compared against the normalized counts of the aggregated sequencing studies. Only genes expressed above background were used for further analysis.
Measurement of cytosolic Zn and imaging analysis.
The NES-ZapCV2 sensor was used to measure cytosolic Zn. This sensor uses the ratio of yellow fluorescent protein (YFP) emission to cyan fluorescent protein (CFP) emission upon CFP excitation (Förster resonance energy transfer [FRET] ratio) to measure labile Zn in cells. The advantage of this genetically encoded sensor is that localization of this sensor is restricted to the cytosol. Briefly, cells were nucleofected with a plasmid encoding NES-ZapCV2 15 h prior to infection with Salmonella Typhimurium. At select time points, postinfection cells were imaged on a widefield fluorescence microscope to measure the resting FRET ratio. For quantification of cytosolic labile Zn in primary macrophages, calibration experiments were performed at the indicated times postinfection on a Nikon Ti-E widefield fluorescence microscope equipped with Nikon elements software, Ti-E perfect focus system, an iXon3 EMCCD camera (Andor), mercury arc lamp, and YFP FRET (434/16 excitation, 458 dichroic, 535/20 emission), CFP (434/16 excitation, 458 dichroic, 470/24 emission), and YFP (495/10 excitation, 515 dichroic, 535/20 emission) filter sets. External excitation and emission filter wheels were controlled by a Lambda 10-3 filter changer (Sutter Instruments), while dichroic mirrors were placed on cubes in the dichroic turret. Channel EM gain and exposure settings for both YFP FRET and CFP were set to be the same. Images were collected using a 60× oil objective (NA 1.40), 200 ms exposure time, EM gain 1 MHz at 16-bit readout mode with an EM gain multiplier of 200, and a neutral density filter with 25% light transmission. Sensor expression level was controlled by selecting cells with YFP intensities between 4,000 to 15,000 fluorescence units under these conditions. Eight fields of view were collected using multipoint acquisition mode and the Perfect Focus System (PFS) set to “ON” in between points. Cells were maintained at 37°C and 5% CO2 in a LiveCell environment chamber (Pathology Devices) during the experiments. Images were collected every minute during RResting and Rmin calibration acquisition phases and every 30 s during the Rmax calibration phase.
Fresh calibration solutions were prepared the day of the experiment and included a 2× solution of Rmin buffer (50 μM TPA in PO43−-free HHBSS) for minimum FRET ratio collection and a 2× solution of Rmax buffer (0.001% saponin + 0.75 μM pyrithione + 23.8 nM buffered Zn in PO43−-, Ca2+-, Mg2+-free HHBSS) for collecting the maximum FRET ratio. The resting FRET ratio of the sensor was collected for 10 min prior to calibration to ensure a stable signal. After 10 min of imaging, 50 μM tetradecanoyl phorbol acetate (TPA) was added to the dish to collect the minimum FRET ratio of the sensor. Once a stable signal had been achieved, cells were then washed with phosphate-, calcium-, and magnesium-free HEPES-buffered HBSS (pH 7.4) to remove the chelate and then treated with pyrithione and Zn with 0.001% (wt/vol) saponin.
All imaging data were analyzed in MATLAB (Mathworks). Images were background corrected by subtracting a local background intensity from each pixel grouped in a certain region of the image. Regions of interest were generated using a segmentation algorithm that segments the image based on the fluorescence intensity of the FRET ratio channel to obtain single cell traces. FRET ratios for each cell trace were calculated by dividing the background-corrected YFP FRET intensity by the background-corrected CFP intensity. The resting, minimum, and maximum FRET ratios for each cell were used to calculate the fractional saturation, the dynamic range, and the reported [Zn] from the sensor in each cell. Resting [Zn] is calculated by where the Kd = 5.3 nM and Hill coefficient = 0.29 for NES-ZapCV2. The dynamic range of the sensor can be impacted by both overexpression and underexpression of the sensor, which has a negative impact on the fidelity of the apparent [Zn]. For this reason, cells were excluded from the analysis if they fell outside of an acceptable dynamic range of 1.6 to 2.3 for the NES-ZapCV2 sensor.
Flow cytometry study of infection outcome with varied zinc availability.
For this study, monocytes from 3 mice were collected and pooled prior to differentiation into macrophages. Four different medium conditions were used for flow cytometry. Standard macrophage growth media (DMEM + 20% FBS) was found to contain 25 μM Zn by ICP-MS and was used as the baseline. The other three media used Chelex-100-treated FBS. Briefly, FBS was treated with Chelex 100 resin (Sigma-Aldrich, St. Louis, MO, USA) for 5 h, stirring at 4°C, followed by sterile filtration using a 0.22-μm PES membrane filter. This Chelex-treated FBS was used to prepare the baseline Chelex medium, which contained DMEM + 20% FBS + l-glutamine (2 mM) + sodium pyruvate (1 mM). Zn-replete medium was made by adding 30 μM ZnCl2 (Sigma-Aldrich, St. Louis, MO, USA) to Chelex medium. Medium with essentially no Zn was made by adding 3 μM 2-([Bis(2-pyridinylmethyl)amino]ethylamino)benzenesulfonic (ZX1, an extracellular Zn chelator [64]) (Strem Chemicals, Inc.) to Chelex medium. ICP-MS measurements were carried out as described previously (65) to quantify the metal content of the media. While Chelex treatment depleted Zn, Ca, and Fe, the Zn-replete, Chelexed medium, and Zn-deficient medium are identical except for the amount of zinc present in the medium (34 μM, 4 μM, no Zn, respectively).
For analysis of intracellular bacterial replication, monocytes from three mice were grown and differentiated to macrophages in 6-well plates. Six days later, Salmonella Typhimurium expressing pDiGc and induced with arabinose was grown to stationary phase with aeration. Arabinose induction of DsRed was halted upon macrophage infection, such that any bacterial division occurring within the macrophage led to dilution of the red fluorescent signal. Macrophages were infected with Salmonella Typhimurium at a multiplicity of infection of 30, and infection proceeded for 45 min before experimental media conditions were introduced. This avoided the confounding variable of Zn’s effect on the phagocytosis of bacteria. At this point, Salmonella Typhimurium medium was removed, cells were washed 3× in PBS, and initial inoculum cells were collected by scraping with a nylon cell lifter (Corning C3008) and homogenized by pipetting gently with a P1000. Homogenized cells were fixed in a gentle fixative for preserving fluorescent protein fluorescence (1% PFA and 1% sucrose) for 15 min and then washed and resuspended in PBS and chilled at 4°C. Medium in all other samples was changed to experimental conditions, which included 10 μg/ml gentamicin. Three wells were treated with each medium, for each time point. At 2, 10, 18, and 24 h postinfection, cells were rinsed, lifted, fixed, and chilled as above. Samples were analyzed on a BD FACSCelesta (BD Biosciences) collecting forward scatter area and width, side scatter area and width, 488 nm excitation with 530/30 nm emission, and 561 nm excitation with 585/15 nm emission. Data were analyzed using FlowJo 10.5.3 software (FlowJo LLC).
The cell gating hierarchy was set as single cells > GFP-positive cells > cells containing replicated bacteria. A total of 25,000 to 30,000 single cells were collected per sample. Single cells were determined first by forward scatter area versus side scatter area, then by side scatter width versus side scatter area. Nonfluorescent uninfected cells were used to set the gate for GFP-positive cells. A ratio of the 488 nm channel to the 561 nm channel was taken by dividing the 488 nm intensity by the 561 nm intensity. Samples collected at 2 h postinfection were used as the “initial inoculum” to determine the fluorescence intensities for cells infected with bacteria that have not undergone replication. Cells containing replicated bacteria were gated as having a 488 nm:561 nm ratio above the initial inoculum.
Data availability.
All raw next-generation sequencing data files and processed data files used to draw conclusions are available at the Gene Expression Omnibus, data series GSE166642.
ACKNOWLEDGMENTS
We thank the following sources for financial support: National Institutes of Health NIH Pioneer Award (DP1 GM114863 to A.E.P.), MIRA (R35 GM139644 to A.E.P.), R01 (GM125871 to R.D.D.), and R01 (HL156475 to M.A.A.). L.N.J. was supported by a National Science Foundation GRF. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Lynn Sanford for technical assistance developing code for analyzing RNA-seq data. We acknowledge the BioFrontiers Institute Advanced Light Microscopy Core and Joseph Dragavon for technical assistance. We acknowledge Theresa Nahreini and the University of Colorado Flow Cytometry Shared Core (BDAria cell sorter, NIH S10OD021601), BioFrontiers Institute Next-Gen Sequencing Core Facility, which performed the Illumina sequencing and library construction, the BioFrontiers high-performance computing resources (NIH 1S10OD012300) supported by BioFrontiers’ IT, and the University of Colorado Biochemistry Cell Culture Core Facility for providing resources and support in cell culture.
Footnotes
Supplemental material is available online only.
Contributor Information
Amy E. Palmer, Email: amy.palmer@colorado.edu.
Manuela Raffatellu, University of California San Diego School of Medicine.
REFERENCES
- 1.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganz T. 2018. Iron and infection. Int J Hematol 107:7–15. 10.1007/s12185-017-2366-2. [DOI] [PubMed] [Google Scholar]
- 4.White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284:33949–33956. 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djoko KY, Ong CY, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961. 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H, Dai W, Zhao L, Min J, Wang F. 2018. The role of zinc and zinc homeostasis in macrophage function. J Immunol Res 2018:6872621. 10.1155/2018/6872621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreini C, Bertini I. 2012. A bioinformatics view of zinc enzymes. J Inorg Biochem 111:150–156. 10.1016/j.jinorgbio.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. 2009. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods 6:737–740. 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin Y, Miranda JG, Stoddard CI, Dean KM, Galati DF, Palmer AE. 2013. Direct comparison of a genetically encoded sensor and small molecule indicator: implications for quantification of cytosolic Zn2+. ACS Chem Biol 8:2366–2371. 10.1021/cb4003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE. 2011. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc Natl Acad Sci USA 108:7351–7356. 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabosseau P, Tuncay E, Meur G, Bellomo EA, Hessels A, Hughes S, Johnson PRV, Bugliani M, Marchetti P, Turan B, Lyon AR, Merkx M, Rutter GA. 2014. Mitochondrial and ER-targeted eCALWY probes reveal high levels of free Zn2+. ACS Chem Biol 9:2111–2120. 10.1021/cb5004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JG, Palmer AE. 2014. Quantitative measurement of Ca2+ and Zn2+ in mammalian cells using genetically encoded fluorescent biosensors, p 29–47. In Zhang J, Ni Q, Newman RH (ed), Fluorescent protein-based biosensors: methods and protocols. Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter KP, Carpenter MC, Fiedler B, Jimenez R, Palmer AE. 2017. Critical comparison of FRET-sensor functionality in the cytosol and endoplasmic reticulum and implications for quantification of ions. Anal Chem 89:9601–9608. 10.1021/acs.analchem.7b02933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratt EPS, Damon LJ, Anson KJ, Palmer AE. 2021. Tools and techniques for illuminating the cell biology of zinc. Biochim Biophys Acta Mol Cell Res 1868:118865. 10.1016/j.bbamcr.2020.118865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eide DJ. 2006. Zinc transporters and the cellular trafficking of zinc. Biochim Biophys Acta 1763:711–722. 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Andrews GK. 2001. Cellular zinc sensors: MTF-1 regulation of gene expression, p 37–51. In Maret W (ed), Zinc biochemistry, physiology, and homeostasis: recent insights and current trends. Springer, Dordrecht, Netherlands. [DOI] [PubMed] [Google Scholar]
- 18.Gaddy JA, Haley KP. 2015. Metalloregulation of Helicobacter pylori physiology and pathogenesis. Front Microbiol 6:911. 10.3389/fmicb.2015.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. 2011. Mycobacterial P1-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10:248–259. 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stocks CJ, Phan M-D, Achard MES, Nhu NTK, Condon ND, Gawthorne JA, Lo AW, Peters KM, McEwan AG, Kapetanovic R, Schembri MA, Sweet MJ. 2019. Uropathogenic Escherichia coli employs both evasion and resistance to subvert innate immune-mediated zinc toxicity for dissemination. Proc Natl Acad Sci USA 116:6341–6350. 10.1073/pnas.1820870116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stafford SL, Bokil NJ, Achard MES, Kapetanovic R, Schembri MA, McEwan AG, Sweet MJ. 2013. Metal ions in macrophage antimicrobial pathways: emerging roles for zinc and copper. Biosci Rep 33:e00049. 10.1042/BSR20130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignesh KS, Figueroa JAL, Porollo A, Caruso JA, Deepe GS. Jr, 2013. Zinc sequestration: arming phagocyte defense against fungal attack. PLoS Pathog 9:e1003815. 10.1371/journal.ppat.1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect Immun 75:5867–5876. 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrarca P, Ammendola S, Pasquali P, Battistoni A. 2010. The Zur-regulated ZinT protein is an auxiliary component of the high-affinity ZnuABC zinc transporter that facilitates metal recruitment during severe zinc shortage. J Bacteriol 192:1553–1564. 10.1128/JB.01310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang K, Wang D, Frederiksen RF, Rensing C, Olsen JE, Fresno AH. 2017. Investigation of the role of genes encoding zinc exporters zntA, zitB, and fieF during Salmonella Typhimurium infection. Front Microbiol 8:2656. 10.3389/fmicb.2017.02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu A, Tymoszuk P, Haschka D, Heeke S, Dichtl S, Petzer V, Seifert M, Hilbe R, Sopper S, Talasz H, Bumann D, Lass-Flörl C, Theurl I, Zhang K, Weiss G. 2017. Salmonella utilizes zinc to subvert antimicrobial host defense of macrophages via modulation of NF-κB signaling. Infect Immun 85:e00418-17. 10.1128/IAI.00418-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapetanovic R, Bokil NJ, Achard MES, Ong CY, Peters KM, Stocks CJ, Phan M-D, Monteleone M, Schroder K, Irvine KM, Saunders BM, Walker MJ, Stacey KJ, McEwan AG, Schembri MA, Sweet MJ. 2016. Salmonella employs multiple mechanisms to subvert the TLR-inducible zinc-mediated antimicrobial response of human macrophages. FASEB J 30:1901–1912. 10.1096/fj.201500061. [DOI] [PubMed] [Google Scholar]
- 28.Campoy S, Jara M, Busquets N, de Rozas AMP, Badiola I, Barbé J. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar Typhimurium virulence. Infect Immun 70:4721–4725. 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal SM, Pinner E, Lepage P, Gauthier S, Gros P. 1996. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains. J Immunol 157:3559–3568. [PubMed] [Google Scholar]
- 30.Brown DE, Libby SJ, Moreland SM, McCoy MW, Brabb T, Stepanek A, Fang FC, Detweiler CS. 2013. Salmonella enterica causes more severe inflammatory disease in C57/BL6 Nramp1G169 mice than Sv129S6 mice. Vet Pathol 50:867–876. 10.1177/0300985813478213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunrath O, Bumann D. 2019. Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 366:995–999. 10.1126/science.aax7898. [DOI] [PubMed] [Google Scholar]
- 32.Saliba A-E, Li L, Westermann AJ, Appenzeller S, Stapels DAC, Schulte LN, Helaine S, Vogel J. 2016. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol 2:16206. 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaffer K, Taylor CT. 2015. The impact of hypoxia on bacterial infection. FEBS J 282:2260–2266. 10.1111/febs.13270. [DOI] [PubMed] [Google Scholar]
- 36.Wrande M, Vestö K, Puiac Banesaru S, Anwar N, Nordfjell J, Liu L, McInerney GM, Rhen M. 2020. Replication of Salmonella enterica serovar Typhimurium in RAW264.7 phagocytes correlates with hypoxia and lack of iNOS expression. Front Cell Infect Microbiol 10:537782. 10.3389/fcimb.2020.537782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Díaz-Bulnes P, Saiz ML, López-Larrea C, Rodríguez RM. 2019. Crosstalk between hypoxia and ER stress response: a key regulator of macrophage polarization. Front Immunol 10:2951. 10.3389/fimmu.2019.02951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugla-Płoskońska G, Kiersnowski A, Futoma-Kołoch B, Doroszkiewicz W. 2009. Killing of Gram-negative bacteria with normal human serum and normal bovine serum: use of lysozyme and complement proteins in the death of Salmonella strains O48. Microb Ecol 58:276–289. 10.1007/s00248-009-9503-2. [DOI] [PubMed] [Google Scholar]
- 39.Dunkelberger JR, Song W-C. 2010. Complement and its role in innate and adaptive immune responses. Cell Res 20:34–50. 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 40.Monack DM, Raupach B, Hromockyj AE, Falkow S. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA 93:9833–9838. 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K, Jayawardena S, De Smaele E, Cong R, Beaumont C, Torti FM, Torti SV, Franzoso G. 2004. Ferritin heavy chain upregulation by NF-κB inhibits TNFα-induced apoptosis by suppressing reactive oxygen species. Cell 119:529–542. 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Nairz M, Ferring-Appel D, Casarrubea D, Sonnweber T, Viatte L, Schroll A, Haschka D, Fang FC, Hentze MW, Weiss G, Galy B. 2015. Iron regulatory proteins mediate host resistance to Salmonella infection. Cell Host Microbe 18:254–261. 10.1016/j.chom.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao X, Yeoh BS, Vijay-Kumar M. 2017. Lipocalin 2: an emerging player in iron homeostasis and inflammation. Annu Rev Nutr 37:103–130. 10.1146/annurev-nutr-071816-064559. [DOI] [PubMed] [Google Scholar]
- 44.Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. 2009. Genetically encoded sensors to elucidate spatial distribution of cellular zinc. J Biol Chem 284:16289–16297. 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiedrowski L. 2011. Cytosolic zinc release and clearance in hippocampal neurons exposed to glutamate—the role of pH and sodium. J Neurochem 117:231–243. 10.1111/j.1471-4159.2011.07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong J, Walker JM, Wang F, Park JG, Palmer AE, Giunta C, Rohrbach M, Steinmann B, Eide DJ. 2012. Promotion of vesicular zinc efflux by ZIP13 and its implications for spondylocheiro dysplastic Ehlers-Danlos syndrome. Proc Natl Acad Sci USA 109:E3530–E3538. 10.1073/pnas.1211775110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiedler BL, Van Buskirk S, Carter KP, Qin Y, Carpenter MC, Palmer AE, Jimenez R. 2017. Droplet microfluidic flow cytometer for sorting on transient cellular responses of genetically-encoded sensors. Anal Chem 89:711–719. 10.1021/acs.analchem.6b03235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanford L, Palmer AE. 2020. Dissociated hippocampal neurons exhibit distinct Zn2+ dynamics in a stimulation-method-dependent manner. ACS Chem Neurosci 11:508–514. 10.1021/acschemneuro.0c00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glassman AB, Rydzewski RS, Bennett CE. 1980. Trace metal levels in commercially prepared tissue culture media. Tissue Cell 12:613–617. 10.1016/0040-8166(80)90016-6. [DOI] [PubMed] [Google Scholar]
- 51.Stapels DAC, Hill PWS, Westermann AJ, Fisher RA, Thurston TL, Saliba A-E, Blommestein I, Vogel J, Helaine S. 2018. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362:1156–1160. 10.1126/science.aat7148. [DOI] [PubMed] [Google Scholar]
- 52.Liu M-J, Bao S, Gálvez-Peralta M, Pyle CJ, Rudawsky AC, Pavlovicz RE, Killilea DW, Li C, Nebert DW, Wewers MD, Knoell DL. 2013. ZIP8 regulates host defense through zinc-mediated inhibition of NF-κB. Cell Rep 3:386–400. 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monack DM, Bouley DM, Falkow S. 2004. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med 199:231–241. 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Núñez G, Sakamoto K, Soares MP. 2018. Innate nutritional immunity. J Immunol 201:11–18. 10.4049/jimmunol.1800325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian Vignesh K, Landero Figueroa JA, Porollo A, Caruso JA, Deepe GS. 2013. Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity 39:697–710. 10.1016/j.immuni.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayadi A, Nguyen A-T, Bard FA, Bard-Chapeau EA. 2013. Zip14 expression induced by lipopolysaccharides in macrophages attenuates inflammatory response. Inflamm Res 62:133–143. 10.1007/s00011-012-0559-y. [DOI] [PubMed] [Google Scholar]
- 57.Cerasi M, Liu JZ, Ammendola S, Poe AJ, Petrarca P, Pesciaroli M, Pasquali P, Raffatellu M, Battistoni A. 2014. The ZupT transporter plays an important role in zinc homeostasis and contributes to Salmonella enterica virulence. Metallomics 6:845–853. 10.1039/c3mt00352c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galván-Peña S, O’Neill LAJ. 2014. Metabolic reprograming in macrophage polarization. Front Immunol 5:420. 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wemyss MA, Pearson JS. 2019. Host cell death responses to non-typhoidal Salmonella infection. Front Immunol 10:1758. 10.3389/fimmu.2019.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orecchioni M, Ghosheh Y, Pramod AB, Ley K. 2019. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS−) vs. alternatively activated macrophages. Front Immunol 10:1084. 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado J de D, Popovich PG, Partida-Sanchez S, Guerau-de-Arellano M. 2015. Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10:e0145342. 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM. 2013. Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14:171–182. 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. 2007. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23:3251–3253. 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 64.Pan E, Zhang X, Huang Z, Krezel A, Zhao M, Tinberg CE, Lippard SJ, McNamara JO. 2011. Vesicular zinc promotes presynaptic and inhibits postsynaptic long-term potentiation of mossy fiber-CA3 synapse. Neuron 71:1116–1126. 10.1016/j.neuron.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y, Sanford L, Simpson DM, Dowell RD, Palmer AE. 2020. Remodeling of Zn2+ homeostasis upon differentiation of mammary epithelial cells. Metallomics 12:346–362. 10.1039/c9mt00301k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI.00273-21-s0001.pdf, PDF file, 2.6 MB (2.6MB, pdf)
Supplemental material. Download IAI.00273-21-s0002.xlsx, XLSX file, 1.5 MB (1.5MB, xlsx)
Supplemental material. Download IAI.00273-21-s0003.xlsx, XLSX file, 0.01 MB (13.4KB, xlsx)
Supplemental material. Download IAI.00273-21-s0004.xlsx, XLSX file, 0.03 MB (29.5KB, xlsx)
Supplemental material. Download IAI.00273-21-s0005.xlsx, XLSX file, 2.4 MB (2.4MB, xlsx)
Supplemental material. Download IAI.00273-21-s0006.xlsx, XLSX file, 0.05 MB (51.7KB, xlsx)
Data Availability Statement
All raw next-generation sequencing data files and processed data files used to draw conclusions are available at the Gene Expression Omnibus, data series GSE166642.