ABSTRACT
Liver transplantation (LT) is a life-saving strategy for patients with end-stage liver disease, hepatocellular carcinoma, and acute liver failure. LT success can be hampered by several short-term and long-term complications. Among them, bacterial infections, especially those due to multidrug-resistant germs, are particularly frequent, with a prevalence between 19 and 33% in the first 100 days after transplantation. In the last decades, a number of studies have highlighted how the gut microbiota (GM) is involved in several essential functions to ensure intestinal homeostasis, becoming one of the most important virtual metabolic organs. The GM works through different axes with other organs, and the gut-liver axis is among the most relevant and investigated ones. Any alteration or disruption of the GM is defined as dysbiosis. Peculiar phenotypes of GM dysbiosis have been associated with several liver conditions and complications, such as chronic hepatitis, fatty liver disease, cirrhosis, and hepatocellular carcinoma. Moreover, there is growing evidence of the crucial role of the GM in shaping the immune response, both locally and systemically, against pathogens. This paves the way to the manipulation of the GM as a therapeutic instrument to modulate infectious risk and outcome. In this minireview, we provide an overview of the current understanding of the interplay between the gut microbiota and the immune system in liver transplant recipients and the role of the former in infections.
KEYWORDS: Clostridium difficile infections, fecal microbiota transplantation, gut microbial dysbiosis, gut microbiota, human microbiome, immune dysfunction, liver failure, liver immunology, liver transplantation, multidrug resistance
THE SYSTEMIC INFLUENCE OF THE GUT MICROBIOTA, THE RELEVANCE OF LIVER TRANSPLANTATION, AND THE ROLE OF INFECTIONS IN LIVER TRANSPLANTATION
Liver transplantation (LT) is the only curative treatment for patients with end-stage liver disease (ESLD), acute liver failure (ALF), and hepatocellular carcinoma (HCC). LT has revolutionized the prognosis of chronic and ALF, increasing survival rates from 10 to 20% (all causes combined) to 75 to 80% at 1 year and 70% at 5 years (1). In recent years, an extension of indications for LT has been observed, as high as 7,614 procedures performed in Europe in 2017 according to the European Liver Transplant Registry (2). One of the most significant complications of LT is represented by infections, especially those due to bacteria originating from the gastrointestinal (GI) tract, with major bacterial infection rates ranging from 36% to 69% in the posttransplant period (3–5). The importance of the gut microbiota (GM) in liver disease has been highlighted lately, with several works dissecting the so-called gut-liver axis. Intestinal commensals and their product can reach the liver, where they modulate both innate and adaptive immunity, regulate liver inflammation, and promote liver fibrosis, among others (6). Moreover, the prevalence of specific bacterial populations has been associated with the development of bloodstream infections (BSI) in the pre- and posttransplant period (7). LT significantly impacts the GM, with relevant changes in its composition. LT seems able to partially improve the GM diversity and dysbiosis that characterize patients with ESLD, even though residual dysbiosis remains (8–10). Interestingly, dysbiosis after LT seems associated with colonization by multidrug-resistant bacteria, suggesting that procedures aiming to modulate GM composition can have relevant clinical consequences (11). In this review, we provide an overview of the GM in liver disease and its correlation with disease severity. We analyzed the data currently available about GM changes after LT and their impact on the immune system. Finally, we highlighted some manipulation strategies of GM already in place and suggested some future directions for research and therapeutic approaches involving GM in LT recipients.
GUT MICROBIOTA: FEATURES AND FUNCTION
The human microbial ecosystems comprise bacteria, archaea, eukaryotes, and viruses, which colonize different body habitats, including the gut, the skin, the vagina, and the respiratory tract. At the gut level, the total number of prokaryotes located in ecological niches includes 14 × 1013 to 14 × 1014 bacteria, renamed the microbiota; the gut microbiota contains 3 × 106 genes, which is 100 times greater than the number of genes in the human genome (3 × 104). For 1,000 bacterial species, with at least 160 species per individual, the microbiota is renamed the microbiome; although the two terms are different, they are often used interchangeably by the scientific community.
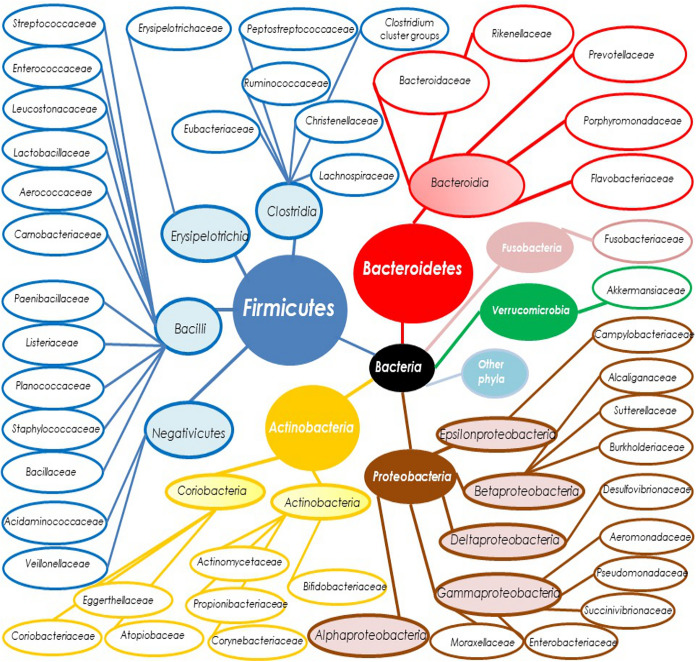
Principal human gut taxa are described in Fig. 1. The human microbiota is individual specific and relatively stable during the adult period but with strong differences according to age, from childhood (higher Clostridium spp. and Bifidobacteria spp.) to elderly age (higher Firmicutes/Bacteroidetes ratio) (12). Moreover, other important factors could influence the diversity of microbiota: both gender and sexual behaviors, body mass index (BMI) (with different microbiota profiles in obese individuals), the kind of diet, the geographic area, and, of course, the use of antibacterial agents or pre- or probiotics. Other conditions that could also modify the composition of microbiota are the development of different gastrointestinal disorders like intestinal inflammatory diseases, metabolic diseases like diabetes, and autoimmune disorders like celiac disease (13).
FIG 1.
Gut microbiota taxa. Gut microbiota composition for the main 6 taxa (at phylum and class level for higher divisions; at family level for lower divisions), which belong to the Bacteria kingdom (black circle); not represented are order, genus, and species taxa. Phyla are represented by big circles, colored backgrounds, and white characters. Classes are presented by smaller circles, colored backgrounds, and black characters. Families are represented by ellipsoid circles, no colored backgrounds, and black characters. Taxa from the phyla Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, and Fusobacteria are represented in blue, red, yellow, brown, green, and turquoise, respectively. “Other phyla” (in sky blue) not represented include Tenericutes, Lentisphaerae, Spirochetes, Synergistetes, and other minor phyla. Taxa from the kingdoms Archaebacteria and Eukaryotes are not represented here.
The gut microbiota serves an essential role in maintaining intestinal homeostasis. Indeed, the vast majority of these bacteria in the colon are strict anaerobes, which are able to perform, essentially, saccharolytic fermentation of nondigestible substrates from dietary fibers. Two of the most important metabolic patterns are gut microbiota related: short-chain fatty acid (SCFA) production and bile acid metabolism, both essential for gut health. Regarding the first aspect, the diet carbohydrates are fermented to SCFAs, i.e., acetate (C2), propionate (C3), and butyrate (C4), in the colon. SCFAs are the major metabolic products of anaerobic fermentation, with total concentrations of 50 to 200 mM in the colon, that can bind specific immunologic receptors and, subsequently, be used as sources of energy, as regulators of gene expression for epithelial integrity, and for immunologic interactions. If acetate is a fermentation product for most gut bacteria, the other two SCFAs, propionate and butyrate, are considered healthy SCFAs, with a different subset of bacterial producers (13, 14). Priopionate is metabolized through two pathways involving specific taxa (mainly Bacteroidetes, Negativicutes class of Firmicutes, and Lachnospiraceae). Conversely, for the final metabolic step toward butyrate, species are distributed among different taxa (Ruminococcaceae, Lachnospiraceae, and other Firmicutes families like Erysipelotrichaceae and Clostridiaceae) (15).
The second significant function of the GM is linked to the secondary bile acid production of lithocholic acid (LCA) and deoxycholic acid (DCA) through 7α-dehydroxylation of the primary bile acids chenodeoxycholic (CDCA) and cholic acid (CA), respectively (16), performed by Clostridium taxa as well as the Eubacterium genus, playing a key role in the enterohepatic circulation of bile acids: secondary bile acids produced by 7α-dehydroxylation are passively absorbed in the large intestine and returned to the liver. Furthermore, with regard to bile metabolism, the gut microbiota is involved in bile salt hydrolases, an activity characterized by several gut microbiota taxa like the genus Bifidobacterium (from the phylum Actinobacteria), the genus Bacteroides (from the phylum Bacteroidetes), and the genera Clostridium, Lactobacillus, and Listeria (from the phylum Firmicutes): it was postulated that this function could play a “detoxification” role. Through these peculiar functions, the gut microbiota plays a significant role both in metabolic function (energy metabolism related to the bile acid metabolites) and anti-infectious activity (antimicrobial effects related to bile acid metabolites, i.e., inhibition of Clostridioides difficile activity performed by secondary bile acids). These specific features determine the concept of the gut-liver axis: an anatomical and functional bidirectional relationship between the gut microbiota and the liver, essentially using portal circulation (17). The alteration of this crucial balance is often associated with both intestinal integrity alterations and gut microbial dysbiosis. It is important to note that dysbiosis represents a change in the numbers of microbes (with an unbalance among taxa) and/or a change in the diversity of the microbiota due to specific diseases. Yet a clear definition of dysbiosis is difficult to provide, because there is disagreement about the term “eubiosis” due to intraindividual differences among distinct body sites and interindividual variations among healthy individuals (18, 19). In the pre-next-generation-sequencing (pre-NGS) era, studies on fecal gut microbial differences were based merely on culture-dependent methods and/or reduced power detection technology through preselected quantitative PCR methods. After the introduction of NGS technology, it was possible to analyze the gut microbial composition through all taxa, from phylum to species, as well as to distinguish some variants among the same species. These results can be analyzed through LEfSe analysis (linear discriminant analysis effect size, an algorithm which emphasizes the differences between two groups) and/or a rough relative abundance comparison at each taxon level. Other appropriate methodologies are related to two different ecological measurements: α- and β-diversity. In humans, α-diversity indexes measure the level of diversity within individual samples: its comparison represents a measure of richness and/or evenness of bacterial taxa. Nevertheless, β-diversity measures the level of diversity (or dissimilarity) between samples, using permutational multivariate analysis of variance (PERMANOVA) analysis. In the end, other methodologies are linked to function studies, instead of composition analyses, from a prediction algorithm (Picrust) to metagenomics and/or metabolomics investigations (20–22).
GUT MICROBIOTA DYSBIOSIS AND CIRRHOSIS DISEASE
End-stage liver disease (ESLD) is a condition characterized by advanced liver disease, decompensated cirrhosis, and liver failure, associated with significant morbidity and mortality. ESLD progressively develops under a continuous inflammatory condition that leads to fibrosis and alteration of liver architecture and function (23). The only available cure is liver transplantation (LT), and the primary disease of LT recipients changed over time. Cirrhosis due to viral infections (hepatitis C virus [HCV], HBV, and HDV), alcoholic liver disease (ALD), and nonalcoholic fatty liver disease (NAFLD) is the most important clinical setting leading to ESLD (24–27). As described by Belli et al. (28), after the advent of direct-acting antivirals (DAA) in 2014, a dramatic decline was observed in the number of liver transplants performed both in patients with decompensated cirrhosis due to HCV and in those with hepatocellular carcinoma associated with HCV, with a 60% and 40% reduction in the need for transplants, respectively. In contrast, transplants for chronic HBV infection have remained stable (even if uncommon) over time in Europe since the introduction of nucleoside analogues (28). The decline in the number of LTs due to HCV infection in Europe and the United States has been outweighed by nonviral causes, particularly non-alcoholic-steatohepatitis (NASH) and ALD (28–30). NAFLD commonly develops in the liver of patients affected by metabolic syndrome: it comprises different clinical manifestations, from simple steatosis to NASH to a more advanced disease (31). Given the rise in obesity and metabolic syndrome in high-income countries, NASH is expected to become the leading indication for LT globally (27, 32) and is now considered one of the growing causes of HCC among patients listed for LT (33).
Each liver disease and stage seem to be characterized by specific gut microbiota dysbiosis profiles; in this review, we will focus on gut microbiota phenotypes linked to cirrhosis and their correlation with severity indexes.
Indeed, cirrhosis represents the final step for LT and plays a crucial role in the pathogenesis of complications, like hepatic encephalopathy (HE), infections, or acute on chronic liver failure (ACLF) as well as spontaneous bacterial peritonitis (SBP) through bacterial translocation and/or their products like lipopolysaccharide (LPS) from Gram-negative bacteria. A considerable number of studies have described gut microbiota alterations in cirrhosis patients compared with healthy controls (Table 1).
TABLE 1.
Gut microbiota dysbiosis during cirrhosis diseasea
| Study | Country | Population | Alpha/beta-diversity | Microbiome modifications based on relative abundance analyses | Correlation of MELD/CTP score and other findings |
|---|---|---|---|---|---|
| Chen et al., 2011 (34) | China | 36 patients with liver cirrhosis (F.sex, 44%; HBV > ALD etiologies; CTP, 8; MELD, 12) –24 healthy controls Material: stool High-throughput 454 pyrosequencing of the 16S rRNA V3 region |
Alpha-diversity (observed, Chao1, Shannon diversity index): no significant reduction compared to controls Beta-diversity: separation according to the unweighted UniFrac index |
Cirrhosis group vs controls: ↓ Bacteroidetes phylum; Lachnospiraceae and Bacteroidaceae families ↑ Proteobacteria and Fusobacteria phyla, Enterobacteriaceae, Veillonellaceae, and Streptococcaceae families Note: ↑ Prevotellaceae in alcoholic subgroup |
CTP + Streptococcaceae − Lachnospiraceae |
| Bajaj et al., 2011 (35) | USA | 25 patients with cirrhosis (F.sex, 8%; ALD > HCV etiologies; MELD, 16) –17 patients with liver cirrhosis + HE (F.sex, 5.8%; MELD, 17) –8 patients with cirrhosis without HE (F.sex, 12.5%; MELD, 12) –10 controls Prospective arm analysis on 7 patients after lactulose withdrawal Material: stool MTPS |
Beta-diversity: according to Bray-Curtis index, a separation between cirrhosis and control groups | Cirrhosis group vs controls: ↓Ruminococcaceae, Lachnospiraceae, and Clostridium incertae sedis XIV ↑ Enterobacteriaceae, Alcaligeneceae, Fusobacteriaceae, Lactobacillaceae, Leuconostocaceae Cirrhosis with HE vs controls: ↑ Enterobacteriaceae, Alcaligenaceae, Lactobacillaceae, Streptococcaceae Cirrhosis with HE vs cirrhosis without HE groups: ↑ Veillonellaceae in HE vs non-HE subgroups Prospective analysis after lactulose withdrawal: ↓Faecalibacterium genus (trend in Veillonella genus) |
MELD Cirrhosis group (HE + non-HE subgroups): + Enterobacteriaceae − Ruminococcaceae (− Prevotellaceae, trend) In non-HE subgroup: − Ruminococcaceae + Veillonellaceae; + Porphyromonadaceae Other findings, inflammation: + Fusobacteriaceae, Veillonellaceae, Enterobacteriaceae − Ruminococcaceae HE subgroup, inflammation: + Leuconostocaceae, Eubacteriaceae, Erysipelotrichaceae, Moraxellaceae, Streptophyta, Streptococcaceae, Fusobacteriaceae, Prevotellaceae Cirrhosis group (and HE subgroup alone), correlation with cognitive impairment + Alcaligeneceae, Porphyromonadaceae |
| Bajaj et al., 2012 (39) | USA | Fecal samples: 60 patients (36 HE, 24 non-HE) –HE subgroup, 36 patients (F.sex, 16.6%; CTP, 9; MELD, 17) –Non-HE subgroup, 24 patients (F.sex, 16.6%; CTP, 6; MELD, 10 –Colonic mucosal (biopsy) samples, 36 patients (19 HE, 17 non-HE) –17 controls Material: stool and rectosigmoid mucosal biopsy specimens MTPS |
Beta-diversity: separation among colonic mucosa groups (controls, HE and non-HE subgroups) according to the Bray Curtis index | Mucosal cirrhosis group (HE and non-HE) vs control mucosa: ↓Dorea, Subdoligranulum, incertae sedis XIV (Clostridia), Lachnospiraceae (other) genera ↑Burkholderia, Ralstonia, Clostridium, Clostridiaceae (other), Proteus, Enterococcus, and Acidaminococcus genera Intracirrhosis group (HE and non-HE) analysis: mucosal vs fecal samples: ↓Roseburia, Blautia, Vibrio, Propionibacterium, Streptomyces, Clostridia incertae sedis XI genera ↑Clostridia incertae sedis XIV, Veillonella, Leuconostoc, Bacteroidales incertae sedis genera Intramucosal cirrhosis analysis: HE vs non-HE: ↓Roseburia genus ↑ Veillonella, Megasphaera, Streptomyces, Bifidobacterium, Enterococcus, Fusobacteriaceae “other”, Burkholderiaceae “other” genera No differences from fecal analyses between patients with rifaximin plus lactulose vs lactulose alone |
MELD and poor cognitive ability, higher inflammation and endothelial activation, mucosal HE subgroup: + Megasphaera, Veillonella, Burkholderia, and Bifidobacterium genera Other findings: less inflammation, endothelial activation, and better cognition, mucosal non-HE subgroup: + Roseburia Other findings: mucosal HE samples vs mucosal non-HE groups: Blautia, Faecalibacterium, Roseburia, Dorea: + good cognition − inflammation Other findings: HE subgroup vs non-HE subgroup comparison, fecal samples: no differences Significant relative abundance differences at mucosal level between patients with rifaximin plus lactulose vs lactulose alone: ↑Propionibacterium ↓Blautia, Roseburia, Alistipes, Veillonellaceae “other” |
| Bajaj et al., 2013 (41) | USA | 219 cirrhotic patients (F.sex, 80.3%; HCV > ALD > NASH) –121 compensated outpatients (F.sex, 76%; MELD, 9.5) –54 decompensated outpatients (F.sex, 74%; MELD, 14.2) –44 inpatients (F.sex, 70%; MELD, 19.4) –25 controls Material: stool MTPS |
Beta-diversity: according to Bray-Curtis index, cluster separation between controls vs all groups (and far from inpatients with infection) | Cirrhotic groups vs controls: ↓Clostridiales XIV taxa, Ruminococcaceae, Veillonellaceae, and Lachnospiraceae families ↑ Enterococcaceae, Staphylococcaceae, and Enterobacteriaceae families (trend Leuconostocaceae) Patients with worsening liver disease vs controls: ↓Veillonellaceae and Porphyromonadaceae |
MELD − Clostridiales XIV, Lachnospiraceae, Ruminococcaceae, Rikenellaceae + Staphylococcaceae, Enterococcaeae, Enterobacteriaceae Other findings: + CDR: a ratio of Lachnospiraceae/Ruminococcaceae/Clostridiales incertae sedis XIV to Enterobacteroidaceae/Bacteroidaceae (a low no. indicates dysbiosis) |
| Qin et al., 2014 (44) | China | Discovery phase: –98 cirrhosis patients (F.sex, 33%) –83 controls Validation phase: –25 cirrhosis patients (F.sex, 36%) –31 controls Material: stool Quantitative metagenomics metaHIT |
Cirrhosis group vs controls: ↓ Bacteroidetes phylum, Bacteroides, Eubacterium, Alistipes, Faecalibacterium, Roseburia, Parabacteroides, Odoribacter, Ruminococcus, Dorea, Bilophila, Coprococcus, Tannerella, Subdoligranulum, Holdemania, and Phascolarctobacterium genera ↑Proteobacteria and Fusobacteria phyla, Veillonella, Streptococcus, Prevotella, Haemophilus, Lactobacillus, Fusobacterium, and Megasphaera genera |
CTP Significant differences between highest and lowest patient-enriched species abundance according to CTP score |
|
| Kakiyama et al., 2013 (37) | USA | 47 cirrhosis patients (F.sex, 21.2%; HCV > ALD etiologies; MELD, 12) –24 advanced cirrhosis patients (CTP class B or C; MELD, 8) –23 early cirrhosis patients (MELD, 16) –14 controls Prospective analysis on 6 patients before and after 8 wks of rifaximin Material: stool MTPS |
Cirrhosis group (early and advanced cirrhosis): ↓ Ruminococcaceae, Lachnospiraceae, Rikenellaceae (more specifically, Blautia genus for Lachnospiraceae family) especially in advanced cirrhosis ↑ Enterobacteriaceae, Veillonellaceae Prospective study: ↓Veillonellaceae family |
Other findings, primary BA, CDCA: + Enterobacteriaceae − Bacteroidaceae Secondary BA, DCA: + Ruminococcaceae DCA/CA ratio: + Ruminococcaceae LCA/CDCA ratio: + Blautia |
|
| Chen et al., 2015 (40) | China | 79 patients with ACLF (F.sex, 21.5%; MELD, 22; CTP, 10; HBV > ALD > HCV) –Survivor group: 43 patients (F.sex, 18.6%; MELD, 19.6; CTP, 10.2) –Nonsurvivor group: 31 patients (F.sex, 22.5%; MELD, 24.7; CTP, 11) Prospective study: 42 patients during 4 wks of follow-up –50 controls Material: stool Illumina 16S rRNA gene V4 |
Alpha-diversity (observed, Chao1, Shannon, and Simpson indexes): ↓alpha-diversity indexes, both richness and evenness indexes, in ACLF vs controls Beta-diversity (unweighted UniFrac index): difference between the ACLF group and controls |
ALCF group vs controls: ↓Bacteroidetes phylum; Bacteroidia class; Bacteroidaceae, Ruminococcaceae, Lachnospiraceae, and Porphyromonadaceae families ↑Proteobacteria and Firmicutes phyla; Bacilli and Gammaproteobacteria classes; Veillonellaceae, Pasteurellaceae, Streptococcaceae, and Enterococcaceae families HE subgroup: ↓ Lachnospiraceae vs non-HE patients |
MELD: higher MELD score and higher Pasteurellaceae family prevalence: significant death hazard (univariate logistic regression) Pasteurellaceae and MELD: independent factors predicting mortality rate (multivariate logistic regression) Other findings: Stability during longitudinal substudy during 4 wks: Gammaprotebacteria class and Pasteurellaceae family higher in nonsurvival patients Lachnospiraceae family lower in nonsurvival patients |
| Ahluwalia et al., 2016 (38) | USA | 147 cirrhotic patients (F.sex, 57.8%; HCV> ALD > NASH etiologies) –85 patients with HE on lactulose and rifaximin use (F.sex, 81%; MELD, 15.5) –62 patients without a previous HE event (F.sex, 25.8%; MELD, 11) –40 controls Material: stool MTPS |
Beta-diversity: reported UniFrac separation among groups | Cirrhotic vs control groups: ↓Bifidobacteriaceae, Incertae sedis XI (Clostridia), incertae sedis XIV (Clostridia), Lachnospiraceae, Peptostreptococcaceae, Ruminococcaceae, Erysipelotrichaceae HE subgroup vs non-HE: ↓ Lachnospiraceae ↑Aerococcaceae |
Other findings, HE subgroup vs controls: worse cognitive performance, systemic inflammation, dysbiosis, and hyperammonemia –correlation between Lachnospiraceae, Ruminococcaceae, and Clostridiales XIV taxa and brain glial MRS manifestations of ammonia + correlations between Streptococcaceae, Enterobacteriaceae, Lactobacillaceae, and Peptostreptococcaceae taxa and ammonia, MELD score, and brain MRS manifestations |
| Santiago et al., 2016 (42) | Spain | 27 cirrhotic patients (F.sex, 29.6%; ALD > virus etiologies) –13 with ascites (F.sex, 31%; MELD, 11.6; CTP, 7.7) –14 without ascites (F.sex, 28%; MELD, 9.5; CTP, 6.1) –17 controls Material: stool Illumina V4 region of the 16S rRNA gene |
Alpha-diversity: ↑ in controls compared to cirrhotic patients, according to the Chao1 index; no differences between ascites and no-ascites subgroups Beta-diversity: unweighted UniFrac and weighted UniFrac separation between cirrhosis group and controls |
Cirrhosis group (with and without ascites) vs controls: ↓Clostridiales “unknown,” Roseburia faecis, Alistipes putredinis species, Oscillospira and Dehalobacterium genera, Mogibacteriaceae family ↑Peptostreptococcaceae family Ascites group vs controls: ↓Clostridiales order, Ruminococcaceae and Peptostreptococcaceae families, Roseburia faecis and Alistipes putredinis species ↑Veillonella dispar |
Other findings, cirrhotic ascites patients + LPB serum |
| Bajai et al., 2017 (9) | USA | 45 cirrhotic patients (pre-LT, HCV >NAFLD > ALD; MELD, 26) –45 controls Material: stool Multitag sequencing V1-V2 16s rRNA Prospective study: see Table 2 |
Alpha-diversity: ↑Chao1 in controls vs. patients pre-LT |
Cirrhotic group vs controls: ↓ Lachnospiraceae, Ruminococcaceae ↓ Clostridiales cluster XIV, Streptococcaceae, Synergisticeae, Desulfovibrionaceae ↑Bacteroidaceae, Enterobacteriaceae (especially Klebsiella and Enterobacter genera), Lactobacillaceae, Veillonellacae |
See Table 2 |
ALD, alcoholic liver disease; HBV, hepatitis B virus; CTP, Child-Turcotte-Pugh index; MELD, model for end-stage liver disease; HE, hepatic encephalopathy; HCV, hepatitis C virus; MTPS, multitag pyrosequencing; F.sex, female sex; NASH, nonalcoholic steatohepatitis; BA, bile acids; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; CDR, cirrhosis dysbiosis ratio; LT, liver transplantation; ACLF, acute-on-chronic liver failure; LPB, lipopolysaccharide-binding protein; MRS, magnetic resonance spectroscopy; upward arrows (↑), increase; downward arrows (↓), decrease. In “Correlation of MELD/CTP score and other findings” column, + means positive correlation, and − means negative correlation.
Chen et al. (34) described for the first time a distinct gut microbiota pattern, showing in patients with cirrhosis a trend in reduction in α-diversity according to the Shannon index. Interestingly, β-diversity analysis described a separation between cirrhotic patients and controls, according to the unweighted UniFrac index. The relative abundance analyses showed a different gut microbiota core characterized by an unbalance in both phylum (i.e., a decrease in Bacteroidetes and an increase in Proteobacteria-Fusobacteria) and family taxa (increases in Enterobacteriaceae, Veillonellaceae, and Streptococcaceae and decreases in Lachnospiraceae family and Bacteroidaceae taxa). In parallel, Bajaj et al. in 2011 (35) observed an increase in Enterobacteriaceae and Fusobacteriaceae families and also Alcaligenaceae, Leuconostocaceae, and Lactobacillaceae, as well as a reduction in Ruminococcaceae, Lachnospiraceae, and Clostridium incertae sedis XIV in the cirrhosis group (with and without HE) in comparison to controls. These findings were consistent with the results of Chen et al. although vaster in number of taxa involved. These significant differences occurred across all taxa, from phylum level to genus level, suggesting that a deep gut microbiota dysbiosis exists in the cirrhotic setting, irrespective of the underlying etiology of liver disease. An exception could be ALD, where a limited influence on gut microbial dysbiosis seems to be performed by this pathology through the involvement of Prevotellaceae in some analyses (34) or the Veillonellaceae-rich and Bacteroidaceae-Porphyromonadaceae-poor profile in others (36).
Interestingly, in another work, Kakiyama et al. in 2013 (37) observed a higher relative abundance of Enterobacteriaceae and Veillonellaceae taxa and a lower relative abundance of Ruminococcaceae, Lachnospiraceae, and Rikenellaceae taxa (more specifically, the genus Blautia of the Lachnospiraceae family) in the cirrhotic group than in the controls; however, these findings were even stronger in advanced cirrhosis according to the Child-Turcotte-Pugh (CTP) score (i.e., classes B and C [see below]). Interestingly, in this work, a positive correlation between Ruminococcaceae and the DCA/CA ratio as well as between Blautia (a 7α-dehydroxylating bacterium) and the LCA/CDCA ratio has been found. From these data, we could infer that a reduction in conversion of primary to secondary bile acids is associated with an abundance of key gut taxa in decompensated cirrhosis. As a conclusion, from these works we can argue that gut microbial dysbiosis during cirrhosis is unbalanced, essentially characterized by a reduction in Firmicutes taxa involved in several metabolic functions (Clostridium incertae sedis XIV, Lachnospiraceae, Ruminococcaceae) in association with an increase in Proteobacteria (essentially Enterobacteriaceae) taxa, often rich in potential pathogens, irrespective of the etiology of cirrhosis, with a peculiar involvement of bile acid metabolism. Other peculiar aspects have yet to be investigated to better understand this complex scenario.
GUT MICROBIOTA DYSBIOSIS AND COMPLICATIONS OF CHRONIC LIVER DISEASE
Another important aspect worth mentioning consists of the link between GM variation and the development of complications in cirrhotic patients. Since their early work, Bajaj et al. (35) have shown how HE, a frequent complication of cirrhosis, is characterized by a GM alteration similar, but not equal, to that in healthy controls. In this study, patients in the HE subgroup had a higher prevalence of Veillonellaceae than the group of cirrhosis patients without HE. In contrast, Enterobacteriaceae, Alcaligenaceae, Lactobacillaceae, and Streptococcaceae were more highly represented in the GM of HE patients than in that of controls. Interestingly, a different dysbiosis profile was reported by Ahluwalia et al. (38), who described a higher expression of Aerococcaceae and a lower representation of Lachnospiraceae in the HE subgroup than in the subgroup without HE.
In line with the previous study, Bajaj et al. (39) did not report substantial differences in fecal gut microbiota between the HE group of cirrhotic patients and the non-HE group of cirrhotic patients, corroborating the idea that during cirrhosis, even in non-HE patients, the gut alterations have begun. Consistent with this, in another work, Chen et al. in 2015 (40) identified a similar pattern, with a reduction only in the Lachnospiraceae family in the HE subgroup of patients compared to the non-HE patients, always from fecal samples.
Despite differences that could be related to the material investigated (16S DNA versus 16S RNA analyses), the etiologies (viral and ALD etiology but not the NAFLD group), and the study site (mucosal versus fecal samples), Bajaj et al. at the end of 2013 (41) also showed how fecal gut microbiota remained unchanged during the follow-up period in cirrhotic patients with no complications. In contrast, when a complication occurs, the gut microbiota could change, with an increase in dysbiosis, which is also expressed by the cirrhosis/dysbiosis ratio (CDR) value, a ratio of “autochthonous” to “no-autochthonous” taxa (Lachnospiraceae plus Ruminococcaceae plus Clostridiales incertae sedis XIV/Enterobacteriaceae plus Bacteroidaceae), irrespective of the introduction of confounding factors like the start of lactulose therapy or the onset of antibacterial agent treatment.
What occurs during the transition from a compensated cirrhosis setting (without complications) to a decompensated cirrhosis setting (with or without complications) remains unknown. In the same study (41), Bajaj et al. showed how fecal gut microbiota differs between compensated and decompensated cirrhosis. For the first subgroup (compensated cirrhosis), they confirmed a typical cirrhotic pattern with a Ruminococcaceae-, Clostridiales incertae sedis XIV-, and Veillonellaceae-poor profile and a Porphyromonadaceae-, Bacteroidaceae-, and Enterobacteriaceae-rich profile, while in the second subgroup (decompensated cirrhosis), an increase in the families Enterobacteriaceae, Enterococcaceae, and Staphylococcaceae associated with a decrease in the family Bacteroidaceae was observed, suggesting that a specific role exists for these taxa in the progression to end-stage liver disease.
Another significant complication of cirrhotic disease is the development of ACLF. Chen et al. in 2015 (40), using Illumina technology, described deep gut microbiota alterations during ACLF consisting of a decrease in Bacteroidaceae, Ruminococcaceae, Porphyromonadaceae, and Lachnospiraceae taxa and an increase in Veillonellaceae, Pasteurellaceae, Streptococcaceae, and Enterococcaceae in the ACLF group compared to controls. In this setting, profound alterations in the GM were observed through the α-diversity indexes for both richness and evenness, as well as a β-diversity separation between the ACFL group and controls.
With regard to SBP, an interesting work by Santiago et al. (42) in 2016 confirmed a reduced α-diversity (according to the Chao1 index) in the cirrhotic group compared to controls and a β-diversity separation (according to the UniFrac indexes) between patients with ascites and patients without ascites. However, they did not see significant differences in intracirrhotic group analysis (patients with ascites versus patients without ascites) in α- and β-diversity analyses, but they found a stronger significant difference in relative abundance analysis, with enrichment in Veillonella dispar and a reduction in Ruminococcaceae, Clostridiales, Peptostreptococcaceae, Roseburia faecis, and Alistipes putredinis taxa. Moreover, a higher level of plasmatic LBP (LPS binding protein), a marker of microbial translocation, was detected in patients with cirrhosis complicated by ascites, suggesting that a potential role of gut microbial dysbiosis also exists in this setting of this complication.
SEVERAL KEY POINTS MUST BE CONSIDERED FOR THE ANALYSIS OF GUT MICROBIOTA IN THIS FIELD
First, it is noteworthy to consider the concomitant treatment taken, in order to have correct data analysis and comparison between studies. In fact, as previously described for instances in which patients are administered omeprazole (Bajaj et al., 2014 [43]), some differences in microbiota composition may be extremely accentuated.
In addition, other differences in GM results also depend on the methodology used. In fact, metagenomic analyses may have higher accuracy and resolution than 16S-related NGS technology (44).
Lastly, some studies highlighted a difference between GM analyzed from intestinal mucosal biopsy specimens and GM analyzed from feces (39, 44, 45). Indeed, lower Roseburia abundance and higher Enterococcus, Veillonella, Megasphaera, and Burkholderia abundance were found in mucosal samples than in fecal samples in both non-HE and HE groups (39). Further analyses are needed to address these research questions.
GUT MICROBIAL DYSBIOSIS AND DISEASE SEVERITY INDEXES
Prognostic models are useful for estimating disease severity and survival prediction and can serve as helpful medical decision-making tools for guiding patient care. Two models, the Child-Turcotte-Pugh (CTP) classification and the model for end-stage liver disease (MELD), are commonly used in classifying and caring for patients with chronic liver disease (CLD). The CTP classification has been used to assess the risk of nonshunt operations in patients with cirrhosis. It includes parameters like serum albumin, bilirubin, prothrombin time (international normalized ratio [INR]), and presence of ascites and encephalopathy, with a score ranging from 5 to 15 in 3 classes: CTP class A (well-compensated cirrhosis) with a score between 5 and 6, CTP class B (significant functional deterioration) with a score between 7 and 9, and CTP class C (decompensated cirrhosis) with a score between 10 and 15. These categories were generated to predict mortality rate in patients with cirrhosis undergoing abdominal surgery, but in several studies they showed a good correlation with survival in nonsurgical settings (46, 47).
MELD was originally developed to predict survival following elective placement of a transjugular intrahepatic portosystemic shunt (TIPS) (48). The model was subsequently validated as a predictor of survival in several cohorts of patients, including patients on the waiting list for LT, hospitalized patients with liver decompensation, ambulatory patients with noncholestatic liver disease, and patients with primary biliary cholangitis. MELD incorporates three laboratory values: INR, serum creatinine, and serum bilirubin.
Implementation of MELD led to a reduction in the number of patients entering the liver transplant waiting list due to its accuracy in prediction of short-term mortality in the vast majority of wait-listed candidates (49).
Furthermore, it has been shown to be an accurate predictor of survival in several settings, such as a predictor of long-term mortality in patients with compensated cirrhosis and in patients with chronic hepatitis B (with or without cirrhosis), as well as in patients with liver complications (50, 51).
Both models, MELD and CTP classification, have been shown to directly correlate with reduced microbial diversity (52); furthermore, a reduced Bifidobacterium/Enterococcus ratio was significantly associated with an increased MELD score (Table 1). Regarding relative abundance analyses, Alcaligenaceae and Porphyromonadaceae taxa (35) were positively correlated with cognitive impairment in both cirrhotic patients and HE patients (35). Furthermore, during cirrhosis (with and without HE), the MELD score positively correlated with Enterobacteriaceae and negatively with Ruminococcaceae with a trend toward lower Prevotellaceae abundance; in parallel, in cirrhosis patients without HE, the MELD score positively correlated with Veillonellaceae and Porphyromonadaceae and negatively correlated with Ruminococcaceae, suggesting that the microbiota composition during cirrhosis is not a simple epiphenomenon but a complex system that could play a significant role in the scenario (35). Interestingly, in another work (41), the MELD score positively correlated with Staphylococcaceae, Enterococcaceae, and Enterobacteriaceae and negatively correlated with Clostridiales incertae sedis XIV, Lachnospiraceae, Ruminococcaceae, and Rikenellaceae. With the exception of Ruminococcaceae, hospitalized and decompensated patients with a higher median MELD score showed distinct gut microbiota core involvement, suggesting that these taxa might play a role in the development of complications too. In the end, the study of Chen et al. (40) showed that a higher MELD score and a higher abundance of the family Pasteurellaceae were significantly associated with a higher risk of death, both resulting as independent factors able to predict mortality by multivariate logistic regression.
In line with these data, the CTP score positively correlated with Streptococcaceae and was negatively associated with Lachnospiraceae members (34). In conclusion, these results seem to be consistent with MELD data and corroborate the theory that gut microbial dysbiosis is strongly interconnected with clinical outcomes.
THE GUT-LIVER AXIS AND ITS IMPACT ON THE IMMUNE RESPONSE TO INFECTIONS AFTER LT
Infections are a major cause of death in patients following LT, with a Canadian cohort estimating that 19% of deaths occurring in a 5-year period after LT were due to infections. Predominant infections are intra-abdominal infections, primary bacteremia, and pneumonia, with coagulase-negative staphylococci, Enterococcus spp., and Escherichia coli being the most frequently identified microorganisms (11). Regarding fungi, LT recipients after invasive fungal infection (IFI) had the lowest survival rate in comparison with recipients of other types of solid organ transplants, with Candida spp. the most frequently identified microorganism (53). Higher MELD scores before LT, the type of biliary anastomosis, presence of biliary complications, and pretransplant infections were identified as independently associated with a higher risk for infections after living-donor LT (54). Other relevant factors are colonization by multidrug-resistant bacteria (MDRB) and degree of immunosuppression after LT (55).
The liver is constantly exposed to bacterial products of gut microbiome origin because of the direct anatomical connection via the portal venous circulation and the biliary tree. Intestinal commensals and their products were repeatedly reported to translocate from the intestinal lumen to the liver, where they may impact the immune responses (6). An alteration of the immune response, defined as cirrhosis-associated immune dysfunction syndrome (CAIDS), is a well described feature in patients with CLD and leads to a decreased ability to clear cytokines, bacteria, and LPS from the circulation. Monocyte spreading, chemotaxis, bacterial phagocytosis, and bacterial killing are significantly reduced in cirrhotic patients compared with controls. Patients with ACLF have reduced expression of the antigen-presenting HLA-DR molecules on monocytes, which may result in decreased monocyte activation and cytokine secretion. In addition to reticuloendothelial system dysfunction, patients with cirrhosis have decreased neutrophil mobilization and phagocytic activity, a phenomenon that correlates with the severity of liver disease (56). In both humans and mice, microbial translocation induces type I interferon production in the liver, leading myeloid cells to produce interleukin-10 (IL-10), which consequently depresses, with negative feedback, their response to antigen stimulation and leads to a loss of infection control and to infection-associated mortality (57). Lymphocytes are also affected; indeed, in cirrhosis, peripheral CD27+ memory B cells exhibit increased sensitivity to Fas-induced apoptosis in an activation-dependent manner to which bacterial LPS contributes, which is associated with a reduced frequency of circulating memory B cells (58).
Currently, there is a paucity of data on how the gut-liver axis changes over the LT pathway and its impact on the immune response to infections. Wu et al. (77) observed high levels of plasma endotoxin and IL-6 among cirrhotic patients, and the values of these molecules were correlated with peculiar phenotypes of the GM. In that study, LT was able to partially restore these alterations detected in the GM and substantially reduced both plasma endotoxin and IL-6 levels, which have been directly associated with the development of infection in the posttransplant period (59). Therefore, it is possible to speculate that the restoration of eubiosis after LT can also reduce the risk of infections through the reduction of microbial translocation and consequent inflammation (Fig. 2). Overall, despite being a promising field, only a few data concerning the immune response modulation by the GM after LT are currently available, supporting the need for further research in this area.
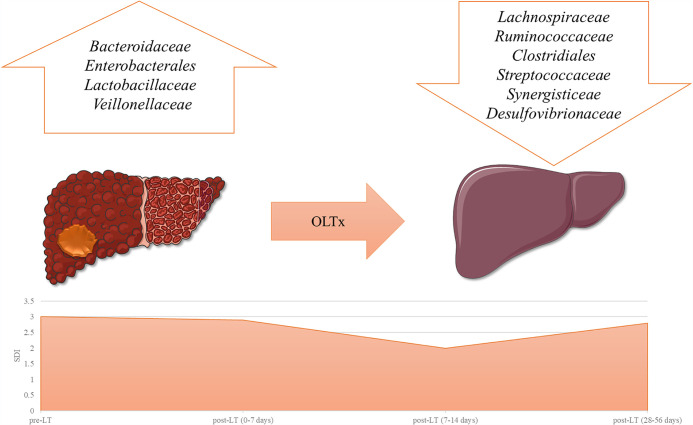
FIG 2.
Model of gut microbiota dysbiosis changes after LT. The figure is a representation of the main alterations occurring after LT, inspired by a report by Kato et al. (7). Top, relative abundance of taxa involved during liver disease. Bottom, an α-diversity index, represented by the SDI (Shannon diversity index) trend between pre-LT and post-LT during a 56-day period of observation, with an initial reduction during the first 2 weeks, followed by a partial subsequent recovery. OLTx, orthotopic liver transplantation.
HOW GUT MICROBIOTA CHANGES AFTER LT: EVIDENCE FROM THE LITERATURE AND DYSBIOSIS AND ITS ASSOCIATION WITH MULTIDRUG-RESISTANT BACTERIA
To date, scarce evidence from a few longitudinal studies is available on how the gut microbiota changes after LT (Table 2). In a study enrolling 45 subjects, Bajaj et al. (9) assessed the characteristics and changes in the intestinal microbiome in patients with cirrhosis, before and after LT. GM diversity, analyzed using fecal samples, was found to be lower in patients undergoing LT than in healthy controls and to increase significantly within 6 months after LT. Interestingly, LT was also associated in 7 of 10 patients with an improvement in cognitive function. These findings were associated with changes in microbial composition, with a relative increase in Ruminococcaceae and Lachnospiraceae and a decrease in Enterobacteriaceae after LT. Similar distributions of taxa were also found in patients with improved cognitive function.
TABLE 2.
Gut microbiota changes after LTa
| Study | Location | Population | Material | Timeline | Major findings | Summary of microbial changes |
|---|---|---|---|---|---|---|
| Bajaj et al., 2017 (9) | USA | 45 patients with cirrhosis listed for LT | Stool | Samples acquired at enrollment (pre-LT) and 6 mo post-LT (7 ± 2 mo) | Shannon microbial diversity index significantly higher post-LT than pre-LT Significantly lower diversity in pre-LT patients than healthy controls, improvement in diversity within 6 mo post-LT Improved cognitive function (according to PHES) post-LT in 71% of patients |
Pre-LT vs post-LT: ↑ Ruminococcaceae, Lachnospiraceae ↓ Enterobacteriaceae (E. coli, Shigella, Salmonella) Patients with improved cognitive function post-LT: ↑ Lachnospiraceae ↓ Enterobacteriaceae |
| Kato et al., 2017 (7) | Japan | 38 patients undergoing LT | Stool | Samples acquired within 2 wks before LT and every 7 days after LT during the first and second mo. Additional samples were acquired when adverse events, such as fever, diarrhea, reoperation, ACR, and infection, occurred |
Shannon microbial diversity index initially decreased (first 3 wks, but no significant changes during postoperative days 0 to 7) and then gradually increased over the course of post-LT Parallel trend according to the observed, Chao1, and whole-tree analysis (PD) indexes Significantly lower diversity in patients with high MELD and CTP scores pre-LT Significantly lower diversity in post-LT patients who experienced ACR Significantly lower diversity in post-LT patients who experienced BSI |
Patients with high pre-LT MELD: ↑ Enterobacteriaceae, Enterococcaceae ↓ Bacteroidaceae, Ruminococcaceae, Lachnospiraceae Patients with high pre-LT CPT score: ↑ Enterococcaceae, Lactobacillaceae ↓ Bacteroidaceae, Ruminococcaceae, Lachnospiraceae ACR patients: ↑ Bacteroidaceae, Enterobacteriaceae, Streptococcaceae, Bifidobacteriaceae ↓ Lachnospiraceae |
| Sun et al., 2017 (10) | China | 24 subjects (9 patients with end-stage liver disease undergoing LT, 15 healthy controls) | Stool | Samples acquired at enrollment (pre-LT) and 3 mo after LT | Different gut microbiota (based on OTU composition) in pre-LT patients and controls Significant difference in gut microbiota pre-LT and post-LT in the same patient No significant difference in gut microbiota post-LT and in controls Metabolic pathway analysis: 15 functional models were enriched and 21 functional modules were less represented in post-LT than pre-LT |
Pre-LT vs post-LT: ↑ Akkermansia, Micromonosporaceae, Desulfobacterales, Eubacteriaceae, Sarcina, Corynebacterium spp., Chitinophagaceae, Coriobacteriaceae ↓ Enterobacteriaceae (Actinobacillus, Escherichia, Shigella), Anaerolineaceae, Fusobacteriales, Clostridium, Fusobacteriaceae, Aeromonas |
| Annavajhala et al., 2019 (11) | USA | 177 patients undergoing LT | Stool | Samples acquired at enrollment (pre-LT), every 7 days during transplant hospitalization, and 2, 3, 6, 9, and 12 mo after LT | ARLD patients and patients with MELD of >18 or CTP class C score showed the lowest α-diversity pre-LT (both Shannon and Chao diversity indexes) MDRB colonization associated with reduction in α-diversity (VRE-colonized patients showing the most drastic reduction) and altered β-diversity; low pre-LT α-diversity significant marker for CRE colonization post-LT Effect of antibiotic exposure dependent on antibiotic class: exposure to group 2 β-lactams (3GCS, first-generation β-lactamase inhibitor combinations), glycopeptides, carbapenems, and fluoroquinolones associated with greater reduction of α-diversity Altered gut microbiome even 2 wks after the most recent antibiotic course Post-LT complications (i.e., postoperative bleeding within 1 wk of transplant, bile leak, and biliary stricture) independently associated with decreased microbiome diversity |
Peri-LT (wk 1 to 3): ↑ Clostridiales, Streptococcus, Enterococcus, Lactobacillus zeae Early post-LT (mo 1 to 3): ↑ Clostridiales ↓ Lactobacillus, Bacterioides Late post-LT (mo 6 to 12): ↑ Ruminococcus bromii, Butyricicoccus pullicaecorum, Blautia producta ↓ Enterococcus, Streptococcus Microbial signature of MDRB colonization: ↑ Enterococcus, Klebsiella, Blautia obeum, Dorea formicigenerans ↓ Bacteroides, Lachnospiraceae, Faecalibacterium Microbial signature of non-MDRB-colonized patients: ↑ Faecalibacterium prausnitzii, Bacterioides, Bifidobacterium, Parabacteroides distasonis, Prevotella copri, Prevotella stercorea Pre-LT marker of disease severity: ↑ Enterococcus casselflavus, Lactobacillus zeae, Enterococcus, Enterobacteriaceae ↓ F. prausnitzii, Ruminococcaceae, Lachnospiraceae, Bacteroidaceae |
LT, liver transplant; ACR, early-phase acute cellular rejection; BSI, bloodstream infection; PHES, psychometric hepatic encephalopathy score; OTU, operational taxonomic units; CRE, carbapenem-resistant Enterobacteriaceae; VRE, vancomycin-resistant enterococci; Ceph-RE, Enterobacteriaceae resistant to third-generation cephalosporins; 3GCS, third-generation cephalosporins; FU, follow-up; ARLD, alcohol-related liver disease; MELD, model for end-stage liver disease; CPS, Child-Pugh score; MDRB, multidrug-resistant bacteria; upward arrows (↑), increase; downward arrows (↓), decrease.
In a similar study, Kato et al. (7) analyzed the gut microbiota of 38 patients undergoing LT. They showed an increase in the microbial diversity index after LT, with a statistically significant increase after the first 7 days post-LT. Moreover, the authors associated a compromised pre-LT hepatic function, assessed by MELD and CTP scores, with a microbial population characterized by a prevalence of Enterobacteriaceae and Enterococcaceae over Ruminococcaceae and Lachnospiraceae. Lower diversity after LT was also significantly associated with postoperative complications such as acute graft rejection and bloodstream infections.
Changes in gut microbiota diversity after LT were also confirmed by Sun et al. (10), who showed a significant difference in microbial composition in the same patients before and after LT, with the post-LT group demonstrating a profile similar to that of healthy controls. Additionally, analysis of metabolic pathways revealed how LT modified 36 functional models (either by enriching or reducing their activity), suggesting that more complex variation is present in the gut ecosystem caused by the procedure.
A large study on 177 patients undergoing LT (11) analyzed GM changes for a period up to 1 year after LT. In addition to confirming previous data on the impact of (high) MELD and CPS scores on (low) pre-LT diversity, the authors found certain gut microbial profiles (decreased presence of the genus Bacteroides and the family Lachnospiraceae and prevalence of the genus Enterococcus) to be associated with an increased risk of post-LT colonization by multidrug-resistant bacteria (MDRB). A decreased pre-LT microbial diversity was significantly linked to MDRB colonization (particularly with vancomycin-resistant enterococci) and to post-LT complications, such as postoperative bleeding, bile leak, and strictures. Of note, exposure to group 2 β-lactams (third-generation cephalosporins and first-generation β-lactamase inhibitor combinations), glycopeptides, carbapenems, and fluoroquinolones was associated with a greater reduction in α-diversity and with the GM alteration persisting even more than 2 weeks after the most recent antibiotic course.
The current knowledge on how the gut microbiota is shaped by LT and its potential role in postoperative complications (namely, MDRB infections) is still limited, and the need for further studies is of utmost importance. A better understanding of these aspects can provide new tools for early recognition of patients at higher risk of developing infections, in addition to other clinically adopted models (60). Moreover, greater expertise regarding the composition of a healthy gut microbiome and of the relationships within the different microbial populations can guide a “tailor-made” modulation, aimed at favoring specific protective taxa.
MANIPULATION OF GUT MICROBIOTA: STRATEGIES AGAINST MDR GRAM-NEGATIVE BACTERIA AND CLOSTRIDIOIDES IN LT
Disruption of normal gut microbiota, known as dysbiosis, can impair host health and cause loss of protection against potentially pathogenic bacterial colonization.
Interesting data are available in the literature about the therapeutic role of gut microbiota manipulation. The two main fields in which it has been studied are the treatment of Clostridioides difficile infection (CDI) and multidrug-resistant bacterial colonization.
In patients with multiple recurrences of CDI, per the latest guidelines of the Infectious Diseases Society of America (IDSA) and the Society for Healthcare Epidemiology of America (SHEA) (61), fecal microbiota transplantation (FMT) is a recommended therapeutic option when appropriate antibiotic treatments have failed.
CDI is a known complication following solid organ transplantation (SOT), with an estimated risk up to 5 times higher than that of the general population (62), and is linked to increased morbidity and mortality (63). Liver transplant recipients are among the populations at higher risk, with a prevalence of CDI estimated at 9.1% (64). CDI prevalence in SOT recipients is highest within the first 3 months after transplantation, when patients are exposed to well-known risk factors such as the start of immunosuppressive therapy (generally at the highest dose), prolonged hospitalization, and antibiotic exposure. Moreover, SOT has also been established as a risk factor for CDI recurrence. Very few data have been gathered about FMT as a treatment for CDI in LT recipients. Schneider et al. (65) reported successful FMT in a patient with therapy-refractory CDI undergoing liver transplantation, with no safety issues and a good clinical response at follow-up evaluations up to 8 weeks after the procedure.
A single-center retrospective study on FMT to treat CDI recurrences in a population comprising SOT recipients described its usage in one patient undergoing kidney and liver transplantation (66). No adverse events were reported, and clinical success was achieved, as the patients remained symptom-free for 8 weeks post-FMT. Furthermore, Bilal et al. (67) reported the case of a liver and kidney transplant recipient with a history of recurrent CDI treated with FMT, with resolution of symptoms accomplished and no further recurrences for the following 6 months of clinical follow-up.
A multicenter retrospective series including 75 adult and 5 pediatric patients treated with FMT for recurrent, refractory, and severe CDI, including 19 SOT recipients, showed an overall cure rate of 89%. Of note, no disaggregated data were available to assess the impact of FMT on SOT patients, and 15% of the subjects had a severe adverse event (SAE) within 12 weeks after FMT. There were no related infectious complications, but several procedure-related complications occurred, such as aspiration during sedation for FMT and superficial mucosal tear caused by the colonoscopy performed to administer FMT (68).
In a larger multicenter retrospective study enrolling 94 SOT patients treated with FMT for recurrent, severe, or fulminant CDI (69), the cure rate at 1 month after the first FMT was 63.8%, while the overall cure rate was 91.3% when additional FMTs and CDI antibiotic treatments were included. Predictors of failure of a single FMT included inpatient status, severe and fulminant CDI, presence of pseudomembranous colitis, and use of non-CDI antibiotics at the time of FMT. Additionally, the study confirmed the good safety profile of FMT in SOT patients, with both nonserious and serious adverse events occurring at rates comparable to those reported in the immunocompetent population (70), and no infectious complications, including bacteremia, related to the fecal transplant material.
A meta-analysis of 44 studies investigated the efficacy of treating CDI and the safety of FMT in 303 immunocompromised patients (71): the pooled success rate was estimated at 88% and 93% with single and multiple FMTs, respectively, and rates of serious adverse events similar to those of immunocompetent patients were reported.
Even if this evidence is encouraging, further studies are needed to assess the efficacy and safety of FMT in cases of CDI recurrences, given its potential therapeutic usefulness in fragile patients, such as SOT recipients, that are exposed to multiple risk factors for CDI (i.e., antibiotic exposure, surgical procedures, prolonged hospitalization).
Few data have been gathered on the use of FMT for MDR Gram-negative bacteria (MDRGNB) decolonization in SOT patients (mostly derived from kidney transplant recipients), with no evidence for LT patients.
The only randomized clinical trial available on the matter enrolled immunocompetent patients and investigated the effectiveness of a sequential approach of 5 days of colistin plus neomycin, followed by FMT, in eradicating carbapenemase-producing Enterobacterales colonization, with no significant decrease found at 6 to 8 weeks in comparison to untreated controls (72).
A recent systematic review (73) comprising 21 studies and 151 patients receiving FMT to prevent MDR bacterial colonization showed eradication rates between 37.5% and 87.5%, with no significant adverse effects reported.
Evidence regarding SOT recipients can be found in a prospective study by Singh et al. (74) assessing the efficacy of FMT in fighting intestinal colonization by extended-spectrum beta-lactamase (ESBL)-producing Enterobacterales. In this report, 5 out of 15 patients enrolled (33%) were renal transplant recipients using immunosuppressive drugs. Whereas the overall eradication rate was 40% (6 out of 15 patients), the efficacy was reduced by half when only the immunocompromised patient cohort was considered, with 1 out of 5 participants achieving decolonization (20%); the remaining 4 maintained gut colonization, despite two FMTs. Of note, no serious adverse event was reported.
Further studies are necessary to assess the efficacy and safety of exogenous gut modulation in immunocompromised patients such as SOT recipients. The current body of evidence suggests that there are two main issues regarding possible adverse events associated with FMT. First, as reported by DeFilipp et al. (75) in 2019, thorough screening of fecal matter donors is crucial to minimize the risk of infections directly related to the procedure. Second, it is important to emphasize procedural caution, especially with patients at high risk of aspiration: two cases of death have been reported due to pneumonia and aspiration related to colonoscopy (68, 76).
CONCLUSIONS AND FUTURE DIRECTIONS: THE MANIPULATION OF GUT MICROBIOTA BEFORE LT?
LT is the only curative treatment for patients with ESLD. Even if in recent years an improvement in LT results has been obtained with satisfactory long-term survival, a relevant issue in terms of lives and costs is still represented by infections occurring in the posttransplant period. During the last decades, the gut-liver axis has been one of the most captivating metabolic organs discovered, and significant evidence has highlighted its multiple influxes also at the systemic level. At the hepatic level, intestinal commensals and their products can reach the liver, where they modulate both innate and adaptive immunity, regulate liver inflammation, and promote liver fibrosis, through a direct anatomical connection via the porta venous circulation and the biliary tree. These effects could be reversed by LT, although without a complete “restitutio ad integrum.” Intriguingly, a high risk of infection due to MDRB persists even after liver transplantation, the mechanism of which remains unknown. The immunosuppressive therapies and the wide-spectrum antibiotics administered in the peri-LT period are probably some of the detrimental factors affecting the above-mentioned events; however, they cannot explain why some MDRB-related infections develop after liver transplantation. The manipulation of the GM with therapeutic strategies like FMT seems a viable tool to impact the infectious risk in LT recipients. Modulating the gut microbiota might be a solution for both the prevention and treatment of CDI and diseases related to MDRB harbored in the gastrointestinal tract. More studied are needed to confirm the safety and applicability of these strategies on a wide scale.
In conclusion, the GM in LT recipients has a crucial role in both the evolution of liver disease before transplantation and the metabolic and infectious profile after transplantation. Promising strategies regarding GM manipulation with significant therapeutic consequences are already available and need to be validated before being implemented in clinical routines.
Contributor Information
Andrea Gori, Email: andrea.gori@policlinico.mi.it.
Anthony R. Richardson, University of Pittsburgh
REFERENCES
- 1.European Association for the Study of the Liver. 2016. EASL clinical practice guidelines: liver transplantation. J Hepatol 64:433–485. 10.1016/j.jhep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 2.European Liver Transplant Registry. 2021. Evolution of LTs in Europe. European Liver Transplant Registry, Villejuif, France. http://www.eltr.org/Evolution-of-LTs-in-Europe.html.
- 3.George DL, Arnow PM, Fox AS, Baker AL, Thistlethwaite JR, Emond JC, Whitington PF, Broelsch CE. 1991. Bacterial infection as a complication of liver transplantation: epidemiology and risk factors. Rev Infect Dis 13:387–396. 10.1093/clinids/13.3.387. [DOI] [PubMed] [Google Scholar]
- 4.Paya CV, Hermans PE, Washington JA, Smith TF, Anhalt JP, Wiesner RH, Krom RAF. 1989. Incidence, distribution, and outcome of episodes of infection in 100 orthotopic liver transplantations. Mayo Clin Proc 64:555–564. 10.1016/S0025-6196(12)65561-X. [DOI] [PubMed] [Google Scholar]
- 5.Markin RS, Stratta RJ, Woods GL. 1990. Infection after liver transplantation. Am J Surg Pathol 14(Suppl 1):64–78. [PubMed] [Google Scholar]
- 6.Zheng D, Liwinski T, Elinav E. 2020. Interaction between microbiota and immunity in health and disease. Cell Res 30:492–506. 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato K, Nagao M, Miyamoto K, Oka K, Takahashi M, Yamamoto M, Matsumura Y, Kaido T, Uemoto S, Ichiyama S. 2017. Longitudinal analysis of the intestinal microbiota in liver transplantation. Transplant Direct 3:e144. 10.1097/TXD.0000000000000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj JS, Khoruts A. 2020. Microbiota changes and intestinal microbiota transplantation in liver diseases and cirrhosis. J Hepatol 72:1003–1027. 10.1016/j.jhep.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Bajaj JS, Fagan A, Sikaroodi M, White MB, Sterling RK, Gilles HC, Heuman D, Stravitz RT, Matherly SC, Siddiqui MS, Puri P, Sanyal AJ, Luketic V, John B, Fuchs M, Ahluwalia V, Gillevet PM. 2017. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl 23:907–914. 10.1002/lt.24754. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Yang Y, Qu W, Zhu Z, Wei L, Ye Z, Zhang J-R, Sun X-Y, Zeng Z-G. 2017. Gut microbiota of liver transplantation recipients. Sci Rep 7:3762. 10.1038/s41598-017-03476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annavajhala MK, Gomez-Simmonds A, Macesic N, Sullivan SB, Kress A, Khan SD, Giddins MJ, Stump S, Kim GI, Narain R, Verna EC, Uhlemann A. 2019. Colonizing multidrug-resistant bacteria and the longitudinal evolution of the intestinal microbiome after liver transplantation. Nat Commun 10:4715. 10.1038/s41467-019-12633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajilić-Stojanović M, de Vos WM. 2014. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38:996–1047. 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenhalgh K, Meyer KM, Aagaard KM, Wilmes P. 2016. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ Microbiol 18:2103–2116. 10.1111/1462-2920.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim CH, Park J, Kim M. 2014. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw 14:277–288. 10.4110/in.2014.14.6.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19:29–41. 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 16.Ridlon JM, Kang D-J, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Kho ZY, Lal SK. 2018. The human gut microbiome—a potential controller of wellness and disease. Front Microbiol 9:1835. 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. 2013. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14:329–339. 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iebba V, Totino V, Gagliardi A, Santangelo F, Cacciotti F, Trancassini M, Mancini C, Cicerone C, Corazziari E, Pantanella F, Schippa S. 2016. Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol 39:1–12. [PubMed] [Google Scholar]
- 20.Hill TCJ, Walsh KA, Harris JA, Moffett BF. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 43:1–11. 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 21.Marcon E, Scotti I, Hérault B, Rossi V, Lang G. 2014. Generalization of the partitioning of Shannon diversity. PLoS One 9:e90289. 10.1371/journal.pone.0090289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potosek J, Curry M, Buss M, Chittenden E. 2014. Integration of palliative care in end-stage liver disease and liver transplantation. J Palliat Med 17:1271–1277. 10.1089/jpm.2013.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. 2018. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 15:397–411. 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen T-CD, Pyrsopoulos N, Rustgi VK. 2018. Microbiota and the liver. Liver Transpl 24:539–550. 10.1002/lt.25008. [DOI] [PubMed] [Google Scholar]
- 26.Beste LA, Leipertz SL, Green PK, Dominitz JA, Ross D, Ioannou GN. 2015. Trends in burden of cirrhosis and hepatocellular carcinoma by underlying liver disease in US veterans, 2001–2013. Gastroenterology 149:1471–1482.e5. 10.1053/j.gastro.2015.07.056. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg D, Ditah IC, Saeian K, Lalehzari M, Aronsohn A, Gorospe EC, Charlton M. 2017. Changes in the prevalence of hepatitis C virus infection, nonalcoholic steatohepatitis, and alcoholic liver disease among patients with cirrhosis or liver failure on the waitlist for liver transplantation. Gastroenterology 152:1090–1099.e1. 10.1053/j.gastro.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belli LS, Perricone G, Adam R, Cortesi PA, Strazzabosco M, Facchetti R, Karam V, Salizzoni M, Andujar RL, Fondevila C, De Simone P, Morelli C, Fabregat-Prous J, Samuel D, Agarwaal K, Moreno Gonzales E, Charco R, Zieniewicz K, De Carlis L, Duvoux C. 2018. Impact of DAAs on liver transplantation: major effects on the evolution of indications and results. An ELITA study based on the ELTR registry. J Hepatol 69:810–817. 10.1016/j.jhep.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Cholankeril G, Ahmed A. 2018. Alcoholic liver disease replaces hepatitis C virus infection as the leading indication for liver transplantation in the United States. Clin Gastroenterol Hepatol 16:1356–1358. 10.1016/j.cgh.2017.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haldar D, Kern B, Hodson J, Armstrong MJ, Adam R, Berlakovich G, Fritz J, Feurstein B, Popp W, Karam V, Muiesan P, O'Grady J, Jamieson N, Wigmore SJ, Pirenne J, Malek-Hosseini SA, Hidalgo E, Tokat Y, Paul A, Pratschke J, Bartels M, Trunecka P, Settmacher U, Pinzani M, Duvoux C, Newsome PN, Schneeberger S. 2019. Outcomes of liver transplantation for non-alcoholic steatohepatitis: a European Liver Transplant Registry study. J Hepatol 71:313–322. 10.1016/j.jhep.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nistal E, Sáenz de Miera LE, Ballesteros Pomar M, Sánchez-Campos S, García-Mediavilla MV, Álvarez-Cuenllas B, Linares P, Olcoz JL, Arias-Loste MT, García-Lobo JM, Crespo J, González-Gallego J, Jorquera Plaza F. 2019. An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity. Rev Esp Enferm Dig 111:275–282. 10.17235/reed.2019.6068/2018. [DOI] [PubMed] [Google Scholar]
- 32.Flegal KM, Kruszon-Moran D, Carroll MD, Fryar CD, Ogden CL. 2016. Trends in obesity among adults in the United States, 2005 to 2014. JAMA 315:2284–2291. 10.1001/jama.2016.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Younossi Z, Stepanova M, Ong JP, Jacobson IM, Bugianesi E, Duseja A, Eguchi Y, Wong VW, Negro F, Yilmaz Y, Romero-Gomez M, George J, Ahmed A, Wong R, Younossi I, Ziayee M, Afendy A. 2019. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clin Gastroenterol Hepatol 17:748–755.e3. 10.1016/j.cgh.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. 2011. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54:562–572. 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 35.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. 2012. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol 302:G168–175. 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, Takei H, Nittono H, Ridlon JM, Fuchs M, Gurley EC, Wang Y, Liu R, Sanyal AJ, Gillevet PM, Bajaj JS. 2014. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol 306:G929–937. 10.1152/ajpgi.00315.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. 2013. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58:949–955. 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahluwalia V, Betrapally NS, Hylemon PB, White MB, Gillevet PM, Unser AB, Fagan A, Daita K, Heuman DM, Zhou H, Sikaroodi M, Bajaj JS. 2016. Impaired gut-liver-brain axis in patients with cirrhosis. Sci Rep 6:26800. 10.1038/srep26800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. 2012. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol 303:G675–G685. 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Guo J, Qian G, Fang D, Shi D, Guo L, Li L. 2015. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol 30:1429–1437. 10.1111/jgh.12932. [DOI] [PubMed] [Google Scholar]
- 41.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. 2014. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 60:940–947. 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santiago A, Pozuelo M, Poca M, Gely C, Nieto JC, Torras X, Román E, Campos D, Sarrabayrouse G, Vidal S, Alvarado-Tapias E, Guarner F, Soriano G, Manichanh C, Guarner C. 2016. Alteration of the serum microbiome composition in cirrhotic patients with ascites. Sci Rep 6:25001. 10.1038/srep25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajaj JS, Cox IJ, Betrapally NS, Heuman DM, Schubert ML, Ratneswaran M, Hylemon PB, White MB, Daita K, Noble NA, Sikaroodi M, Williams R, Crossey MME, Taylor-Robinson SD, Gillevet PM. 2014. Systems biology analysis of omeprazole therapy in cirrhosis demonstrates significant shifts in gut microbiota composition and function. Am J Physiol Gastrointest Liver Physiol 307:G951–G957. 10.1152/ajpgi.00268.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. 2014. Alterations of the human gut microbiome in liver cirrhosis. Nature 513:59–64. 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 45.Bajaj JS, Betrapally NS, Gillevet PM. 2015. Decompensated cirrhosis and microbiome interpretation. Nature 525:E1–E2. 10.1038/nature14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Infante-Rivard C, Esnaola S, Villeneuve JP. 1987. Clinical and statistical validity of conventional prognostic factors in predicting short-term survival among cirrhotics. Hepatology 7:660–664. 10.1002/hep.1840070408. [DOI] [PubMed] [Google Scholar]
- 47.Albers I, Hartmann H, Bircher J, Creutzfeldt W. 1989. Superiority of the Child-Pugh classification to quantitative liver function tests for assessing prognosis of liver cirrhosis. Scand J Gastroenterol 24:269–276. 10.3109/00365528909093045. [DOI] [PubMed] [Google Scholar]
- 48.Malinchoc M, Kamath PS, Gordon FD, Peine CJ, Rank J, ter Borg PC. 2000. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology 31:864–871. 10.1053/he.2000.5852. [DOI] [PubMed] [Google Scholar]
- 49.Wiesner R, Edwards E, Freeman R, Harper A, Kim R, Kamath P, Kremers W, Lake J, Howard T, Merion RM, Wolfe RA, Krom R, United Network for Organ Sharing Liver Disease Severity Score Committee. 2003. Model for end-stage liver disease (MELD) and allocation of donor livers. Gastroenterology 124:91–96. 10.1053/gast.2003.50016. [DOI] [PubMed] [Google Scholar]
- 50.Yoo HY, Thuluvath PJ. 2005. Short-term postliver transplant survival after the introduction of MELD scores for organ allocation in the United States. Liver Int 25:536–541. 10.1111/j.1478-3231.2005.01011.x. [DOI] [PubMed] [Google Scholar]
- 51.Onaca N, Levy MF, Ueno T, Martin AP, Sanchez EQ, Chinnakotla S, Randall HB, Dawson S, Goldstein RM, Davis GL, Klintmalm GB. 2006. An outcome comparison between primary liver transplantation and retransplantation based on the pretransplant MELD score. Transplant Int 19:282–287. 10.1111/j.1432-2277.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 52.Grąt M, Hołówko W, Wronka KM, Grąt K, Lewandowski Z, Kosińska I, Krasnodębski M, Wasilewicz M, Gałęcka M, Szachta P, Zborowska H, Patkowski W, Krawczyk M. 2015. The relevance of intestinal dysbiosis in liver transplant candidates. Transpl Infect Dis 17:174–184. 10.1111/tid.12352. [DOI] [PubMed] [Google Scholar]
- 53.Neofytos D, Fishman JA, Horn D, Anaissie E, Chang CH, Olyaei A, Pfaller M, Steinbach WJ, Webster KM, Marr KA. 2010. Epidemiology and outcome of invasive fungal infections in solid organ transplant recipients. Transplant Infectious Dis 12:220–229. 10.1111/j.1399-3062.2010.00492.x. [DOI] [PubMed] [Google Scholar]
- 54.Abad CLR, Lahr BD, Razonable RR. 2017. Epidemiology and risk factors for infection after living donor liver transplantation. Liver Transpl 23:465–477. 10.1002/lt.24739. [DOI] [PubMed] [Google Scholar]
- 55.Hand J, Patel G. 2016. Multidrug-resistant organisms in liver transplant: mitigating risk and managing infections. Liver Transpl 22:1143–1153. 10.1002/lt.24486. [DOI] [PubMed] [Google Scholar]
- 56.Bonnel AR, Bunchorntavakul C, Reddy KR. 2011. Immune dysfunction and infections in patients with cirrhosis. Clin Gastroenterol Hepatol 9:727–738. 10.1016/j.cgh.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 57.Hackstein CP, Assmus LM, Welz M, Klein S, Schwandt T, Schultze J, Förster I, Gondorf F, Beyer M, Kroy D, Kurts C, Trebicka J, Kastenmüller W, Knolle PA, Abdullah Z. 2017. Gut microbial translocation corrupts myeloid cell function to control bacterial infection during liver cirrhosis. Gut 66:507–518. 10.1136/gutjnl-2015-311224. [DOI] [PubMed] [Google Scholar]
- 58.Chang LY, Li Y, Kaplan DE. 2016. Endotoxemia contributes to CD27+ memory B-cell apoptosis via enhanced sensitivity to Fas ligation in patients with cirrhosis. Sci Rep 6:36862–36811. 10.1038/srep36862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Függer R, Hamilton G, Steininger R, Mirza D, Schulz F, Mühlbacher F. 1991. Intraoperative estimation of endotoxin, TNFα, and IL-6 in orthotopic liver transplantation and their relation to rejection and postoperative infection. Transplantation 52:302–306. 10.1097/00007890-199108000-00022. [DOI] [PubMed] [Google Scholar]
- 60.Giannella M, Trecarichi EM, De Rosa FG, Del Bono V, Bassetti M, Lewis RE, Losito AR, Corcione S, Saffioti C, Bartoletti M, Maiuro G, Cardellino CS, Tedeschi S, Cauda R, Viscoli C, Viale P, Tumbarello M. 2014. Risk factors for carbapenem-resistant Klebsiella pneumoniae bloodstream infection among rectal carriers: a prospective observational multicentre study. Clin Microbiol Infect 20:1357–1362. 10.1111/1469-0691.12747. [DOI] [PubMed] [Google Scholar]
- 61.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Sammons JS, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48. 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cózar-Llistó A, Ramos-Martinez A, Cobo J. 2016. Clostridium difficile infection in special high-risk populations. Infect Dis Ther 5:253–269. 10.1007/s40121-016-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsu JL, Enser JJ, Mckown T, Leverson GE, Pirsch JD, Hess TM, Safdar N. 2014. Outcomes of Clostridium difficile infection in recipients of solid abdominal organ transplants. Clin Transplant 28:267–273. 10.1111/ctr.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paudel S, Zacharioudakis IM, Zervou FN, Ziakas PD, Mylonakis E. 2015. Prevalence of clostridium difficile infection among solid organ transplant recipients: a meta-analysis of published studies. PLoS One 10:e0124483. 10.1371/journal.pone.0124483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider KM, Wirtz TH, Kroy D, Albers S, Neumann UP, Strowig T, Sellge G, Trautwein C. 2018. Successful fecal microbiota transplantation in a patient with severe complicated Clostridium difficile infection after liver transplantation. Case Rep Gastroenterol 12:76–84. 10.1159/000481937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alrabaa S, Jariwala R, Zeitler K, Montero J. 2017. Fecal microbiota transplantation outcomes in immunocompetent and immunocompromised patients: a single-center experience. Transplant Infect Dis 19:e12726. 10.1111/tid.12726. [DOI] [PubMed] [Google Scholar]
- 67.Bilal M, Khehra R, Strahotin C, Mitre R. 2015. Long-term follow-up of fecal microbiota transplantation for treatment of recurrent clostridium difficile infection in a dual solid organ transplant recipient. Case Rep Gastroenterol 9:156–159. 10.1159/000430491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, Gordon S, Gluck M, Hohmann EL, Kao D, Kao JY, McQuillen DP, Mellow M, Rank KM, Rao K, Ray A, Schwartz MA, Singh N, Stollman N, Suskind DL, Vindigni SM, Youngster I, Brandt L. 2014. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol 109:1065–1071. 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng YW, Phelps E, Ganapini V, Khan N, Ouyang F, Xu H, Khanna S, Tariq R, Friedman-Moraco RJ, Woodworth MH, Dhere T, Kraft CS, Kao D, Smith J, Le L, El-Nachef N, Kaur N, Kowsika S, Ehrlich A, Smith M, Safdar N, Misch EA, Allegretti JR, Flynn A, Kassam Z, Sharfuddin A, Vuppalanchi R, Fischer M. 2019. Fecal microbiota transplantation for the treatment of recurrent and severe Clostridium difficile infection in solid organ transplant recipients: a multicenter experience. Am J Transplant 19:501–511. 10.1111/ajt.15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S, Xu M, Wang W, Cao X, Piao M, Khan S, Yan F, Cao H, Wang B. 2016. Systematic review: adverse events of fecal microbiota transplantation. PLoS One 11:e0161174. 10.1371/journal.pone.0161174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shogbesan O, Poudel DR, Victor S, Jehangir A, Fadahunsi O, Shogbesan G, Donato A. 2018. A systematic review of the efficacy and safety of fecal microbiota transplant for Clostridium difficile infection in immunocompromised patients. Can J Gastroenterol Hepatol 2018:1394379. 10.1155/2018/1394379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huttner BD, de Lastours V, Wassenberg M, Maharshak N, Mauris A, Galperine T, Zanichelli V, Kapel N, Bellanger A, Olearo F, Duval X, Armand-Lefevre L, Carmeli Y, Bonten M, Fantin B, Harbarth S, Colle L, Kloosterman F, van Bentum-Puijk W, Vlooswijk J, Andremont A, Ben Hayoun M, Canoui E, Chabrol A, Gamany N, Lafaurie M, Lefort A, Lepeule R, Louis Z, Rondinaud E, Sadou-Yaye H, Sarfati L, Zarrouk V, Brossier C, Carrez L, Lazarevic V, Renzi G, von Dach E, Cohen Percia S, Shvartz R, Lellouche J, R-Gnosis WP3 Study Group. 2019. A 5-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect 25:830–838. 10.1016/j.cmi.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 73.Saha S, Tariq R, Tosh PK, Pardi DS, Khanna S. 2019. Faecal microbiota transplantation for eradicating carriage of multidrug-resistant organisms: a systematic review. Clin Microbiol Infect 25:958–963. 10.1016/j.cmi.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 74.Singh R, De Groot PF, Geerlings SE, Hodiamont CJ, Belzer C, Berge IJMT, De Vos WM, Bemelman FJ, Nieuwdorp M. 2018. Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing Enterobacteriaceae: a proof of principle study. BMC Res Notes 11:190. 10.1186/s13104-018-3293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen Y-B, Hohmann EL. 2019. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med 381:2043–2050. 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 76.Fischer M, Kao D, Kelly C, Kuchipudi A, Jafri SM, Blumenkehl M, Rex D, Mellow M, Kaur N, Sokol H, Cook G, Hamilton MJ, Phelps E, Sipe B, Xu H, Allegretti JR. 2016. Fecal microbiota transplantation is safe and efficacious for recurrent or refractory Clostridium difficile infection in patients with inflammatory bowel disease. Inflamm Bowel Dis 22:2402–2409. 10.1097/MIB.0000000000000908. [DOI] [PubMed] [Google Scholar]
- 77.Wu Y, Wang M, Zhu Y, Lin S. 2016. Serum interleukin-6 in the diagnosis of bacterial infection in cirrhotic patients: a meta-analysis. Medicine 95:e5127. 10.1097/MD.0000000000005127. [DOI] [PMC free article] [PubMed] [Google Scholar]