Abstract
Background
Glioblastoma remains a deadly brain cancer with dismal prognosis. Genetic alterations, including IDH mutations, 1p19q co-deletion status and MGMT promoter methylation have been proven to be prognostic and predictive to response to treatment in gliomas. In this manuscript, we aimed to correlate other mutations and genetic alterations with various clinical endpoints in patients with IDH-wild-type (IDHwt) glioblastoma.
Methods
We compiled a comprehensive clinically annotated database of IDHwt GBM patients treated at the Ohio State University Wexner Medical Center for whom we had mutational data through a CLIA-certified genomic laboratory. We then added data that is publicly available from Memorial Sloan Kettering Cancer Center through cBioPortal. Each of the genetic alterations (mutations, deletions, and amplifications) served as a variable in univariate and multivariate Cox proportional hazard models.
Results
A total of 175 IDHwt GBM patients with available MGMT promoter methylation data from both cohorts were included in the analysis. As expected, MGMT promoter methylation was significantly associated with improved overall survival (OS). Median OS for MGMT promoter methylated and unmethylated GBM was 26.5 and 18 months, respectively (HR 0.45; P = .003). Moreover, EGFR/ERBB alterations were associated with favorable outcome (HR of 0.37 (P = .003), but only in MGMT promoter unmethylated GBM. We further found that patients with EGFR/ERBB alterations who also harbored PDGFRA amplification had a significantly worse outcome (HR 7.89; P = .025).
Conclusions
Our data provide further insight into the impact of genetic alterations on various clinical outcomes in IDHwt GBM in 2 cohorts of patients with detailed clinical information and inspire new therapeutic strategies for IDHwt GBM.
Keywords: EGFR, GBM, IDH, MGMT, next-generation sequencing, PDGFR
Key Points.
In our analysis of 2 cohorts of IDHwt GBM, EGFR/ERBB alterations were associated with favorable outcome in MGMT promoter unmethylated GBM.
Patients with EGFR/ERBB alterations who also harbored PDGFRA amplification had significantly worse outcome.
Importance of the Study.
In the era of personalized medicine, it has become routine practice to sequence tumors to aide in decisions making regarding treatment options, either for the sake of clinical trials inclusion, or for salvage treatment for patients who have exhausted standard-of-care options. In this paper, we report our experience with CLIA-certified molecular sequencing tests for patients with IDH-wild-type glioblastoma at the Ohio State University. We correlate mutations and genetic alterations with various clinical endpoints in patients from our cohort as well as a publically available cohort from Memorial-Sloan Kettering Cancer Center.
Gliomas are tumors that arise from the glial cells in the central nervous system. Glioblastoma (GBM) is the most common malignant glioma and represents astrocytoma grade IV.1 Despite our deeper understanding of the genomic alterations that precipitate gliomagenesis, only a few genetic and epigenetic modifications have been identified to be meaningful in clinical practice: At the chromosomal level, simultaneous copy number losses of chromosomes 1p and 19q—based on the World Health Organization 2016 classification of brain tumors—define oligodendrogliomas.2 Oligodendrogliomas have better survival outcomes compared to astrocytomas.3,4 Similarly, it is well established that isocitrate dehydrogenase (IDH) 1 and 2 mutations carry favorable outcomes in patients with astrocytomas and secondary GBM.5 Finally, the promoter methylation status of the gene encoding for the repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) predicts response to the standard-of-care alkylating chemotherapy agent used in glioma: temozolomide.6
Besides tumor-treating fields, little progress has been made in the management of GBM over the past 2 decades despite enormous research efforts. In fact, GBM was the first cancer type to be analyzed by The Cancer Genome Atlas (TCGA) project which revealed several genomic subtypes of the tumor.7 Genomic alterations—including copy number variations (CNV) and mutations—lead to activation of oncogenes and inactivation of tumor suppressor genes. Hence, we now better understand the various cellular mechanisms and pathways utilized by gliomas for growth and survival. The most common altered pathways in GBM include: receptor tyrosine kinase (RTK/RAS) pathway (eg, via amplification of epidermal growth factor receptor [EGFR] and platelet-derived growth factor receptor [PDGFR]), phosphatidylinositol 3-kinase (PI3K) pathway (eg, via deletion of the tumor suppressor [PTEN]), cell cycle pathway (eg, via mutations in CDKN2A/B and RB1), P53 pathway (eg, via mutations in P53 and MDM2), and telomere length maintaining pathways (eg, TERT promoter mutations).8
Targeted therapies emerged to tackle specific pathways utilized by cancer cells. While these treatments have had successes in cancers such as melanoma and non-small-cell lung cancer, clinical trials of small molecule inhibitors, antibodies, vaccines, and kinase inhibitors targeting these pathways have not improved overall survival (OS) in patients with GBM. A key reason for failure of target inhibition in gliomas appears to be tumor heterogeneity and cancer cell plasticity leading to redundant inputs that maintain the downstream signaling pathways allowing the cancer cells to survive even if one upstream signaling receptor is blocked.9
In this study, we aimed to correlate genomic alterations with clinical patient outcomes in 2 cohorts of IDHwt GBM and to assess interactions among the various alterations.
Methods
Under an IRB-approved protocol, we compiled a comprehensive clinically annotated database of adult patients with GBM at The Ohio State University (OSU) for whom we had next-generation sequencing (NGS) data through CLIA-certified commercial platforms (Foundation One and Tempus). The database included information detailing the pathologic diagnosis, age, race, gender, performance status, tumor location, treatments utilized, occurrence of complications (radiation necrosis, leptomeningeal spread, or thromboembolic disease defined as deep vein thrombosis and/or pulmonary embolism at any time during the disease course), molecular classifications (IDH mutations, MGMT promoter methylation, 1p19q co-deletion), programmed death ligand-1 (PD-L1) protein expression on immunohistochemistry, and survival data, in addition to NGS data.
Furthermore, we added publicly available data from a cohort of IDH-wild-type (IDHwt) GBM from Memorial Sloan Kettering Cancer Center (MSKCC)10 through cBioPortal.11,12 We also acquired MGMT promoter methylation data for this cohort from MSKCC as this data was not available through cBioPortal. The aim was to correlate mutational data with clinical outcomes, namely OS. Each of the mutations/alterations served as a variable in univariate and multivariate analyses.
We grouped certain alterations under one variable based on the core signaling pathway altering functions.8 The molecular variables included are listed in Table 1.
Table 1.
Variables Included in the Univariate and Multivariate Survival Models Grouped Based on Function in the Core Signaling Pathways
| Variable | Mutations/Alterations |
|---|---|
| MDM/TP53 | MDM2, MDM4 or TP53 mutations |
| CDKN2A/B | CDKN2A/B/C deletions or mutations (rare) |
| CDK/CCND | CDK4/CDK6 amplifications, or CCND1/CCND2 amplifications |
| RB1 | RB1 mutations/deletions |
| EGFR/ERBB | EGFR amplifications/mutations or ERBB (2-4) mutations. |
| FGFR | FGFR mutations/amplifications |
| PDGFRA/KIT | PDGFRA mutations/amplifications |
| NF1 | NF1 mutations |
| PTEN | PTEN mutations/deletions |
| PIK3R1 | PIK3R1 mutations/deletions |
| PI3K gain | PIK3CA mutations, mTOR mutations or AKT amplifications |
| MYC | MYC: MYC/MYCN amplifications |
| TERT | TERT promoter mutations |
Statistical Analysis
A Cox’s proportional hazards model was used for univariate and multivariate survival analyses. Backward stepwise regression models were used when looking at subgroups to decrease the number of variables in smaller cohorts. Chi-square tests were used for categorical variables. A nonparametric test was used to compare the median age for the 2 cohorts as age was not normally distributed P values less than .05 were considered significant. IBM SPSS Statistics 26 was used.
Results
Patient Characteristics
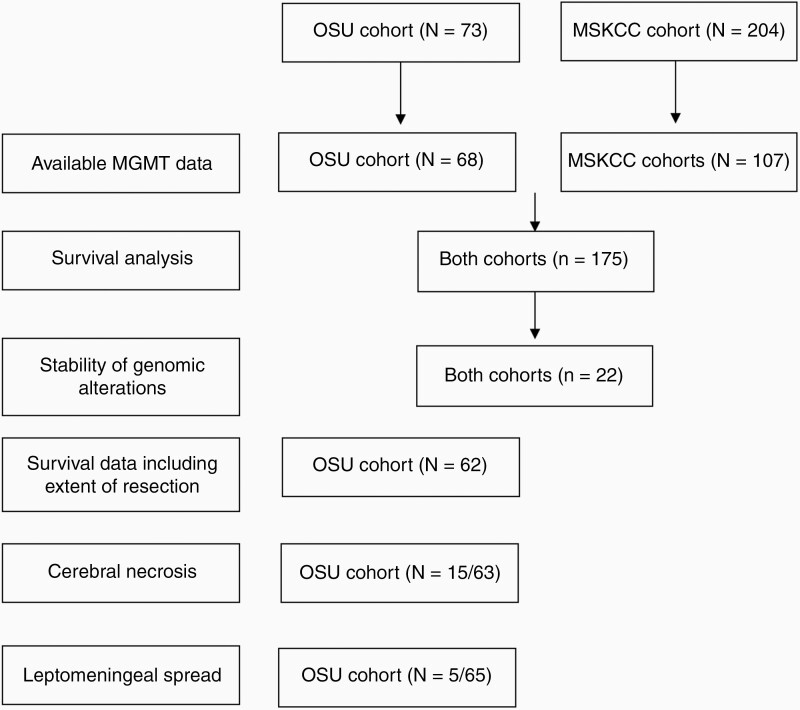
Of the 73 patients with IDHwt GBM from the OSU cohort included in the study, MGMT promoter methylation data was available on 68/73 patients and these patients were included in the survival analysis. Similarly, we were able to obtain MGMT promoter methylation data on 107/204 patients from the MSKCC cohort. Only patients with known MGMT promoter methylation status were included in this analysis as we figured that any model that does not include MGMT would be inaccurate. Figure 1 shows a flow diagram of the analyses performed on each or both cohorts and number of patients included in each analysis.
Figure 1.
A flow diagram showing the analyses performed on the OSU and MSKCC cohorts and number of patients included in each analysis.
The median age for the OSU cohort was 60 years (20–78) and the median age for the MSKCC cohort was 61 years (22–91; P = .649). The median OS for the OSU cohort of 20.88 months (95% CI, 15.88–25.87) was similar to that for the MSKCC cohort of 18.77 months (95% CI, 16.46–21.08; P = .464).
Genomic Alterations and Overall Survival (both cohorts)
A total of 175 patients were included in this analysis. Results of the univariate and multivariate analyses are shown in Table 2. As expected, MGMT promoter methylation was significantly associated with improved OS. Median OS for methylated and unmethylated MGMT promoter GBM were 26.5 and 18 months, respectively (multivariable HR 0.45; P = .003). Unexpectedly, EGFR/ERBB alterations were also associated with significantly improved OS. OS for EGFR/ERBB altered GBM and EGFR/ERBBwt GBM were 24.95 and 16.86 months, respectively (multivariable HR 0.31; P < .001). EGFR amplification, specifically, was also associated with improved OS (multivariable HR 0.41; P = .009).
Table 2.
Univariate and Multivariate Analyses Correlating the Various Genomic Alterations With Overall Survival
| OSU+MSK with MGMT (N = 175) | Univariate Analysis P value (HR) |
Multivariate Model (0.001)- P value (HR) |
|---|---|---|
| Age | .619 | .793 (1.003) |
| MGMT | .008 (0.53) | .003 (0.45) |
| MDM/P53 | .507 | .023 (0.49) |
| CDKN2A/B | .765 | .388 |
| CDK/CCND | .706 | .985 |
| RB1 | .145 | .06 (0.367) |
| EGFR/ERBB | .003 (0.49) | .0002 (0.31) |
| FGFR | .568 | .378 |
| PDGFRA/KIT | .941 | .974 |
| NF1 | .208 | .641 |
| PTEN | .026 (1.66) | .513 |
| PIK3R1 | .936 | .067 (0.38) |
| PI3K gain | .386 | .281 |
| MYC | .988 | .183 |
| TERT promoter | .043 (1.93) | .074 (2.07) |
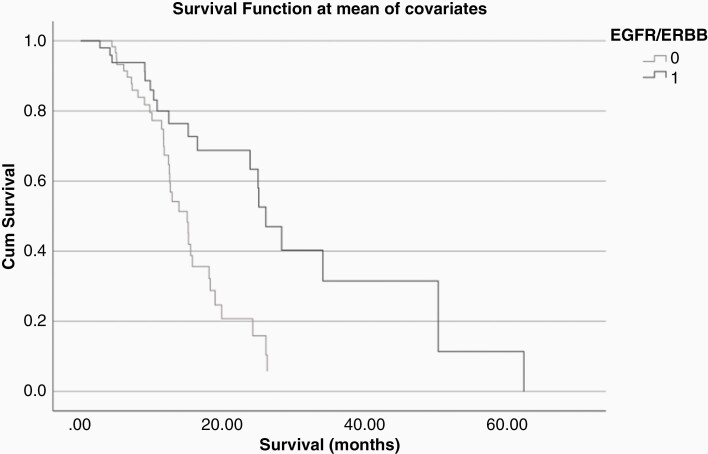
Favorable survival related to EGFR/ERBB alterations appeared to hold true only when MGMT promoter was unmethylated: in a backward stepwise regression model (P = .003) of the 112 unmethylated MGMT promoter cohort, EGFR/ERBB alterations were associated with favorable HR of 0.37 (P = .003; Figure 2). Median OS for MGMT unmethylated, EGFR/ERBB altered GBM was 25.05 months compared to 15.12 months for MGMT unmethylated, EGFR/ERBBwt GBM. In patients with unmethylated MGMT promoter GBM, MDM/P53 alterations were also associated with favorable outcome (HR 0.51; P = .027). On the other hand, when MGMT promoter was methylated (N = 63), a backward stepwise model (P = .001) revealed FGFR and PTEN as markers of worse survival (HR 10.23; P = .001 and HR 3.02; P = .012, respectively).
Figure 2.
The presence of EGFR/ERBB alterations was associated with favorable outcome in a cohort of 112 patients with unmethylated MGMT promoter GBM.
Association Between EGFR/ERBB and PDGFRA Amplification
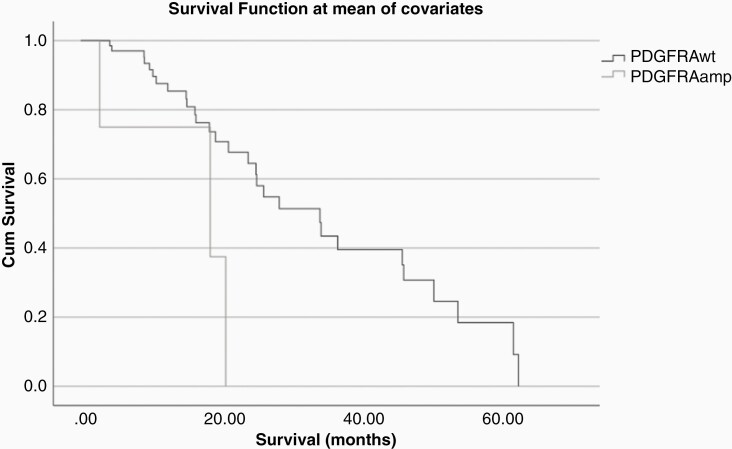
The multivariable analysis hinted at an association between EGFR/ERBB and PDGFRA. We therefore performed separate analyses for patients who were positive for EGFR/ERBB alterations (N = 78; Table 3) and those were not. In patients with EGFR/ERBB alterations (or those who specifically had EGFR amplification), univariate and multivariate analyses showed that those who also had PDGFRA (N = 5) amplification had significantly worse survival (multivariate HR 7.89; P = .025; Figure 3). Median OS for patients who were positive for EGFR/ERBB alterations and PDGFRA amplification was 18.44 m compared to 34.06 m for patients positive for EGFR/ERBB alterations without PDGFRA amplification.
Table 3.
Univariate and Multivariate Analyses Correlating the Various Genomic Alterations With Overall Survival Within EGFR/ERBB Altered Tumors
| OSU+MSK with MGMT (EGFR/ERBB altered) (N = 78) | Univariate Analysis P value (HR) |
Multivariate Model (0.023)- P value (HR) |
|---|---|---|
| Age | .261 | .120 |
| MGMT | .224 | .241 |
| MDM/P53 | .520 | .317 |
| CDKN2A/B | .434 | .505 |
| CDK/CCND | 0.742 | .976 |
| RB1 | .686 | .878 |
| FGFR | .495 | .981 |
| PDGFRA/KIT | .034 (3.86) | .025 (7.89) |
| NF1 | .361 | .877 |
| PTEN | .01 (2.60) | .454 |
| PI3K gain | .112 | .175 |
| MYC | .196 | .721 |
| TERT promoter | .031 (5.17) | .139 |
Figure 3.
The presence of PDGFRA amplification in EGFR/ERBB altered GBM was associated with worse survival in a cohort of 78 patients.
In patients without EGFR/ERBB alterations, a backward stepwise model (P = .008), revealed only MGMT promoter methylation as a favorable marker (HR 0.45; P = .023). Age was associated with worse outcome (HR 1.02; P = .047).
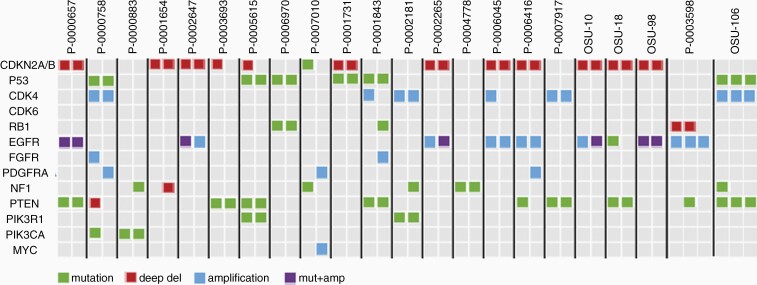
Stability of Genomic Alterations Across Recurrent Samples
Four patients from the OSU cohort and 22 patients from the MSKCC cohort had DNA sequencing performed on recurrent samples. CDKN2A/B, EGFR, TP53, and PTEN alterations were most consistent between the primary and recurrent samples (Figure 4).
Figure 4.
Mutations and copy number variations in primary and recurrent samples for each patient are shown in pairs. MSKCC patient IDs are as identified in cBioPortal.
Genomic Alterations and Clinical Outcomes (OSU cohort)
Overall survival
Looking at the OSU cohort alone, we were able to incorporate extent of resection into the model (N = 62). A strongly significant backward stepwise regression model (P < .001) revealed that extent of resection (P = .023), MGMT (HR 0.54; P = .086), and EGFR/ERBB alterations (HR 0.31; P = .003) correlate with significant favorable outcomes. Biopsy only compared to gross total resection carried significantly worse outcome (HR 4.97; P = .006). Subtotal was not significantly different from gross total resection in the multivariate model. Also, of note, presence of thromboembolic disease was also associated with worse OS in the OSU cohort in univariate but not multivariate analysis (univariate HR 2.26; P = .009).
Progression-free survival
A strongly significant backward stepwise model (P < .001) revealed extent of resection (P = .022), MGMT promoter methylation (HR 0.29; P < .001), EGFR/ERBB alterations (HR 0.40; P = .011), CDKN2A/B loss (HR 0.39; P = .012), and PIK3R1 mutations (HR 0.04; P = .003) as significant favorable outcomes. Biopsy only as opposed to gross total resection carried significantly worse risk of progression (HR 3.627; P = .007).
Cerebral necrosis
Treatment-related cerebral necrosis is a potential complication in patients with gliomas. Information about cerebral necrosis was available on 63 patients. Nine out of 24 (37.5%) of patients with methylated MGMT promoter GBM developed RN versus 6/39 (15.4%) in the unmethylated group. Chi-square test (P = .045).
Leptomeningeal spread
Five out of 65 (7.7%) patients (with available data) developed leptomeningeal disease (LMD). All tumors appeared to have extended into the ventricles on brain imaging by the time of development of LMD. Four tumors had evidence of subependymal spread prior (range 45–93 days) to development of LMD. Four patients had available CSF studies, 2 of whom had evidence of atypical or malignant cells. CSF protein was elevated in all 4 samples (range 113–492 mg/dL). CSF glucose was low in all 4 samples (range < 10–46 mg/dL). MGMT promoter was methylated in 2 tumors and unmethylated in 2 and unknown in 1. The genomic alterations varied among samples and included EGFR V765M mutation (N = 1), PDGFRA amplification (N = 1), PDGFRA Y849C subclonal mutation (N = 1), CDKN2A/B loss (N = 2), TP53 mutations (N = 3), and TERT promoter mutations (N = 3). No case had EGFR amplification or EGFRvIII mutation. Other alterations observed included DNMT3A mutation, KIT, MYC, and MDM2 amplifications and RB1 losses. PD-L1 expression in tumor samples ranged from 5% to 40%. One intracranial sample had a 20% PD-L1 expression with LMD from this tumor exhibiting 40% PD-L1 expression. Median OS was 11.67 months (range 6.05–18.31). Median OS after LMD diagnosis was 2.76 months (range 1.91–7.39).
Discussion
We compiled a comprehensive clinically annotated database of with detailed demographic and clinical data of 73 patients with IDHwt GBM. We then added data that is publicly available from MSKCC through cBioPortal. This cohort included 204 patients with IDHwt GBM. The aim was to correlate different mutational data with clinical outcomes, namely survival. Each of the mutations/alterations served as a variable in univariate and multivariate analyses. Survival analyses were performed on 175 patients for whom MGMT promoter methylation status is known.
As expected, MGMT promoter methylation was significantly associated with better outcome HR 0.45; P = .003. Furthermore, GBM with EGFR/ERBB alterations (and specifically EGFR amplification) had better outcome compared to EGFR/ERBBwt GBM. However, this appeared to be true only in MGMT promoter unmethylated GBM. Previous literature is unclear in prognostic role of EGFR in GBM, as some studies suggested favorable outcome and others suggested worse outcome.13,14 However, to our knowledge this has not been looked at previously in the setting of multivariable analysis and specifically MGMT promoter methylation. This finding will need to be validated in bigger datasets.
In the OSU cohort alone, more aggressive surgical resection, MGMT promoter methylation and EGFR/ERBB alterations yielded favorable OS and PFS outcomes. Moreover, focusing on patients with EGFR/ERBB alterations, in both cohorts, univariate and multivariate analysis showed an association between EGFR and PDGFRA. More specifically, patients with an EGFR/ERBB alteration who also exhibited PDGFRA amplification had significantly worse survival (HR 7.89; P = .025) hinting to a potential interaction between the 2 receptors that needs further evaluation.
PDGFRα and PDGFRβ (encoded by PDGFRA and PDGFRB, respectively) are both expressed as transmembrane receptors on GBM cell surface and are drivers of glioma growth. PDGFRA amplification is the more common alteration and occurs in 13.1% of GBM.7 While concurrent amplification of EGFR and PDGFRA has been reported in up to 5% of GBM,7 EGFR and PDGFRA co-expression at the mRNA level is seen in 37% of GBM sphere lines as reported by Chakravarty et al.15 The paper also showed functional transactivation of PDGFRα by EGFR: EGF stimulation in this setting can result both in EGFR-EGFR homodimerization as well as EGFR-PDGFRα heterodimerization which can drive proliferation.15 Further supporting this interaction, Hegi et al. observed that the expression of p-EGFR correlated with p-PDGFRβ in a window of opportunity study of 22 patients with recurrent GBM who were treated with at least 5 days of gefitinib (an EGFR inhibitor) prior to re-resection.16 We believe that the interaction between EGFR and PDGFRA alterations may have therapeutic implications and escaping to PDGFR signaling may in part explain the failure of EGFR targeted therapy in GBM.
Tumor heterogeneity and multiple RTK pathway activation have rendered GBM highly resistant to targeted treatment,17 particularly in the recurrent setting. GBM has been reported to change methylation subclass upon recurrence, which may further point to emergence of adaptive resistance mechanisms to therapy.18 Likewise, EGFR amplification has been previously reported to be lost in 16–27% of recurrent samples.18,19 We illustrate how CDKN2A/B, EGFR, TP53, and PTEN alterations were most consistent between the primary and recurrent samples, however, none of the mutations/copy number variations appear to be invariably consistent between the primary and recurrent samples.
As previously described,20MGMT promoter methylation was associated with increased risk of treatment related cerebral necrosis: (37.5%) versus (15.4%) in the methylated and unmethylated groups, respectively, in the OSU cohort. Furthermore, GBM patients who developed leptomeningeal disease had no one consistent genetic alteration. Rather, in all instances, the lesions had extended toward the ventricles prior to development of LMD.
In summary, NGS provides valuable information when caring for patients with GBM. The study is limited by the small sample size, especially for the analyses performed on the OSU cohort alone, leading to low power to make definitive statistical conclusions. The study is also limited by the fact that glioblastoma was diagnosed solely based on pathologic evaluations; new diagnostic entities have been established based on methylation testing.21 The study is rather hypothesis generating and our findings will need validation from other bigger cohorts. We find that MGMT unmethylated GBM that harbors EGFR/ERBB alterations appear to have better prognosis in comparison to MGMT unmethylated EGFR/ERBBwt tumors. Moreover, there appears to be a significant interaction between EGFR and PDGFRA. This may in part explain EGFR inhibition resistance in GBM and highlights the plasticity of GBM cells. Combining targeted treatments against these 2 receptors may be an attractive therapeutic target.
Acknowledgment
We thank Dr. Ingo Mellinghoff and the MSKCC team for providing MGMT methylation data for the cBioPortal dataset.
Funding
This project did not receive any funding support.
Conflict of interest statement. The authors have no conflicts of interest to disclose.
Authorship Statement
I.A., A.R., M.G.P.: Project design, data collection, data analysis, manuscript writing. S.O., P.G., V.P.: Project design, data analysis, manuscript writing.
References
- 1. Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erdem-Eraslan L, Gravendeel LA, de Rooi J, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol. 2013;31(3):328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Eng J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 7. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Touat M, Idbaih A, Sanson M, Ligon KL. Glioblastoma targeted therapy: updated approaches from recent biological insights. Ann Oncol. 2017;28(7):1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hobbs J, Nikiforova MN, Fardo DW, et al. Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas. Am J Surg Pathol. 2012;36(8):1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J, Liang R, Song C, Xiang Y, Liu Y. Prognostic significance of epidermal growth factor receptor expression in glioma patients. Onco Targets Ther. 2018;11:731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chakravarty D, Pedraza AM, Cotari J, et al. EGFR and PDGFRA co-expression and heterodimerization in glioblastoma tumor sphere lines. Sci Rep. 2017;7(1):9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hegi ME, Diserens A-C, Bady P, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol Cancer Ther. 2011;10(6):1102–1112. [DOI] [PubMed] [Google Scholar]
- 17. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Körber V, Yang J, Barah P, et al. Evolutionary trajectories of IDHWT glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell. 2019;35(4):692-704. e612. [DOI] [PubMed] [Google Scholar]
- 19. van den Bent MJ, Gao Y, Kerkhof M, et al. Changes in the EGFR amplification and EGFRvIII expression between paired primary and recurrent glioblastomas. Neuro Oncol. 2015;17(7):935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brandes AA, Franceschi E, Tosoni A, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26(13):2192–2197. [DOI] [PubMed] [Google Scholar]
- 21. Bender K, Perez E, Chirica M, et al. High-grade astrocytoma with piloid features (HGAP): the Charité experience with a new central nervous system tumor entity. J Neurooncol. 2021;153(1):109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]