Abstract
Plant-growth-promoting bacteria (PGPB) are used to improve plant health and promote crop production. However, because some PGPB (including Bacillus subtilis) do not maintain substantial colonization on plant roots over time, it is unclear how effective PGPB are throughout the plant growing cycle. A better understanding of the dynamics of plant root community assembly is needed to develop and harness the potential of PGPB. Although B. subtilis is often a member of the root microbiome, it does not efficiently monoassociate with plant roots. We hypothesized that B. subtilis may require other primary colonizers to efficiently associate with plant roots. We utilized a previously designed hydroponic system to add bacteria to Arabidopsis thaliana roots and monitor their attachment over time. We inoculated seedlings with B. subtilis and individual bacterial isolates from the native A. thaliana root microbiome either alone or together. We then measured how the coinoculum affected the ability of B. subtilis to colonize and maintain on A. thaliana roots. We screened 96 fully genome-sequenced strains and identified five bacterial strains that were able to significantly improve the maintenance of B. subtilis. Three of these rhizobacteria also increased the maintenance of two strains of B. amyloliquefaciens commonly used in commercially available bioadditives. These results not only illustrate the utility of this model system to address questions about plant–microbe interactions and how other bacteria affect the ability of PGPB to maintain their relationships with plant roots but also may help inform future agricultural interventions to increase crop yields.
Keywords: bacteriology, microbiome, plants, rhizosphere, phyllosphere
As criticism around environmentally costly chemical fertilizers and pesticides increases (Aloo et al. 2019; Busby et al. 2017), scientists are focusing on developing microbe-based agricultural treatments (Keswani et al. 2019; Li et al. 2019). Bacterial species that increase plant growth or crop yield, regardless of mechanism (Cawoy et al. 2014; Ongena and Jacques 2008; Tahir et al. 2017; Yi et al. 2016), have been defined as plant-growth-promoting bacteria (PGPB) (Ferreira et al. 2019). In laboratory settings, these bacteria colonize what is collectively known as the rhizosphere (root surfaces and the areas directly surrounding plant roots) (Bulgarelli et al. 2013; Reinhold-Hurek et al. 2015). Still, some studies indicate that PGPB may not reproducibly maintain their root associations over extended periods of time, in spite of indications that such maintenance may be required for consistent beneficial effects on plants (Gadhave et al. 2018; Mendis et al. 2018).
Several species of Bacillus are currently used as agricultural PGPB (Aloo et al. 2019; Asari et al. 2016; El-Daim et al. 2019; Radhakrishnan et al. 2017; Shafi et al. 2017), in part due to their ability to form hardy spores, which allow commercial formulations to remain shelf stable for over a year (Kröber et al. 2014). However, similar to other PGPB, one concern surrounding the use of Bacillus PGPB is that they may not attach to the root or maintain their colonization at the levels needed to elicit beneficial effects on the plant (Gadhave et al. 2018). Indeed, because the beneficial effects of Bacillus subtilis on plants appear to rely on biofilm formation (Chen et al. 2013), prolonged plant–root interactions may be required for Bacillus’s PGPB effects. Thus, developing mechanisms to explicitly enhance the maintenance of Bacillus spp. on plant roots over time may enable us to improve the beneficial impacts of PGPB on crops.
Previous work has indicated that B. subtilis biofilm formation can be influenced by interactions with neighboring microbes (Powers et al. 2015; Shank et al. 2011). Documented examples of biofilm synergy (i.e., mixes of bacteria increasing biofilm biomass beyond simply additive effects) (Ren et al. 2015) led us to consider the possibility that B. subtilis (and other Bacillus PGPB) might form more stable associations with plant roots when in the presence of additional plant-derived microbes.
Here, we tested the hypothesis that native rhizosphere bacteria can promote maintenance of B. subtilis on Arabidopsis thaliana roots. We chose A. thaliana as our plant host because similar studies using Bacillus PGPB have been conducted with this model plant (Asari et al. 2017). To test this hypothesis, we used our previously developed hydroponic plant-growth system (Harris et al. 2019) to screen a library of rhizosphere-derived bacterial isolates to identify strains that increase B. subtilis maintenance on A. thaliana seedling roots. We identified five bacterial isolates that significantly increased B. subtilis colonization and maintenance on A. thaliana roots. In addition, coculture with three of these strains (either alone or in combination) increased maintenance of agriculturally relevant species of Bacillus. Taken together, these data suggest that mixed bacterial interactions can prolong colonization of Bacillus on plant roots and that application of multispecies inoculants might further improve PGPB-based agricultural interventions (Wu et al. 2015).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All Bacillus and rhizosphere isolates used in this article are listed in Supplementary Table S1. Overnight cultures of B. subtilis ES748 and ES749 were grown on Lysogeny Broth (LB)-Lennox medium (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter) at 30°C. For inoculation, bacteria were suspended in 10 mM MgCl2 + 15% glycerol to a concentration of approximately 5 × 107 CFU/ml. To select for growth of Bacillus strains for CFU determinations from mixed bacterial samples, bacterial suspensions were grown on agar plates containing 80% mannitol salt agar medium and 20% LB-Lennox medium. When needed, chloramphenicol was used at 5 μg/ml and erythromycin-lincomycin was used at 1 and 25 μg/ml.
The rhizobacterial strains screened were 96 bacterial strains previously isolated from the roots of A. thaliana grown in natural soil (Lebeis et al. 2015; Lundberg et al. 2012). Working stocks of these strains were made as described above and dispensed into 100-μl aliquots that were frozen at −80°C before use. Aliquots were thawed at room temperature prior to inoculation.
Colonization of A. thaliana roots in a hydroponic growth system.
A. thaliana ecotype Col-0 seed were grown hydroponically, essentially as described by Harris et al. (2019). Sterilized mesh disks (Stretchable High-Temperature PTFE Plastic Mesh; 0.045-by-0.025-in. opening, 18 in. wide; McMaster-Carr 1100T43) of 0.5 cm in diameter were cut using a standard hole punch, sterilized by autoclaving, and distributed in a single layer across the surface of agar plates containing 0.5× Murashige-Skoog (MS) salts (with morpholineethanesulfonic acid buffer and ethylenediaminetetraacetic acid ferric sodium [NaFe-EDTA] constituting a stock solution at 5 ml/liter containing 5.57 g of FeSO4 · 7H2O and 7.45 g of Na2-EDTA) at 50 mg/liter. Two surface-sterilized seeds of A. thaliana previously stratified at 4°C were placed on each mesh disk and the entire plate was sealed with gas-permeable tape (BS-25 Aeraseal Excel Scientific) before being placed in a Conviron incubator set for long-day plant growth (16 h of light, 21°C day and 18°C night temperatures). Seed were germinated for between 8 and 12 days.
A. thaliana Col-0 seedlings were colonized with bacterial isolates as previously described (Harris et al. 2019), with some modifications. Germinated seedlings embedded in sterilized mesh were transferred to 24-well plates containing 0.1× LB liquid medium. Bacterial suspensions (10 μl each) of approximately 5 × 107 CFU/ml were added to the liquid medium. After covering with a gas-permeable membrane and replacing the lid, plates were incubated in a Conviron incubator set for short-day plant growth (9 h of light, 21°C day and 18°C night temperatures). In the incubator, the plates were placed on a shaker set at 200 rpm to prevent anoxia of the medium (Haney et al. 2015; Harris et al. 2019).
Maintenance of colonization of A. thaliana roots in a hydroponic growth system.
Following 20 h of incubation with bacteria, the mesh disks containing seedlings were removed from the inoculation wells and transferred to wells of a 24-well plate containing 1 ml of 10 mM MgCl2 and allowed to stand at room temperature for 10 min to remove any bacteria not tightly associated with the roots. At this point, seedlings were either used for A. thaliana hydroponic maintenance assays or collected for quantification of colonization CFU from A. thaliana seedlings. To assay hydroponic maintenance, after the 10 min “rinse”, mesh with seedlings were transferred to a 24-well plate filled with 1.0 ml of 0.5× MS liquid media. After covering with a gas-permeable Aeraseal and the 24-well plate lid, the 24-well plate was transferred to an orbital shaker set to 220 rpm in a Conviron incubator set for short-day plant growth.
Quantification of bacterial CFU from A. thaliana seedlings.
Following either A. thaliana hydroponic colonization or maintenance, mesh disks and their embedded seedlings were removed from wells and transferred to wells of a 24-well plate containing 1 ml of 10 mM MgCl2 and allowed to stand at room temperature for 10 min to remove bacteria not tightly associated with the roots. Both seedlings were removed from their mesh using EtOH-flame-sterilized forceps and transferred to wells of a 24-well plate containing 1 ml of 10 mM MgCl2. Each plate was sealed with a gas-impermeable AlumaSeal and sonicated (Qsonica Sonicator q700) three times with 20-s pauses between each round using the following program: amplitude 40, 12-s process time, 2-s pulse on, 1-s pulse off.
It was empirically determined that Mannitol-LB agar plates enable B. subtilis and B. amyloliquefaciens to grow while minimizing or eliminating the growth of the other 22 cocultured strains. Serial dilutions (10-fold) in 10 mM MgCl2 were plated on 0.1× LB with and without the antibiotics erythromycin at 6 μg/ml + chloramphenicol at 5 μg/ml to monitor overall bacterial growth across wells, as well as on Mannitol-LB agar plates with or without the antibiotics erythromycin at 6 μg/ml + chloramphenicol at 5 μg/ml to select for Bacillus strains.
Plates for CFU per milliliter determinations were incubated at room temperature for 2 to 4 days after plating. Serial dilutions were plated on half-strength LB and full-strength LB with erythromycin at 6 μg/ml + chloramphenicol at 5 μg/ml and were counted after 1 to 2 days. Centrifuged samples plated on Mannitol-LB plates were counted 3 to 5 days later. All results represent at least three replicates performed on separate days, each of which contained at least three seedling-floats per sample type.
Imaging of bacteria on A. thaliana seedlings.
For microscopy imaging, colonization and maintenance of A. thaliana were performed as described above but using a B. subtilis strain containing a constitutive transcriptional mYPet reporter. Regions of the roots were selected for imaging based on their approximate locations relative to either end of the seedling to avoid biasing of collected images. Root attachment images were taken with a Zeiss-710 laser-scanning microscope (Zeiss, Oberkochen, Germany) and were processed and linearly adjusted using ImageJ (Schneider et al. 2012). All results represent at least three biological replicates performed on separate days, each containing at least three technical replicates.
Phylogenetic trees.
To build phylogenetic trees, we followed the protocol established by Finkel et al. (2019). Briefly, we used a supermatrix approach to build the phylogenetic tree of the rhizobacterial isolates as described by Levy and colleagues (2018). We scanned 120 previously defined marker genes across the rhizosphere isolate collection genomes using the hmmsearch tool from the hmmer version 3.1b2 (Wheeler and Eddy 2013). We selected 47 markers that were present as single-copy genes in 100% of our isolates and aligned each individual marker using MAFFT (Katoh and Standley 2013). Low-quality columns in the alignment were filtered using trimAl (Capella-Gutiérrez et al. 2009). Afterward, all filtered alignments were concatenated into a superalignment. Phylogeny was inferred using FastTree version 2.1 (Price et al. 2010) via the WAG model of evolution.
Statistical analysis.
Calculation of variance (one-way analysis of variance [ANOVA]) and mean comparison between treatments were carried out based on the Dunnett’s multiple comparison test at the 0.05 probability level using Graphpad PRISM version 8.3.1 for Windows (GraphPad Software, La Jolla, CA U.S.A.).
RESULTS
Monitoring loss of B. subtilis root colonization in a hydroponic system.
To begin investigating the kinetics of B. subtilis colonization and maintenance, we utilized a hydroponic growth system (Fig. 1) previously published by our laboratory (Harris et al. 2019). Briefly, we germinated sterile A. thaliana seedlings on agar on top of small pieces of sterile mesh; when germinated in this manner, the roots migrated to the opposite side of the mesh from the cotyledons. The mesh allowed the seedlings to be easily transferred to liquid wells containing various bacterial strains. After 20 h of inoculation to permit bacteria to colonize the roots, we gently washed and transferred the plants and their mesh to 0.5× MS, a plant minimal salts medium. Following transfer to 0.5× MS, we removed and rinsed the plants at either 0, 12, 24, 48, 72, or 96 h post-transfer; we then removed bacteria from the surface of the roots through sonication and measured the relative number of B. subtilis CFU. Within our system, B. subtilis NCIB3610 (hereafter simply B. subtilis) repeatedly colonized A. thaliana seedlings to approximately 1 × 104 CFU/seedling (Fig. 2A). However, following transfer to the plant minimal salts liquid medium, B. subtilis CFU declined sharply for the first 24 h, at which point the titer leveled out at approximately two log-fold below the initial colonization levels (Fig. 2A). Therefore, we set out to determine whether bacterial isolates from the native rhizosphere of A. thaliana could individually colonize and maintain on roots or were able to affect the association of B. subtilis with plant roots.
Fig. 1.
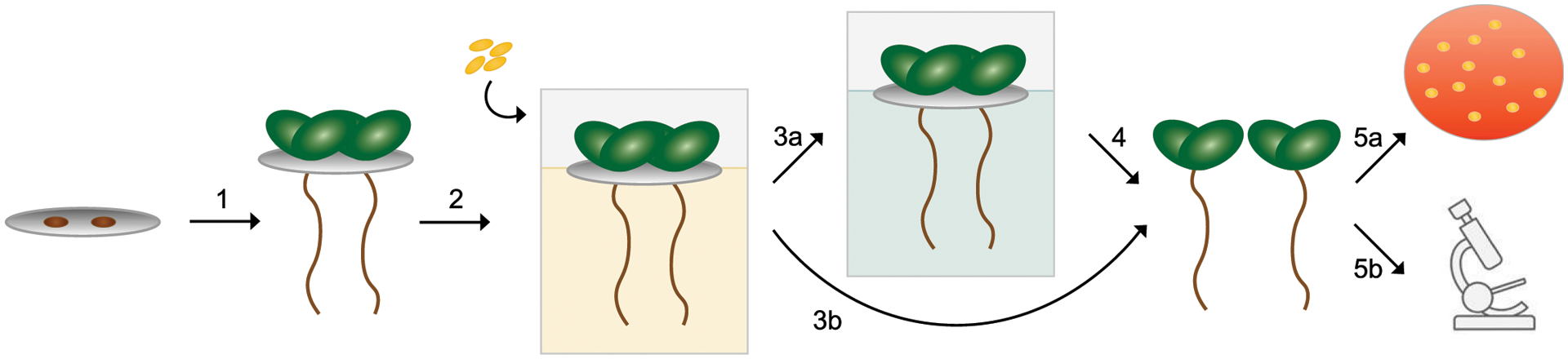
Schematic of hydroponic growth system for quantification and imaging of bacterial associations with the roots of Arabidopsis thaliana seedlings. (1) Two surface-sterilized seeds of A. thaliana were germinated on discs of sterile Teflon mesh for 7 to 10 days on top of agar. (2) Mesh discs embedded with two seedlings were transferred to liquid colonization medium, into which bacterial strains were inoculated. (3) After 20 h of colonization, the mesh and seedlings were rinsed and either (3a) the seedling was transferred to the liquid maintenance medium or (3b) the seedlings were immediately (4) removed from the multiwell plate for sample collection, to be either (5a) sonicated in liquid to resuspend bacteria for plating for CFU determinations or (5b) transferred to microscope slides for fluorescence microscopy imaging.
Fig. 2.
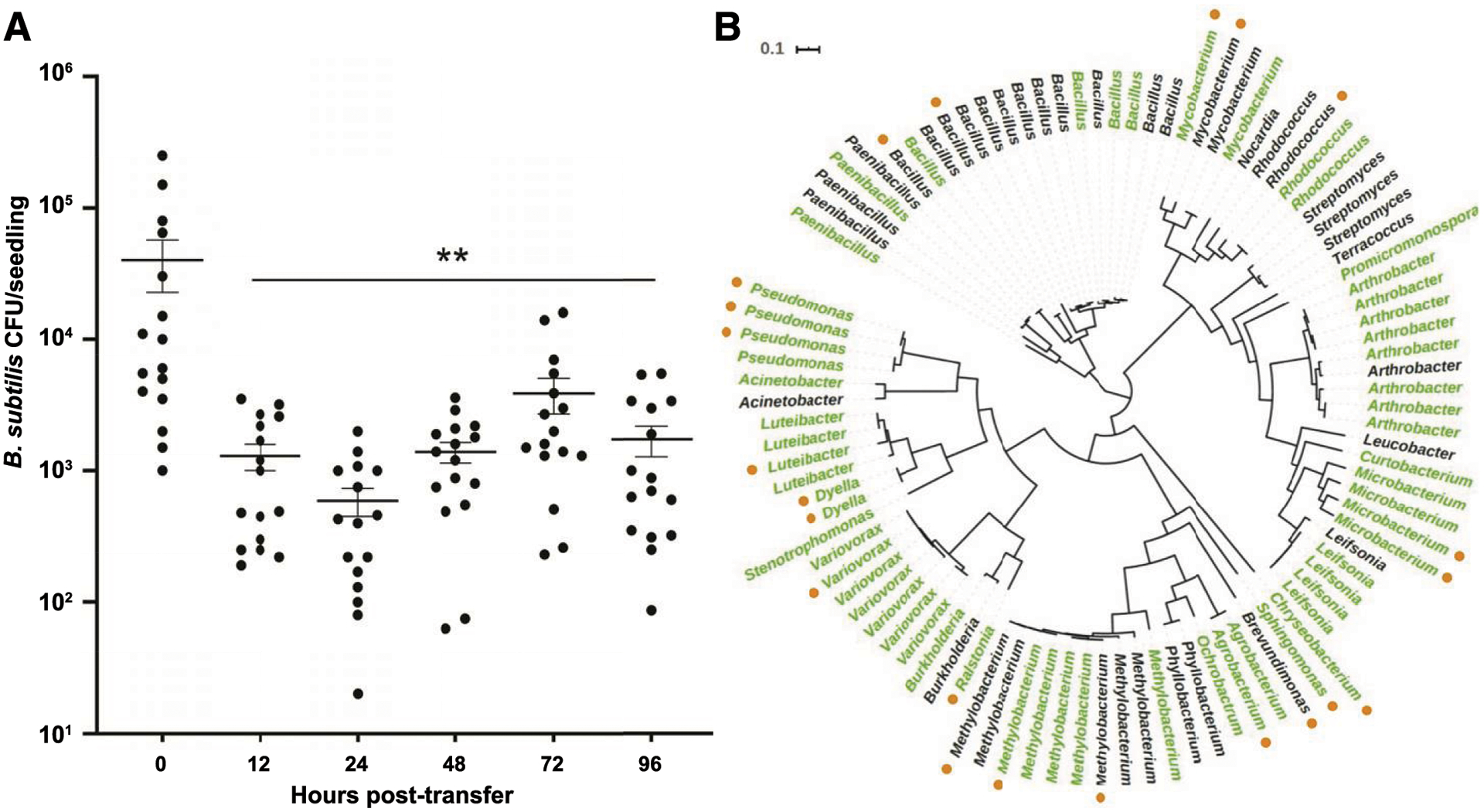
A, Bacillus subtilis colonizes the hydroponic roots of Arabidopsis thaliana seedlings but does not maintain its association when seedlings are transferred to minimal salts medium for continued incubation. Error bars = standard error of the mean; ** indicates P < 0.01. B, An unrooted phylogenetic tree showing the relationships between the 96 rhizosphere bacterial strains tested for their hydroponic association with A. thaliana roots. Isolates found to maintain their associations with A. thaliana roots at a level at least one log-fold higher than B. subtilis alone are shown in green. The outer ring of orange dots indicates the 22 isolates selected in the primary screen for further study.
Maintenance of root colonization by native rhizosphere isolates.
During the design of our hydroponic bacterial-plant-root colonization and maintenance system (Fig. 1), we discovered that some bacteria were better able to colonize and maintain on roots than others (Harris et al. 2019). Therefore, we wanted to more broadly identify which members of a natural plant microbiome were able to maintain their association with root seedlings over time. We aimed to not only identify individual strains as potential PGPB but also look for phylogenetic patterns of the bacteria and their behaviors to determine whether particular bacterial clades were more consistently able to reassociate with plant roots.
To do so, we screened 96 fully genome-sequenced bacterial strains (Fig. 2B) originally isolated from the rhizosphere of A. thaliana ecotype Col-0 (Lebeis et al. 2015; Lundberg et al. 2012) for isolates that were able to maintain their colonization over 24 h. We elected to set the cutoff for “maintenance” as being at least one log-fold higher than the number of B. subtilis CFU in the same experimental replicate after 24 h of maintenance. Our initial screen identified 51 of these 96 bacterial isolates that were able to maintain on the root at least one log-fold higher titer than B. subtilis (Fig. 2B). Thus, 61% of the tested isolates were classified as “maintainers” and 39% (including B. subtilis) were classified as “nonmaintainers.” These designations did not show any obvious correlation to the phylogeny of the bacteria (Fig. 2B; Supplementary Fig. S1).
Identification of strains from the rhizosphere that promote B. subtilis maintenance on roots.
We were intrigued that not all of the native A. thaliana rhizosphere isolates were able to maintain their associations with roots over time in monoculture, despite being originally isolated from the rhizosphere of the same A. thaliana ecotype. Based on existing literature, which indicates that bacterial interactions can increase biofilm biomass (Ren et al. 2015), we wondered whether the presence of a maintainer could increase persistence of a nonmaintainer such as B. subtilis. To examine this question, we coinoculated each of the 96 rhizosphere isolates with a strain of B. subtilis engineered to express a constitutive fluorescent protein that is also resistant to multiple antibiotics (ES749 B. subtilis NCIB3610 amyE::PspacC-mTurq [cmR]; lacA::PtapA-mYpet [ermR]) (Yannarell et al. 2019) at similar initial CFU per milliliter on A. thaliana seedlings. Following 1 day of incubation in 0.5× MS, we compared B. subtilis CFU per seedling when inoculated alone or with one of the rhizobacteria. Using this method, we flagged 22 of the 96 strains as putative helper bacteria: in coculture, these 22 strains appeared to increase the CFU of B. subtilis maintenance by at least a log-fold relative to when B. subtilis was inoculated alone (Fig. 3). Not all of these strains were initially identified as maintainers on their own (Fig. 2B), suggesting that the ability of individual strains to “help” B. subtilis maintain may not be dependent on their own ability to persist on roots. It is important to note that, for technical reasons, we did not quantify the CFU of these rhizosphere strains but only examined their effects on B. subtilis.
Fig. 3.
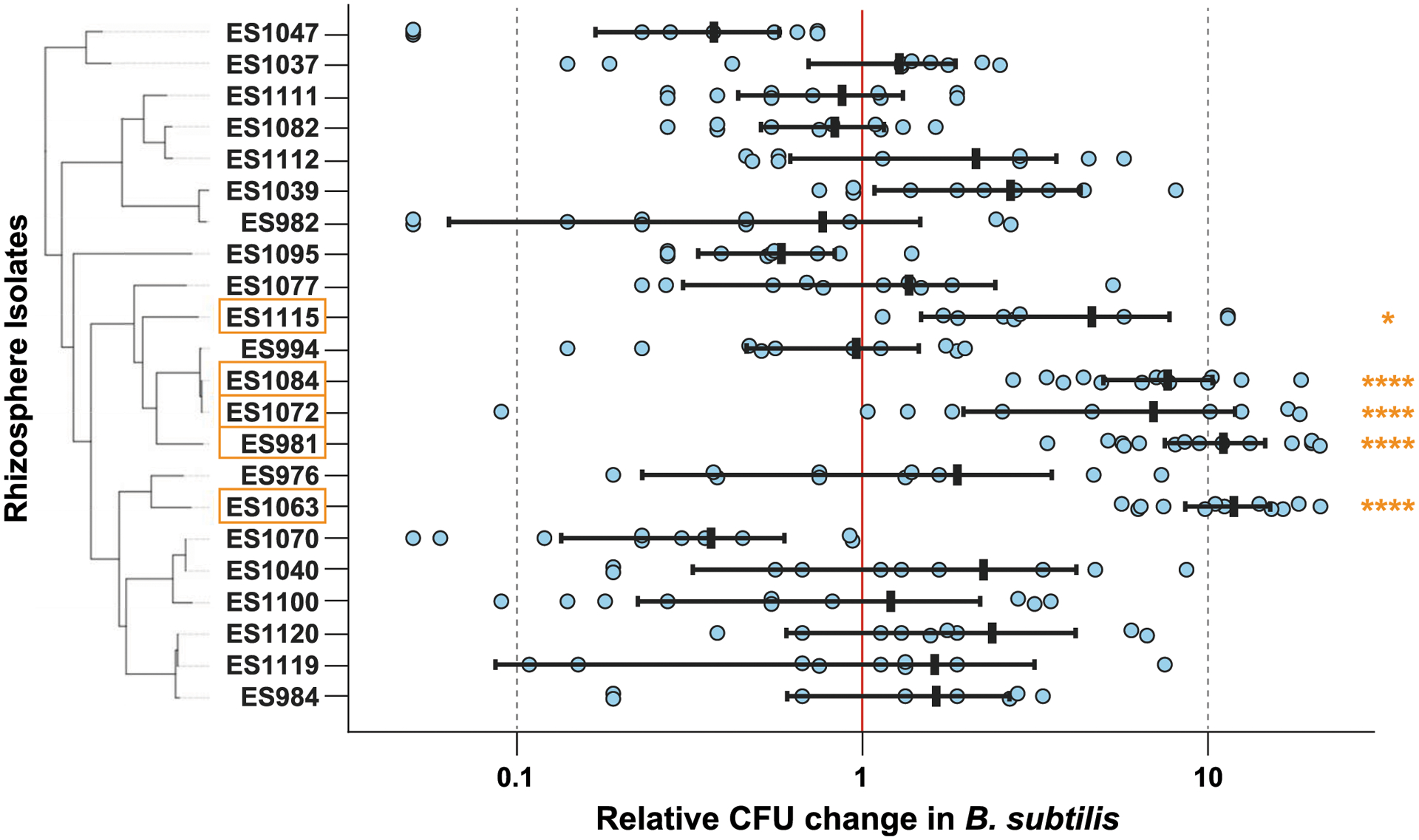
We extensively retested the 22 rhizosphere isolates initially implicated to increase Bacillus subtilis maintenance when in coculture. Of these strains, we identified five strains that reproducibly increased B. subtilis maintenance, which are outlined in orange; four of these strains increased B. subtilis by one log-fold (ES981, ES1063, ES1084, and ES1115). Differences in B. subtilis CFU per seedling when coinoculated with an isolate are reported as log-fold changes compared with the average B. subtilis CFU per seedling in the same biological experiment. Error bars = standard error of the mean; * and **** indicate P < 0.05 and 0.0001, respectively.
To validate and confirm which of these 22 strains could reliably increase maintenance of B. subtilis on the roots of A. thaliana, we then quantified the effect of these strains in a secondary screen across multiple days and with additional replicates relative to our initial screen (specifically, testing at least three mesh floats containing two seedlings grown in individual wells per sample type per day) (Fig. 3). Values above the line of neutrality (Fig. 3, the middle “1” line on the y-axis) indicated that coculturing had a positive effect on B. subtilis adherence. Coculture with five strains (Agrobacterium sp. ES981, Variovorax sp. ES1063, Methylobacterium sp. ES1072, Methylobacterium sp. ES1084, and Brevundimonas sp. ES1115) significantly increased B. subtilis maintenance (P < 0.001 for ES981, ES1063, ES1072, and ES1084; P < 0.05 for ES1115). Although two Methylobacterium spp. are within this group, the overall phylogenetic diversity of these five helper strains is broad and appears unconserved across phylogeny. The other 17 strains that passed our initial screen but not our more rigorous secondary screen were likely artifacts from the high levels of biological variability observed for these strains. Of the five helper strains, we chose three to focus on during subsequent experiments (Agrobacterium sp. ES981, Variovorax sp. ES1063, and Methylobacterium sp. ES1084) due to their lower sample variance compared with ES1072 and their stronger significance compared with ES1115 (Fig. 3). These three strains were also found to be maintainers when in monoculture association with the root (Fig. 2B).
Effects of cocolonization on the spatial distribution of B. subtilis on the root surface.
Bacillus spp. show preferences in the location of their attachment along the length of plant roots, presumably based on the zone of root cell differentiation (Fan et al. 2012); furthermore, the colonization preferences of some Bacillus spp. have been shown to be affected by the presence of other bacteria (Massalha et al. 2017). Finally, bacterial biofilms found on natural surfaces often comprise multiple species (Flemming et al. 2016). Thus, we wondered whether and how these three helper strains were affecting the spatial localization patterns of B. subtilis on A. thaliana roots, and whether different bacterial species were in close contact on the roots or whether they were spatially segregated.
To address this question, we performed the maintenance assay as before but now also collecting seedlings following the colonization step (prior to the transfer to 0.5× MS). In addition, for these experiments, we used a B. subtilis strain engineered to constitutively express the mYPet fluorescent protein (ES748 B. subtilis NCIB3610 lacA::PtapA-mYPet [ermR]) (Yannarell et al. 2019). Other than the fluorescent gene construct (mYPet versus mTurq) and a different antibiotic resistance gene, this strain was genotypically identical to the parental B. subtilis strain used in our original screening assay; however, its brighter fluorescence allowed us to better visualize the bacteria along the root using microscopy.
Using confocal laser fluorescence microscopy, we imaged the roots at four sections along their length during colonization and maintenance: the crown (where the root meets the shoot), the upper middle (in the half closest to the crown) and lower middle (in the half closest to the root tip) sections of the main root, and the tip of the main root. We saw the most consistent differences across seedling roots at the lower-middle section of the root (Fig. 4A) (representative images from all four locations along the root length are shown in Supplementary Figure S2). Following colonization with B. subtilis alone, only a few B. subtilis cells were visible along the surface of the root; in comparison, slightly more fluorescent bacteria were found when B. subtilis was coinoculated with the Methylobacterium spp., and clumps of fluorescent bacteria were visible when B. subtilis was coinoculated with either the Agrobacterium or the Variovorax sp. (Fig. 4A).
Fig. 4.
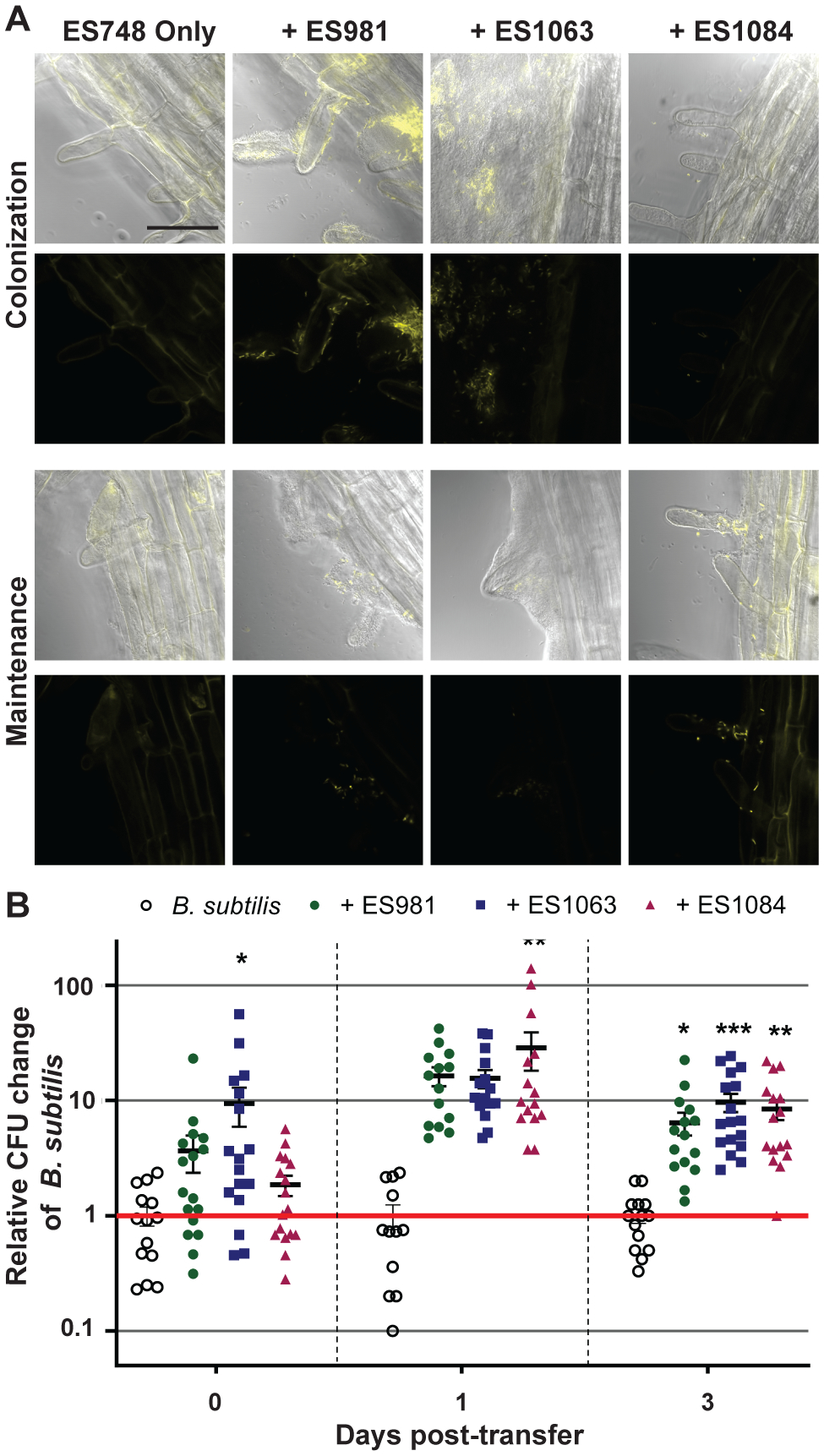
A, Distributions of Bacillus subtilis on the lower regions of plant roots following colonization (top) and maintenance (bottom) when inoculated either alone or with the indicated strains. Fluorescent cells were false colored yellow in both the DIC-fluorescent image overlays and the fluorescent images. Images from each section were collected from at least two technical replicates of three independent biological replicates of these experiments. Bar = 50 μm. B, Plant root attachment of B. subtilis following colonization (0 days) and maintenance (1 and 3 days) is increased by cocolonization with either ES981, ES1063, or ES1084. Differences in B. subtilis CFU per seedling when coinoculated with another strain are reported as log-fold changes compared with the average B. subtilis CFU per seedling of three replicates in the same biological experiment. Error bars = standard error of the mean; *, **, and *** indicate P < 0.05, 0.01, and 0.001, respectively.
Following maintenance, we also observed differences in B. subtilis abundance when in association with A. thaliana alone or with cocolonizing bacteria: it was rare to visualize any fluorescent cells on roots inoculated with B. subtilis alone; in contrast, fluorescent B. subtilis cells were always apparent when B. subtilis was coinoculated with either the Agrobacterium sp. or the Variovorax sp. There appeared to be fewer B. subtilis cells at the lower-midroot location following colonization with the Variovorax sp. as compared with coinoculation with the other two helper strains; however, B. subtilis cells were more prevalent following maintenance when coinoculated with the Variovorax sp. compared with when coinoculated with either the Agrobacterium sp. or the Methylobacterium sp. (Fig. 4A).
Because none of these three helper strains were engineered to express a fluorescent protein and they did not exhibit appreciable autofluorescence, we could not determine their exact locations on the root. However, based on DIC images, it appeared that these bacteria were often in direct contact with the B. subtilis cells. For example, during cocolonization with the Agrobacterium sp., it appears that small clumps of B. subtilis cells are encased in a larger colony of nonfluorescent cells, which most likely are Agrobacterium cells. This apparent mixing is more obvious when visualized through three-dimensional reconstructions of confocal microscopy images taken at multiple Z-planes (Supplementary Videos S1, S2, S3, and S4).
The most promising strains increase colonization and maintenance of B. subtilis.
To determine how these three strains affected the kinetics and dynamics of the association of B. subtilis with the plant root, we performed coinoculation assays as before but now removed plants for determination of bacterial CFU at multiple timepoints (at the initial transfer after colonization, after 1 day of maintenance, and after 3 days of maintenance). Coinoculation with each of the three helper strains increased B. subtilis’s colonization by at least one log-fold as compared with B. subtilis’s colonization alone (Fig. 4B). Due to high variance between technical replicates and biological experiments, however, some of the day 1 results do not achieve statistical significance (see Discussion for more about our use of an ANOVA versus an unpaired t test here and below). Nevertheless, a clear trend is apparent. Importantly, this increase in colonization was not due to a general increase in B. subtilis growth in the coculture colonization media itself, because B. subtilis CFU counts from the liquid medium showed no effect of coinoculation (Supplementary Fig. S3). In addition, although the overall titer of B. subtilis CFU/root decreased between 1 and 3 days of incubation in maintenance medium, we observed approximately the same trends at both time points. Thus, based on these results, in future screening efforts to identify isolates able to increase of B. subtilis maintenance, 1 day of maintenance incubation may be sufficient to identify additional helper strains. It is worth noting that our assays require destructive sampling; thus, we could not compare the colonization and subsequent maintenance on individual plants over time.
Maintenance of agriculturally relevant Bacillus PGPB also improved by strains that affect B. subtilis both individually and in combination.
Up to this point, we used the undomesticated B. subtilis NCIB3610 strain (Nye et al. 2017) for our screens due to the wealth of information about its behavior (Vlamakis et al. 2013) and range of available genetic tools, as well as the observation that it inhibits invasion by the pathogenic Ralstonia solanacearum on tomato plants (Chen et al. 2013). However, we next wanted to determine whether the three helper strains that increased the maintenance of B. subtilis NCIB3610 were also able to promote the maintenance of other Bacillus spp., including Bacillus strains used as PGPB additives in agricultural interventions. We tested whether these helper strains or their combination could affect the maintenance of B. amyloliquefaciens strains GB03 and FZB42, which are widely used as PGPB additives in agriculture (Chowdhury et al. 2015; Wu et al. 2015), and had been previously compared with B. subtilis 3610 in terms of their biofilm-forming capabilities (albeit in a slightly different growth format) (Beauregard et al. 2013). Interestingly, over time, FZB42 also exhibited decreasing CFU on lettuce plants compared with its levels at colonization (Kröber et al.2014).
Based on the results from our microscopy imaging (Fig. 4A), it appeared that the three helper strains we had identified affected localization of B. subtilis differently. Therefore, we considered whether each individual strain might affect localization of B. subtilis through distinct mechanisms that could be mutually beneficial if combined. This concept is consistent with the idea that multispecies communities can exhibit emerging properties that arise through complex multiway interactions (Coyte et al. 2015). Therefore, we wanted to determine both how each of the three strains individually would affect the maintenance of these Bacillus spp. as well as whether their combination would have additive or synergistic impacts on the Bacillus PGPB.
We inoculated plants with each separate Bacillus strain and the three helper strains either alone or in a 1:1:1 mix. After 1 day of maintenance, we plated sonicated root samples on selective mannitol/LB agar plates without antibiotics and counted Bacillus spp. CFU (note that none of the helper strains grew on mannitol/LB, whereas all of the Bacillus strains did). We then compared the number of the Bacillus CFU in the presence of the other strains compared with the Bacillus-only inoculation (Fig. 5). Due to biological variability, in some cases, our analysis of these data did not indicate statistical significance; however, once again there was a clear trend in the data: the helpers improved Bacillus maintenance on roots relative to monoculture inoculation.
Fig. 5.
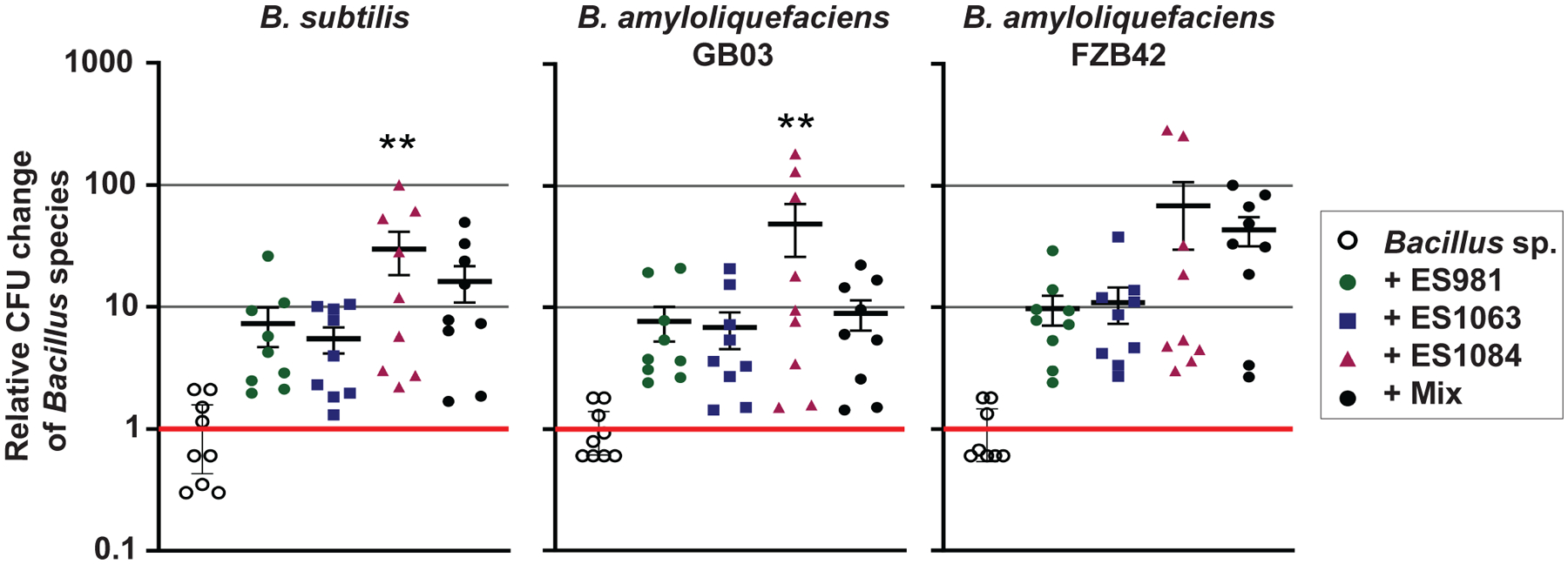
We inoculated three Bacillus spp. (Bacillus subtilis ES749, B. amyloliquefaciens GB03, and B. amyloliquefaciens FZB42) either alone or with the addition of ES981, ES1063, and ES1084, either with each strain alone or all together (Mix). Graphs show the differences in Bacillus CFU per seedling when coinoculated with the other bacteria, reported as log-fold changes compared with the average Bacillus sp. CFU per seedling of three monoculture replicates in the same biological experiment. Error bars = standard error of the mean; * and ** indicate P < 0.05 and 0.01, respectively.
We found that coinoculation with each of the three strains individually did increase the maintenance of B. subtilis and both B. amyloliquefaciens strains (Fig. 5). These results indicate that screening for maintenance effects of coinoculation of B. subtilis 3610 with bacterial isolates may be used to identify helper strains that can also affect other agriculturally relevant Bacillus spp. Unexpectedly, however, when all three helper strains were collectively coinoculated, they did not increase the maintenance of the Bacillus strains more than any of the individual helpers did (i.e., there was no synergy of their interactions or additional benefits of combining them) (Fig. 5). This lack of observed benefit could have been the result of helper-helper antagonism (e.g., due to the helpers exerting their effects through similar and, thus, not additive pathways) or due to a physical or structural limit on the number of bacterial or Bacillus cells the plant root can support. These findings suggest that, in at least some cases, using a combination of strains might not supplement (or detract) from the effects of the presence of other bacteria.
DISCUSSION
To leverage the potential power of microbe-based agricultural interventions, we must first understand how diverse biotic and abiotic environmental factors mediate plant productivity. This includes studying how mixed microbial species affect phytobiome health and stability. The rhizosphere microbiomes of plants grown in natural soils frequently comprise hundreds of species (Lundberg et al. 2012). The assembly of these complex plant microbiomes often requires interspecies bacterial interactions, where the presence of certain species facilitates the subsequent growth, maintenance, and succession of other species (Bradáčová et al. 2019; Niu et al. 2017; Y. Zhang et al. 2019). Indeed, recent studies found that B. subtilis NCIB3610 colonization of melon roots is affected by Pseudomonas chlororaphis (Molina-Santiago et al. 2019), while peptidoglycan from B. cereus facilitates rhizospheric bacterial commensalism (Peterson et al. 2006). Therefore, we reasoned that interbacterial interactions could play a role in the persistence and behavior of Bacillus PGPB on plant roots.
B. subtilis is commonly utilized as a PGPB; it has been shown to increase root growth of melon seedlings (Magno-Pérez-Bryan et al. 2015) and inhibit plant pathogen invasion through production of specialized metabolites (Chen et al. 2013). Although B. subtilis strains have been included in commercial agricultural bioadditives, other strains of Bacillus such as B. amyloliquefaciens FZB42 and GB03 are more widely used for such purposes (Aloo et al. 2019; Wu et al. 2015). That said, Bacillus spp. added to plant growth systems as PGPB do not always maintain their initial colonization on roots over time (Gadhave et al. 2018; Kröber et al. 2014).
Using our hydroponic growth assay to measure bacterial presence on plant roots, we identified rhizosphere bacteria that could maintain root colonization on their own as well as those that could promote maintenance of the model bacterium B. subtilis and PGPB strains of B. amyloliquefaciens on A. thaliana. Despite all tested bacterial coinoculants being originally isolated from the rhizosphere of A. thaliana grown in natural soil (Lundberg et al. 2012), only 51 of the 96 rhizobacteria tested were able to maintain their independent associations with A. thaliana roots over time. (It is important to note that some of the 44 strains classified as nonmaintainers may potentially still be present on plant roots but simply at lower levels than our cutoff.) Even so, the overall low percentage of rhizobacteria able to maintain on plant roots on their own suggests that additional microbe–microbe or microbe–environment interactions may be important to promote bacterial association with roots.
We next identified five rhizosphere isolates that significantly (P < 0.05) increased the CFU of B. subtilis associated with roots (single strains each of Agrobacterium, Variovorax, and Brevundimonas and two Methylobacterium strains). Again, a relatively small percentage of the tested isolates affected B. subtilis root association, even though all of the rhizosphere bacteria examined were originally isolated from native soil rhizospheres. These results are consistent with strain specificity being crucial for interbacterial interactions, an idea supported by the fact that multiple phylogenetically close relatives of the identified helper strains were not able to increase B. subtilis maintenance in our assay. When we more deeply explored three of these helper strains (ES981, ES1063, and ES1084), we saw that they were able to increase the association of B. subtilis with roots over time, while also generally improving the root associations of B. amyloliquefaciens. Bacteria belonging to the genera these helpers are a part of (the Agrobacterium, Variovorax, and Methylobacterium clades) have all previously been found in association with plants; however, minimal existing literature explicitly discusses the potential interactions of bacteria from these groups with Bacillus spp. What is known is that some strains of B. subtilis and B. amyloliquefaciens reduce the incidence of disease by pathogenic Agrobacterium tumefaciens, which causes crown gall on eudicots, when cocolonized (Frikha-Gargouri et al. 2017). Notably, although Agrobacterium spp. are most commonly considered plant pathogens, some species have been shown to have a phytostimulatory effect (Walker et al. 2013). That said, work clearly remains to understand the specifics of the mechanisms by which these particular helper strains either directly or indirectly affect the association of Bacillus with plant roots.
The cell–cell interactions between plant-associated microbes such as the helpers identified here and Bacillus PGPB may be reliant on chemical or physical mechanisms. Microbial chemical communication that alters bacterial behavior is well established (Abisado et al. 2018; Mukherjee and Bassler 2019; Straight and Kolter 2009), and some strains of Variovorax, Agrobacterium, and Methylobacterium have been shown to have the potential to make or degrade the quorum-sensing molecules N-acyl homoserine-lactone or autoinducer-2 (AI-2) (Daniels et al. 2004; Dourado et al. 2013; Han et al. 2011; Leadbetter and Greenberg 2000). Whether and how B. subtilis responds to such quorum signals (or other microbial chemical cues) in ways that might increase their ability to maintain their association with plant roots is not yet clear, although there is evidence that AI-2 may affect biofilm formation in some B. subtilis strains (Duanis-Assaf et al. 2016; Lombardía et al. 2006). Directed experiments using mutant helper strains lacking genes known to be involved in these microbial signaling systems could provide information about the underlying mechanism of the interactions described here. In addition, metabolomics of these plant–microbe systems or comparative transcriptomics of B. subtilis associated with the plant root either with or without helpers could also provide additional mechanistic insights. It is also possible that the helpers’ ability to enhance B. subtilis maintenance could be the indirect result of the plant’s response to microbial inoculation. As an example, the addition of root exudates into bacterial growth agar affects the interactions between B. cereus and other plant commensal bacteria (Peterson et al. 2006). Thus, exploring how patterns of root secretions change due to inoculation with helper strains in future work may provide metabolic insights into how B. subtilis is better able to remain on the root over time. Complementary studies testing how mutagenized plant lines, or plant lines containing fluorescent transcriptional reporters for specific candidate genes of interest, respond to differences in microbial maintenance could also potentially reveal relevant plant–microbe signals.
There also may be physical mechanisms at work to alter B. subtilis plant root maintenance, either directly or in response to chemical cues such as those just discussed. Indeed, our confocal fluorescence microscopy images of fluorescently labeled B. subtilis coinoculated with the three helper strains corroborates the idea that plant-associated bacterial biofilms may be affected by coinoculation. Our images and movies clearly show that these different species are often in direct contact within the root-associated biofilms. Future work with all bacterial strains being fluorescently labeled either using genetic (Downie et al. 2014; Massalha et al. 2017) or chemical (Welch et al. 2016) means would enable their physical relationships on the root to be more precisely described. These approaches would be particularly beneficial for visualizing mixtures of more than two bacteria; in part, because inoculation of Bacillus spp. has previously been shown to modify the root endophytic bacterial diversity (Qiao et al. 2017), it would be interesting to see how B. subtilis affects the localization of the helpers along the root rather than just how the helpers affect B. subtilis’s localization. In addition, gaining an understanding of whether direct physical interaction between these bacteria is required for increased PGPB association with roots (or whether diffusion of their metabolites may be sufficient) may inform methods for future agricultural interventions.
It is interesting to note that, although the three helper strains we identified consistently demonstrate strong trends toward increasing the association of Bacillus spp. with plant roots, there is significant biological variability observed in the data. This is consistent with published articles demonstrating that plants inoculated with a Bacillus PGPB led to increased variance of plant growth, rather than a consistent increase across all plants (Gadhave et al. 2018). However, this increase in variance was only apparent when the individual data points were examined (and the data points were not obscured by box-and-whisker plots or bar graphs) (Weissgerber et al. 2015). Our data demonstrate clear trends that coinoculation with these strains increases the presence of Bacillus spp. on Arabidopsis thaliana roots, even though an ANOVA analysis did not indicate statistical significance in all cases. Because of this, we further analyzed these data using multiple unpaired t test comparisons and found that, using this approach, all of the helpers increased the association of B. subtilis with the roots (at both days 1 and 3, with a significance of P < 0.05 or less). Furthermore, with the exception of the combination of FZB42 with ES1084, the other Bacillus strains also demonstrated significant increases on the root with all helper strains by t test (again, with P < 0.05). However, although t test comparisons are commonly used in the field, the ANOVA analysis we present is more stringent and appropriate for this type of data, which is why we focus on that approach. Regardless, altogether, our data and both statistical analyses indicate that these helper strains increase the association of Bacillus PGPB with A. thaliana roots.
Previous studies have examined the effects of mixed PGPB consortia on plant growth and microbial composition (Hashmi et al. 2019; Jha et al. 2013; L.-N. Zhang et al. 2019) but, here, we were interested in the (related) question of whether multiple potential helper strains showed additive or synergistic impacts on a single PGPB when coinoculated. Multispecies communities are typically more stable that single strains are, and emergent properties can arise in complex multiway interactions (Coyte et al. 2015). However, in this case, we found that combining helper strains did not increase the maintenance of the PGPB bacilli over that of the most-effective helper alone. Even so, in more complex natural environments, the benefit of applying multiple strains simultaneously might stabilize their impact across different environments; indeed, adjustments to our assay could be made to directly scan a variety of possible environmental perturbations (nutrient availability, salinity, and so on) to identify suites of helper strains able to benefit PGPB under a range of growth conditions. At this point it is also unclear whether the bacterium–root associations we have described translate into beneficial plant phenotypes. The timeframe of our assays (up to 96 h) did not allow any differences in A. thaliana growth or morphology to be detected. Future work using either longer incubation periods or other, larger plant hosts will be required to better monitor the consequences of these bacterium–root associations on plant phenotypes of interest.
It remains unclear whether our findings in this hydroponic growth system will translate into soil settings. Previous work indicates that, in at least some cases, the essential elements of native growth environments can be sufficiently recapitulated in lab systems to provide important insights. For example, Chen et al. (2013) demonstrated that the ability of wild B. subtilis isolates to form biofilms correlated with the ability of these strains to colonize roots in soil and inhibit the growth of the pathogen R. solanacearum. A comparative study using confocal microscopy showed that FZB42 associated with root hairs in both gnotobiotic and soil growth systems, despite the root morphology of plants grown on agar differing from those grown in soil and the initial bacterial inoculum needing to be higher in soil systems for bacteria to be observable (Fan et al. 2012). In addition, some Bacillus and Pseudomonas spp. that inhibit pathogens on agar plates were able to suppress disease in soil pot assays, suggesting that these bacteria may have similar metabolic activity in both growth settings (Asaka and Shoda 1996; Khabbaz et al. 2015). Such metabolic similarities are not universal, however: a B. amyloliquefaciens antifungal compound displayed different concentration kinetics on the wheat spikes of plants grown in the greenhouse compared with those grown in the field (Crane et al. 2013), while the antifungal activity of a bacterial biocontrol agent on plates did not correlate with its ability to inhibit disease on olive plants (Mercado-Blanco et al. 2004). In sum, these results speak to the complexity of natural plant–microbe interaction systems. Nevertheless, evidence indicates that at least some plant–microbe interactions are robustly conserved. Although we are not yet capable of making reliable predictions about which relationships will display consistency across lab and field systems, we are enthusiastic about the potential translation of the hydroponic screen results described here to soil studies.
The complexity of plant–microbe interactions underscores the importance of building our knowledge from simple to more challenging growth settings. Novel technical approaches are needed to study phytobiome systems so we can better manipulate and manage phytobiomes in agricultural ecosystems across rapidly changing conditions around the world (Herrmann and Lesueur 2013). Although here we focused on the interactions between Bacillus spp. and A. thaliana, our hydroponic assay enables many different plant and bacterial species to be studied (Haney et al. 2015) under a variety of environmental conditions. This simplified and high-throughput screening pipeline was designed to investigate the role of different bacterial isolates on B. subtilis maintenance on plant roots, and provides a fast, straightforward assay that lays the foundation for future work in more natural settings (e.g., sand or soil or greenhouse experiments). Identifying microbes that affect the associations of PGPB with roots could enhance the efficacy of microbial agricultural interventions as well as improve our understanding of interkingdom interactions (Wu et al. 2015). Our work complements that of ongoing studies elucidating the effects of plant inoculation with mixed-species communities (Aleklett et al. 2018; Lozano et al. 2019; Marschner et al. 2004), especially in the context of soils (Finkel et al. 2019; Kremer et al. 2018). Integrating findings across these different efforts will be essential for transferring basic research results to future field and agricultural applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Dangl (University of North Carolina at Chapel Hill [UNC-CH]) for providing the library of rhizosphere isolates, I. S. González (UNC-CH) for constructing the phylogenetic trees, M. Fischbach (Stanford University) for providing the B. amyloliquefaciens strains, J. Winshell and the laboratory of J. Kieber (UNC-CH) for providing sterilized A. thaliana seed, C. McGehee for assistance with bacterial stock preparation, T. Perdue and the UNC Biology Department Microscopy Core for use of and support for a Zeiss 710 laser-scanning microscope, and C. Darnell for ongoing advising.
Funding: This work was supported by research funds from the National Institute of General Medical Sciences (GM112981) and the Department of Energy Biological and Environmental Research (DE-SC0013887 and DE-SC0019012) to E. A. Shank. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The e-Xtra logo stands for “electronic extra” and indicates that supplementary figures, one supplementary table, and four supplementary videos are published online.
The author(s) declare no conflict of interest.
LITERATURE CITED
- Abisado R, Benomar S, Klaus J, Dandekar A, and Chandler J 2018. Bacterial quorum sensing and microbial community interactions. MBio 9: e02331–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleklett K, Kiers ET, Ohlsson P, Shimizu TS, Caldas VEA, and Hammer EC 2018. Build your own soil: Exploring microfluidics to create microbial habitat structures. ISME J. 12:312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloo BN, Makumba BA, and Mbega ER 2019. The potential of Bacilli rhizobacteria for sustainable crop production and environmental sustainability. Microbiol. Res 219:26–39. [DOI] [PubMed] [Google Scholar]
- Asaka O, and Shoda M 1996. Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol 62:4081–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asari S, Matzén S, Petersen MA, Bejai S, and Meijer J 2016. Multiple effects of Bacillus amyloliquefaciens volatile compounds: Plant growth promotion and growth inhibition of phytopathogens. FEMS Microbiol. Ecol 92:fiw070. [DOI] [PubMed] [Google Scholar]
- Asari S, Tarkowská D, Rolčídk J, Novák O, Palmero DV, Bejai S, and Meijer J 2017. Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 245:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard PB, Chai Y, Vlamakis H, Losick R, and Kolter R 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U.S.A 110:E1621–E1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradáčová K, Sittinger M, Tietz K, Neuhäuser B, Kandeler E, Berger N, Ludewig U, and Neumann G 2019. Maize inoculation with microbial consortia: Contrasting effects on rhizosphere activities, nutrient acquisition, and early growth in different soils. Microorganisms 7:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, and Schulze-Lefert P 2013. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol 64:807–838. [DOI] [PubMed] [Google Scholar]
- Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Mustafa M, Eisen JA, Leach JE, and Dangl JL 2017. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15:e2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, and Gabaldón T 2009. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy H, Mariutto M, Henry G, Fisher C, Vasilyeva N, Thonart P, Dommes J, and Ongena M 2014. Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant-Microbe Interact 27:87–100. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yan F, Chai Y, Liu H, Kolter R, Losick R, and Guo J 2013. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol 15:848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury SP, Hartmann A, Gao X, and Borriss R 2015. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol 6:780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, and Foster KR 2015. The ecology of the microbiome: Networks, competition, and stability. Science 350:663–666. [DOI] [PubMed] [Google Scholar]
- Crane J, Gibson D, Vaughan R, and Bergstrom G 2013. Iturin levels on wheat spikes linked to biological control of Fusarium head blight by Bacillus amyloliquefaciens. Phytopathology 103:146–155. [DOI] [PubMed] [Google Scholar]
- Daniels R, Vanderleyden J, and Michiels J 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev 28:261–289. [DOI] [PubMed] [Google Scholar]
- Dourado M, Bogas A, Pomini A, Andreote F, Quecine M, Marsaioli A, and Luiz A 2013. Methylobacterium-plant interaction genes regulated by plant exudate and quorum sensing molecules. Braz. J. Microbiol 44: 1331–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie HF, Valentine TA, Otten W, Spiers AJ, and Dupuy LX 2014. Transparent soil microcosms allow 3D spatial quantification of soil microbiological processes in vivo. Plant Signal. Behav 9:e970421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanis-Assaf D, Steinberg D, Chai Y, and Shemesh M 2016. The LuxS based quorum sensing governs lactose induced biofilm formation by Bacillus subtilis. Front. Microbiol 6:1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Daim IAA, Bejai S, and Meijer J 2019. Bacillus velezensis 5113 induced metabolic and molecular reprogramming during abiotic stress tolerance in wheat. Sci. Rep 9:16282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Borriss R, Bleiss W, and Wu X 2012. Gram-positive rhizobacterium Bacillus amyloliquefaciens FZB42 colonizes three types of plants in different patterns. J. Microbiol 50:38–44. [DOI] [PubMed] [Google Scholar]
- Ferreira CMH, Soares HMVM, and Soares EV 2019. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ 682: 779–799. [DOI] [PubMed] [Google Scholar]
- Finkel OM, Salas-González I, Castrillo G, Spaepen S, Law TF, Teixeira PJPL, Jones CD, and Dangl JL 2019. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol 17:e3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, and Kjelleberg S 2016. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol 14:563–575. [DOI] [PubMed] [Google Scholar]
- Frikha-Gargouri O, Ben Abdallah D, Bhar I, and Tounsi S 2017. Antibiosis and bmyB gene presence as prevalent traits for the selection of efficient Bacillus biocontrol agents against crown gall disease. Front. Plant Sci 8: 1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadhave KR, Devlin PF, Ebertz A, Ross A, and Gange AC 2018. Soil inoculation with Bacillus spp. modifies root endophytic bacterial diversity, evenness, and community composition in a context-specific manner. Microb. Ecol 76:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J-I, Choi H-K, Lee S-W, Orwin PM, Kim J, LaRoe SL, Kim T. g., O’Neil J, Leadbetter JR, Lee SY, Hur C-G, Spain JC, Ovchinnikova G, Goodwin L, and Han C 2011. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol 193:1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney CH, Samuel BS, Bush J, and Ausubel FM 2015. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 1:15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Pelaez CA, and Shank EA 2019. Monitoring bacterial colonization and maintenance on Arabidopsis thaliana roots in a floating hydroponic system. J. Vis. Exp 147:e59517. [DOI] [PubMed] [Google Scholar]
- Hashmi I, Paul C, Al-Dourobi A, Sandoz F, Deschamps P, Junier T, Junier P, and Bindschedler S 2019. Comparison of the plant growth-promotion performance of a consortium of Bacilli inoculated as endospores or as vegetative cells. FEMS Microbiol. Ecol 95:fiz147. [DOI] [PubMed] [Google Scholar]
- Herrmann L, and Lesueur D 2013. Challenges of formulation and quality of biofertilizers for successful inoculation. Appl. Microbiol. Biotechnol 97: 8859–8873. [DOI] [PubMed] [Google Scholar]
- Jha M, Chourasia S, and Sinha S 2013. Microbial consortium for sustainable rice production. Agroecol. Sustain. Food Syst 37:340–362. [Google Scholar]
- Katoh K, and Standley DM 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswani C, Prakash O, Bharti N, Vílchez JI, Sansinenea E, Lally RD, Borriss R, Singh SP, Gupta VK, Fraceto LF, de Lima R, and Singh HB 2019. Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ 690:841–852. [DOI] [PubMed] [Google Scholar]
- Khabbaz SE, Zhang L, Cáceres LA, Sumarah M, Wang A, and Abbasi PA 2015. Characterisation of antagonistic Bacillus and Pseudomonas strains for biocontrol potential and suppression of damping-off and root rot diseases. Ann. Appl. Biol 166:456–471. [Google Scholar]
- Kremer JM, Paasch BC, Rhodes D, Thireault C, Froehlich JE, Schulze-Lefert P, Tiedje JM, and He SY 2018. FlowPot axenic plant growth system for microbiota research. bioRxiv doi:10.1101/25495310.1101/254953https://www.biorxiv.org/content/10.1101/254953v1https://www.biorxiv.org/content/10.1101/254953v1 [Google Scholar]
- Kröber M, Wibberg D, Grosch R, Eikmeyer F, Verwaaijen B, Chowdhury SP, Hartmann A, Pühler A, and Schlüter A 2014. Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front. Microbiol 5:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter JR, and Greenberg EP 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol 182: 6921–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, del Rio TG, Jones CD, Tringe SG, and Dangl JL 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. [DOI] [PubMed] [Google Scholar]
- Levy A, Gonzalez IS, Mittelviefhaus M, Clingenpeel S, Paredes SH, Miao J, Wang K, Devescovi G, Stillman K, Monteiro F, Alvarez BR, Lundberg DS, Lu T-Y, Lebeis S, Jin Z, McDonald M, Klein AP, Feltcher ME, Rio TG, Grant SR, Doty SL, Ley RE, Zhao B, Venturi V, Pelletier DA, Vorholt JA, Tringe SG, Woyke T, and Dangl JL 2018. Genomic features of bacterial adaptation to plants. Nat. Genet 50:138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Cai X, Gong J, Xu T, Ding G, and Li J 2019. Long-term organic farming manipulated rhizospheric microbiome and Bacillus antagonism against pepper blight (Phytophthora capsici). Front. Microbiol 10:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardía E, Rovetto A, Arabolaza A, and Grau R 2006. A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J. Bacteriol 188:4442–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano GL, Bravo JI, Diago MFG, Park HB, Hurley A, Peterson SB, Stabb EV, Crawford JM, Broderick NA, and Handelsman J 2019. Introducing THOR, a model microbiome for genetic dissection of community behavior. MBio 10:e02846–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, and Dangl JL 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magno-Pérez-Bryan MC, Martínez-García PM, Hierrezuelo J, Rodríguez-Palenzuela P, Arrebola E, Ramos C, de Vicente A, Pérez-García A, and Romero D 2015. Comparative genomics within the Bacillus genus reveal the singularities of two robust Bacillus amyloliquefaciens biocontrol strains. Mol. Plant-Microbe Interact 28:1102–1116. [DOI] [PubMed] [Google Scholar]
- Marschner P, Crowley D, and Yang CH 2004. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition, and soil type. Plant Soil 261:199–208. [Google Scholar]
- Massalha H, Korenblum E, Malitsky S, Shapiro OH, and Aharoni A 2017. Live imaging of root–bacteria interactions in a microfluidics setup. Proc. Natl. Acad. Sci. U.S.A 114:4549–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis HC, Thomas VP, Schwientek P, Salamzade R, Chien J-T, Waidyarathne P, Kloepper J, and de la Fuente L 2018. Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I-1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions. PLoS One 13:e0193119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Blanco J, Rodríguez-Jurado D, Hervás A, and Jiménez-Díaz RM 2004. Suppression of verticillium wilt in olive planting stocks by root-associated fluorescent Pseudomonas spp. Biol. Control 30:474–486. [Google Scholar]
- Molina-Santiago C, Pearson JR, Navarro Y, Berlanga-Clavero MV, Caraballo-Rodriguez AM, Petras D, García-Martín ML, Lamon G, Haberstein B, Cazorla FM, de Vicente A, Loquet A, Dorrestein PC, and Romero D 2019. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat. Commun 10:1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, and Bassler BL 2019. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol 17: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Paulson JN, Zheng X, and Kolter R 2017. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A 114:E2450–E2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye TM, Schroeder JW, Kearns DB, and Simmons LA 2017. Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announce 5:e00364–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M, and Jacques P 2008. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125. [DOI] [PubMed] [Google Scholar]
- Peterson SB, Dunn AK, Klimowicz AK, and Handelsman J 2006. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl. Environ. Microbiol 72:5421–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ, Sanabria-Valentín E, Bowers AA, and Shank EA 2015. Inhibition of cell differentiation in Bacillus subtilis by Pseudomonas protegens. J. Bacteriol 197:2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J, Yu X, Liang X, Liu Y, Borriss R, and Liu Y 2017. Addition of plant-growth-promoting Bacillus subtilis PTS-394 on tomato rhizosphere has no durable impact on composition of root microbiome. BMC Microbiol. 17: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan R, Hashem A, and Abd_Allah EF 2017. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol 8:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, and Hurek T 2015. Roots shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol 53:403–424. [DOI] [PubMed] [Google Scholar]
- Ren D, Madsen JS, Sørensen SJ, and Burmølle M 2015. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 9:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW 2012. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi J, Tian H, and Ji M 2017. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip 31: 446–459. [Google Scholar]
- Shank EA, Klepac-Ceraj V, Collado-Torres L, Powers GE, Losick R, and Kolter R 2011. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc. Natl. Acad. Sci. U.S.A 108:E1236–E1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight P, and Kolter R 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol 63:99–118. [DOI] [PubMed] [Google Scholar]
- Tahir HAS, Gu Q, Wu H, Niu Y, Huo R, and Gao X 2017. Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci. Rep 7:40481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R, and Kolter R 2013. Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol 11:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker V, Bruto M, Bellvert F, Bally R, Muller D, Prigent-Combaret C, Moënne-Loccoz Y, and Comte G 2013. Unexpected phytostimulatory behavior for Escherichia coli and Agrobacterium tumefaciens model strains. Mol. Plant-Microbe Interact 26:495–502. [DOI] [PubMed] [Google Scholar]
- Weissgerber TL, Milic NM, Winham SJ, and Garovic VD 2015. Beyond bar and line graphs: Time for a new data presentation paradigm. PLoS Biol. 13:e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JLM, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG 2016. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A 113:E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, and Eddy SR 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29:2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wu H-J, Qiao J, Gao X, and Borriss R 2015. Novel routes for improving biocontrol activity of Bacillus based bioinoculants. Front. Microbiol 6:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarell SM, Grandchamp GM, Chen S-Y, Daniels KE, and Shank EA 2019. A dual-species biofilm with emergent mechanical and protective properties. J. Bacteriol 201:e00670–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H-S, Ahn Y-R, Song GC, Ghim S-Y, Lee S, Lee G, and Ryu C-M 2016. Impact of a bacterial volatile 2,3-butanediol on Bacillus subtilis rhizosphere robustness. Front. Microbiol 7:993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-N, Wang D-C, Hu Q, Dai X-Q, Xie Y-S, Li Q, Liu H-M, and Guo J-H 2019. Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol 10:1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Shen Z, Zhu C, Jiao Z, Li R, and Shen Q 2019. Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant Soil 439:553–567. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


