Abstract
Background:
Bipolar disorder is highly heritable and polygenic. The polygenic risk for bipolar disorder overlaps with that of schizophrenia, and polygenic scores are normally distributed in the population. Bipolar disorder has been associated with structural brain abnormalities, but it is unknown how these are linked to genetic risk factors for psychotic disorders.
Methods:
We tested whether polygenic risk scores for bipolar disorder and schizophrenia predict structural brain alterations in 98 patients with bipolar disorder and 81 healthy controls. We derived brain cortical thickness, surface area and volume from structural MRI scans. In post-hoc analyses, we correlated polygenic risk with functional hub strength, derived from resting-state functional MRI and brain connectomics.
Results:
Higher polygenic risk scores for both bipolar disorder and schizophrenia were associated with a thinner ventromedial prefrontal cortex (vmPFC). We found these associations in the combined group, and separately in patients and drug-naive controls. Polygenic risk for bipolar disorder was correlated with the functional hub strength of the vmPFC within the default mode network.
Limitations:
Polygenic risk is a cumulative measure of genomic burden. Detailed genetic mechanisms underlying brain alterations and their cognitive consequences still need to be determined.
Conclusion:
Our multimodal neuroimaging study linked genomic burden and brain endophenotype by demonstrating an association between polygenic risk scores for bipolar disorder and schizophrenia and the structure and function of the vmPFC. Our findings suggest that genetic factors might confer risk for psychotic disorders by influencing the integrity of the vmPFC, a brain region involved in self-referential processes and emotional regulation. Our study may also provide an imaging–genetics vulnerability marker that can be used to help identify individuals at risk for developing bipolar disorder.
Introduction
Bipolar disorder is a highly heritable1 and severe psychiatric disorder characterized by recurrent episodes of (hypo)mania and depression.2 Psychotic symptoms similar to those seen in schizophrenia are commonly observed during manic episodes and, consistent with their overlapping diagnostic criteria and symptoms,3 bipolar disorder and schizophrenia share at least half of their genetic risk.4
Findings from neuroimaging case–control studies suggest that bipolar disorder is associated with abnormalities primarily in brain regions involved in emotional regulation, including the temporal and prefrontal cortices.5–9 However, such morphological brain alterations might be secondary to disease progression, psychiatric comorbidity or drug treatment. Furthermore, patient–control comparisons in adults with established illness cannot conclusively reveal brain abnormalities that preceded the development of bipolar disorder. We propose that brain regions that play a pivotal role in the development of bipolar disorder should not only show structural and functional abnormalities in case–control comparisons, but also relate to genetic risk for the disorder, because genetic factors likely contribute to bipolar disorder–related brain abnormalities.10 Although cortical abnormalities have been elucidated previously by us and by others,5–7,11,12 the effect of genetic factors has been addressed mainly in family studies of bipolar disorder and schizophrenia, where the degree of genetic risk is only indirectly inferred;13 it has been addressed only rarely in studies in which genetic risk has been assessed by direct genotyping.
Genome-wide association studies of bipolar disorder and schizophrenia have found multiple risk alleles with small effects that additively increase the risk for these disorders.14,15 The cumulative genetic risk for a disorder is reflected in the polygenic risk score.16 Given the shared genetic risk of bipolar disorder and schizophrenia,4 we investigated the polygenic risk scores for both.17 Polygenic risk scores for traits are normally distributed in the general population, and although on average healthy people show lower polygenic risk scores for bipolar disorder and schizophrenia than patients, there is a distributional overlap of polygenic risk scores for patients and healthy people.18 Therefore, potential correlations between polygenic risk score and brain morphology should be present not only in patients, but also in the general population. Importantly, associations found in healthy people would not be confounded by secondary effects of the disorder or drug treatment.
The primary aim of this study was to investigate whether higher polygenic risk scores for bipolar disorder and schizophrenia predicted lower cortical thickness in patients with bipolar disorder and healthy controls. We expected to find such associations in frontotemporal regions of interest that have been shown to be affected in bipolar disorder. We computed associations with other structural brain phenotypes (surface area and volume) and whole brain analyses for completeness. In post-hoc secondary analyses, we tested the associations between genetic risk and such brain regions’ functional integrity (functional hub strength) using resting-state functional MRI and brain connectomics.
Methods
Participants
Patients with bipolar disorder were recruited from the St. Göran Bipolar project, which is a long-term follow-up program at the affective disorder outpatient unit at the Northern Stockholm psychiatric clinic, Stockholm, Sweden. Details on exclusion and inclusion criteria and diagnostic tools can be found in Appendix 1, available at jpn.ca/200165-a1. In brief, we included MRI and genetic data from 98 patients with bipolar disorder and 81 matched healthy controls who had no family history of bipolar disorder or schizophrenia in first-degree relatives. Bipolar disorder types I and II were included in this study. All participants were Swedish and consented to participate verbally and in writing. The study was approved by the ethics committee of the Karolinska Institutet, Stockholm, Sweden.
MRI acquisition
We acquired MRI scans at the MR Research Centre, Karolinska University Hospital, Stockholm, Sweden, using a 1.5 T Signa Excite MRI medical scanner equipped with an 8-channel head coil. We acquired T1-weighted images with a spoiled gradient echo recall sequence using a repetition time of 21 ms, an echo time of 6 ms, a field-of-view of 18 cm, a flip angle of 30°, an acquisition matrix of 256 × 256 × 128 and a voxel size of 0.7 × 0.7 × 1.8 mm3. We acquired additional T2-weighted images for examination by senior radiologists to ensure that the investigated sample was free of clinically significant pathologies. We collected structural MRI data between December 2007 and January 2010.
In post-hoc analyses, we analyzed data obtained from resting-state functional MRI (fMRI), which were added to the scan protocol at a follow-up that occurred 6 to 7 years after the baseline scan. For resting-state fMRI, we acquired 180 volumes; each volume comprised 39 blood-oxygen-level-dependent sensitive T2*-weighted axial echo-planar images, with a resolution of 3.79 mm × 3.79 mm and a slice thickness of 4 mm covering the whole brain. Volumes were acquired with a repetition time of 2.5 s, an echo time of 40 ms, a field of view of 24.3 cm and a flip angle of 85° (total duration 7.5 min). The first 4 (dummy) volumes of each run were discarded to allow for T1 equilibration. Resting-state fMRI data were available for 45 patients and 45 controls. The fMRI data were collected between September 2012 and March 2017. All data were analyzed in 2018 and 2019.
Structural MRI processing
Details on imaging acquisition and processing are described in Appendix 1. In short, we obtained measures of regional cortical volume, thickness and surface area for 34 regions of interest per hemisphere (Desikan atlas19) from structural T1-weighted images using the semi-automated cortical surface reconstruction and parcellation methods provided by Free-Surfer.20–24 These measures were defined in the participant’s native space. The primary analysis was a hypothesis-driven region-of-interest approach, focused on brain areas that have consistently been reported to be affected in patients with bipolar disorder.5–7,25 These included regions involved in the core symptoms of bipolar disorder according to current disease models8 and known to be structurally influenced by genetic factors.26–28 This approach resulted in a total of 16 cortical regions (listed in Appendix 1), including 11 frontal regions, 4 temporal regions and the insular cortex. Additional rationales for adopting this region-of-interest approach were to increase statistical power and to enable comparisons with previous studies that used the same parcellation method, including large-scale multi-centre ENIGMA studies of bipolar disorder5 and major depression.29 In complementary secondary analyses, we also tested the relationships between polygenic risk score and brain structure in other regions of interest (Appendix 1) as well as at the vertex level across the whole brain.
Resting-state fMRI processing for secondary analyses
We performed resting-state fMRI preprocessing using fMRIPrep version 1.0.11.30 We conducted post-processing using nilearn (version 0.5.0)31 and teneto (version 0.4.4).32 See Appendix 1 for fMRI preparatory methods and further details.
Quantification of vmPFC functional-connectivity-based hub strength in the default mode network
Brain connectomics have revealed altered connectivity patterns in bipolar disorder,33 and connectomics-based methods have revealed altered ventromedial prefrontal cortex (vmPFC) hub properties in bipolar disorder.34 We used brain connectomics and resting-state fMRI in post-hoc secondary analyses to test the functional relevance of our main findings by calculating an individual’s vmPFC functional hub strength, which we then related to polygenic risk scores.
We extracted time series of 400 brain regions following the Schaefer parcellation35 by averaging over voxels in each brain region. We used Pearson correlation to quantify the statistical relationship between the time series for each pair of nodes, creating a 400 × 400 adjacency matrix. We assigned the 400 nodes to 1 of 7 communities according to the node network template of Yeo and colleagues36 and the highly spatially overlapped 400-area cortical parcellation procedure of Schaefer and colleagues.35 For each participant, we calculated the within-module degree z-score per node to evaluate its hub strength and assess a node’s role as a possible “provincial hub” within its community. This measure is the z-scored strength (i.e., the sum of all its weights) of a node when considering only the nodes assigned to the same community.
We focused this secondary analysis on the within-module degree z-score for the vmPFC, based on our primary observation of an association between polygenic risk score and cortical thickness in the vmPFC, as well as on a previous finding that bipolar disorder is associated with a missing hub in the vmPFC.34 This z-score reflects the vmPFC functional hub strength within the default mode network (DMN), referred to hereafter as “DMN hubiness.”
We selected the vmPFC nodes of interest based on the polygenic risk score–cortical thickness relationship observed in the vertex-wise follow-up analysis (most significant vertex at Montreal Neurological Institute coordinates x, y, z = 11, 54, −6; right PFCm_1, parcel 379 in the 400-parcel and 7-network version of the Schaefer atlas35). For completeness, we investigated the functional DMN hubiness of the corresponding region in the left hemisphere (parcel 168). We also calculated participation coefficients quantifying the number of between-community connections a node had (see Guimera and colleagues37 for more information about both measures). This allowed us to distinguish between provincial hubs and connector hubs (see Results for a description). We used BCTpy38 to calculate the network measures.
Polygenic risk scores
Participants provided blood samples for genotyping. A score reflecting the aggregated polygenic risk for bipolar disorder and schizophrenia was generated for each participant. Genome-wide association studies used to create the polygenic risk scores were performed in people of European ancestry (excluding genetically identified non-Europeans). See Appendix 1 for methodological details.
Statistical analyses
We tested differences between groups in terms of demographic variables using t tests or Fischer exact χ2 tests.
Associations between polygenic risk scores and cortical measures
In the main analysis (region-of-interest approach), we tested the effects of polygenic risk score (independent variable, variable of interest) on regional cortical thickness (dependent variables) using general linear regression, controlling for age and sex (independent variables; covariates of no interest) using SPSS version 25 (IBM). We also tested how the inclusion of other covariates influenced the findings.
Cortical thickness is a widely used measure of cortical integrity in controls and in pathological conditions,39–42 including bipolar disorder.5–7,43,44 For this reason, and because the largest cross-sectional study on bipolar disorder to date specifically found abnormalities in cortical thickness but not surface area,5 we used cortical thickness as the primary structural measure in the present study. Given that cortical volume is a function of cortical thickness and surface area — 2 phenotypically and genetically distinct measures45,46 — we performed separate analyses on cortical surface area and cortical volume for completeness.
We performed the primary analysis on the combined sample of patients and controls. The rationale was to increase statistical power and test if a relationship between genetic risk and brain alterations could be detected regardless of categorical diagnostic label. Although a correlation driven by case–control differences can reflect a genuine relationship between polygenic risk score and brain structure, we also ran analyses controlling for patient or control status, and separately within the patient and control groups, to test whether observed correlations were driven by case–control differences. For each imaging phenotype, we corrected for multiple testing using Bonferroni Dubey Armitage–Parmar/Sidak adjustment of α-level, which considers the number of regions (n = 16) performed and their intercorrelation.47 We tested distributions of dependent variables for normality using the 1-sample Kolmogorov–Smirnov test.
Sensitivity analyses for potential confounding effects
In sensitivity tests, we repeated the main analysis while controlling for clinical and demographical variables, including medication use, bipolar subtype and comorbidity. Although participants were Swedish and the summary statistics for generation of polygenic risk scores were derived from genome-wide association studies performed on European samples, we also adjusted for principal components derived from linkage disequilibrium–pruned genome-wide data to account for population substructure and potential differences in ancestry. See Appendix 1 for methodological details.
Exploratory analyses
We performed exploratory analyses on regions of interest located outside the 16 regions of interest included in the study, and at a high-resolution vertex-level across the whole brain. In addition to post-hoc analyses, in which we tested the functional relevance of our structural findings by investigating whether polygenic risk score predicted vmPFC DMN hubiness (dependent variable), we also explored correlations between vmPFC DMN hubiness and cortical thickness. All statistical tests were 2-tailed. See Appendix 1 for methodological details.
Results
Participants
Table 1 displays clinical and demographic characteristics. We found no differences between patients and controls with respect to educational level or intracranial volume. As expected, patients with bipolar disorder had higher polygenic risk scores for bipolar disorder and schizophrenia than healthy controls, had a higher mean body mass index, were more likely to smoke, and showed higher depression ( Montgomery–Åsberg Depression Rating Scale) and mania (Young Mania Rating Scale) ratings. Because body mass index and smoking have frequently been associated with bipolar disorder,48 we did not include these variables in the main analysis, but we did correct for these variables in sensitivity analyses, which did not indicate that smoking or body mass index confounded the findings.
Table 1.
Participant demographic and clinical characteristics*
| Characteristic | Patients (n = 98) | Controls (n = 81) | p value† |
|---|---|---|---|
| Age, yr | 40 ± 12 | 39 ± 15 | NS |
| Male/female | 32/66 | 38/43 | NS |
| Polygenic risk score for bipolar disorder | −3.07 ± 3.07 | −6.06 ± 4.05 | < 0.001 |
| Polygenic risk score for schizophrenia | −19.52 ± 3.06 | −21.18 ± 2.27 | < 0.001 |
| Bipolar disorder subtype (I/II) | 55/43 | — | — |
| Education level‡ | 3 ± 1 | 3 ± 1 | NS |
| Montgomery–Åsberg Depression Rating Scale score | 7 ± 7 | 1 ± 2 | < 0.001 |
| Young Mania Rating Scale score | 1 ± 2 | 0 ± 1 | 0.001 |
| Intracranial volume, L | 1.6 ± 0.2 | 1.6 ± 0.2 | NS |
| Smoker | 35 | 15 | 0.007 |
| Snuff user | 17 | 11 | NS |
| Body mass index | 25.7 ± 4.7 | 24.1 ± 3.9 | 0.017 |
| Age at onset of bipolar disorder, yr | 19.3 ± 11.2 | — | — |
| Manic episodes, lifetime | 2 ± 3 | 0 | < 0.001 |
| Depressive episodes, lifetime | 12 ± 17 | 0 | < 0.001 |
| Comorbidities | |||
| Panic disorder | 33 | 0 | < 0.001 |
| Social phobia | 13 | 0 | < 0.001 |
| Alcohol misuse | 10 | 0 | 0.002 |
| Drug misuse | 9 | 0 | 0.004 |
| Eating disorder | 12 | 0 | 0.001 |
| History of psychosis | 47 | 0 | < 0.001 |
| Obsessive–compulsive disorder | 8 | 0 | 0.006 |
| Generalized anxiety disorder | 12 | 0 | 0.001 |
| Post-traumatic stress disorder | 3 | 0 | NS |
| Attention-deficit/hyperactivity disorder | 8 | 0 | 0.006 |
| Medication use | |||
| Antidepressants | 38 | 0 | < 0.001 |
| Antiepileptics | 32 | 0 | < 0.001 |
| Antipsychotics | 22 | 0 | < 0.001 |
| Lithium | 53 | 0 | < 0.001 |
NS = nonsignificant.
Significance of group differences. Comparisons were performed using t tests and χ2 tests.
Values are mean ± standard deviation or n.
Educational achievement was categorized as follows: 1 pre-high school; 2 high school; 3 university (less than 3 years); and 4 university (3 years or longer).
Associations between polygenic risk score and cortical measures
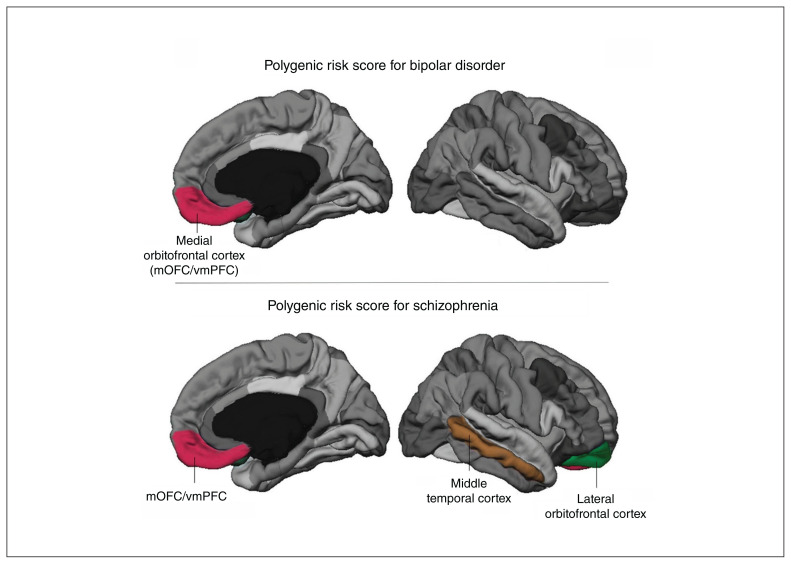
In the primary analysis, higher polygenic risk scores for schizophrenia (p < 0.001, t = −4.55, β = −0.31) and bipolar disorder (p = 0.014, t = −2.48, β = −0.17) were significantly associated with lower cortical thickness in the medial orbitofrontal cortex (mOFC) region of interest. We also observed significant main effects of age (polygenic risk score for schizophrenia: β = −0.30, p < 0.001; polygenic risk score for bipolar disorder: β = −0.28, p < 0.001) and sex (β = 0.24, p < 0.001; β = 0.23, p = 0.001). The mOFC region of interest also contains the vmPFC when generated in FreeSurfer. Therefore, we refer to this region of interest as the mOFC/vmPFC. Further, polygenic risk score for schizophrenia was negatively associated with thickness in the lateral orbitofrontal and the middle temporal cortex regions of interest (Figure 1 and Appendix 1, Table S1). We found no correlations between polygenic risk scores and cortical surface area (Appendix 1, Table S2). Results of regression analyses are presented in Appendix 1, Tables S1 to S3.
Figure 1.
Region-of-interest approach (main analysis). Regions of interest in which we observed significant correlations between thickness and polygenic risk score in the combined cohort are labelled according to the corresponding Desikan atlas19 label and are highlighted in colour. Cortical thickness in the medial orbitofrontal cortex region of interest (mOFC/vmPFC) was significantly inversely correlated with polygenic risk score for bipolar disorder (top). Polygenic risk score for schizophrenia predicted cortical thickness in the same region, and in the lateral orbitofrontal cortex and middle temporal cortex (bottom). The correlation between polygenic risk score for schizophrenia and mOFC/vmPFC thickness remained when correcting for group status and when analyzing patients and controls separately (see Figure 2). mOFC = medial orbitofrontal cortex; vmPFC = ventromedial prefrontal cortex.
The correlation between polygenic risk score for schizophrenia and cortical thickness in the mOFC/vmPFC remained significant when controlling for patient or control status (p = 0.005, t = −2.869, β = −0.182, R2 = 0.357, F4,173 = 25.558; p[model] < 0.001), as well as when analyzed separately in patients (p = 0.035, t = −2.146, β = −0.206, R2 = 0.185, F3,93 = 7.034; p[model] < 0.001) and controls (p = 0.040, t = −2.093, β = −0.225, R2 = 0.141, F3,77 = 4.213; p[model] < 0.008). In contrast, polygenic risk score for bipolar disorder was not correlated with mOFC/vmPFC cortical thickness when controlling for patient or control status (p = 0.91, t = 0.116, β = 0.008; p[group] < 0.001, t = −4.75, β = −0.475), or when tested within patients (p = 0.45, t = 0.763, β = 0.076) and controls separately (p = 0.58, t = −0.556, β = −0.061).
Secondary whole brain analyses at the vertex level confirmed the results we obtained in the region-of-interest analysis (Figure 2 and Appendix 1). The results of whole brain region-of-interest analyses are presented in Appendix 1, Tables S5 to S7. Controlling for demographic and clinical variables, including body mass index, smoking status, comorbidity, bipolar subtype, medication use and ancestry (principal components derived from genomic data) did not change the findings. Detailed results of such tests are not shown because of the large number of sensitivity tests performed.
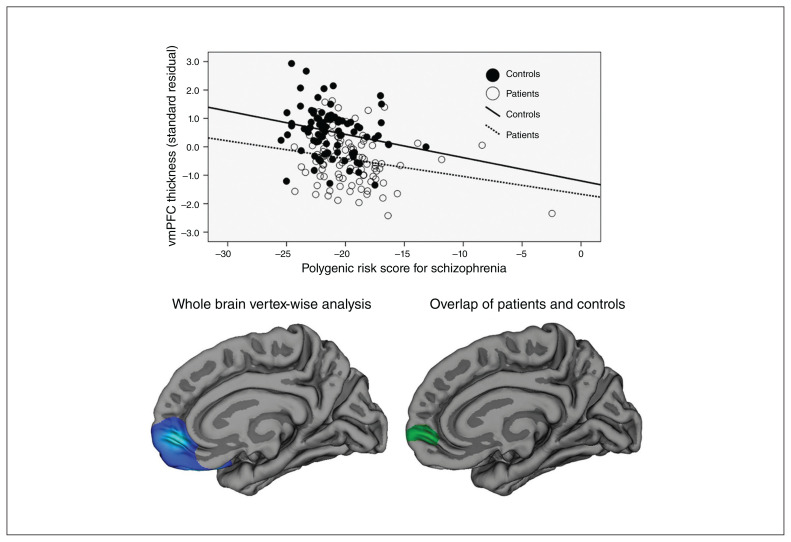
Figure 2.
Correlations between polygenic risk score for schizophrenia and cortical thickness. (Top) Scatter plot displaying the significant correlations between polygenic risk score for schizophrenia and thickness of the mOFC/vmPFC for patients (dashed line) and controls (solid line) observed in the region-of-interest analysis (after regressing out the effects of age and sex on cortical thickness). (Bottom left) Vertex-wise whole brain analysis in the combined group (correcting for age and sex). The blue cluster depicts the brain area with a significant correlation between polygenic risk score for schizophrenia and cortical thickness after correction for multiple comparisons across the whole brain (medial view). Colour gradient in this cluster corresponds to vertex-wise significance levels. We observed the most significant vertex in the right vmPFC (lightest blue colour; Montreal Neurological Institute coordinates: x, y, z = 11, 54, −6). We observed similar correlations in patients and controls, and when correcting for patient–control status. See Appendix 1, including Figures S1 and S3, for detailed statistical results from these follow-up tests. (Bottom right) Cortical region in which the correlations in patients and controls overlapped (green cluster). The correlation was still significant after removing participants with high polygenic risk scores for schizophrenia. mOFC = medial orbitofrontal cortex; vmPFC = ventromedial prefrontal cortex.
Associations between polygenic risk score and vmPFC functional hub strength
Post-hoc analyses revealed that patients showed significantly lower vmPFC DMN hubiness than controls in the right hemisphere (controls [mean ± SD] 0.17 ± 0.62, patients −0.27 ± 0.78; p = 0.004, F1,88 = 8.53), but we observed no group differences in the left hemisphere (controls 0.47 ± 0.60, patients 0.33 ± 0.83, p = 0.37, F1,88 = 0.42; Appendix 1, Table S7). The results did not change when we corrected for age and sex. The mean of right vmPFC DMN hubiness was negative in patients and positive in controls.
Polygenic risk scores for bipolar disorder correlated negatively with right vmPFC DMN hubiness in the combined cohort (n = 90; Pearson r = −0.324, p = 0.002, Dubey adjusted α = 0.020), and this finding remained significant when we corrected for age and sex (partial r86 = −0.353, p = 0.001) and for age, sex and patient/control status (partial r85 = −0.252, p = 0.018). The correlations were not significant when we analyzed them separately in patients (n = 45; Pearson r = −0.246, p = 0.10) and controls (n = 45; Pearson r = −0.165, p = 0.17). Polygenic risk scores for schizophrenia did not correlate with vmPFC DMN hubiness. Associations between vmPFC hubiness and cortical thickness are presented in Figure 3 and Appendix 1.
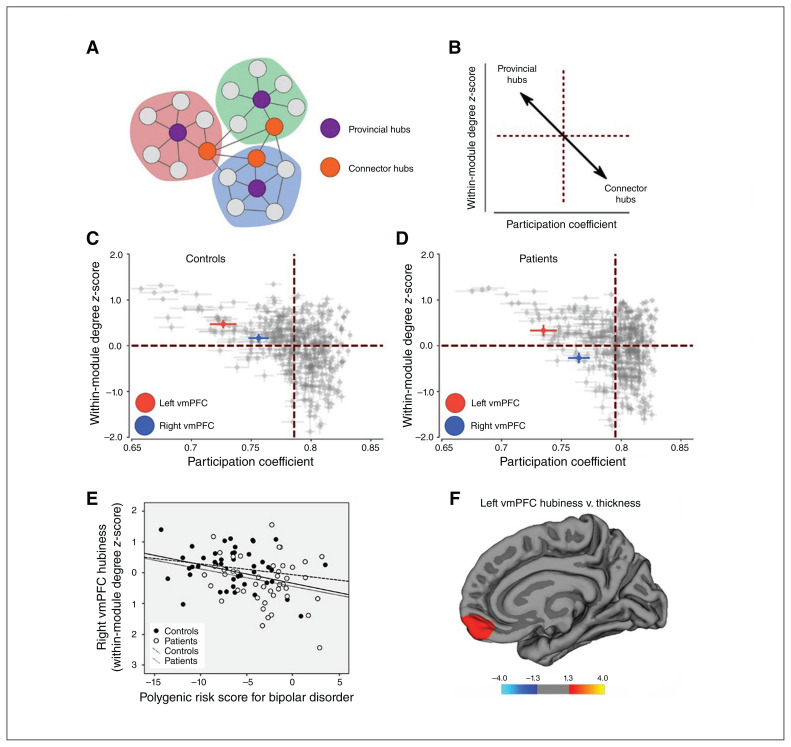
Fig. 3.
Resting-state functional MRI–based vmPFC hub strength (DMN hubiness). (A) Sketch displaying 2 types of hubs: provincial hubs (purple) exhibit connections to many other nodes within a network; connector hubs (orange) show connections across networks. (B) Nodes exhibiting high within-module degree z-scores and low participation coefficients are more likely to be provincial hubs (upper left quadrant). Nodes with low within-module degree z-scores (labelled DMN hubiness in the main text) and low participation coefficients are more likely to be connector hubs (bottom right quadrant). (C and D) Plots of within-module degree z-scores versus participation coefficients of all 400 nodes of the Schaefer atlas (Appendix 1) in controls and patients. Within-module degree z-scores for the 2 vmPFC nodes investigated in this study are indicated in colour (left vmPFC, red; right vmPFC, blue). Patients and controls differed significantly in terms of DMN hubiness in the right vmPFC (Appendix 1, Table S7). (E) Scatter plot displaying the correlation between polygenic risk score for bipolar disorder and right vmPFC hubiness in the combined cohort (solid line). Regression lines within patients (dotted) and controls (dashed) are shown. Correlations in the separate groups were weak (nonsignificant), but the correlation in the full cohort was still significant when controlling for patient–control status. Note: the correlation was still significant after removing the participant with the lowest vmPFC hubiness. (F) Correlations between left vmPFC DMN hubiness and cortical thickness in patients with bipolar disorder at the vertex level across the whole brain. Coloured areas represent brain regions in which we observed significant correlations between vmPFC hubiness and cortical thickness (after correcting for multiple comparisons). Warm colours represent positive correlations. We observed no negative correlations. Significance is displayed on a log(p) scale. See Appendix 1, Figure S4, for a lateral view and correlations with right vmPFC hubiness, where we observed no correlations. DMN = default mode network; vmPFC = ventromedial prefrontal cortex.
Discussion
Previous case–control studies suggested bipolar disorder–related cortical abnormalities mainly in prefrontal and temporal regions.5–7 The causes of these abnormalities are still unknown. Given the high heritability of bipolar disorder, the integrity of brain regions that are related to the development of bipolar disorder should relate to genetic risk for psychotic disorders. We therefore combined multimodal MRI techniques with genomic data to identify brain regions affected by a person’s polygenic risk scores for bipolar disorder and schizophrenia. Our main finding was that polygenic risk scores for bipolar disorder and schizophrenia were associated with a thinner vmPFC. We also found that vmPFC functional hub strength was correlated with polygenic risk scores for bipolar disorder. Taken together, these results indicate that genetic factors affect the structure and functional role of the vmPFC.
We found that higher polygenic risk scores for bipolar disorder and schizophrenia were significantly associated with thinner mOFC/vmPFC. We made these observations using a hypothesis-driven region-of-interest approach, as well as a high-resolution vertex-wise whole brain analysis. Notably, the result we obtained for polygenic risk score for bipolar disorder (p = 0.014) was at the threshold for significance after correcting for multiple testing and should be interpreted with caution. However, structural vmPFC abnormalities have been reported previously in bipolar disorder,5–7 and our results link those findings to genetic factors. Interestingly, we did not observe correlations between polygenic risk score and cortical surface area. This was in line with the notion that genetic factors that influence cortical thickness are believed to be distinct from those that influence surface area.45,46
The relationship between polygenic risk score for schizophrenia (but not bipolar disorder) and vmPFC thickness remained when adjusting for patient or control status, and when controls and patients were analyzed separately. The lower predictive power of polygenic risk score for bipolar disorder in such follow-up tests may be explained by the fact that the polygenic risk score for schizophrenia was derived from larger genome-wide association studies than the polygenic risk score for bipolar disorder, or that correcting for group status could disguise relationships of interest because patient status relates to polygenic risk score. However, the results of sensitivity tests and the fact that the gene-structure associations were replicated in controls indicate that medication use, psychiatric comorbidity and long-term disease progression did not confound the main results in the combined cohort. Thus, the observed link between higher polygenic risk score and thinner prefrontal cortex suggests that genetically mediated vmPFC alterations could be a risk factor for developing psychotic disorders. The polygenic effects on brain structure reported here — in patients with bipolar disorder and controls — have not been demonstrated previously. Previous studies attempting to examine the effects of polygenic risk score on brain structure most likely lacked anatomic resolution, lacked polygenic risk score accuracy and/or excluded high-risk individuals.49–53 Prefrontal areas, especially the orbitofrontal cortex, are also prone to imaging artifacts and require thorough quality control, which is not feasible in large-scale studies.51 However, our findings resembled those of prefrontal brain alterations in family studies on schizophrenia and bipolar disorder, where the degree of genetic risk was indirectly assessed,13 and support a recent review article on associations between polygenic risk score and functional MRI outcomes, indicating that the genetic load for schizophrenia and bipolar disorder affects task-related brain function.54
Given our structural findings, we tested whether genetic factors also affected the functional integrity of the vmPFC. We used brain connectomics and resting-state fMRI, which can reveal dysfunctional connectivity at a network level.33 Previous studies using resting-state fMRI and brain connectomics have shown that the vmPFC is an important hub region (i.e., exhibiting many strong functional connections to other brain regions) in the DMN.55,56 The DMN is essential for processing internal information56,57 and it has been suggested that it is implicated in bipolar disorder.33 Indeed, the vmPFC has been found to be functionally impaired11,12,33 and lose its network hub property in patients with bipolar disorder.34 We found that the functional hub strength of the vmPFC in the DMN (DMN hubiness) was significantly lower in patients than in controls. This finding suggests that the vmPFC has a provincial hub role in the DMN in controls but not in patients with bipolar disorder and aligns with a recent diffusion tensor imaging study reporting that the vmPFC loses its structural hub property in patients with bipolar disorder.34 We further tested whether the decrease in functional DMN hub strength was correlated with individual polygenic risk scores and found that a higher polygenic risk score for bipolar disorder was associated with lower right vmPFC DMN hubiness. Intriguingly, vmPFC hubiness was also correlated contralaterally with vmPFC thickness in patients with bipolar disorder, supporting the functional relevance of cortical thickness measures. This type of structure–function relationship has not been shown previously, and may be of relevance for our general understanding of how these 2 phenotypically distinct measures influence each other.
Taken together, our findings point toward genetically mediated compromised structural and functional integrity of the vmPFC in patients with bipolar disorder and people with an elevated polygenic risk score for bipolar disorder. Previous research has strongly linked the vmPFC to emotional regulation,58,59 in alignment with current neurocognitive models of bipolar disorder suggesting that disruptions of prefrontal and subcortical connections lead to diminished emotional top-down regulation.8,33 Moreover, the vmPFC has a central role as a hub in the DMN55 and is recruited during introspective and self-referential processes.33,56,57,60–62 Self-referential processes have been linked to mania and depression in bipolar disorder.63,64 Speculatively, if the vmPFC is affected, emotional regulation and corresponding self-referential processes could fail and lead to symptoms of bipolar disorder. Our findings could direct future research to test whether the ability to correct emotional states ultimately depends on genetic factors.
Limitations
Polygenic risk is a cumulative measure; the individual risk alleles contributing to our findings and the detailed genetic mechanisms underlying brain alterations need to be investigated. In this Swedish sample, polygenic score calculation was based on European populations, and controlling for potential ancestry differences using genetic information did not indicate a confound by ethnicity. However, it remains unknown how self-reported ethnic background relates to our findings. Although the p value threshold we selected for polygenic risk score analyses best predicted case–control status in previous studies and was a good balance between variance explained and single nucleotide polymorphism inclusion, analyses using different p value thresholds for polygenic risk score extraction may produce different associations. Furthermore, the association between polygenic risk score for bipolar disorder and cortical thickness needs replication in larger samples; we could not determine how genetically influenced brain alterations lead to cognitive impairments and symptom development based on this study. Moreover, the cross-sectional design did now allow us to make conclusions about the relationship between genetic risk and progressive brain changes over time.
Conclusion
The present study provides evidence that polygenic risk scores for bipolar disorder and schizophrenia affect cortical thickness and the functional connectivity of the vmPFC, a brain region essential for self-referential processes and emotional regulation. We suggest that these genetically mediated vmPFC abnormalities may play an important role in risk for developing bipolar disorder.
Acknowledgements
We thank the patients who participated in this study. We also thank the staff at the St. Göran bipolar affective disorder unit, including study coordinators, nurses and data managers.
Footnotes
Data availability: Data access and codes can be obtained on reasonable request.
Funding: This work was supported by the Swedish Medical Research Council (grant no. 2018-02653; to ML), the Swedish Foundation for Strategic Research (grant no. KF10-0039; to ML), the Swedish Brain Foundation (to ML), the Swedish Federal Government under the LUA/ALF agreement (grant nos. ALF20170019 and ALFGBG-71680; to ML), and the Barbro and Bernard Osher Foundation (to MI). Funding sources were not involved in study design, collection, analysis, or interpretation of data, nor in the writing of the report or decision to submit the paper for publication. PP was supported by the Swedish Research Foundation (Vetenskapsrådet; VR; 2019-01253 and 2014-30186-113005-199, ALF Medicine (20160039 and 20140306), the Swedish Brain Foundation (Hjärnfonden; FO2016-0083), Marianne och Marcus Wallenbergs Stiftelse (MMW2014.0065) and Karolinska Institutet (2-70/2014-97).
Competing interests: Dr. Landén reports grants from the Swedish Medical Research Council, the Swedish Foundation for Strategic Research and the Swedish Federal Government during the conduct of the study. No other competing interests declared.
Contributors: P. Petrovic and M. Landén designed the study. C. Sellgren, M. Ingvar and M. Landén acquired the data, which C. Abé, P. Petrovic, W. Ossler, W. Thompson, B. Liberg, J. Song, S. Bergen and P. Fransson analyzed. C. Abé, P. Petrovic, W. Ossler, B. Liberg, J. Song and M. Landén wrote the article, which P. Petrovic, W. Thompson, B. Liberg, S. Bergen, C. Sellgren, P. Fransson, M. Ingvar and M. Landén reviewed. All authors approved the final version to be published and can certify that no other individuals not listed as authors have made substantial contributions to the paper.
References
- 1.Craddock N, Sklar P. Genetics of bipolar disorder. Lancet 2013; 381:1654–62. [DOI] [PubMed] [Google Scholar]
- 2.Merikangas KR, Jin R, He JP, et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch Gen Psychiatry 2011;68:241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearlson GD. Etiologic, phenomenologic, and endophenotypic overlap of schizophrenia and bipolar disorder. Annu Rev Clin Psychol 2015;11:251–81. [DOI] [PubMed] [Google Scholar]
- 4.Lee SH, Ripke S, Neale BM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013;45:984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibar DP, Westlye LT, Doan NT, et al. Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol Psychiatry 2018; 23:932–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe C, Ekman CJ, Sellgren C, et al. Cortical thickness, volume and surface area in patients with bipolar disorder types I and II. J Psychiatry Neurosci 2016;41:240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanford LC, Nazarov A, Hall GB, et al. Cortical thickness in bipolar disorder: a systematic review. Bipolar Disord 2016;18:4–18. [DOI] [PubMed] [Google Scholar]
- 8.Phillips ML, Swartz HA. A critical appraisal of neuroimaging studies of bipolar disorder: toward a new conceptualization of underlying neural circuitry and a road map for future research. Am J Psychiatry 2014;171:829–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ching CRK, Hibar DP, Gurholt TP, et al. What we learn about bipolar disorder from large-scale neuroimaging: findings and future directions from the ENIGMA Bipolar Disorder Working Group. Hum Brain Mapp 2020. July 29 [Epub ahead of print]. 10.1002/hbm.25098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med 2015;45:2461–80. [DOI] [PubMed] [Google Scholar]
- 11.Vargas C, Lopez-Jaramillo C, Vieta E. A systematic literature review of resting state network–functional MRI in bipolar disorder. J Affect Disord 2013;150:727–35. [DOI] [PubMed] [Google Scholar]
- 12.Chen CH, Suckling J, Lennox BR, et al. A quantitative meta-analysis of fMRI studies in bipolar disorder. Bipolar Disord 2011; 13:1–15. [DOI] [PubMed] [Google Scholar]
- 13.McDonald C, Bullmore ET, Sham PC, et al. Association of genetic risks for schizophrenia and bipolar disorder with specific and generic brain structural endophenotypes. Arch Gen Psychiatry 2004; 61:974–84. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M, Saito T, Kondo K, et al. Genome-wide association studies of bipolar disorder: a systematic review of recent findings and their clinical implications. Psychiatry Clin Neurosci 2018;72:52–63. [DOI] [PubMed] [Google Scholar]
- 15.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray NR, Lee SH, Mehta D, et al. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 2014;55:1068–87. [DOI] [PubMed] [Google Scholar]
- 17.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460:748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranlund S, Calafato S, Thygesen JH, et al. A polygenic risk score analysis of psychosis endophenotypes across brain functional, structural, and cognitive domains. Am J Med Genet B Neuropsychiatr Genet 2018;177:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–80. [DOI] [PubMed] [Google Scholar]
- 20.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999; 9:179–94. [DOI] [PubMed] [Google Scholar]
- 21.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II. Inflation, flattening, and a surface-based coordinate system. Neuroimage 1999;9:195–207. [DOI] [PubMed] [Google Scholar]
- 22.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 2000; 97:11050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischl B, van der Kouwe A, Destrieux C, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex 2004;14:11–22. [DOI] [PubMed] [Google Scholar]
- 24.Fischl B, Salat DH, van der Kouwe AJ, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage 2004; 23(Suppl 1):S69–84. [DOI] [PubMed] [Google Scholar]
- 25.Hibar DP, Westlye LT, van Erp TG, et al. Subcortical volumetric abnormalities in bipolar disorder. Mol Psychiatry 2016;21:1710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi AA, Lepore N, Joshi SH, et al. The contribution of genes to cortical thickness and volume. Neuroreport 2011;22:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimol LM, Panizzon MS, Fennema-Notestine C, et al. Cortical thickness is influenced by regionally specific genetic factors. Biol Psychiatry 2010;67:493–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt JE, Neale MC, Fassassi B, et al. The dynamic role of genetics on cortical patterning during childhood and adolescence. Proc Natl Acad Sci U S A 2014;111:6774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmaal L, Hibar DP, Samann PG, et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry 2017; 22: 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esteban O, Markiewicz CJ, Blair RW, et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods 2019; 16:111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham A, Pedregosa F, Eickenberg M, et al. Machine learning for neuroimaging with scikit-learn. Front Neuroinform 2014;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson WH, Brantefors P, Fransson P. From static to temporal network theory: applications to functional brain connectivity. Netw Neurosci 2017;1:69–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry A, Roberts G, Mitchell PB, et al. Connectomics of bipolar disorder: a critical review, and evidence for dynamic instabilities within interoceptive networks. Mol Psychiatry 2019;24:1296–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandes HM, Cabral J, van Hartevelt TJ, et al. Disrupted brain structural connectivity in pediatric bipolar disorder with psychosis. Sci Rep 2019;9:13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer A, Kong R, Gordon EM, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex 2018;28:3095–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo BT, Krienen FM, Sepulcre J, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 2011;106:1125–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guimera R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature 2005;433:895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 2010;52:1059–69. [DOI] [PubMed] [Google Scholar]
- 39.Dickerson BC, Wolk DA. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology 2012;78:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durazzo TC, Mon A, Gazdzinski S, et al. Chronic cigarette smoking in alcohol dependence: associations with cortical thickness and N-acetylaspartate levels in the extended brain reward system. Addict Biol 2013;18:379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almeida Montes LG, Prado Alcantara H, Martinez Garcia RB, et al. Brain cortical thickness in ADHD: age, sex, and clinical correlations. J Atten Disord;2013;17:641–54. [DOI] [PubMed] [Google Scholar]
- 42.Burzynska AZ, Nagel IE, Preuschhof C, et al. Cortical thickness is linked to executive functioning in adulthood and aging. Hum Brain Mapp 2012;33:1607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elvsashagen T, Westlye LT, Boen E, et al. Bipolar II disorder is associated with thinning of prefrontal and temporal cortices involved in affect regulation. Bipolar Disord 2013;15:855–64. [DOI] [PubMed] [Google Scholar]
- 44.Abé C, Liberg B, Song J, et al. Longitudinal cortical thickness changes in bipolar disorder and the relationship to genetic risk, mania, and lithium use. Biol Psychiatry 2020;87:271–81. [DOI] [PubMed] [Google Scholar]
- 45.Panizzon MS, Fennema-Notestine C, Eyler LT, et al. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex 2009;19:2728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkler AM, Kochunov P, Blangero J, et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 2010;53:1135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sankoh AJ, Huque MF, Dubey SD. Some comments on frequently used multiple endpoint adjustment methods in clinical trials. Stat Med 1997;16:2529–42. [DOI] [PubMed] [Google Scholar]
- 48.Goldstein BI, Liu SM, Zivkovic N, et al. The burden of obesity among adults with bipolar disorder in the United States. Bipolar Disord 2011;13:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tesli M, Egeland R, Sonderby IE, et al. No evidence for association between bipolar disorder risk gene variants and brain structural phenotypes. J Affect Disord 2013;151:291–7. [DOI] [PubMed] [Google Scholar]
- 50.Doan NT, Kaufmann T, Bettella F, et al. Distinct multivariate brain morphological patterns and their added predictive value with cognitive and polygenic risk scores in mental disorders. Neuroimage Clin 2017;15:719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neilson E, Shen X, Cox SR, et al. Impact of polygenic risk for schizophrenia on cortical structure in UK Biobank. Biol Psychiatry 2019;86:536–44. [DOI] [PubMed] [Google Scholar]
- 52.Reus LM, Shen X, Gibson J, et al. Association of polygenic risk for major psychiatric illness with subcortical volumes and white matter integrity in UK Biobank. Sci Rep 2017;7:42140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neilson E, Bois C, Gibson J, et al. Effects of environmental risks and polygenic loading for schizophrenia on cortical thickness. Schizophr Res 2017;184:128–36. [DOI] [PubMed] [Google Scholar]
- 54.Dezhina Z, Ranlund S, Kyriakopoulos M, et al. A systematic review of associations between functional MRI activity and polygenic risk for schizophrenia and bipolar disorder. Brain Imaging Behav 2019;13:862–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Persem A, Verhagen L, Amiez C, et al. The human ventromedial prefrontal cortex: sulcal morphology and its influence on functional organization. J Neurosci 2019;39:3627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raichle ME. The brain’s default mode network. Annu Rev Neurosci 2015;38:433–47. [DOI] [PubMed] [Google Scholar]
- 57.Mak LE, Minuzzi L, MacQueen G, et al. The default mode network in healthy individuals: a systematic review and meta-analysis. Brain Connect 2017;7:25–33. [DOI] [PubMed] [Google Scholar]
- 58.Etkin A, Buchel C, Gross JJ. The neural bases of emotion regulation. Nat Rev Neurosci 2015;16:693–700. [DOI] [PubMed] [Google Scholar]
- 59.Petrovic P, Castellanos FX. Top-down dysregulation—from ADHD to emotional instability. Front Behav Neurosci 2016;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amodio DM, Frith CD. Meeting of minds: the medial frontal cortex and social cognition. Nat Rev Neurosci 2006;7:268–77. [DOI] [PubMed] [Google Scholar]
- 61.Wagner DD, Haxby JV, Heatherton TF. The representation of self and person knowledge in the medial prefrontal cortex. Wiley Interdiscip Rev Cogn Sci 2012;3:451–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kable JW, Glimcher PW. The neurobiology of decision: consensus and controversy. Neuron 2009;63:733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pavlickova H, Varese F, Turnbull O, et al. Symptom-specific self-referential cognitive processes in bipolar disorder: a longitudinal analysis. Psychol Med 2013;43:1895–907. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, Fletcher K, Whitehead R, et al. Toward new therapeutic mechanisms in bipolar disorder: analog investigation of self-compassion and nonattachment to self. Front Psychol 2018;9:1848. [DOI] [PMC free article] [PubMed] [Google Scholar]