The integral membrane acyltransferase PatA uses a molecular ruler mechanism for acyl chain length recognition and transfer.
Abstract
Glycolipids are prominent components of bacterial membranes that play critical roles not only in maintaining the structural integrity of the cell but also in modulating host-pathogen interactions. PatA is an essential acyltransferase involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIMs), key structural elements and virulence factors of Mycobacterium tuberculosis. We demonstrate by electron spin resonance spectroscopy and surface plasmon resonance that PatA is an integral membrane acyltransferase tightly anchored to anionic lipid bilayers, using a two-helix structural motif and electrostatic interactions. PatA dictates the acyl chain composition of the glycolipid by using an acyl chain selectivity “ruler.” We established this by a combination of structural biology, enzymatic activity, and binding measurements on chemically synthesized nonhydrolyzable acyl–coenzyme A (CoA) derivatives. We propose an interfacial catalytic mechanism that allows PatA to acylate hydrophobic PIMs anchored in the inner membrane of mycobacteria, through the use of water-soluble acyl-CoA donors.
INTRODUCTION
Bacterial membranes play fundamental roles not only in the physiology of the cell but also in host–pathogen interactions (1, 2). They are primarily used as physiochemical barriers allowing bacterial cells to be functionally constituted and differentiated from the environment (3). Phospholipids and glycolipids are the fundamental building blocks, whereas proteins, the other major components of bacterial membranes, associate permanently or temporarily with them, performing a diverse set of key functions in the cells (4, 5). Specifically, they regulate the selective transport of molecules across the bacterial membranes and, as enzymes, they catalyze a myriad of biochemical reactions. The bacterial membrane constituents are represented by a substantial number of species, the amount and distribution of which also varies in space and time (6–12). Therefore, the structural diversity of membranes provides identity to bacterial cells, which strongly influence the development and maintenance of a large number of bacteria life forms.
Acyltransferases are enzymes that catalyze the transfer of fatty acid chains from activated fatty acid donors to a variety of acceptor molecules of different chemical structure and complexity, playing key roles in the biosynthesis of phospholipids and glycolipids (4). The fatty acid chains can be activated by esterification of their carboxyl groups with the thiol group of coenzyme A (CoA) or that of the acyl carrier protein (ACP), yielding acyl-thioesters. Acyltransferases, as many other enzymes, are very often required to access both hydrophobic and hydrophilic substrates that reside within distinct chemical environments defined by the phospholipid/glycolipid-based membrane and the aqueous cytoplasm and periplasm, respectively (5, 6). The molecular and mechanistic aspects by which membrane enzymes, including acyltransferases, govern substrate recognition and catalysis represent a major challenge and are, compared to their soluble counterparts, poorly understood.
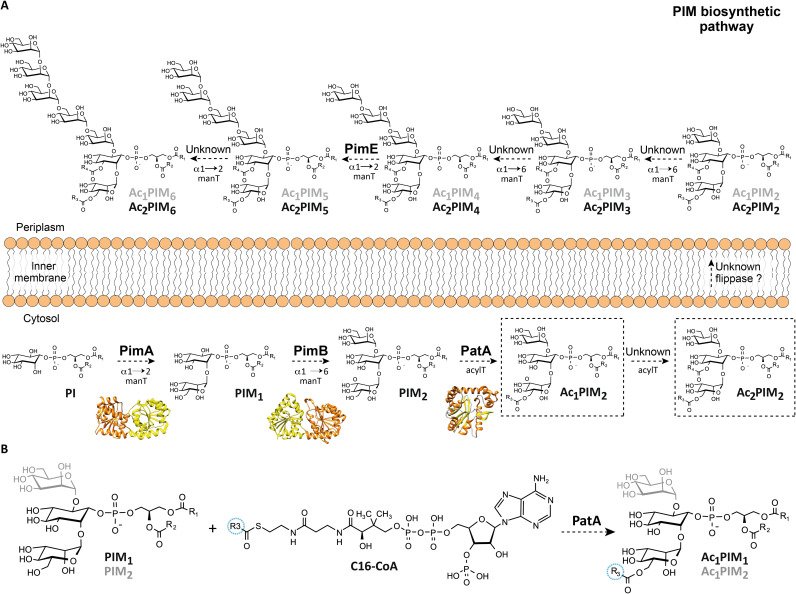
PatA is an essential acyltransferase involved in the biosynthesis of phosphatidyl-myo-inositol mannosides (PIMs) and the metabolically related lipomannan (LM) and lipoarabinomannan (LAM). These species are glycolipids/lipoglycans noncovalently anchored through their phosphatidyl-myo-inositol (PI) moiety to the inner and outer membranes of the mycobacterial cell envelope (2, 7–14). PIMs, LM, and LAM are thought to be important virulence factors during of Mycobacterium tuberculosis infection (15). The PIM family of glycolipids is composed of PI mono-, di-, tri-, tetra-, penta-, and hexamannosides with different degrees of acylation (7, 8). PIM2 and PIM6 are the two most abundant classes found in Mycobacterium bovis bacillus Calmette-Guérin (BCG), M. tuberculosis H37Rv, and Mycobacterium smegmatis 607 (Fig. 1A) (16). According to the currently accepted model, the PIM biosynthetic pathway takes place on both sides of the mycobacterial inner membrane (IM) (8, 17). The early steps occur in the cytosolic face of the IM and involve the consecutive action of three enzymes. The phosphatidyl-myo-inositol mannosyltransferase PimA (Rv2610c in M. tuberculosis H37Rv), which catalyzes the transfer of Manp residue from guanosine diphosphate–mannose to the 2-position of the myo-inositol ring of PI, to form phosphatidyl-myo-inositol monomannoside (PIM1; Fig. 1A) (7, 18–20). PimA was found essential for M. smegmatis mc2155 and M. tuberculosis growth in vitro and in vivo (18, 21). The second mannosylation step involves the action of another essential enzyme PimB (Rv2188c in M. tuberculosis H37Rv), which transfers a Manp residue to the 6-position of the myo-inositol ring of PIM1 to form PIM2 (20, 22). The PIM acyltransferase A, PatA (Rv2611c in M. tuberculosis H37Rv), catalyzes the transfer of a palmitoyl moiety from palmitoyl-CoA to the 6-position of the mannose ring linked to the 2-position of inositol in PIM1 or PIM2, to obtain Ac1PIM1 or Ac1PIM2 (Fig. 1B) (20, 23). PatA was found important for the optimal growth of M. smegmatis mc2155 and an essential enzyme for the growth of M. tuberculosis in vitro and in vivo (11, 23). Two models were originally proposed for the biosynthesis of Ac1PIM2 in mycobacteria. In the first model, PI is mannosylated to form PIM1. PIM1 is then mannosylated to form PIM2, which is acylated to form Ac1PIM2. In the second model, PIM1 is first acylated to Ac1PIM1 and then mannosylated to Ac1PIM2. Recent evidence indicates that although both pathways might coexist in mycobacteria, the sequence of events PI-PIM1-PIM2-Ac1PIM2 is favored (Fig. 1, A and B) (20). Last, Ac1PIM2 can be further acylated on position 3 of the myo-inositol ring to form Ac2PIM2. However, this acyltransferase and most of the mannosyltransferases that catalyze the formation of higher PIMs still remains to be identified (Fig. 1A) (7).
Fig. 1. Proposed PIM biosynthetic pathway in mycobacteria.
(A) The PIM biosynthetic pathway in mycobacteria. According to the currently accepted model, the mannosylation steps of the PIM derivatives start in the cytosolic face of the IM with the consecutive mannosylation of PI, catalyzed by PimA and PimB to form PIM2. PIMs are further acylated by PatA and by a not yet identified acyltransferase, to form Ac1PIM2 and Ac2PIM2, at the cytosolic face of the IM, respectively. It is worth noting that the inner leaflet of the inner mycobacterial membrane is likely composed nearly entirely of Ac2PIM2 (79). Although Ac1PIM2/Ac2PIM2 and Ac1PIM6/Ac2PIM6 are the most abundant forms of PIMs in M. bovis BCG, M. tuberculosis H37Rv, and M. smegmatis 607, all forms of PIM2 could eventually translocate to the outer leaflet of the IM by an unknown mechanism, which could involve the action of a flippase. PimE (Rv1159), an integral membrane glycosyltransferase family C (GT-C) enzyme that use polyprenyl-linked sugars as donors, is involved in the biosynthesis of higher-order PIMs in the periplasmic phase of the IM (17). Other mannosyltransferases involved in the biosynthesis of polar PIMs remain to be identified. (B) Specifically, PatA catalyzes the transfer of a palmitoyl moiety from palmitoyl-CoA to the 6-position of the mannose ring linked to 2-position of myo-inositol in PIM1 and PIM2.
A close interaction of the enzyme with the cytosolic face of the mycobacterial plasma membrane might be a strict requirement for PIM1 or PIM2 modification by PatA. Supporting this notion, PatA was found to colocalize in mycobacterial membrane fractions (20, 23, 24). PatA is the paradigm of a large family of acyltransferases that work at the membrane-water interface and for which the understanding of the molecular mechanism of substrate recognition and membrane association still remains a challenge. Here, surface plasmon resonance (SPR), electron spin resonance (ESR), circular dichroism (CD), differential scanning calorimetry (DSC), and structural and molecular dynamics (MD) data are used to define the membrane association mechanism of PatA from M. smegmatis. Moreover, chemical synthesis, enzymatic activity, isothermal titration calorimetry (ITC), and x-ray crystallography are used to describe the molecular ruler mechanism of PatA. Together, our results support a model of interfacial catalysis, allowing PatA to recognize both hydrophobic and hydrophilic substrate molecules, that facilitates its reaction at the membrane surface.
RESULTS
PatA preferentially binds to anionic phospholipids
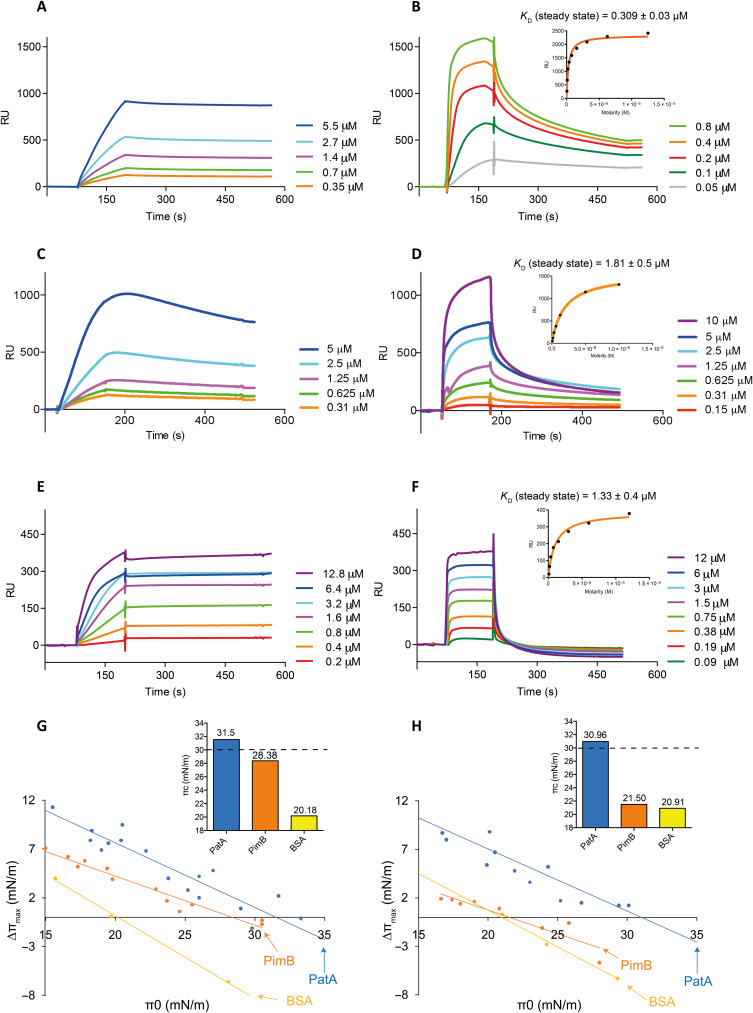
SPR is a powerful technique to study the mechanisms of membrane protein association and integration (25–27). To determine whether PatA binds to anionic [1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG)] or zwitterionic [1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)] phospholipids, we immobilized small unilamellar vesicles (SUVs) composed of DOPG:DOPC (60:40, w/w) (DOPG-SUVs) or DOPC (DOPC-SUVs) into a L1 chip and injected different concentrations (micromolar range) of purified full-length PatA (figs. S1 and S2) to determine the apparent dissociation constants (KD) (see Materials and Methods for details). It is worth noting that the SPR experiments with DOPG-SUVs and DOPC-SUVs were performed at 25°C, temperature at which these lipids are in the fluid phase (phase transition temperature for DOPG and DOPC are −18° and − 17°C, respectively). PatA did not bind to DOPC-SUVs (fig. S2B); however, the enzyme showed binding to the anionic DOPG-SUVs. Thus, we decided to use the zwitterionic DOPC-SUVs as the negative control. As depicted in Fig. 2A, PatA binding to anionic DOPG-SUVs is consistent with the strict requirement that a negative charge needs to be present in the phospholipid for interaction with PatA to occur. It is worth noting that it was not possible to measure the KD value for PatA, because the equilibrium was not reached during the injection phase. However, as PatA is injected, the SPR signal increases and when the protein injection is stopped, the SPR signal remains constant, indicating that the protein is firmly associated with the DOPG-SUVs (Fig. 2A). We determined that PimB, the enzyme that carries out the second step of the PIM biosynthetic pathway (20), also preferentially binds anionic DOPG-SUVs. However, in contrast to PatA, once the injection of PimB is stopped, the SPR signal decreased suggesting that the interaction with the lipid bilayer is temporarily (Fig. 2B and fig. S2A), typical of peripheral membrane proteins (28). It was possible to measure the binding affinity of PimB for DOPG-SUVs (KD), which is in the low micromolar range (0.309 ± 0.03 μM). We also determined the binding of PatA and PimB to 1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DMPG):1,2-dimyristoyl-sn-glycero-phosphatidylcholine (DMPC) (60:40, w/w) (DMPG-SUVs) or DMPC-SUVs at 30°C, temperature at which these lipids are also in the fluid phase (the phase transition temperature for DMPG and DMPC are 23° and 24°C, respectively; Fig. 2, C and D). DMPG, an anionic saturated phospholipid, is one of the most widely used lipid system mimicking bacterial membranes. PatA and PimB showed a similar binding interaction with DMPG-SUVs than DOPG-SUVs (Fig. 2, C and D). Specifically, PatA remains bound to the DMPG-SUVs after stopping the injection while PimB dissociates rapidly after the injection ends (Fig. 2, C and D). To further explore the differences between PatA and PimB in an environment more similar to its natural one, we analyzed the binding of both enzymes using SUVs generated with membrane lipids isolated from M. smegmatis mc2155 cells (Fig. 2, E and F; see Materials and Methods). The behavior of the two enzymes was reminiscent of that observed with the DOPG-SUVs; however, the association rates were faster for both enzymes, and the dissociation rate was faster for PimB, while that for PatA did not change. Together, the SPR data indicate that (i) PatA and PimB preferentially bind anionic phospholipids and (ii) PatA firmly associates with the lipid bilayer, while (iii) PimB associates temporarily with the lipid bilayer.
Fig. 2. PatA interaction and insertion with different lipid membrane models.
SPR sensograms of increasing concentrations of PatA with captured DOPG-SUVs at 25°C (A), DMPG-SUVs at 30°C (C), and M. smegmatis membrane lipid SUVs at 25°C (E); and PimB with captured DOPG-SUVs at 25°C (B), DMPG-SUVs at 30°C (D), and M. smegmatis membrane lipid SUVs (F). (G and H) Surface pressure increase (Δπ) plotted as a function of the initial pressure (Δπ0) to determine the critical pressure of PatA and PimB in the presence of DOPG and DOPC, respectively. Critical pressure of protein insertion (πc) of the three proteins is represented on top-right corner of (G) and (H). The lateral biological membrane pressure is marked with a dashed black line (30 mN/m). Bovine serum albumin (BSA) (in yellow) surface pressure data are also shown as a negative control.
We also investigated the interaction between PatA and membranes by CD and DSC (25, 29, 30). CD spectroscopy was used to examine the effect of phospholipids binding to the secondary structure and thermal stability of PatA and PimB. Because both proteins preferentially associate with anionic membranes, we analyzed protein conformational changes in the presence of DOPG-SUVs and the saturated DMPG-SUVs. The far-ultraviolet CD spectra of the enzymes in solution display spectral features characteristic of α-helical–rich proteins, whose helical content is only slightly altered upon binding to the negatively charged SUVs (fig. S3A). The thermal stability of both proteins was investigated by following the ellipticity at 222 nm upon heating (fig. S3B). Denaturation curves were fitted to a two-state model of protein unfolding, providing the melting temperature (Tm) and apparent enthalpy (ΔHapp) change of the transition (table S1) (31). Both enzymes are more thermally stable in the presence of lipids at high concentrations, with PimB (ΔTm ~ 2.6° to 3.4°C) being more perturbed than PatA (ΔTm ~ 1.1° to 1.8°C). The unfolding ΔHapp of membrane-bound PatA is larger than that of the free enzyme, whereas the unfolding enthalpy of PimB is comparatively reduced in the presence of the lipids. This result indicates a different interaction mechanism for each enzyme with lipid membranes. The increase in the Tm and the unfolding enthalpy of PatA bound to the lipid bilayer suggests an enhancement of both intraprotein and protein-lipid interactions that contribute to the stabilization of the enzyme’s native state.
To evaluate the impact of the enzymes on the thermotropic phase behavior of lipid vesicles, we carried out DSC experiments using DMPG-containing large multilamellar vesicles (LUVs). Binding of both PimB and PatA to DMPG-LUVs (140-nm size; Materials and Methods for details) affects the thermodynamics of the lipids in a concentration-dependent manner (fig. S4A and table S2). The enzymes broaden the heat capacity profile of the membrane, reducing both the calorimetric enthalpy change and the transition cooperativity of the protein-associated membranes. The DSC thermograms of the protein-containing samples display two components: The narrow peak corresponds to the protein-free LUVs, while the broader one can be attributed to protein-associated membranes. The gel–to–liquid crystalline phase transition temperature of the narrow peak remains virtually unperturbed of protein concentration, whereas the broad peak is shifted to higher temperatures and becomes more enthalpic with the addition of increasing enzyme concentrations. Moreover, both proteins alter the pretransition of the DMPG-LUVs (low enthalpic peak at about 14°C; fig. S4A), suggesting structural changes in the polar head group of the lipids (32). The thermograms of the membrane registered after PimB denaturation are very different from those recorded before protein melting. Upon protein denaturation, the broad, PimB-associated membrane peak vanishes, suggesting that PimB is released into solution. In contrast, the PatA-associated LUV peak remains, suggesting that upon protein denaturation, PatA is still attached to the phospholipids (fig. S4B).
PatA inserts into anionic phospholipid monolayers
The insertion of PatA into phospholipid membranes was evaluated using Langmuir films at various initial pressure (π0) values. PatA was injected underneath DOPG:DOPC (60:40, w/w) (DOPG) and DOPC monolayers prepared at the air-water interface, and the interaction was quantified by surface pressure measurements. The critical pressure of insertion (πc), determined by the abscissa intercept of the surface pressure increase versus π0 plot, indicates the protein propensity for membrane insertion. Hence, a larger πc indicates a higher propensity for protein insertion into monolayers. The lateral pressure of biological membranes corresponds to a surface pressure of 30 to 35 mN m−1 in the Langmuir monolayers experiments (33). Therefore, it is generally accepted that πc values between 30 and 35 mN m−1 strongly suggest insertion of the protein into the lipid monolayer (34–36). PatA displayed a πc of 31.54 mN m−1 in DOPG (R2 = 0.92) phospholipid monolayer and a πc of 30.96 mN m−1 (R2 = 0.89) in DOPC monolayer experiments, supporting the notion that the enzyme inserts into both types of phospholipid monolayers but preferably into the DOPG monolayer. PimB showed a πc value of 28.38 mN m−1 (R2 = 0.95) in DOPG experiments and 21.5 mN m−1 (R2 = 0.85) in DOPC experiments, consistent with a peripheral localization, while the soluble bovine serum albumin (BSA) has a πc value of 20.18 mN m−1 (R2 = 0.99) and 20.91 mN m−1 (R2 = 0.99) in DOPG and DOPC experiments, respectively (Fig. 2, G and H).
PatA insertion modifies the dynamics of anionic phospholipid bilayers
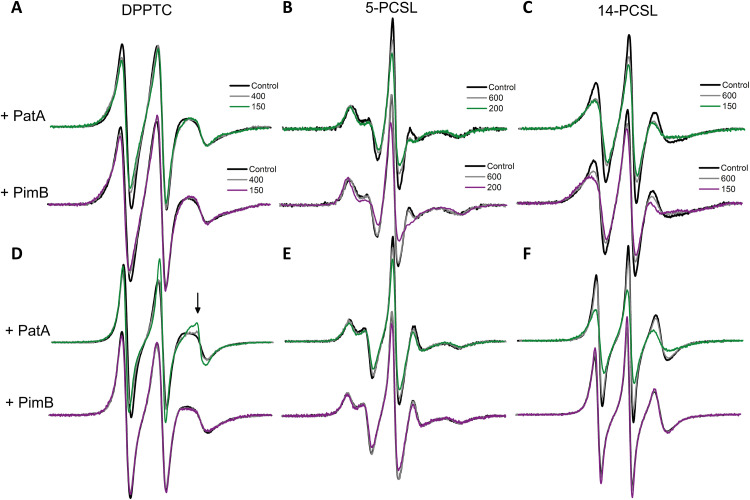
We applied ESR spectroscopy to probe the effects of protein binding on the ordering and dynamics of phospholipid bilayer membranes. Spin-labeling ESR is a powerful spectroscopic tool to study changes in the structural dynamics of lipids and proteins during protein-lipid interactions (37–39). ESR experiments were performed with DMPG-SUVs at different lipid-to-protein (L/P) molar ratios, using nitroxide labels attached to both the head group region [1,2-dipalmitoyl-sn-glycero-3-phospho(tempo)choline (DPPTC)] and the acyl chain region (5-PCSL and 14-PCSL) of phosphatidylcholines, allowing to monitor protein-induced changes from the membrane surface (DPPTC) to the hydrophobic core of the phospholipid bilayer (5-PCSL and 14-PCSL) (39).
In the DMPG-SUVs ripple gel phase, remarkable line broadening was obtained in the presence of the enzymes for all spin labels (Fig. 3, A and C). The observed spectral changes suggest that both PimB and PatA reduce lipid mobility and increase the membrane packing from the lipid-water interface down to the center of the bilayer. In particular, the effective order parameter (Szz) of the 5-PCSL spectra (see Materials and Methods) increased by 0.06 and 0.09 in the presence of PatA and PimB, respectively, at an L/P of 200. Moreover, PimB is slightly more effective than PatA at reducing the parameter h+1/h0, the ratio between the intensity of the low (h+1) and the central (h0) field lines of the 14-PCSL spectra (14.6% versus 12.8%, respectively). The lower the h+1/h0, the more packed or less dynamic are the spin probes. These results indicate that the binding of PimB to DMPG-SUVs makes the gel phase of the protein-lipid system more packed/ordered than the PatA/lipid gel phase. In contrast, the head group region is more perturbed by PatA than by PimB (Fig. 3A and fig. S5B), suggesting different membrane interaction mechanisms for both enzymes. The protein-induced perturbation of the ordering and mobility of the head group region and acyl chains in the DMPG gel phase may explain the reduced intermolecular cooperativity of the lipid phase transition and the effect of the proteins on the lipid pretransition, as seen by DSC. However, the most interesting results were obtained in the fluid phase of DMPG-SUVs. PatA substantially increased the lipid ordering and decreased the lipid mobility (fluidity) of the acyl chain region of the membrane, as evidenced by the broadening of the 5-PCSL and 14-PCSL spectra (Fig. 3, E and F). The Szz values for the 5-PCSL increased by 0.07 in the presence of PatA at an L/P of 200, while the parameter h+1/h0 for the 14-PCSL was reduced by about 9% at an L/P of 150. In contrast, the effect of PimB on the acyl chain region of the bilayer was practically negligible, yielding virtually similar ESR spectra for the 5-PCSL and 14-PCSL (Fig. 3, E and F).
Fig. 3. PatA changes the structural dynamics of DMPG lipid bilayers.
ESR spectra of DPPTC (A and D), 5-PCSL (D and E), and 14-PCSL (C and F) obtained in the gel phase (20°C; A to C) and in the fluid phase (30°C; D to F) of DMPG-SUVs in the absence (black lines) and presence of PatA (spectra on the top of each panel) and PimB (spectra on the bottom of each panel) at different L/P molar ratios (colored lines). The arrow in (D) indicates a sharp spectral component. The spectra were normalized by the number of spins. Spectral width is 100 Gauss.
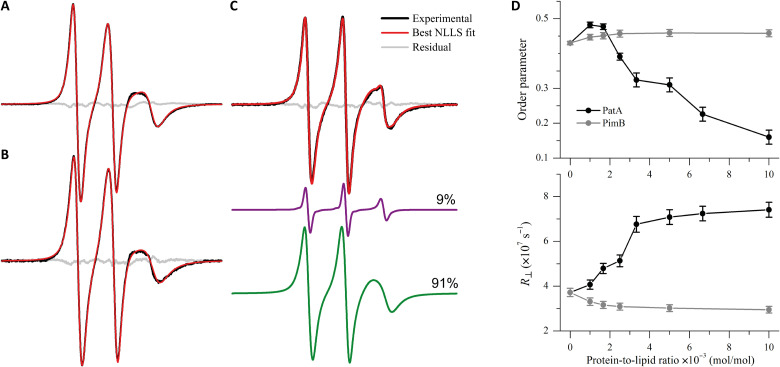
On the other hand, binding of PatA and PimB to DMPG-SUVs in the fluid phase yields opposing effects in the head group region, despite both proteins decreasing the h+1/h0 values for the DPPTC (fig. S5B). While PatA induces extensive line narrowing in a concentration-dependent manner, PimB slightly broadens the DPPTC ESR spectra (Fig. 3D). The interaction of PatA with the DMPG-SUV in the fluid phase gives rise to a second, narrow component in the DPPTC spectra (arrow in Fig. 3D) superimposed with a broad signal. The relative population of the narrow component grows in intensity with the addition of PatA (arrow in the fig. S5A). To quantify the order parameter (S0), the rotational diffusion rate (R⊥), rotational correlation times (τ), and the populations of both spectral components, nonlinear least-squares (NLLS) simulations of the DPPTC ESR spectra were performed, and the best-fit parameters are summarized in the table S3.
NLLS spectral fitting successfully reproduced the two-component feature of the DPPTC ESR signal from the PatA/DMPG system (Fig. 4C) along with the typical one-component spectra of DPPTC in the protein-free (Fig. 4A) and PimB-decorated membranes (Fig. 4B). Spectral simulations indicate that PatA insertion into DMPG-SUVs induces a much less packed (lower S0) and more dynamic lipid head group than the protein-free membranes, as indicated by the dependence of S0 and R⊥ of the DPPTC broad component as a function of PatA concentration (Fig. 4D). In contrast, the peripheral interaction of PimB with DMPG-SUVs in the fluid phase only slightly increases the ordering and decreases the rotational diffusion of the head group region (Fig. 4D).
Fig. 4. PatA promotes extensive disordering of the DMPG head group region.
NLLS simulations of the DPPTC ESR spectra acquired at 30°C in the absence (A) and presence of 0.5 mole percent (mol %) (L/P = 200) of PimB (B) or PatA (C). Experimental spectra are shown in black, the best fit in red, and the residuals in gray. The spectra in (A) and (B) are typical one-component spectra, whereas the spectrum in (C) was simulated with two spectral components. The sharp component (purple) of the spectrum in (C) accounts for 9% of the total spin population. Spectral width is 100 Gauss. (D) Dependence of the order parameter (top) and the rotational diffusion rate (bottom) of DPPTC as a function of the protein-to-lipid molar ratio obtained from the NLLS spectral simulations. For PatA, both S0 and R⊥ are those determined from the broad spectral component.
Moreover, the sharp component is characterized by much faster motion (τ of about 0.32 to 0.41 ns) and lower packing (S0 ~ 0.13) than those obtained from the broad signals (τ of 1.08 to 2.24 ns and S0 ~ 0.16 to 0.48). The relative population of the sharp component roughly linearly increases with PatA addition (from 0.9% at L/P = 1000 to about 22.5% at L/P = 100; table S3), suggesting an additive effect. The appearance of sharp signals in the ESR spectra of nitroxide-labeled lipids upon protein-membrane interactions has been attributed to defect formation in the lipid bilayer (40). In particular, a defect-like structure in the membrane hydrophobic core has been observed for the Escherichia coli dihydroorotate dehydrogenase. The authors concluded that the insertion of the enzyme N-terminal helices into the lipid bilayer acts as a spacer of the lipid acyl chains, generating sharp spectral components in the ESR spectra (41). Therefore, the emergence of a narrow component in the DPPTC spectra in the PatA-embedded DMPG-SUV is consistent with the formation of local defects in the head group region likely due to the insertion of the PatA N-terminal α helices into the lipid bilayer.
Our ESR results are summarized as follows. In the gel phase, PatA and PimB remarkably alter the ordering and mobility of the DMPG-SUVs lipids, exerting a differential effect in the head group and in the acyl chain regions. In the fluid phase, (i) the enzymes promote opposing effects in the structural dynamics of the DMPC/DMPG head group region; (ii) the effect of PimB is restricted to the bilayer surface, consistent with a peripheral membrane attachment mechanism; and (iii) PatA increases the packing of the acyl chains and enhances defects in the membrane head group region, supporting a model of PatA insertion into the bilayer hydrophobic core.
A two-helix motif positions PatA active site for catalysis within the membrane bilayer
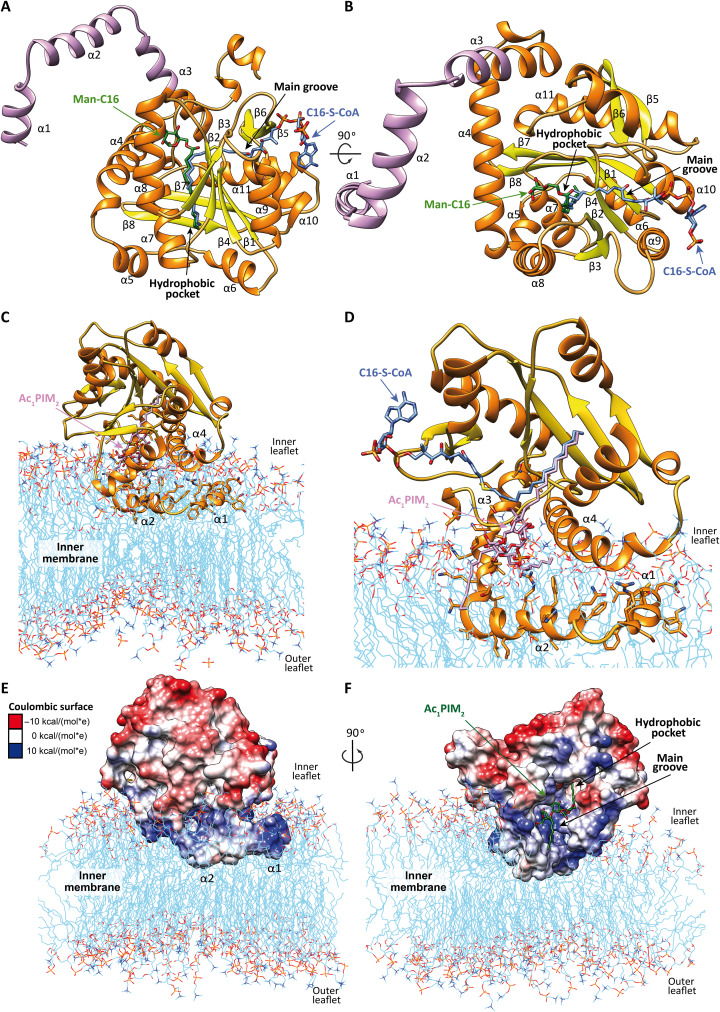
Analysis of the amino acid sequence of PatA revealed the lack of a signal peptide or hydrophobic transmembrane segments, suggesting that PatA associates with only one side of the lipid bilayer, a feature typical of peripheral or monotopic integral membrane proteins (42). We have recently solved crystal structures of PatA from M. smegmatis mc2155 (MSMEG_2934; 304 residues) in complex with (i) its naturally occurring palmitoyl donor (PatA-C16) (43), (ii) a nonhydrolyzable palmitoyl-CoA analog (PatA-S-C16-CoA) (43), and (iii) the 6-O-palmitoyl-α-d-mannopyranoside product (PatA-ManC16) (44). The high quality of the electron density maps allowed the trace of residues ca. 40 to 303. The structures revealed an α/β architecture, with the palmitoyl residue deeply buried into a hydrophobic pocket that runs perpendicular to a long groove where the active site is located (Fig. 5A). The 4-phosphopantetheinate moiety of S-C16-CoA is located at the entrance of the main groove, in close contact with a highly conserved region flanked by the β2-α8 (residues 149 to 153), β3-α9 (residues 174 to 180) and β4-β5 (residues 199 to 207) loops, and two α helices, α9 (residues 181 to 190) and α10 (residues 221 to 230). The adenosine 3′,5′-diphosphate (3′,5′-ADP) moiety of the ligand sticks out from the core domain of the enzyme and is exposed to the bulk solvent (43, 44). In contrast, the Manp ring of ManC16 is located within a cavity located at the end of the main groove and comprising helices α4 and α8 and the connecting loops β1-α7 (residues 83 to 90), β2-α8 (residues 148 to 154), and α11-α12 (residues 282 to 291; Fig. 5B). Specifically, the Manp ring is mainly stabilized by van der Waals interactions with the side chains of four aromatic residues Tyr83, Trp84, Trp130, and Phe160. In addition, the O6 atom of the Manp ring makes a hydrogen bond with the NE2 atom of the imidazole moiety in His126, whereas the O2 atom makes a weak hydrogen bond with the NH2 atom of Arg164 (Fig. 5B). It is worth noting that both PIM2 acyl chains are essential for PatA enzymatic activity, supporting the notion that they play a prominent structural role in the arrangement of the sugar moiety/moieties into the catalytic center. The first 40 residues of PatA predict the formation of two amphipathic helices, α1 and α2 (Fig. 5, A to D). However, the positioning and involvement of these helices in the binding of the PIM1/PIM2 lipid acceptors and membrane interaction remain unknown.
Fig. 5. PatA overall structure and membrane association.
(A and B) Two views of the x-ray crystal structures of PatA in complex with nonhydrolyzable S-C16-CoA [Protein Data Bank (PDB) code 5F34]. The Man-C16 product was placed by superposition of the crystal structure of PatA in complex with the Man-C16 product (PDB code 5OCE). S-C16-CoA is shown in blue, Man-C16 product in green, PatA’s β-strands in yellow, and α helices in orange. The modeled α1, α2, and partial α3 are highlighted in purple. (C) Model of PatA and membrane interaction result from MD simulations. The membrane bilayer apolar face is shown in light blue, phospholipids external heads are shown in red and orange, the PatA β strands are shown in yellow, and α helices are shown in orange. The reaction product, Ac1PIM2, is modeled in purple. (D) A closer snapshot derived from 0.5-μs MD simulations superimposed with the nonhydrolyzable S-C16 CoA (in blue). (E and F) Two views of Coulombic surface representation of PatA and the membrane interaction model obtained by MD calculations. The polar character of the enzyme is shown in three colors; negatively charged patches in red (−10 kcal mol−1), hydrophobic zones in white (0 kcal mol−1), and positively charged residues in blue (10 kcal mol−1). The modeled Ac1PIM2 reaction product is colored in green. The palmitate chain is deeply buried in the hydrophobic pocket, the inositol and mannose rings are located in the main groove, and the acyl chains are located close to the membrane.
The electrostatic potential of the PatA surface revealed a clear solvent-exposed area bordering the main groove containing several hydrophobic and aromatic residues interspersed with positively charged residues (Fig. 5, E and F). Specifically, this region comprises the amphipathic helices α3, α4, and α8, and the connecting loop β2-α8 (residues 148 to 154; Fig. 5C). In contrast, the opposite side of PatA exhibits a negatively charged surface, which would generate significant electrostatic repulsion with the anionic phospholipid bilayer. Therefore, the polar character of PatA seems to determine the correct orientation of the enzyme into the membrane (Fig. 5C). To establish the preferred binding orientation of full-length PatA within the phospholipid bilayer and the involvement of the helices α1 and α2, in PIM1/PIM2 acceptor and membrane association, we modeled helices α1, α2, and partially α3 and carried out 0.5-μs MD simulations of PatA embedded in one leaflet of the bilayer (see Materials and Methods). To this end, we used the coordinates of PatA containing (i) the C16-CoA donor molecule based on the PatA-S-C16-CoA crystal structure and (ii) the PIM2 glycolipid molecule docked into the acceptor binding site based on the PatA-ManC16 structure (Fig. 5, C and D). The calculations show that the complex is stable in the membrane throughout the simulation time and that helices α1 and α2 interact with the polar fragments of the inner layer via several hydrogen bonds (fig. S6A). In addition, the inositol moiety, one of the mannose, and the phosphate group of the PIM2 glycolipid are involved in hydrogen bonds (fig. S6B) and hydrophobic contacts with the protein.
The ruler mechanism for acyl chains recognition
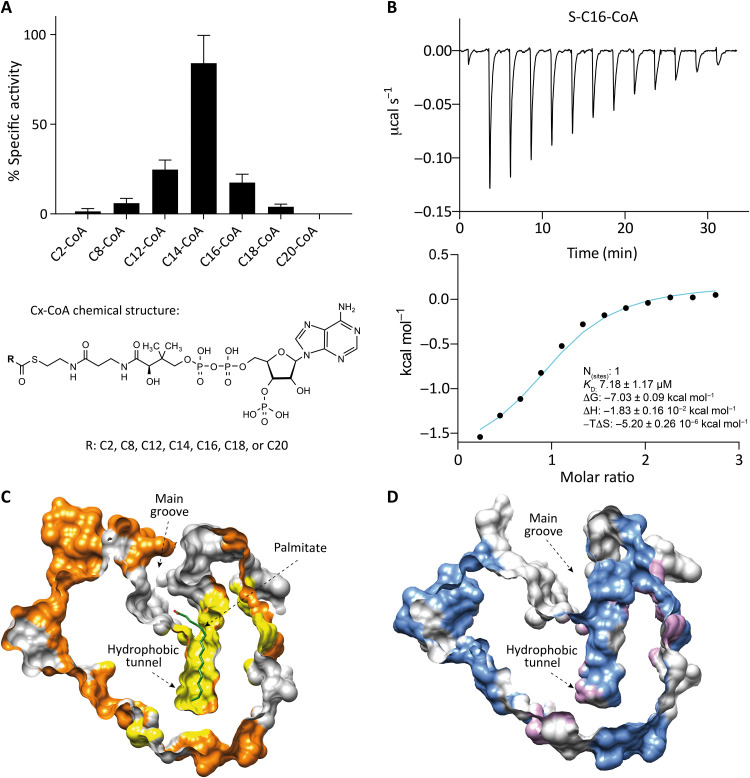
In the crystal structures previously reported, we observed that PatA has a hydrophobic cavity where the acyl chain of the palmitoyl-CoA (C16) donor is deeply buried (Fig. 5, A and B). The length of this cavity might determine the ability of the enzyme to recognize a certain number of carbon atoms in the acyl chain of the acyl-CoA substrates. To this end, we measured the specific activity of PatA against acyl-CoA derivatives with different lengths of the acyl chain, including C2-CoA, C8-CoA, C12-CoA, C14-CoA, C16-CoA, C18-CoA, and C20-CoA, by a spectrophotometric assay using 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB; Fig. 6A). The assay measures the absorption of the released TNB+2 produced by reaction between DTNB and the CoA released when PatA catalyzes the transfer of the acyl chain from an acyl-CoA derivative to PIM2. All reactions were performed in the presence of an artificial membrane model DOPC-SUVs, which is required for the PatA reaction (44). The assays showed that the highest activity corresponds to C14-CoA, followed by the C12-CoA and C16-CoA derivatives; the last was previously described as the preferred acyl-CoA donor substrate of the enzyme (16, 45). The formation of the reaction products was detected and confirmed by mass spectrometry (MS) in all the reactions (fig. S7 and table S4). The increase or decrease in the acyl chain length in the CoA substrate derivatives, compared to C14-CoA, resulted in a gradual reduction of enzymatic activity, with almost no activity with C2-CoA and C20-CoA. The activity measurements showed a clear Gaussian-like curve profile, strongly supporting the notion that PatA can recognize subtle differences in acyl chain lengths in CoA derivatives. The C14-CoA derivative unexpectedly showed the highest specific activity compared to C16-CoA. This could be due to a different binding mode between the enzyme and the C14-CoA, with respect to the natural substrate C16-CoA. With this in mind, we studied the impact of acyl chain length on the binding of different acyl-CoA derivatives to PatA, using ITC. To this end, we chemically synthesized the nonhydrolyzable ether substrates S-lauroyl-CoA (S-C12-CoA), S-stearoyl-CoA (S-C18-CoA), and S-arachidoyl-CoA (S-C20-CoA). Specifically, the nonhydrolyzable ether substrates were obtained by conjugation of CoA-SH with the corresponding alkyl iodide following described protocols (46). In addition, S-tetradecyl-CoA (S-C14-CoA) and S-hexadecyl-CoA (S-C16-CoA) were purchased in Avanti Polar Lipids. We detected binding exclusively with S-C16-CoA (KD = 7.18 ± 1.17 μM), the derivative that was previously described as the preferred donor substrate of the enzyme (Fig. 6B and fig. S8) (47). Together, the experimental data support a fatty acid ruler as the mechanism by which PatA specifically recognizes the donor acyl chain of CoA.
Fig. 6. Acyl chain length recognition activity and binding experiments.
(A) PatA-specific hydrolytic activity measured by spectrophotometric DTNB assay with different acyl-CoA derivatives (C2-CoA, C8-CoA, C12-CoA, C14-CoA, C16-CoA, C18-CoA, and C20-CoA) in the presence of DOPC-SUVs as an artificial membrane model. The activity results of PatA for each compound are plotted in reference to the most active one, the C14-CoA derivative. (B) ITC raw and integrated data for the binding interaction of PatA and nonhydrolyzable C16-CoA derivative. The experimental points are represented as filled squares, and the best fit of these points to a one-site binding model is represented as a solid curve. (C) The hydrophobic pocket surface shape of PatA crystal structure in complex with palmitate (in green) (PDB code 5F2Z). (D) The empty hydrophobic pocket surface shape of the crystal structure PDB code 7OJT.
To better understand the recognition mechanism of the donor substrate, we attempted to obtain the cocrystal structure of PatA in complex with the nonhydrolyzable substrates S-C12-CoA, S-C14-CoA, S-C18-CoA, and S-C20-CoA. We could obtain good-diffracting crystals for PatA in the presence of the S-C12-CoA derivative and solved the structure at 3.67-Å resolution in the space group P21 (Fig. 6D, fig. S9, and table S5). Structural comparison of this new crystal structure with the previously reported PatA-S-C16-CoA complex [Protein Data Bank (PDB) code 5F34] (43) revealed that the overall protein structure is essentially preserved [root mean square deviation (r.m.s.d.) of 0.36 Å for 245 residues; Fig. 6, C and D]. We could not find clear electron density to model the S-C12-CoA substrate in the hydrophobic cavity. It is worth noting that C16 was present in the hydrophobic cavity in all reported crystal structures of PatA (PDB codes 5OCE, 5F2Z, 5F2T, and 5F34). Unexpectedly, the hydrophobic cavity maintains its conformation although it is empty. These data suggest that the S-C12-CoA derivative can access both the main grove and the hydrophobic cavity and displaces the palmitic acid coming from the purification (43). However, since it displays a short acyl chain, the S-C12-CoA is released from the active site, as it is not sufficiently stabilized.
DISCUSSION
Acyltransferases play a central role in the biosynthesis of bacterial phospholipids and glycolipids by catalyzing the transfer of activated acyl chains to acceptor molecules of different chemical structure and complexity. To this end, fatty acids are usually activated for subsequent reactions by esterification of their carboxyl groups with the thiol of CoA or ACP, resulting in acyl-thioesters (4). Similar to many other membrane acyltransferases, PatA catalyzes a reaction between substrates of completely different nature: (i) the PIM2 glycolipid, which is deeply anchored in the mycobacterial IM, and (ii) water soluble palmitoyl-CoA. How does the active site of PatA access these two chemically different substrates? Our experimental data support a model in which the reaction mediated by PatA occurs at the lipid-water (mycobacterial IM–cytosol) interface. PatA is permanently attached to the lipid bilayer. This interaction is mainly mediated by the insertion of N-terminal helices α1 and α2 into anionic lipid bilayers (Fig. 5, C and D). In addition, PatA displays several hydrophobic patches interspersed with clusters of positively charged residues adjacent to the main groove that contain the catalytic site. These hydrophobic and negatively charge residues certainly maintain and stabilize PatA into the bilayer through electrostatics and London dispersion forces with other bilayer components. In addition, the enzyme displays a negatively charged region at the opposite side of the enzyme, oriented toward the cytosol. Therefore, the polar character of PatA also contributes to determine the correct orientation of the enzyme into the membrane (Fig. 5, E and F) (43, 44). In that context, PatA recognizes the polar head group of the PIM1/PIM2 glycolipid acceptor molecule in a region of the major groove mainly flanked by helix α4. The fatty acid chains of PIM1/PIM2 are essential for the activity of the enzyme, contributing to the mannose/s and inositol residues into the binding site (44). Notably, PatA can sequester the acyl chain of the acyl-CoA donor substrate from the cytosol and incorporate it into a hydrophobic tunnel that extends entirely through the core of the enzyme. The adenosine 3′,5′-ADP moiety of the acyl-CoA protrudes away from the groove and is exposed to bulk solvent, as observed in other acyl-CoA modifying enzymes. Thus, PatA transfers the C16 acyl group to PIM1/PIM2 in a two-step reaction mechanism in which His126 acts as a general base to deprotonate the hydroxyl group of a mannose ring in PIM1/PIM2, facilitating nucleophilic attack on the thioester of the acyl-CoA. The reaction is assisted by Glu200, which modulates the pKa of the central histidine as a general base or acid during the catalytic cycle, similar to the charge-relay system of serine proteases.
Brennan and Ballou (47) were the first to study the fatty acid donor specificity of PatA, in the context of the acylation of PIMs. By using radioactivity experiments, they showed that palmitic (C16), myristic (C14), and oleic (C18, cis-9) acid could be incorporated into phospholipids isolated from Mycobacterium phlei. More recently, different acyl-forms of PIM2, and among them, Ac1PIM2, the product of the reaction catalyzed by PatA, were isolated from M. bovis BCG and characterized (45). Matrix-assisted laser desorption/ionization tandem MS analysis have notably revealed that the Manp is selectively acylated by a C16 acyl appendage (48). Our hydrolytic experiments, DNTB assays and direct liquid chromatography (LC)–MS detection, showed that PatA could transfer all the acyl-CoA derivatives used (C2, C8, C12, C14, C16, C18, and C20) to PIM2. Although the enzyme showed the highest activity using C14-CoA, and not C16-CoA, we only detected binding of PatA to the nonhydrolyzable S-C16-CoA derivative by ITC, supporting the notion that C16-CoA is the natural donor substrate of PatA (45) The high activity of PatA in the presence of C14-CoA, a non-natural substrate of the enzyme, and the low binding affinity of the enzyme to the nonhydrolyzable homolog derivative with the same chain length might be explained by an easier release of the C14 product facilitating reaction turnover.
Our activity measurements showed a clear Gaussian-type curve profile of PatA activity against acyl-CoA derivatives of different lengths (Fig. 6A). These data strongly support the notion that PatA can recognize acyl-CoA derivatives differently, based on acyl chain length. Specifically, the hydrophobic tunnel acts as a ruler that measures the length of the acyl chains and therefore determines the specific recognition of the substrate. The acyltransferase LpxM from Acinetobacter baumannii (AbLpxM), a structural homolog of PatA (49) (PDB codes 5KN7 and 5KNK; Z score of 26.7; r.m.s.d. value of 2.6 Å for 243 aligned residues), transfers C12 length acyl chains instead other acyl chains length tested (C10, C14, and C16). We showed that the C16-CoA derivative was too long to fit in the hydrophobic groove of AbLpxM, which is much shorter than the one present in PatA (44). This hydrocarbon ruler mechanism has evolved in other enzymes that need to distinguish between different fatty acyl chain lengths in CoA and ACP derivatives, including (i) the pimeloyl-CoA synthetase (BioW) from Bacillus subtilis involved in the biosynthesis of biotin; (ii) the acyltransferase LpxA from E. coli; and (iii) the outer membrane acyltransferase PagP from E. coli, (50, 51), which are both involved in the biosynthesis of Lipid A; (iv) the 1-acyl-glycerol-3-phosphate acyltransferase from Thermotoga maritime (TmPlsC), involved in membrane phospholipids biosynthesis (52); and (v) the eukaryotic S-acyltransferase (human DHHC20 and zebrafish DHHC15) that catalyzes protein palmitoylation (53). Notably, acyl chain length specificity could be modified by modulating the length corresponding acyl chain recognition grooves using point mutations (51–53). This ruler mechanism is a general substrate recognition strategy, widespread in enzymes that catalyze several reaction types, including soluble and membrane proteins (51, 54) such as glycosyltransferases, amylomaltases, ribonucleases, and aminopeptidases (51, 54–57).
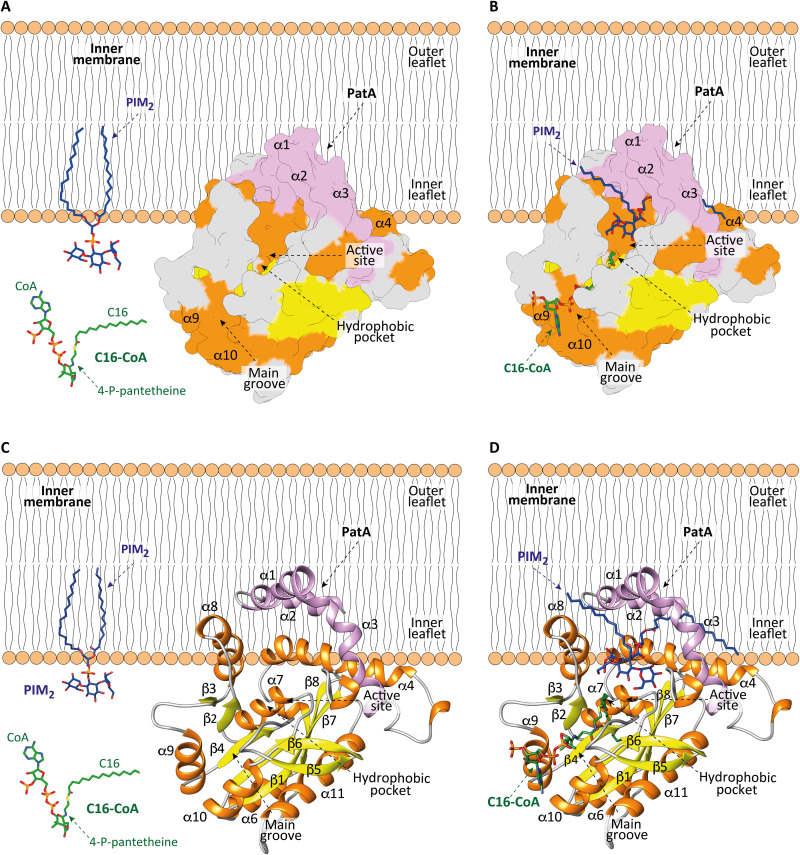
Evolution has led enzymes to acquire exquisite molecular mechanisms to access lipophilic substrates located in membranes and then catalyze their chemical reaction with polar, water-soluble compounds (5). When enzymes have an active site embedded in the membrane, the hydrophobic substrate laterally diffuses from the plane of the membrane and into the transmembrane region through hydrophilic channels. These channels are present in enzymes with different overall folds. When enzymes show an active site not embedded in a membrane, the hydrophobic substrate is extracted by a desorption mechanism from the membrane and binds to a channel or cavity located within the main core of the protein. Last, some enzymes have the active site located exactly at the membrane surface, where both hydrophobic and hydrophilic substrates may stay in their original environment with the reaction occurring at the membrane-water interface (5). PatA follows a different mechanism. The enzyme displays an active site located in a major groove facing the membrane surface. PatA sequesters the C16 acyl group from the soluble C16-CoA and incorporates it into a hydrophobic tunnel, deeply buried in the core of the enzyme. The polar head of PIM1/PIM2 enters into a cavity located in the major groove, whereas the fatty acid chains of the glycolipid remain attached or partially sequestered from the membrane (Fig. 7).
Fig. 7. Model of membrane association mechanism of PatA.
(A to C) The N-terminal helices α1 and α2 and the polarity of PatA determines the right orientation of the enzyme on the membrane. PIM2 is anchored into the inner leaflet of the mycobacterial IM. C16-CoA is localized in the cytosol. (B to D) PatA sequesters the acyl chain of the acyl-CoA donor substrate from the cytosol and incorporates it into a hydrophobic tunnel that extends entirely through the core of the enzyme. In this orientation, the polar head of PIM2 enters into a cavity located in the major groove to react with the acyl-CoA, at the protein-membrane interphase. The Ac1PIM2 product is formed and CoA-SH is released to the cytosol.
In summary, we showed that the interaction and insertion of the acyltransferase PatA within anionic lipid bilayers are typical of an integral membrane protein. This mode of interaction, as well as the architecture of the substrates binding sites and the catalytic site, determines the ability of the enzyme to facilitate the reaction of molecules with different chemical characteristics. Moreover, our enzymatic and binding experiments showed that PatA can transfer acyl groups from acyl-CoAs of different chain lengths via a hydrocarbon ruler mechanism. Many structural, chemical, and mechanistic aspects of this type of membrane enzymology remain intriguing and major challenge. From a drug discovery perspective, we hypothesize that the presence of clear and deep pockets might facilitate the discovery of potent inhibitors against the enzyme.
MATERIALS AND METHODS
Materials
The phospholipids DOPC, DOPG, DMPC, and DMPG; the spin labels 1-palmitoyl-2-stearoyl(n-doxyl)-sn-glycero-3-phosphocholine (n-PCSL; where n = 5 and 16) and DPPTC; the CoA derivatives, 12:0 CoA (C12-CoA), 14:0 CoA (C14-CoA), 16:0 CoA (C16-CoA), 18:0 CoA (C18-CoA), and 20:0 CoA (C20-CoA) ammonium salts; and the nonhydrolyzable CoA derivatives 16:0 ether CoA and 14:0 ether CoA ammonium salts, were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). The 02:0 acetyl-CoA lithium salt (C2-CoA) and 08:0 octanoyl-CoA ammonium salt (C8-CoA) were purchased from Sigma-Aldrich (Missouri, USA). All reagents were used without further purification. PIM2 was chemical synthetized as previously described (44).
Expression and purification of PatA from M. smegmatis mc2155
The full length patA gene from M. smegmatis mc2155 (MSMEG_2934; UniProt code A0QWG5) was inserted into the pJAM3 plasmid, using the Nde I and Xho I sites (pJAM3-patA). The pJAM3 plasmid is a derivative of pJAM2 plasmid, where an Nde I site was generated in the multiple cloning site. M. smegmatis mc2155 cells, transformed with pJAM3-patA, were grown in MM63 medium [15 mM (NH4)2SO4 and 100 mM KH2PO4] supplemented with 0.5 mM MgSO4, 0.025% (v/v) tyloxapol, and 17 mM succinic acid, adjusted to pH 7.0 with KOH, supplemented with kanamycin (20 μg ml−1; fig. S1) (44). When the culture reached optical density at 600 nm (OD600) = 0.6, the expression of PatA was induced by adding 0.2% acetamide. After 30 hours at 37°C, cells were harvested at 5000g for 20 min at 4°C and resuspended in 20 mM tris-HCl (pH 7.5) containing protease inhibitors (Complete EDTA-free, Roche) and culture of benzonase (0.5 μl liter−1; Sigma-Aldrich). Cells were disrupted by sonication in 36 cycles of 10-s pulses, with 60-s cooling intervals between the pulses, and 60% of amplitude. To solubilize the protein from the mycobacterial membrane, we added a final concentration of 2 mM CHAPS, 300 mM NaCl, and 10% glycerol, to the previous disrupted cells mixture. The suspension was gently stirred during 1 hour at 4°C and centrifuged at 59,000g for 30 min at 4°C. The supernatant was filtered by 0.22-μm pore-size Merck Millipore Durapore polyvinylidene difluoride (PVDF) membrane filters and applied to a Co2+ Talon Resin (GE Healthcare; ratio 1 ml of resin:1000 ml of culture volume) equilibrated in 20 mM tris-HCl (pH 7.5), 300 mM NaCl, and 10% glycerol and incubated with gently stirring on rotator for 1 hour at 4°C. The resin was then transferred to an empty column and washed with 25 column volumes (CV) of 20 mM tris-HCl (pH 7.5), 300 mM NaCl, 10 mM imidazol, and 10% glycerol. The proteins were then eluted by gravity flow with a step gradient of 20, 50, 75, 100, 200, 300, and 500 mM imidazole, in 20 mM tris-HCl (pH 7.5), 300 mM NaCl, and 10% glycerol, three CV each. The fractions of interest were pooled and dialyzed against 5 liters of 50 mM tris-HCl (pH 7.5), 150 mM NaCl, and 10% glycerol overnight at 4°C. The protein was loaded onto a Superdex 200 10/300 GL (GE Healthcare) equilibrated in the 20 mM tris-HCl (pH 7.5) and 150 mM NaCl. The resulting preparation displayed a single-protein band when analyzed by SDS–polyacrylamide gel electrophoresis (PAGE). The purified full-length PatA was stored at −80°C. We obtained 1.6 mg of purified PatA per liter of culture.
Expression and purification of PimB from M. smegmatis mc2155
E. coli BL21(DE3) cells transformed with pET29a-pimB were grown in 2xYT medium supplemented with kanamycin (50 μg ml−1) at 37°C (29). When the culture reached OD600 = 0.6, PimB expression was induced by adding 1.0 mM isopropyl-β-d-thiogalactopyranoside. After 20 hours at 18°C, cells were harvested at 5000g for 20 min at 4°C and resuspended in 50 mM tris-HCl (pH 8.0), 500 mM NaCl, 10 mM imidazol, and 25% glycerol containing protease inhibitors (cOmplete EDTA-free, Roche) and culture of benzonase (0.5 μl liter−1; Sigma-Aldrich). Cells were then disrupted by sonication in 12 cycles of 10-s pulses, with 60-s cooling intervals between the pulses, and 60% of amplitude, and the suspension was centrifuged for 30 min at 59,000g at 18°C. The supernatant was filtered by 0.22-μm pore-size Merck Millipore Durapore PVDF membrane filters and subjected to Ni2+-affinity chromatography using a HisTrap Chelating column (1 ml; GE Healthcare) equilibrated in 50 mM tris-HCl (pH 8.0), 500 mM NaCl, 10 mM imidazole, and 25% glycerol. The elution was performed with a linear gradient of 20 to 250 mM imidazole at 1 ml min−1. The fractions of interest were pooled and dialyzed overnight at 4°C against 50 mM tris-HCl (pH 7.5), 350 mM NaCl, and 25% glycerol. The protein was loaded onto a Superdex 200 10/300 GL (GE Healthcare) equilibrated in 50 mM tris-HCl (pH 7.5), 350 mM NaCl, and 25% glycerol. The resulting preparation displayed a single protein band when analyzed by SDS-PAGE. The purified PimB was aliquoted and stored at −80°C. We obtained 6.25 mg of purified PimB per liter of growth culture.
Isolation of native lipids from M. smegmatis mc2155
M. smegmatis mc2155 strain was cultured in 200 ml of Middlebrook 7H9 supplemented with 10% (v/v) OADC (oleic acid, albumin, dextrose and catalase) enrichment medium [NaCl (8.8 g liter−1), albumin bovine fraction V (Amresco), dextrose (20 g liter−1), 0.05% (v/v) of 1% stock solution of oleic acid, catalase (40 mg liter−1), and 0.5% (v/v) glycerol] for 3 days at 37°C in absence of agitation. Bacterial cultures were centrifuged at 5000g for 10 min at 4°C. The resulting pellet was washed with 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4 [phosphate-buffered saline (PBS)] at 4°C. The cells were resuspended in PBS, 250 mM sucrose, and 1 mM EDTA, supplemented with protease inhibitors, and disrupted in a FastPrep-24 5G bead beater using zirconia beads (0.1 mm zirconia/silica; BioSpec Products), nine beating cycles of 30 s, with 60-s cooling intervals between the pulses. The lysed cells were centrifuged at 10,000g for 10 min at 4°C to remove the beads and unbroken cells. The supernatant was centrifuged at 100,000g for 1 hour at 4°C. The isolated M. smegmatis membranes were resuspended in PBS, 250 mM sucrose, 20 mM tris-HCl (pH 7.5), 300 mM NaCl, and 10% glycerol and quickly frozen at −80°C.
Liquid-liquid extraction technique was used to purify the membrane’s lipids. To this end, 2.4 ml of CHCl3:CH3OH (2:1) were added to 0.3 m of the M. smegmatis mc2155 membranes. The mixture was vigorously mixed, and the organic and aqueous phases were separated because of the solvents immiscibility at 4°C. The organic phase was washed with water and 2 M NaCl, and the polar phase was washed with CHCl3:CH3OH (2:1) twice. All the saved organic phases were collected in a tube, dried under a N2 flow, and centrifuged for 1 hour under vacuum in a RVC 2-25 CD plus concentrator at 37°C to remove traces of organic solvents. Two milligrams of dry lipid film was obtained.
Preparation of SUVs
For SPR measurements, a dry lipidic film [DOPC, DOPG:DOPC (60:40, w/w), or M. smegmatis mc2155 membrane lipids] was prepared from a CHCl3:CH3OH (2:1) solution by removing the solvent under a stream of N2 and ultracentrifuged under vacuum for 1 hour to remove traces of organic solvent. SUVs of each lipid (DOPC films to create DOPC-SUVs, DOPG:DOPC mixture film to create DOPG-SUVs, and M. smegmatis mc2155 membrane lipids films to create M. smegmatis mc2155-SUVs) were prepared by dispersion of the lipids 20 mM tris-HCl (pH 7.5) and 150 mM NaCl by vigorous vortexing and following sonication at 4°C with a probe microtip sonicator [MSE Soniprep 150 (MSE, UK)] of two cycles of 5 min (5-s on and off cycles) at 100-μm amplitude and a break of 5 min in between cycles.
For CD, DSC, and ESR measurements, appropriate amounts of phospholipids and nitroxide-labeled lipids [0.5 mol percent (mol %) for ESR] dissolved in CHCl3:CH3OH (2:1) were mixed in a round-bottom glass tube, dried under a nitrogen stream, and kept under vacuum overnight to remove traces of organic solvent. The lipid film was hydrated with the appropriate buffer at a temperature ~10°C higher than the main phase transition temperature (Tm) of the lipid components, and submitted to intense vortexing. Samples were then subjected to five freeze-thaw cycles. SUVs were prepared by sonicating the resulting multilamellar vesicles (MLVs) on a bath-type sonicator at T > Tm + 10°C until the solution became clear. LUVs were obtained by subjecting the MLVs to 13 to 17 passes through polycarbonate membranes of 100-nm pore size (Nuclepore Corp., Cambridge, CA) using an Avanti Mini Extruder (Alabaster, AL, USA). The size distribution of the resultant LUVs and SUVs was confirmed by dynamic light scattering using a Zetasizer μV (Malvern Instruments).
The preparation of the lipid/protein samples for CD, DSC, and ESR was the following. An aliquot of the protein stock solutions was added into preformed SUVs or LUVs and gently vortexed for a few seconds. The samples were kept equilibrating at a temperature T > Tm of the lipids (22°C for DOPG/DOPC or 30°C for DMPG/DMPC) for 20 min before the experiments.
SPR experiments
SPR experiments were performed in a BIAcore 3000 system (GE Healthcare). The running buffer for PatA and PimB experiments was 50 mM tris-HCl (pH 7.5), 150 mM NaCl, 50 mM tris-HCl (pH 7.5), 350 mM NaCl, and 25% glycerol, respectively. The chip surfaces were first washed with 50 mM NaOH:isopropanol (2:3) and 20 mM CHAPS, and then the lipid vesicles were immobilized on the chip’s surface. DOPC-SUVs and DMPC-SUVs were immobilized at 5 μl min−1 to reach a density of 2000 RU (resonance unit) in flow cell 1 as negative control because we did not observe binding of PatA neither PimB to DOPC-SUVs in preliminary experiments. DOPG-SUVs, DMPG-SUVs, and M. smegmatis mc2155 lipids SUVs were immobilized at 2 μl min−1 to reach the same density in flow cell 2. After lipid vesicle immobilization, BSA (1 mg ml−1) was injected at 30 μl min−1 to fill any possible noncovered chip surface. After baseline stabilization, twofold concentration series (from 12 μM up to 0.05 μM) of 60 μl of the corresponding protein was injected at 30 μl min−1 for 1 min. After 5 min of dissociation time, the surface was regenerated with 50 mM NaOH:isopropanol (2:3) and 20 mM CHAPS between binding cycles. Experimental data were corrected for instrumental and bulk artifacts by referencing to the flow cell 1, using the BIAevaluation 4.1 software. The affinity constants (KD) were calculated using a general steady-state equilibrium model and the BIAevaluation 4.1 software.
Monolayer studies by Langmuir films
Protein-lipid interactions were studied with the Langmuir film balance technique using a Kibron microtensiometer (μTROUGH SX, Kibron Inc. Helsinki, Finland). We have prepared a solution of 1 mM of the DOPC- and DOPG-lipids [DOPG:DOPC (60:40, w/w)]. Monomolecular films of the indicated lipid were spread on 20 mM tris-HCl (pH 7.5) and 150 mM NaCl for PatA and 50 mM tris-HCl (pH 7.5), 350 mM NaCl, and 25% glycerol for PimB subphases (volume of 1250 μl). The desired lipidic monolayer was created spreading over the aqueous subphase of the DOPC or DOPG with a 10-μl Hamilton syringe inducing different superficial tension values (π0) between 15 and 40 mN m−1. After baseline equilibration, a final concentration of 100 μM protein (PatA, PimB, and BSA) was injected. PimB and BSA were used as negative controls. Superficial pressure changes were continuously recorded as a function of time, and the achieved pressure was saved when a plateau was reached (πprotein). The data were analyzed with the FilmWareX 3.57 program (Kibron Inc.). The experiment results were plotted following a linear regression between the monolayer pressure difference before and after the protein addition (Δπmax = πprotein − π0) versus the initial monolayer superficial tension values (π0). The critical pressure of the protein insertion (πc) is the value where, in the plot, the abscissa axis is crossed.
Continuous wave ESR assays
Lipid and L/P samples transferred to glass capillaries (1.5 mm I.D) were placed in a quartz tube. X-band (9.5 GHz) ESR experiments were carried out on a Varian E-109 spectrometer using 10-mW microwave power and 0.5- to 1.0-G field modulation amplitude. The center field was set to 3365 G, and the scan width was 100 to 140 G. The spectra were recorded with an acquisition time of 240 s using a time constant of 128 ms. The temperature was controlled by a homemade temperature control unit coupled to the spectrometer. ESR spectral analysis were carried out as previously described (39) using the Multicomponent LabVIEW (National Instruments) software developed by C. Altenbach (University of California, Los Angeles, CA). The effective order parameter, Szz, was calculated from the outer (2Amax) and inner (2Amin) hyperfine splitting values extracted from the 5-PCSL ESR spectra (58).
DSC assays
The effect of PatA and PimB on the thermotropic phase behavior of DMPG/DMPC (60:40, mol/mol) liposomes was recorded in a VP-DSC MicroCal MicroCalorimeter (Microcal, Northampton, MA, USA) using a heating rate of 1.0°C min−1. The samples were firstly degassed, loaded into the calorimeter cells, and equilibrated for 10 min at 5°C before the measurements. Lipid concentration ranged between 0.6 and 0.8 mM, whereas the protein-to-lipid molar ratios were 1:400, 1:200, and 1:100. The lipid vesicles were prepared in 20 mM tris-HCl and 150 mM NaCl (pH 7.5). The analyses of the heat capacity profiles of the protein-free and protein-bound lipid vesicles were performed using Microcal Origin software (OriginLab Corporation, Northampton, MA).
CD analysis
CD experiments were performed on a Jasco J-715 spectropolarimeter using a 1-mm path-length quartz cell. The bandwidth and response time were set, respectively, to 1 nm and 2 s. The spectra of PatA and PimB (3 to 5 μM) in the absence and presence of DOPG/DOPC (60:40, mol/mol) and DMPG/DMPC (60:40, mol/mol) SUVs were recorded at 20°C over the wavelength range of 200 to 270 nm and at 100 nm min−1. The resultant spectra were averaged over six spectral accumulations, each corresponding to an average of six scans, and subtracted by the spectra of the protein-free control samples acquired under the same conditions. The final spectra were converted to mean residue molar ellipticity. Thermal unfolding of PatA and PimB in the absence and presence of the lipid vesicles was recorded at 222 nm using a heating rate of 1°C min−1 in the 15° to 80°C range. The temperature was controlled using a Peltier thermal device. The data were fitted to a two-state equilibrium model between folded and unfolded states without heat capacity changes, as described elsewhere (31).
Molecular modeling
First, helices α1, α2, and partially α3 were modeled for PatA using the crystal structure of lipid A secondary acyltransferase LpxM as a template (PDB code 5KN7) (49) and with PyMOL 2.4 software. Subsequently, the PIM2 glycolipid was docked to the PatA-ManC16 molecule, whose binding pose with the protein was previously revealed using x-ray crystallography by our group (PDB code 5OCE) (44).
Bilayers of 1,2-distearoyl-sn-glycero-3-phosphocholine lipid, together with the modeled complex, H2O molecules, and Cl− to neutralize the system, were constructed using the CHARMM Membrane Builder GUI (www.charmm-gui.org) (59) and converted to Lipid17 PDB format with the charmmlipid2amber.x script included in Amber20 (60). MD simulations were performed using the ff14SB force field, which is an evolution of the Stony Brook modification of the Amber 99 force field force field (ff99SB) (61). Lipid17 force field (62) was used to properly simulate the conformational behavior of the lipid membrane. Parameters for the PIM2 glycolipid were generated with the antechamber module of Amber20 using the general Amber force field (63) with partial charges set to fit the electrostatic potential generated with HF/6-31G(d) by RESP (64). The charges were calculated according to the Merz-Singh-Kollman scheme using Gaussian 09 (65). The TIP3P water model was used for H2O (66). The full system was minimized for 10,000 steps, of which the first 5000 steps used the steepest descent method and the remaining steps used the conjugate gradient method (67). The system was then heated from 0 to 100 K using Langevin dynamics (68) for 5 ps at constant volume, with weak restraints on the lipid (force constant, 10 kcal mol−1 Å−2). Following this, the volume was allowed to change freely and the temperature increased to 300 K with a Langevin collision frequency of γ = 1.0 ps−1 and anisotropic Berendsen regulation (69) (1 atm) with a time constant of 2 ps for 100 ps. The same weak restraint of 10 kcal mol−1 Å−2 was maintained on the lipid molecules. Constant pressure and constant temperature (NPT) runs were performed using the AMBER 20. Bonds involving hydrogen were constrained using the SHAKE algorithm, allowing a 2-fs time step. Structural data were recorded every 10 ps. PME (particle mesh Ewald) was used to treat all electrostatic interactions with a real space cutoff of 10 Å. A long-range analytical dispersion correction was applied to the energy and pressure. All simulations were performed at constant pressure of 1 atm and constant target temperature. Temperature was controlled by the Langevin thermostat, with a collision frequency of γ = 1.0 ps−1. Pressure was regulated by the anisotropic Berendsen method (1 atm) with a pressure relaxation time of 1.0 ps. The system was simulated for 0.5 μs.
PatA palmitoyl-CoA hydrolytic activity assay
The specific activity of full-length PatA using different acyl-CoA derivatives as acyl donors (C2-CoA, C8-CoA, C12-CoA, C14-CoA, C16-CoA, C18-CoA, and C20-CoA) was measured by the spectrophotometric disulfide DTNB assay, a method described for other acyltransferases (43, 44, 70, 71). Specifically, PatA transfers the acyl chain from the acyl-CoA compound to PIM2, producing CoA from the hydrolysis of the acyl donor. Thus, CoA-SH liberated in the reaction reacts with DTNB, releasing TNB−2 as a product. TNB−2 absorbance is measured at 412-nm wavelength in a NanoDrop One spectrophotometer (Thermo Fisher Scientific). The reaction contained 20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM DOPC-SUV, 200 μM PIM2 mixture, 200 μM of the acyl-CoA derivative, 600 μM DTNB, and 1 μM full-length PatA. DOPC-SUV and PIM2 were vortexed together in 20 mM tris-HCl (pH 8.3), in two cycles of 2 min with a 2-min time lapse in between. The protein was added after the blank was measured. After the reaction started, TNB−2 absorbance was measured every 3 s for the first 180 s and then every 7 s until 1600 s was reached. The absorbance values during the first 60 s were converted into the amount of hydrolyzed acyl-CoA derivative per milligram of protein in 1 min and compared with the most active one activity (C14-CoA in our scenario). All enzymatic activity measurements were determined in duplicates.
PatA acyltransferase activity assay
PatA transferase activity was measured by high-resolution LC coupled to MS. A typical reaction contained 20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM DOPC-SUV, 200 μM PIM2 mixture, 200 μM of the acyl-CoA derivative, 600 μM DTNB, and 3.5 μM full-length PatA. DOPC-SUV and PIM2 are vortexed together in 20 mM tris-HCl (pH 8.3), in two cycles of 2 min with a 2-min time lapse in between. The reaction was incubated for 3 hours at room temperature. The reaction was quenched by adding 60% (v/v) final concentration of acetonitrile. In negative control reactions, full-length PatA was added after 1 hour and immediately quenched. The quenched samples were sonicated for 10 min. To 350 μl of sonicated mixture, 350 μl of chloroform were added, and the solution was incubated for 30 min at room temperature under shaking, followed by centrifugation for 5 min at 4°C and 21,000g for phase separation. One hundred microliters of the aqueous phase was dried at −4°C in a rotary vacuum concentrator (Labconco). Before LC-MS analysis, samples were resuspended and vortexed in 150 μl of a CH3OH:H2O (2:1) mixture.
LC-MS measurement
Relative quantification of target compounds was performed using an Agilent 1290 LC coupled to an Agilent 6560 quadrupole time-of-flight (Q-TOF) MS system equipped with a Dual Agilent Jet Stream ESI (electrospray ionization) source. The column (Waters ACQUITY UPLC BEH C18, 100 × 1 mm, 1.7-μm particle size) used in this study was maintained at 40°C. The autosampler was kept at 4°C, and the injection volume was 10 μl. The flow rate was set to 0.12 ml min−1, and the mobile phases were consisted of 10 mM ammonium formate in 60:40 CH3CN:H2O (Eluent A, pH unadjusted) and 10 mM ammonium formate in 90:10 IPN:CH3CN (Eluent B, pH unadjusted). The run consisted of a linear gradient from 40 to 99% Eluent B over 6 min, followed by an isocratic delivery of 99% Eluent B over 7 min and a reequilibration phase on starting conditions with 40% Eluent B for 7 min. Total run time was 20 min per sample.
MS experiments were performed using ESI in negative mode with a capillary voltage of 2000 V and a nozzle voltage of 500 V. The deprotonated target molecules were monitored in high-resolution mode (slicer position: 5) and Extended Dynamic Range (2 GHz) with the following Q-TOF MS conditions: drying gas temperature, 325°C; drying gas flow, 10 liters min−1 (nitrogen); nebulizer, 35 psig; sheath gas temperature, 350°C; sheath gas flow, 12 liters min−1; fragmentor, 380 V; and Oct RF Vpp, 750 V. Full-scan spectra were acquired from mass/charge ratio (m/z) 50 to 1600 (1 spectrum s−1). External mass calibration was performed before measurement of each set of samples. A reference solution (m/z 112.9855 and m/z 966.0007) was used online mass correction during the acquisition. All data were acquired with Agilent Mass Hunter LC/MS Data Acquisition (ver B.09.00, Build 9.0.9044.0) and analyzed with Agilent Mass Hunter Profinder (ver 10.0 SP1, Build 10.0.10142.1). Target compounds were identified by exact mass (mass error ± 5 parts per minute), isotopic pattern, and retention time (± 0.15 min) matching (Batch Targeted Feature Extraction; table S4). Semiquantification was based on integrated peak area of the deprotonated target compound.
Chemical synthesis of nonhydrolyzable C12-CoA and C18-CoA derivatives
To a stirred solution of CoA-SH (1.0 equivalent) in a 1.0 M aqueous buffer solution of triethylammonium bicarbonate (1 ml, pH 8.1 to 8.5) was added the corresponding alkyl iodide (20 equivalent) at room temperature. The reaction mixture was heated at 50° to 60°C for 16 hours before being cooled to room temperature, and then water was added. The mixture was extracted with CH2Cl2 to remove the unreacted alkyl iodide. The aqueous phase was concentrated, and the residue was purified by C18 chromatography (CH3OH:H2O from 1:1 to 1:3). The collected fractions were concentrated in vacuum and lyophilized to give the final products as fluffy white solids. See the Supplementary Materials for details.
PatA crystallization and data collection
PatA was crystallized by mixing 0.2 μl of a protein solution at 5.22 mg ml−1 in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl in presence of 1 mM S-C12-CoA derivative, with 0.2 μl of 100 mM imidazole/MES monohydrate (pH 8.5), 60 mM divalent cations mixture (magnesium chloride hexahydrate and calcium chloride dihydrate), and 50% (w/v) of precipitant mix based on 20% (w/v) PEG 4000 and 40% (w/v) glycerol (Morpheus protein crystallization screen). The crystals grew in 7 days. The crystals were transferred to a cryo-protectant solution containing 30% glycerol and frozen under liquid nitrogen. Complete x-ray diffraction datasets for both crystal forms were collected at beamline i03 (Diamond Light Source, Oxfordshire, UK). PatA crystallized in the monoclinic space group P21 with four molecules in the asymmetric unit and diffracted to a maximum resolution of 3.67 Å (table S5). All datasets were integrated and scaled with XDS following standard procedures (72).
PatA structure determination and refinement
Structure determination of PatA was carried out by molecular replacement methods implemented in Phaser (73) and the PHENIX suite (74) and using the PDB code 5F2Z as a search template. The final manual building was performed with Coot (75) and refinement with phenix.refine (76). The structure was validated by MolProbity (77). Data collection and refinement statistics are presented in table S5. The atomic coordinates and structure factors have been deposited with the PDB code 7OJT. Molecular graphics and structural analyses were performed with the UCSF Chimera package (78).
ITC measurements
ITC essays were performed on a MicroCal PEAQ-ITC (Malvern) at 25°C. Two hundred microliters of 50 μM full-length PatA in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl was degasified for 30 min and placed into the cell. Forty microliters of 0.716 mM nonhydrolyzable acyl-CoA derivatives in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl (S-C12-CoA, S-C14-CoA, S-C16-CoA, and S-C18-CoA) were added in 13 injections (0.4 μl of first injection and 3 μl of the rest of them with 150-s time lapse between each injection) at 750 rpm. Before data analysis was carried out, dilution experiments were performed and subtracted (buffer versus buffer, buffer versus full-length PatA, and CoA NH derivatives versus buffer) to the raw data. Subtracted experimental data were fitted, and KD parameters were calculated with MicroCal PEAQ-ITC Analysis software (Malvern Instruments Ltd.) following One Set of Sites-Single Injection fitting model.
Acknowledgments
We acknowledge Diamond Light Source (proposals mx20113), ALBA synchrotron beamline BL13-XALOC (mx2018093013), and iNEXT (proposals 1618/2538) for providing access to synchrotron radiation facilities. We thank LCSB Metabolomics Platform for technical support. We thank the Biophysics and Structural Biology Group from the São Carlos Physics Institute at the University of São Paulo, Brazil for providing access to the Biophysical Facility.
Funding: This work was supported by the MINECO/FEDER EU contracts BFU2016-77427-C2-2-R, BFU2017-92223-EXP, PID2019-105649RB-I00, Severo Ochoa Excellence Accreditation SEV-2016-0644; the Basque Government contract KK-2019/00076; NIH R01AI149297 (to M.E.G.); the MINECO/FEDER EU contract BFU-2015-68981-P and the Basque Government (IT1264-19) (to F.-X.C.); MINECO/FEDER EU contract RTI2018-099592-B-C21 (to F.C.); the Canadian Glycomics Network Grant SD-1 (to T.L.L.). European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 844905 (to B.T.); the Basque Government (to I.A.); CONACyT fellowship no. 291276 (to E.D.P.-P.).
Author contributions: B.T. and M.E.G. conceived the project. I.A., L.G.M.B., L.W., A.M., E.D.P.-P., C.J., F.G., M.T., S.P., J.P., and B.T. performed the experiments. I.A., L.G.M.B., L.W., A.M., E.D.P.-P., C.J., F.G., M.T., S.P., F.-X.C., J.P., M.G., C.L.L., F.C., T.L.L., B.T., and M.E.G. analyzed the results. I.A., L.G.M.B., B.T., and M.E.G. wrote the original draft. I.A., L.G.M.B., L.W., A.M., E.D.P.-P., C.J., F.G., M.T., S.P., F.-X.C., J.P., M.G., C.L.L., F.C., T.L.L., B.T., and M.E.G. reviewed and edited the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S9
Tables S1 to S5
REFERENCES AND NOTES
- 1.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angala S. K., Belardinelli J. M., Huc-Claustre E., Wheat W. H., Jackson M., The cell envelope glycoconjugates of Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 49, 361–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strahl H., Errington J., Bacterial membranes: Structure, domains, and function. Annu. Rev. Microbiol. 71, 519–538 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Rottig A., Steinbuchel A., Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev. 77, 277–321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forneris F., Mattevi A., Enzymes without borders: Mobilizing substrates, delivering products. Science 321, 213–216 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Dufrisne M. B., Petrou V. I., Clarke O. B., Mancia F., Structural basis for catalysis at the membrane-water interface. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1368–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerin M. E., Korduláková J., Alzari P. M., Brennan P. J., Jackson M., Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J. Biol. Chem. 285, 33577–33583 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morita Y. S., Fukuda T., Sena C. B. C., Yamaryo-Botte Y., McConville M. J., Kinoshita T., Inositol lipid metabolism in mycobacteria: Biosynthesis and regulatory mechanisms. Biochim. Biophys. Acta, Gen. Subj. 1810, 630–641 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Jackson M., The mycobacterial cell envelope-lipids. Cold Spring Harb. Perspect. Med. 4, a021105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batt S. M., Minnikin D. E., Besra G. S., The thick waxy coat of mycobacteria, a protective layer against antibiotics and the host’s immune system. Biochem. J. 447, 1983–2006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boldrin F., Anso I., Alebouyeh S., Sevilla I. A., Geijo M., Garrido J. M., Marina A., Mazzabò L. C., Segafreddo G., Guerin M. E., Manganelli R., Prados-Rosales R., The phosphatidyl-myo-inositol dimannoside acyltransferase PatA is essential for Mycobacterium tuberculosis growth in vitro and in vivo. J. Bacteriol. 203, e00439-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalscheuer R., Palacios A., Anso I., Cifuente J., Anguita J., Jacobs W. R. W. R., Guerin M. E., Prados-Rosales R., The Mycobacterium tuberculosis capsule: A cell structure with key implications in pathogenesis. Biochem. J. 476, 1995–2016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jankute M., Cox J. A. G., Harrison J., Besra G. S., Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Sancho-Vaello E., Albesa-Jove D., Rodrigo-Unzueta A., Guerin M. E., Structural basis of phosphatidyl-myo-inositol mannosides biosynthesis in mycobacteria. Biochim. Biophys. Acta 1862, 1355–1367 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Maeda N., Nigou J., Herrmann J. L., Jackson M., Amara A., Lagrange P. H., Puzo G., Gicquel B., Neyrolles O., The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278, 5513–5516 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Gilleron M., Quesniaux V. F. J. J., Puzo G., Acylation state of the phosphatidylinositol hexamannosides from Mycobacterium bovis bacillus Calmette Guérin and Mycobacterium tuberculosis H37Rv and its implication in Toll-like receptor response. J. Biol. Chem. 278, 29880–29889 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Morita Y. S., Sena C. B. C., Waller R. F., Kurokawa K., Sernee M. F., Nakatani F., Haites R. E., Billman-Jacobe H., McConville M. J., Maeda Y., Kinoshita T., PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J. Biol. Chem. 281, 25143–25155 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Korduláková J., Gilleron M., Mikuová K., Puzo G., Brennan P. J., Gicquel B., Jackson M., Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis: PimA is essential for growth of mycobacteria. J. Biol. Chem. 277, 31335–31344 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M., Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282, 20705–20714 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Guerin M. E., Kaur D., Somashekar B. S., Gibbs S., Gest P., Chatterjee D., Brennan P. J., Jackson M., New insights into the early steps of phosphatidylinositol mannoside biosynthesis in Mycobacteria. J. Biol. Chem. 284, 25687–25696 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boldrin F., Ventura M., Degiacomi G., Ravishankar S., Sala C., Svetlikova Z., Ambady A., Dhar N., Kordulakova J., Zhang M., Serafini A., Vishwas V. G., Kolly G. S., Kumar N., Palù G., Guerin M. E., Mikusova K., Cole K. M., Manganelli R., The phosphatidyl-myo-inositol mannosyltransferase PimA is essential for Mycobacterium tuberculosis growth in vitro and in vivo. J. Bacteriol. 196, 3441–3451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lea-Smith D. J., Martin K. L., Pyke J. S., Tull D., McConville M. J., Coppel R. L., Crellin P. K., Analysis of a new mannosyltransferase required for the synthesis of phosphatidylinositol mannosides and lipoarbinomannan reveals two lipomannan pools in corynebacterineae. J. Biol. Chem. 283, 6773–6782 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Kordulakova J., Gilleron M., Puzo G., Brennan P. J., Gicquel B., Mikusova K., Jackson M., Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of mycobacterium species. J. Biol. Chem. 278, 36285–36295 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Svetlíková Z., Baráth P., Jackson M., Korduláková J., Mikušová K., Purification and characterization of the acyltransferase involved in biosynthesis of the major mycobacterial cell envelope glycolipid - Monoacylated phosphatidylinositol dimannoside. Protein Expr. Purif. 100, 33–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sastre D. E., Basso L. G. M., Trastoy B., Cifuente J. O., Contreras X., Gueiros-Filho F., de Mendoza D., Navarro M. V. A. S., Guerin M. E., Membrane fluidity adjusts the insertion of the transacylase PlsX to regulate phospholipid biosynthesis in Gram-positive bacteria. J. Biol. Chem. 295, 2136–2147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Šakanovič A., Hodnik V., Anderluh G., Surface plasmon resonance for measuring interactions of proteins with lipids and lipid membranes. Methods Mol. Biol. 2003, 53–70 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Li L., Storm P., Karlsson O. P., Berg S., Wieslander Å., Irreversible binding and activity control of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii at an anionic lipid bilayer surface. Biochemistry 42, 9677–9686 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Batt S. M., Jabeen T., Mishra A. K., Veerapen N., Krumbach K., Eggeling L., Besra G. S., Fütterer K., Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis. J. Biol. Chem. 285, 37741–37752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerin M. E., Schaeffer F., Chaffotte A., Gest P., Giganti D., Korduláková J., van der Woerd M., Jackson M., Alzari P. M., Substrate-induced conformational changes in the essential peripheral membrane-associated mannosyltransferase PimA from mycobacteria: Implications for catalysis. J. Biol. Chem. 284, 21613–21625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giganti D., Albesa-Jove D., Urresti S., Rodrigo-Unzueta A., Martinez M. A., Comino N., Barilone N., Bellinzoni M., Chenal A., Guerin M. E., Alzari P. M., Secondary structure reshuffling modulates glycosyltransferase function at the membrane. Nat. Chem. Biol. 11, 16–18 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Greenfield N. J., Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 1, 2527–2535 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janiak M. J., Small D. M., Shipley G. G., Nature of the thermal pretransition of synthetic phospholipids: Dimyristoyl- and dipalmitoyllecithin. Biochemistry 15, 4575–4580 (1976). [DOI] [PubMed] [Google Scholar]
- 33.Bernardino De La Serna J., Hansen S., Berzina Z., Simonsen A. C., Hannibal-Bach H. K., Knudsen J., Ejsing C. S., Bagatolli L. A., Compositional and structural characterization of monolayers and bilayers composed of native pulmonary surfactant from wild type mice. Biochim. Biophys. Acta Biomembr. 1828, 2450–2459 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Marsh D., Components of the lateral pressure in lipid bilayers deduced from HII phase dimensions. Biochim. Biophys. Acta Biomembr. 1279, 119–123 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Söderlund T., Alakoskela J. M. I., Pakkanen A. L., Kinnunen P. K. J., Comparison of the effects of surface tension and osmotic pressure on the interfacial hydration of a fluid phospholipid bilayer. Biophys. J. 85, 2333–2341 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H., Sood R., Jutila A., Bose S., Fimland G., Nissen-Meyer J., Kinnunen P. K. J., Interaction of the antimicrobial peptide pheromone Plantaricin A with model membranes: Implications for a novel mechanism of action. Biochim. Biophys. Acta Biomembr. 1758, 1461–1474 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Marsh D., Horváth L. I., Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim. Biophys. Acta Rev. Biomembr. 1376, 267–296 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Borbat P. P., Costa-Filho A. J., Earle K. A., Moscicki J. K., Freed J. H., Electron spin resonance in studies of membranes and proteins. Science 291, 266–269 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Basso L. G. M., Mendes L. F. S., Costa-Filho A. J., The two sides of a lipid-protein story. Biophys. Rev. 8, 179–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge M., Cohen J. S., Brown H. A., Freed J. H., ADP ribosylation factor 6 binding to phosphatidylinositol 4,5-bisphosphate-containing vesicles creates defects in the bilayer structure: An electron spin resonance study. Biophys. J. 81, 994–1005 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Couto S. G., Cristina Nonato M., Costa-Filho A. J., Defects in vesicle core induced by escherichia coli dihydroorotate dehydrogenase. Biophys. J. 94, 1746–1753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blobel G., Walter P., Gilmore R., Intracellular protein topogenesis. Prog. Clin. Biol. Res. 168, 3–10 (1984). [PubMed] [Google Scholar]
- 43.Albesa-Jové D., Svetlíková Z., Tersa M., Sancho-Vaello E., Carreras-González A., Bonnet P., Arrasate P., Eguskiza A., Angala S. K., Cifuente J. O., Korduláková J., Jackson M., Mikušová K., Guerin M. E., Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA. Nat. Commun. 7, 10906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tersa M., Raich L., Albesa-Jové D., Trastoy B., Prandi J., Gilleron M., Rovira C., Guerin M. E., The molecular mechanism of substrate recognition and catalysis of the membrane acyltransferase PatA from mycobacteria. ACS Chem. Biol. 13, 131–140 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Gilleron M., Ronet C., Mempel M., Monsarrat B., Gachelin G., Puzo G., Acylation state of the phosphatidylinositol mannosides from Mycobacterium bovis bacillus Calmette Guérin and ability to induce granuloma and recruit natural killer T cells. J. Biol. Chem. 276, 34896–34904 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Zheng W., Cole P. A., Novel bisubstrate analog inhibitors of serotonin N-acetyltransferase: The importance of being neutral. Bioorg. Chem. 31, 398–411 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Brennan P., Ballou C. E., Biosynthesis of mannophosphoinositides by Mycobacterium phlei. Enzymatic acylation of the dimannophosphoinositides. J. Biol. Chem. 243, 2975–2984 (1968). [PubMed] [Google Scholar]
- 48.Gilleron M., Lindner B., Puzo G., MS/MS approach for characterization of the fatty acid distribution on mycobacterial phosphatidyl-myo-inositol mannosides. Anal. Chem. 78, 8543–8548 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Dovala D., Rath C. M., Hu Q., Sawyer W. S., Shia S., Elling R. A., Knapp M. S., Metzger L. E. IV, Structure-guided enzymology of the lipid a acyltransferase LpxM reveals a dual activity mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, E6064–E6071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bishop R. E., The lipid A palmitoyltransferase PagP: Molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Ahn V. E., Lo E. I., Engel C. K., Chen L., Hwang P. M., Kay L. E., Bishop R. E., Privé G. G., A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 23, 2931–2941 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson R. M., Yao J., Gajewski S., Kumar G., Martin E. W., Rock C. O., White S. W., A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer. Nat. Struct. Mol. Biol. 24, 666–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rana M. S., Kumar P., Lee C.-J., Verardi R., Rajashankar K. R., Banerjee A., Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 359, eaao6326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarshis L. C., Proteau P. J., Kellogg B. A., Sacchettini J. C., Poulter C. D., Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. U.S.A. 93, 15018–15023 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramírez A. S., Boilevin J., Mehdipour A. R., Hummer G., Darbre T., Reymond J. L., Locher K. P., Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase. Nat. Commun. 9, 445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth C., Weizenmann N., Bexten N., Saenger W., Zimmermann W., Maier T., Sträter N., Amylose recognition and ring-size determination of amylomaltase. Sci. Adv. 3, e1601386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Y.-H., Lavoie M., Comeau M.-A., Elela S. A., Ji X., Structure of a eukaryotic RNase III postcleavage complex reveals a double-ruler mechanism for substrate selection. Mol. Cell 54, 431–444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schorn K., Marsh D., Extracting order parameters from powder EPR lineshapes for spin-labelled lipids in membranes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 53, 2235–2240 (1997). [Google Scholar]
- 59.Lee J., Patel D. S., Ståhle J., Park S. J., Kern N. R., Kim S., Lee J., Cheng X., Valvano M. A., Holst O., Knirel Y. A., Qi Y., Jo S., Klauda J. B., Widmalm G., Im W., CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J. Chem. Theory Comput. 15, 775–786 (2019). [DOI] [PubMed] [Google Scholar]
- 60.D. A. Case, K. Belfon, I. Y. Ben-Shalom, S. R. Brozell, D. S. Cerutti, T. E. Cheatham, III, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, G. Giambasu, M. K. Gilson, H. Gohlke, A. W. Goetz, R. Harris, S. Izadi, S. A. Izmailov, K. Kasavajhala, A. Kovalenko, R. Krasny, T. Kurtzman, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, V. Man, K. M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, A. Onufriev, F. Pan, S. Pantano, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. L. Simmerling, N. R. Skrynnikov, J. Smith, J. Swails, R. C. Walker, J. Wang, L. Wilson, R. M. Wolf, X. Wu, Y. Xiong, Y. Xue, D. M. York, P. A. Kollman, AMBER 2020 (2020).
- 61.Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dickson C. J., Madej B. D., Skjevik Å. A., Betz R. M., Teigen K., Gould I. R., Walker R. C., Lipid14: The amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A., Development and testing of a general Amber force field. J. Comput. Chem. 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Bayly C. I., Cieplak P., Cornell W. D., Kollman P. A., A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 97, 10269–10280 (1993). [Google Scholar]
- 65.M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, D. J. Fox, Gaussian 09, Revision A.02 (2016).
- 66.Kiyohara K., Gubbins K. E., Panagiotopoulos A. Z., Phase coexistence properties of polarizable water models. Mol. Phys. 94, 803–808 (1998). [Google Scholar]
- 67.W. H. Press, S. A. Teukolsky, W. T. Vettering, B. P. Flannery, NUMERICAL RECIPES The Art of Scientific Computing Third Edition (2007).
- 68.Pastor R. W., Brooks B. R., Szabo A., An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 65, 1409–1419 (1988). [Google Scholar]
- 69.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R., Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
- 70.Rodgers J. B. Jr., Assay of acyl-CoA:monoglyceride acyltransferase from rat small intestine using continuous recording spectrophotometry. J. Lipid Res. 10, 427–432 (1969). [PubMed] [Google Scholar]
- 71.Ellman G. L., Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 (1959). [DOI] [PubMed] [Google Scholar]
- 72.Kabsch W., XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen V. B., Arendall W. B., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Bansal-Mutalik R., Nikaido H., Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. U.S.A. 111, 4958–4963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S9
Tables S1 to S5