Full-length structures of the Lon protease complex reveal a tensegrity helix triangle for substrate selection and unfolding.
Abstract
Lon is an evolutionarily conserved proteolytic machine carrying out a wide spectrum of biological activities by degrading misfolded damaged proteins and specific cellular substrates. Lon contains a large N-terminal domain and forms a hexameric core of fused adenosine triphosphatase and protease domains. Here, we report two complete structures of Lon engaging a substrate, determined by cryo–electron microscopy to 2.4-angstrom resolution. These structures show a multilayered architecture featuring a tensegrity triangle complex, uniquely constructed by six long N-terminal helices. The interlocked helix triangle is assembled on the top of the hexameric core to spread a web of six globular substrate-binding domains. It serves as a multipurpose platform that controls the access of substrates to the AAA+ ring, provides a ruler-based mechanism for substrate selection, and acts as a pulley device to facilitate unfolding of the translocated substrate. This work provides a complete framework for understanding the structural mechanisms of Lon.
INTRODUCTION
Lon is an evolutionarily conserved adenosine 5′-triphosphate (ATP)–dependent protease found in prokaryotic cytoplasm and eukaryotic organelles. Lon plays a major role in cellular protein homeostasis by degrading damaged or misfolded proteins. Lon also participates in the regulation of diverse biological processes by degrading specific regulatory proteins (1). Lon has been implicated in the virulence of many pathogenic bacteria (2–4); mutations and dysregulation of Lon determine severe diseases and pathologic conditions in humans (5).
Lon is a large multidomain protein that assembles into a homohexamer (6). The N-terminal region contains a bilobal globular domain (7–10), which recognizes and binds protein substrates (11–13), followed by a 40-residue C-terminal linker with unknown structural and functional roles. The fused AAA+ and the protease domains form a hexameric core complex (14–18), which carries out ATP-dependent substrate degradation (1). Recent structures of substrate-bound Lon composed of the AAA+ and the protease domains form a close-ring hexamer in which the substrate-engaged adenosine triphosphatase (ATPase) domains show a spiral staircase arrangement; these results have suggested a conserved processive rotary mechanism for substrate translocation (19–21). So far, none of the available structures of the hexameric Lon complex contain the N-terminal region. Consequently, current knowledge and understanding of the structure of Lon and its mode of action are fragmented. The mechanisms by which Lon selects protein targets as the substrates, controls access of the substrates to the AAA+ ring, and mediates the unfolding of the substrates remain poorly understood. A complete structure of the full-length Lon including the functionally indispensable N-terminal region would be key to answer these fundamental questions.
Here, we report complete three-dimensional (3D) structures of the Lon protease from Meiothermus taiwanensis (MtaLon), in two substrate-bound states, determined to 2.4-Å resolution by cryo–electron microscopy (cryo-EM). This work has revealed previously unknown structural features and new roles of the N-terminal domain; the structures also show the path of a translocating substrate engaged by the N-terminal and the AAA+ domains. This study has provided answers to the mechanistic questions described above.
RESULTS AND DISCUSSION
A tensegrity helix triangle assembled by six N-terminal helices
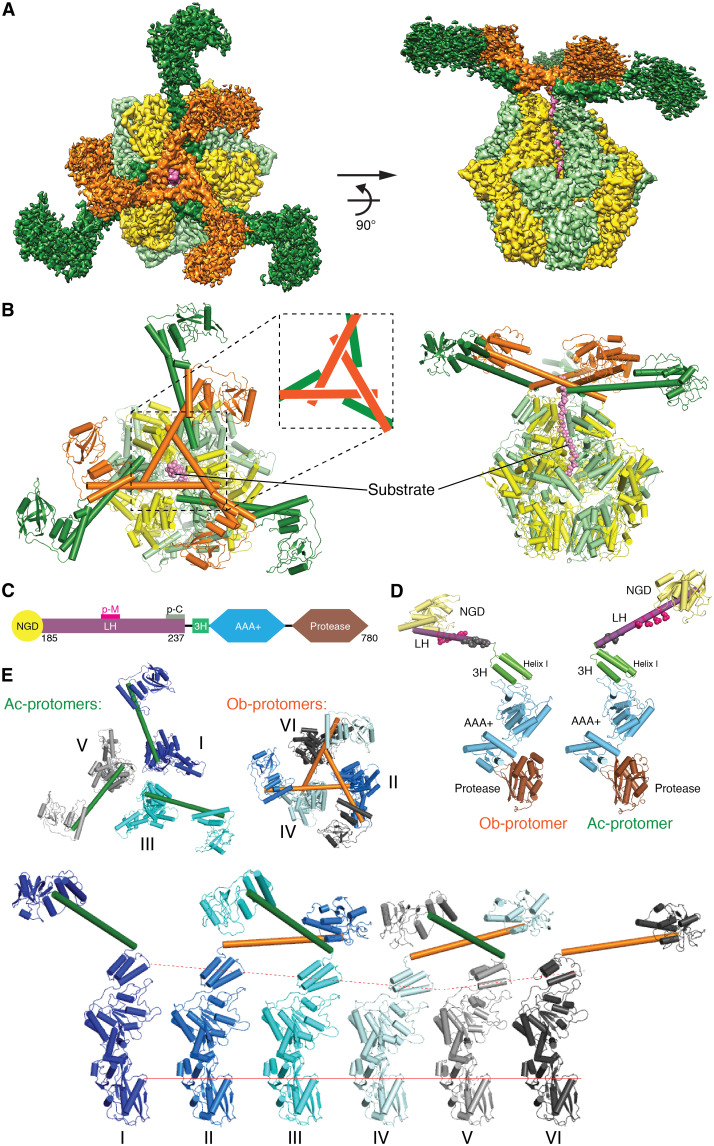
In our previous work, MtaLon was purified by size exclusion chromatography in the presence of excess amounts of adenosine 5′-[γ-thio]-triphosphate (ATP-γ-S) and the model substrate Ig2; the cryo-EM data yielded a substrate-bound structure with six protomers, of which only the ATPase domain and the protease domain (residues 241 to 780) were resolved in the map (21). However, when the cryo-EM specimen was prepared with α-casein, a common model substrate for Lon, processing of the acquired micrographs yielded cryo-EM maps with interpretable density emerging on the top of the spirally arranged ring of the ATPase domains. On the basis of the initial result, more micrograph images were collected, and the box size was increased for particle extraction (see Materials and Methods). Image processing with 3D classification eventually yielded two high-resolution maps to the resolution of 2.4 Å (Fig. 1A and figs. S1 and S2), in which some water molecules could be identified (fig. S3). These maps (map 1 and map 2) revealed three 78-Å-long shafts crossing over each other to form a triangular structure mounted on the top of the hexameric core; each of the three intercrossed sticks is adjoined by a globular domain structure, similar in shape to a golf club (Fig. 1A). The density for the three clubheads is not well resolved; nevertheless, the overall map fits well with the crystal structure of the globular N-terminal fragment (residues 1 to 206) (13). At a lower threshold, the map further reveals a weak density for three more golf club–shaped structures. Later, particle subtraction and focused local refinement were applied to improve the reconstruction for the N-terminal region. The focused refined maps, with the resolutions of 3.7 and 4.3 Å (figs. S1 and S2), showed interpretable side-chain features for the helices forming the triangular structure (residues 207 to 240) and allowed us to build the complete structure of Lon.
Fig. 1. Complete structure of the Lon complex.
(A) Reconstructed cryo-EM map of MtaLon in the top (left) and side (right) views. One group of nonadjoining protomers is colored in orange for the N-terminal and in yellow for the C-terminal regions, and the other group is colored in forest green for the N-terminal and in lime for the C-terminal regions. The substrate is colored in pink. For better display, different contour levels in Chimera are used: N-terminal regions of the protomers, colored in forest green, 0.1; N-terminal regions of the protomers, in orange, 0.3; C-terminal regions of all protomers, 0.4; substrate, 0.2. (B) The structure of MtaLon in the top and side views, in the same coloring scheme as (A). The inset shows a tensegrity triangle containing three α helices as rods. (C) Domain organization of the full-length MtaLon. The locations of patch-M and patch-C residues, which form the crossover contacts in the tensegrity triangle structure, are marked as p-M and p-C, respectively. (D) Side view of two representative protomers (chains A and B) in the same orientation, one with the LH forming an obtuse angle with helix I of 3H (Ob-protomer; shown by chain B, left) and the other with an acute-angled LH (Ac-protomer; shown by chain A, right). Each of the composed domains is colored in the same scheme as (C). The p-M/C residues are shown in spheres. (E) Structures of the full-length Lon protomers. The top row shows the top-view arrangements of three Ac-protomers (left; with forest green LHs) and three Ob-protomers (right; with orange LHs). The bottom row shows the side view of the six protomers with distinct conformational states (P1 to P6); these protomers are aligned on the basis of the protease domain.
We refer to the two determined structures based on map 1 and map 2 as Con1 and Con2, respectively. Because map 1 shows better quality overall, hereafter, the presentation and description are mainly based on the structure of Con1 (movie S1), unless mentioned otherwise. The full-length structure of Lon contains three interlocked long helices (hereafter termed LHs), extended from three non-neighboring protomers in the hexameric complex, forming a tensegrity triangle structure (22, 23). The tensegrity helix triangle of Lon consists of three long α helices; each helix intersects with two other helices, and each of the vertices is connected by crossing over the C-terminal end of one helix by another helix (Fig. 1B). The LH in each Lon protomer bridges the N-terminal globular domain (NGD) and the first helix I of the three-helix bundle (3H) of the AAA+ module via a flexible glycine-rich hinge loop (Fig. 1C). Each of the three interlocked LHs joins the AAA+ module by forming an obtuse angle with helix I of the 3H; by contrast, those of the remaining three form an acute angle (Fig. 1, D and E). The protomers with acute-angled LH (Ac-protomers) and those with obtuse-angled LH (Ob-protomers) are not adjoining in the hexameric complex (Fig. 1E). The tensegrity triangle structure is therefore formed by the LHs of the Ob-protomers; the crossovered LHs at every corner are further supported by additional interhelical contacts with another LH from each of the three Ac-protomers (Fig. 1, B and E).
Two conserved hydrophobic patches mediating crossover helix triangle formation
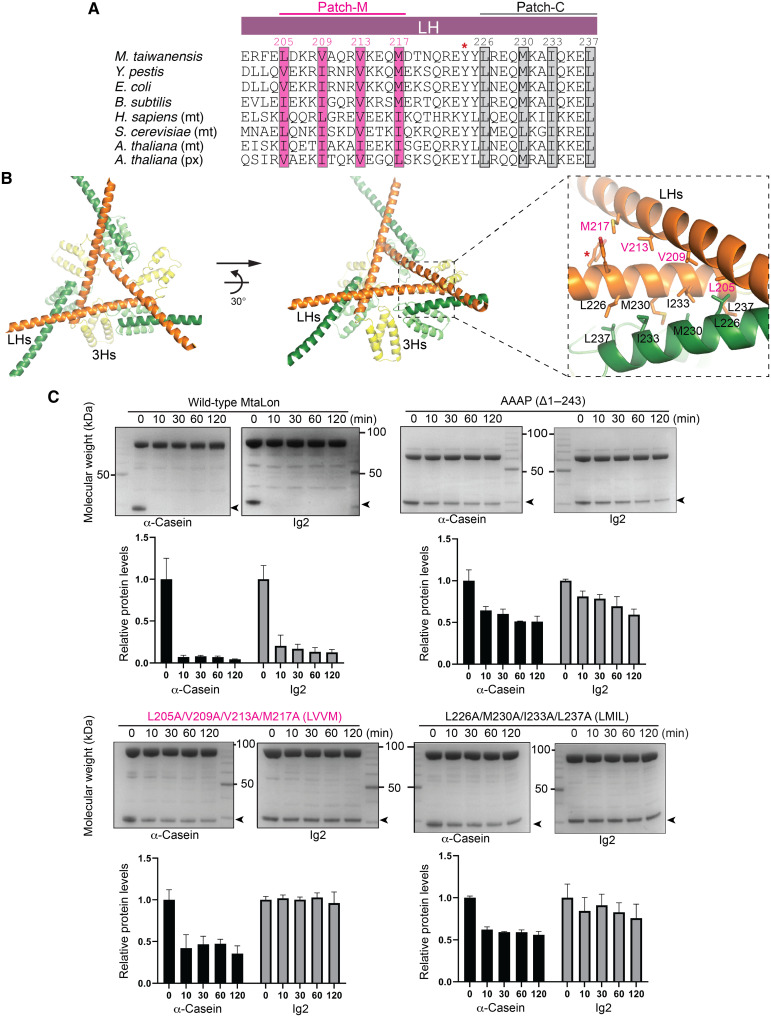
The LHs are involved in two types of interactions in the Lon hexamer: one making three crossovers of the helix triangle and the other binding to the crossovered helices. The interhelical interactions are made possible by the presence of two universally conserved hydrophobic patches, patch-M and patch-C, located in the middle and the C-terminal portions of the LH, respectively (Figs. 1C and 2A). Patch-M consists of L205, V209, V213, and M217; patch-C consists of L226, M230, I233, and L237. The crossover interaction involves patch-M and patch-C residues from the LHs of two non-neighboring Ob-protomers; the crossovered helices are further bound by patch-C residues from the LH of an Ac-protomer (Fig. 2B).
Fig. 2. Analysis of the interhelical interactions forming a tensegrity helix triangle.
(A) Structure-based sequence alignment of LH. The conserved hydrophobic residues constituting patch-M and patch-C are shaded. The conserved Y224 making contact with the substrate is indicated by the asterisk. The UniProt IDs of the aligned Lon proteins are as follows: A0A059VAZ3 for M. taiwanensis, A0A5P8YJ65 for Y. pestis, P0A9M0 for Escherichia coli, P37945 for Bacillus subtilis, P36776 for Homo sapiens, P36775 for Saccharomyces cerevisiae, and P93655 and O64948 for Arabidopsis thaliana. Mt and px are abbreviations for mitochondrion and peroxisome, respectively. (B) Two views showing the interlocked triangle complex of six LHs and their C-terminally linked 3Hs. The dashed box on the right shows the zoom-in view of the crossover interhelical interaction sites by LHs from two Ob-protomers (orange; 3Hs colored in yellow) and one Ac-protomer (forest green; 3Hs colored in lime), mediated by the hydrophobic residues of patch-M and patch-C. Y224 is indicated by the asterisk. (C) Mutational analysis showing the essential role of the hydrophobic residues in patch-M and patch-C on the activity of Lon. The substrate bands in the gels are indicated by arrowheads, and the names are shown below each gel image. Histograms of the substrate bands, which were scanned and quantified, are plotted below as means ± SD. Each gel assay was repeated three times.
We made two quadruple mutants with the four patch-M residues (LVVM) and patch-C residues (LMIL) replaced by alanines and tested their degradation activity against two substrates: Ig2 and α-casein (Fig. 2C) (17, 18). Ig2 is partially unfolded at 55°C, the preferred assay temperature for thermophilic MtaLon (17, 18); by contrast, α-casein is intrinsically disordered. Degradation of Ig2 by MtaLon strictly requires the N-terminal domain. Some loss of α-casein was observed by an MtaLon mutant lacking the entire N-terminal domain (Δ1–243) at the initial time point but no further degradation (Fig. 2C). Both LVVM and LMIL mutants completely lost the activity to degrade the substrate Ig2. Degradation of α-casein by the quadruple mutants was severely inhibited to a degree comparable to that by MtaLon Δ1–243 mutant (Fig. 2C). The results on α-casein may suggest that only a single round of degradation of α-casein occurs due to the loss of the N-terminal tensegrity structure, which is required for further rounds of α-casein degradation. These results suggest that patch-M-C and patch-C-C interactions, which mediate the formation of the prominent triangular structure, are critical for substrate-degrading activity.
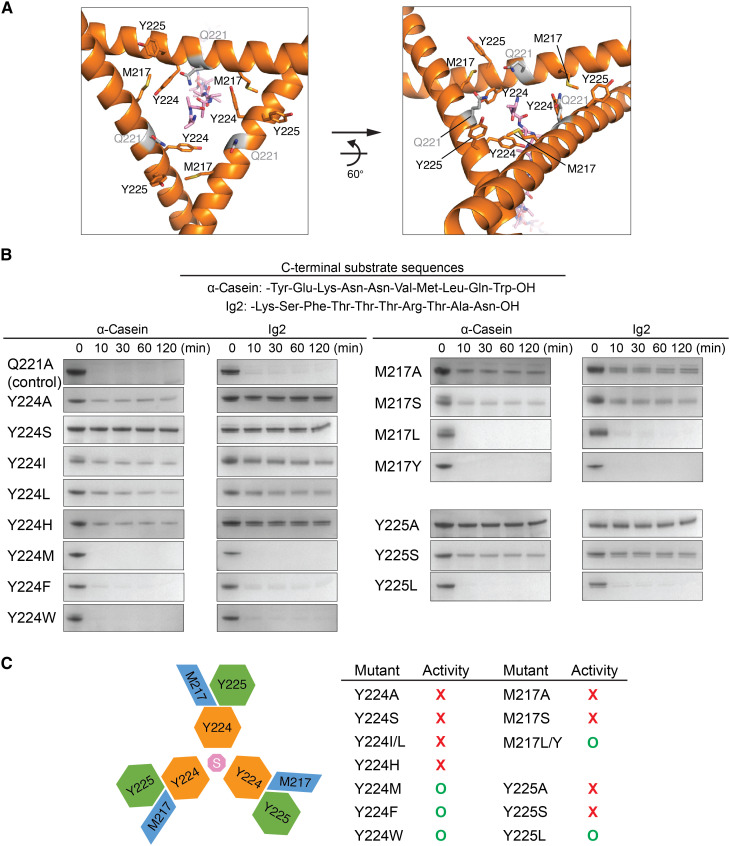
A conserved hydrophobic triad presenting a tyrosine gate to engage substrate
The helix triangle structure forms an open pore lined with three copies of the gate residue Y224, whose side chains are each packed by the neighboring Y225 and M217 from patch-M of a crossing LH (Figs. 2B and 3A). Y224 and Y225 are also highly conserved in prokaryotic and eukaryotic Lon proteins (Fig. 2A); the aromatic side chains of Y224 make contact with the substrate bound at the center of the pore (Fig. 3A). Therefore, the gate residue Y224 may play an essential role to control substrate entry. We conducted mutational analysis to test the role of each of the triad residues M217, Y224, and Y225. The results showed that the Y224A and Y224S mutants could not degrade Ig2 and α-casein, two model substrates with distinct C-terminal sequences (Fig. 3, B and C). Y224I and Y224L, with shorter and less bulky hydrophobic side chains, did not show the activity either. However, the Y224M, Y224F, and Y224W mutants displayed an activity similar to wild-type MtaLon or the control mutant Q221A (Fig. 3, B and C). These results suggest that long hydrophobic or bulky aromatic side chains of the gate residue are essential and sufficient for engaging the substrate. Moreover, we found that M217A, M217S, Y225A, and Y225S mutations all lost the substrate-degrading activity; by contrast, M217L, M217Y, and Y225L are active (Fig. 3, B and C). These results suggest that hydrophobic stacking interactions among the triad residues are also important, perhaps for positioning the hydrophobic side chain of the gate residue Y224 for effective substrate interaction; a flexible side-chain conformation of Y224 in the pore may instead impede entry of substrate polypeptide. In line with this notion, Y224H was not active; in the mutant, the charged aromatic side chain of H224 may not stack with M217 and Y225 (Fig. 3, B and C).
Fig. 3. Substrate-entry gate and mutational analysis.
(A) Structure of the substrate-entry gate formed by three LHs from Ob-protomers (orange), with key residues shown in sticks. The bound substrate is shown in pink sticks. (B) Mutational analysis of the hydrophobic triad M217, Y224, and Y225. The C-terminal sequences of two tested substrates are listed. Q221, shown in gray sticks in (A), is used as a mutation control. (C) Summary of the substrate-degrading activity of the mutants. The top-view diagram on the left illustrates the side-chain packing interaction of the triad residues M217, Y224, and Y225. The letter “S” indicates the bound substrate. Each gel assay was repeated three times.
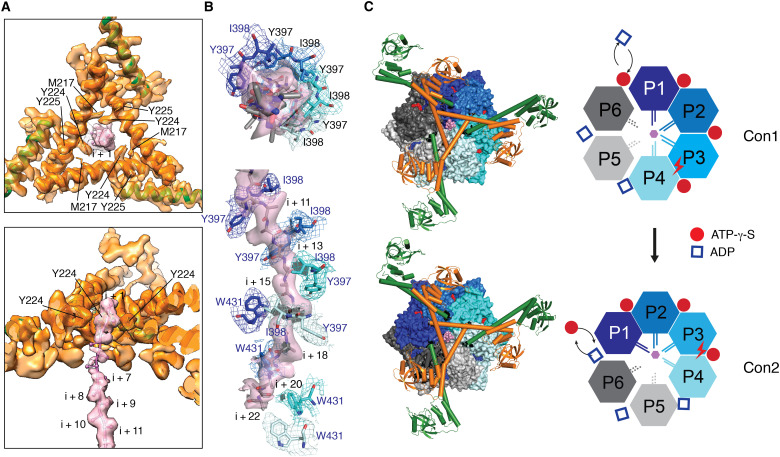
A translocation pathway delineated by the interlocked helices and the AAA+ pore loops
At the center of the triangular gate encircled by Y224, the maps reveal density for the translocating substrate, which extends down the central axis of the hexamer (Fig. 4A). After residue i + 3, the density becomes weaker and visible only at lower threshold. After residue i + 5, the density for the translocating polypeptide backbone becomes stronger and remains continuous until residue i + 22 (Fig. 4B). The translocating substrate is engaged by pore-loop-I residues Y397 and I398, the two acting as a pincer gripping the peptide bond moieties of every other residue of the substrate polypeptide; the substrate backbone is also captured by pore-loop-II residue W431. These pore-loop-I and pore-loop-II residues are contributed by four protomers P1 to P4 arranged spirally in a clockwise descending order; together, they demarcate the full path for a translocating substrate engaged by the AAA+ domains (Fig. 4B). The protomers P5 and P6 serve as two ascending nonengaging “seam” protomers, an arrangement similar to human mitochondrial Lon but not Yersinia pestis Lon (19–21). In Con1, the P1 substrate-engaging protomer is an Ac-protomer, with an acute-angled LH, whereas in Con2, the P1 protomer is an Ob-protomer; therefore, Con1 and Con2 may be regarded as two sequential conformational states in the ATP-hydrolysis cycle, which proceeds counterclockwise in each of the six protomers (Fig. 4C). On the basis of Con1 and Con2 structures, it was possible to extrapolate additional conformational states associated with the subsequent steps in the ATPase cycle (movie S2). In both Con1 and Con2 structures, the substrate-engaging protomers P1, P2, and P3 are bound to ATP-γ-S; the engaging P4 and nonengaging P5 are bound to adenosine 5′-diphosphate (ADP). Notably, the nonengaging protomer P6 is bound to ATP-γ-S in Con1 but ADP in Con2 (Fig. 4C and fig. S4). This result suggests that the seam protomer P6 is in a conformational state permitting ADP-to-ATP exchange, which lends experimental support to the proposed processive rotary model for substrate translocation (24).
Fig. 4. Engagement of Lon with α-casein and the nucleotides.
(A) Focused refined map and molecular model of the triangular gate formed by interlocked LHs, shown in the top (top) and cutaway side (bottom) views. The map is contoured at 6.5 σ. The density for the bound substrate (pink) is modeled with poly-Ala, with the residues denoted by i + n, where n is a positive integer. The LHs are colored in the same scheme as Figs. 1A and 2B. (B) Cryo-EM map and poly-Ala model of the substrate, shown in the top (top) and side (bottom) views, which is bound by the pore-loop-I residues Y397 and I398 and by pore-loop-II residue W431, shown in sticks, of four engaging protomers (colored in four different shades of blue). The maps with matching colors were contoured at 4.0 σ for both the substrate and pore-loop-I residues and at 3.5 σ for pore-loop-II residues. (C) Top view of the Con1 and Con2 structures and cartoon illustrations. On the left, the structures are shown in spheres for the AAA+ and protease domains. The N-terminal portions are shown in ribbons and cylindrical helices in the same coloring scheme as Fig. 1B. Substrate, ATP-γ-S, and ADP are colored in pink, red, and blue, respectively. On the right, cartoons show the protomers in four substrate-engaging conformational states (P1 to P4, in four different shades of blue) and two nonengaging conformational states (P5 and P6, in two shades of gray). ATP-γ-S and ADP are represented by red circles and blue squares, respectively. The substrate is represented by a pink hexagon. The nucleotide assumed to undergo hydrolysis is marked by a lightning symbol.
Implication of the tensegrity helix triangle on the assembly process
The unique interlocked helix triangle structure assembled in a homohexameric complex may presume a specific assembly process. One possibility is to involve a specific protomer dimer pair as the building block (fig. S5), joined by an Ac-protomer and an Ob-protomer, of which their LHs interact with each other via patch-C residues (Fig. 2B). A previous study has shown the monomers and dimers of Lon in solution (25). An Ac-Ob dimer may be bound by the second Ac-Ob dimer, joining counterclockwise to form a tetramer, thereby forming a LH crossover joint. Forming the final hexamer may be completed by adding the third Ac-Ob dimer or, alternatively, by a monomer binding next to the exposed Ac-protomer to form an Ob-protomer, completing the triangular LH crossovers, followed by the sixth monomer joining as the final Ac-protomer for completion. This model is consistent with previous chemical cross-linking results, suggesting a dimer ↔ tetramer ↔ hexamer assembly model (26).
A triangular platform for substrate binding, selection, and unfolding
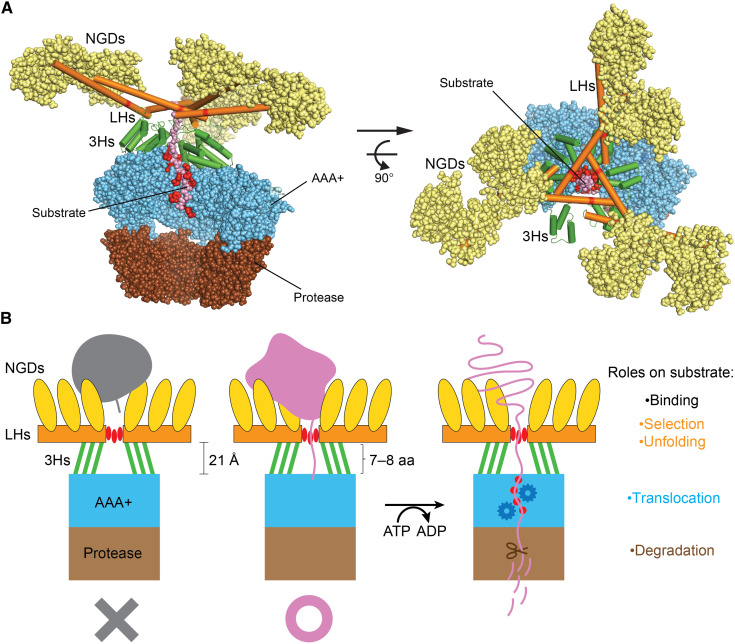
This work uncovers a tensegrity helix triangle previously not described in proteins. The unique interlocked helix structure is mounted on the top of the AAA+ rings, serving as a scaffold to spread a web of six substrate-binding NGDs (Figs. 1B and 5A). NGDs show different degrees of flexibility (fig. S6); some of them tumble independently of the hexameric complex (13). These NGDs have been shown to selectively bind to the exposed hydrophobic regions of the folded or unfolded protein substrates (13). Here, the NGDs further serve as the compression elements for the tensegrity structure, allowing the multiple LHs to be stably mounted on the top of the AAA+ ring for interacting with diverse protein substrates with different folding states and molecular sizes. The open gate formed in the helix triangle is located at the central axis of the hexamer where the gate residue Y224 is ~21 Å above the pore-loop-I I398 of the substrate-engaging P1 protomer. The substrate polypeptide chain captured by Y224 and I398 encompasses a segment of seven to eight residues. Many characterized Lon substrates are large or incompletely synthesized, folded or partially unfolded proteins or polypeptides carrying an exposed region (or degron) of at least seven residues at the C terminus rich in hydrophobic and aromatic residues (27–31). A protein with a shorter C-terminal tail, with less than seven to eight residues, would unlikely be degraded, as the short tail would not be able to reach beyond the triangular gate to be engaged by the pore-loop-I residue. Therefore, the structural features of the mounted multihelix platform suggest that it may function as a ruler to measure and target diverse protein substrates with sufficiently long C-terminal tails (Fig. 5B).
Fig. 5. Structural features and mechanism of Lon.
(A) Side (left) and top (right) views of the structure of Lon highlighting the multitier architectural features. To reveal the engaged substrate (colored in pink) inside the core chamber, the AAA+ and protease domains of the nonengaging protomers E and F are removed for clarity. From the top down, the NGDs are shown in wheat spheres, the LHs (in orange) and 3Hs (in green) are shown in cylindrical helices, and the core complex of the AAA+ and protease domains are shown as sky blue and brown spheres, respectively. The substrate-engaging Y224 and all the pore-loop residues are highlighted in red. (B) Schematic diagram of cutaway view illustrating the structural features and molecular mechanism of Lon. The NGDs (colored in yellow) bind substrates with exposed hydrophobic region. The LH-3H framework (in orange and green, respectively) carries out selection of substrates with a sufficiently long (>7 to 8 residues) exposed tail rich in aromatic or hydrophobic residues, which interact with the gate residue Y224 (shown in three red ovals). The LH platform mounted on the top of the AAA+ ring (in sky blue) with a triangular gate forms a pulley-like structure to trigger substrate unfolding by translocation of substrate polypeptide, which requires the four sets of pore-loop residues (denoted by red dots) and ATP-powered conformational changes of the AAA+ ring. In the core chamber, the translocated polypeptide is degraded by the protease domains (in brown) by a mechanism not depicted here. aa, amino acid.
According to the processive rotatory model, the translocating substrate polypeptide chain is pulled by pore-loop residues of the AAA+ domains in processive movement powered by sequential rotary ATP hydrolysis around the hexameric ring. The gated triangular platform may form a pulley-like structure functionally coupled to the motile AAA+ ring. A substrate undergoing translocation may be regarded as in an engaged state that its exposed tail threading through the gate is gripped by the spiraling AAA+ pore loops below the triangular platform, while the other exposed hydrophobic regions are bound by the multiple NGDs protruding above the triangular platform. The pulley-like feature may thus serve to convert the directional pull of the translocating substrate into mechanical force to facilitate unfolding of the substrate (Fig. 5B). The global stability and plasticity of the pulley-like platform are strategically provided by the tensegrity structure formed by the N-terminal helices. In light of the present work, the full-length structures of other AAA+ protease complexes with analogous activities may show similar substrate selection and pulley-like molecular features; this speculation awaits experimental evidence from future structural studies.
MATERIALS AND METHODS
Cloning and mutagenesis
Gene encoding wild-type MtaLon with a C-terminal 6xHis-tag was cloned into pET21a(+) vector, which was used as the template for site-directed mutagenesis, as previously described (17). All mutants were constructed by site-directed mutagenesis (the primers are listed in table S2). All constructs in this study were sequenced before use by the DNA Sequencing Core Facility of Academia Sinica (AS-CFII-108-115).
Protein expression and purification
Cells were grown in LB medium at 37°C until the optical density reached 0.8. Isopropyl-β-d-thiogalactopyranoside was then added to a final concentration of 1 mM to the culture and incubated for another 4 hours at 25°C, allowing expression of the full-length MtaLon. Cells were harvested by centrifugation and suspended in lysis buffer containing 50 mM tris-HCl (pH 8.0) and 500 mM NaCl. Cell lysate was ruptured by a French press (Avestin) and centrifuged at 35,000g. The supernatant was collected and incubated with Ni-nitrilotriacetic acid resins (Qiagen) at 4°C for 2 hours. The resins were loaded into an open column, washed with 20 mM imidazole, and lastly eluted with 250 mM imidazole. The eluted protein was dialyzed against a buffer containing 20 mM tris-HCl (pH 8.0), 100 mM NaCl, and 2 mM dithiothreitol (DTT).
Purified MtaLon was incubated with 10 mM MgCl2 for 1 hour and then incubated with 1 mM bortezomib, 5 mM ATP-γ-S, and α-casein (Sigma-Aldrich; in fivefold molar excess) overnight. Before cryo-sample preparation, the mixture was treated by adding an additional 1 mM ATP-γ-S.
Cryo-EM data acquisition
The substrate-engaged MtaLon sample was diluted at a final concentration of around 0.5 mg/ml. Three microliters of the sample was applied onto glow-discharged 200-mesh R1.2/1.3 Quantifoil copper grids. The grids were blotted for 4 s and rapidly cryocooled in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4°C and 100% humidity. The sample was screened using a Talos Arctica cryo–electron microscope (Thermo Fisher Scientific) operated at 200 kV. It was then imaged in a Titan Krios cryo–electron microscope (Thermo Fisher Scientific) at a magnification of ×96,000 (corresponding to a calibrated sampling of 0.82 Å per pixel). Micrographs were recorded by EPU software (Thermo Fisher Scientific) with a Falcon 4 detector, where each image was composed of 40 individual frames in gain-normalized mrc format with an exposure time of 5.8 s and an exposure rate of 8.28 electrons/s per square angstrom. A total of 6479 movie stacks were collected.
Single-particle image processing and 3D reconstruction
All micrographs were first imported into Relion (32) for image processing. The motion correction was performed using MotionCor2 (33), and the contrast transfer function was determined using CTFFIND4 (34). Then, the micrographs with “rlnMotionEarly < 10” and “rlnCtfMaxResolution < 5” were selected using the “subset selection” option in Relion. All particles were autopicked using the NeuralNet option (threshold 1 = 0; threshold 2 = −5) in EMAN2 (35). Then, particle coordinates were imported to Relion, where the poor 2D class averages were removed by several rounds of 2D classification. Notably, we initially extracted the particles with a box size of 256 pixels and found some extra density on the top of the hexamer core from 2D analysis; therefore, the box size was increased to 336 pixels to include all possible densities from N termini (fig. S1). A total of 692,834 particles were transferred to cryoSPARC (36) for ab initio map generation. Then, a good class with apparent N termini and better-resolved AAA+ and protease domains, containing 317,318 particles, was subjected to homogeneous refinement, and a 2.3-Å map was obtained. However, the features of N termini were not clearly visible, indicating their heterogeneity. Therefore, another round of heterogeneous refinement was performed to further classify the particles, and two good classes (Con1 and Con2) having distinct N termini were generated. Homogeneous refinement and local refinement were then conducted to achieve the final maps. A 2.4-Å map from 158,553 particles for Con1 and a 2.4-Å map from 156,330 particles for Con2 were lastly obtained. To better resolve the N-terminal region, particle subtraction and local refinement on this region were applied, resulting in two locally refined maps of the N-terminal region with the resolutions of 3.7 and 4.3 Å, respectively. Resolutions for the final maps were estimated with the 0.143 criterion of the Fourier shell correlation curve. Resolution maps were calculated in cryoSPARC using the “Local Resolution Estimation” option. The figures were prepared using Chimera (37) and PyMOL (38) (see more information in figs. S1 and S2 and table S1).
Model building
Model building was first performed on the 2.4-Å-resolution cryo-EM map of Con1. The crystal structure of a hexameric Lon protease (residues 244 to 780) (Protein Data Bank ID: 4YPL) from our previous work (18) was rigidly fitted into the cryo-EM map. As the six protomers were conformationally different in the cryo-EM density, molecular dynamics flexible fitting (MDFF) (39) was used. The MDFF was completed in three runs, where each run included 104 minimization steps and 105 molecular dynamics steps. After no noticeable deformation, the MDFF was stopped. The resultant model was refined using phenix.real_space_refine (40).
To model the N-terminal domain of Con1, a locally refined map, at 3.7-Å resolution, was used together with the crystal structure of the N-terminal fragment (residues 1 to 206) (13). The region (residues 207 to 237) that was previously unresolved was modeled using SWISS-MODEL (41). These models were combined and rigidly fitted into the locally refined map and subjected to one round of MDFF, followed by optimization by Coot (42) and phenix.real_space_refine. The resulting model was then docked into the 2.4-Å full map. The linker (residues 238 to 243) between the N-terminal domain and the AAA+ module was manually built using Coot to complete the full-length model of Con1. Then, phenix.real_space_refine and Coot were applied for model optimization.
The type of the bound nucleotides, ADP or ATP-γ-S, was determined by LigandFit in Phenix, with an overall correlation coefficient of the ligand to the map of more than 0.7. The bound substrate (α-casein) was manually built with Coot. The complete model consisting of the full-length MtaLon, bound nucleotide, and substrate was optimized using phenix.real_space_refine.
The same modeling procedures were also conducted on the 2.4-Å full map and 4.3-Å locally refined map of Con2. The final models were evaluated by MolProbity (43). Statistics of the map reconstruction and model building are summarized in table S1. All figures were prepared using PyMOL and Chimera.
Substrate degradation assay
Ig2 as a model substrate of MtaLon has been reported previously (17). Ig2 (4 μM) or α-casein (8 μM) was incubated with 0.8 μM MtaLon (hexamer concentration) in the reaction buffer containing 50 mM tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, and 5 mM ATP at 55°C. At different time points, reaction aliquots were stopped by adding 5× SDS–polyacrylamide gel electrophoresis (SDS-PAGE) loading dye and heated at 95°C for 5 min. Substrate degradation was assessed by SDS-PAGE (SurePAGE gel, 4 to 20% Bis-Tris, Genscript) and Coomassie Blue staining. The substrate band intensities were scanned by ImageJ (National Institutes of Health); data were analyzed using Prism 5 (GraphPad Software Inc., San Diego, CA, USA) and presented as means ± SD of three independent experiments.
Acknowledgments
We thank W. Chiu at Stanford University for the support of cryo-EM data collection and Academia Sinica Cryo-EM Facility for the support of cryo-grid preparation. We also thank C. Chang for manuscript editing.
Funding: This work was supported by Academia Sinica and MOST grant 108-2320-B-001-011-MY3 (to C.-I.C.) and Start-up funding from the University of Science and Technology of China (KY9100000032 and KJ2070000080 to K.Z.).
Author contributions: K.Z. and C.-I.C. conceived the study. K.Z. and C.-I.C. designed experiments. S.L. and K.Z. collected data, solved the structures, and built the models. S.L., C.-I.K., S.-H.L., K.-Y.H., and K.Z. performed experiments. S.L., G.D.P., K.Z., and C.-I.C. analyzed data. S.L., K.Z., and C.-I.C. wrote the manuscript with inputs from all other authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Cryo-EM maps of MtaLon bound to α-casein, with their associated atomic models, have been deposited in the Electron Microscopy Data Bank (EMD-31534 and EMD-31535) and the Protein Data Bank (7FD4 and 7FD5). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for movies S1 and S2
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Gur E., The Lon AAA+ protease. Subcell. Biochem. 66, 35–51 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Breidenstein E. B. M., Janot L., Strehmel J., Fernandez L., Taylor P. K., Kukavica-Ibrulj I., Gellatly S. L., Levesque R. C., Overhage J., Hancock R. E. W., The Lon protease is essential for full virulence in Pseudomonas aeruginosa. PLOS ONE 7, e49123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddicker J. D., Jones B. D., Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 72, 2002–2013 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takaya A., Suzuki M., Matsui H., Tomoyasu T., Sashinami H., Nakane A., Yamamoto T., Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect. Immun. 71, 690–696 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibellini L., De Gaetano A., Mandrioli M., Van Tongeren E., Bortolotti C. A., Cossarizza A., Pinti M., The biology of Lonp1: More than a mitochondrial protease. Int. Rev. Cell Mol. Biol. 354, 1–61 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Rotanova T. V., Botos I., Melnikov E. E., Rasulova F., Gustchina A., Maurizi M. R., Wlodawer A., Slicing a protease: Structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 15, 1815–1828 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M., Rasulova F., Melnikov E. E., Rotanova T. V., Gustchina A., Maurizi M. R., Wlodawer A., Crystal structure of the N-terminal domain of E. coli Lon protease. Protein Sci. 14, 2895–2900 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duman R. E., Löwe J., Crystal structures of Bacillus subtilis Lon protease. J. Mol. Biol. 401, 653–670 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Li M., Gustchina A., Rasulova F. S., Melnikov E. E., Maurizi M. R., Rotanova T. V., Dauter Z., Wlodawer A., Structure of the N-terminal fragment of Escherichia coli Lon protease. Acta Crystallogr. D Biol. Crystallogr. 66, 865–873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Zhang S., Bi F., Guo C., Feng L., Wang H., Yao H., Lin D., Crystal structure of the N domain of Lon protease from Mycobacterium avium complex. Protein Sci. 28, 1720–1726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wohlever M. L., Baker T. A., Sauer R. T., Roles of the N domain of the AAA+ Lon protease in substrate recognition, allosteric regulation and chaperone activity. Mol. Microbiol. 91, 66–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng I., Mikita N., Fishovitz J., Frase H., Wintrode P., Lee I., Identification of a region in the N-terminus of Escherichia coli Lon that affects ATPase, substrate translocation and proteolytic activity. J. Mol. Biol. 418, 208–225 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Tzeng S.-R., Tseng Y.-C., Lin C.-C., Hsu C.-Y., Huang S.-J., Kuo Y.-T., Chang C.-I., Molecular insights into substrate recognition and discrimination by the N-terminal domain of Lon AAA+ protease. eLife 10, e64056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botos I., Melnikov E. E., Cherry S., Tropea J. E., Khalatova A. G., Rasulova F., Dauter Z., Maurizi M. R., Rotanova T. V., Wlodawer A., Gustchina A., The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site. J. Biol. Chem. 279, 8140–8148 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Cha S.-S., An Y. J., Lee C. R., Lee H. S., Kim Y.-G., Kim S. J., Kwon K. K., De Donatis G. M., Lee J.-H., Maurizi M. R., Kang S. G., Crystal structure of Lon protease: Molecular architecture of gated entry to a sequestered degradation chamber. EMBO J. 29, 3520–3530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao J.-H., Ihara K., Kuo C.-I., Huang K.-F., Wakatsuki S., Wu S.-H., Chang C.-I., Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors. Acta Crystallogr. D Biol. Crystallogr. 69, 1395–1402 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Su S.-C., Lin C.-C., Tai H.-C., Chang M.-Y., Ho M.-R., Babu C. S., Liao J.-H., Wu S.-H., Chang Y.-C., Lim C., Chang C.-I., Structural basis for the magnesium-dependent activation and hexamerization of the Lon AAA+ protease. Structure 24, 676–686 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Lin C.-C., Su S.-C., Su M.-Y., Liang P.-H., Feng C.-C., Wu S.-H., Chang C.-I., Structural insights into the allosteric operation of the Lon AAA+ protease. Structure 24, 667–675 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Shin M., Watson E. R., Novick S. J., Griffin P., Luke Wiseman R., Lander G. C., Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Nat. Commun. 12, 3239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin M., Puchades C., Asmita A., Puri N., Adjei E., Wiseman R. L., Karzai A. W., Lander G. C., Structural basis for distinct operational modes and protease activation in AAA+ protease Lon. Sci. Adv. 6, eaba8404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K., Li S., Hsieh K.-Y., Su S.-C., Pintilie G. D., Chiu W., Chang C.-I., Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease. Cold Spring Harb. Lab. 10.1101/2020.04.29.068361 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D., Wang M., Deng Z., Walulu R., Mao C., Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 126, 2324–2325 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Liedl T., Högberg B., Tytell J., Ingber D. E., Shih W. M., Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 5, 520–524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puchades C., Sandate C. R., Lander G. C., The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol. 21, 43–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S.-C., Jia B., Yang J.-K., Van D. L., Shao Y. G., Han S. W., Jeon Y.-J., Chung C. H., Cheong G.-W., Oligomeric structure of the ATP-dependent protease La (Lon) of Escherichia coli. Mol. Cells 21, 129–134 (2006). [PubMed] [Google Scholar]
- 26.Lee A. Y.-L., Hsu C.-H., Wu S.-H., Functional domains of Brevibacillus thermoruber lon protease for oligomerization and DNA binding: Role of N-terminal and sensor and substrate discrimination domains. J. Biol. Chem. 279, 34903–34912 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Ge Z., Karzai A. W., Co-evolution of multipartite interactions between an extended tmRNA tag and a robust Lon protease in Mycoplasma. Mol. Microbiol. 74, 1083–1099 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gur E., Sauer R. T., Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 22, 2267–2277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gur E., Sauer R. T., Evolution of the ssrA degradation tag in Mycoplasma: Specificity switch to a different protease. Proc. Natl. Acad. Sci. U.S.A. 105, 16113–16118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishii Y., Amano F., Regulation of SulA cleavage by Lon protease by the C-terminal amino acid of SulA, histidine. Biochem. J. 358, 473–480 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puri N., Karzai A. W., HspQ functions as a unique specificity-enhancing factor for the AAA+ Lon protease. Mol. Cell 66, 672–683.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Rigsby R. E., Parker A. B., Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 44, 433–437 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Trabuco L. G., Villa E., Mitra K., Frank J., Schulten K., Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L.-W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, P. H. Zwart, in International Tables for Crystallography (2012), pp. 539–547. [Google Scholar]
- 41.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., Schwede T., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for movies S1 and S2
Tables S1 and S2
Movies S1 and S2