Abstract
The significance of canonical DNA non-homologous end-joining (c-NHEJ) for DNA double strand break (DSB) repair has increased from lower organisms to higher eukaryotes, and plays the predominant role in human cells. Ku, the c-NHEJ end-binding component, binds DSBs with high efficiency enabling c-NHEJ to be the first choice DSB repair pathway, although alternative pathways can ensue after regulated steps to remove Ku. Indeed, radiation-induced DSBs are repaired rapidly in human cells. However, an important question is the fidelity with which radiation-induced DSBs are repaired, which is essential for assessing any harmful impacts caused by radiation exposure. Indeed, is compromised fidelity a price we pay for high capacity repair. Two subpathways of c-NHEJ have been revealed; a fast process that does not require nucleases or significant chromatin changes and a slower process that necessitates resection factors, and potentially more significant chromatin changes at the DSB. Recent studies have also shown that DSBs within transcriptionally active regions are repaired by specialised mechanisms, and the response at such DSBs encompasses a process of transcriptional arrest. Here, we consider the limitations of c-NHEJ that might result in DSB misrepair. We consider the common IR-induced misrepair events and discuss how they might arise via the distinct subpathways of c-NHEJ.
Introduction
Early studies on DNA double strand break (DSB) repair in prokaryotes and lower eukaryotes demonstrated that homologous recombination (HR) is the major DSB repair pathway and HR-deficient mutants display high radiation (IR)-sensitivity.1 Surprisingly, IR-sensitive and DSB repair defective mammalian cell lines proved to be proficient for HR but deficient in a pathway that did not require extensive homology.1 Subsequently, identification of the genetic defects in such mutants revealed the process of DNA canonical non-homologous end-joining (c-NHEJ), and its functional distinction to HR. Here, we define this process as c-NHEJ to distinguish it from other end-joining pathways, such as Alt-NHEJ. The prevalence of a pathway distinct to HR pathway in mammalian cells was unexpected given that DNA repair pathways are evolutionarily well-conserved, that HR’s ability to restore DNA integrity using an undamaged template is exquisitely elegant and that diploid mammalian cells were anticipated to be able to use HR regardless of cell-cycle phase. However, HR in mammalian cells was shown later to only utilise the sister homologue as a template, suggesting that homologous chromosomes have insufficient homology to drive HR. Consequently, HR usage is restricted to late S/G2 where a sister homologue exists.2 The large mammalian genome was also proposed to hinder homology searching, providing a further factor restricting HR usage in mammalian cells. Strikingly, however, even in G2, the majority of DSBs are repaired by c-NHEJ,3 although transcription-associated DSBs (TA-DSBs) preferentially undergo repair by HR.4,5
The question, therefore, is what has driven the preference for c-NHEJ and what are its limitations. The processes of c-NHEJ have been reviewed previously and only salient points will be described here.6 Ku, the DSB end-binding factor of c-NHEJ, is highly abundant in human cells, binding DSB ends in a non-sequence dependent manner with extraordinary efficiency. Current studies support the notion that Ku is the first responder to most, and likely, all DSBs, protecting them from nucleolytic degradation although end-degradation can ensue to allow alternative pathway usage in an appropriate, controlled manner.6 Although other pathways can function even after Ku end-binding, most IR-induced DSBs in human cells are repaired by c-NHEJ, even after high doses.7 Loss of c-NHEJ proteins confers extreme IR-sensitivity. These and additional findings argue that c-NHEJ is highly efficient at rejoining DSBs, and suggest that, despite often being called error-prone, it is likely to be predominantly accurate. However, there is evidence that its robust ligation activity can result in some misrepair, potentially a price paid for its rapid and high rejoining capacity.8 Here, we focus on how misrepair might arise during c-NHEJ. Importantly, we focus on errors generated during c-NHEJ end-joining and not those arising following cell cycle progression or replication. To achieve this, we consider the fidelity of repair in G0/G1 cells where c-NHEJ is the sole or major DSB repair pathway. We review our mechanistic understanding of c-NHEJ from the perspective of its accuracy and discuss findings assessing fidelity. Although our focus lies on repair processes in G1 phase, understanding mechanisms of repair in G1 emerged through examining G2 cells. Thus, we commence by reviewing DSB repair in G2 prior to discussing G0/G1 cells. We then evaluate the potential classes of, and evidence for, misrepair events, evaluating them in the context of our mechanistic understanding.
DSB repair in G2 phase
Analysis of DSB repair kinetics in different genetic backgrounds using γH2AX foci enumeration and cell cycle markers has provided insight into the relative contribution of c-NHEJ versus HR to DSB repair.3 In G2, c-NHEJ repairs ~70–75% of IR-induced DSBs with fast kinetics whilst 25–30% of DSBs are repaired more slowly by HR. Consistent with this notion, RPA and RAD51 foci only form at DSBs repaired with slow kinetics. Recent studies have delineated the DSBs repaired by HR into two subclasses. One-third of these DSBs are located within transcriptionally active regions, since their repair can be influenced by drugs inhibiting the initiation of transcription or over expression of RNASE H1.5 This component of HR (TA-HR) requires RAD52 and XPG to process R-loops, which form at these DSBs. Importantly, dysfunction of TA-HR promotes repair by c-NHEJ with a strikingly high level of rearrangements. These and other findings suggest that active transcription directs DSB repair towards HR and failure to progress HR results in chromosomal rearrangements, possibly due to the open structure at these DSBs.4 Although there is some insight, the precise mechanism promoting HR at TA-DSBs remains unclear.9 In contrast, 70–75% of the DSBs repaired by HR appear to represent those within heterochromatic or compacted DNA regions (HC-DSBs), in part because their repair can be influenced by depletion of compacting factors such as KAP1 or HP1.10 The current working model is that Ku binds to most X-ray induced DSBs, promoting fast c-NHEJ. Resection ensues in the presence of Ku via an ordered process involving MRE11 endo- and exonuclease activities, CtIP, EXO1, and BRCA1 relieving a barrier posed by 53BP1.11–15 Although proteolytic and nucleolytic removal of Ku has been proposed, the precise mechanism underlying Ku removal at these DSBs remains unclear.16,17
In summary, three classes of DSB repair can be identified in G2 phase; fast c-NHEJ which repairs ~70–75% of DSBs with HR repairing 25–30% of DSBs with slower kinetics. HR can be subdivided into TA-HR representing ~30% of all HR events and HC-HR representing the remaining 70% of HR events.
DSB repair in G0/G1 phase
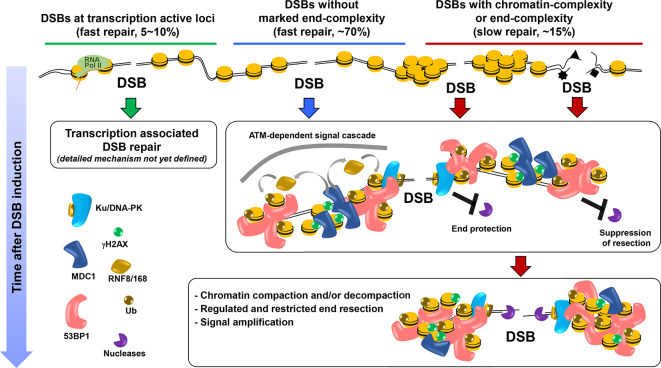
DSB repair studies in G0/G1 phase cells have revealed similar, although distinct, kinetics to those in G2, which is perhaps surprising given the inability of HR to function in G0/G118 (Figure 1). As in G2, c-NHEJ repairs ~75% of DSBs with fast kinetics. However, the slow DSB repair process in G0/G1 also requires c-NHEJ proteins, as well as ATM signalling proteins, including 53BP1 and factors required for its DSB localisation, Polo-like kinase 3 (PLK3), CtIP, the nuclease Artemis, and additional factors required for resection including MRE11 exonuclease and EXO1.21,22 MRE11 endonuclease is not required, however. Thus, there are distinctions yet overlap between the slow process in G0/G1 vs G2. Interestingly, the kinetics of slow repair in G1 vs G2 differ subtly, consistent with the notion that they occur via distinct pathways. A further distinction is that the slow process is approximately one-third smaller in G1 vs G2 and does not appear to encompass TA-DSBs.23 The slowly repaired DSBs are, however, affected by HP1 or KAP1 depletion, similar to the HC-DSBs in G2.10 RAD51 foci are not observed at this DSB subset, consistent with the process representing c-NHEJ rather than HR.21,22 RPA foci are not formed at X-ray induced DSBs in G0/G1, which initially suggested that resection does not arise. However, the requirement for resection factors argues otherwise. A plausible explanation is that the length of resection is restricted in G1 and is insufficient to promote RPA binding. The slow repair process in G0/G1 has been called Resection and Artemis-dependent c-NHEJ to distinguish it from resection-independent c-NHEJ, the fast process.21 It is noteworthy that depletion of CtIP, EXO1, or loss of MRE11 exonuclease activity does not confer a slow component repair defect but rather rescues the defect observed in Artemis-deficient cells. Such genetic “rescue” suggests that the role of resection factors can be masked by the use of alternative pathways (including resection-independent c-NHEJ), an important phenomenon to consider when evaluating the requirement for DSB repair factors.21 It is also noteworthy that the number of DSBs repaired with slow kinetics is similar following exposure to X-rays, neocarzinostatin, calchiamycin or H2O2.24 However, high LET radiation has a greater requirement for Resection and Artemis-dependent DSB repair. These findings argue that DSB end complexity enhances the usage of the slow repair process but another factor must also be significant. We suggest that chromatin complexity also influences repair pathway usage. The similar fraction of DSBs repaired with slow kinetics after X-rays, neocarzinostatin or calchiamycin suggests that DSB end-complexity induced by X-rays may not substantially influence pathway choice.
Figure 1.
Repair processes in G1. Three distinct forms of DSB repair have been identified to function in G1 cells. The majority of DSBs are repaired with fast kinetics via a process involving c-NHEJ factors (central part of the figure shown by blue line and blue arrow head). The presence of the Ku/DNA-PK complex acts as a first step providing protection from resection (shown by the inhibitory arrow to Ku/DNA-PK complex). ATM and 53BP1 likely also contribute to the suppression of resection at this stage. Briefly, ATM phosphorylates the histone variant, H2AX, at the DSB site, which causes recruitment of MDC1, a mediator protein, followed by recruitment of two ubiquitin ligases, RNF8/168. H2AK15 is also ubiquitylated and H4K20Me2 residues become exposed, causing the recruitment of 53BP1. Following ATM-dependent phosphorylation of 53BP1 in its N-terminus, RIF1 is also recruited (see refs1,19 for reviews). Collectively these co-ordinated reactions promote a chromatin environment suppressive for resection (shown by the inhibitory arrow to 53BP1). At a later stage, e.g. 8-24 h after low LET IR, 10–15% of DSBs are repaired with slow kinetics via the Resection and Artemis-dependent process involving c-NHEJ factors (right side of the figure shown by red line and red arrow head). The initiation of this process seems to be promoted by chromatin-complexity and DSB end-complexity, although the precise regulation is unclear. DSBs within transcriptionally active (TA) regions undergo ultrafast repair, although precise details remain to be clarified (left side of figure presented by green line and green arrow)20 ; Shibata abstract report at http://icrr2019manchester.com}. c-NHEJ, canonical non-homologous end-joining; DSB, double strand break; TA, transcriptionally active.
How are TA-DSBs repaired in G0/G1 cells? Damage-induced transcriptional repression (DITR) is an ATM- and PBAF-dependent process inhibiting transcription within the DSB vicinity. Failure to activate DITR via siBAF180, a DITR essential factor, confers a subtle DSB repair defect at early times post IR, suggesting that in G0/G1 TA-DSBs might be repaired with ultrafast kinetics.20 This possibility has been substantiated and extended in recent work suggesting that TA-DSB undergo repair with fast kinetics in G1 and are prone to translocation formation when the fast repair process is dysfunctional (Shibata abstract report at http://icrr2019manchester.com).
Collectively, these findings reveal: (i) slow DSB repair occurs via HR in G2 but Resection and Artemis-dependent c-NHEJ in G1, (ii) TA-DSBs are repaired distinctly in G1 vs G2 phase, (iii) the slow process in G1 phase involves restricted end-resection and (iv) chromatin and end complexity influence repair pathway choice. These points are important for evaluating the fidelity of DSB repair since the distinct pathways are likely to differ in their accuracy.
A consideration of DNA misrepair
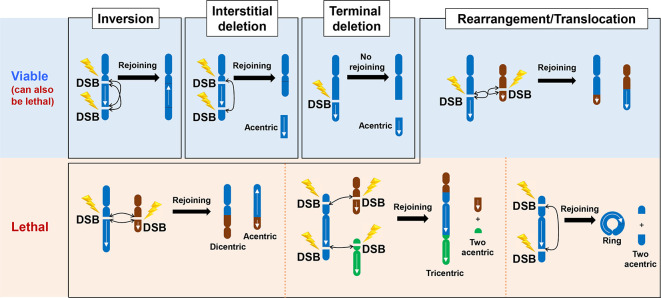
Mutational and recent sequencing analysis suggests that IR does not efficiently induce missense mutations.25,26 However, two categories of misrepair events do arise, namely large-scale chromosome rearrangements (Figure 2) and short insertion or deletion (indel) events (Figure 3). Chromosome rearrangements are predominantly lethal events. Balanced translocations, however, need not be lethal and are a signature change in radiation-induced tumours.27 Genetic and cell cycle analysis has shown that chromosome aberrations can arise via c-NHEJ (see below). Thus, the ability of c-NHEJ to rejoin ends without extensive end-homology appears to allow rejoining of the wrong ends if correct-end synapsis is not maintained. ATM might play a significant role in promoting correct end-synapsis considering its major impact on the chromatin environment around the DSB (see further discussion below).23
Figure 2.
Mis-repair of the incorrect DNA ends by c-NHEJ in G1 phase to generate chromosome rearrangements. Since we consider mis-repair events arising from c-NHEJ we have focused on chromosome rearrangements in G0/G1 phase cells, where c-NHEJ is the major, and possibly the sole, DSB repair process. Thus, we have shown chromosomes with a single chromatid, which can be visualised using PCC techniques. Failure to repair DSBs in G1 phase can result in terminal or interstitial deletions, translocation events including dicentrics and ring chromosomes. These events are depicted in the figure. Generally, significant loss of chromosome material leads to lethality whilst inversion events or balanced translocations can allow viability. However, some smaller deletions events need not confer lethality, including small interstitial or terminal deletions. The formation of dicentric or ring chromosomes and larger acentric fragments are normally lethal events after cell division. c-NHEJ, canonical non-homologous end-joining; DSB, double strand break.
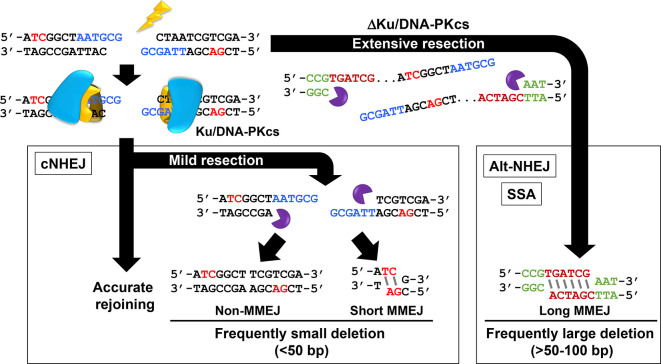
Figure 3.
Mis-repair of the correct DNA ends causing indel formation. The figure shows a DSB within a construct. Accurate rejoining (black arrow on the right hand site) reconstitute the sequence. A small level of resection involving loss of the sequences in blue from both strands followed by rejoining without MH usage can result in small deletions (non-MMEJ). The use of short MH for rejoining (sequences in red) can also lead to short deletions (short MMEJ). Such small deletions can arise by c-NHEJ. Alt-NHEJ involves larger deletions and longer regions of MH. The green sequences lie some regions away from the original break site. Thus, c-NHEJ rejoining can arise causing small deletions (and occasionally insertions), which are usually very short. Such indels may arise without any MH usage or involve a few base pairs of microhomology. Alt-NHEJ does not arise frequently in normal cells unless Ku or DNA-PKcs are absent. Note that MMEJ, any rejoining that involves regions of microhomology, can arise via resection-mediated c-NHEJ or Alt-NHEJ (as well as via single strand annealing, which is not shown here). c-NHEJ, canonical non-homologous end-joining; DSB, double strand break; PCC, premature chromosome condensation.
Indels, effectively, represent erroneous rejoining of the correct ends suggesting that synapsis is maintained but with sequence loss. Small deletions, the most common event, can involve microhomology (MH) usage. Shorter deletions (<around 50 bp), which often involve only a few base pairs of MH, are generally attributed to c-NHEJ misrepair. Deletions of >50 bp are usually attributed to Alt-NHEJ and involve greater MH (Figure 3). Since we are considering misrepair during c-NHEJ, we will focus on deletion events involving shorter deletions with no or short MH usage.
c-NHEJ mediated misrepair in G0/G1
To focus on c-NHEJ mediated misrepair, as opposed to misrepair following replication or mitosis with DSBs, we will consider misrepair in primary or hTERT immortalised fibroblasts following irradiation and maintenance in G0/G1, where c-NHEJ is the major, and likely the sole, DSB rejoining process in normal cells. Analysis using premature chromosome condensation (PCC) of G0/G1 cells following fusion with mitotic cells has shown that chromosome rearrangements can arise in G0/G1 after IR via c-NHEJ since they are not affected by deficiency of Alt-NHEJ factors.8,21 Indeed, although Alt-NHEJ promotes IR-induced translocations in mouse cells, translocations arise predominantly via c-NHEJ in human cells.28 Insightful early studies revealed that rearrangements in G0/G1 cells arise with dose squared kinetics, although they appear to be linear at low doses.8 Additionally, they can be reduced by dose splitting (a phenomenon termed sparing); thus a single dose of, e.g. 8 Gy confers a higher aberration yield than two consecutive exposures to 4 Gy or 4 exposures to 2 Gy.29 Detailed analysis assessing optimal doses and intervals suggested a model whereby mis-repair is diminished when DSBs are separated in time and space arguing that the interaction between DSBs is dependent on the number present at any time. Assuming 1 Gy induces 30 DSBs, 8 Gy will yield 240 DSBs, which is still a small number of DSBs per genome. It is, arguably, surprising that detectable misrejoining and dose sparing arises following 8 Gy (and its fractionation) given that Ku is highly abundant and protects DSBs. This begs the question: how is synapsis promoted? ATM and 53BP1 play important roles in compacting chromatin at DSBs, which is likely to protect against aberration formation.23,30 Significantly, rearrangement events in G0/G1 fibroblasts are enhanced in A-T fibroblasts.8 This impact cannot be attributed to impaired cell cycle arrest but could arise from a failure to arrest transcription in the DSB vicinity or to correctly organise chromatin structure at DSBs, known impacts of ATM loss.20,23 Since the DSBs repaired with slow kinetics remain unrepaired in the absence of ATM, the enhanced aberrations in A-T fibroblasts must arise from those DSBs normally repaired with faster kinetics.18 The role of 53BP1 in regulating rearrangements specifically in G1 has not been carefully examined.
Using PCC analysis in G0/G1 cells, we examined whether the fast and slow processes differ in their ability to generate chromosome aberrations. We found that approximately one-half of the exchange events formed with fast kinetics; the other half arose with slower kinetics.21,31 Further, the exchange events arising with slow kinetics required Artemis and CtIP, factors specifically required for the slow repair process. Thus, the kinetics and genetic requirements for aberration formation reflect the identified DSB repair processes. Given that 80% of DSBs are repaired with fast kinetics, these findings suggest that the slow process is 3–4 times more prone to translocations than the fast process, potentially due to greater “end-opening” to effect Resection and Artemis-dependent c-NHEJ.
Another question is whether the fast and slow c-NHEJ processes generate small indels at similar frequencies. Addressing this requires careful sequence analysis, which has not been undertaken, although indel formation has been observed by signature sequencing after IR exposure.26 Assuming that small deletion events involve resection, it is possible that they only arise via Resection and Artemis-dependent c-NHEJ, that is, the slow repair process. Constructs harbouring two closely located I-Sce1 sites have been used as a model system to examine this following selection for rejoining events that lose the intervening sequence.21 This system does not model incorrect rejoining of the correct ends, but has provided useful insight. Importantly, 83% of the selected mis-repair events, harbour sequence loss at their junctions (i.e. only 17% of rejoining events involving loss of the intervening fragment arise without end-resection). Further, such rejoining requires CtIP and Artemis, i.e. the factors required for slow repair, suggesting that Resection and Artemis-dependent c-NHEJ might frequently generate indels. Although this construct may not represent most DSBs repaired with slow kinetics, it is possible that it reflects those DSBs where rejoining by fast c-NHEJ is precluded promoting a switch to the resection-dependent process. It is possible, however, that some form of templating, e.g. use of an RNA sequence, might overcome this limitation or another process restricting sequence loss to one DNA strand. Another question is whether indel formation can also be diminished following dose splitting.
Finally, we should consider the fidelity of rejoining TA-DSBs, which are an important although minor DSB subset, particularly those involving interaction between the transcription and DSB repair machinery. Recent studies have shown that such DSBs are repaired by HR in G2 phase and failure to undergo TA-HR enhances translocation formation.4,5 Thus, an important question is how such DSBs are repaired in G0/G1 phase. Of note, DITR arrests transcription in the DSB vicinity and impaired DITR causes the slowing of DSBs normally repaired with superfast kinetics.20 It is tempting to speculate that such DSBs could contribute to IR-induced rearrangements.
Summary
Now that we have gained significant insight into the pathways of DSB repair, including the subpathways of c-NHEJ, it is important to establish the fidelity of these pathways since erroneously repaired DSBs can cause both lethality and carcinogenesis. Indeed, efficient radiation protection necessitates an assessment of the fidelity of DSB repair. Additionally, radiotherapy regimes have involved dose fractionation to limit normal tissue effects and understanding the underlying mechanism could enhance the efficacy. Here, we discuss the potential limitations of c-NHEJ leading to chromosomal rearrangements and/or indel formation. An important tool to drive the work forward is the exploitation of state-of-the-art sequencing procedures to assess the accuracy of the c-NHEJ sub pathways, the genomic location of DSB misrepair events and how they respond to dose, dose rate and the quality of the radiation.
Footnotes
Funding: JSPS KAKENHI grant JP17H04713
Contributor Information
Atsushi Shibata, Email: shibata.at@gunma-u.ac.jp.
Penny A Jeggo, Email: P.A.Jeggo@sussex.ac.uk.
REFERENCES
- 1.Jeggo P, Lavin MF. Cellular radiosensitivity: how much better do we understand it? Int J Radiat Biol 2009; 85: 1061–81. doi: 10.3109/09553000903261263 [DOI] [PubMed] [Google Scholar]
- 2.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J 2000; 19: 3398–407. doi: 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. Embo J 2011; 30: 1079–92. doi: 10.1038/emboj.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marnef A, Cohen S, Legube G. Transcription-Coupled DNA double-strand break repair: active genes need special care. J Mol Biol 2017; 429: 1277–88. doi: 10.1016/j.jmb.2017.03.024 [DOI] [PubMed] [Google Scholar]
- 5.Yasuhara T, Kato R, Hagiwara Y, Shiotani B, Yamauchi M, Nakada S, et al. Human RAD52 promotes XPG-Mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 2018; 175: 558: e11–70. doi: 10.1016/j.cell.2018.08.056 [DOI] [PubMed] [Google Scholar]
- 6.Shibata A, Jeggo P, Löbrich M. The pendulum of the Ku-Ku clock. DNA Repair 2018; 71: 164–71. doi: 10.1016/j.dnarep.2018.08.020 [DOI] [PubMed] [Google Scholar]
- 7.Rothkamm K, Löbrich M. Misrejoining of DNA double-strand breaks in primary and transformed human and rodent cells: a comparison between the HPRT region and other genomic locations. Mutat Res 1999; 433: 193–205. doi: 10.1016/S0921-8777(99)00008-7 [DOI] [PubMed] [Google Scholar]
- 8.Cornforth MN, Bedford JS. A quantitative comparison of potentially lethal damage repair and the rejoining of interphase chromosome breaks in low passage normal human fibroblasts. Radiat Res 1987; 111: 385–405. doi: 10.2307/3576926 [DOI] [PubMed] [Google Scholar]
- 9.Pfister SX, Ahrabi S, Zalmas L-P, Sarkar S, Aymard F, Bachrati CZ, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014; 7: 2006–18. doi: 10.1016/j.celrep.2014.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodarzi AA, Kurka T, Jeggo PA. Kap-1 phosphorylation regulates Chd3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 2011; 18: 831–9. doi: 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- 11.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois M-M, et al. Dna double-strand break repair pathway choice is directed by distinct Mre11 nuclease activities. Mol Cell 2014; 53: 7–18. doi: 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008; 455: 770–4. doi: 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 2011; 25: 350–62. doi: 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isono M, Niimi A, Oike T, Hagiwara Y, Sato H, Sekine R, et al. Brca1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep 2017; 18: 520–32. doi: 10.1016/j.celrep.2016.12.042 [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z, Chung W-H, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008; 134: 981–94. doi: 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng L, Chen J. The E3 ligase RNF8 regulates Ku80 removal and NHEJ repair. Nat Struct Mol Biol 2012; 19: 201–6. doi: 10.1038/nsmb.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myler LR, Gallardo IF, Soniat MM, Deshpande RA, Gonzalez XB, Kim Y, et al. Single-Molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Mol Cell 2017; 67: 891–8. doi: 10.1016/j.molcel.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riballo E, Kühne M, Rief N, Doherty A, Smith GCM, Recio María-José, Recio MJ, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol Cell 2004; 16: 715–24. doi: 10.1016/j.molcel.2004.10.029 [DOI] [PubMed] [Google Scholar]
- 19.Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol 2013; 14: 661–72. doi: 10.1038/nrm3659 [DOI] [PubMed] [Google Scholar]
- 20.Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Künzel J, et al. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell 2014; 55: 723–32. doi: 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biehs R, Steinlage M, Barton O, Juhász S, Künzel J, Spies J, et al. Dna double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol Cell 2017; 65: 671–84. doi: 10.1016/j.molcel.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton O, Naumann SC, Diemer-Biehs R, Künzel J, Steinlage M, Conrad S, et al. Polo-Like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J Cell Biol 2014; 206: 877–94. doi: 10.1083/jcb.201401146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata A, Jeggo P. A historical reflection on our understanding of radiation-induced DNA double strand break repair in somatic mammalian cells; Interfacing the past with the present. Int J Radiat Biol 2019; 95: 945–56. doi: 10.1080/09553002.2018.1564083 [DOI] [PubMed] [Google Scholar]
- 24.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, et al. Atm signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 2008; 31: 167–77. doi: 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- 25.Singh B, Gupta RS. Mutagenic response to ultraviolet light and x-rays at five independent genetic loci in Chinese hamster ovary cells. Environ Mutagen 1982; 4: 543–51. doi: 10.1002/em.2860040505 [DOI] [PubMed] [Google Scholar]
- 26.Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, et al. A compendium of mutational signatures of environmental agents. Cell 2019; 177: 821–36. doi: 10.1016/j.cell.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behjati S, Gundem G, Wedge DC, Roberts ND, Tarpey PS, Cooke SL, et al. Mutational signatures of ionizing radiation in second malignancies. Nat Commun 2016; 7: 12605. doi: 10.1038/ncomms12605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghezraoui H, Piganeau M, Renouf B, Renaud J-B, Sallmyr A, Ruis B, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell 2014; 55: 829–42. doi: 10.1016/j.molcel.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedford JS, Cornforth MN. Relationship between the recovery from sublethal X-ray damage and the rejoining of chromosome breaks in normal human fibroblasts. Radiat Res 1987; 111: 406–23. doi: 10.2307/3576927 [DOI] [PubMed] [Google Scholar]
- 30.Ochs F, Karemore G, Miron E, Brown J, Sedlackova H, Rask M-B, et al. Stabilization of chromatin topology safeguards genome integrity. Nature 2019; 574: 571–4. doi: 10.1038/s41586-019-1659-4 [DOI] [PubMed] [Google Scholar]
- 31.Löbrich M, Jeggo P. A process of Resection-Dependent nonhomologous end joining involving the Goddess ARTEMIS. Trends Biochem Sci 2017; 42: 690–701. doi: 10.1016/j.tibs.2017.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]