Abstract
A mouse cell variant carrying in heteroplasmic form a nonsense mutation in the mitochondrial DNA-encoded ND5 subunit of the respiratory NADH dehydrogenase has been isolated and characterized. The derivation from this mutant of a large number of cell lines containing between 4 and 100% of the normal number of wild-type ND5 genes has allowed an analysis of the genetic and functional thresholds operating in mouse mitochondria. In wild-type cells, ∼40% of the ND5 mRNA level was in excess of that required for ND5 subunit synthesis. However, in heteroplasmic cells, the functional mRNA level decreased in proportion to the number of wild-type ND5 genes over a 25-fold range, pointing to the lack of any compensatory increase in rate of transcription and/or stability of mRNA. Most strikingly, the highest ND5 synthesis rate was just sufficient to support the maximum NADH dehydrogenase-dependent respiration rate, with no upregulation of translation occurring with decreasing wild-type mRNA levels. These results indicate that, despite the large excess of genetic potential of the mammalian mitochondrial genome, respiration is tightly regulated by ND5 gene expression.
One of the most striking features of the mitochondrial genomes of both higher and lower eukaryotes is the discrepancy between the large number of copies of these genomes and the relatively low rate of expression of the mitochondrial genes (3). This “copy number paradox” is most clearly illustrated by the observation that, in HeLa cells, the ratio of rRNA molecules synthesized per cell generation to rRNA genes is 2 orders of magnitude lower in the mitochondrial compartment than in the cytoplasmic compartment (3). Very little is known about the regulation of gene expression in mammalian mitochondria and its adaptation to the ATP demands of the cell. In particular, no information is available as to whether, and under which conditions, the apparent excess of mitochondrial genetic potential is utilized by the cell. The observation in HeLa cells that both the mitochondrial mRNAs and rRNAs are metabolically unstable (21) suggested that the basal rate of transcription in these cells is in great excess over the cell requirements for protein synthesis. On the other hand, in both African green monkey cells (14) and mouse cells (32), a large increase in mitochondrial mRNA stability has been observed under conditions where the synthesis of the organelle RNA was blocked. Regulation of mitochondrial RNA stability has also been suggested to play an important role during rat liver development (42). While the large excess of both mitochondrial DNA (mtDNA) and its transcriptional activity could, in principle, allow a rapid adaptation to increased respiratory and ATP synthesis demands, it is intriguing that, in some developmental and physiological situations, an increased level of mitochondrial gene expression is frequently accompanied, and possibly determined, by an increase in the level of mtDNA (9, 49, 50). Furthermore, there is well-documented evidence of transcriptional regulation of mitochondrial gene expression in rat liver mitochondria by thyroid hormones (16) and during early embryogenesis in Xenopus laevis (1).
There is also very little information concerning the thresholds operating at the level of mitochondrial translation. Thus, it is not known how much the rate of mitochondrial protein synthesis exceeds the requirements for the assembly of the enzyme complexes capable of supporting a normal rate of oxidative phosphorylation and whether it can be upregulated in case of need.
Answers to the issues discussed above would be essential for understanding how different cells or even different subcellular compartments adapt their respiratory and ATP-producing capacity in various developmental and physiological situations. Furthermore, the discovery of disease-causing mtDNA mutations, affecting either components of the translation apparatus or subunits of the oxidative phosphorylation complexes, and the increasing evidence of progressive damage to the oxidative phosphorylation activities associated with aging and neurodegenerative diseases have raised important questions concerning the genetic and functional thresholds controlling gene expression and oxidative phosphorylation in mammalian mitochondria.
In the present work, the isolation of a nonsense heteroplasmic mutation in the mitochondrial gene for ND5, an essential subunit of the mouse respiratory NADH dehydrogenase (complex I), and the application of specific technologies for the manipulation of the mitochondrial genome (5, 29, 30) have allowed the construction of a set of transmitochondrial cell lines carrying, in a constant nuclear background, various copy numbers of the wild-type ND5 gene, from ∼4 to 100% of the normal level. Analysis in these transformant cell lines of the total and wild-type mRNA levels and of the rates of mRNA translation and complex I-dependent respiration have revealed a stringent regulation of ND5 gene expression and respiration. These findings have given novel insights into the regulation of mitochondrial function in mammalian cells and provided a paradigm of tight thresholds which are likely to apply to other cellular processes.
MATERIALS AND METHODS
Cell lines and media.
All the cell lines used in the present work were grown in monolayer culture. The cell line A9 (ATCC CCL-1.4) is a derivative of the L mouse fibroblast cell line deficient in hypoxanthine-guanine phosphoribosyl transferase and is, thus, resistant to 8-azaguanine and 6-thioguanine (34) and incapable of growing in hypoxanthine-aminopterin-thymidine (HAT) medium (19). This cell line was grown in Dulbecco modified Eagle medium with 4.5 mg of glucose/ml (DMEM) supplemented with 10% calf serum and 3 μg of 8-azaguanine per ml. The A9-derived rotenone-resistant clone 3A, isolated as previously described (5), was grown in the above-described medium supplemented with 1.2 μM rotenone. The mouse cell line LL/2, derived from Lewis lung carcinoma (7; ATCC CRL-1642), was grown in DMEM supplemented with 10% calf serum. 3A clone-derived transformants (see below) were also grown in the same medium. The mtDNA-less (ρ°) LL/2-m21 cell line, a derivative of LL/2 cells (see below), was grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 50 μg of uridine per ml.
Isolation of mtDNA-less LL/2 derivatives and mitochondrion-mediated transformation.
The mtDNA-less ρ° LL/2-m21 cell line was isolated by a modification of a method described earlier (13, 29), which involved treatment of LL/2 cells with high concentrations of ethidium bromide (5). In particular, LL/2 cells were exposed to 5 μg of ethidium bromide per ml for 11 to 12 weeks in medium supplemented with 50 μg of uridine per ml. Two clones were isolated and tested for the presence of mtDNA by Southern blot hybridization of a FokI-digested total cell DNA with a mouse mtDNA probe [clone MumX7.6, carrying the mtDNA fragment between positions 953 and 8529 in pBluescript II KS(+)], which was digested with EcoRV and 32P labeled by random priming (17). Both clones showed complete absence of mtDNA. One of these clones, ρ° LL/2-m21, was utilized in the present work. Mitochondrion-mediated transformation of ρ° cells by cytoplast fusion was carried out as described previously (29) by fusing 3A mutant cells, which had been enucleated by centrifugation in the presence of cytochalasin B, with ρ° LL/2-m21 cells in the presence of 40% polyethylene glycol 1500 (BDH). Mitochondrial transformants were isolated in DMEM supplemented with HAT medium components (i.e., hypoxanthine, aminopterin, and thymidine) (19) and 10% FBS. The transformant clones were subsequently cultured in DMEM medium with 10% calf serum. In order to increase the proportion of mutant mtDNA in one of the ρ° LL/2-m21 transformants (3A20), this was cultured in DMEM supplemented with 10% FBS and 50 μg of uridine and 1 μg of ethidium bromide per ml for 10 days (30). The cells were then trypsinized and replated at low density (0.5 cell per well) in a 96-well dish in the same medium without ethidium bromide. Ten days later, individual colonies were selected, transferred to 100-mm plates, and grown further in medium lacking uridine.
Chromosome analysis.
To distinguish cybrids from hybrids among the ρ° LL/2-m21 transformants, cells were arrested in metaphase by treatment with 0.05 μg of colchicine per ml for 3 h. Karyotype analysis was carried out as described previously (40).
O2 consumption measurements.
The medium of the cell lines to be analyzed was changed with fresh medium (rotenone free in the case of the original 3A mutant cell line) 24 h before the measurements. The total O2 consumption rate was determined on cells in DMEM lacking glucose, supplemented with 5% dialyzed FBS, in a YS oxygraph (model 5300 Biological Oxygen Monitor), as previously described (29). After the measurement, 100 nM rotenone was added to the chamber, and the rotenone-insensitive O2 consumption rate was measured. This rate was subtracted from the total respiration to calculate the rotenone-sensitive rate. For measurements of O2 consumption in digitonin-permeabilized cells (27), about 2 × 106 cells were resuspended in 1 ml of buffer (20 mM HEPES [pH 7.1], 10 mM MgCl2, 250 mM sucrose), and then 100 μg of digitonin (1 μl of a 10% dimethyl sulfoxide solution) in 1 ml of buffer was added. After incubation for 1 min at room temperature, the cell suspension was diluted with 8 ml of buffer. The cells were rapidly pelleted and then resuspended in respiration buffer (20 mm HEPES [pH 7.1], 250 mM sucrose, 2 mM KPi, 10 mM MgCl2, and 1.0 mM ADP). The measurements were carried out in the chamber of the YSI oxygraph. The substrates (adjusted to a pH of ∼7.0 with NaOH) and inhibitors were added with Hamilton syringes. The final concentrations were as follows: malate, 5 mM; glutamate, 5 mM; succinate, 5 mM; glycerol-3-phosphate, 5 mM; ascorbate, 10 mM; N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), 0.2 mM; rotenone, 100 nM; antimycin, 20 nM; and KCN, 1 mM.
Enzymatic tests.
The mitochondrial fraction of the desired cell type was isolated from 0.5 to 1.0 ml of packed cells as described previously (44), resuspended in 8 ml of 50 mM Tris-HCl buffer (pH 7.5 at 25°C), and sonicated with a Branson Sonifier for 40 s (four 10-s pulses separated by 15-s intervals) on ice. Mitochondrial membranes were pelleted by centrifugation at 39,000 rpm in a Beckman Ty65 fixed-angle rotor for 60 min and resuspended in 500 μl of the above-described buffer. The oxidoreductase activities were measured, at protein concentrations of 120 μg/ml for Q1 reduction and 25 μg/ml for K3Fe(CN)6 reduction, in medium containing 20 mM Tris-HCl (pH 7.5 at 25°C), 1 mM KCN, 100 μM NADH, and either 50 μM Q1 (Eisai Co., Tokyo, Japan) or 1 mM K3Fe(CN)6. The reaction was monitored by absorbance measurements at 275 nm for the reduction of Q1 (ɛ = 12,250 M−1 cm−1) and at 410 nm for the reduction of K3Fe(CN)6 (ɛ = 1,000 M−1 cm−1). The NADH-Q1 oxidoreductase activity of the LL/2 control samples was >98% sensitive to 100 nM rotenone.
Mitochondrial protein synthesis analysis.
To measure the rate of mitochondrial protein synthesis, pulse-labeling experiments with [35S]methionine were performed according to the method of Chomyn et al. (11). Samples of 2 × 106 cells of the desired type were plated on 10-cm petri dishes, incubated overnight, washed with methionine-free DMEM, and then incubated for 7 min at 37°C in 4 ml of the same medium containing 50 μg of the cytoplasmic translation inhibitor emetine per ml. Thereafter, [35S]methionine (0.2 mCi [1,175 Ci/mmol] per plate) was added, and the cells were incubated for 30 min. The labeled cells were trypsinized, washed, and lysed in 1% sodium dodecyl sulfate (SDS). Samples containing 40 μg of protein were electrophoresed through an SDS–15 to 20% exponential polyacrylamide gradient gel. The intensities of the bands were quantified by laser densitometry of appropriately exposed fluorograms or by phosphorimager analysis. To measure the protein labeling as a function of time, four samples of 5 × 105 cells of each of the LL/2 and 3A33 cell lines were plated on 10-cm petri dishes. Labeling conditions were the same as those described above, except that a greater amount of [35S]methionine (1.2 mCi per plate) was used. Protein synthesis was stopped by the addition of 4 μM puromycin at 10, 20, 30, and 45 min. The intensities of the bands were quantified by phosphorimager analysis after electrophoresis.
Immunoprecipitation experiments.
For immunoprecipitation of complex I, cells were subjected to pulse-chase labeling (12). For this purpose, about 8 × 107 cells were grown for 22 h in the presence of 40 μg of the mitochondrial translation inhibitor chloramphenicol (CAP) per ml in order to allow the accumulation of the nucleus-encoded subunits of the oxidative phosphorylation apparatus and, therefore, facilitate the incorporation into these complexes of the mtDNA-encoded subunits synthesized after the removal of CAP. Labeling was carried out as described in the previous section, except that emetine was replaced with the reversible protein synthesis inhibitor cycloheximide, and the incubation time with [35S]methionine was extended to 2 h; thereafter, the cells were washed and subjected to a 19-h chase in complete unlabeled medium in the absence of cycloheximide in order to allow the incorporation of the labeled mtDNA-encoded subunits into the complexes. The pulse-chase-labeled cells were then collected and pelleted to yield ∼0.2 ml of packed cells. The mitochondrial fraction was isolated from these cells by homogenization and differential centrifugation and then lysed with 0.5% Triton X-100 (39). Samples of 120 μg of protein were incubated at 4°C with 72 μg of gamma globulins from an antiserum against the COOH-terminal heptapeptide of the human mtDNA-encoded subunit ND4L (38) or from normal rabbit serum. Immunocomplexes were bound to formaldehyde-fixed Staphylococcus aureus (Zysorbin; Zymed Laboratories, San Francisco, Calif.) (11), pelleted, and washed repeatedly by centrifugation and resuspension. The protein was eluted from the immunoadsorbant in the final pellets with 1% SDS, 5 mM Tris-HCl (pH 8) (25°C), and 1 mM phenylmethylsulfonyl fluoride and then electrophoresed through an SDS–15 to 20% exponential polyacrylamide gradient gel.
DNA analysis.
For ND5 mtDNA sequencing, total DNA samples were isolated from cells with an Applied Biosystems 340A DNA extractor and then subjected to PCR amplification of the ND5 gene by using the primers ND5-5′-1 and ND5-3′-1 (see below). DNA sequencing of the gel-purified (QIAEXII; Qiagen) product was carried out by using the ABI PRISM Dye Terminator Cycle Sequencing Core (Perkin-Elmer) with the primers ND5-5′-2, ND5-5′-3, ND5-5′-5′4, ND5-5′5, ND5-5′6, ND5-3′-2, ND5-3′-3, ND5-3′-4, and ND5-3′-5 (see below).
The mtDNA content of the various cell lines was determined at the time of O2 consumption measurements by DNA transfer hybridization of total cell DNA carried out with a slot blot apparatus (29). For this purpose, samples of 2 × 105 cells were lysed in PCR buffer containing 1% NP40 and 100 μg of proteinase K per ml, incubated for 1 h at 55°C and then for 10 min at 95°C, treated with 0.5 M NaOH for 16 h, blotted in triplicate, and hybridized with a mixture of three mouse mtDNA-specific probes 32P labeled by random priming (plasmid MumX1.9, containing the mouse mtDNA sequence from positions 8984 to 10907; plasmid MumX5.1, containing the mtDNA sequence from positions 10907 to 15973; and plasmid MumX7.6, containing the mtDNA sequence from positions 953 to 8529 [8]). Quantification of the intensities of the bands was done by using a PhosphorImager (Molecular Dynamics) and the ImageQuant program.
Quantification of the mtDNA mutation (a C-to-A point mutation, which destroys a ClaI site) was carried out by analysis of the products of a restriction digestion reaction. For this purpose, a 465-bp fragment of the ND5 gene was amplified by PCR with the primers ND5-5′-2 and ND5-3′-4 in a 50-μl volume. To avoid the errors arising from resistance to enzyme digestion of heteroduplexes of wild-type and mutant mtDNA, the “last cycle hot” PCR was performed (41). For this purpose, 5 μCi of [α-32P]dATP was added to a sample of the reaction mixture before the last cycle. A 5-μl sample of the final PCR mixture was then subjected to ClaI digestion (5 U) in a 20-μl reaction volume at 37°C overnight: under these conditions, the wild-type ND5 fragment was cut into two small fragments of 297 and 168 bp, while the mutant ND5 fragment remained intact. A 5-μl sample of the above-described reaction mixture was subsequently electrophoresed on a 6% polyacrylamide gel. Quantification of the intensity of the bands was done by phosphorimager analysis.
The sequences of the primers used in this study were as follows: ND5-5′-1 (positions 11613 to 11630), GATTGCAAGAACTGCTTA; ND5-5′-2 (positions 11785 to 11802), CCCCAATCCTAATTTCAA; ND5-5′-3 (positions 12968 to 12986), ACACTAATCGCCACTTCT; ND5-5′-4 (positions 12186 to 12203), TCTTTCCTACTAATTGGA; ND5-5′-5 (positions 12761 to 12778), CTCTGGCTCAATCATTCA; ND5-5′-6 (positions 13381 to 13398), CCCTAACTCTCCTAGACT; ND5-3′-1 (positions 13609 to 13592), GGTAGTAGCTGGGTGATC; ND5-3′-2 (positions 13556 to 13539), CTCGAGATTAATTGAGTA; ND5-3′-3 (positions 12912 to 12895), CTTTTGAGTAGAACCCTG; ND5-3′-4 (positions 12248 to 12231), TGCTTGTAGGGCTGCAGT; and ND5-3′-5 (positions 11917 to 11900), TATTCTATATTATTGTGG.
The sequences of the ND1, ND4, and ND6 genes were determined by PCR amplification of appropriate overlapping fragments and sequencing of the gel-purified products. For this purpose, 18- to 20-nucleotide primers were used with the starting positions of the 5′ primers being 2653, 2679, 2951, and 3231 for the ND1 gene, 10124, 10176, 10451, and 10781 for the ND4 gene, and 13433 and 13496 for the ND6 gene and the starting positions of the 3′ primers being 3835, 3796, and 3520 for the ND1 gene, 11644, 11610, 11320, and 11030 for the ND4 gene, and 14202 and 14164 for the ND6 gene.
RNA analysis.
Total cell RNA was isolated by the RNAzol B procedure (Tel-Test, Inc., Friendswood, Tex.), which is based on a modification of the single-step method by acid guanidinium thiocyanate-phenol-chloroform extraction (10). RNA extracted from 1 × 106 to 3 × 106 cells was treated with 30 U of RNase-free DNase I (Boehringer Mannheim) at 37°C overnight in Taq buffer supplemented with 3 mM MgCl2 and 100 U of RNasin (Promega). The analysis of the genotypes of the transcripts was carried out by reverse transcription (RT)-PCR (6) and restriction digestion by ClaI. The RT was carried out in a 20-μl reaction mixture containing 1 μg of RNA (dissolved in water), 0.4 μg of random hexamer mixture, 10 mM dithiothreitol, 40 U of RNasin, 0.5 mM concentrations of each of the four deoxynucleoside triphosphates (dNTPs), 4 μl of 5× RT buffer and 200 U of mouse mammary leukemia virus reverse transcriptase (dNTPs were from Amersham; all other products were from Promega). The reaction was carried out at 37°C for 1 h, followed by 5 min of heating at 95°C to inactivate the enzyme. Then, 4-μl samples of each cDNA mixture and, as a control, of reaction mixtures from which reverse transcriptase had been omitted were used directly as substrates for PCR. The PCR and the ClaI digestion were carried out as for DNA analysis. For quantification of the ND5 mRNA, an RNA transfer hybridization analysis was carried out as follows. First, 20 μg of the total cell RNA was fractionated by electrophoresis through a 1.4% agarose–2.2 M formaldehyde gel; then it was transferred onto a Zeta-probe membrane (Bio-Rad) and hybridized to a mouse mtDNA-specific probe 32P labeled by random priming (plasmid MumX5.1, containing the mtDNA sequence from positions 10907 to 15973, which include the ND5, ND6 and CYTb genes and part of the ND4 gene [8]). Quantification of the intensity of the bands was done by phosphorimager analysis.
RESULTS
Isolation of a mouse ND5 gene mutant.
In previous work (5), the mouse fibroblast cell line A9 (deficient in hypoxanthine-guanine phosphoribosyltransferase [HGPRT]) was screened for mutants defective in one or another of the mtDNA-encoded subunits of complex I by following an approach based on the cell resistance to high concentrations of rotenone, a specific inhibitor of the enzyme (5, 24, 25). Eleven clones, which had been adapted to grow in the presence of 1.2 μM rotenone by exposing them to increasing concentrations of the drug (5), all showed a variable decrease, relative to the A9 level, in malate-glutamate-dependent respiration, which usually reflects the rate-limiting activity of complex I, whereas the succinate–glycerol-3-phosphate (G-3-P)-driven respiration, which usually reflects the activity of ubiquinone-cytochrome c oxidoreductase (complex III), and the TMPD-ascorbate-driven respiration, which reflects the activity of cytochrome c oxidase (COX; complex IV), were not significantly decreased. The clone most severely affected, clone 4A, was analyzed further and shown to carry a near-homoplasmic frameshift mutation in the ND6 subunit gene (5). In the present work, another of the 11 clones, clone 3A, was tested and found not to exhibit any decrease, relative to the level in the parental A9 cells, in overall respiration (Fig. 1a), but a significant reduction (∼34%) in malate-glutamate-dependent O2 consumption (Fig. 1b). This reduction contrasted with the lack of any decrease in succinate–G-3-P-dependent respiration and a slight increase in TMPD-ascorbate-driven respiration.
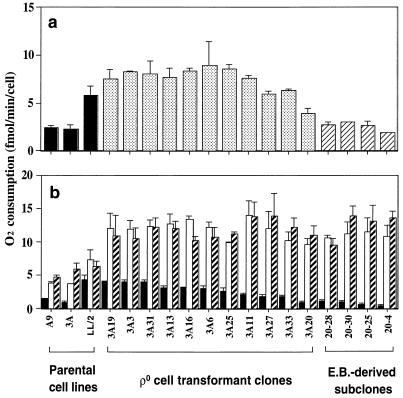
FIG. 1.
Total respiration rate (a) and activities of the enzymes of the mitochondrial respiratory chain (b) in the three parental cell lines (A9, 3A, and LL/2), 11 ρ° LL/2-m21 cell transformants, and 4 3A-20 ethidium bromide (E.B.)-treated derivatives. In both panels, the data are displayed in the following order: first for the A9, 3A, and LL/2 cell lines; then for the transformants; and, finally, the 4 3A-20 derivatives. The cell lines in the latter two groups are arranged in order of decreasing malate-glutamate-dependent respiration. In panel a, the total respiration rate was measured on ∼2 × 106 cells. In panel b, the activities of the various components of the respiratory chain were determined on 2 × 106 cells as respiration dependent on malate-glutamate (solid bars), succinate–G-3-P (open bars), and TMPD-ascorbate (hatched bars). Three to five determinations were made for each cell line. The error bars indicate the standard error of the mean (SEM).
In order to investigate the genetic origin of the rotenone resistance and of the respiratory defect in the 3A cells and the relationship between the two phenomena, advantage was taken of the mtDNA-less (ρ°) cell repopulation approach (29). The ρ° LL/2-m21 cell line had been isolated in this laboratory from the mouse LL/2 cell line after long-term treatment with ethidium bromide (5; see also Materials and Methods). Mitochondria from 3A cells were transferred into the mouse ρ° cells by fusion of the latter with a population of predominantly enucleated cells (cytoplasts) derived from the mutant cells. The cybrids and hybrids were selected for in DMEM without the addition of uridine (a medium in which ρ° cells cannot survive [29]) and supplemented with HAT medium components in order to kill any nonenucleated HGPRT-deficient 3A cells. Eleven transformants, named 3A3, 3A6, 3A11, 3A13, 3A16, 3A19, 3A20, 3A25, 3A27, 3A31, and 3A33, were selected for further analysis.
Karyotype analysis of the 3A transformants revealed a range of chromosome numbers (43 to 54) similar to those of the LL/2 and ρ° LL/2-m21 cell lines (i.e., ∼42), as expected for cybrids. None of the transformants was able to grow in the presence of 1.2 μM rotenone (data not shown). These results strongly suggested that the resistance to rotenone was due to a mutation in a nuclear gene, as previously observed for the rotenone-resistant human and mouse cells (5, 24, 25).
In confirmation of earlier findings (5), the rates of overall O2 consumption (Fig. 1a) and of malate-glutamate-dependent respiration (Fig. 1b) in A9 cells were found to be significantly lower than in LL/2 cells. The previous experimental evidence strongly suggested that this was due to some difference in the nuclear backgrounds of A9 and LL/2 cells (5). Accordingly, in the present work, the O2 consumption rates of the ρ° LL/2-m21 transformants were compared with those of the LL/2 cells, which had the same nuclear background as the ρ° cells. It was found that, in contrast to what occurred in rotenone resistance, the defect in complex I activity of the clone 3A cells was transferred into ρ° cells with their mtDNA, pointing to an mtDNA mutation as its cause. Thus, the rates of malate-glutamate-dependent respiration in the 11 transformants analyzed were reduced, relative to that in LL/2 cells, by 7 to 78%, as shown in decreasing order in Fig. 1b. By contrast, the rate of overall respiration was increased by 2 to 54% in all transformants, except in 3A20, in which it was decreased by ∼32% (Fig. 1a). A plausible explanation of the increase in overall respiratory capacity in nearly all of the transformants was provided by the observation that the succinate–G-3-P-dependent respiration rate was increased in all transformants by 31 to 91%, relative to the LL/2 rate, while the TMPD-ascorbate-dependent respiration rate was increased by 66 to 119% (Fig. 1b).
Among the 11 ρ° LL/2-m21 transformants analyzed, 3A20 showed both the lowest overall respiration rate (3.9 fmol/min/cell; 67% of the LL/2 rate) (Fig. 1a) and the lowest malate-glutamate-dependent respiration rate (0.95 fmol/min/cell; 22% of the LL/2 rate) (Fig. 1b). In order to increase further the proportion of the putative mutant mtDNA in this transformant, an approach based on treatment of the cells with a low concentration of the mtDNA replication selective inhibitor ethidium bromide (30) was used. The goal was to reduce the mtDNA level to an average of one molecule per cell, a state in which the majority of the cells would be expected to be homoplasmic for mutant or wild-type mtDNA. At this time, withdrawal of ethidium bromide would allow the single mtDNA molecule to repopulate the cell. Previous work had, in fact, shown that cells partially depleted of mtDNA by treatment with ethidium bromide rapidly regain a normal level of mtDNA upon removal of the drug (51). Accordingly, 3A20 cells were grown for 10 days in DMEM supplemented with 50 μg of uridine per ml and containing 1 μg of ethidium bromide per ml. Single cells were cloned, as detailed in Materials and Methods, and among 32 clones isolated, the four which had the lowest overall and malate-glutamate-dependent respirations were selected (Fig. 1). Among them, the 3A20-4 clone exhibited the most significant reduction in total respiration rate (1.9 fmol/min/cell; 32% of the LL/2 rate) and in malate-glutamate-dependent respiration rate (0.44 fmol/min/cell; 10% of the LL/2 rate).
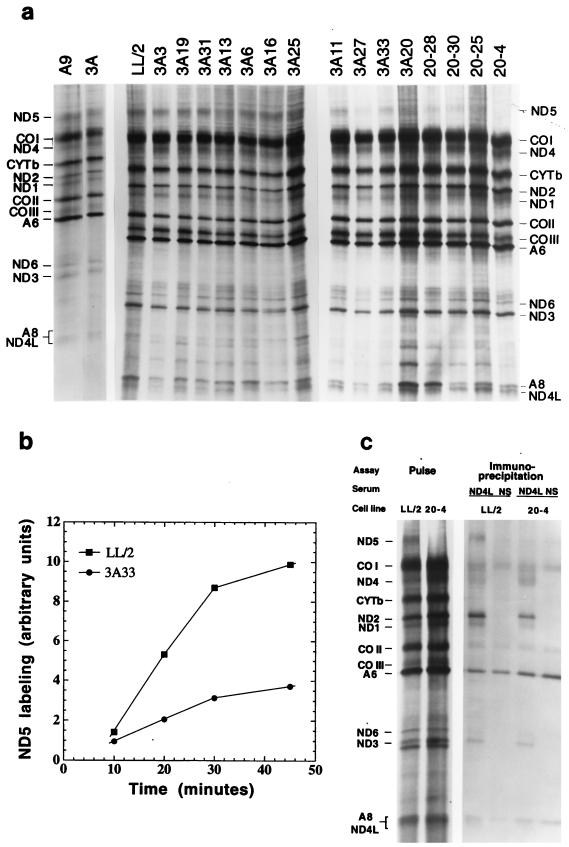
The transfer of the complex I defect into ρ° cells with 3A cell mtDNA pointed strongly to the presence in this DNA of a mutation in one of the genes encoding subunits of the respiratory NADH dehydrogenase. In order to obtain some indication of the possible site of this mutation, the mitochondrial translation products of the various cell lines were labeled with [35S]methionine for 30 min in the presence of the inhibitor of cytoplasmic protein synthesis emetine. As can be seen in Fig. 2a, in the electrophoretic patterns from the 11 transformants and the four ethidium bromide-derived 3A20 subclones, the only obvious difference from the wild-type patterns of A9 and LL/2 cells is the reduction in the relative labeling of the polypeptide identified from its electrophoretic mobility as the ND5 gene product. A control experiment utilizing different [35S]methionine pulses verified that the labeling of the ND5 polypeptide (Fig. 2b), as well as the overall labeling of the mitochondrial translation products (not shown), was linear over the 30-min period in both the parental line LL/2 and the 3A33 transformant. Therefore, the 30-min labeling data reflect the rates of synthesis of the various polypeptides. In Fig. 2a, the electrophoretic patterns for the transformant and ethidium bromide-derived clones are displayed, with minor deviations, in order of decreasing malate-glutamate-dependent respiration rate of the cell lines. After correction for the obvious differences in loading of lanes 3A25, 3A27, and 3A20, a general trend toward a progressive decrease in the relative intensity of the ND5 band is clear, indicating a decrease in the rate of synthesis of the ND5 protein. These observations pointed to the ND5 gene as the best candidate for carrying the putative mutation.
FIG. 2.
Electrophoretic analysis of SDS mitochondrial lysates from the [35S]methionine 30-min-pulse-labeled 3 parental lines, 11 ρ° LL/2-m21 transformants, and 4 ethidium bromide-derived 3A20 subclones (a), kinetics of the ND5 labeling in LL/2 and 3A33 cells (b), and electrophoretic patterns of immunoprecipitates obtained from Triton X-100 mitochondrial lysates of pulse-chased LL/2 and 3A20-4 cells with gamma globulins from an antiserum against the human ND4L subunit (ND4L) or from normal rabbit serum (NS) (c). For details, see Materials and Methods. In panel a, the patterns for the transformants and 3A20-4 are arranged, with minor deviations, in the order of decreasing malate-glutamate-dependent respiration rate.
The ND5 mutation is a C-to-A transversion creating a mitochondrial stop codon.
On the basis of the above findings, the PCR-amplified ND5 gene was sequenced by the chain termination method (43) in both the wild-type cell line LL/2 and the mutant cell line 3A20-4. As shown below, a C-to-A transversion was found at position 12081 (8), which changed the arginine codon CGA to the mitochondrial stop codon AGA (2):
 |
The mouse ND5 gene encodes a 607-amino-acid polypeptide. The C-to-A mutation resulted in a 115-amino-acid truncated polypeptide. It seems very likely that this truncated protein is an unstable product, as previously shown for the prematurely terminated human ND4 and ND5 polypeptides (24, 25) and the mouse ND6 polypeptide (5). However, the near identity in size of this truncated product with the ND3 subunit (114 amino acids) prevented its identification in the electrophoretic patterns of newly synthesized proteins from the mutant cell lines.
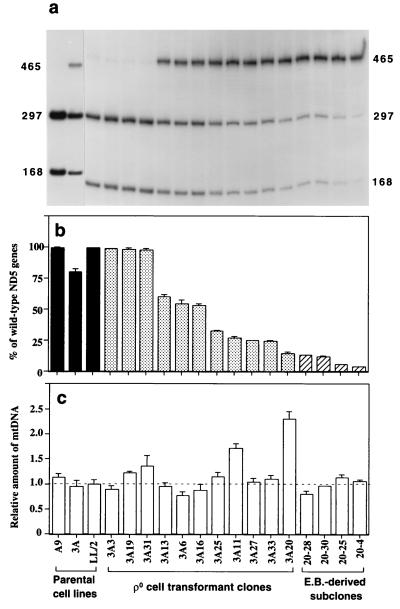
The sequence data showed that the C-to-A mutation at position 12081 also destroyed a ClaI restriction site (ATCGAT, underlined in the sequence shown above), and this was confirmed by the ClaI digestion of a PCR-amplified ND5 fragment (Fig. 3a). The quantitative analysis of the ClaI digestion patterns revealed that, in the original 3A mutant clone, there was about 20% of ND5 mutation-carrying mtDNA. In the transformants 3A3, 3A19, and 3A31, mutant mtDNA was not detectable above a very weak background signal observed also in LL/2 and, in clones 3A20 and 3A20-4, the mutant mtDNA had increased up to 85 and ∼96%, respectively (Fig. 3b). To exclude any additional structural alteration in any of the subunits of NADH dehydrogenase, the genes for the subunits which have been found to be affected by disease-causing mutations, i.e., ND1, ND4, and ND6 (47), were completely sequenced. No sequence difference from the corresponding genes in A9 mtDNA was found.
FIG. 3.
Quantification of the A12081C mutation, by ClaI digestion of a PCR-amplified ND5 fragment (a and b), and of total mtDNA content, by slot blot hybridization analysis (c), in the parental cell lines, transformants, and ethidium bromide (E.B.)-derived 3A20 subclones. In panel c, the mtDNA content of the various cell lines is expressed relative to the value in LL/2 cells. Two determinations of the proportion of wild-type ND5 genes and three to six determinations of mtDNA content were made for each cell line. The error bars indicate the SEM.
A quantification of the mtDNA content of the transformant cell lines, carried out by hybridization with mouse mtDNA probes, failed to reveal any significant difference in mtDNA level from the LL/2 control value (the deviations being ≤36%, mostly in the range of 4 to 25%), except in clones 3A11 and 3A20 (Fig. 3c). The latter two clones exhibited 71 and 130% increases in mtDNA content, respectively. These increases presumably reflected a compensatory amplification of mtDNA, a phenomenon which was previously observed (5, 53).
Further characterization of the 3A20-4 ND5 mutant cells.
To further characterize this ND5 mutant cell, a biochemical analysis of the activity of complex I was carried out by enzymological tests on partially purified mitochondrial membranes (24). The NADH oxidoreductase activity was determined with a water-soluble ubiquinone analog (Q1) in both LL/2 wild-type cells and 3A20-4 mutant cells. The NADH-K3Fe(CN)6 oxidoreductase activity of the membranes, which is catalyzed by the nucleus-encoded flavoprotein fragment of the enzyme (20), was also measured in the same cells.
As shown in Table 1, the NADH-Q1 oxidoreductase activity of the 3A20-4 clone mitochondrial membranes was reduced to less than ∼1.4% of the LL/2 activity. By contrast, the NADH-K3Fe(CN)6 oxidoreductase activity did not appear to be significantly affected in the 3A20-4 clone membrane preparation. It has previously been shown with Neurospora crassa that the flavoprotein fragment is assembled independently of the membrane fragment, which contains the mtDNA-encoded subunits (48), and the same is probably true in mammalian cells (5, 24, 25). Therefore, the NADH-K3Fe(CN)6 oxidoreductase activity was used to correct for differences in mitochondrial content among the crude mitochondrial membrane preparations from the two cell lines tested. The corrected activities are shown in Table 1. It appears that the normalized NADH-Q1 oxidoreductase activity was reduced in 3A20-4 cells to less than 1.7% of the LL/2 activity.
TABLE 1.
Measurements of NADH-Q1 and NADH-K3Fe(CN)6 oxidoreductase activities in mitochondrial membranes isolated from LL/2 and 3A20-4 cellsa
| Cell line | Avg activity (nmol/min/mg, ± 2 SEM)b
|
NADH-Q1/NADH-K3Fe(CN)6 (103)c | |
|---|---|---|---|
| NADH-Q1 oxidoreductase | NADH-K3Fe(CN)6 oxidoreductase | ||
| LL/2 | 11.0 (±1.2) | 2,079 (±245) | 5.3 |
| 3A20-4 | <0.15 | 1,693 (±150) | <0.089 (<1.7) |
The values for NADH-Q1 represents the total activity, with >98% being rotenone sensitive.
Determinations were made in triplicate.
Value in parentheses represents the percentage of normalized NADH-Q1 oxidoreductase activity in 3A20-4 relative to that in LL/2.
In order to investigate whether, in the cell lines carrying a nonsense mutation in ND5, the assembly of the other mtDNA-encoded subunits is affected, immunoprecipitation experiments were carried out by using antibodies against the COOH-terminal synthetic heptapeptide of the mtDNA-encoded subunit ND4L of human complex I, which is located in the membrane arm (38). These antibodies had previously been shown to be able to precipitate all mtDNA-encoded subunits and, therefore, presumably the whole mouse complex I, from a 0.5% Triton X-100 mitochondrial lysate (5), and thus were expected to reveal whether the mtDNA-encoded subunits of the mutant cell lines are assembled into the membrane arm of complex I. For the purpose of these immunoprecipitation experiments, as detailed in Materials and Methods, LL/2 and 3A20-4 cells were labeled for 2 h with [35S]methionine in the presence of the reversible inhibitor of cytoplasmic protein synthesis cycloheximide and then chased for 19 h in complete unlabeled medium in the absence of the inhibitor. As shown in Fig. 2c, when a 0.5% Triton X-100 mitochondrial lysate from pulse-chased LL/2 cells was incubated with gamma globulins from the anti-ND4L antiserum, the antibodies precipitated several mtDNA-encoded complex I subunits, in particular, clearly recognizable, ND1, ND2, ND3, ND4, ND5, and ND6. By contrast, no evidence of these subunits could be seen in the corresponding immunoprecipitate obtained with normal serum gamma globulins. A nonspecific precipitate of COI, COII, A6, and A8 polypeptides was observed, for unknown reasons, in the precipitate obtained when gamma globulins of either normal serum or anti-ND4L antiserum were used. In the mitochondrial lysate from mutant 3A20-4 cells, which lacked almost completely the ND5 product, all the recognizable remaining mtDNA-encoded complex I subunits, i.e., ND1, ND2, ND3, ND4, and ND6, were also immunoprecipitated, although to a somewhat reduced extent compared to levels produced by the lysate from LL/2 cells. The above results indicated that the lack of ND5 subunit does not prevent the assembly of the membrane arm of complex I, as was previously shown for human cells lacking ND5 (25), although it reduces the efficiency of this process or affects the stability of the membrane arm.
Effect of ND5 gene dosage alteration on mutant and wild-type ND5 mRNA levels.
The availability of a set of ρ° LL/2-m21 transformants and ethidium bromide-derived 3A20 subclones carrying, in a constant nuclear background, a content of functional ND5 genes which varied over a wide range, from ∼4 to 100% of the wild-type LL/2 level, allowed an analysis of the genetic and functional thresholds operating in the expression of this gene. In particular, the effects of the dosage of ND5 genes on the ND5 mRNA level, the ND5 synthesis rate, and the assembly of a functional NADH dehydrogenase were investigated in detail. In this analysis, the cell line 3A3, which had virtually 100% wild-type ND5 genes and an mtDNA content very close to that of the LL/2 parental line (Fig. 3b and c) and exhibited a nuclear background and mtDNA haplotype identical to those of all LL/2-m21 transformants and ethidium bromide-derived 3A20 subclones, was used as a reference for comparison purposes.
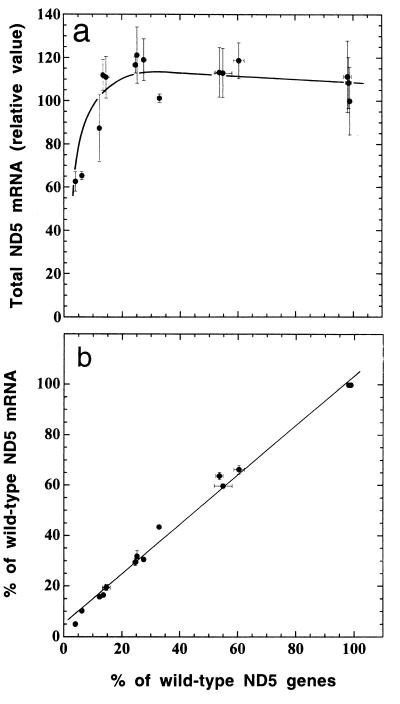
RNA transfer hybridization experiments, in which a mouse mtDNA fragment carrying the sequences of the ND5, ND6, and CYTb genes and of part of the ND4 gene was used as a probe, and the data of ND5 hybridization to the ND4, ND6, and CYTb sequences were utilized for normalization showed that almost all transformants and ethidium bromide-derived 3A20 subclones had a total ND5 mRNA level that was approximately constant, independent of the proportion of mutant genes. Only two ethidium bromide-derived subclones (20-25 and 20-4), which are nearly homoplasmic for mutant mtDNA (containing 6.2 and 4.0% wild-type mtDNA, respectively), exhibited a significantly reduced mRNA level (by 35 to 40% relative to the reference 3A3 level) (Fig. 4a). In other experiments, the proportions of wild-type and mutant ND5 mRNAs were analyzed by taking advantage of the destruction of the ClaI site produced by the C12081A transversion. Thus, by RT-PCR it was shown that, in every transformant or ethidium bromide-derived subclone, the proportion of wild-type ND5 mRNA was very close to the proportion of wild-type ND5 genes (Fig. 4b). Control experiments, in which reverse transcriptase was omitted, failed to yield any PCR products, as expected.
FIG. 4.
Quantification of total ND5 mRNA in the ρ° LL/2-m21 transformants and ethidium bromide-derived 3A20 subclones by RNA transfer hybridization (a) and relationship of the proportion of wild-type ND5 mRNA, as determined by RT-PCR, to the proportion of wild-type ND5 genes (b). The average total ND5 mRNA content per cell was normalized as detailed in the text and expressed relative to the value for the transformant 3A3, which has virtually 100% wild-type ND5 genes (Fig. 3b). Three to five determinations of total ND5 mRNA content and three determinations of the proportion of wild-type ND5 mRNA by RT-PCR were made for each cell line. The error bars represent the SEM; the error bars that fall within the individual data symbols are not shown.
Effect of wild-type ND5 mRNA level alteration on ND5 synthesis rate.
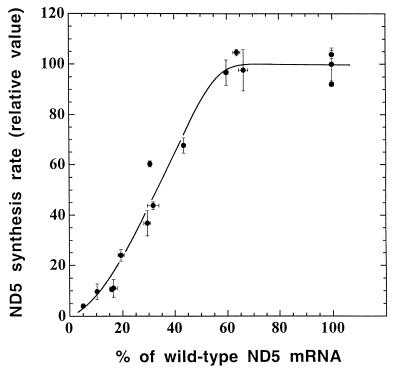
In order to investigate how the rate of ND5 protein synthesis varied in the 15 ρ° LL/2-m21 transformants and ethidium bromide-derived subclones as a function of the amount of wild-type ND5 mRNA, the ND5 labeling data of the various samples analyzed in the experiments shown in Fig. 2a were normalized for variation in overall mitochondrial protein labeling associated with differences in lane loading or other experimental factors. As shown in Fig. 5, where the normalized rates of ND5 synthesis in the various transformants and ethidium bromide-derived clones are expressed relative to the reference 3A3 transformant rate and plotted versus the percentage of wild-type ND5 mRNA, a threshold phenomenon was observed. It appears, in fact, that the relative rate of ND5 synthesis starts declining below the control rate when the percentage of wild-type ND5 mRNA, which is equivalent to the proportion of wild-type ND5 genes (Fig. 4b), becomes lower than 60% of the control. This means that 60% of the normal level of functional ND5 mRNA is adequate to maintain a normal rate of ND5 protein synthesis. The same threshold was observed when the rates of ND5 protein synthesis were normalized relative to the labeling of the CYTb and/or ND2 band (data not shown).
FIG. 5.
Relationship between the ND5 synthesis rate, expressed relative to the rate in the 3A3 transformant, and the proportion of wild-type ND5 mRNA in the ρ° LL/2-m21 transformants and ethidium bromide-derived 3A20 subclones. The individual values for the rate of ND5 protein labeling, as determined by laser densitometry of appropriately exposed fluorograms, were normalized to the overall protein labeling. Two independent labeling experiments and two electrophoretic analyses of the mitochondrial translation products after each labeling were performed for each cell line. Almost identical curves were obtained by measuring the intensities of the ND5 bands by phosphorimager analysis and/or normalizing the data to the intensities of the CYTb and/or ND2 band (not shown). Symbols are as defined for Fig. 4.
Dependence of a functional complex I assembly on ND5 protein synthesis.
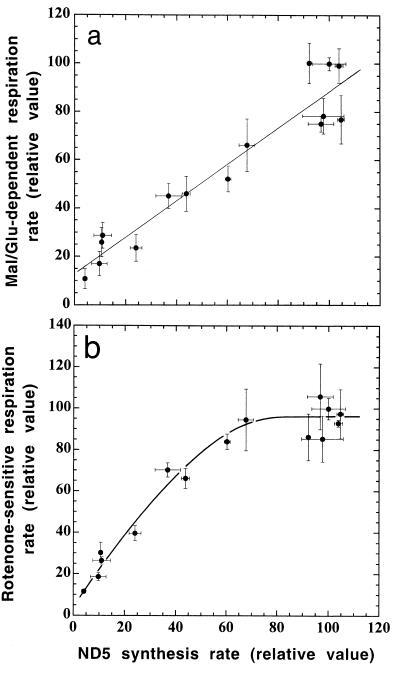
The evidence obtained previously (25) and in the present work has clearly indicated that the ND5 subunit is essential for complex I activity in human cells and mouse cells. In order to investigate the control that the synthesis of the ND5 subunit plays on the assembly of a functional NADH dehydrogenase, the malate-glutamate-dependent respiration rates determined in digitonin-permeabilized transformants and ethidium bromide-derived subclones (Fig. 1b) and expressed relative to the 3A3 reference value were plotted versus the corresponding rates of ND5 subunit synthesis, as determined in the experiments of Fig. 2a. The latter values had been normalized to the corresponding overall mitochondrial protein synthesis rates and also expressed relative to the 3A3 reference value. It appears that, with decreasing protein synthesis rate, the malate-glutamate-dependent respiration rate decreases nearly in parallel (Fig. 6a). This surprising result clearly indicates that there is very little excess of ND5 protein synthesis capacity over that required to maintain a normal rate of assembly of a functional complex I.
FIG. 6.
Relationship between rate of malate-glutamate-dependent respiration (a) or rate of rotenone-sensitive respiration (b), as expressed relative to the rate in the 3A3 transformant, and relative ND5 synthesis rate in the ρ° LL/2-m21 transformants and ethidium bromide-derived 3A20 subclones. Symbols are as defined for Fig. 4.
In order to have an estimate of the control that the ND5 synthesis rate plays on complex I activity in intact cells, the rotenone-sensitive endogenous respiration rates of the various cell lines, which reflect the contribution of complex I to the total respiration, were plotted versus the corresponding rates of ND5 synthesis. The proportions of the endogenous respiration rate which was rotenone sensitive had been previously determined to be ∼85, ∼67, and ∼50% in the cell lines which had, respectively, 30 to 100, ∼10, and ∼5% wild-type mtDNA (data not shown). As shown in Fig. 6b, the rotenone-sensitive respiration rates, expressed relative to the 3A3 reference value, remain fairly constant with a decreasing rate of ND5 synthesis, until this value reaches ca. 80% of the control value, and then declines progressively to nearly zero.
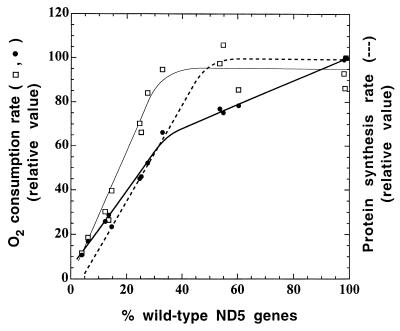
To obtain an estimate of the threshold for the capacity of ND5 genes to support respiration, the malate-glutamate-dependent and rotenone-sensitive respiration rates, expressed relative to the 3A3 reference values, were plotted against the percentage of wild-type genes in the transformant and ethidium bromide-derived cell lines analyzed in the present work. Figure 7 shows the minimum number of wild-type ND5 genes required to maintain a normal rate of rotenone-sensitive respiration or malate-glutamate-dependent respiration. The difference in behavior between the two parameters is discussed below.
FIG. 7.
Relationship between malate-glutamate-dependent respiration rate (●) or rotenone-sensitive respiration rate (□), as expressed relative to the rate in the 3A3 transformant, and proportion of wild-type ND5 genes in the ρ° LL/2-m21 transformants and ethidium bromide-derived 3A20 subclones. For comparison, the relative rate of ND5 protein synthesis (dashed line) is also shown.
DISCUSSION
In the present work, the structural alteration of the ND5 gene in 3A, a derivative of the mouse cell line A9, was found to be a nonsense mutation converting an arginine codon to the mitochondrial stop codon AGA. This is in contrast to the other complex I-defective mutants previously isolated by the rotenone resistance method (5, 24, 25), which exhibited frameshifts corresponding to homopolymeric tracts, presumably due to stuttering of the γ DNA polymerase. As discussed for these mutations, also the 12081 mutation analyzed here is likely to have preexisted in the A9 cell population and to have then been selected for by a combination of a replicative advantage of the mutant mtDNA molecules (33, 52) and of a progressive adaptation to an exclusively glycolytic energy metabolism. This adaptation would have made the cells complex I independent and rotenone insensitive (25). No mutation was detected in any of the other mtDNA genes encoding subunits of NADH dehydrogenase in which disease-causing mutations have been previously identified (ND1, ND4, and ND6) (47). Furthermore, in clone 3A and in the ρ° LL/2-m21 transformants and ethidium bromide-derived subclones, no change was observed in the rate or pattern of mitochondrial translation other than the decrease in the rate of synthesis of the ND5 subunit. These observations clearly indicated that the ND5 mutation detected here was responsible for the physiological changes observed in these cell lines. Further support for this conclusion came from the close similarity in phenotype of the mouse ND5 mutant analyzed here and from the human ND5 frameshift mutant investigated earlier (25). Particularly important is the confirmation of the previous finding that the ND5 subunit is essential for the activity of complex I but is not required for the assembly of the other mtDNA-encoded subunits of the complex (25).
Transfer of mitochondria from clone 3A to the mtDNA-less ρ° LL/2-m21 generated a large set of transformants which exhibited a proportion of mutant mtDNA varying between 0 and 85% compared with the 20% mutant mtDNA proportion of the original clone 3A. This large heterogeneity in mutation content among the transformants presumably reflected mainly the original intercellular variation in the 3A cell population. A surprising finding in the analysis of the transformants and the ethidium bromide-derived subclones was a general increase in the rates of both succinate–G-3-P-dependent and TMPD-ascorbate-dependent respiration, which showed no correlation with the decrease in malate-glutamate-dependent respiration or an increase in mtDNA content. This phenomenon presumably reflected a better functionality of complex III and complex IV carrying 3A mtDNA-encoded subunits in the LL/2 nuclear background. These observations illustrate clearly the influence of the nuclear background on the biochemical phenotype of the transmitochondrial cell lines, with obvious implications for future therapeutic approaches based on mitochondria transfer.
Lack of upregulation of transcription and mRNA stabilization with decreasing wild-type ND5 gene copy number.
The present work has yielded the surprising findings that, in all transformants and ethidium bromide-derived subclones, the total ND5 mRNA had an almost constant level, except for a significant decrease (35 to 40%) in two transformants nearly homoplasmic for the mutation (Fig. 4a), and that the proportions of wild-type ND5 gene and wild-type ND5 mRNA were almost identical in all cell lines (Fig. 4b). These observations have clearly pointed to the lack of a compensatory upregulation of transcription of the ND5 gene and/or to an increase in stability of the ND5 mRNA under conditions where the decrease in wild-type ND5 gene copy number caused a significant decrease in rates of ND5 synthesis and of complex I-dependent respiration. In particular, the absence of evidence of any increase in metabolic stability of the ND5 mRNA stands in striking contrast to previous observations indicating a pronounced stabilization of the mitochondrial mRNAs in African green monkey cells (14) and in mouse cells (32) under conditions where mtDNA transcription was severely blocked. Since, in the present experiments, no evidence of transcription inhibition was found, one would have to conclude that either the increase in mitochondrial mRNA stability in mammalian cells is a compensatory phenomenon intimately connected with a block in transcription activity or that the ND5 mRNA escapes this regulation. Furthermore, the near identity of the mutant-to-wild-type ratios in the ND5 genes and the ND5 mRNA indicated that nonsense mutation-activated degradation of mRNA, which has been well established for cytoplasmically translated mRNAs in eukaryotic cells (37), does not operate effectively in the case of the mitochondrial ND5 mRNA.
ND5 mRNA in wild-type cells is in significant excess over the requirement for normal protein synthesis but lacks the capacity of translational upregulation.
The observation that transformants carrying ≥60% of the wild-type ND5 mRNA level were capable of carrying out a normal rate of ND5 synthesis (Fig. 5) indicated clearly that, in wild-type cells, ∼40% of the ND5 mRNA is in excess of the level required for the maximal rate of ND5 synthesis. This finding should be correlated with previous observations indicating that, in cultured human cells, the amounts of different mitochondrial tRNAs investigated are in two- to threefold excess of the levels required to support a normal rate of protein synthesis (15, 22, 23). This suggests that the rate of assembly of the mitochondrial translation apparatus exceeds to a significant extent the requirements for normal protein synthesis. On the other hand, the observation made in the present work that, when the concentration of functional ND5 mRNA falls below the thresholds required for normal malate-glutamate-dependent or rotenone-sensitive respiration, the mitochondrial protein synthesis rate continues to decline progressively to zero with decreasing mRNA levels indicates that the cells investigated here have no capacity to upregulate translation of this mRNA. It is significant, in this connection, to mention the previous observations that, among the mtDNA-encoded proteins synthesized in isolated rat brain synaptosomes (35) and in isolated rat quadriceps muscle fibers (4), the ND5 protein was apparently specifically missing or strongly underrepresented, despite the presence of a normal level of its mRNA (35). These findings may point to the occurrence of an ND5 translation factor(s) with tissue-specific regulation. In Saccharomyces cerevisiae, several nucleus-encoded mitochondrial mRNA-specific translational activators have been identified that may play a rate-limiting role in modulating mitochondrial gene expression in response to environmental conditions (18).
The ND5 synthesis rate is rate limiting for complex I-dependent respiration.
The most striking observation of the present analysis has been that the rate of ND5 protein synthesis in ρ° LL/2-m21 transformants and ethidium bromide-derived cell lines is rate limiting, or nearly so, for the assembly of a functional complex I. A difference was observed between malate-glutamate-dependent respiration and rotenone-sensitive respiration, as concerns the rate of ND5 protein synthesis required to support full respiration, which was found to correspond to 100 and ∼80%, respectively, of the mitochondrial synthetic capacity for this protein. This difference is presumably accounted for by the fact that the malate-glutamate-dependent respiration rate in permeabilized cells measures the maximum capacity of complex I-dependent respiration under conditions where NADH and ubiquinone are not limiting. By contrast, the rotenone-sensitive respiration rate in intact cells gives an estimate of the contribution of complex I to the endogenous respiration in various experimental situations. It is reasonable to expect that, in different in vivo situations, the ND5 protein synthesis rate required to support full endogenous respiration may vary and may reach, under certain conditions, the maximum synthetic capacity for this protein. Considering that the malate-glutamate-dependent respiration is usually the rate-limiting step in respiration (27, 45), the surprising conclusion of these experiments is that the synthesis of ND5 is nearly rate limiting for respiration. Whether this conclusion also applies to the synthesis of other mtDNA-encoded subunits of NADH dehydrogenase remains to be determined.
Implications for general mitochondrial function control and for mitochondrial diseases.
The picture of the regulation of ND5 gene expression and its effect on respiration which has emerged from the present work is one involving very low reserve capacities at the level of transcription, translation, and respiration. This conclusion is in striking contrast to the large excess of genetic potential of the eukaryotic mitochondrial genome (3). Whether this tightness of control is unique for the ND5 gene and possibly related to its translational regulation needs to be established. However, it is pertinent to mention here that the conclusions of these studies are consistent with and extend previous findings from this laboratory. These observations had indicated that, in a considerable variety of human cell types, the COX capacity, measured in intact uncoupled cells or in digitonin-permeabilized cells in state 3, is in low excess or nearly limiting, respectively, relative to the capacity needed to support the endogenous or the glutamate-malate-dependent respiration (45, 46). In a general sense, it is reasonable to assume that an effective and fine modulation of activity of a given biochemical pathway, such as to match the different cellular needs in various physiological or developmental situations, would require a limiting level of at least one of the components of the pathway. Therefore, the tightness of control observed for ND5 gene expression in the present work may be a paradigm of what occurs in many physiological and developmental pathways.
In another context, the present observations have relevance for understanding the pathogenetic role of disease-causing mtDNA mutations that produce only a moderate decrease in the activity of a given component of the respiratory chain (26, 28, 31, 36). Furthermore, they may help elucidate the basis for the striking tissue specificity of the mutation-associated defects and for the late onset of some of these disorders. Thus, it is perfectly plausible that tissue-specific or age-related variations in the rate of mitochondrial protein synthesis, which is under the control of nuclear genes, could, by affecting the expression of a critical gene like ND5 and consequently the rate of respiration, be one of the critical factors underlying the tissue specificity and the time of appearance of the disease phenotype.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM11726 to G.A. and by Eisai Co., Ltd., Tokyo, Japan.
We are very grateful to Anne Chomyn for providing the anti-ND4L antibodies and for critical reading of the manuscript and to Gaetano Villani for helpful discussions. We also thank R. Zedan, A. Drew, and B. Keeley for expert technical assistance.
REFERENCES
- 1.Ammini C V, Hauswirth W W. Mitochondrial gene expression is regulated at the level of transcription during early embryogenesis of Xenopus laevis. J Biol Chem. 1999;274:6265–6271. doi: 10.1074/jbc.274.10.6265. [DOI] [PubMed] [Google Scholar]
- 2.Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int Rev Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 3.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 4.Attardi G, Chomyn A, Loguercio P. Evidence for translational control of mitochondrial gene expression in rat muscle and brain synaptosome mitochondria. In: Benzi G, editor. Advances in myochemistry. Vol. 2. London, England: John Libbey Eurotext, Ltd.; 1989. pp. 55–64. [Google Scholar]
- 5.Bai Y D, Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y D, Lee D, Yu T D, Chasin L A. Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 1999;27:1126–1134. doi: 10.1093/nar/27.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram J S, Janik P. Establishment of a cloned line of Lewis lung carcinoma cells adapted to cell culture. Cancer Lett. 1980;11:63–73. doi: 10.1016/0304-3835(80)90130-5. [DOI] [PubMed] [Google Scholar]
- 8.Bibb M J, Van Etten R A, Wright C T, Walberg M W, Clayton D A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 9.Cantatore P, Polosa P L, Fracasso F, Flagella Z, Gadalenta M N. Quantitation of mitochondrial RNA species during rat liver development: the concentration of cytochrome oxidase subunit I (COI) messenger RNA increases at birth. Cell Differ. 1986;19:125–132. doi: 10.1016/0045-6039(86)90069-2. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Chomyn A, Mariottini P, Cleeter M W J, Ragan C I, Matsuno-Yagi A, Hatefi Y, Doolittle R F, Attardi G. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory chain NADH dehydrogenase. Nature. 1985;314:592–597. doi: 10.1038/314592a0. [DOI] [PubMed] [Google Scholar]
- 12.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 13.Desjardins P, Frost E, Morais R. Ethidium bromide induced loss of mitochondrial DNA from primary chicken embryo fibroblasts. Mol Cell Biol. 1985;5:1163–1169. doi: 10.1128/mcb.5.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.England J, Constantino P, Attardi G. Mitochondrial RNA and protein synthesis in enucleated African green monkey cells. J Mol Biol. 1978;119:455–462. doi: 10.1016/0022-2836(78)90226-7. [DOI] [PubMed] [Google Scholar]
- 15.Enriquez J A, Chomyn A, Attardi G. mtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNALys and premature translation termination. Nat Genet. 1995;10:47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- 16.Enriquez J A, Fernandez-Silva P, Garrido-Perez N, Montoya J. Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol Cell Biol. 1999;19:657–670. doi: 10.1128/mcb.19.1.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 18.Fox T D. Translational control of endogenous and recorded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia. 1996;52:1130–1135. doi: 10.1007/BF01952112. [DOI] [PubMed] [Google Scholar]
- 19.Freshney R I. Culture of animal cells. New York, N.Y: Wiley-Liss, Inc.; 1994. [Google Scholar]
- 20.Galante Y M, Hatefi Y. Purification and molecular and enzymatic properties of mitochondrial NADH dehydrogenase. Arch Biochem Biophys. 1979;192:559–568. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- 21.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan M X, Enriquez J A, Fischel-Ghodsian N, Puranam R S, Lin C P, Maw M A, Attardi G. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi J-I, Ohta S, Kikuchi A, Takemitsu M, Goto Y-I, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofhaus G, Attardi G. Lack of assembly of mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase and loss of enzyme activity in a human cell mutant lacking the mitochondrial ND4 gene product. EMBO J. 1993;12:3043–3048. doi: 10.1002/j.1460-2075.1993.tb05973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofhaus G, Attardi G. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol Cell Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofhaus G, Johns D R, Hurko O, Attardi G, Chomyn A. Respiration and growth defects in transmitochondrial cell lines carrying the 11778 mutation associated with Leber's hereditary optic neuropathy. J Biol Chem. 1996;271:13155–13161. doi: 10.1074/jbc.271.22.13155. [DOI] [PubMed] [Google Scholar]
- 27.Hofhaus G, Shakeley R M, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 1996;264:476–483. doi: 10.1016/s0076-6879(96)64043-9. [DOI] [PubMed] [Google Scholar]
- 28.Jun A S, Trounce I A, Brown M D, Shoffner J M, Wallace D C. Use of transmitochondrial cybrids to assign a complex I defect to the mitochondrial DNA-encoded NADH dehydrogenase subunit 6 gene mutation at nucleotide pair 14459 that causes Leber hereditary optic neuropathy and dystonia. Mol Cell Biol. 1996;16:771–777. doi: 10.1128/mcb.16.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King M P, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 30.King M P. Use of ethidium bromide to manipulate ratio of mutant and wild-type mitochondrial DNA in cultured cells. Methods Enzymol. 1996;264:339–344. doi: 10.1016/s0076-6879(96)64032-4. [DOI] [PubMed] [Google Scholar]
- 31.Kuznetsov A V, Clark J F, Winkler K, Kunz W S. Increase of flux control of cytochrome c oxidase in copper-deficient mottled brindled mice. J Biol Chem. 1996;271:283–288. doi: 10.1074/jbc.271.1.283. [DOI] [PubMed] [Google Scholar]
- 32.Lansman R A, Clayton D A. Mitochondrial protein synthesis in mouse L cells: effect of selective nicking of mitochondrial DNA. J Mol Biol. 1975;99:777–793. doi: 10.1016/s0022-2836(75)80184-7. [DOI] [PubMed] [Google Scholar]
- 33.Larsson N G, Holme E, Kristiansson B, Oldfors A, Tulinius M. Progressive increase of the mutated mitochondrial DNA fraction in Kearns-Sayre syndrome. Pediatr Res. 1990;28:131–136. doi: 10.1203/00006450-199008000-00011. [DOI] [PubMed] [Google Scholar]
- 34.Littlefield J W. The inosinic acid pyrophosphorylase activity of mouse fibroblasts partially resistant to 8-azaguanine. Proc Natl Acad Sci USA. 1963;50:568–576. doi: 10.1073/pnas.50.3.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loguercio Polosa P, Attardi G. Distinctive pattern and translational control of mitochondrial protein synthesis in rat brain synaptic endings. J Biol Chem. 1991;266:10011–10017. [PubMed] [Google Scholar]
- 36.Majander A, Finel M, Savontaus M L, Nikoskelainen E, Wikstrom M. Catalytic activity of complex I in cell lines that possess replacement mutations in the ND genes in Leber's hereditary optic neuropathy. Eur J Biochem. 1996;239:201–207. doi: 10.1111/j.1432-1033.1996.0201u.x. [DOI] [PubMed] [Google Scholar]
- 37.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1988;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 38.Mariottini P, Chomyn A, Riley M, Cottrell B, Doolittle R F, Attardi G. Identification of the polypeptides encoded in the unassigned reading frame-2, frame-4, frame-4L and frame-5 of human mitochondrial DNA. Proc Natl Acad Sci USA. 1986;83:1563–1567. doi: 10.1073/pnas.83.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mariottini P, Chomyn A. Immunoprecipitation of human NADH:ubiquinone oxidoreductase and cytochrome c oxidase with single subunit-specific antibodies. Methods Enzymol. 1996;260:202–210. doi: 10.1016/0076-6879(95)60138-4. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell C H, Attardi G. Cytoplasmic transfer of chloramphenicol resistance in a human cell line. Somat Cell Genet. 1978;4:737–744. doi: 10.1007/BF01543161. [DOI] [PubMed] [Google Scholar]
- 41.Moraes C T, Ricci E, Bonilla E, DiMauro S, Schon E A. The mitochondrial tRNALeu(UUR) mutation in mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am J Hum Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
- 42.Ostronoff L K, Izquierdo J M, Cuezva J M. mt-mRNA stability regulates the expression of the mitochondrial genome during liver development. Biochem Biophys Res Commun. 1995;217:1094–1098. doi: 10.1006/bbrc.1995.2881. [DOI] [PubMed] [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storrie B, Attardi G. Expression of the mitochondrial genome in HeLa cells. XIII. Effect of selective inhibition of cytoplasmic or mitochondrial protein synthesis on mitochondrial nucleic acid synthesis. J Mol Biol. 1972;71:177–199. doi: 10.1016/0022-2836(72)90345-2. [DOI] [PubMed] [Google Scholar]
- 45.Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in wild-type and mitochondrial DNA mutation-carrying human cells. Proc Natl Acad Sci USA. 1997;94:1166–1171. doi: 10.1073/pnas.94.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J Biol Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- 47.Wallace D C, Shoffner J M, Trounce I, Brown M D, Ballinger S W, Corral-Debrinski M, Horton T, Jun A S, Lott M T. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochem Biophys Acta. 1995;127:141–151. doi: 10.1016/0925-4439(95)00021-u. [DOI] [PubMed] [Google Scholar]
- 48.Weiss H, Friedrich T, Hofhaus G, Preis D. The respiratory chain NADH dehydrogenase (complex I) of mitochondria. Eur J Biochem. 1991;197:563–576. doi: 10.1111/j.1432-1033.1991.tb15945.x. [DOI] [PubMed] [Google Scholar]
- 49.Williams R S. Mitochondrial gene expression in mammalian striated muscle: evidence that variation in gene dosage is the major regulatory event. J Biol Chem. 1986;261:12390–12394. [PubMed] [Google Scholar]
- 50.Williams R S, Salmons S, Newsholme E A, Kaufman R E, Mellor J. Regulation of nuclear and mitochondrial gene expression by contractile activity in skeletal muscle. J Biol Chem. 1986;261:376–380. [PubMed] [Google Scholar]
- 51.Wiseman A, Attardi G. Reversible ten-fold reduction in mitochondrial DNA content of human cells treated with ethidium bromide. Mol Gen Genet. 1978;167:51–63. doi: 10.1007/BF00270321. [DOI] [PubMed] [Google Scholar]
- 52.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci USA. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoneda M, Miyatake T, Attardi G. Complementation of mutant and wild-type human mitochondrial DNAs coexisting since the mutation event and lack of complementation of DNAs introduced separately into a cell within distinct organelles. Mol Cell Biol. 1994;14:2699–2712. doi: 10.1128/mcb.14.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]