Abstract
Arginine/serine-rich (RS) domain-containing proteins and their phosphorylation by specific protein kinases constitute control circuits to regulate pre-mRNA splicing and coordinate splicing with transcription in mammalian cells. We present here the finding that similar SR networks exist in Schizosaccharomyces pombe. We previously showed that Dsk1 protein, originally described as a mitotic regulator, displays high activity in phosphorylating S. pombe Prp2 protein (spU2AF59), a homologue of human U2AF65. We now demonstrate that Dsk1 also phosphorylates two recently identified fission yeast proteins with RS repeats, Srp1 and Srp2, in vitro. The phosphorylated proteins bear the same phosphoepitope found in mammalian SR proteins. Consistent with its substrate specificity, Dsk1 forms kinase-competent complexes with those proteins. Furthermore, dsk1+ gene determines the phenotype of prp2+ overexpression, providing in vivo evidence that Prp2 is a target for Dsk1. The dsk1-null mutant strain became severely sick with the additional deletion of a related kinase gene. Significantly, human SR protein-specific kinase 1 (SRPK1) complements the growth defect of the double-deletion mutant. In conjunction with the resemblance of dsk1+ and SRPK1 in sequence homology, biochemical properties, and overexpression phenotypes, the complementation result indicates that SRPK1 is a functional homologue of Dsk1. Collectively, our studies illustrate the conserved SR networks in S. pombe consisting of RS domain-containing proteins and SR protein-specific kinases and thus establish the importance of the networks in eucaryotic organisms.
Arginine/serine-rich (RS) domain-containing proteins are among the best-characterized non-snRNP proteins participating in pre-mRNA splicing (for reviews, see references 8 and 19). Members of the protein superfamily are involved in constitutive splicing and are specific modulators of alternative splicing (15, 19). Mammalian serine/arginine-rich (SR) proteins are featured by one or more RNA recognition motifs at the NH2 terminus and by an RS domain at the COOH terminus. Other RS domain-containing proteins are relatively less defined with respect to the arrangement of the two structural elements in a protein (8, 11, 19, 35).
SR proteins are heavily phosphorylated, predominantly in the RS domain (4, 5, 12, 41). Several kinases have been reported to phosphorylate RS domain-containing splicing factors (5, 12, 30, 39, 50, 53), including SR protein-specific kinase (SRPK) and Cdc28/Cdc2-like kinase (Clk/Sty). Based on studies in mammalian nuclear extracts, both phosphorylation and dephosphorylation of SR proteins are required for pre-mRNA splicing. Phosphorylation of SR proteins may promote spliceosome assembly by facilitating specific protein interactions while preventing SR proteins from binding randomly to RNA (54). Once a functional spliceosome has formed, dephosphorylation of SR proteins is necessary to allow the transesterification reaction to occur (3, 23). Recently, human type 2C Ser/Thr phosphatase PP2Cγ was reported to be required during early stages of spliceosome assembly and to be physically associated with the spliceosome in vitro (29). Therefore, the sequential phosphorylation and dephosphorylation of SR proteins may mark the transition between stages in one round of splicing reaction.
The phosphorylation state of SR proteins not only regulates their functional properties in splicing reaction but also modulates their subnuclear distribution in vivo (5, 12, 26, 50). The phosphorylation of the serine residues in the RS domain is a prerequisite for the release of splicing factors from the storage loci, nuclear speckles, to the sites of transcription and splicing, suggesting that protein phosphorylation functions as a control switch for spatially linking transcription with splicing in vivo (24). In a simplified scenario, the ability of the splicing machinery to respond to mRNA synthesis in the cell may be conferred by the differential phosphorylation of SR proteins, so that sufficient splicing factors can be recruited to the sites of transcription as gene expression is activated.
In addition to transcription, pre-mRNA splicing is closely coordinated in space and time with other nuclear events, including 5′ capping, and the 3′ processing of RNA (25). Gene expression is also synchronized with the cell division cycle, such that it is active during interphase and repressed upon entry into mitosis (9). Therefore, intricate interplay exists among pre-mRNA splicing, transcription, and cell cycle. RS domain-containing proteins and SR protein-specific kinases may constitute a protein relay or networks to regulate the coupling of splicing, transcription, and cell cycle in mammalian cells (6, 25).
The fission yeast Schizosaccharomyces pombe, as a genetically tractable system, has been widely used to investigate cell cycle control (14, 31). S. pombe also bears resemblance to mammalian systems with respect to the high content and structure of introns in protein-encoding genes (13, 36, 48). An increasing body of evidence suggests the interplay between pre-mRNA splicing and cell cycle in fission yeast. A splicing defect is coupled with a cdc phenotype at a restrictive temperature in 10 of 14 prp ts mutants identified in fission yeast, i.e., prp1, prp2, prp5 through prp8, and prp11 through prp14 (33, 45, 48, 49). Defects in nuclear division, cytokinesis, and particularly G2/M transition were observed in those 10 prp mutants. These cell cycle defects are not simply a result of malfunction in splicing since not all prp mutants impose a block on mitotic progression. Since reorganization of nuclear architecture, including splicing machinery, occurs at the onset and the exit of mitosis (25), it is possible that defects in some splicing factors may affect the proper reorganization of nuclear architecture and cell cycle progression.
Protein components similar to elements of the mammalian SR networks exist in S. pombe. First, several RS domain-containing proteins have been identified. The Prp2 protein, also named spU2AF59 due to its homology to the large subunit of human U2AF (35), is essential for pre-mRNA splicing in vivo (34, 35). Another prp2 mutant allele, mis11-453, affects chromosome segregation and leads to minichromosome loss (45). In addition to Prp2/Mis11 protein, Srp1 and Srp2 are two proteins containing RS repeats recently found in S. pombe (11). The srp2+ gene is essential for viability, while the srp1+ gene is not. Overexpression of Srp1 protein with a mutant RS domain or the RNA-binding domain alone inhibits splicing in fission yeast, suggesting a role for Srp1 in pre-mRNA splicing (11). Second, kinases that phosphorylate RS domain-containing proteins have been discovered. Dsk1 is an S. pombe protein kinase that specifically phosphorylates Prp2 in vitro (47). Although initially described as a mitotic regulator (46), Dsk1 has also been implicated in pre-mRNA splicing according to its sequence homology to human SRPK1 (12). Another protein kinase, Prp4 (38), is reported to phosphorylate human SF2/ASF protein in vitro (10).
In further investigating the kinase activity of Dsk1 and its interaction with RS domain-containing proteins, we show here for the first time that phosphorylation of S. pombe RS domain-containing proteins by Dsk1 produces the same phosphoepitope found in mammalian SR proteins. We also obtained in vivo evidence to support the kinase-substrate relationship between Dsk1 and Prp2. The dsk1-null mutant became severely sick with additional deletion of a related kinase. Significantly, human SRPK1 protein expressed in fission yeast is capable of compensating for the loss of Dsk1 in vivo. Consistent with the notion that SRPK1 is a functional homologue of Dsk1, the overexpression phenotype of SRPK1 resembles that of dsk1+ in S. pombe. Taken together, our studies document the conservation of the SR protein-specific kinases through evolution and the importance of the SR networks in eucaryotic organisms.
MATERIALS AND METHODS
S. pombe strains.
The following haploid strains of S. pombe were used: 1913 (h− leu1), B8 (h− leu1 ura4 dsk1::ura4+) (46), 2A5 (h− leu1 ura4 kic1::ura4+ his2), 2D4 (h− leu1 ura4 kic1::ura4+ dsk1::ura4+ his2). Standard genetic procedures and media for growing S. pombe strains are described elsewhere (1, 27).
Plasmid construction.
Fission yeast srp1+ gene was obtained by PCR (42) from the S. pombe cDNA library (Clontech) by using two primers complementary to the 5′ and 3′ sequence of the gene, respectively: 5′-GCGCGCGGATCCATGAGTCGCAGAAGCCTTCGT-3′, including a BamHI site, and 5′-GCCGGATAGTCGACATTAACTGTGTTACGG-3′, including a SalI site. The BamHI-SalI fragment of ∼900 bp was then inserted into pET-28a (Novagen) to generate pET-28a srp1+. To construct pET-28bGST-srp1+, a BamHI-SalI fragment was produced by PCR by using plasmid pET-28a srp1+ as a template with two primers: 5′-GGTCGGGATCCGATGAGTCGCAGAAGC-3′, including a BamHI site, and 5′-GCTTGTCGACATTAACTGTGTTACG-3′, including a SalI site. Plasmids pET-28bGST, pET-28adsk1+, and pET-28bGST-prp2+ have been described (47). Plasmid pGADGH srp2+ DNA was isolated from the S. pombe cDNA library by using srp1+ gene as bait (unpublished data). An EcoRI fragment containing the coding sequence of the srp2+ gene was ligated to vector pET-28b and pET-28bGST to produce pET-28bsrp2+ and pET-28bGSTsrp2+. To generate pREP1prp2+, a 1.4-kb NdeI-BamHI DNA fragment encoding the Prp2 protein was inserted into pREP1 vector (20, 21). The pREP1SRPK1 plasmid was constructed by inserting a SalI-BamHI fragment containing the open reading frame of human SRPK1 into the same sites of pREP1. The SRPK1 SalI-BamHI fragment was synthesized by PCR by using pcDNA3-FLAG-SRPK1 (from X. D. Fu University of California at San Diego) as template and two oligonucleotides (5′-AGCTGCCTGTCGACAATGGACTACAAAGACGAT-3′ and 5′-TGTGGGATCCCTGCTGTGGTGCTG-3′) as primers.
Production of recombinant proteins.
Recombinant proteins GST-Srp1, Srp1, GST-Srp2, Srp2, GST-Prp2, GST-SF2/ASF, and Dsk1 were expressed in Escherichia coli BL21(DE3)pLysS as described earlier (47). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, instead of 0.4 mM, to assure a full induction of a T7lac promoter (Novagen) in bacteria. Bacterial lysate preparations and histidine-tagged Dsk1 protein purification have been described (47). The relative amounts of recombinant proteins in lysates were estimated based on Coomassie blue-stained gel by using bovine serum albumin as a standard, or the intensity of protein bands was visualized on immunoblots.
GST pulldown assay.
Bacterial lysates containing glutathione S-transferase (GST) or GST fusion proteins were incubated with or without various non-GST- tagged proteins in lysates to allow complex formation at 23°C for 30 min. The mixture was then incubated with glutathione beads at 4°C for 1 h with constant agitation. After pulldown by microcentrifugation at 7,000 rpm for 1 min at room temperature, the beads were washed in TBS (10 mM Tris-HCl, pH 7.4; 150 mM NaCl) two to three times with 0.1% NP-40 and four times without NP-40. The beads were resuspended in TBS as a 50% suspension, aliquoted, frozen in liquid nitrogen, and stored at −80°C until use. All steps were performed in the presence of protease inhibitors: 5 mg of pepstatin, 5 mg of chymostatin, and 5 mg of leupeptin per ml plus 1 mM phenylmethylsulfonyl fluoride.
Kinase assay.
Purified or bead-bound Dsk1 was incubated at 23°C for 30 min with RS domain-containing proteins in bacterial lysates, purified, or bound to glutathione beads in a total volume of 20 to 60 μl in a kinase buffer (50 mM Tris-HCl, pH 7.4; 10 mM MgCl2; 1 mM dithiothreitol) with 50 μM ATP and 0.1 μCi of [γ-32P]ATP per μl. When a bead-bound protein was present in a kinase reaction, an end-to-end rotor was used to mix the sample during incubation. The kinase reaction was terminated by boiling in sodium dodecyl sulfate (SDS) sample buffer, and the samples were resolved on an SDS–10% polyacrylamide gel. Protein phosphorylation was detected by autoradiography. For Western blot analysis, the kinase reaction was performed by employing an ATP-regenerating system (10 mM creatine phosphate, 1 mM ATP, and 0.1 mg of creatine phosphokinase per ml) without radioisotopes. Immunoblotting in most experiments was performed as previously described (47). When 3C5 monoclonal antibody was used, 25 mM NaF and 1 mM NaVO3 were present as phosphatase inhibitors to prevent dephosphorylation.
Antibodies.
The anti-Dsk1 peptide polyclonal antibodies were generated and affinity purified as described earlier (47). Monoclonal antibody (MAb) 3C5 was obtained from mouse ascites and was used in a ×500 dilution. Anti-GST polyclonal antibodies were from Santa Cruz Biotechnology. Anti-T7-Tag monoclonal antibody was purchased from Novagen.
Transformation of S. pombe.
Transformation of fission yeast was accomplished by using the lithium acetate method (1) with modifications. A 3- to 5-ml culture in YES medium (27) was grown at 33°C for about 5 h with shaking at 225 rpm. A 100- to 200-ml culture was then started by adding a calculated amount of cells from the small culture so that cell density would reach 0.5 × 107 to 1.5 × 107 cells/ml overnight. Cells were harvested and resuspended at a density of approximately 109 cells/ml in 0.1 M lithium acetate in TE buffer. After 1 h of incubation at 30°C with shaking at 170 to 200 rpm, 1 μg of plasmid DNA in 15 μl of TE was mixed with 100 μl of the cell resuspension, followed by the addition of 290 μl of 50% polyethylene glycol. Samples were incubated for 1 h with occasional gentle vortexing. After heat shock at 42°C for 15 min, cells were incubated at room temperature for 10 min. Cells were then collected, washed, and resuspended in 200 μl of EMM2 (minimal medium) (1). Finally those transformed cells were spread on EMM2 plates in the presence of 2 μM thiamine and incubated at 33°C until colonies appeared.
Expression of fission yeast prp2+ gene and human SRPK1 gene in S. pombe.
The plasmid pREP1prp2+ was transformed into fission yeast wild-type (strain 1913), dsk1-null (Δdsk1), and kic1-null (Δkic1) strains. The human SRPK1 gene was introduced as plasmid pREP1SRPK1 into wild-type (strain 1913) and dsk1 kic1 double-null (Δdsk1Δkic1) strains of S. pombe. The expression of prp2+ gene and human SRPK1 gene under the control of nmt+ promoter was induced according to procedures described elsewhere (46).
DAPI staining.
Methods for DAPI (4′,6′-diamidino-2-phenylindole) staining were modified from Alfa et al. (1) and the Fission Yeast Handbook (www.bio.uva.nl/pombe/handbook/). Cells were fixed on a slide at 70°C for 1 min on a hot plate. Then, 3 to 4 μl of a freshly diluted 1× DAPI solution (1 μg of DAPI per ml, 1 mg of antifade per ml, 45% glycerol) was added to the fixed cells. Slides were kept in the dark to prevent fading before they were observed under a microscope.
RESULTS
Dsk1-mediated phosphorylation of fission yeast Srp1, Srp2, and Prp2 proteins generates the same phosphoepitope as in mammalian SR proteins.
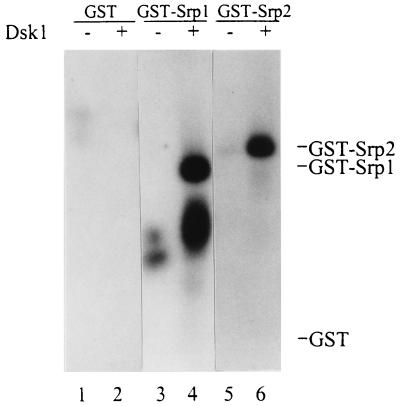
We showed previously that Dsk1 protein specifically phosphorylates fission yeast Prp2/Mis11, a U2AF65 homologue, in vitro (47). To extend our studies of the SR networks in S. pombe we investigated whether Srp1 and Srp2 proteins are also substrates for Dsk1 in vitro. Full-length Srp1 and Srp2 proteins fused at the NH2 terminus to GST, designated GST-Srp1 and GST-Srp2, were isolated on glutathione-agarose beads and incubated with or without purified Dsk1 in the presence of [γ-32P]ATP. As shown in Fig. 1, 32P-labeled proteins with apparent molecular sizes of ∼56 kDa (lane 4) and ∼66 kDa (lane 6) were detected, matching the predicted sizes of GST-Srp1 and GST-Srp2 proteins, respectively. These bands were not detected in the samples without Dsk1 protein (Fig. 1, lanes 3 and 5). The lower-molecular-size band observed in lane 4 of Fig. 1 was probably a degradation product of GST-Srp1. The GST portion of the fusion proteins did not contribute to the phosphorylation by Dsk1, since GST alone was not phosphorylated by Dsk1 (Fig. 1, lane 2). Therefore, in addition to Prp2, Dsk1 phosphorylates Srp1 and Srp2 proteins in vitro.
FIG. 1.
Dsk1 phosphorylates fission yeast Srp1 and Srp2 proteins in vitro. GST fusion proteins were isolated from bacterial lysates by binding to glutathione beads. After a washing, the bound GST (lanes 1 and 2), GST-Srp1 (lanes 3 and 4), and GST-Srp2 (lanes 5 and 6) were individually incubated with purified Dsk1 (lanes 2, 4 and 6) in the presence of [γ-32P]ATP at 23°C for 30 min. Samples were resolved on an SDS–10% polyacrylamide gel and visualized with X-ray film. The expected positions of GST, GST-Srp1, and GST-Srp2 proteins on the gel are indicated on the right. Truncated forms of GST-Srp1 protein were observed as a lower-molecular-size band (lane 4).
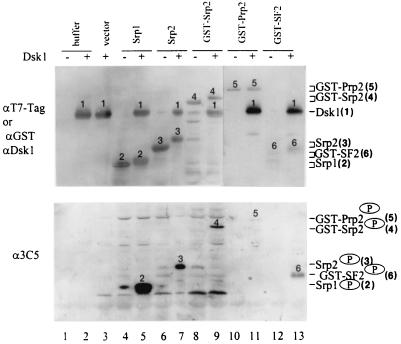
To assess the specificity of phosphorylation, we probed the Dsk1-phosphorylated proteins with SR protein-specific MAbs. Mammalian SR proteins share common phosphoepitopes, which specifically react to two MAbs, MAb 104 (40) and MAb 3C5 (2). Since MAb 3C5 is more sensitive and specific for detecting phosphorylated SR proteins than MAb 104 in some studies (2), we included MAb 3C5 in our experiments. Bacterial lysates containing recombinant Srp1, Srp2, or GST-Srp2 were incubated with purified Dsk1 protein in the presence of an ATP regenerating system. Samples were split, resolved on SDS–10% polyacrylamide gels, and transferred to Immobilon membrane to generate duplicate blots. One blot was used to monitor the amount of the recombinant proteins in each sample (Fig. 2, top panel), while the other blot was probed with MAb 3C5 for the phosphorylation of those proteins (bottom panel). Srp1, Srp2, and GST-Srp2, as well as Dsk1, were detected by anti-T7-Tag MAb, since they possessed a T7-Tag (Fig. 2, top panel, lanes 2 to 9). The mobility of Srp1, Srp2, and GST-Srp2 observed in samples with Dsk1 was slower (Fig. 2, top panel, lanes 5, 7, and 9) than that of those same proteins in samples without Dsk1 (lanes 4, 6, and 8), suggesting that the proteins were phosphorylated. Consistent with the mobility changes the slower-migrating bands were recognized by MAb 3C5 (Fig. 2, bottom panel, lanes 5, 7, and 9). Thus, phosphorylation of Srp1 and Srp2 by Dsk1 produced 3C5-reactive epitope, regardless of whether the substrate was fused with GST or not. Purified GST-Prp2 and GST-SF2/ASF were analyzed similarly (Fig. 2, top panel, lanes 10 to 13). Both proteins were detected by anti-GST polyclonal antibodies (Fig. 2, top panel, lanes 10 to 13), and after phosphorylation by Dsk1, they were recognized by MAb 3C5 (bottom panel, lanes 11 and 13). In these samples Dsk1 protein was monitored by anti-Dsk1 polyclonal antibodies (top panel, lanes 11 and 13). As shown in this experiment, all three RS domain-containing proteins, Srp1, Srp2, and Prp2, were recognized by MAb 3C5 after Dsk1 action, reflecting a general feature of Dsk1-mediated phosphorylation. The weaker signal of Prp2 is likely due to the lower amount of Prp2 protein present in the reaction mixture as well as the presence of fewer RS repeats in Prp2 than in the other proteins. The results provide the first biochemical evidence that fission yeast RS domain-containing proteins phosphorylated by Dsk1 share the same phosphoepitope with the mammalian SR proteins. Therefore, Dsk1 behaves similarly to its mammalian counterparts at the molecular level. The conserved phosphorylation of RS domain-containing proteins from distinctive organisms implicates its importance in eucaryotic systems.
FIG. 2.
Dsk1-mediated phosphorylation of Srp1, Srp2, and Prp2 proteins generates a phosphoepitope specifically recognized by MAb 3C5. Purified GST-Prp2 (lanes 10 and 11), purified GST-SF2/ASF (lanes 12 and 13), or a bacterial lysate containing individual recombinant proteins (lanes 4 to 9) as indicated at the top of each lane was incubated with (lanes 5, 7, 9, 11, and 13) or without (lanes, 4, 6, 8, 10, and 12) purified Dsk1 protein in the presence of an ATP regenerating system for 30 min at 23°C. Buffer (lanes 1 and 2) and lysate from bacteria with the pET28a vector alone (lane 3) were used as negative controls. The samples were then processed for immunoblotting with anti-T7-Tag MAb (top panel, lanes 1 to 9) or anti-GST and anti-Dsk1 polyclonal antibodies in successive order (top panel, lanes 10 to 13) or MAb 3C5 monoclonal antibody (bottom panel). Alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit (top panel) or goat anti-mouse (bottom panel) IgM antibodies were used as secondary antibodies. The identity of the proteins is marked above each band with numbers 1 to 6 representing Dsk1, Srp1, Srp2, GST-Srp2, GST-Prp2, and GST-SF2, respectively, as indicated on the right side of the figure. The same amount of Srp1 and Srp2 was used, while GST-Srp2 at 1/4 of the amount and GST-Prp2 and GST-SF2/ASF at <1/10 of the amount were added to the indicated samples.
Dsk1 forms kinase-competent complexes with RS domain-containing proteins.
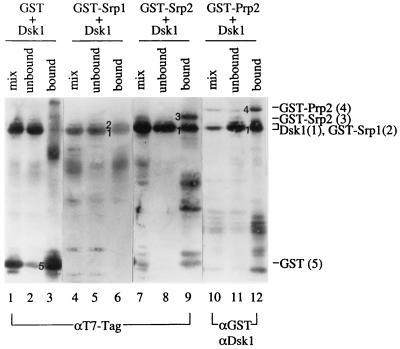
As kinase-substrate pairs, physical association of Dsk1 with the RS domain-containing proteins must take place during the phosphorylation process. If these interactions are stable, Dsk1 protein should coisolate with GST fusion substrates in a GST pulldown assay. Lysates containing similar amounts of GST fusion proteins were incubated with a lysate containing Dsk1 protein. Glutathione-agarose beads were then added to bind the GST fusions. Portions of the mixed lysates, unbound fractions, and bound fractions from each sample were analyzed by Western blots by using anti-T7-Tag MAb (for detecting GST-Srp1, GST-Srp2, and Dsk1), anti-GST (for detecting GST-Prp2), and anti-Dsk1 polyclonal antibodies (Fig. 3). The different appearance of protein bands in lanes 10 to 12 from that in lanes 1 to 9 may reflect the different sensitivities of the antibodies used in these experiments. As displayed in Fig. 3, Dsk1 protein was brought down with GST-Srp1, GST-Srp2, or GST-Prp2 (Fig. 3, lanes 6, 9, and 12). GST-Srp1 protein has a molecular mass very similar to that of Dsk1 and was distinguished from Dsk1 as a protein with a slightly slower mobility on the gel (Fig. 3, lane 6). The interaction observed was specific between Dsk1 and the yeast RS domain-containing proteins, since Dsk1 did not bind GST in this assay (Fig. 3, lane 3). Therefore, Dsk1 protein forms a complex with its substrates.
FIG. 3.
Srp1, Srp2, and Prp2 proteins individually form a complex with Dsk1 in vitro. A bacterial lysate containing Dsk1 protein was incubated with a lysate containing GST (lanes 1 to 3) or GST fusion (lanes 4 to 12) proteins as indicated at the top of each lane to allow complex formation at 23°C for 30 min. Glutathione beads were then added to pulldown bound proteins at 4°C as described in Materials and Methods. Portions of mixed lysates, unbound fractions, and bound fractions from each sample were analyzed by SDS-polyacrylamide gel electrophoresis. Some samples were processed for immunoblotting by using anti-T7-Tag MAb (lanes 1 to 9), which detects GST, GST-Srp1, GST-Srp2, and Dsk1. Other samples were processed for immunoblotting first with anti-GST and subsequently with anti-Dsk1 polyclonal antibodies (lanes 10 to 12). Dsk1 protein was pulled down by each of the four RS domain-containing proteins (lanes 6, 9, and 12) but not by GST protein (lane 3). Numbers 1 to 5 on the left of the protein bands in the bound fraction of each sample represent Dsk1, GST-Srp1, GST-Srp2, GST-Prp2, and GST, respectively, as indicated on the right side of the figure.
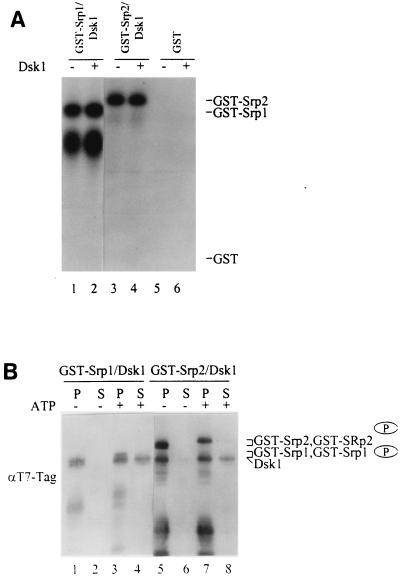
We next examined the kinase activity of the bound Dsk1 on its associated substrates. The GST-Srp1/Dsk1 and GST-Srp2/Dsk1 complexes isolated by the GST pulldown procedure were incubated with [γ-32P]ATP (Fig. 4A). GST-Srp1 (Fig. 4A, lane 1) or GST-Srp2 (lane 3) protein became phosphorylated in the complex containing Dsk1, whereas no phosphorylation was detected in the GST control (lane 5). Dsk1 protein added exogenously did not substantially increase the phosphorylation of GST-Srp1 (Fig. 4A, compare lanes 1 and 2) or GST-Srp2 (compare lanes 3 and 4). The result indicates that the bound Dsk1 protein was active and sufficient to phosphorylate the substrate in each complex. Therefore, Dsk1 binds Srp1 or Srp2 protein in a kinase-competent conformation.
FIG. 4.
Dsk1 is dissociated from the complex after phosphorylation of Srp1 or Srp2 protein. (A) The bound Dsk1 phosphorylates Srp1 and Srp2 in the complex in the presence of ATP. The pulldown complexes GST-Srp1/Dsk1 (lanes 1 and 2) and GST-Srp2/Dsk1 (lanes 3 and 4), as described in Fig. 3, were incubated with (lanes 2 and 4) or without (lanes 1 and 3) purified Dsk1 protein in the presence of [γ-32P]ATP. GST protein was also used in place of the GST fusion proteins as a negative control (lanes 5 and 6). Samples were resolved on an SDS–10% polyacrylamide gel and visualized by autoradiography. The bound Dsk1 phosphorylated Srp1 and Srp2 in the complex (lanes 1 and 3). (B) After the kinase reaction, Dsk1 is released from the Srp1/Dsk1 and Srp2/Dsk1 complexes. GST-Srp1/Dsk1 and GST-Srp2/Dsk1 protein complexes were incubated individually with (lanes 3, 4, 7, and 8) or without (lanes 1, 2, 5, and 6) an ATP regenerating system for 30 min at 23°C. Following the kinase reaction, protein-bound beads were pelleted by centrifugation. The supernatant (S) and bead (P) portions of each sample were resolved on an SDS–10% polyacrylamide gel and subsequently processed for immunoblotting with anti-T7-Tag MAb. Dsk1 was released from the complex to the supernatant in the presence of ATP (lanes 4 and 8), but it is not dissociated from the complex in the absence of ATP (lanes 2 and 6). Note the phosphorylated Srp proteins (indicated with a circled P) have slower mobility than that of their nonphosphorylated forms.
Does the binding of Dsk1 with its substrates change upon phosphorylation by Dsk1? To address this question, GST-Srp1/Dsk1 and GST-Srp2/Dsk1 protein complexes bound to glutathione-agarose beads were incubated with an ATP regenerating system. Following the kinase reaction, samples were centrifuged to pellet the beads, and proteins released from the complexes should be retained in the supernatant. Both the bound (P) and the released (S) fractions were analyzed by immunoblotting (Fig. 4B). Dsk1 was released from the complex to the supernatant after incubation with ATP (Fig. 4B, lanes 4 and 8), while no Dsk1 was released in the absence of ATP (Fig. 4B, lanes 2 and 6). Note that GST-Srp1 (Fig. 4B, lane 3) and GST-Srp2 (lane 7) migrated more slowly on the gel upon phosphorylation. Therefore, Dsk1 was dissociated from the complex after phosphorylating the GST-Srp1 or GST-Srp2 protein. Some Dsk1 protein was retained in the pellet fraction of the samples with ATP (Fig. 4B, lanes 3 and 7). This may be due to trapping of some released Dsk1 molecules in the pellet fraction since, once separated from the supernatant, the beads were not washed following the kinase reaction. Based on these results the Dsk1 reaction is dissected into three distinct steps: substrate binding, substrate phosphorylation, and release of the kinase from the product. Quantitative measurement for the percentage and rate of Dsk1 release from the complex has not been carried out.
Genetic interaction between prp2+ and dsk1+.
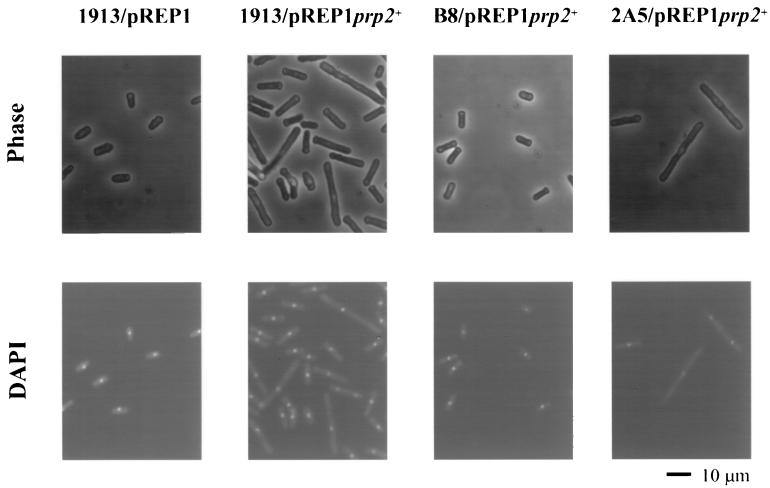
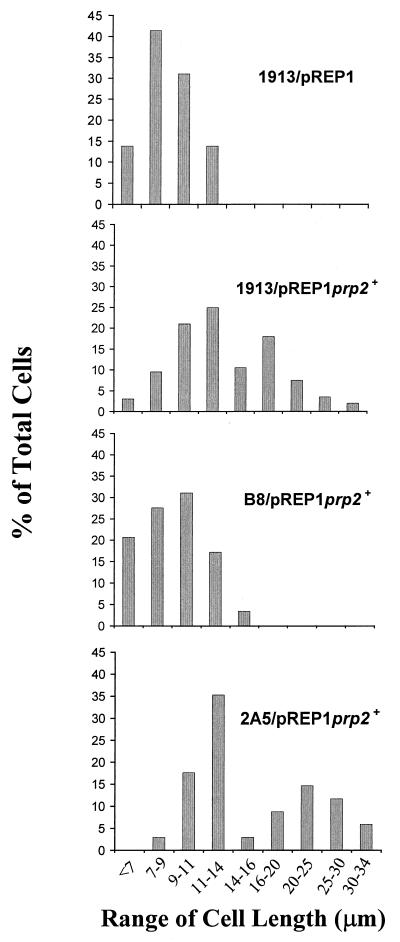
To understand the biological functions of Dsk1 protein kinase, it is necessary to investigate interactions of Dsk1 with the RS domain-containing proteins in vivo. For example, if Prp2 protein is an in vivo target of Dsk1, overexpression of prp2+ may confer a phenotype, which is only apparent in the strain with the dsk1+ gene. To test this, we placed prp2+ gene under the control of a thiamine-repressible nmt1+ (no message in thiamine) promoter of S. pombe (20, 21), so that Prp2 protein could be produced at a high level by growing cells in medium without thiamine. Consistent with a recent report (37), induction of prp2+ expression from the nmt1+-driven plasmid, pREP1prp2+, in wild-type cells leads to smaller colonies than those transformed with the vector pREP1 alone (data not shown). Exponentially growing cells in liquid culture were transferred to thiamine-depleted medium and grown for 21 h. The cells were then stained with DAPI and examined by phase-contrast (Fig. 5, top panels) and fluorescence (bottom panels) microscopy. Elongated cells were observed when the expression of the plasmid-borne prp2+ gene was induced in the wild-type strain 1913; the average cell length increased about 60% (from 8.8 to 14.2 μm) compared to that of the cells harboring the vector pREP1 (Table 1). In addition, more than 40% of the cells had a cell length exceeding the regular range for 1913/pREP1 cells (Fig. 6). Although multiple nuclei were observed in some cells, many elongated cells seemed to have a single nucleus (Fig. 5, second column, bottom panel). In contrast, overexpression of prp2+ gene in a dsk1-null strain (Δdsk1), B8, did not display any elongation phenotype (Fig. 5, third column) under the same condition; the average size of the cells (9.2 μm) remained similar to that of the wild-type strain, i.e., 1913/pREP1 (8.8 μm) (Table 1). Moreover, the “elongated” population as seen in 1913/pREP1prp2+ disappeared in strain B8/pREP1prp2+ (Fig. 6). Therefore, the elongation characteristic of prp2+ overexpression requires the presence of dsk1+ gene.
FIG. 5.
The cell elongation phenotype resulting from Prp2 overproduction is dependent on the dsk1+ gene. Strains 1913 (wild type), B8 (Δdsk1), and 2A5 (Δkic1) were transformed with pREP1prp2+. Strain 1913 containing pREP1 vector was also generated as a negative control. Cells were first grown at 32°C to midlogarithmic phase in minimal medium (EMM2) with thiamine, and Prp2 overproduction was then induced for 21 h in the absence of thiamine. Cells were fixed by heating them on slides, and they were then stained with DAPI. Cell images obtained by phase-contrast (top panel) and fluorescence (bottom panel) microscopy were indicated. Magnification is ×400 in all panels. The elongated cells were observed in strain 1913 (wild type, second column) and 2A5 (Δkic1, fourth column) but not in strain B8 (Δdsk1, third column). The scale bar represents 10 μm.
TABLE 1.
Measurements of cell length
| Strain | No. of cells | Avg (μm) (range) |
|---|---|---|
| 1913/pREP1 | 29 | 8.82 (5.70–13.57) |
| 1913/pREP1prp2+ | 200 | 14.21 (5.28–32.56) |
| B8/pREP1prp2+ | 29 | 9.16 (5.44–15.60) |
| 2A5/pREP1prp2+ | 34 | 16.81 (7.01–33.20) |
FIG. 6.
Size distribution of cell population in strains with prp2+ gene overexpressed. The cell length of the four samples in Fig. 5 was measured. The cell populations with a size range as indicated for 1913/pREP1, 1913/pREP1prp2+, B8/pREP1prp2+, and 2A5/pREP1prp2+ are displayed as histograms. A population of cells longer than 16 μm was observed in 1913/pREP1prp2+ and 2A5/pREP1prp2+. The distribution pattern of 2A5/pREP1prp2+ is similar to that 1913/pREP1prp2+, while the pattern of B8/pREP1prp2+ resembles that of the negative control, 1913/pREP1.
To address the specificity of the genetic interaction between dsk1+ and prp2+, we examined the prp2+ overexpression phenotype in another kinase-deletion strain. The S. pombe katb+ gene (GenBank accession number Q10156) encodes a protein closely related in sequence to mammalian Clk/Sty. Interestingly, overexpression of the katb+ gene in S. pombe leads to branched cells with multiple septa and nuclei, which was different from the phenotype conferred by dsk1+ overexpression (unpublished data). Since the name katb+ is not conventional nomenclature for a S. pombe gene, we changed it to kic1+ for “kinase in Clk” family. The kic1+ gene was disrupted, and a haploid strain with a null allele was found to be viable (unpublished data). The pREP1prp2+ plasmid was transformed into a kic1-null mutant strain (Δkic1), 2A5. Similar to wild type, overexpression of prp2+ gene in the kic1-null mutant strain, 2A5 (Δkic1), resulted in elongated cells (Fig. 5, fourth column). The average size of the cells in 2A5/pREP1 prp2+ was 16.8 μm, increased approximately 90% compared to 8.8 μm in 1913/pREP1 (Table 1). An “elongated” population representing more than 40% of the cells was again observed (Fig. 6). Thus, cell elongation caused by Prp2 overproduction is specifically dependent on the presence of the dsk1+ gene but does not require the kic1+ gene. These in vivo results substantially support the notion that Prp2 protein is a target of Dsk1 action in fission yeast and reinforce the in vitro data demonstrating the binding of the two proteins and phosphorylation of Prp2 protein by Dsk1.
The prp2+ overexpression phenotype in strains 1913 and 2A5 displayed two distinct populations, one with normal length distribution and the other elongated. This is perhaps due to the leakiness of the prp2+ overexpression phenotype. This is consistent with the observation that overexpressing prp2+ did not kill the cell but instead produced smaller colonies. The dual population phenomenon indicates that the prp2+ overexpression may block cell cycle progression only part of the time. Alternatively or additionally, it suggests that the prp2+ overexpression may affect multiple steps of the cell cycle. Since plasmid in S. pombe is not stable, cells that lost the pREP1-prp2+ plasmid might also contribute to the population with apparently normal cell sizes. Finally, because the prp2+ overexpression was induced in an asynchronous cell population, a portion of the cells may already pass the elongation phase and resume the normal cell cycle.
Human SRPK1 protein is a functional homologue of fission yeast Dsk1 protein in vivo.
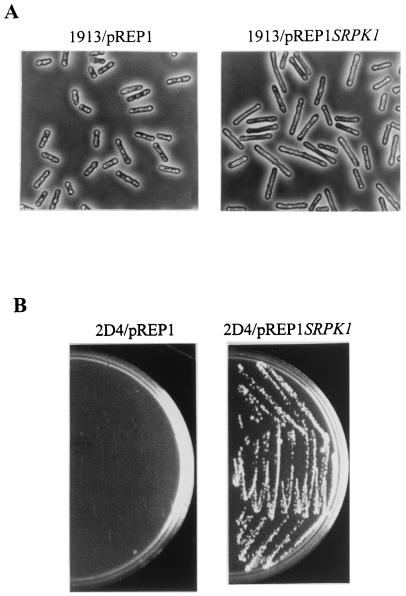
Sequence analysis and kinase assays indicate that S. pombe Dsk1 is homologous to human SRPK1 (12). We here tested the functional similarity between Dsk1 and SRPK1 in vivo. When the dsk1+ gene is overexpressed, it results in highly elongated cells with a delay in the progression from G2 to M phase (46). Thus, we first examined whether overexpression of the human SRPK1 gene in fission yeast would produce a phenotype similar to that of dsk1+ overexpression. Plasmid pREP1SRPK1 was introduced into a wild-type S. pombe strain, and the expression of the SRPK1 gene was induced in the absence of thiamine. The majority of the SRPK1-overproducing cells became elongated (Fig. 7A, right panel) compared to cells containing only the pREP1 vector (left panel). Therefore, like dsk1+ overexpression, overexpression of the human SRPK1 gene in S. pombe leads to elongated cells that are indicative of a delay at the G2/M-phase transition.
FIG. 7.
Human SRPK1 is a functional homologue of fission yeast Dsk1. (A) Overexpression of human SRPK1 gene in S. pombe results in elongated cells similar to the cells with dsk1+ overexpression. Strain 1913 (wild type) was transformed with either pREP1 or pREP1SRPK1. Exponentially grown cells were induced for 16 to 18 h in the absence of thiamine and fixed for microscopy. Left panel and right panel show at a magnification of ×400 the phase-contrast micrographs of cells harboring pREP1 and pREP1SRPK1, respectively. Fission yeast cells with human SRPK1 gene overexpressed were elongated (right panel) compared to those carrying pREP1 vector alone (left panel) under the same condition. (B) Expression of human SRPK1 gene complements the growth defect of Δdsk1Δkic1 double-deletion strain (2D4). Strain 2D4 (Δdsk1Δkic1) was transformed with either pREP1 or pREP1SRPK1. The transformants were subsequently analyzed on minimal medium plates in the presence of thiamine and incubated for 4 days at 33°C. Cells carrying pREP1SRPK1 formed healthy colonies (right panel), whereas cells harboring pREP1 hardly grew (left panel) under the same condition.
One stringent evaluation for functional homology is complementation of the loss of one gene by another gene. We anticipated that if human SRPK1 is a true functional homologue of Dsk1, it should compensate for the loss of Dsk1 in the cell. The genetic complementation test had not been accomplished because dsk1+ is not essential for the viability of the cell. Interestingly, dsk1-null mutant yeast cells became very sick when a related kinase gene in the cell, kic1+, was also disrupted. With the double deletions, cells grew extremely slowly and formed microcolonies (see Fig. 7B). Taking advantage of this recent finding, we transformed the dsk1 kic1 double-null mutant, 2D4 (Δdsk1Δkic1), with either pREP1 or pREP1SRPK1. Since the nmt1+ promoter is leaky, a considerable amount of expression occurs even in the presence of thiamine (28). Transformants were first selected and subsequently restreaked for growth analysis on thiamine-containing plates (repressed condition). Cells carrying pREP1SRPK1 formed healthy colonies (Fig. 7B, right panel), whereas cells containing the pREP1 vector alone did not grow (left panel). Therefore, expression of human SRPK1 gene complemented the growth defect of the double-null mutant. In conjunction with the results that SRPK1 has sequence homology closer to Dsk1 than Kic1 and that the SRPK1 overexpression produces elongated but not branched cells, SRPK1 is more likely to compensate for the loss of dsk1+ than that of kic1+ in S. pombe. These data indicate that human SRPK1 protein is an in vivo functional homologue of fission yeast Dsk1 protein and further demonstrate the conservation of the SR protein-specific kinases from fission yeast to human.
DISCUSSION
Together with our previous studies (47), we show in this report that Dsk1 protein specifically phosphorylates S. pombe RS domain-containing proteins Prp2, Srp1, and Srp2 in vitro. Consistent with its substrate specificity, Dsk1 forms kinase-competent complexes with those RS domain-containing proteins. The kinase-substrate interaction is supported by the in vivo evidence for the dependency of prp2+ overexpression phenotype on dsk1+ gene. Despite the evolutionary gap separating fission yeast and human, SRPK1 not only shares similar biochemical properties with Dsk1 but also compensates for the loss of Dsk1 in fission yeast cells. The functional conservation of these kinases at the molecular and cellular level illustrates the importance of the SR protein-specific kinases in eucaryotic systems.
The evidence accumulated in recent years indicates that SR networks exist in the fission yeast S. pombe which consist of RS domain-containing proteins and their kinases. Our studies suggest that the phosphorylation patterns and interactions of the SR networks are conserved from fission yeast to mammals. We have shown for the first time that all four S. pombe RS domain-containing proteins, including Prp2, Srp1, Srp2, and Rsd1 (T.-L. Tseng and A. R. Krainer, personal communication), are phosphorylated by Dsk1 in vitro, and these phosphorylated proteins are recognized by 3C5 MAb (this report; Z. Tang, R.-J. Lin, T.-L. Tseng and A. R. Krainer, unpublished data), indicating that the kinase reaction generates a phosphoepitope identical to that found in mammalian SR proteins. In agreement with the in vitro observation, Rsd1 isolated from wild-type fission yeast is also recognized by MAb 3C5 (T.-L. Tseng and A. R. Krainer, personal communication), providing in vivo evidence for the conservation of phosphorylation specificity.
Here we dissected the Dsk1-mediated kinase reaction in vitro into three discrete steps: substrate binding, substrate phosphorylation, and release of the Dsk1 from the complex after phosphorylating its substrate. Thus, Dsk1 forms transient complexes with RS domain-containing proteins in the presence of ATP. Similar kinase-substrate complexes were recently observed between human SRPKs and SR proteins. Both SRPK1 and SRPK2 bind and subsequently phosphorylate GST-SF2/ASF. The expression of a kinase-deficient mutant SRPK2 leads to trapping SF2/ASF in the cytoplasm, possibly by forming a stable complex between the two proteins (16).
The phosphorylation by Dsk1 may affect the interactions between and/or the activity of these proteins in splicing. In agreement, pre-mRNA splicing is partially impaired in dsk1 deletion strain of S. pombe (unpublished data), and the interaction of Srp1 and Srp2 proteins is inhibited by Dsk1-mediated phosphorylation in vitro (2; Tang et al., unpublished data). It has been shown that the phosphorylation status of SF2-ASF exerts distinct effects on its association with various protein targets in vitro (55). Additionally, changes in SR protein phosphorylation play a role in the activation of pre-mRNA splicing during early development in the nematode (43). It was also established in Drosophila that SR protein phosphorylation is essential for developmentally regulated alternative splicing (7). Dsk1 influences the activity of Prp2 in vivo. Overexpression of prp2+ in different strains of S. pombe demonstrated that the ability of Prp2 to cause cell elongation is Dsk1-dependent (Fig. 5 and 6). Moreover, the observation that kic1+ gene does not have obvious effect on the phenotype of prp2+ overexpression substantiates the specific interaction between Dsk1 and Prp2. The effect on Prp2 probably is through phosphorylation of Prp2 by Dsk1, especially that Dsk1 displays high activity in phosphorylating Prp2 in vitro (47). It will be very interesting to investigate whether Dsk1 is indeed required for the phosphorylation of Prp2, Srp1, and Srp2 in vivo. The phosphorylation levels of these target proteins can be determined by in vivo 32P labeling of wild-type and dsk1-null mutant cells followed by immunoprecipitation of individual proteins with antibodies specifically against each protein. Alternatively or additionally, it can be done by using 3C5 MAb, which is specific to the SR-phosphoepitope, to probe these target proteins isolated from wild-type and dsk1-null mutant cells. We plan to address this important issue in the future.
It was reported that SRPK1 and Clk/Sty also phosphorylate human U2AF65 protein in vitro (4), although the consequence of the phosphorylation on the function of U2AF65 is not known. Perhaps Dsk1-mediated phosphorylation changes the ability of Prp2/Mis11 protein to interact with other splicing factors, such as the fission yeast homologue of human U2AF35, spU2AF23 (51). Therefore, as in mammalian systems, phosphorylation and/or dephosphorylation of RS domain-containing proteins may regulate the properties of these proteins and the organization of the protein relay in fission yeast.
We performed the first cross-species test for viability complementation of SR protein-specific kinases. Since the discovery of human SRPK1 (12), members of SRPK and Clk/Sty families were identified from various eucaryotic organisms, including mammals, Drosophila, and yeasts, based on sequence analysis and kinase specificity (4, 5, 18, 44, 47, 50, 57). Recently, the SRPK homologue in Saccharomyces cerevisiae, Sky1, was shown to phosphorylate Npl3, a budding yeast RNA binding protein containing SR/RS dipeptide repeats (44) and several mammalian SR proteins in vivo (56). The phosphorylation by Sky1 affects the cellular localization and protein interactions of these mammalian SR proteins in yeast cells (56). Interestingly, mammalian SRPK1 and Clk/Sty specifically substitute the activity of Sky1 in mediating RS domain interactions in vivo (56). However, the viability complementation strategy had not been applied to measure the functional similarity of these protein kinases prior to this study, perhaps partly due to their redundancy in cells, so that single mutation in one protein kinase lacks the apparent phenotype. Our genetic result exhibited that human SRPK1 compensates for the loss of the S. pombe Dsk1 in vivo and thus is a functional homologue of Dsk1. Collectively, these data provide both in vitro and in vivo evidence for the conservation of the SR networks through evolution.
A common feature shared between Dsk1 and Prp2 is their dual functional potential. Dsk1 protein plays a role in mitotic control (46) and is an SR protein-specific kinase involved in pre-mRNA splicing (12, 47). Prp2 is essential for pre-mRNA splicing (34, 35) and also affects chromosome segregation (45). A similar type of dual functional feature is also found in other proteins such as Ran, a small guanosine triphosphatase. It was recently shown that Ran functions to trigger the formation of the mitotic spindle, in addition to its well-characterized role in nuclear trafficking (32, 52). Another example is fission yeast Cdc5 protein, which is required for G2/M transition and is a component of a 40S snRNP-containing complex essential for pre-mRNA splicing (22). The putative dual functions of Dsk1 and Prp2 in both splicing and the cell cycle may be fulfilled through the action of Dsk1 on Prp2/Mis11, since we demonstrated that Dsk1 and Prp2/Mis11 proteins genetically interact with each other. It is conceivable that differential phosphorylation of Prp2/Mis11 may regulate its ability to either participate in chromosome segregation or to be engaged in splicing. Phosphorylation by Dsk1 may also modulate the activity of Prp2/Mis11 through altering its cellular or subnuclear localization. In agreement with its connection to the cell cycle, the phosphorylation state, cellular localization, and kinase activity of Dsk1 all change in a cell-cycle-dependent fashion (46). Supporting the model in the differential effects of phosphorylation, distinct phosphorylation sites on budding yeast transcription factor Pho4 play separable roles in altering its subcellular localization and interaction with another transcription factor, providing multiple levels of regulation to control the activity of Pho4 (17). Thus, studying the S. pombe RS domain-containing proteins and their kinases may help determine the regulatory pathways that link pre-mRNA splicing with the cell division cycle.
Our studies provide novel information about the fission yeast SR networks. The functional conservation of SRPKs from fission yeast to human makes S. pombe a valuable system for studying the biological roles of the kinase family. The powerful genetics of S. pombe will facilitate the elucidation of functions of the SR networks in eucaryotic gene expression.
ACKNOWLEDGMENTS
We thank Xiang-Dong Fu (University of California at San Diego) for providing the human SRPK1 gene, Mitsuhiro Yanagida (Kyoto University, Kyoto, Japan) for dsk1-null mutant strain, and Paul Salvaterra for advice in microscopic analysis. We also thank Xiang-Dong Fu, David Horowitz, Adam Bailis, and Glenn Manthey for critical reading of the manuscript and for constructive suggestions. We thank the reviewers for their valuable suggestions for improving the manuscript.
This work was supported by City of Hope Beckman Endowment Grant.
REFERENCES
- 1.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 2.Bridge E, Xia D X, Carmo-Fonseca M, Cardinali B, Lamond A I, Pettersson U. Dynamic organization of splicing factors in adenovirus-infected cells. J Virol. 1995;69:281–290. doi: 10.1128/jvi.69.1.281-290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao W, Jamison S F, Garcia-Blanco M A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 4.Colwill K, Feng L L, Yeakley J M, Gish G D, Caceres J F, Pawson T, Fu X D. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- 5.Colwill K, Pawson T, Andrews B, Prasad J, Manley J L, Bell J C, Duncan P I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 6.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 7.Du C, McGuffin M E, Dauwalder B, Rabinow L, Mattox W. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol Cell. 1998;2:741–750. doi: 10.1016/s1097-2765(00)80289-0. [DOI] [PubMed] [Google Scholar]
- 8.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesfeld J M, Forbes D J. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 10.Gross T, Lutzelberger M, Weigmann H, Klingenhoff A, Shenoy S, Kaufer N F. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 1997;25:1028–1035. doi: 10.1093/nar/25.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross T, Richert K, Mierke C, Lutzelberger M, Kaufer N F. Identification and characterization of srp1, a gene of fission yeast encoding a RNA binding domain and a RS domain typical of SR splicing factors. Nucleic Acids Res. 1998;26:505–511. doi: 10.1093/nar/26.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gui J F, Lane W S, Fu X D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 13.Kaufer N F, Simanis V, Nurse P. Fission yeast Schizosaccharomyces pombe correctly excises a mammalian RNA transcript intervening sequence. Nature. 1985;318:78–80. doi: 10.1038/318078a0. [DOI] [PubMed] [Google Scholar]
- 14.Kirschner M. The cell cycle then and now. Trends Biochem Sci. 1992;17:281–285. doi: 10.1016/0968-0004(92)90435-c. [DOI] [PubMed] [Google Scholar]
- 15.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 16.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer A R, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J Biol Chem. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- 17.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 18.Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- 19.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 20.Maundrell K. nmt1 of fission yeast: a highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 21.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 22.McDonald W H, Ohi R, Smelkova N, Frendewey D, Gould K L. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol Cell Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermoud J E, Cohen P T, Lamond A I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misteli T, Caceres J F, Clement J Q, Krainer A R, Wilkinson M F, Spector D L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misteli T, Spector D L. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 26.Misteli T, Spector D L. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 28.Murone M, Simanis V. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 1996;15:6605–6616. [PMC free article] [PubMed] [Google Scholar]
- 29.Murray M V, Kobayashi R, Krainer A R. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 1999;13:87–97. doi: 10.1101/gad.13.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolakaki E, Simos G, Georgatos S D, Giannakouros T. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J Biol Chem. 1996;271:8365–8372. doi: 10.1074/jbc.271.14.8365. [DOI] [PubMed] [Google Scholar]
- 31.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 32.Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 33.Potashkin J, Kim D, Fons M, Humphrey T, Frendewey D. Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr Genet. 1998;34:153–163. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- 34.Potashkin J, Li R, Frendewey D. Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J. 1989;8:551–559. doi: 10.1002/j.1460-2075.1989.tb03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- 36.Prabhala G, Rosenberg G H, Kaufer N F. Architectural features of pre-mRNA introns in the fission yeast Schizosaccharomyces pombe. Yeast. 1992;8:171–182. doi: 10.1002/yea.320080303. [DOI] [PubMed] [Google Scholar]
- 37.Romfo C M, Lakhe-Reddy S, Wise J A. Molecular genetic analysis of U2AF59 in Schizosaccharomyces pombe: differential sensitivity of introns to mutational inactivation. RNA. 1999;5:49–65. doi: 10.1017/s1355838299981323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberg G H, Alahari S K, Kaufer N F. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol Gen Genet. 1991;226:305–309. doi: 10.1007/BF00273617. [DOI] [PubMed] [Google Scholar]
- 39.Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou J F, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- 40.Roth M B, Murphy C, Gall J G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth M B, Zahler A M, Stolk J A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Sanford J R, Bruzik J P. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siebel C W, Feng L, Guthrie C, Fu X. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc Natl Acad Sci USA. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi M, Yanagida M. A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle stage dependent phosphorylation and localization. Mol Biol Cell. 1993;4:247–260. doi: 10.1091/mbc.4.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Z, Yanagida M, Lin R-J. Fission yeast mitotic regulator is an SR protein-specific kinase. J Biol Chem. 1998;273:5963–5969. doi: 10.1074/jbc.273.10.5963. [DOI] [PubMed] [Google Scholar]
- 48.Urushiyama S, Tani T, Ohshima Y. Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol Gen Genet. 1996;253:118–127. doi: 10.1007/s004380050304. [DOI] [PubMed] [Google Scholar]
- 49.Urushiyama S, Tani T, Ohshima Y. The prp1+ gene required for pre-mRNA splicing in Schizosaccharomyces pombe encodes a protein that contains TPR motifs and is similar to Prp6p of budding yeast. Genetics. 1997;147:101–115. doi: 10.1093/genetics/147.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H Y, Lin W, Dyck J A, Yeakley J M, Songyang Z, Cantley L C, Fu X D. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wentz-Hunter K, Potashkin J. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 1996;24:1849–1854. doi: 10.1093/nar/24.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 53.Woppmann A, Will C L, Kornstadt U, Zuo P, Manley J L, Luhrmann R. Identification of an snRNP-associated kinase activity that phosphorylates arginine/serine rich domains typical of splicing factors. Nucleic Acids Res. 1993;21:2815–2822. doi: 10.1093/nar/21.12.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 55.Xiao S H, Manley J L. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeakley J M, Tronchere H, Olesen J, Dyck J A, Wang H Y, Fu X D. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun B, Farkas R, Lee K, Rabinow L. The Doa locus encodes a member of a new protein kinase family and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev. 1994;8:1160–1173. doi: 10.1101/gad.8.10.1160. [DOI] [PubMed] [Google Scholar]