Abstract
Current coronavirus (CoV) vaccines primarily target immunodominant epitopes in the S1 subunit, which are poorly conserved and susceptible to escape mutations, thus threatening vaccine efficacy. Here, we use structure-guided protein engineering to remove the S1 subunit from the Middle East respiratory syndrome (MERS)-CoV spike (S) glycoprotein and develop stabilized stem (SS) antigens. Vaccination with MERS SS elicits cross-reactive β-CoV antibody responses and protects mice against lethal MERS-CoV challenge. High-throughput screening of antibody-secreting cells from MERS SS-immunized mice led to the discovery of a panel of cross-reactive monoclonal antibodies. Among them, antibody IgG22 binds with high affinity to both MERS-CoV and severe acute respiratory syndrome (SARS)-CoV-2 S proteins, and a combination of electron microscopy and crystal structures localizes the epitope to a conserved coiled-coil region in the S2 subunit. Passive transfer of IgG22 protects mice against both MERS-CoV and SARS-CoV-2 challenge. Collectively, these results provide a proof of principle for cross-reactive CoV antibodies and inform the development of pan-CoV vaccines and therapeutic antibodies.
Keywords: coronaviruses (CoV), spike (S) glycoprotein, stem stabilization, cross-reactivity, protective antibodies, protective antigens, cryo-EM structure, stem-helix antibodies
Graphical abstract
Hsieh et al. generate MERS-CoV spike stabilized stem (SS) antigens using a structure-guided approach. Mice immunized with MERS SS are protected against MERS-CoV challenge and used for isolation of cross-reactive monoclonal antibodies, including IgG22, which protects mice against MERS-CoV and SARS-CoV-2 challenges. Structures of Fab22-spike complexes reveal a conserved epitope.
Introduction
Coronaviruses (CoVs) are a genetically diverse group of enveloped viruses containing a large approximately 30-kb, positive-sense, single-stranded RNA genome (Li, 2016). Four endemic (or “seasonal”) human CoVs (HCoVs)—HCoV-OC43, -HKU1, -NL63, and −229E—circulate globally and produce primarily mild upper respiratory illness in otherwise healthy individuals. CoVs exhibit adaptation to alternative host species, and spillover of three novel β-CoVs—Middle East respiratory syndrome (MERS)-CoV, severe acute respiratory syndrome (SARS)-CoV, and SARS-CoV-2—from bats directly or indirectly into humans within the past two decades underscores the serious and persistent threat to public health that CoVs hold. Most recently, SARS-CoV-2 has led to 200 million cases worldwide and has left in its wake devastating socioeconomic consequences. Effective vaccines not only are crucial for mitigating overall morbidity and mortality of SARS-CoV-2 infection but also may enable populations to achieve herd immunity and temper the emergence and spread of new viral variants. Many of the coronavirus disease 2019 (COVID-19) vaccines in the global portfolio are based on an engineered version of the full-length viral attachment protein, spike (S), stabilized in its prefusion conformation ((Walls et al., 2020); (Wrapp et al., 2020)). In particular, mRNA-1273 and BNT162b2 showed >94% efficacy against COVID-19 in adult phase III clinical trials (Baden et al., 2020; Polack et al., 2020). However, recent emergence of numerous SARS-CoV-2 variants harboring diverse changes in S substantiate concerns about the breadth of this efficacy ((Choi et al., 2020); (Grubaugh et al., 2021); (Kupferschmidt, 2021); (Tang et al., 2021))
All CoV S proteins are composed of an S1 subunit—a major determinant for host cell tropism—and an S2 subunit, which contains the machinery that drives virus-cell fusion (Li, 2016; (Siebert et al., 2003)). The S1 subunit, which directly interacts with the host receptor, is composed of an N-terminal domain (NTD), a receptor-binding domain (RBD), and subdomains (SDs). For SARS-CoV-2, transient hinging of the RBD into an “up” conformation allows for host-cell receptor engagement. In the S2 subunit, a helix-loop region spanning from the fusion peptide (FP) to heptad repeat 1 (HR1) constitutes a metastable structure that transitions to a long stable α-helix in the postfusion conformation during viral entry. The FP, HR1, central helix (CH), and connector domain (CD) compose the globular head of the S2 subunit. In contrast, the helical stalk region connecting the globular head to the viral membrane is elongated and highly flexible ((Ke et al., 2020); (Turoňová et al., 2020)). The stalk region can be further divided into a hip, knee, and ankle, in which the knee functions as a hinge, providing a considerable degree of flexibility and promoting movement of S on the viral surface. As the S1 is responsible for interacting directly with the host receptor, the majority of vaccine-induced antibodies and antibodies elicited upon natural infection target this subunit, which contains the immunodominant RBD (Corbett et al., 2020b, 2020a; (Jackson et al., 2020)). In addition, a portion of neutralizing monoclonal antibodies (mAbs) isolated from convalescent COVID-19 patients recognize the NTD ((Chi et al., 2020); (Liu et al., 2020)). Amino acid changes are concentrated in the S1 subunit among the majority of SARS-CoV-2 variants described thus far. Additionally, recent data suggest that antibodies in convalescent sera that target neutralization-sensitive epitopes in the S1 domain create a selective pressure that yields escape mutations in these epitopes ((Andreano et al., 2021)).
Several new SARS-CoV-2 “variants of concern (VOC),” distinguished by one to many unique or recurring changes in S, recently emerged. These variants have spread at disproportionately fast rates relative to predecessor variants in certain geographic regions, consistent with an augmented replication capacity, greater transmissibility, and/or immune evasion. Among recurring changes, multiple amino acid deletions in the NTD (del 69-70, del 144, and del 242-244) and K417N/T, E484K, and N501Y substitutions in the RBD have garnered special attention due to an enhanced affinity of S for the SARS-CoV-2 receptor angiotensin converting enzyme 2 (ACE2) and reduced sensitivity to neutralization by convalescent polyclonal sera and anti-RBD therapeutic mAbs ((Gu et al., 2020); (Starr et al., 2020); (Wang et al., 2021); (Weisblum et al., 2020); (Wu et al., 2021); (Zhao et al., 2021)). The del 69-70 and N501Y alterations have evolved both together and separately in independent VOC lineages, as has the E484K mutation ((Ku et al., 2021)). Altogether, these data indicate that selective pressures on the neutralization-sensitive epitopes of the S1 subunit ultimately result in escape variants.
Although most neutralizing antibodies target the S1 subunit, the S2 subunit is more conserved, likely constrained by the complex and precisely timed refolding events essential for productive viral entry. Nevertheless, antibodies targeting this region may be neutralizing and protective against infection and pathology in vivo ((Chi et al., 2020); (Ng et al., 2020); (Zhang et al., 2021)). Notably, it has been observed that SARS-CoV-2-infected children and young adolescents recently infected with an endemic HCoV develop higher titers of S2 antibodies than SARS-CoV-2-infected adults without a recognized antecedent endemic HCoV infection (Ng et al., 2020). Memory B cells trained on conserved S2 epitopes and stimulated by SARS-CoV-2 infection may account for the skewed neutralizing antibody response to the SARS-CoV-2 S2 subunit in young individuals. In individuals not previously infected with SARS-CoV-2, a majority of the anti-SARS-CoV-2 immunoglobulin G (IgG) repertoire targets the S2 subunit, underscoring the potential for cross-reactivity directed at conserved epitopes on HCoVs ((Nguyen-Contant et al., 2020)). Thus, the S2 subunit serves as an attractive target for the development of broad-spectrum vaccines and therapeutic antibodies for current and future novel HCoVs.
In this report, we describe the design and study of two MERS-CoV stabilized stem (SS) constructs, namely, MERS SS.V1 and MERS SS.V2, both of which lack the S1 subunit (Figure 1 A). We show that both antigens are highly immunogenic in mice, inducing broadly cross-reactive antibodies to MERS-CoV, HCoV-HKU1, SARS-CoV, and SARS-CoV-2 S, and confer protection in a lethal murine MERS-CoV challenge model. Two mAbs, namely, IgG22 and IgG72, isolated using microfluidic single B cell screening technology from MERS SS.V1-immunized mice, neutralized authentic MERS-CoV. Strikingly, the passive transfer of IgG22 protected mice from both MERS-CoV and SARS-CoV-2 lethal challenge. Single-particle cryoelectron microscopy (cryo-EM) studies revealed that IgG22 binds a region in the S2 stalk of MERS-CoV and SARS-CoV-2 S proteins that undergoes substantial changes during the pre-to-postfusion conformational transition. High-resolution crystal structures of the stalk peptide–antibody complexes further provided a basis for binding of this stalk-targeting antibody to highly conserved sequences in human β-CoVs. Our findings establish the feasibility of rationally designing stabilized S2 constructs as protective vaccine immunogens against emerging HCoVs, which upon further development may exhibit efficacy against divergent HCoV species. In addition, stem cross-reactive antibodies may serve as valuable tools for prospective serological surveillance of future CoV spillover into humans.
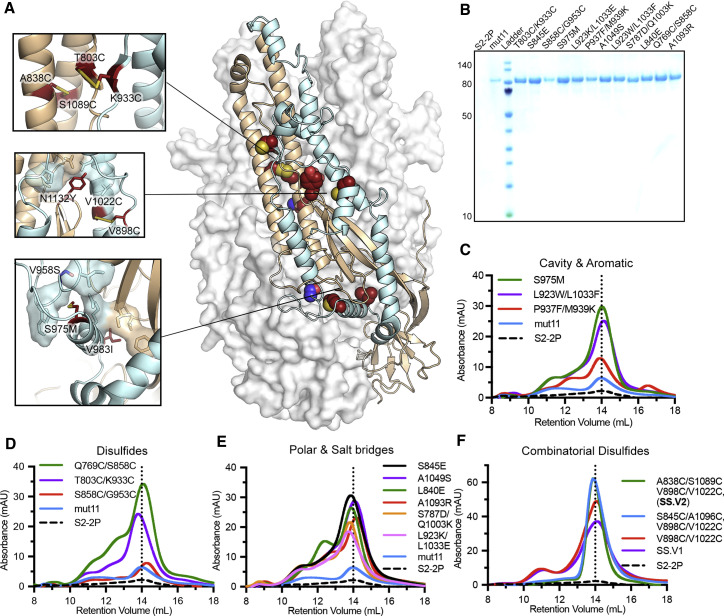
Figure 1.
Characterization of MERS stem stabilized (SS) spike (S) variants
(A) Side view of the trimeric MERS-CoV globular S ectodomain in a prefusion conformation (PDB: 5W9I) with S1 subunits omitted. One protomer of the S2 subunit is shown as a ribbon diagram with the other two protomers shown in a transparent molecular surface. The regions that refold during the pre-to-post-fusion transition are colored cyan with the rest of S2 in tan. Each inset corresponds to the location of beneficial substitutions in the MERS SS.V1 construct.
(B) SDS-PAGE of MERS-CoV S2-2P and individual S variants on mut11 backbone. Molecular weight standards in kDa are indicated at the left.
(C–E) Size-exclusion chromatography (SEC) traces of purified S variants, grouped by type (C, cavity and aromatic; D, disulfide; E, polar and salt bridge). A vertical dotted line indicates the peak retention volume for S2-2P.
(F) SEC traces for combinatorial disulfide-substituted S variants. One or two additional disulfide substitutions are introduced on the MERS SS.V1 backbone.
See Figures S1 and S2.
Results
Structure-based vaccine design of MERS-CoV SS antigens
To stabilize the stem region (S2 subunit) of S, we first used the Protein Repair One-Stop Shop (PROSS) algorithm to computationally design stabilizing mutations based on a prefusion-stabilized structure of the MERS-CoV S ectodomain (PDB: 5W9I) (Goldenzweig et al., 2016; Pallesen et al., 2017). Among 53 designs, 11 substitutions were selected and introduced in various combinations into the S2 subunit of MERS-CoV S. The protein expression level of the mutant containing all 11 substitutions (mut11) was substantially higher than that of MERS S2-2P (base construct) (Figure 1B). To further improve the protein expression and thermostability of S2, we used a variety of stabilization strategies to design 12 different substitutions, which were added onto the mut11 background. The strategies we used include using hydrophobic residues to fill loosely packed internal cavities such as S975M substitution (Figures 1A and 1C), disulfide bonds to lock the regions that move substantially during the pre-to-postfusion transition such as T803C/K933C substitution (Figures 1A and 1D), polar residues to counter internal charge imbalance such as V958S substitution (Figures 1A and 1E), and aromatic side chains to favor pi-pi or cation-pi interactions with positively charged residues such as L923W/L1033F substitution (Figures 1C and S1A). Except for the S858C/G953C variant, all other single substitutions increased the protein expression to various extents compared with mut11. Particularly, S975M, L923W/L1033F, Q769C/S858C, T803C/K933C, S845E, A1049S, and A1093R substitutions exhibited a more than 10-fold higher expression compared to their parental construct mut11 (Figures 1B–1E). The size-exclusion chromatography (SEC) traces of all variants showed a major trimeric peak with some minor shoulder peaks, with the retention volumes of the trimeric variants being similar to the base construct and mut11.
Next, we sought to examine the contribution of the individual substitutions comprising mut11 to protein expression and thermostability by reverting each of the substitutions back to the wild-type (WT) residue. Reverting K816R, H1020Q, H1146Y, or V1150T increased the protein expression relative to mut11, meaning these four substitutions in mut11 are dispensable. Thus, a new base construct containing 7 substitutions (mut7) was used for subsequent designs (Figure S1B). To test whether our structure-based designs (previously tested individually on the mut11 background) have additive effects, we added a disulfide design (T803C/K933C), a cavity filling design (S975M), or a combination of both designs (S975M, T803C/K933C) on top of mut7. However, unlike the increase in expression for mut11, T803C/K933C decreased the protein expression relative to mut7 but increased the melting temperature (Tm) by 1.7°C (Figure S1C). S975M substitution not only boosted protein expression relative to mut7, but also enhanced the thermostability. Combining both T803C/K933C and S975M on the mut7 backbone restored the expression to a level similar to mut7 and increased Tm by 4.0°C. After iterative screening of WT reversions in mut7 S975M/T803C/K933C, we found that A918P and V1139F had adverse effects on protein expression and thermostability (Figures S1D and S1E). Removal of A918P and V1193F resulted in our first SS antigen, MERS SS.V1 (Figures S1B and S1C), containing 7 substitutions in addition to the original 2P substitutions (S975M, T803C/K933C, V958S, V983I, S1091E, L1094Q, N1132Y, V1060P, and L1061P).
To evaluate the viability of MERS SS.V1 as an immunogen, we investigated large-scale production in FreeStyle 293-F cells, thermostability, and epitope integrity. After two consecutive runs of SEC, MERS SS.V1 exhibited a monodispersed trimeric peak, with a yield of 2.2 mg from 1 L of cell culture. The Tm of MERS SS.V1 is 59.9°C, which is a 6.3°C increase relative to mut11 (Figure S1F and S1G). mAb G4 is one of very few S2-directed antibodies showing neutralizing activity to MERS-CoV (Wang et al., 2015). The binding kinetics of MERS SS.V1 to G4 IgG were comparable to those of the MERS S-2P ectodomain, with apparent affinities of 8.5 nM and 13.2 nM, respectively (Figures S2A and S2B). We also examined the conformation of MERS SS.V1 complexed with G4 Fab by negative stain electron microscopy (nsEM) analysis (Figure S2C). Although the images from 2D class averages of SS.V1 did not resemble an elongated postfusion conformation, it was not clear that our SS antigens maintained a fully prefusion state. Taken together, these results suggest that MERS SS.V1 retains the integrity of the known neutralization epitope and should be amenable to large-scale production.
Introducing disulfide linkages to lock viral glycoproteins in the prefusion conformation has proven effective in class I viral fusion proteins such as respiratory syncytial virus (RSV) F, human immunodeficiency virus type 1 (HIV-1) Env, and Ebola GP ((McLellan et al., 2013); (Rutten et al., 2020); (Sanders et al., 2013)). Although it was not successfully applied to the S2 subunit of the SARS-CoV-2 S (Hsieh et al., 2020), it was effective to lock the RBD from the S1 subunit to the S2 subunit and stabilize a closed S trimer conformation (Henderson et al., 2020; McCallum et al., 2020; Xiong et al., 2020). We thus explored the feasibility of the disulfide design on MERS SS.V1. Three disulfide designs, namely, A838C/S1089C, S845C/A1096C, and V898C/V1022C, were introduced individually or in combination to stabilize the region proximal to the FP and CH. With the addition of V898C/V1022C, the protein expression level increased 1.3-fold relative to MERS SS.V1 and the Tm increased by 4.0°C (Figures 1F and S1F). Upon combination with either A838C/S1089C or S845C/A1096C substitutions, the protein expression levels increased 1.7-fold relative to MERS SS.V1 and the Tm increased by 6.3°C. Notably, both constructs exhibited a sharp monodisperse trimeric peak on SEC without any prominent shoulder peaks. This led to our best construct, namely, MERS SS.V2, which contains A838C/S1089C and V898C/V1022C added onto the MERS SS.V1 background. The large-scale expression of MERS SS.V2 outperformed MERS SS.V1, yielding 9.5 mg from 1 L of FreeStyle 293-F cells (Figure S1G).
MERS SS immunization elicits cross-reactive β-CoV antibodies
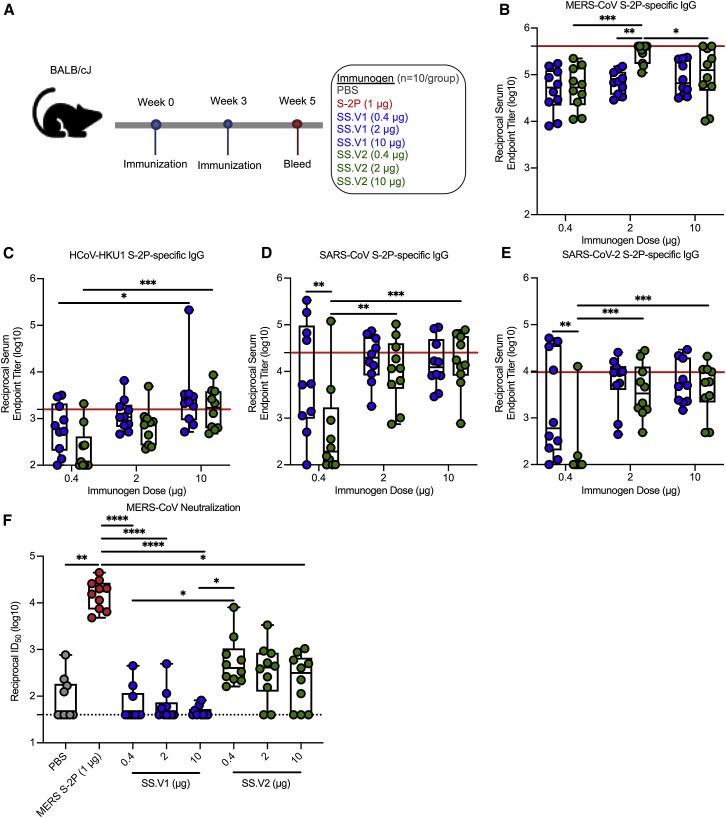
To evaluate the cross-reactive immunogenicity of MERS SS immunogens, we immunized BALB/cJ mice with 0.4, 2, or 10 μg of MERS SS.V1 or SS.V2 or the maximally effective dose of 1 μg of stabilized MERS S ectodomain (MERS S-2P) (Pallesen et al., 2017; Figure 2 A). All doses of both immunogens elicited ∼105 MERS S-2P-specific reciprocal endpoint binding titers at 2 weeks post-boost (Figure 2B). Both MERS SS.V1 and SS.V2 elicited HCoV-HKU1, SARS-CoV, and SARS-CoV-2 S-2P-specific IgG that trended toward dose dependency; most notably at the 0.4 μg dose, heterotypic SARS- and SARS-2 S-2P-specific binding IgG responses were more significant in mice immunized with SS.V1 than in mice immunized with SS.V2 (Figures 2C–2E). Conversely, neutralizing antibody responses against a pseudotyped lentivirus reporter expressing MERS-CoV S were not dose dependent. As expected, both SS immunogens elicited significantly fewer neutralizing antibodies than MERS S-2P given the lack of the S1 subunit in the SS immunogens. Although MERS SS.V1 elicited only modest baseline neutralizing antibody responses (up to a geometric mean reciprocal ID50 titer, geometric mean titer [GMT] = 156), SS.V2 induced stronger neutralization responses with a GMT of 550 ID50 (Figure 2F). Altogether, these data corroborate that the introduction of stabilizing disulfide bonds into the SS.V1 backbone promotes the production of more potently neutralizing MERS-CoV S2-specific antibodies. Additionally, they underscore the importance of targeting conserved epitopes to induce broad binding antibody responses.
Figure 2.
Immunogenicity of MERS SS immunogens in mice
(A) BALB/cJ mice (N = 10/group) were inoculated with PBS (gray); 1 μg of MERS S-2P (red); or 0.4, 2, or 10 μg of MERS SS.V1 (blue) or SS.V2 (green) adjuvanted with Sigma Adjuvant System (SAS). Mice were immunized at weeks 0 and 3 and bled at week 5 (2 weeks post-boost) for serological assessments.
(B–E) ELISAs to quantify S-2P specific binding IgG titers for MERS-CoV (B), HCoV-HKU1 (C), SARS-CoV (D), and SARS-CoV-2 (E). Red line respresents the GMT of MERS S-2P immunized group.
(F) Neutralizing antibody titers against MERS-CoV England1 pseudovirus. The dotted line represents the lower limit of detection.
In (B) to (F), each circle represents and individual mouse. Box and whisker plots are shown. Boxes and horizontal bars denote the interquartile range (IQR) and medians, respectively; whisker end points are equal to the maximum and minimum values. In (B) to (E), two-way ANOVA with multiple comparisons post-tests were used to compare doses within immunogen group and groups at each dose. In (F), groups were compared by one-way ANOVA with Kruskal-Wallis post-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See Figures S3 and S4.
MERS SS protects humanized-DPP4 mice against lethal MERS-CoV challenge
In order to assess the ability of MERS SS immunogens to protect against lethal MERS-CoV challenge, we immunized 288/330+/+ mice (Corbett et al., 2020a) with 10 μg of MERS SS.V1 or S-2P, adjuvanted with Sigma Adjuvant System (SAS) at weeks 0, 3, and 9—inclusive of an additional boost to further promote cross-reactive humoral responses. Mock-immunized mice received PBS (Figure 3 A). In 288/330+/+ mice, the Dpp4 gene has been modified to encode two amino acids on positions 288 and 330 to match the human gene, rendering them susceptible to MERS-CoV infection and replication. Serial passaging led to a mouse-adapted version of MERS-CoV (maM35c4) ((Douglas et al., 2018)) that not only replicated efficiently in mouse lungs but also caused symptoms like weight loss, decreased pulmonary function, and histopathological manifestations of severe lung disease (Figure 3A). First, ELISA revealed MERS SS.V1 elicits similar levels (∼105) of homotypic binding IgG as MERS S-2P (Figure 3B). However, stem-specific binding antibodies were sub-neutralizing as MERS SS.V1-immunized mice had significantly reduced homotypic pseudovirus neutralizing antibody responses (geometric mean reciprocal ID50 titer, GMT = 185) as compared to mice receiving MERS S-2P (GMT = 5,758), similar to BALB/cJ mice in Figure 2F. In fact, only 5 out of 20 MERS SS.V1-immunized 288/330+/+ mice had detectable neutralizing antibody responses (Figure 3C).
Figure 3.
Efficacy of SS.V1 against lethal MERS-CoV challenge in mice
(A) 288/330+/+ mice (N = 20/group) were immunized at weeks 0, 3, and 9 with PBS (gray) or 10 μg of MERS-CoV S-2P (red) or SS.V1 (blue) adjuvanted with SAS.
(B–C) At week 11 (two weeks post-boost 2), mice were bled for analysis of MERS-CoV S-2P-specific IgG (B) and MERS-CoV maM35c4 pseudovirus neutralizing antibodies (C).
(D) Following challenge, mice were monitored for weight loss. The mean of each group is represented by a circle, error bars represent SEM.
(E–H) At days 3 and 5 post-challenge, a subset of mouse lungs (n = 5/time point) was harvested for analysis of viral titers (E and F) and tissue discoloration (G and H) (0 = no discoloration, 4 = severe discoloration in all lobes). (B, C, and E–H) Each circle represents an individual mouse. Box and whisker plots are shown. Boxes and horizontal bars denote the interquartile range (IQR) and medians, respectively; whisker end points are equal to the maximum and minimum values. Dotted lines represent lower limit of detection.
In (B) to (C) and (E) to (H)experimental groups were compared to the PBS control group by one-way ANOVA with Kruskal-Wallis post-test. In (D), experimental groups were compared to the PBS control group at each day post challenge by one-way ANOVA with Kruskal-Wallis post test. Statistics account for varability in mouse numbers following lung harvest. ND indicates statistical analysis is not determined due to limited number of PBS control mice. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Following challenge, 4 weeks post-boost, mice immunized with either MERS S-2P or MERS SS.V1 demonstrated no weight loss throughout the course of infection. Conversely, mice in the PBS control group demonstrated severe clinical presentations and succumbed to weight loss by 1 week post-challenge (Figure 3D). At day 3 post-challenge, the day of peak lung viral titers in this model, MERS S-2P-immunized mice had no detectable lung viral load, and lung viral titers in MERS SS.V1-immunized mice hovered about the viral detection limit (geometric mean PFU per lung lobe = 167) (Figure 3E). By day 5 post-challenge, MERS SS.V1- and S-2P-immunized mice alike had cleared viral replication, contrasting against PBS-immunized mice, which had 2 × 106 geometric mean PFU per lung lobe (Figure 3F). In line with trends of viral replication among SS.V1- and S-2P-immunized mice, at both days 3 and 5 post-challenge, there was little to no distinguishable pulmonary damage, as evidenced by low discoloration scores, indicating that both vaccines effectively protected against lung viral replication and disease (Figures 3G and 3H). These findings emphasize the utility of eliciting protective immunity by targeting epitopes outside immunodominant and neutralization-sensitive S1 sites.
Discovery of pan-CoV antibodies from MERS SS-immunized mice
Next, we used AbCellera’s single B cell technology to isolate cross-reactive mAbs from the spleen, thymus, and lymph nodes of mice immunized with the MERS SS.V1 antigen. Using HCoV-HKU1 S-2P, SARS-CoV S-2P, MERS-CoV S-2P, and MERS SS.V1 as probes, we discovered 146 antibodies with a diverse spectrum of specificities to the β-CoV S proteins. We selected for antibodies exhibiting broader S reactivities, higher frequencies, and unique CDR3s in both heavy and light chains in order to cover a wide distribution of the germline family. The selected IgGs were then expressed recombinantly in vitro and characterized by binding specificities to β-CoV prefusion-stabilized S, namely, HCoV-HKU1 S-2P, MERS-CoV S-2P, and SARS-CoV S-2P. IgG20, IgG23, and IgG62 demonstrated cross-reactive binding to MERS- and SARS-CoV S with higher apparent affinity to the former (Figure S3A). Intriguingly, IgG21, IgG22, and IgG72 were also MERS and SARS-specific, but the association rates of these mAbs to SARS-CoV S were nearly 5-fold higher than those to MERS-CoV S (Figure S3B). In contrast, IgG12, IgG19, and IgG42 only bound to MERS-CoV S (Figure S3C). We also performed competitive binding experiments with G4, the only MERS S2-targeted antibody with a structurally defined epitope (Pallesen et al., 2017). One-third of selected mAbs (IgG20, IgG23, and IgG62) competed with G4 binding to MERS-CoV S (Figures S4A and S4B). By contrast, IgG12, IgG19, IgG21, IgG22, IgG42, and IgG72 appeared to bind to regions outside the G4 epitope. Although the initial screening from single B cell technology showed that IgG7 reacted with HKU1 S-2P, IgG7 was shown to target a foldon trimerization motif that was artificially linked to all of the S antigens and the control RSV F protein. Finally, we performed a nsEM analysis for one of the non-G4 competing antibodies, Fab20, complexed with MERS-CoV S. In line with competitive binding data, the low-resolution reconstruction of the complex indicated that Fab20 bound to a different site from the G4-binding loop at the bottom of the S2 subunit (Figure S4C).
IgG22 binds to multiple CoV S proteins and neutralizes authentic MERS-CoV
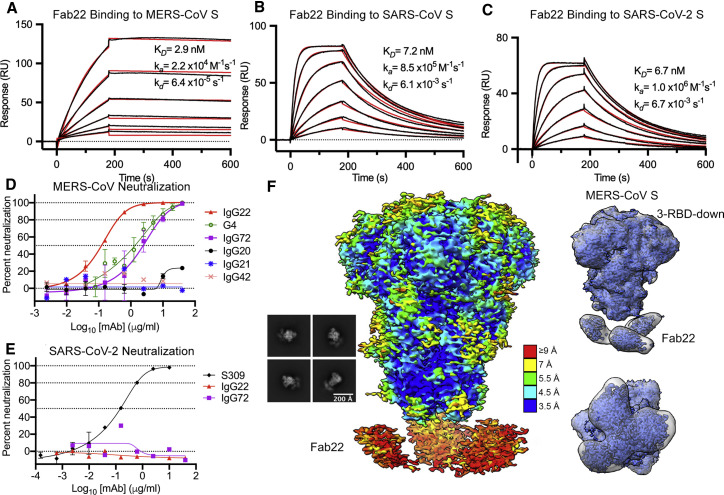
IgG22 and the clonally related IgG72 are two of the cross-reactive antibodies that exhibited faster association to SARS-CoV S than MERS-CoV S (Figure S3B). To determine the kinetics of their interactions, we performed surface plasmon resonance experiments for which we flowed Fab over S ectodomain immobilized to a sensor chip. The binding affinity of Fab22 to MERS-CoV S was comparable to those of SARS-CoV S and SARS-CoV-2 S, with equilibrium dissociation constant (K D) values of 2.9 nM, 7.2 nM, and 6.7 nM, respectively (Figures 4 A–4C). However, Fab22 exhibited distinct binding kinetics to MERS-CoV S, featuring a much slower on-rate and off-rate than SARS-CoV or SARS-CoV-2 S. We examined the neutralization activities of selected mAbs against authentic MERS-CoV. IgG22 and IgG72 exhibited potent neutralizing activity, with IC50 values of 0.12 μg/ml and 2.45 μg/ml, respectively (Figure 4D). On the other hand, IgG20, IgG21, and IgG42 showed no neutralization activity against MERS-CoV. Interestingly, IgG22 and IgG72 failed to neutralize authentic SARS-CoV-2 (Figure 4E), which may be due to the faster off-rate of IgG22 from SARS-CoV-2. However, we cannot rule out other possibilities, such as differences in S density between the two viruses.
Figure 4.
MERS stem-targeted mAbs neutralize authentic MERS-CoV
(A–C) Binding of Fab22 to MERS-CoV S (A), SARS-CoV S (B), and SARS-CoV-2 S (C) assessed by surface plasmon resonance. Binding data are shown as black lines, and the best fit to a 1:1 binding model is shown as red lines.
(D) IgG22 and IgG72 demonstrated concentration-dependent neutralization of MERS-CoV infectivity. Other SS.V1 elicited mAbs shown lacked detectable neutralizing activity against MERS-CoV. mAbs IgG20, IgG22, and IgG72 were tested in duplicate, and remaining mAbs were tested once. Neutralizing mAb G4 was included as a positive control and tested in triplicate.
(E) Neither IgG22 nor IgG72 exhibited detectable neutralization of SARS-CoV-2. mAb S309, which neutralizes SARS-CoV-2, was included as a positive control. IgG22 and IgG72 were tested once, and S309 was tested in duplicate.
(F) Representative 2D class averages and cryo-EM map of Fab22 bound to MERS S-2P. The map is colored by local resolution. The side view and top-down view of the Gaussian filtered maps indicate distinct Fab volumes.
Cryo-EM structure of Fab22 bound to the MERS-CoV S
To investigate Fab22 and Fab72 binding sites on S, we performed nsEM analysis for the Fab-S complexes. 2D classification showed three distinct densities for Fab22 attached to the S2 stalk region of MERS-CoV S-2P, a region that undergoes substantial conformation changes during the pre-to-postfusion transition (Figure S5A). Interestingly, Fab72 also binds to a similar region of S2 in proximity to HR2 (Figure S5B). Given that the binding of HR2 could potentially prevent virus-cell membrane fusion, we sought to determine a cryo-EM structure of Fab22 bound to MERS S-2P. Similar to the nsEM analysis results, we were able to see three distinct Fab densities from multiple 2D classes (Figure 4F). The initial 3D reconstruction exhibited a single conformation of the S, with all three RBDs in the down conformation. A total of 183,556 particles led to a 3.3-Å reconstruction with well-defined side chains densities at the core of S2. Three Fab volumes could be visualized at the bottom stalk of S2, although one of the Fabs displayed a broader than usual Fab volume. It is possible that the highly flexible nature of the stalk-HR2 region could lead to broadened Fab volumes. Therefore, the initial set of particles was subjected to two additional rounds of heterogeneous refinement to sort out 3D classes with diverse Fab orientations. We were able to obtain a 3D reconstruction (3.3 Å) from about one-half of the total particles (48.7%) demonstrating three distinct Fabs bound to the S. Interestingly, both reconstructions displayed three-RBD-down conformations. Focused refinement on the Fab22 volumes was attempted, but the Fab-S interface could not be resolved.
Cryo-EM structure of Fab22 bound to the SARS-CoV-2 S
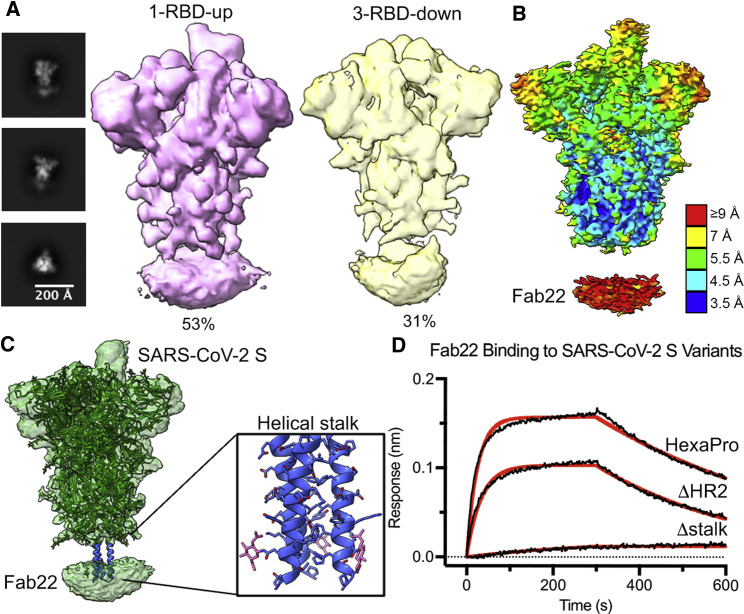
Similar to the Fab22-MERS-CoV S complex, nsEM analysis showed that Fab22 binds to the stalk region of SARS-CoV-2 S (Figure S5C). To ascertain the discrepancy between the MERS-CoV and SARS-CoV-2 neutralization by IgG22, we attempted to determine a high-resolution structure of Fab22 bound to SARS-CoV-2 S HexaPro, a prefusion-stabilized S (Hsieh et al., 2020). The initial 3D reconstruction exhibited a disc-shaped Fab volume beneath the helical stalk of the globular ectodomain. After two additional rounds of heterogeneous refinement, we were able to sort out two different conformations of S, as follows: one-RBD-up at 4.0 Å (53% of the total particles) and three-RBD-down at 4.4 Å (31% of the total particles) (Figures 5 A and 5B). Although Fab22 volumes were not well defined, the Fab binding site on the S could be located to the helical stalk, which is only resolved in the full-length S model (PDB: 6XR8) (Figure 5C). Sequence alignments of the helical stalk show that, among the β-CoVs, HCoV-HKU1 is the least conserved in this region. This finding could explain why IgG22 has nearly no affinity to the HCoV-HKU1 S. To further narrow down the binding site, we constructed the following two C-terminal truncations of the S variants: ΔHR2 (1–1160) and Δstalk (1–1142). In line with the cryo-EM map, Fab22 retained affinity to HexaPro-ΔHR2 but completely lost binding to HexaPro-Δstalk (Figure 5D). Collectively, these data demonstrate that the conserved helical stalk region between residues 1142 and 1160 is an epitope with the potential to elicit cross-reactive S2-targeted antibodies.
Figure 5.
Stem-targeted IgG22 binds to helical stalk region of SARS-CoV-2 S
(A) Representative 2D class averages and 3D reconstructions of Fab22 bound to SARS-CoV-2 S.
(B) Side view of the cryo-EM map of Fab22 bound to SARS-CoV-2 S. The map is colored by local resolution. Fab22 binds to the base of the S.
(C) Model of full-length wild-type SARS-CoV-2 S (PDB: 6XR8) docked into EM map of Fab22 bound to SARS-CoV-2 S. The S protein is colored green, and the helical stalk is colored blue. Zoomed view of the helical stalk has side chains shown in blue and N-linked glycans shown in pink.
(D) Binding of Fab22 to three SARS-CoV-2 S variants captured by RBD-directed IgG S309 is assessed by biolayer interferometry (BLI). Binding data are shown as black lines, and the best fit to a 1:1 binding model is shown as red lines.
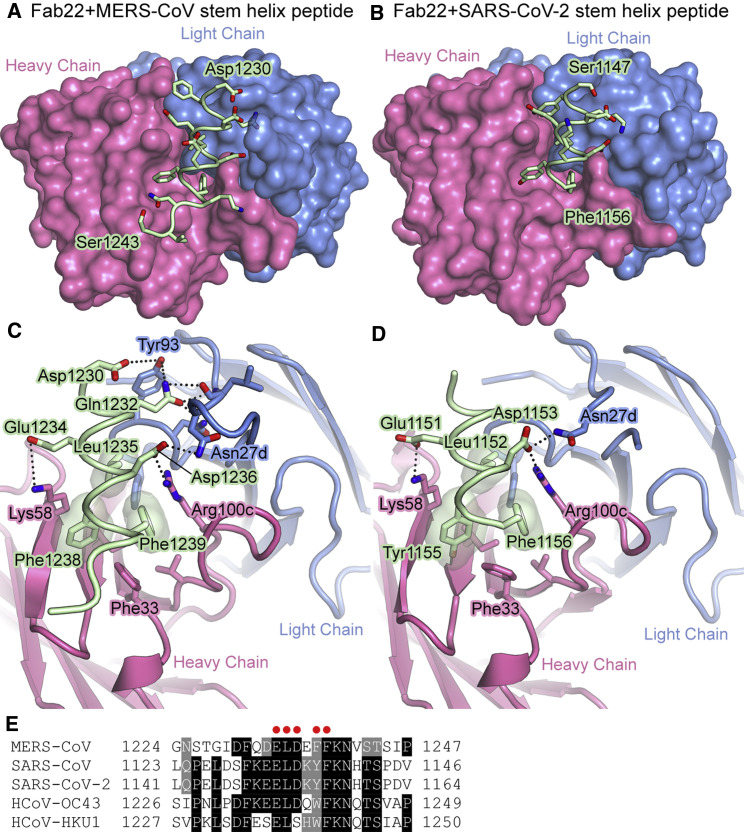
Crystal structures of Fab22 in complex with MERS-CoV S and SARS-CoV-2 S stem helix peptides
To gain molecular insight into the ability of Fab22 to bind both MERS-CoV S and SARS-CoV-2 S, we determined crystal structures of Fab22 bound to either a MERS-CoV S peptide or a SARS-CoV-2 S peptide containing the putative S2 epitope (Sauer et al., 2021; Figures 6 A, 6B, and S6). These structures revealed that Fab22 can bind both MERS-CoV and SARS-CoV-2 S by engaging residues that are conserved between both viruses. The Fab22–peptide interfaces each contain 3 key hydrophobic interactions, formed by Leu1235, Phe1238, and Phe1239 in MERS-CoV S and by Leu1152, Tyr1155, and Phe1156 in SARS-CoV-2. These conserved residues occupy the same position in both structures, with Phe1239/Phe1156 sandwiched between CDRH3 Arg100c and CDRH1 Phe33, and with Leu1235/Leu1152 and Phe1238/Tyr1155 packing into identical hydrophobic pockets of Fab22 (Figures 6C and 6D). In addition, the conserved Asp1236/Asp1152 forms a salt bridge with CDRH3 Arg100c, and the conserved Glu1234/Glu1151 forms a salt bridge with CDRH2 Lys58 (Figures 6C and 6D). The conservation of these five positions in the Fab22 epitope provides a molecular basis for its cross-reactivity (Figure 6E).
Figure 6.
IgG22 binds to a conserved region in S2
(A and B) Crystal structure of Fab22 bound to the MERS-CoV (A) and SARS-CoV-2 (B) stem helix peptides. Heavy and light chains of the Fab are shown as molecular surfaces, and the main chain of the peptides is shown as a tube, with the side chains shown as sticks. Oxygen atoms are colored red and nitrogen atoms are colored blue. The N-terminal and C-terminal residues of the peptides are labeled.
(C and D) Zoomed-in view of the interface between Fab22 and the MERS-CoV (C) and SARS-CoV-2 (D) stem helix peptides. Fab22 is shown as a ribbon, and the main chain of the peptides is shown as a tube, with the side chains shown as sticks.
(E) Alignment of the stem helix region of S proteins from multiple β-CoVs. Fab22 interface residues conserved between MERS-CoV and SARS-CoV-2 are denoted with a red dot.
IgG22 protects mice against lethal MERS-CoV and SARS-CoV-2 challenge
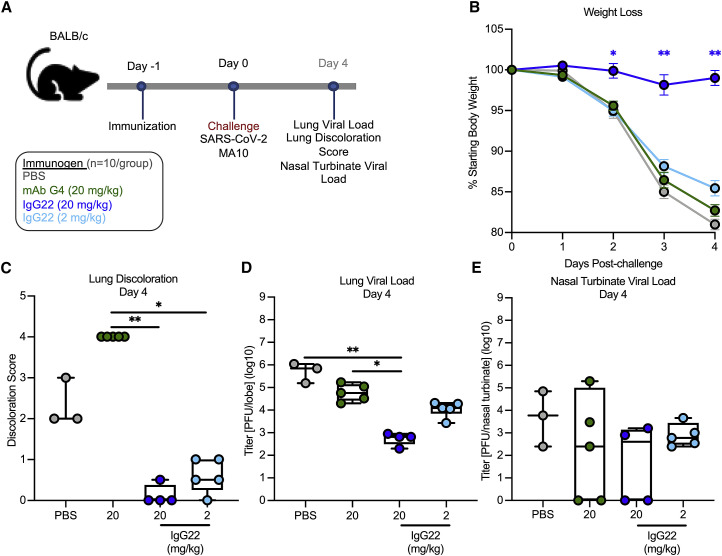
The observed cross-reactivity of IgG22 to MERS-CoV and SARS-CoV-2 S suggested its ability to protect against both viruses in vivo. We first assessed protection against a lethal dose of MERS-CoV using mouse-adapted strain maM35c4 ((Douglas et al., 2018)) in 288/330+/+ mice ((Corbett et al., 2020b); Figure S7A), and subsequently, for SARS-CoV-2, we used a BALB/c model (Dinnon et al., 2020; Leist et al., 2020; Figure 7 A). In both models, 2 or 20 mg/kg of IgG22 or 20 mg/kg of MERS-CoV S2-directed mAb G4 ((Wang et al., 2018)) was passively transferred into mice and compared to mock-immunized, PBS control mice (Figures S7A and 7A). IgG22 protected against homotypic MERS-CoV challenge in a dose-dependent fashion; 20 mg/kg of IgG22 and G4 significantly reduced weight loss as compared to PBS control mice, which demonstrated an average loss of 14.1% of the initial starting weight by day 5 post-challenge (Figure S7B). In contrast, there was no difference comparing PBS control mice and mice immunized with 2 mg/kg of IgG22 (Figure S7B). These results were mirrored in assessments of lower airway viral replication and disease. On day 5 post-challenge, PBS control mice had a geometric mean of 5.0 × 104 PFU/lobe and the 2- and 20-mg/kg IgG22 group had 70.5-fold and 6.7-fold less lung-resident MERS-CoV, respectively (Figure S7D). Additionally, discoloration of lung tissues was completely inhibited in both the 20-mg/kg IgG22 and G4 groups, but evidence of lung discoloration was present in the 2-mg/kg IgG22-immunized and PBS-control mice by the fifth day post-challenge (Figure S7C). In the upper airway, by day 5, there was significantly less (8.7-fold) MERS-CoV in nasal turbinates of mice that received 20 mg/kg IgG22 as compared to PBS control mice; however 2-mg/kg IgG22 did not prevent upper airway MERS-CoV replication (Figure S7E). Notably, across all measurements, there were similarities in the ability of mAbs G4 and IgG22 to protect against MERS-CoV-2, further suggesting these two S2-directed potently neutralizing antibodies provide homotypic protection by a similar mechanism.
Figure 7.
Passive transfer of IgG22 protects mice from lethal SARS-CoV-2 challenge
(A) BALB/c mice (N = 10/group) were treated with 20 mg/kg of mAb G4 (green), 20 mg/kg of IgG22 (blue), or 2 mg/kg (light blue) intraperitoneally on the day before challenge.
(B) Following challenge, mice were monitored for weight loss.The mean of each group is represented by a circle, error bars represent SEM.
(C–E) On day 4 post-challenge, lungs were harvested for analysis of tissue discoloration (C) and viral titers (D). Nasal turbinates were harvested for analysis of viral titers (E). Each circle represents an individual mouse. Box and whisker plots are shown. Boxes and horizontal bars denote the interquartile range (IQR) and medians, respectively; whisker end points are equal to the maximum and minimum values. Dotted lines represent lower limit of detection.
In (B), experimental groups were compared to the PBS control group at each day post-challenge by one-way ANOVA with Kruskal-Wallis post test. In (C) to (E), groups were compared to the PBS control by one-way ANOVA with Kruskal-Wallis post-test. = ∗p < 0.05, ∗∗p < 0.01.
See Figure S7.
For SARS-CoV-2 challenge, mice were infected with 104 PFU SARS-CoV-2 MA10 (Leist et al., 2020) at 24 hours following intraperitoneal (i.p.) treatments. Mice immunized with 20 mg/kg of mAb G4, 2 mg/kg of IgG22, and PBS all demonstrated similar trends in weight loss kinetics—losing on average 17.3%, 14.6%, or 19.0% of the initial starting weight, respectively—by day 4 post-challenge (Figure 7B). In contrast, mice in the 20-mg/kg IgG22 group maintained their weight throughout the challenge, only losing 1% of the starting body weight on average by day 4 post-challenge (Figure 7B). On day 4 post-challenge, mice that received 20-mg/kg IgG22 showed significantly lower (10.5-fold) lung SARS-CoV-2 than the PBS control mice. In a dose-dependent fashion, there was a modest 1.7-fold lower SARS-CoV-2 titer in lungs of mice immunized with 2-mg/kg IgG22 than in PBS control mice (Figure 7D). There were negligible differences in nasal turbinate SARS-CoV-2 titers across all groups, suggesting that IgG22, at up to 20 mg/kg, was insufficient at preventing viral replication in the upper airway (Figure 7E). In all, the ability of IgG22 to prophylactically protect against both homotypic MERS-CoV and heterotypic SARS-CoV-2 in mice shows promise for the use of SS antigens, such as MERS SS, to induce not only cross-reactive but also protective antibody responses against related HCoVs. The efficacy of IgG22 in heterotypic challenge additionally emphasizes the utility of such cross-reactive S2-directed antibodies in acute prevention of CoV lower airway disease.
Discussion
The S2 subunit of CoV S, analogous to HA2 of influenza virus hemagglutinin (HA) and gp41 of HIV-1 Env, functions as a fusogen by bringing viral and host cell membranes together, thus enabling viral entry. To mediate fusion, the S2 subunit transitions from a metastable prefusion to a highly stable post-fusion conformation (Li, 2016). We previously reported that the introduction of stabilizing mutations into the S2 subunit of MERS-CoV, SARS-CoV, and SARS-CoV-2 permitted the production of large quantities of prefusion-stabilized trimeric full-length S retaining well-folded 3D structures resembling native S on the virion surface (Hsieh et al., 2020; (Ke et al., 2020); (Kirchdoerfer et al., 2018); Pallesen et al., 2017; (Turoňová et al., 2020); (Wrapp et al., 2020)). Importantly, neutralization-sensitive epitopes and host-receptor-binding sites in the S1 subunit were completely preserved. Collectively, the strategy of stabilizing S through engineered prevention of S2 refolding has been successful for generating vaccine antigens that protect animals in CoV-challenge models and humans from SARS-CoV-2 infection (Baden et al., 2020; Corbett et al., 2020a, 2020b; Polack et al., 2020).
Efforts toward a universal influenza A virus (IAV) vaccine yielded the first proof of concept for exploiting antigenic conservation in the HA stem region (functionally analogous to the CoV S2 subunit) to elicit broad protection in animals. Specifically, two independent research groups rationally engineered the HA stem by introducing disulfide bridges, increasing surface hydrophilicity, stabilizing the hydrophobic core, optimizing the protease cleavage site, and/or displaying the HA stem on nanoparticles ((Impagliazzo et al., 2015); (Yassine et al., 2015)). The HA stem trimers stabilized in the prefusion conformation induced neutralizing antibodies targeting stems of various group 1 HA subtypes and protected animals from heterotypic IAV challenge. A similar strategy based on structure-guided vaccine design was successfully applied to more distantly related group 2 IAVs ((Boyoglu-Barnum et al., 2020); (Corbett et al., 2019)).
In this study, we extended our structure-guided design strategy to generate MERS-CoV-S2-only antigens, with the intention of eliciting cross-reactive antibodies against conserved S2 epitopes. Using the PROSS server (Goldenzweig et al., 2016) to identify candidate stabilizing mutations and biochemical analyses to down select expressed S2 mutant proteins, we arrived at two S2 subunit constructs optimized for stability, conformational fidelity, and product yields as immunogens. Intra-protomer disulfide bonds were the most effective means of enhancing protein expression and thermostability. The Cys803-Cys933 pair was introduced to cross-link regions of mobility and immobility during the pre- to post-fusion transition in a manner similar to the Cys155-Cys290 substitution in RSV F (DS-Cav1) ((McLellan et al., 2013)). We also aimed to make a covalent linkage across the S2′ protease cleavage site, by the formation of a Cys838-Cys1089 bridge, akin to the Cys93(HA2)-Cys310(HA1) substitution in the HA stem ((Impagliazzo et al., 2015)). Finally, Cys898 and Cys1022 substitutions were placed in the FP and HR1 to prevent the helix-loop region from transitioning to a single, elongated helix. Both Cys898 and Cys1022 substitutions were at regions that move substantially, and a similar approach was used to increase the trimer fraction of HA stem (Cys68-Cys76) and to keep PIV3 F in a prefusion conformation (Cys162-Cys168) ((Impagliazzo et al., 2015); (Stewart-Jones et al., 2018)). Disulfide engineering has also been used to restrict local flexibility of secondary structures in SARS-CoV-2 S (Hsieh et al., 2020) or to trap the RBDs in a closed configuration (Henderson et al., 2020; McCallum et al., 2020; Xiong et al., 2020). Most importantly, MERS SS V2 elicited 10-fold higher neutralizing antibodies against MERS-CoV than SS V1 in a single prime-boost immunization regimen. Each disulfide substitution significantly improved the thermostability of the MERS SS antigens, consistent with disulfide designs in other class I viral fusion proteins.
The cavity-filling approach proved to be successful in stabilizing loosely packed regions of RSV F in its prefusion conformation ((Krarup et al., 2015); (McLellan et al., 2013)). We applied this tactic in the optimization of a stable S2-only MERS immunogen by substituting Ser975 and Val983 with Met and Ile, respectively, at the base of MERS-CoV SS. These changes neatly filled a hydrophobic pocket between HR1 and amino acids that remain stationary during the pre- to post-fusion transition. A lone S975M change proved to be the most effective, leading to more than a 10-fold increase in protein yields relative to the parental molecule. Similarly, the N1132Y substitution seems to pack against Pro937, Val1206, and Pro1131 and possibly forms H bonds with Gln800 and Asn1029. Other modifications intended to enhance polar interactions—for instance, V958S, S1091E, and L1094Q substitutions—were considered as beneficial additions to our best construct. Alternative strategies, such as stabilizing proline substitutions, reducing hydrophobicity at the S1-S2 interface, and nanoparticle display, are viable options to further improve our MERS SS.V2 antigens in efforts to induce cross-neutralizing antibodies.
MERS SS immunization stimulated the production of antibodies that were cross-reactive to multiple CoV S proteins. To date, our study is the first to show protection against lethal MERS-CoV challenge with a stem-only immunogen—protection that comes without elicitation of potent neutralizing antibody responses. Previous studies suggest that in vivo activity of influenza virus stem-specific antibodies rely on Fc-mediated functions (DiLillo et al., 2014, 2016). In fact, only a handful of S2-specific neutralizing antibodies against MERS-CoV have been defined (Wang et al., 2015; Widjaja et al., 2019). One of these antibodies, murine mAb G4, protects against challenge in a murine lethal model of MERS-CoV infection and binds a hypervariable loop containing a unique N-glycosylation site in the CD (Pallesen et al., 2017; (Wang et al., 2018)). Roughly one-third of MERS-SS-reactive antibodies compete with G4 Fab for the loop epitope, which suggests at least two types of neutralizing antibodies (G4-like and stalk-targeting) were elicited by MERS-stem-only antigens. Deletion of the highly immunogenic loop from an S2 subunit vaccine might be advantageous to prevent natural immunofocusing on an epitope poorly conserved among even closely related lineage C β-HCoVs.
Our study also describes a conserved epitope near the base of S, distinct from the G4-binding site. Two antibodies that recognize this epitope, namely, IgG22 and IgG72, display heterotypic binding to all highly pathogenic β-HCoVs. Both antibodies neutralize authentic MERS-CoV and at low nM concentrations in the case of IgG22. However, IgG72 demonstrated markedly reduced neutralizing activity against MERS-CoV as compared to IgG22 (approximately a 10-fold reduction in inhibitory titer). Cryo-EM structures of Fab22 complexed with MERS S-2P and prefusion-stabilized SARS-CoV-2 S (HexaPro) revealed the Fab binding site to be in close proximity to the helical portion of the stem, a region that is highly conserved within β-CoVs. Although the binding interface was incompletely resolved in the cryo-EM maps, we were able to obtain high-resolution crystal structures of stalk peptide–Fab22 complexes, which revealed a conserved binding modality of the stalk-targeting antibody. The Fab22 binding site is within a stem helix that must dramatically refold to form a six-helix bundle with HR1, which leads to virus-cell fusion. Sequence alignments of Fab22 and Fab72 reveal only four amino acid differences—one in CDR-L3 and three in CDR-H2—and none of them directly interact with the stem helix, plausibly explaining shared binding profiles for HCoV S proteins (Figure S6C). The faster (nearly 100-fold) dissociation rate of Fab22 for HexaPro (and SARS-CoV S-2P) than that for MERS-CoV S-2P provides an explanation for the undetectable heterotypic neutralizing activity by IgG22 (and IgG72). We note that similar antibodies have been recently described (Pinto et al., 2021; Sauer et al., 2021; Zhou et al., 2021), including some isolated from humans.
We explore the potential function of the cross-reactive antibody IgG22 by assessing its ability to provide protection in lethal challenge models of MERS-CoV and SARS-CoV-2. Although mAb G4 has been shown to induce protection at 20 mg/kg in mouse models ((Wang et al., 2018)), here, we confirm its specificity for MERS-CoV and demonstrate that IgG22 at the equivalent dose protects against not only homologous challenge but also a SARS-CoV-2 lethal challenge. The specificity of mAb G4 toward a highly variable glycosylated loop in the generally well-conserved S2 domain of MERS-CoV (Pallesen et al., 2017) may help explain its failure to both cross-react and to protect against SARS-CoV-2 challenge. Our studies reveal that unlike G4, IgG22 recognizes a highly conserved site in S2 that drives virus-cell fusion, which provides a rationale for why targeting this site protects against other β-CoVs. Although our studies demonstrated that IgG22 fails to neutralize authentic SARS-CoV-2, it significantly reduced tissue damage and lung discoloration scores compared to untreated mice and promoted rapid clearance of virus from the lower respiratory tract. Passive administration of IgG22 did not reduce virus replication in the upper respiratory tract, corroborating recent findings that the threshold for IgG-mediated inhibition of SARS-CoV-2 replication is lower for the lung than upper airways (Corbett et al., 2021). These findings emphasize the importance of determining other functional mechanisms by which non-neutralizing antibodies protect, especially in the context of cross-protection. Identifying the conserved IgG22 binding site may inform future vaccine designs needed not only to manage the ongoing pandemic but also for the development of broader vaccines to combat future spillovers of novel CoVs into humans. Identification of sites, like IgG22’s binding epitope that cannot freely accumulate substitutions to avoid loss-of-function mutations, enables vaccine development efforts to target sites less likely to escape antibody-mediated protection. As current vaccines use full-length SARS-CoV-2 S antigens, the immunodominant RBD and NTD skew humoral responses upon immunization—which may have reduced activity against SARS-CoV-2 VOCs harboring conformational changes in the S1 domain. The use of stem antigens may lessen the selective pressure for advantageous substitutions in the regions that are responsible for interacting directly with host cells.
In summary, this study provides a proof of concept that the CoV S stem can be rationally engineered as a stabilized immunogen to elicit antibodies cross-reactive with all three epidemic β-HCoVs and to provide complete protection in animals against a lethal MERS-CoV challenge. Using a SS construct, we induced the production of antibodies that enabled the identification of a site of vulnerability in the S2 subunit, which will inform the development of next-generation CoV vaccines and fortify CoV pandemic preparedness.
Limitations of the study
One limitation of this study is the lack of structural data showing the extent to which the stabilizing mutations in the MERS-CoV S2 subunit favor the prefusion conformation. Further structure-based designs focused on stabilizing the trimer interface could potentially lead to more native-like S2 immunogens. In addition, optimization of the helical stalk regions where the cross-protective antibodies bound could be an alternative approach to design next-generation pan-CoV vaccines. Another limitation is that the immunogen did not provide cross-protection for animals against SARS-CoV-2 infection. Elicitation of broadly neutralizing or protective sera is desirable, but these first-generation immunogens were unable to meet that standard. Although IgG22 provided protection in mice against both MERS-CoV and SARS-CoV-2 challenge, additional immunogenicity studies conducted in non-human primates need to be performed to strengthen this conclusion.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-MERS-CoV S antibody, G4 | PMID: 28807998 | N/A |
| Goat anti-mouse IgG-horseradish peroxidase (HRP) conjugate | SigmaAldrich | Cat # G-21040 |
| Anti-MERS-SS antibody, IgG22 | This manuscript | N/A |
| Anti-MERS-SS antibody, IgG72 | This manuscript | N/A |
| Anti-SARS-CoV S antibody, S309 | PMID: 32422645 | N/A |
| Bacterial and virus strains | ||
| MERS-CoV EMC/2012 | Scobey et al., 2013 | N/A |
| DH5α competent cells | ThermoFisher | Cat# 18258012 |
| SARS-CoV-2/human/USA/WA-CDC-WA1/2020 | CDC | GenBank: MN985325.1 |
| Mouse-adapted MERS-CoV EMC derivative, maM35c4 | PMID: 29277291 | N/A |
| SARS-CoV-2 MA10 | PMID: 33031744 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 3,5,3′5’-tetramethylbenzidine (TMB) | KPL | Cat# 5150-0021 |
| MERS-CoV S-2P protein | PMID: 28807998 | N/A |
| SARS-CoV-1 S-2P protein | PMID: 28807998 | N/A |
| SARS-CoV-2 S-2P protein | PMID: 32075877 | N/A |
| SARS-CoV-2 S-HexaPro protein | PMID: 32703906 | N/A |
| DS-Cav1 protein | PMID: 24179220 | N/A |
| T4 foldon protein | PMID: 28807998 | N/A |
| MERS-CoV SS.V1 protein | This manuscript | N/A |
| MERS-CoV SS.V2 protein | This manuscript | N/A |
| MERS peptide- DFQDELDEFFKNVST | This manuscript | N/A |
| SARS peptide- DSFKEELDKYFKNHT SPDVD |
This manuscript | N/A |
| Bovine serum albumin | Sigma-Aldrich | Cat# A8327 |
| Anti-Human IgG Fc Capture (AHC) Biosensors | FortéBio | Cat# 18-5063 |
| SYPRO Orange Protein Gel Stain | ThermoFisher | Cat# S6651 |
| FreeStyle 293 Expression Medium | GIBCO | Cat# 12338002 |
| OPTI-MEM, Reduced Serum Medium | ThermoFisher | Cat# 11058021 |
| Kifunensine | GlycoSyn | Cat# FC-034 |
| 25 kDa linear polyethylenimine | Polysciences | Cat# 3966-2 |
| Critical commercial assays | ||
| Luciferase Assay System | Promega | Cat# E1500 |
| Luciferase Cell Culture Lysis 5X Reagent | Promega | Cat# E1531 |
| FuGENE® HD Transfection Reagent | Promega | Cat# E2311 |
| Deposited data | ||
| Crystal structure of Fab22 + MERS-CoV S peptide | This manuscript | PDB ID: 7S3M |
| Crystal structure of Fab22 + SARS-CoV-2 S peptide | This manuscript | PDB ID: 7S3N |
| EM map of Fab22 + MERS-CoV S | This manuscript | EMDB ID: 25072 |
| EM map of Fab22 + SARS-CoV-2 S | This manuscript | EMDB ID: 25073 |
| Experimental models: Cell lines | ||
| Huh7.5 cells | Provided by Dr. Deborah R. Taylor of the US FDA | N/A |
| Freestyle 293F cells | ThermoFisher Scientific | Cat# R7007 |
| Vero E6 cells | PMID: 14569023 | N/A |
| Vero 81 cells | ATCC | Cat# CCL-81 |
| Experimental models: Organisms/strains | ||
| Mus musculus, strain BALB/cJ | Jackson Laboratories | JAX stock #000651 |
| Mus musculus, strain BALB/cAnNHsD (BALB/c) | Envigo | Stock # 047 |
| Mus musculus, strain C57BL/6J | Jackson Laboratories | JAX stock #000664 |
| Mus musculus, strain 288/330+/+ | PMID: 31883094 | N/A |
| Recombinant DNA | ||
| pCG1-SARS-2-S | PMID: 32142651 | N/A |
| pCG1-SARS-S | PMID: 24023659 | N/A |
| pαH expression plasmid | McLellan Laboratory | N/A |
| pαH-MERS-SS.V1 | This manuscript | N/A |
| pαH-MERS-SS.V1 | This manuscript | N/A |
| pαH-SARS-CoV-1 S-2P | PMID: 28807998 | N/A |
| pαH-MERS-CoV S-2P | PMID: 28807998 | N/A |
| pαH-SARS-CoV-2 S-HexaPro | PMID: 32703906 | N/A |
| pVRC8400-VH22 | This manuscript | N/A |
| pVRC8400-VL22 | This manuscript | N/A |
| pVRC8400-VH72 | This manuscript | N/A |
| pVRC8400-VL72 | This manuscript | N/A |
| pHR′ CMV-Luc | Graham Laboratory | N/A |
| CMV/R-MERS-CoV S | Graham Laboratory | N/A |
| CMV/R-SARS-CoV-1 S | Graham Laboratory | N/A |
| Software and algorithms | ||
| Protein Repair One-Stop Shop (PROSS) server | PMID: 27425410 | https://pross.weizmann.ac.il/step/pross-terms/ |
| Octet Data Analysis software | FortéBio | v11.1 |
| GraphPad Prism | (Motulsky and Brown, 2006) | V9.0.2 |
| Biacore X100 Evaluation Software | GE Healthcare | V2.0.1 |
| iMOSFLM | Battye et al., 2011 | https://www.mrc-lmb.cam.ac.uk/harry/imosflm/ver721/downloads.html |
| Aimless | Evans and Murshudov, 2013 | www.ccp4.ac.uk/download/ |
| Phaser | McCoy et al., 2007 | www.ccp4.ac.uk/download/ |
| COOT | Emsley and Cowtan, 2004 | http://bernhardcl.github.io/coot/ |
| Phenix | Adams et al., 2002; Afonine et al., 2018 | https://phenix-online.org/ |
| ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| cisTEM | Grant et al., 2018 | https://cistem.org/ |
| cryoSPARC | Punjani et al., 2017 | v2.15.0 |
| Other | ||
| Strep-Tactin Superflow resin | IBA Lifesciences | Cat# 2-1206-010 |
| Pierce™ Protein A Agarose | ThermoFisher Scientific | Cat# 20334 |
| Biacore X100 Sensorchip NTA | GE Healthcare | Cat# BR100407 |
| Superose 6 increase 10/300 GL | GE Healthcare | Cat# 29091596 |
| Sigma Adjuvant System | Sigma-Aldrich | Cat# S6322-1VL |
| Carbon Film 400 Mesh, Cu | Electron Microscopy Sciences | Cat# CF400-Cu-50 |
| C-Flat Holey Carbon Grid CF-1.2/1.3-4C, 400 mesh Cu | Electron Microscopy Sciences | Cat# CF413-50 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jason McLellan (jmclellan@austin.utexas.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell lines
FreeStyle293F mammalian cells (ThermoFisher) were maintained in FreeStyle293 expression medium (ThermoFisher) in a 37°C shaker supplied with 8% CO2 and 80% humidity. Huh7.5, Vero81, and Vero E6 cells were incubated in DMEM (ThermoFisher) at 37°C and 5% CO2.
Mouse strains
Female BALB/cJ mice and C57BL/6J mice aged 6- to 8-weeks were obtained from Jackson Laboratory. Female 10-week-old BALB/c were obtained from Envigo (BALB/cAnNHsD; stock # 047). Sixteen to 20-week-old male and female 288/330+/+ mice were generated in the Baric Laboratory. All mouse experiments were carried out in compliance with all pertinent US National Institutes of Health regulations and approval from the Animal Care and Use Committee (ACUC) of the Vaccine Research Center, from Institutional Animal Care and Use Committee at University of North Carolina at Chapel Hill to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the U.S. Department of Agriculture, or from ACUC of Abcellera Biologics. All infection experiments were done in animal biosafety level 3 (BSL-3) facilities at the University of North Carolina at Chapel Hill.
Method details
Design scheme for MERS SS variants
The base construct used for the S2 subunit of MERS-CoV S-2P variant contained residues 762-1291 of MERS-CoV S (GenBank ID: AFY13307) with proline substituted at residues 1060 and 1061, the foldon trimerization motif of T4 fibritin, an HRV3C protease recognition site, an octa-histidine tag, and a tandem Twin-Strep-tag, cloned into the mammalian expression plasmid pαH. All mutations in subsequent designs were introduced into this base construct. The initial design was conducted via the PROSS server (Goldenzweig et al., 2016), and a total of 11 substitutions were picked (yielding the ‘mut11’ construct) from 53 computational designs based on the biochemical property of the residues and steric effect on the prefusion-stabilized MERS S-2P structure (PDB ID: 5W9I). Using structure-based design, additional substitutions that were aimed at favoring the stability of the prefusion structure were introduced into the mut11 backbone. Pairs of core-facing residues less than 5 Å apart were replaced with aromatic sidechains or pairs of aromatic and positively charged sidechains to favor pi-pi or pi-cation interactions, respectively. Alternatively, residues were replaced with extended or bulkier hydrophobic sidechains in efforts to fill pre-existing internal cavities. Disulfide bonds were designed to increase overall stability or prevent formation of the postfusion conformation. The charged or polar substitutions were aimed to establish hydrogen bonds or salt bridges with the native residues that were predicted to be within 4.0 Å. To examine the effect of the substitutions in the mut11 backbone, the mutations were individually or combinatorially reverted back to the wild-type sequence. Three designs, each containing 5–7 of the beneficial substitutions from the PROSS server (mut5, mut6 and mut7) were chosen to serve as backgrounds for subsequent rounds of design. These were then combined with individual substitutions or combinations of substitutions from the structure-based designs that were shown to be beneficial, for subsequent assessment of improved monodispersity and thermostability. Substitutions predicted to potentially interfere with one another or clash with native residues were avoided.
Expression and purification of MERS SS
Plasmids encoding MERS SS variants were transiently transfected into FreeStyle293F cells (Thermo Fisher) using polyethylenimine, with 5 μM kifunensine being added 3h post-transfection. Cultures were grown for 6 days, and culture supernatant was separated via centrifugation and passage through a 0.22 μm filter. Protein was purified from supernatants using StrepTactin resin (IBA). MERS SS variants were further purified by size-exclusion chromatography (SEC) using a Superose 6 increase 10/300 column (GE Healthcare) in a buffer composed of 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3. For initial purification and characterization, single-substitution and combinatorial variants were purified from 40 mL cultures. The SS.V1 and SS.V2 variants were further tested in large-scale expression and purification from 1 L cultures. The protein purity, monodispersity and expression level were determined by SDS-PAGE and SEC.
Differential scanning fluorimetry
MERS SS variants were prepared at a concentration of 1.5 μΜ with a final concentration 5X SYPRO Orange Protein Gel Stain (ThermoFisher) in a white, opaque 96-wellplate. Continuous fluorescence measurements (λex = 465 nm, λem = 580 nm) were performed using a Roche LightCycler 480 II, with a temperature ramp rate of 4.4°C/minute, and a temperature range of 25°C to 95°C. Data were plotted as the derivative of the melting curve.
Mouse Experiments
All mouse experiments were carried out in compliance with all pertinent US National Institutes of Health regulations and approval from the Animal Care and Use Committee (ACUC) of the Vaccine Research Center, from Institutional Animal Care and Use Committee at University of North Carolina at Chapel Hill to guidelines outlined by the Association for the Assessment and Accreditation of Laboratory Animal Care and the U.S. Department of Agriculture, or from ACUC of Abcellera Biologics. All infection experiments were done in animal biosafety level 3 (BSL-3) facilities at the University of North Carolina at Chapel Hill. For immunogenicity studies, female BALB/cJ mice aged 6- to 8-weeks (Jackson Laboratory) were used. Per the experimental design schema outlined in Figure 3A, mice were inoculated intramuscularly with protein immunogens adjuvanted with SAS as previously described (Pallesen et al., 2017) and bled for serological assays. For challenge studies to evaluate MERS-CoV vaccines, 16- to 20-week-old male and female 288/330+/+ mice (Cockrell et al., 2016) were immunized, bled, and challenged, as detailed in Figure 4A. Mice were challenged with 5 × 105 PFU of a mouse-adapted MERS-CoV EMC derivative, maM35c4 (Douglas et al., 2018). On days 3 and 5 post-challenge, lungs were collected from selected mice to assess viral titers and discoloration, using previously published methods. Briefly, caudal right lung lobes were harvested for analysis of viral load by plaque assay. Lung lobes were homogenized in 1mL of PBS and glass beads. Clarified homogenates were used to inoculate monolayers of Vero E6 cells for SARS-CoV-2 MA10 or Vero CCL81 cells for MERS-CoV maM35c4 which were stained with Neutral Red dye after 72 hours to visualize plaques. For S-reactive monoclonal antibody isolation, female C57BL/6J mice aged 4- to 8-weeks (Jackson Laboratory) were used. Mice were immunized intramuscularly with 10 μg MERS SS.V1 + SAS at weeks 0, 3, and 9. At week 13, mice were euthanized and spleens, thymuses, and lymph nodes were harvested for single B cell technology for mAb isolation. For passive transfer studies to consider the protective efficacy of IgG22 in MERS- and SARS-CoV-2 challenges, male and female 288/330+/+ mice aged 16–20 weeks (Cockrell et al., 2016) and 10-week-old female BALB/c mice (Dinnon et al., 2020) (Envigo), respectively, received either 20 mg/kg or 2 mg/kg of antibody diluted appropriately in 1X PBS via intraperitoneal injection one day prior to challenge. On the day of challenge, mice were infected intranasally with 5 × 105 PFU MERS-CoV maM35c4, or 1 x 104 PFU SARS-CoV-2 MA10 (Leist et al., 2020). For 288/330+/+ mice infected with MERS-CoV, mice were monitored up to 5 days post-challenge, and were euthanized and lungs were collected to investigate lung discoloration, viral burden, and nasal turbinate was collected from select mice to determine viral burden in the upper airway as described above. For BALB/c mice infected with SARS-CoV-2, mice were monitored up to 4 days post-challenge, and were euthanized and lungs were collected to investigate lung discoloration, viral burden, and nasal turbinate was collected from select mice to determine viral burden in the upper airway as described above. Sample size for animal experiments was determined on the basis of criteria set by institutional ACUC. Experiments were neither randomized nor blinded.
Serum IgG Measurement
HCoV S-2P-specific IgG in immunized mouse sera were quantified via enzyme-linked immunosorbent assay (ELISA). Briefly, Nunc MaxiSorp 96-well plates (ThermoFisher) were coated with either MERS S-2P, SARS S-2P, SARS-CoV-2 S-2P, or HKU1 S-2P at 1 μg/mL in 1X PBS at 4°C for 16h. Sera dilutions were prepared in blocking buffer, which consisted of PBS-Tween 20 + 5% non-fat dairy milk, and diluted serially at 1:100, four-fold, 8x. To eliminate foldon-specific binding, 50 μg/mL of foldon was added to the dilutions and incubated for an hour at room temperature. After standard washes and blocks, goat anti-mouse IgG-horseradish peroxidase (HRP) conjugate (SigmaAldrich) was used as secondary antibody. Plates were reacted with 3,5,3′5’-tetramethylbenzidine (TMB) (KPL) to detect binding responses. Plates were read at OD450/650 using SpectraMax Paradigm (Molecular Devices). Endpoint titers were calculated as the reciprocal serum dilution that yielded a signal 4x greater than that of the background signal (secondary antibody alone).
MERS-CoV Pseudovirus Neutralization Assay
Neutralization activity was assessed as previously described (Pallesen et al., 2017). Briefly, Huh7.5 cells were seeded at 10,000 cells/well in 96-well black/white Isoplates (PerkinElmer) 24-h prior to infection. Sera were serially diluted (1:40, 4-fold dilutions, 8x) in DMEM (GIBCO) + 1% penicillin/streptomycin, and mixed with a pseudotyped MERS-CoV England1 lentivirus reporter that was previously titrated to 104 RLU, and incubated for 30 minutes at room temperature. The sera + pseudovirus mixture was then added to Huh7.5 cells in duplicate, and incubated at 37°C and 5% CO2 for 2h. Then, 100 μL of DMEM supplemented with 10% FBS, 2 mM glutamine, and 1% penicillin/streptomycin was added to each well, and incubated for 72h. Cells were then lysed, and luciferase substrate (Promega) was added. Luciferase activity was measured as relative luciferase units (RLU) at 570 nm, using a SpectraMaxL (Molecular Devices). Sigmoidal curves, taking averages of triplicates at each dilution, were generated from RLU readings; 50% neutralization (ID50) titers were calculated considering uninfected cells as 100% neutralization and cells transduced with only virus as 0% neutralization, by fitting RLU readings to a log(agonist) versus normalized-response (variable slope) nonlinear regression model in Prism v9 (GraphPad).
Single-cell screening and recovery
Immunized mice exhibiting elevated serum titers were sacrificed and plasma cells from lymph node, spleen, and bone marrow tissues were isolated using standard protocols. Samples were screened with AbCellera’s high-throughput single-cell microfluidic platform using a multiplexed microbead assay on devices containing individual nanoliter-volume reaction chambers (Lecault et al., 2011). The multiplexed assay employed multiple optically-encoded beads, each conjugated to one of the following unique antigens: full-length pre-fusion stabilized S proteins of MERS-CoV, SARS-CoV, or HKU1-CoV, or the S2 subunit of MERS-CoV S. Bead-conjugated bovine serum albumin (BSA) His-tag and T4 foldon trimerization domain were used as negative controls. Beads were flowed into microfluidic screening devices and incubated with single antibody-secreting cells, and monoclonal antibody binding to cognate antigens was detected via a fluorescently labeled anti-human IgG secondary antibody. Positive hits were identified using machine vision and recovered using automated robotics-based protocols.
Single-cell sequencing and cloning
For recovery of paired heavy and light chain sequences, single-cell polymerase chain reaction (PCR) and next-generation sequencing (MiSeq, Illumina) were performed using automated workstations (Bravo, Agilent) and custom molecular biology protocols. Sequences were analyzed using a custom bioinformatics pipeline to yield paired heavy and light chain sequences for each recovered antibody-secreting cell. Each sequence was assigned the closest germline (V(D)J) genes, degree of somatic hypermutation, and potential sequence liabilities.
Expression and purification of antibodies
Twenty monoclonal antibodies discovered by single B cell technology were selected for characterization based on their binding specifies to HKU1-S, MERS-S and SARS-1-S (monospecific, bi-specific or tri-specific), diversity of heavy and light chain CDR3s, and high frequency rates in the B cell repertoire (independently isolated by the probes more than 2 times). The individual VH sequence of selected IgGs was cloned into a mammalian expression plasmid pVRC8400 containing HRV 3C cleavage site in the hinge, and human IgG1 Fc domain. VL sequences were also cloned into pVRC8400 with human CL. Paired VH and VL in a 1:1 ratio were co-transfected transiently into FreeStyle293F cells as previously described. The supernatant was harvested six days post-transfection and IgGs were purified with Protein A agarose (ThermoFisher). IgGs were eluted with 100 mM glycine, pH 3 into 1/10th volume 1 M Tris-HCl pH 8.0. IgGs were then buffer exchanged into PBS pH 7.4. Fabs were generated by digesting the IgGs with HRV 3C protease at 4°C. Fc was removed by passing digests over fresh Protein A agarose, leaving the Fab in the flowthrough, which was further purified by SEC using a Superdex 200 increase 10/300 column (GE Healthcare) in PBS buffer, pH 7.4.
Biolayer interferometry
The binding affinity of purified IgGs to HKU1-CoV S, MERS-CoV S or SARS-CoV S was characterized by BLI using an Octet RED96e (FortéBio). Briefly, anti-human Fc (AHC) sensors (FortéBio) with captured IgG were dipped into the wells containing 100 nM of CoV S in a BLI buffer composed of 10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% v/v Tween 20 and 1 mg/ml BSA. After 600 s association step, the dissociation step was carried out in the wells containing only BLI buffer for 600 s. RSV F-foldon was also included in the experiments as negative control. The binding affinity of purified mAb G4 to MERS-CoV S and MERS SS was also characterized by BLI using a similar approach. AHC sensors with captured mAb G4 were dipped into the wells containing serial dilutions of MERS-CoV S or MERS SS at concentrations ranging from 100 to 1.56 nM in a BLI buffer. The data were aligned to a baseline prior to association step and fit to a 1:1 binding model to calculate rate and dissociation constants using Octet Data Analysis software v11.1. The IgGs exhibiting binding to MERS-CoV S were further examined for the ability to compete with mAb G4, the only S2-targeted MERS-S antibody with a defined epitope (Pallesen et al., 2017). Four-fold molar excess of G4 Fab was preincubated with 100 nM MERS-S at room temperature for 10 min. The IgG being tested was loaded on AHC sensors and then dipped into either 100 nM apo MERS-S or 100 nM G4-presaturated MERS-S. G4 IgG was also included in one set of the experiments as a control. The data were plotted as the difference between the binding level of G4-saturated and apo MERS-S, normalized to the binding level of apo MERS-S to the IgG of interest.
Surface Plasmon Resonance
To accurately determine the binding kinetics, His-tagged S variants (MERS-CoV S-2P, SARS-CoV S-2P and SARS-CoV-2 S-HexaPro) were immobilized to a Ni-NTA sensorchip (GE Healthcare) to a level of ∼600 response units (RUs) using a Biacore X100 (GE Healthcare) and running buffer composed of 10 mM HEPES pH 8.0, 150 mM NaCl and 0.05% Tween 20. Serial dilutions of purified Fab22 were injected at concentrations ranging from 400 to 6.25 nM over immobilized MERS-CoV S-2P. For the SARS-CoV S-2P and SARS-CoV-2 S-HexaPro binding experiments, Fab22 concentrations ranging from 100 to 3.13 nM were used instead. The Ni-NTA sensorchip was regenerated between each cycle with 0.35 M EDTA, 50 mM NaOH and followed by 0.5 mM NiCl2. Response curves were double-reference subtracted and fit to a 1:1 binding model using Biacore X100 Evaluation Software (GE Healthcare).
Plaque-reduction neutralization test (PRNT)
Four-fold serial dilutions of mAbs were combined with an average of 124 plaque-forming units of MERS-CoV (HCoV-EMC/2012) or SARS-CoV-2 (SARS-CoV-2/human/USA/USA-WA1/2020) in 200 μL gelatin saline (0.3% [wt/vol] gelatin in phosphate-buffered saline supplemented with CaCl2 and MgCl2) for 20 min at 37°C, and 100 μL of virus-mAb mixture was applied to each of two confluent Vero 81 or Vero E6 cell monolayers, respectively, in 6-well (10-cm2) plates. Monolayers were overlaid with Dulbecco’s modified Eagle’s medium (DMEM) containing 1% agar following virus adsorption for 30 min at 37°C, and plaques were enumerated at 72 h or 96 h post-infection. Percent plaque reduction resulting from mAb treatment (relative to untreated virus control) was plotted as a function of log10 mAb concentration. Neutralization data were subjected to five-parameter logistic regression modeling using PRISM 9 (GraphPad). Minimum mAb concentrations resulting in 50% and 80% virus neutralization were interpolated from fitted dose-response curves.
Negative stain EM for S-Fab complexes
Purified MERS-CoV S-2P or SARS-CoV-2 S-HexaPro were incubated with 2-fold molar excess of Fab22 or Fab72 or Fab20 in 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3 at room temperature for 30 min. The S-Fab complexes were diluted to a concentration of 0.06 mg/mL in 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3. Each protein complex was deposited on a CF-400-CU grid (Electron Microscopy Sciences) that had been plasma cleaned for 30 s in a Solarus 950 plasma cleaner (Gatan) with a 4:1 ratio of O2/H2 and stained using methylamine tungstate (Nanoprobes). Grids were imaged at a magnification of 92,000X (corresponding to a calibrated pixel size of 1.63 Å/pix) in a Talos F200C TEM microscope equipped with a Ceta 16M detector (Thermo Fisher Scientific). The CTF-estimation and particle picking were performed in cisTEM (Grant et al., 2018). Particles were then imported into cryoSPARC v2.15.0 for 2D classification (Punjani et al., 2017).
Cryo-EM
Purified MERS-CoV S-2P at 1 mg/mL was incubated with 2-fold molar excess of Fab22 in 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3 at room temperature for 30 min. Sample was then deposited on a plasma-cleaned CF-400 1.2/1.3 grid before being blotted for 5 s with +1 force in a Vitrobot Mark IV (ThermoFisher) and plunge-frozen into liquid ethane. Similarly, purified SARS-CoV-2 S (HexaPro variant) complexed with 2-fold molar excess of Fab22 was diluted to a concentration of 0.37 mg/mL in 2 mM Tris pH 8.0, 200 mM NaCl, 0.02% NaN3 and applied to plasma-cleaned CF-400 1.2/1.3 grids before being blotted for 3.5 s with −4 force in a Vitrobot Mark IV and plunge-frozen into liquid ethane. For the Fab22-MERS S-2P sample, 4330 micrographs were collected from a single grid. For the Fab22-HexaPro sample, 2,013 micrographs were collected from a single grid. FEI Titan Krios (ThermoFisher) equipped with a K3 direct electron detector (Gatan) was used for imaging. Data were collected at a magnification of 22,500x, corresponding to a calibrated pixel size of 1.07 Å/pix. A full description of the data collection parameters can be found in Table S1. Warp was used for motion correction, CTF estimation, and particle picking ((Tegunov and Cramer, 2019)). The particle stack was then imported into cryoSPARC v2.15.0, which was used to curate the particles via iterative rounds of 2D classification (Punjani et al., 2017). The final reconstructions were then arrived at via ab initio reconstruction, heterogeneous refinement, and subsequently non-uniform homogeneous refinement of final classes. The structure validation can be found in Figure S5 .
X-ray crystallography
To crystallize the Fab22–MERS-stem-helix-peptide complex, MERS stem helix peptide (DFQDELDEFFKNVST, GenScript) was added in 4-fold molar excess to Fab22, resulting in a final concentration of 14 mg/ml Fab22 and 2 mg/ml peptide in 2 mM Tris pH 8, 200 mM NaCl, 0.02% NaN3. Peptide complex (0.1 μl) was mixed with 0.1 μL mother liquor (10% PEG1000, 10% PEG8000) using an NT8 (Formulatrix), and the 0.2 μL drop was spotted onto an MRC2 crystallization tray to allow for vapor diffusion, with 60 μL mother liquor in the well. For the SARS-CoV-2 stem helix peptide complex, a modified version of Fab22 containing a mutation in the light chain constant Ig domain (CL) that favors Fab crystallization was used (LCmod-Fab22, HQGLSSP to QGTTS) (Lieu et al., 2020). SARS-CoV-2 stem helix peptide (DSFKEELDKYFKNHTSPDVD, GenScript) was added in 5-fold molar excess to LCmod-Fab22, resulting in a final concentration of 11 mg/ml LCmod-Fab22 and 2.1 mg/ml peptide in 2 mM Tris pH 8, 200 mM NaCl, 0.02% NaN3. Peptide complex (0.1 μl) was mixed with 0.05 μL mother liquor (0.1 M sodium citrate pH 5.5, 23% PEG8000) using an NT8 (Formulatrix), and the 0.15 μL drop was spotted onto an MRC2 crystallization tray to allow for vapor diffusion, with 60 μL mother liquor in the well. Crystals were rapidly soaked in their respective mother liquors supplemented with 30% ethylene glycol, looped, and flash-frozen in liquid nitrogen. Remote data collection was performed at the SBC beamline 19ID (Advanced Photon Source, Argonne National Laboratory). Diffraction data were indexed and integrated in iMOSFLM (Battye et al., 2011), before being merged and scaled (to 2.4 Å for MERS peptide complex and to 1.9 Å for SARS-CoV-2 peptide complex) using Aimless (Evans and Murshudov, 2013). Molecular replacement was performed in Phaser (McCoy et al., 2007), and models were then subjected to multiple rounds of model building and refining in Coot and Phenix, respectively (Adams et al., 2002; Emsley and Cowtan, 2004). Data collection and refinement statistics can be found in Table S2.
Quantification and statistical analysis
Figure legends detail all quantification and statistical analyses, inclusive of animal numbers (n), dispersion and precision measures, and statistical tests. For comparisons of antibody responses and challenge outcomes between experimental groups, one-way ANOVA with Kruskal-Wallis post-tests were used. Serum IgG endpoint titers and pseudovirus neutralizing antibody titers were transformed so that all values were on a log(10) scale prior to statistical analyses. All one-way ANOVA analyses did not assume Gaussian distribution and were performed as non-parametric tests. One-way ANOVA analyses with Kruskal-Wallis post-tests corrected for multiple comparisons using statistical hypothesis testing via Dunn’s tests. For comparisons of serum IgG endpoint titers (Figures 2B–2E), two-way ANOVA analyses were performed between groups receiving the same dose of different immunogens, and between groups receiving the same immunogen at different doses. Two-way ANOVA analyses with multiple comparisons compared values within each row across columns; i.e., each cell mean was compared with every other cell mean within their respective row. Corrections for multiple comparisons using statistical hypothesis testing were performed via Sidak tests for all two-way ANOVA analyses. p values are summarized within figures as: ∗ < 0.05, ∗∗ < 0.01, ∗∗∗ < 0.001, ∗∗∗∗ < 0.0001. All statistical analyses were conducted using Graphpad Prism v9.0.2, i.e., geometric mean titers (GMT), means, and SEM (standard error of the mean) for serum IgG endpoint titers, pseudovirus neutralization titers, plaque reduction neutralization titers, body weight loss of mice in challenge assessments, mouse lung viral loads, and discoloration scores.
Acknowledgments
We would like to thank Dr. Sasha Dickinson from the Sauer Structural Biology Laboratory at the University of Texas at Austin for his assistance with microscope data collection. We thank members of the VRC Translational Research Program for technical and administrative assistance with animal experiments; Kaitlyn Morabito and Gabriela Alvarado for project management support; Seyhan Byogolu-Barnum for technical assistance with animal experiments; and Cynthia Ziwawo, Dr. Lingshu Wang, and Dr. John Mascola for technical assistance with assays. X-ray Crystallography results shown in this report are derived from work performed at Argonne National Laboratory (ANL), Structural Biology Center (SBC) at the Advanced Photon Source (APS), under U.S. Department of Energy, Office of Biological and Environmental Research contract DE-AC02-06CH11357. This work was supported in part by a National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) grant awarded to J.S.M. (R01-AI127521) and by the Intramural Research Program of the Vaccine Research Center (VRC), NIAID, NIH (B.S.G.). PRNT assays were funded under NIH contract HHSN261200800001E agreement 17x198 (to J.D.C.), furnished through Leidos Biomedical Research. MERS-CoV mouse challenge studies were funded under NIH contract HHSN272201700036I task order no. 75N93019F00132 requisition no. 5494549 and U01 AI149644 (to R.S.B.). K.S.C. is the recipient of a research fellowship that was partially funded by the Undergraduate Scholarship Program, Office of Intramural Training and Education, Office of the Director, NIH. This study was also funded by the US Department of Defense, Defense Advanced Research Projects Agency (DARPA) - Pandemic Prevention Platform, agreement no. D18AC00002.
Author contributions
C.-L.H. and J.S.M. preformed the antigen design. C.-L.H., A.P.W., K.S.C., and J.S.M. conceptualized the study. A.P.W., S.R.L., O.M.A., A.W., E.J.F., K.H.D., K.S.C., and A.S. conducted animal experiments. A.P.W. conducted pseudovirus neutralization assays. L.J.S. and J.D.C. conducted live virus neutralization assays. E.F., K.W., and K.M. isolated MERS SS murine antibodies. C.-L.H. and J.A.G. performed the structural studies. C.-L.H., A.P.W., C.-W.C., and O.M.A., characterized the antigen-antibody interactions. C.-L.H., J.A.G., and A.P.W. drafted the manuscript. M.R.D., J.D.C., R.S.B., B.S.G., K.S.C., and J.S.M. revised and polished the manuscript.
Declaration of interests
C-L.H. and J.S.M. are inventors on U.S. patent application no. 63/188,813 (“Stabilized S2 Beta-coronavirus Antigens”). This research was, in part, funded by the U.S. Government. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the U.S. Government. S.R.L., K.H.D., and R.S.B. have a pending patent for recombinant SARS-CoV-2 MA10 used in this study.
Published: October 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109929.
Supplemental information
Data and code availability
-
•
Atomic coordinates for the Fab22–MERS-CoV S peptide and the Fab22–SARS-CoV-2 S peptide structures have been deposited into the Protein Data Bank and assigned PDB IDs 7S3M and 7S3N, respectively. Cryo-EM maps of Fab22–MERS-CoV S-2P and Fab22–SARS-CoV-2 S-HexaPro have been deposited in the EMDB and assigned codes EMD-25072 and EMD-25073.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta. Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnsonv N., Paciello I., Dal Monego S., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2103154118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., el Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyoglu-Barnum S., Hutchinson G.B., Boyington J.C., Moin S.M., Gillespie R.A., Tsybovsky Y., Stephens T., Vaile J.R., Lederhofer J., Corbett K.S., et al. Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat. Commun. 2020;11:791. doi: 10.1038/s41467-020-14579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Moin S.M., Yassine H.M., Cagigi A., Kanekiyo M., Boyoglu-Barnum S., Myers S.I., Tsybovsky Y., Wheatley A.K., Schramm C.A., et al. Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. MBio. 2019;10 doi: 10.1128/mBio.02810-18. e02810–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Nason M.C., Flach B., Gagne M., O’ Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune Correlates of Protection by mRNA-1273 Immunization against SARS-CoV-2 Infection in Nonhuman Primates. bioRxiv. 2021 doi: 10.1101/2021.04.20.440647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., III, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M.G., Kocher J.F., Scobey T., Baric R.S., Cockrell A.S. Adaptive evolution influences the infectious dose of MERS-CoV necessary to achieve severe respiratory disease. Virology. 2018;517:98–107. doi: 10.1016/j.virol.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenzweig A., Goldsmith M., Hill S.E., Gertman O., Laurino P., Ashani Y., Dym O., Unger T., Albeck S., Prilusky J., et al. Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Mol. Cell. 2016;63:337–346. doi: 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T., Rohou A., Grigorieff N. cisTEM, user-friendly software for single-particle image processing. Elife. 2018;7 doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Hodcroft E.B., Fauver J.R., Phelan A.L., Cevik M. Public health actions to control new SARS-CoV-2 variants. Cell. 2021;184:1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:eabd0826. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A., Milder F., Kuipers H., Wagnerv M., Zhu X., Hoffman R.M.B., van Meersbergen R., Huizingh J., Wanningen P., Verspuij J., et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- Krarup A., Truan D., Furmanova-Hollenstein P., Bogaert L., Bouchier P., Bisschop I.J.M., Widjojoatmodjo M.N., Zahn R., Schuitemaker H., McLellan J.S., et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku Z., Xie X., Davidson E., Ye X., Su H., Menachery V.D., Li Y., Yuan Z., Zhang X., Muruato A.E., et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021;12:469. doi: 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. Fast-spreading U.K. virus variant raises alarms. Science. 2021;371:9–10. doi: 10.1126/science.371.6524.9. [DOI] [PubMed] [Google Scholar]
- Lecault V., Vaninsberghe M., Sekulovic S., Knapp D.J.H.F., Wohrer S., Bowden W., Viel F., McLaughlin T., Jarandehei A., Miller M., et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., III, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu R., Antonysamy S., Druzina Z., Ho C., Kang N.R., Pustilnik A., Wang J., Atwell S. Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing. PLoS One. 2020;15:e0232311. doi: 10.1371/journal.pone.0232311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Bowen J.E., Corti D., Veesler D. Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation. Nat. Struct. Mol. Biol. 2020;27:942–949. doi: 10.1038/s41594-020-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky H.J., Brown R.E. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B.E., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A., et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio. 2020;11 doi: 10.1128/mBio.01991-20. e01991–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rutten L., Gilman M.S.A., Blokland S., Juraszek J., McLellan J.S., Langedijk J.P.M. Structure-Based Design of Prefusion-Stabilized Filovirus Glycoprotein Trimers. Cell Rep. 2020;30:4540–4550.e3. doi: 10.1016/j.celrep.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Derking R., Cupo A., Julien J.-P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J., et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M.M., Tortorici M.A., Park Y.-J., Walls A.C., Homad L., Acton O., Bowen J., Wang C., Xiong X., de van der Schueren W., et al. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D., et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert D.N., Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones G.B.E., Chuang G.-Y., Xu K., Zhou T., Acharya P., Tsybovsky Y., Ou L., Zhang B., Fernandez-Rodriguez B., Gilardi V., et al. Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4. Proc. Natl. Acad. Sci. USA. 2018;115:12265–12270. doi: 10.1073/pnas.1811980115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bülow S., Gecht M., Bagola K., Hörner C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Toovey O.T.R., Harvey K.N., Hui D.D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 2021;82:e8–e10. doi: 10.1016/j.jinf.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N., et al. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J. Virol. 2018;92 doi: 10.1128/JVI.02002-17. e02002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M., et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. ELife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja I., Wang C., van Haperen R., Gutiérrez-Álvarez J., van Dieren B., Okba N.M.A., Raj V.S., Li W., Fernandez-Delgado R., Grosveld F., et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg. Microbes Infect. 2019;8:516–530. doi: 10.1080/22221751.2019.1597644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. BioRxiv. 2021 doi: 10.1101/2021.01.25.427948. [DOI] [Google Scholar]
- Xiong X., Qu K., Ciazynska K.A., Hosmillo M., Carter A.P., Ebrahimi S., Ke Z., Scheres S.H.W., Bergamaschi L., Grice G.L., et al. CITIID-NIHR COVID-19 BioResource Collaboration A thermostable, closed SARS-CoV-2 spike protein trimer. Nat. Struct. Mol. Biol. 2020;27:934–941. doi: 10.1038/s41594-020-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine H.M., Boyington J.C., McTamney P.M., Wei C.-J., Kanekiyo M., Kong W.-P., Gallagher J.R., Wang L., Zhang Y., Joyce M.G., et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wu Q., Liu Z., Wang Q., Wu J., Hu Y., Bai T., Xie T., Huang M., Wu T., et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat. Microbiol. 2021;6:51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- Zhao S., Lou J., Cao L., Zheng H., Chong M.K.C., Chen Z., Chan R.W.Y., Zee B.C.Y., Chan P.K.S., Wang M.H. Quantifying the transmission advantage associated with N501Y substitution of SARS-CoV-2 in the United Kingdom: An early data-driven analysis. J. Travel Med. 2021;28:taab011. doi: 10.1093/jtm/taab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yuan M., Song G., Beutler N., Shaabani N., Huang D., He W.-T., Zhu X., Callaghan S., Yong P., et al. A protective broadly cross-reactive human antibody defines a conserved site of vulnerability on beta-coronavirus spikes. bioRxiv. 2021 doi: 10.1101/2021.03.30.437769. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Atomic coordinates for the Fab22–MERS-CoV S peptide and the Fab22–SARS-CoV-2 S peptide structures have been deposited into the Protein Data Bank and assigned PDB IDs 7S3M and 7S3N, respectively. Cryo-EM maps of Fab22–MERS-CoV S-2P and Fab22–SARS-CoV-2 S-HexaPro have been deposited in the EMDB and assigned codes EMD-25072 and EMD-25073.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.