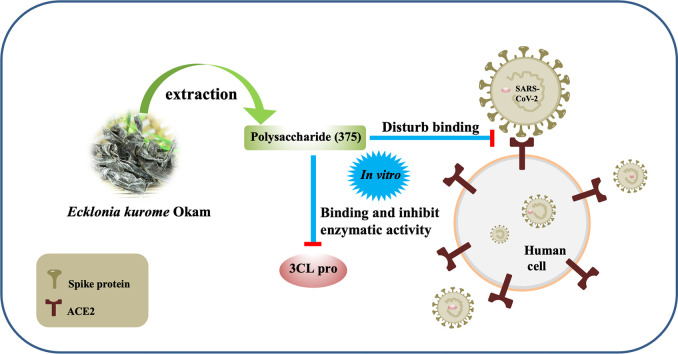
Abstract
Previous researches suggested that polysaccharides from brown algae had anti-virus activity. We hypothesized that nature polysaccharide from marine plants might have the effect on anti-SARS-CoV-2 activity. By high throughput screening to target 3CLpro enzyme using polysaccharides library, we discover a crude polysaccharide 375 from seaweed Ecklonia kurome blocked 3CLpro enzymatic activity and shows good anti-SARS-CoV-2 infection activity in cell. Further, we show that homogeneous polysaccharide 37502 from the 375 may bind to 3CLpro well and disturb spike protein binding to ACE2 receptor. The structure characterization uncovers that 37502 is alginate. These results imply that the bioactivities of 375 on SARS-CoV-2 may target multiple key molecules implicated in the virus infection and replication. The above results suggest that 375 may be a potential drug candidate against SARS-CoV-2.
Chemical compounds studied in this article: Alginate, Mannuronic acid, Guluronic acid, Heparan sulfate, Fucan, Sodium borohydride, Dimethyl sulfoxide, Deuterium oxide, Acetone, 1-Cyclo-hexyl-3-(2-mopholinoethyl) carbodiimidemetho-p-toluenesulfonate
Keywords: COVID-19, SARS-CoV-2, Angiotensin converting enzyme 2 (ACE2), 3CL protease, Ecklonia kurome Okam, Polysaccharide
Graphical abstract
1. Introduction
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has made a pandemic of Coronavirus Disease 2019 (COVID-19) cross the globe. Up to now, this virus has spread more than 200 countries. 78 million individuals have been infected and over 1.7 million deaths to date (Gaebler et al., 2021). Vaccination and keeping social distancing are functional strategies to slow down the infection during the coming vaccine injection period and effective medicine. Currently, although some potential new drugs are on the clinical trial, only one anti-SARS-CoV-2 agent, remdesivir, has been approved by FDA for the treatment of adult COVID-19 patients (Beigel et al., 2020). Scientific research institutions and pharmaceutical companies are trying to understand the mechanism of SARS-CoV-2 infection and potential antivirus drug to treat COVID-19. A chymotrypsin-like cysteine protease called 3C-like protease (3CLpro) and papain-like protease (PLpro) are required to process polyproteins into mature nonstructural proteins such as RNA-dependent RNA polymerase (RdRp) and helicase, which are essential for viral transcription and replication (Su et al., 2020). Kumar et al. found that SARS-CoV-2 had only 12.8% of difference with SARS-CoV in S protein and has 83.9% similarity in minimal receptor-binding domain with SARS-CoV (Kumar et al., 2020). In 2003, it had been identified that angiotensin-converting enzyme 2 (ACE2) could efficiently bind to the S1 domain of the SARS-CoV S protein (Li et al., 2003). The S protein is a heavily glycosylated protein, which possess 22 potential N-glycosylation sites and facilitates attachment, entry and membrane fusion (Shajahan et al., 2020). Previous researchers showed that several viruses interacted with sialic acids located on the ends of glycans in glycolipids and glycoproteins surrounding the cells. Some other viruses might interact with heparan sulfate (HS) that is attached to cell membrane (Milewska et al., 2014) or extracellular matrix proteoglycans (Lindahl et al., 2015).
Latest research found that SARS-CoV-2 entry the human cell through binding of the virus spike (S) protein to angiotensin-converting enzyme 2 (ACE2) and cellular HS on the surface of the host cell (Clausen et al., 2020). Hence, blocking the transcription and replication of viral and interfering the binding of SARS-CoV-2 and ACE2 are rational strategies to fight SARS-CoV-2 infection. Interestingly, traditional Chinese medicine has been paid more attention for antivirus clinical drug application during SARS-CoV and SARS-CoV-2 spreading (Leung, 2007; H. Luo et al., 2020; Wen et al., 2011). Indeed, baicalin and baicalein, two ingredients of Chinese traditional patent medicine Shuanghuanglian, were characterized as the first noncovalent, nonpeptidomimetic inhibitors of SARS-CoV-2 3CLpro and exhibited potent antiviral activities in a cell-based system (Su et al., 2020). However, the detailed mechanism underlying active components against the virus is still vague.
Traditional Chinese medicine E. kurome, named Kunbu in China, is a seaweed of the Laminariaceae, belonging to the genus Laminariales (Kuda et al., 2007). The high pharmaceutical value of this seaweed is at least partially relied on the biomacromolecule polysaccharide. Different from other general polysaccharide, the polysaccharide in E. kurome is mainly present on cell wall of brown algae plants and has a sulfate group at the end of its molecular chain, which may make it has significance bioactivities (Li et al., 2020). For instance, a modified sulfate polysaccharide extracted from E. kurome has anti-angiogenesis anti-tumor effect (Kuda et al., 2007). Alginate is a naturally acidic polysaccharide which can be extracted from marine brown seaweeds, such as Ectocarpus, Laminaria, Colpomenia, Fucales and Sargassum (Sachan et al., 2009; Szekalska et al., 2016). It is a liner anionic polymer of β-(1–4)-D-mannuronic acid and of its C-5 epimer, α-(1–4)-L-guluronic acid and is a main acid polysaccharide in E. kurome. They consist of alternate homopolymeric blocks of poly-β-(1–4)-D-mannuronic acid (M), and poly-α-(1–4)-L-guluronic acid (G), and of heteropolymeric blocks with random arrangements of both monomers (Fujihara & Nagumo, 1993).
Although alginate could be derived from different seaweed, the biological activities of alginates were closely related to the distribution of M and G residues, the molecular mass (MM) and the uronic acid composition (Stokke et al., 1991). The structural features such as the molecular weight (Mw), M/G ratio, arrangement of M and G residues and acetylation degree of alginates vary depending on species, geographic location, season and other parameters (Goh et al., 2012; Setyawidati et al., 2018). The antimicrobial activities of alginates extracted from Laminaria hyperborean, Laminaria digitata, Laminaria japonica, Ascophyllum nodosum and Macrocystis pyrifera have been reported (Li et al., 2017). Alginates from three genera of Tunisian brown algae (Padina pavonica, Cystoseira compressa and Dictyopteris membranaceae) had gastroprotective activities, but the effect of them depends mainly on their uronic acid composition (Ammar et al., 2018). Besides, previous researches also revealed that alginate had the effect on anti-tumor and immunoenhancement (De Sousa et al., 2007; Luo et al., 2020).
According to previous studies, most of the papers showed that sulfated fucan in brown algae had anti-virus activity and rarely reported about the alginate. For instance, a sulfated fucan from the Laminaria abyssalis marine algae might inhibit the cell-to-cell contact essential for the spreading of the human T cell lymphotropic virus type 1 (HTLV-1) (Romanos et al., 2002). In 2010, sulfated fucoidan and guluronic acid-rich alginate derived from Sargassum tenerrimum showed activity against herpes simplex virus type 1 (HSV-1). The 50% inhibitory concentration (IC50) values were in the range 0.5–15 μg/mL (Sinha et al., 2010). A fucose-containing polysaccharide purified from Sargassum trichophyllum showed anti-HSV-2 properties in vitro (Lee et al., 2011). Two sulfated fucoidans SHAP-1 and SHAP-2 were isolated from Sargassum henslowianum. The antiviral test showed that SHAP-1 at the concentration of 0.89 μg/mL and SHAP-2 at 0.82 μg/mL could inhibit HSV-1 activity by plaque reduction assay, whereas both polysaccharides at concentration of 0.48 μg/mL might also impede HSV-2 activity (Sun et al., 2020). However, the anti-virus activity of alginate was reported in 1999 and the results supported that it could inhibit tobacco mosaic virus infection (Sano, 1999).
In this study, we firstly extracted crude polysaccharide, named as 375 and an alginate 37502 from E. kurome and examine their bioactivities against SARS-CoV-2 virus. The structure of 37502 from E. kurome was further characterized with chemical methods (molecular weight (Mw), monosaccharide composition, infrared spectroscopy (IR) and nuclear magnetic resonance (NMR) spectra). Unlike previous findings on the anti-virus effect of sulfated polysaccharide in brown algae, we firstly reported the alginate in E. kurome with the anti-SARS-CoV-2 bioactivity by targeting and inhibiting 3CLpro and disturbing the binding of S1 protein and ACE2 molecules. These results provide us a good foundation for the development of anti-SARS-CoV-2 virus drug.
2. Experimental
2.1. Materials and reagents
E. kurome was come from Fujian province, China. DEAE Sepharose Fast Flow was obtained from GE healthcare. Dimethyl sulfoxide (DMSO) was from E. Merck. U.S.A. Sodium borohydride (NaBH4) and iodomethane were obtained from Sinopharm Chemical Reagent Co. Ltd. Standard monosaccharides were purchased from Shanghai Aladdin Bio-Chem Technology Co. Ltd. Other reagents were analytical grade and from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China).
2.2. Determination of physicochemical property of polysaccharides
The carbohydrate content was determined by the PhOH-H2SO4 method using glucose as a standard (Dubios et al., 1956). Protein content was evaluated using a BCA protein assay kit (Beyotime Biotechnology, China). Uronic acid content was determined by the meta-hydroxydiphenyl method using galacturonic acid as a standard. All the measurements were repeated three times. Novostar microplate reader was employed to detect the absorbance at OD490 for sugar, OD520 for uronic acid and OD562 for protein.
2.3. Homogeneous polysaccharide preparation
Crude polysaccharide was fractionated on a DEAE Sepharose Fast Flow column. 200 mg polysaccharide was dissolved in 20 mL distilled water and centrifuged for each time. The supernatant was applied to a DEAE Sepharose Fast Flow column and eluted with distilled water, 0.05, 0.1, 0.2 and 0.3 M NaCl solutions stepwise. The solution was pooled according to the elution profile based on phenol-sulfuric acid method. Then the fraction eluted with distilled water, 0.2 M NaCl and 0.3 M NaCl were collected, followed by concentration, dialysis with distill water and freeze-dried.
2.4. Homogeneity and molecular weight
The homogeneity and molecular weight (Mw) were examined by HPGPC (high performance gel permeation chromatography) method on Agilent 1260 HPLC (Santa Clara, CA, USA) system equipped with two series-connected columns (UItrahydrogel™ 2000 and 500). The columns were calibrated by pullulans standards. 0.1 mol/L NaNO3 was used as an eluent and the flow rate was maintained at 0.6 mL/min. The column temperature was maintained at 40.0 °C ± 0.1 °C. The samples were prepared with mobile phase as 0.2% (W/V) solution. 20 μL of sample was injected for analysis.
2.5. Monosaccharide composition analysis
The method of monosaccharide composition was PMP pre-column derivatization based on the previous reported (Dai et al., 2010).
2.6. FT-IR spectrum and NMR analysis
The IR spectra were determined according to the previous report (Cong et al., 2014). 2 mg native polysaccharide was mixed with dried KBr powder and pressed into pellet, then scanned from 4000 to 600 cm−1 for the analysis.
For the NMR analysis, polysaccharide was deuterium-exchanged and dissolved in deuterium oxide (D2O) in 60 mg/mL. 1D-NMR and 2D-NMR spectra were obtained on a Bruker AVANCE III NMR spectrometer operating at 500 MHz at 25 °C with acetone as internal standard.
2.7. Uronic acid reduction
The method was based on the reported method (Taylor & Conrad, 1972). In brief, 40 mg polysaccharide was dissolved in 40 mL H2O. CMC (600 mg) was added and pH was kept at 4.75 with 0.01 M HCl for 2 h. Then 2 M fresh aqueous sodium borohydride (15 mL) was added slowly to the mixture in 45 min and 4 M HCl was added concurrently to keep pH at 7.0. The mixture was stirred for 2 h and dialyzed (1000 mL × 4) for 24 h at room temperature. Then the retentate was lyophilized to achieve carboxyl reduced polysaccharide, followed by monosaccharide composition and linkage pattern analyses.
2.8. Methylation analysis
10 mg sample was methylated with a modified method from Ciucanu and Kerek. Briefly, the sample was dried overnight with P2O5 and dissolved in DMSO (2 mL) and 100 mg powdered NaOH was added into the reaction bulb and stirred at room temperature for 3 h. Iodomethane (0.5 mL) was added dropwise within 45 min in ice-cold water bath. The mixture was stirred for 2.5 h at room temperature in a dark place and then 1 mL deionized water was added to terminate the reaction. The redundant CH3I was removed by evaporation under depressed pressure. The solution was extracted by 15 mL CHCl3 and 15 mL water (1:1, v/v) and the organic phase was washed with deionized water for three times. After removing the residual water by adding Na2SO4, the organic phase was concentrated to get the methylated polysaccharide. Then the product was hydrolyzed with 2 M TFA for 2 h at 110 °C and converted into the partially methylated alditol acetates (PMAA) and analyzed by GC–MS. The GC–MS program designed for methylation analysis was based on the reported method (Cong et al., 2014).
2.9. Enzymatic activity and inhibition assays
The enzyme activity and inhibition assays have been described previously (Xue et al., 2007; Yang et al., 2005). The recombinant SARS-CoV-2 3CLpro (30 nM at a final concentration) was mixed with serial dilutions of each compound in 80 μL assay buffer (50 mM Tris-HCl, pH 7.3, 1 mM EDTA) and incubated for 10 min. The reaction was initiated by adding 40 μL fluorogenic substrate with a final concentration of 20 μM. After that, the fluorescence signal at 320 nm (excitation)/405 nm (emission) was immediately measured every 30 s for 10 min with a Bio-Tek Synergy4 plate reader. The Vmax of reactions added with compounds at various concentrations compared to the reaction added with DMSO were calculated and used to generate IC50 curves. For polysaccharide 375, IC50 values against SARS-CoV-2 3CLpro were measured at 7 concentrations and three independent experiments were performed. All experimental data was analyzed using GraphPad Prism software.
2.10. ELISA
10 μg/mL ACE2 coating buffer were used to treat the 96 well plate at 4 °C overnight following with 200 μL washing buffer for three times. Then the 96-well plate was blocked by 2% BSA at room temperature for 2 h. After that, 100 μL biotinylated S1 protein was added and incubated at room temperature. At the same time, the positive control and the negative control were set. After incubation for 1 h, the plate was washed for three times and each time for 5 min. Following, 100 μL Streptavidin-HRP was added to a final concentration 200 ng/mL and the reaction was incubated for 1 h at room temperature. After 1 h incubation, the plate was washed for three times. Then, 100 μL TMB were added and incubated in the dark for 35 min. Finally, 50 μL stop solution was added to stop the reaction and the value was detected at A450 by microplate reader (BioTek).
2.11. Isothermal titration calorimetry (ITC)
Binding test was performed by ITC as described in previous publication (Su et al., 2020). It was conducted with an iTC200 instrument (Microcal, GE Healthcare) at 25 °C, and the resulting data were processed by the supplied MicroCal Origin software package. The concentration of 3CLpro protein and polysaccharide 37502 was 300 μM and 600 μM, respectively. Titrations were run in triplicate to ensure reproducibility. In all the cases, a single binding site mode was employed and a nonlinear least-squares algorithm was used to obtain best-fit values of the stoichiometry (n), change in enthalpy (ΔH), and binding constant (K). Thermodynamic parameters were subsequently calculated with the formula ΔG = ΔH − T ΔS = −RT ln K, where T, R, ΔG, and ΔS are the experimental temperature, the gas constant, the changes in free energy, and entropy of binding, respectively.
2.12. Antiviral test in vitro
The experiments related to SARS-CoV-2 are completed at National Biosafety Laboratory, Wuhan, Chinese Academy of Sciences.
SARS-CoV-2 (WIV04) was passaged in Vero E6 cells and tittered by plaque assay. Vero E6 cells were treated with drugs at indicated concentration and infected by SARS-CoV-2 virus at MOI 0.01. After 24 h incubation at 37 °C, supernatants were collected and the viral RNAs were extracted by Magnetic Beads Virus RNA Extraction Kit (Shanghai Finegene Biotech, FG438), and quantified by real-time RT-PCR with Taqman probe targeting to the RBD region of S gene.
3. Results
3.1. Isolation, purification and monosaccharide composition analysis of 37502
The crude polysaccharide, 375 (200 mg) was fractioned by DEAE Sepharose™ Fast Flow. 37501 (20 mg, yield: 10%), 37502 (44 mg, yield: 22%) and 37503 (35 mg, yield: 17%) were obtained by elution with distilled water, 0.2 M NaCl and 0.3 M NaCl. 375 contained 65% total carbohydrate, 5% protein and 28% uronic acid. 37501 contained 12% total carbohydrate and 7.7% uronic acid. 37502 contained 16.4% total carbohydrate and 32.0% uronic acid. The content of total carbohydrate, protein and uronic acid of 37503 was 40.3%, 2.3% and 11.9%, respectively. The homogeneity of 37502 was determined by high performance gel permeation chromatography (HPGPC) that showed a single symmetrical peak with the Mw of 27.9 kDa (Fig. S1A, Supplementary data). The monosaccharide compositions of 375, 37501, 37502 and 37503 were analyzed by HPLC after PMP derivatization with PMP in parallel with monosaccharides standards. Compared with standards, the monosaccharide composition of 375, 37501 and 37503 were shown in Fig. S2, the crude polysaccharide 375 contained guluronic acid (3.0%), mannuronic acid (15.3%), mannose (10.9%), rhamnose (2.7%), glucouronic acid (1.6%), galacturonic acid (2.8%), glucose (1.9%), galactose (17.7%), xylose (6.9%) and fucose (36.9%). 37501 mainly contained guluronic acid (1.4%), mannuronic acid (3.6%), mannose (14.6%), rhamnose (3.6%), glucouronic acid (2.0%), galactose (28.1%), xylose (7.37%) and fucose (39.1%). 37503 was composed of mannose (30.1%), rhamnose (9.6%), glucose (3.4%) galactose (7.3%), xylose (10.9%) and fucose (36.9%). Relatively, the monosaccharide composition of 37502 was simpler, it was composed of mannuronic acid (89.3%) and guluronic acid (10.7%). More structure information was revealed by FT-IR and NMR spectra.
3.2. FT-IR spectrum
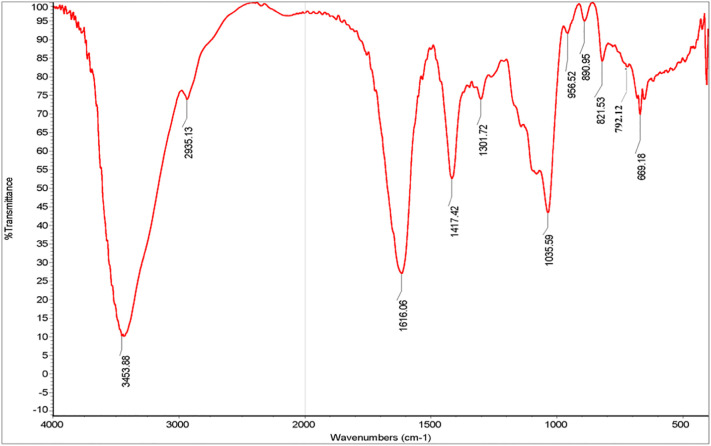
FT-IR spectrum of 37502 showed the typical absorption bands of the polysaccharide (Fig. 1 ). 3453.88 cm−1 was assigned to the O—H stretching bands. 2935.13 cm−1 was from the stretching bands of C—H group. 1616.06 cm−1 and 1417.42 cm−1 could be reassigned to the asymmetrical and symmetrical stretching vibration of COO-, respectively, which confirmed the presence of uronic acid. The absorption at 1035.59 cm−1 showed the existence of the stretching vibration of C—C. 956.52 cm−1 was the stretching vibration of uronic acid residues. 890.95 cm−1 was the variable angular vibration of C1—H of β-mannuronic acid (Mao et al., 2004). 821.53 cm−1 was the special absorption peak of mannuronic acid and 792.12 cm−1 was the special absorption peak of guluronic acid (Lin et al., 2007).
Fig. 1.
FT-IR spectrum of 37502.
3.3. Linkage pattern analysis
In order to determine the linkage types, the methylation method was employed. First, 37502 was reduced with 1-Cyclo-hexyl-3-(2-mopholinoethyl) carbodiimidemetho-p-toluenesulfonate (CMC) and the reduced products were further methylated. Result revealed that 37502 was composed of (1, 4)-linked β-D-mannuronate (M) and (1, 4)-linked α-L-guluronate (G) residues in a molar ratio of 9:1. More structure information will be shown by NMR spectra.
3.4. NMR spectral analysis
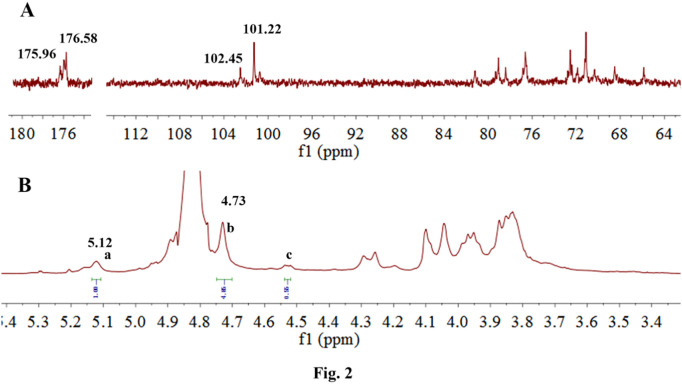
13C NMR spectra of 375, 37502 and 37503 were compared in Fig. S3 and the 1H, 13C NMR spectra of 37502 were shown in Fig. 2. In the 13C NMR spectra of 37502 (Fig. 2A), the signals at δ 102.45 ppm and 101.22 ppm were assigned to C1 of 1, 4-linked α-L-guluronic acid and 1, 4-linked β-D-mannuronic acid, respectively (Heyraud et al., 1996). The strong signals at δ 65.87, δ 78.46 and δ 68.57 were ascribed to C-2, C-3 and C-5 of the 1,4-linked α-L-guluronic acid. The signals at δ 71.12, δ 72.55 and δ 76.69 were ascribed to C-2, C-3 and C-5 of the 1, 4-linked β-D-mannuronic acid. The signals at δ 81.2 and δ 79.06 were assigned to C-4 of 1,4-linked α-L-guluronic acid and 1, 4-linked β-D-mannuronic acid, respectively (Schürks et al., 2002).
Fig. 2.
1H NMR spectrum (A) and 13C NMR spectrum (B) of 37502.
In the 1H NMR (Fig. 2B) and HSQC (Fig. 3B) spectra, δ 5.12 was assigned to H-1 of 1,4-linked α-L-guluronic acid, which correlated to C-1 of 1,4-linked α-L-guluronic acid (102.45 ppm). δ 4.73 was assigned to H-1 of 1, 4-linked β-D-mannuronic acid correlated to C-1 of 1, 4-linked β-D-mannuronic acid (101.22 ppm). In 1H-1H COSY (Fig. 3A) spectra, δ 4.04 was assigned to H-2 of 1,4-linked α-L-guluronic acid, which correlated to H-1 (δ 5.12) of 1,4-linked α-L-guluronic acid. δ 4.10 was assigned to H-2 of 1, 4-linked β-D-mannuronic acid, which correlated to H-1 (δ 4.73) of 1, 4-linked β-D-mannuronic acid. Other signals were listed in Table.1 .
Fig. 3.
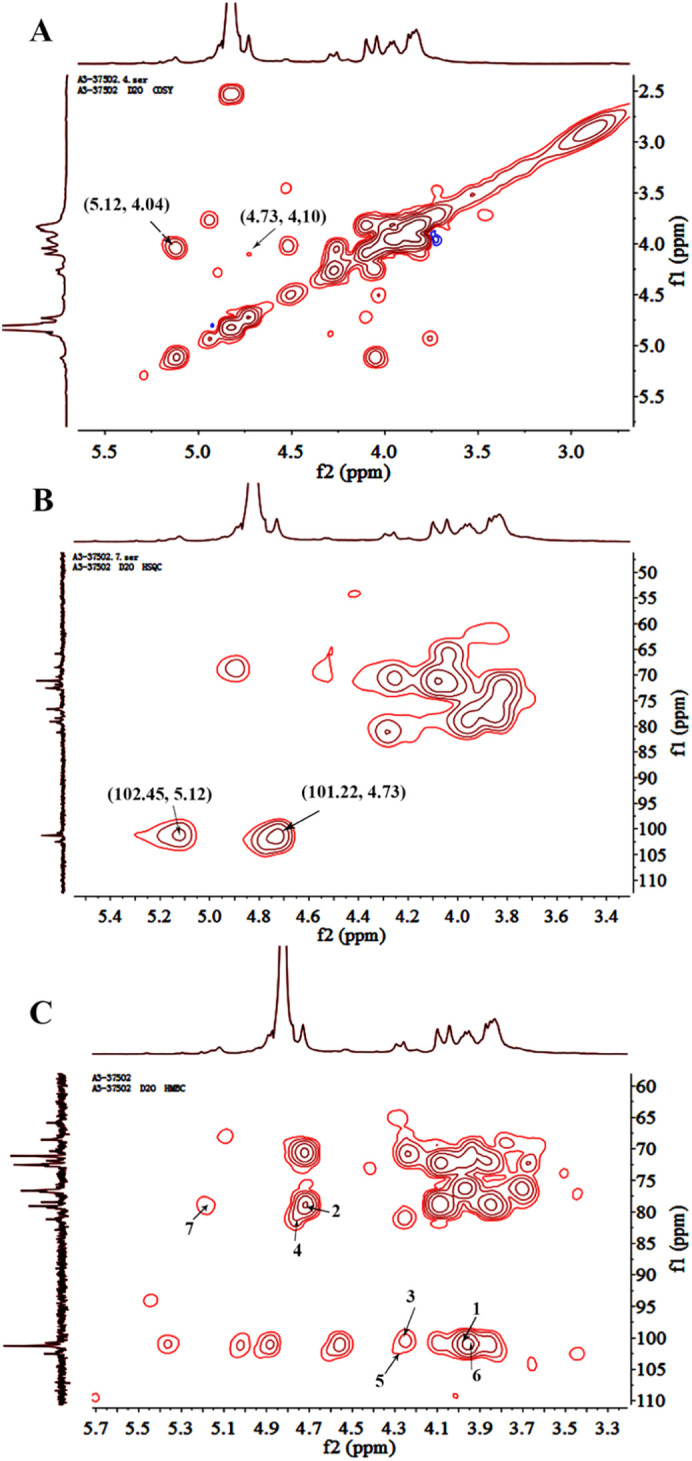
Two dimensional spectra of 37502; A. 1H-1H COSY spectrum; B. HSQC spectrum; C. HMBC of spectrum.
Table. 1.
1H NMR and 13C NMR chemical shifts (ppm) assignments for 37502.
| Residues | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| 1, 4-Linked guluronic acid (G) | C | 102.45 | 65.87 | 78.46 | 81.2 | 68.57 | 176.58 |
| H | 5.12 | 4.04 | 3.95 | 4.29 | 4.53 | ||
| 1, 4-linked mannuronic acid (M) | C | 101.22 | 71.12 | 72.55 | 79.06 | 76.69 | 175.96 |
| H | 4.73 | 4.10 | 3.83 | 3.93 | 3.87 | ||
HMBC spectrum was employed to analyze the structure backbone and the substitution sites. In Fig. 3C, the strong peak 1 (101.22/3.93) represented the correlation between C-1 and H-4 of 1, 4-linked mannuronic acid. The cross peak 2 (79.06/4.73) represented the correlation between C-4 and H-1 of 1, 4-linked mannuronic acid. These two peaks showed the existence of liner 1, 4-linked mannuronic acid. The cross peak 3 (101.22/4.29) showed the correlation between C-1 of 1, 4-linked mannuronic acid and H-4 of the neighboring 1, 4-linked guluronic acid. The peak 4 (81.2/4.73) represent the correlation between C-4 of 1, 4-linked guluronic acid and H-1 of the neighboring 1, 4-linked mannuronic acid. These results suggested that guluronic acid may linked to the liner mannuronic acid. The cross peak 5 (102.45/4.29) showed the correlation between C-1 of 1, 4-linked guluronic acid and H-4 of neighboring 1, 4-linked guluronic acid, which showed the existence of liner 1, 4-linked guluronic acid. The cross peak 6 (102.45/3.93) overlapped by peak 1 showed the correlation between C-1 of 1, 4-linked guluronic acid and H-4 of neighboring 1, 4-linked mannuronic acid. The cross peak 7 (79.06/5.12) also showed the correlation between C-4 of 1, 4-linked mannuronic acid and H-1 of 1, 4-linked guluronic acid. It showed the existence of heteropolymeric blocks with random arrangements of both mannuronic acid and guluronic acid.
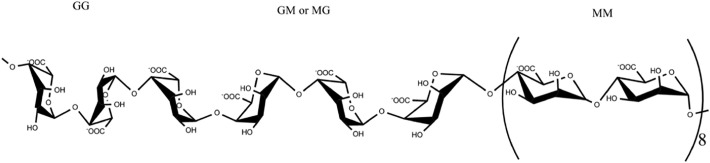
3.5. Putative structure of 37502
Based on the above results, we proposed that 37502 is an alginate consisting of different dimer structures (MM, MG, GM and GG). The molar ratio (F) of dimer structures was speculated according to the integral area (I) of a, b, c in 1H NMR and following formula: FG = I a / (I b + I c), FGG = I c / (I b + I c), FGG + FGM = FG, FMM + FMG = FM, FMG = FGM. In 1H NMR of 37502, the integral area of a, b, c was 1.0, 4.85 and 0.55, respectively. So, FG = 0.19; FM = 0.9; FMM = 0.81; FGG = 0.1; FGM = FGM = 0.09. The molar ratio (0.19: 0.9) of between FG and FM was consisted of the results of monosaccharide composition. The ratio of FMM was highest and the ratio of FGG, FMG and FGM was similar. Finally, the possible repeated unit of 37502 (Mw: 27.9 kDa) was shown in Fig. 4 .
Fig. 4.
Proposed repeated unit of 37502.
3.6. Crude polysaccharide 375 was a potent inhibitor of SARS-COV-2 3CLpro and disturb the interaction between SARS-CoV-2-S1 and ACE2
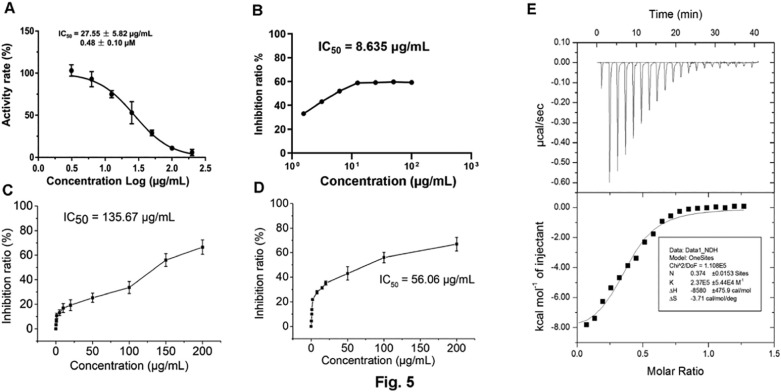
People showed that marine polysaccharide may inhibit SARS-CoV-2 infection (Song et al., 2020), however the convinced targeting molecule and mechanism are still unknown. SARS-CoV-2 includes two open reading frames ORF1a and ORF1b (Yin et al., 2020). ORF1a encodes two cysteine proteases, a papain-like protease (PLpro) and a 3 C-like protease (3CLpro). Scientists have provided evidences that main protease (3CLpro) is one of the good targets to discover new antiviral agents before vaccines are available (Derosa et al., 2020). To explore more potent leading compounds against SARS-CoV-2 by targeting key molecule in RNA synthesis of the virus, recombinant SARS-CoV-2 3CLpro was firstly expressed and purified from Escherichia coli (Xue et al., 2007; Yang et al., 2003). A fluorescently labeled substrate MCA-AVLQSGFR-Lys (Dnp)-Lys-NH2, derived from the N-terminal autocleavage sequence from the viral protease was designed and synthesized for the enzymatic assay. Then the binding test targeting 3CLpro was examined. The results showed that 375 might potently inhibit the activity of 3CLpro of SARS-CoV-2 (Fig. 5 ). Further we used a fluorescence resonance energy transfer (FRET) based cleavage assay to determine the median inhibitory concentration (IC50) values. The results also revealed good inhibitory potency, with IC50 values of 0.48 ± 0.1 μM (Fig. 5A). The spike protein of SARS-CoV-2 shows more than 90% amino acid similarity to that of pangolin and bat CoVs which also use human angiotensin-convert enzyme 2 (ACE2) as a receptor for the virus infection. Receptor binding domain of S protein of SARS-CoV-2 which is processed into two subunits including S1 and S2, can bind with ACE2 to invade target cells (Lan et al., 2020). Thus, S protein is very vital for viral invasion. Interestingly, the N-terminal domain of S1 protein has the glycan binding site (Kirchdoerfer et al., 2016). This implies S protein can bind carbohydrate. Based on these backgrounds, 375 was employed to examine whether it might disturb the binding between S1 protein and ACE2 by ELISA method. The result showed that 375 could impede the binding of S1 protein and ACE2 with the median inhibitory concentration (IC50) 135.67 μg/mL (Fig. 5C). It implies that 375 may contain the bioactive components to inhibit SARS-CoV-2 replication and infection. Since 375 is a crude polysaccharide, the results inspire us to explore which bioactive components from crude polysaccharide 375 may contribute the effect against SARS-CoV-2 and their underlying mechanism.
Fig. 5.
(A) Inhibitory activity profiles of 375 against SARS-CoV-2 3CLpro. (B) Inhibitory activity profiles of 37502 against SARS-CoV-2 3CLpro. (C) Competitive intervention of polysaccharide 375 on S1 protein and ACE2 by ELISA. (D) Competitive intervention of polysaccharide 37502 on S1 protein and ACE2 by ELISA. (E) Binding test of polysaccharide 37502 with SARS-CoV-2 3CLpro by ITC.
3.7. 37502 may bind and inhibit SARS-CoV-2 3CLpro and disturb the interaction between SARS-CoV-2-S1 and ACE2
To explore which compound from 375 may interfere 3CLpro enzyme activity, isothermal titration calorimetry (ITC) method was employed (Su et al., 2020). Result showed that 37502, one homogeneous polysaccharide from 375 could bind SARS-CoV-2 3CLpro protein very well (Fig. 5E) and the Kd value was 4.23 × 10−6 M. This result suggested that 375 or 37502 might interfere the replication of SARS-CoV-2 in some way.
To further understand the bioactivity of 375 against SARS-CoV-2, alginate 37502 from 375 was employed to examine whether it might inhibit 3CLpro activity and disturb the binding between S1 protein and ACE2 by ELISA. The results showed that polysaccharide 37502 could inhibit the 3CLpro activity and the IC50 was 8.635 μg/mL (Fig. 5B). This polysaccharide also effectively impeded the binding of S1 protein with ACE2 and the IC50 was 56.06 μg/mL (Fig. 5D). This result suggested that 37502 might had the potential to inhibit SARS-CoV-2 3CLpro activity and block SARS-CoV-2 infection through disturbing the S protein binding to ACE2 receptor.
3.8. 375 and 37503 exhibit anti-viral effect on SARS-CoV-2
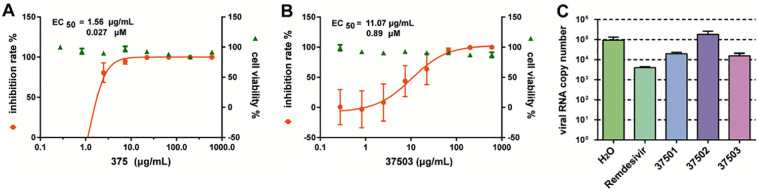
The above results inspired us to explore whether those polysaccharides might really block SARS-CoV-2 replication. Firstly, the inhibition effect of native polysaccharide 375 was examined. Surprisingly, as shown in Fig. 6A, polysaccharides 375 nearly completely inhibit SARS-CoV-2 in vitro, exhibiting very good anti-SARS-CoV-2 infection activity in Vero E6 cells. The EC50 value is 27 nM (or 1.56 μg/mL) (Fig. 6A). However, the toxicity of 375 on mice is low and the LD50 is 136 mg/kg (Table S1, Supplementary data). To further understand which component from native polysaccharide 375 is contributing to inhibit effect on SARS-CoV-2, quantitative real-time polymerase chain reaction (qRT-PCR) was also employed to monitor the antiviral activity of the purified polysaccharide 37501, 37502 and 37503 from 375. The result showed that polysaccharide 37501 and 37503 exhibited antivirus effect on SARS-CoV-2 (Fig. 6C). Although 37502 might attenuate 3CLpro enzymatic activity significantly (Fig. 5B), it had no significant direct effect against the active virus replication (Fig. 6C). To examine whether purified polysaccharide 37503 have more stronger bioactivity than that of native polysaccharide 375, EC50 of 37503 against SARS-CoV-2 was also measured. Interestingly, the bioactivity of 37503 was significant feeble than that of 375, while EC50 of 37503 is 0.89 μM (or 11.07 μg/mL) (Fig. 6B). This indicated the effects of 375 against SARS-CoV-2 were cocktail-like synergistic contribution of combined components of 37501, 37502, and 37503, the impact of individual homogeneous polysaccharide might be weaker although. In brief, although polysaccharide 375 and 37503 are probably the potential drug candidate for inhibiting the SARS-CoV-2 infection, obviously cocktail-liked crude polysaccharide 375 is the best option for anti-SARS-CoV-2 new drug development. Thus, we focused on the 375 and 37503 for further investigation.
Fig. 6.
In vitro inhibition of polysaccharides against SARS-CoV-2. (A) EC50 of crude polysaccharide 375 against SARS-CoV-2; (B) EC50 of homogenous polysaccharide 37503 against of SARS-CoV-2. (C) Viral RNA copy number was detected by qPCR after the treatment of solvent (H2O) control, Remdesivir positive control, crude polysaccharide 375 and their fragmentation 37501, 37502 and 37503.
In conclusion, cocktail polysaccharide from Ecklonia kurome might have better impact against SARS-CoV-2 virus infection than individual homogeneous polysaccharide with different bioactivities on 3CLpro, ACE2 and S1 protein binding.
CRediT authorship contribution statement
All authors have reviewed the final version of the manuscript and approved it for publication. This original manuscript has neither been formally published nor considering for publication in elsewhere. The first co-authors contributed equally to this paper. Among them, Shihai Zhang was responsible for the structure analysis of polysaccharide and preparation, creation of the published work, specifically writing the initial draft of chemical part. Rongjuan Pei was responsible for anti-SARS-Cov-2 infection test. Meixia Li was responsible for data curation, visualization, formal analysis and writing (Original Draft). Chunfan Huang prepared partial polysaccharide extract and separation. Hao Sun was responsible for the RNA extraction and qRT-PCR experiment. Minbo Su was responsible for binding test of crude polysaccharide 375 with 3CL pro. Haixia Su perform the binding test of 37502 with 3CLpro by the isothermal titration calorimetry (ITC). Yaqi Ding conducted competitive binding test of the polysaccharide and ACE2 and spike protein using Elisa method. Xia Chen was responsible for structure part revising (Review & Editing). Zhenyun Du was in charge of daily animal management and examine safety test of polysaccharides on animal. Can Jin and were responsible for preparation of chemical reagents and material and literature research. Yi Zang, Jia Li were in charge of polysaccharide library drug screening against 3CLpro. Yechun Xu were involved in targeting molecule confirmation conceptualization in this paper. Xiwen Chen was in charge of partial funding acquisition and experimental design. Bo Zhang was responsible for the management and coordination responsibility for the research activity planning and execution in center for biosafety Mega-Science. Kan Ding was in charge of supervision, project administration, writing (review & editing), manuscript revision, and funding acquisition.
Acknowledgment
This work was supported by National Key Research and Development Program of China, Ministry of Science and Technology of the People's Republic of China (grant number 2019YFC1711-000), Key New Drug Creation and Manufacturing Program (grant number 2019ZX09735001), COVID-19 Emergency Research Project founded by Zhejiang University (2020XGZX080) and Shanghai Municipal Science and Technology Commission Major Project. In addition, we are particularly grateful to Tao Du and Lun Wang from Zhengdian Biosafety Level 3 Laboratory and the running team of the laboratory for their work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.carbpol.2021.118779.
Appendix A. Supplementary data
Supplementary material
References
- Ammar H.H., Lajili S., Sakly N., Cherif D., Rihouey C., Le Cerf D., Bouraoui A., Majdoub H. Influence of the uronic acid composition on the gastroprotective activity of alginates from three different genus of tunisian brown algae. Food Chemistry. 2018;239:165–171. doi: 10.1016/j.foodchem.2017.06.108. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the treatment of Covid-19 - final report. The New England Journal of Medicine. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4):1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q., Xiao F., Liao W., Dong Q., Ding K. Structure and biological activities of an alginate from Sargassum fusiforme, and its sulfated derivative. International Journal of Biological Macromolecules. 2014;69:252–259. doi: 10.1016/j.ijbiomac.2014.05.056. [DOI] [PubMed] [Google Scholar]
- Dai J., Wu Y., Chen S.-W., Zhu S., Yin H.-P., Wang M., Tang J. Sugar compositional determination of polysaccharides from Dunaliella Salina by modified RP-HPLC method of precolumn derivatization with 1-phenyl-3-methyl-5-pyrazolone. Carbohydrate Polymers. 2010;82(3):629–635. [Google Scholar]
- De Sousa A.P.A., Torres M.R., Pessoa C., Moraes M.O.D., Filho F.D.R., Alves A.P.N.N., Costa-Lotufo L.V. In vivo growth-inhibition of Sarcoma 180 tumor by alginates from brown seaweed Sargassum vulgare. Carbohydrate Polymers. 2007;69(1):7–13. [Google Scholar]
- Derosa G., Maffioli P., D'Angelo A., Di Pierro F. A role for quercetin in coronavirus disease 2019 (COVID-19) Phytotherapy Research. 2020;35(3):1230–1236. doi: 10.1002/ptr.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubios M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. Colorimetric method for determination of sugar and related substances. Analytical Chemistry. 1956;28:250–256. [Google Scholar]
- Fujihara M., Nagumo T. An influence of the structure of alginate on the chemotactic activity of macrophages and the antitumor activity. Carbohydrate Research. 1993;243(1):211–216. doi: 10.1016/0008-6215(93)84094-m. [DOI] [PubMed] [Google Scholar]
- Goh C.H., Heng P.W.S., Chan L.W. Alginates as a useful natural polymer for microencapsulation and therapeutic applications. Carbohydrate Polymers. 2012;88(1):1–12. [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyraud A., Gey C., Leonard C., Rochas C., Girond S., Kloareg B. NMR spectroscopy analysis of oligoguluronates and oligomannuronates prepared by acid or enzymatic hydrolysis of homopolymeric blocks of alginic acid. Application to the determination of the substrate specificity of Haliotis tuberculata alginate lyase. Carbohydrate Research. 1996;289:11–23. doi: 10.1016/0008-6215(96)00060-2. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N.S., Pallesen J, Yassine H.M., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuda T., Kunii T., Goto H., Suzuki T., Yano T. Varieties of antioxidant and antibacterial properties of Ecklonia stolonifera and Ecklonia kurome products harvested and processed in the Noto peninsula, Japan. Food Chemistry. 2007;103(3):900–905. [Google Scholar]
- Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) VirusDisease. 2020;31(1):13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee J.-B., Takeshita A., Hayashi K., Hayashi T. Structures and antiviral activities of polysaccharides from Sargassum trichophyllum. Carbohydrate Polymers. 2011;86(2):995–999. [Google Scholar]
- Leung P.-C. The efficacy of chinese medicine for SARS: A review of chinese publications after the crisis. The American Journal of Chinese Medicine. 2007;35(04):575–581. doi: 10.1142/S0192415X07005077. [DOI] [PubMed] [Google Scholar]
- Li J., He J., Huang Y. Role of alginate in antibacterial finishing of textiles. International Journal of Biological Macromolecules. 2017;94:466–473. doi: 10.1016/j.ijbiomac.2016.10.054. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Qin G., Cheng C., Yuan B., Huang D., Cheng S., et al. Purification, characterization and anti-tumor activities of polysaccharides from ecklonia kurome obtained by three different extraction methods. International Journal of Biological Macromolecules. 2020;150:1000–1010. doi: 10.1016/j.ijbiomac.2019.10.216. [DOI] [PubMed] [Google Scholar]
- Lin C.-Z., Guan H.-S., Li H.-H., Yu G.-L., Gu C.-X., Li G.-Q. The influence of molecular mass of sulfated propylene glycol ester of low-molecular-weight alginate on anticoagulant activities. European Polymer Journal. 2007;43(7):3009–3015. [Google Scholar]
- Lindahl U., Couchman J., Kimata K., Esko J.D. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2015. Proteoglycans and Sulfated Glycosaminoglycans; pp. 207–221. [DOI] [Google Scholar]
- Luo C., Wu W., Lou S., Zhao S., Yang K. Improving the in vivo bioavailability and in vitro anti-inflammatory activity of tanshinone IIA by alginate solid dispersion. Journal of Drug Delivery Science and Technology. 2020;60:101966. doi: 10.1016/j.jddst.2020.101966. [DOI] [Google Scholar]
- Luo H., Tang Q.-L., Shang Y.-X., Liang S.-B., Yang M., Robinson N., Liu J.-P. Can chinese medicine be used for prevention of corona virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chinese Journal of Integrative Medicine. 2020;26(4):243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W., Li B., Gu Q., Fang Y., Xing H. Preliminary studies on the chemical characterization and antihyperlipidemic activity of polysaccharide from the brown alga Sargassum fusiforme. Hydrobiologia. 2004;512(1):263–266. [Google Scholar]
- Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. Journal of Virology. 2014;88(22):13221. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanos M.T.V., Andrada-Serpa M.J., Mourão P.A.S., Yoneshigue-Valentin Y., Costa S.S., Pereira M.S., et al. A sulphated fucan from the laminaria abyssalis inhibits the human T cell lymphotropic virus type 1-induced syncytium formation in HeLa cells. Antiviral Chemistry and Chemotherapy. 2002;13(4):219–221. doi: 10.1177/095632020201300402. [DOI] [PubMed] [Google Scholar]
- Sachan N., Pushkar S., Jha A., Bhattcharya A. Sodium alginate: The wonder polymer for controlled drug delivery. Journal of Pharmacy Research. 2009;2(8):1191–1199. [Google Scholar]
- Sano Y. Antiviral activity of alginate against infection by tobacco mosaic virus. Carbohydrate Polymers. 1999;38(2):183–186. [Google Scholar]
- Schürks N., Wingender J., Flemming H.C., Mayer C. Monomer composition and sequence of alginates from Pseudomonas aeruginosa. International Journal of Biological Macromolecules. 2002;30(2):105–111. doi: 10.1016/s0141-8130(02)00002-8. [DOI] [PubMed] [Google Scholar]
- Setyawidati N.A.R., Puspita M., Kaimuddin A.H., Widowati I., Deslandes E., Bourgougnon N., Stiger-Pouvreau V. Seasonal biomass and alginate stock assessment of three abundant genera of brown macroalgae using multispectral high resolution satellite remote sensing: A case study at Ekas Bay (Lombok, Indonesia) Marine Pollution Bulletin. 2018;131:40–48. doi: 10.1016/j.marpolbul.2017.11.068. [DOI] [PubMed] [Google Scholar]
- Shajahan A., Supekar N., Gleinich A., Azadi P. Deducing the N- and O- glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Astani A., Ghosh T., Schnitzler P., Ray B. Polysaccharides from Sargassum tenerrimum: Structural features, chemical modification and anti-viral activity. Phytochemistry. 2010;71(2):235–242. doi: 10.1016/j.phytochem.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Song S., Peng H., Wang Q.-L., Liu Z.-Q., Dong X.-P., et al. Inhibitory activities of marine sulfated polysaccharides against SARS-CoV-2. Food Function. 2020;11:7415–7420. doi: 10.1039/d0fo02017f. [DOI] [PubMed] [Google Scholar]
- Stokke B.T., Smidsroed O., Bruheim P., Skjaak-Braek G. Distribution of uronate residues in alginate chains in relation to alginate gelling properties. Macromolecules. 1991;24(16):4637–4645. [Google Scholar]
- Su H.-X., Yao S., Zhao W.-F., Li M.-J., Liu J., Shang W.-J., et al. Anti-SARS-CoV-2 activities in vitro of shuanghuanglian preparations and bioactive ingredients. Acta Pharmacologica Sinica. 2020;41(9):1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.-L., Li Y., Ni L.-Q., Li Y.-X., Cui Y.-S., Jiang S.-L., et al. Structural characterization and antiviral activity of two fucoidans from the brown algae sargassum henslowianum. Carbohydrate Polymers. 2020;229 doi: 10.1016/j.carbpol.2019.115487. [DOI] [PubMed] [Google Scholar]
- Szekalska M., Puciłowska A., Szymańska E., Ciosek P., Winnicka K. Alginate: Current use and future perspectives in pharmaceutical and biomedical applications. International Journal of Polymer Science. 2016;8:1–17. [Google Scholar]
- Taylor R.L., Conrad H.E. Stoichiometric depolymerization of polyuronides and glycosaminoglycuronans to monosaccharides following reduction of their carbodiimide-activated carboxyl groups. Biochemistry. 1972;11(8):1383–1388. doi: 10.1021/bi00758a009. [DOI] [PubMed] [Google Scholar]
- Wen C.-C., Shyur L.-F., Jan J.-T., Liang P.-H., Kuo C.-J., Arulselvan P., et al. Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and taxillus chinensis inhibit SARS-CoV replication. Journal of Iraditional and Complementary Medicine. 2011;1(1):41–50. doi: 10.1016/S2225-4110(16)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., et al. Production of authentic SARS-CoV mpro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. Journal of Molecular Biology. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biology. 2005;3(10) doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of AME. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X.D., Shen D.-D., Shen Q., Su H., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material