Abstract
The t(14,18) chromosomal translocation that occurs in human follicular lymphoma constitutively activates the BCL2 gene and disrupts control of apoptosis. Interestingly, 70% of the t(14,18) translocations are confined to three 15-bp clusters positioned within a 150-bp region (major breakpoint region or [MBR]) in the untranslated portion of terminal exon 3. We analyzed DNA-protein interactions in the MBR, as these may play some role in targeting the translocation to this region. An 87-bp segment (87MBR) immediately 3′ to breakpoint cluster 3 was essential for DNA-protein interaction monitored with mobility shift assays. We further delineated a core binding region within 87MBR: a 33-bp, very AT-rich sequence highly conserved between the human and mouse BCL2 gene (37MBR). We have purified and identified one of the core factors as the matrix attachment region (MAR) binding protein, SATB1, which is known to bind to AT-rich sequences with a high propensity to unwind. Additional factors in nuclear extracts, which we have not yet characterized further, increased SATB1 affinity for the 37MBR target four- to fivefold. Specific binding activity within 37MBR displayed cell cycle regulation in Jurkat T cells, while levels of SATB1 remained constant throughout the cell cycle. Finally, we demonstrated in vivo binding of SATB1 to the MBR, strongly suggesting the BCL2 major breakpoint region is a MAR. We discuss the potential consequences of our observations for both MBR fragility and regulatory function.
The t(14,18) translocation of human follicular lymphoma activates the oncogene BCL2 (22, 25) and aborts programmed cell death (11, 12, 18). Although the BCL2 gene is more than 200 kb long, at least 70% of translocations occur within a 150-bp sequence located in the untranslated portion of the terminal exon, designated the major breakpoint region (MBR) (4, 25, 26). We have shown that the targeting of BCL2 translocation within the MBR is considerably more precise than previously appreciated (27). Nearly all breakpoints that we examined (51 of 62) occurred within three 15-bp-wide clusters spaced evenly at approximately 50-bp intervals along the MBR. The breakpoints demonstrated fusion of BCL2 sequences to JH coding sequences in a canonical form suggesting rearrangement into immunoglobulin gene coding joints. Like immunoglobulin heavy-chain (IgH) rearrangements, untemplated nucleotides were usually present at the breakpoints.
Tsujimoto et al. (26) suggested that these recombinations could be explained by the presence of a pseudo-DH element within the MBR. Under this model, aberrant events leading to translocation would involve the V(D)J recombinase recognizing and using immunoglobulin signal sequences within the MBR to target the MBR-JH fusion. The inappropriate use of ectopic immunoglobulin signal sequences has been convincingly demonstrated in the translocation of other genes, such as TTG1 (19). However, the clustering of translocation breakpoints as described in our study is completely inconsistent with the pseudo-D model. In this latter instance, translocations should occur in one cluster—the junction of pseudo-D coding and signal sequences—rather than at the three observed cluster sites. Furthermore, no signal joint has ever been found on the reciprocal product, der 18q-. Therefore, while the V(D)J recombinase probably participates in BCL2 translocation by site-directed cleavage and DNA end generation within the IGH locus, some other determinant(s) must be responsible for the involvement of the MBR.
Two general alternatives to the pseudo-D model, which are not mutually exclusive, may be offered in explanation of BCL2 translocation. The first is that translocations take place through the stochastic intervention of any one of a host of recombination/repair mechanisms (RRM). The hyperrecombination phenotype of several chromosome instability disorders, for example, is likely to be explained by deficiencies of products important in these several pathways; the rare errors of an otherwise intact RRM could, therefore, generate translocations. It should be noted that this hypothesis includes the possible existence of DNA signals that target RRM, creating translocation hot spots. Although such hot spots may be distributed throughout the genome, in those (presumably fortuitous) instances in which hot spots and growth-regulatory genes, such as BCL2, are located in the same region of DNA, rearrangements targeted by the hot spot may create a selectable phenotype through regulatory or coding sequence alterations in the nearby gene.
We have previously shown that an octamer consensus sequence, CC(A/T)CC(A/T)GC, present in 50% of highly recombinogenic minisatellite repeat unit motifs, also occurs at or near the breakpoints of several oncogene translocations, including those involving MYC and BCL2 (16). Of particular interest is the observation that tandem 8-for-8 and 7-for-8 matches of this octamer, designated χ because of its homology to the recognition signal of the RecBCD recombinase of Escherichia coli (23), form the immediate 5′ boundary of the MBR: The first translocation cluster begins at the first MBR base 3′ to the 8/8 χ. On the basis of this association, we have suggested the hypothesis that the consensus is a targeting signal for an RRM and, as such, defines a recombination hot spot for translocation within the MBR.
The alternative model is that BCL2 translocations take place at precise locations within the MBR because such rearrangements disrupt or alter functional configurations that occur there. Other than polyadenylation and message stability signals, however, little is known of the potential functional features of untranslated, terminal exons. In the case of BCL2, mRNA produced from translocated genes shows no half-life alteration (22), although an advantage over wild-type mRNA in posttranscriptional processing has been noted (21). Therefore, a functional basis for targeting rearrangements would necessarily involve a currently unknown (or unexpected) use of DNA sequence.
To begin distinguishing between these possibilities, we have identified and characterized several complexes that form when nuclear extracts are incubated with DNA probes from this region of BCL2. Our initial studies indicated complexes forming with a 279-bp target that included the three breakpoint clusters described by Wyatt et al. (27) and approximately 175 bp of 5′ and 3′ flanking DNA. The most prominent of these was a very slowly migrating complex with a binding site within the AT-rich region immediately flanking breakpoint cluster 3 (7). We now report purification and characterization of a matrix attachment region (MAR) binding protein present in this complex.
MATERIALS AND METHODS
Cell lines.
Jurkat is a human T-cell leukemia cell line. SUDHL-6 is a cell line derived from a human diffuse B-cell lymphoma and carries the t(14,18) translocation characteristic of BCL2-IGH fusions. HL60 is derived from a human acute promyelocytic leukemia, and MCF-7 is a human breast cancer cell line.
Preparation of nuclear extract.
Nuclear extracts were prepared as described by Dignam et al. (8). The nuclear extracts from sorted cells and transient transfections were prepared as described by Andrews and Faller (1). Protease inhibitors phenylmethylsulfonyl fluoride (PMSF) and benzamidine to 1 mM and aprotinin, leupeptin, and pepstatin to 1 μg/ml were used in all nuclear extract preparations.
Preparation of target DNA.
The 279-bp double-stranded DNA probe for gel shift analysis was PCR labeled as described by DiCroce and Krontiris (7), using primers RTW2 and RTW3. The 87MBR (87-bp segment of the MBR) probe was PCR labeled by using oligomers PDC4 (5′AATGATCAGACCTTTGAATGA 3′) and the 3′ oligomer, RTW3. The 37MBR oligomer probe was prepared by annealing 37-bp sense and antisense oligonucleotides in the presence of 50 mM NaCl. The ends of the oligonucleotides containing SalI and XhoI sites were then filled in the presence of [α-32P]dATP (3,000 Ci/mmol), [α-32P]dCTP (3,000 Ci/mmol), dTTP, and dGTP, using the Klenow fragment of DNA polymerase I (New England Biolabs). The radiolabeled 37MBR probe was then purified by using a QIAquick nucleotide removal kit (Qiagen) or probeQuant G-50 microcolumns (Pharmacia). Gel-purified 37MBR oligomers for the DNA affinity column were prepared similarly to the method for gel-purified 279MBR probe preparation (7).
MSA.
DNA-protein binding reactions were carried out in 20 μl of a mixture containing 10 mM Tris HCl (pH 7.5), 40 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 4% glycerol, 0.5 μg of poly(dI-dC), 3 ng of probe, and 2 μg of nuclear extract. The samples were incubated for 30 min at room temperature. The reaction products were separated by electrophoresis through a 4% polyacrylamide gel in 1× TBE (89 mM Tris HCl [pH 8.0], 89 mM boric acid, 2 mM EDTA). In competition experiments, nonradioactive oligonucleotide competitors were added along with labeled probe to the binding reaction prior to the addition of nuclear extract. For Resource Q column fractions, assays were performed with 100 ng of poly(dI-dC) per reaction; for 37MBR and poly(rA)-oligo(dT) columns, 50 ng of poly(dI-dC) and 4 μg of bovine serum albumin (BSA) per reaction were used. Some gel mobility shift assays (MSAs) (Fig. 6 and 7A) were done as described by Dickinson et al. (6), with slight modifications. The reaction volume was 20 μl containing 10 mM HEPES (pH 7.9), 1 mM dithiothreitol (DTT), 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, BSA (0.2 μg/μl), poly(dI-dC) (0.05 to 0.25 μg), and purified SATB1 (special AT-rich binding protein 1) at room temperature for 20 min. The samples were separated over a 6% acrylamide minigel (8 by 10 cm) containing 0.05% bisacrylamide, 5% glycerol, and 0.5× TBE. The gel was run at 120 V at 4°C for 2 h.
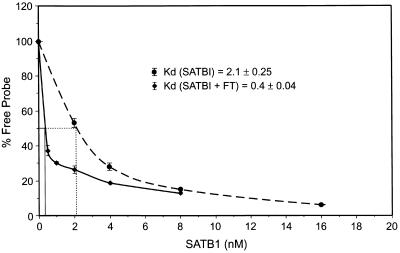
FIG. 6.
Kd determination for SATB1 binding to 37MBR. Increasing amounts of SATB1 were incubated with a constant amount of the 37MBR probe in binding reactions as described by Dickinson et al. (6). The amount of free probe and B1 complex were quantitated with a PhosphorImager. The percentage of free probe was plotted against nanomoles of SATB1 used, and the Kd was calculated as the amount of SATB1 required to bind 50% of probe in the formation of the B1 complex. The dotted line represents three experiments in which only SATB1 was used in the binding reaction; the solid line represents binding performed with purified SATB1 and flowthrough (FT) from 37-mer affinity column.
FIG. 7.
(A) Antibody MSA inhibition assay. The assay was performed by the MSA protocol of Dickinson et al. (6) in the presence of 37MBR probe and Jurkat nuclear extracts. Lane 1, no antibody added; lanes 2 and 3, increasing amounts of anti-SATB1 antiserum incubated with complexes following the binding reaction; lanes 4 and 5, increasing amounts of anti-PCNA antibody similarly incubated with complexes. (B) Western blotting of proteins present in the MBR B1 complex: anti-SATB1 antibody. Preparative-scale MSAs were performed with Jurkat nuclear extracts or with purified eluate (SATB1 and PARP) mixed with the flowthrough from the 37-mer DNA affinity column (FTDAC) in the presence and absence of the 37MBR probe. The B1 complex, or the corresponding region from MSA gels when probe was absent, was excised from the gel, and proteins were eluted and subjected to Western blot analysis. Lane 1, nuclear extract from Jurkat cells demonstrating the position of a positive SATB1 signal; lane 2, protein from B1 complex obtained with Jurkat nuclear extract; lane 3, protein from reconstituted B1 complex formed by mixture of the SATB1 and PARP eluates with the flowthrough from the DNA affinity column; lane 4, protein from the region of the reconstituted complex on MSA gels in the absence of probe. (C) Western blotting of proteins present in the MBR B1 complex: anti-PARP antibody. The Western blot in panel B was reprobed with anti-PARP antibody.
Chromatographic purification of SATB1 from Jurkat cells.
The nuclear extract (5 mg) was passed through a NAP-10 column equilibrated with buffer A (20 mM HEPES [pH 7.9], 0.5 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, 0.5 mM PMSF, 0.1 M NaCl). The extract in buffer A and 0.1 M NaCl was applied onto a Resource Q column (1 ml; Pharmacia) attached to an AKTA purifier system (Pharmacia). The column was washed with 10 ml of buffer A, and protein was eluted with an increasing linear gradient of NaCl in buffer A. Fractions containing complex B1 or B3/B4 were diluted in buffer B (20 mM HEPES [pH 7.9], 12.5 mM MgCl2, 1 mM DTT, 20% glycerol, 0.1% NP-40, 1 mM PMSF, 0.075 M NaCl) and passed over the 37-mer DNA affinity column. The proteins were eluted with an NaCl step gradient. The fraction containing SATB1 and poly(ADP-ribose) polymerase (PARP) (0.3 M NaCl) was diluted in buffer B (without NP-40) and passed through a poly(rA)-oligo(dT)-cellulose column. The bound proteins were eluted with an NaCl step gradient.
Preparation of DNA affinity and poly(rA)-oligo(dT) columns.
Gel-purified oligomers of 37MBR were annealed and ligated as described by Kadonaga and Tjian (13). The oligomers (400 μg) were coupled to 0.8 g of cyanogen bromide-activated Sepharose 4B (Pharmacia). The coupling of the oligomers to the resin was performed as described by Kadonaga and Tjian (13). The resin was stored in column storage buffer (10 mM Tris HCl [pH 7.6], 1 mM EDTA, 0.3 M NaCl, 0.02% sodium azide) at 4°C. The poly(rA)-oligo(dT)-cellulose column was prepared by suspending 1 g of oligo(dT)-cellulose (Pharmacia) in buffer D (50 mM Tris HCl [pH 8.0], 0.5 mM EDTA, 12 mM β-mercaptoethanol, 5 mM MgCl2, 1 mM PMSF, 5% glycerol) for 10 min at room temperature (15). A solution of poly(rA) (10 mg/ml in 10 mM Tris HCl [pH 8.0]–1 mM EDTA) was incubated with the oligo(dT)-cellulose suspension [100 A260 units/g (dry powder) of oligo(dT)] with shaking at room temperature for 30 min. The unbound poly(rA) was removed with equilibration buffer washes.
Western blot analysis.
Protein samples were mixed with equal volumes of 2× sample buffer (100 mM Tris HCl [pH 6.8], 200 mM DTT, 4% sodium dodecyl sulfate (SDS), 0.2% bromophenol blue, 20% glycerol), boiled for 5 min, and resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in 6% minigels. The proteins were electrophoretically transferred to Immobilon P membrane (Millipore) in 25 mM Tris HCl (pH 8.3)–192 mM glycine for 1 h at 60 V. The membrane was blocked in 5% BSA in TST buffer (20 mM Tris HCl [pH 7.4], 0.5 M NaCl, 0.05% Tween 20), washed in TST buffer and incubated with a 1:2,000 dilution of rabbit anti-SATB1 serum (20) at 4°C overnight. The membrane was washed and incubated with anti-rabbit antibody conjugated to horseradish peroxidase as the secondary antibody (1:10,000) for 2 h at room temperature. The blot was extensively washed with TST buffer, incubated with enhanced chemiluminescence reagent solution (Amersham), and exposed to X-ray film for visualization of the SATB1 band. For detection of PARP protein, the primary antibody was goat polyclonal anti-PARP antibody (1:500; Santa Cruz Biotechnology), and the secondary antibody was an anti-goat antibody conjugated to alkaline phosphatase (Bio-Rad). This blot was developed with an Immune-Lite chemiluminescence assay kit (Biorad).
Staining of protein gels.
Protein gels were silver stained by using a Bio-Rad Silver Stain Plus kit.
Transient transfection.
MCF-7 cells were transfected with a total of 30 μg of DNA from pECE (vector alone) or pECH (SATB1 expression plasmid) (6), using a calcium phosphate transfection kit (Promega). Cells were incubated with DNA for 20 h following transfection. The medium was then changed, and the cells were harvested 48 h posttransfection in TEN (40 mM Tris HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl) and washed once with phosphate-buffered saline. The nuclear extracts were prepared by the protocol of Andrews and Faller (1).
FACS.
The Jurkat cells (5 × 106 cells/ml) were incubated with Hoeschst 33342 dye (10 μg/ml) at 37°C for 1 h. The cells were then concentrated to 3 × 107/ml and incubated on ice for 30 min. Cells were sorted into six fractions with a fluorescence-activated cell sorting (FACS) analyzer (MoFlo; Cytomation, Inc.).
Liquid chromatography-MS/MS sequencing analysis.
The purified proteins were separated on an SDS–6% polyacrylamide gel. The protein in the gel was then subjected to the in-gel reduction-alkylation-digestion procedure as described by Hellman et al. (10). The peptides obtained following tryptic digestion were then subjected to liquid chromatography-tandem mass spectrometry (MS) analysis (24) on a MAT TSQ-700 triple-quadrupole mass spectrometer (Finnigan Corp., San Jose, Calif.).
In vivo SATB1 binding assay.
Jurkat cells were grown in RPMI 1640 medium containing 10% fetal bovine serum in a humidified atmosphere of 5% CO2. In vivo cross-linking and immunoprecipitation with anti-SATB1 and control antibodies were performed by the published protocol (5, 15a) except that the cross-linked chromatin was digested with RsaI and HindIII (New England Biolabs). After the reversal of formaldehyde cross-links and DNA purification, DNA was PCR amplified to identify sequences binding SATB1 in vivo. Six oligonucleotides were used as primers to PCR amplify three loci: BCL2 locus (5′CTTTAGAGTTGCTTTACGTTG3′/5′TCCATATTCATCACTTTGACAA3′), SBS3 locus (5′CAAGATTTTGATGGCCCCAAGCA3′/5′GGGCTTTACCAGATGAGAAT 3′), and HBB (5′AGAAACTGCAGATTCTCTGCAT3′/5′CCTTCTACTTAGCCTACTTTTGA3′). SBS3 is a locus isolated from Jurkat cell DNA that served as a positive control for SATB1 binding in vivo (5; GenBank/EMBL/DDBJ accession no. AF051676). The sequence chosen from human β-globin locus (HBB; accession no. U013171, positions 6789 to 7098), in which there is no SATB1 binding site, served as a negative control. The accession number for the MBR region is M13994.
PCR was performed in a 50-μl volume including 2.5 U of Taq polymerase (Display Systems Biotech), 1× manufacturer's buffer, 1.5 mM MgCl2, four (200 μM each), deoxynucleoside triphosphates and 2 μM each primer. Cycling conditions were as follows: 2 cycles of 95°C for 3 min, 53°C for 3 min, and 72°C for 3 min, 38 cycles of 94°C for 1 min, 53°C for 1 min, and 72°C for 1 min, and an additional 5 min at 72°C. PCR products were examined by 2% agarose gel electrophoresis in the presence of ethidium bromide.
RESULTS
Sequence localization, specificity, and preliminary characterization of nuclear complexes binding the MBR.
We previously described the appearance of a slowly migrating nucleoprotein complex when nuclear extracts from particular human leukocyte cell lines, such as Jurkat, were incubated with the 279MBR target (7). A distinct complex that migrated even more slowly than that of Jurkat formed in the presence of nuclear extracts from all other human and mouse cell lines tested, including those of epithelial, fibroblastic, and leukocytic origin. To characterize these complexes further, we performed deletion analysis of the 279MBR region, using nuclear extracts from two cell lines displaying prototypic forms of the slowly migrating complexes, Jurkat and MCF-7, in MSAs. Three targets of successively smaller size were used: 279MBR, 87MBR, and 37MBR (Fig. 1A). The Jurkat complex formed with all targets (Fig. 2A). Interestingly, the more slowly migrating complex seen in MCF-7 and other cell lines was observed only with the larger targets, 279MBR and 87MBR; no complex was visualized with the smaller 37MBR target (Fig. 2A). Therefore, we concluded that the core binding complex in Jurkat extracts required only the AT-rich, 37-bp region just downstream of breakpoint cluster 3 and that the core factor(s) involved was absent or inactive in MCF-7 and related nuclear extracts. Specificity of the core complex present in Jurkat nuclear extracts was verified in MSAs using cold (unlabeled) competitors. With a labeled 87MBR target, cold competitors consisting of either 87MBR or 37MBR effectively competed for binding, whereas 151MBR competitor representing the 5′ region of the MBR did not compete.
FIG. 1.
(A) Schematic diagram of the BCL2 locus. Positions of the BCL2 gene exons, the MBR, the breakpoint clusters (C1, C2, and C3) and the AT-rich 3′ end are shown. The subregions of the MBR (279MBR, 87MBR, 37MBR, and 151MBR) used in the DNA-protein interaction analyses are also shown. (B) DNA sequence of the MBR. Positions of the probes are demarcated with thin arrows (279), thick hatched arrows (87), and thick solid arrow (37). The three breakpoint clusters are underlined; a hatched horizontal line indicates the extent of 37MBR. (C) MSA probes and competitors. The 37-bp oligomer corresponding to 37MBR and its mutations used in gel MSAs are shown. Lowercase boldface letters indicate positions of mutations.
FIG. 2.
(A) Complex formation in the MBR region of the BCL2 gene. Probes corresponding to the various regions of MBR (279, 87, and 37 in Fig. 1B) were used in MSAs with Jurkat and MCF-7 nuclear extracts. The reaction products were electrophoresed through 4% polyacrylamide gels, dried, and subjected to autoradiography as shown. Positions of the specific complexes in Jurkat and MCF-7 are marked B1 and B1′, respectively. (B) Competition MSA with the 87MBR target. The radiolabeled 87MBR probe was incubated with Jurkat nuclear extract in the absence and presence of cold oligonucleotide competitors. Lane 1 has no competitor; lanes 2, 4, and 6 have 5-fold and lanes 3, 5, and 7 have 10-fold molar excesses of the cold competitors indicated (see also Fig. 1A). In lane 8, the binding reaction was done with 37MBR as the probe. The reaction products were electrophoresed through a 6% polyacrylamide gel and processed as described above. (C) Competition MSA with 37MBR as the target. The MSA reaction was carried out as for panel B in the absence (lane 1) or presence (lanes 2 to 7) of 10-fold molar excess of the cold competitors (Fig. 1C). The products were analyzed on a 4% polyacrylamide gel. The DNA-protein complexes, B1 through B4, are indicated with arrows. Wt, wild type; As, single-stranded antisense DNA competitor; Ss, single-stranded sense DNA competitor.
Following localization of core factor binding to the 37MBR fragment, we used this target in all subsequent analyses and purifications. As seen in the final lane of Fig. 2B, as well as in Fig. 2C, four complexes were reproducibly detected with this probe, although their relative intensities varied from experiment to experiment. They were designated B1 through B4 and represented the slowest- to fastest-migrating complexes, respectively. B1 was consistently present in large amounts when the 37MBR target was used, as was B4. There was considerable variation in the amount of B2 complex from experiment to experiment. B3 was always present in very low amounts.
To complete our characterization of the specificity of these complexes, competition MSAs were performed with a variety of mutated targets (depicted in Fig. 1C). The principal complex observed in all MBR MSAs with Jurkat was B1, and this complex was effectively competed only by wild-type oligomer. Several of the mutant oligomers showed diminished effects in competition, as expected. Of note, the Mu2 competitor competed somewhat more effectively than Mu1 and Mu3, indicating that the terminal region of the 37-bp target was more important to the overall interaction with the core factor(s). The nonspecific binding of the B2 complex was competed by both wild-type and mutant competitors. Complex B4 was not abolished by any competitor, and B3 showed a variable pattern of competition suggesting, perhaps, an intermediate degree of specificity; as seen in Fig. 2C, competition from the wild-type oligomer was somewhat more effective than competition with Mu1 to Mu3. Because the 37MBR target was so AT rich, we were concerned that it could denature under our experimental conditions and bind as single-stranded DNA. Lanes 6 and 7 of Fig. 2C indicate no competition of complex formation in the presence of either antisense- or sense-strand single-stranded DNA competitor. Thus, complex formation required a double-stranded target.
Purification of B1 and B3/B4 components from Jurkat nuclear extract.
In preparation for purification of the complexes that we observed with the 37MBR target, we used bromodeoxyuridine-mediated UV cross-linking to determine the number and size of factors present in Jurkat nuclear complexes (data not shown). Following binding to bromodeoxyuridine-substituted and radiolabeled 37MBR probe, complexes B1 through B4 were excised from gels and subjected to SDS-PAGE. The B1 complex consisted of several high-molecular-weight proteins. One major protein band was approximately 300 kDa, while three to four more bands were clustered between 100 and 140 kDa. These bands were abolished when the affinity labeling was performed in the presence of cold wild-type competitor but not in the presence of Mu1. The most prominent of the B1 proteins was approximately 100 kDa. The B2 complex had one major protein in common with B1 at ∼100 kDa. Several proteins appeared in the B4 complex; the most prominent was ∼70 kDa, with minor bands at ∼86 to 90 and >300 kDa. The latter were not reproducibly present. The B3 complex shared the 70- and 86-kDa bands with B4 and had variable amounts of the ∼100-kDa protein. Larger proteins were absent in this complex.
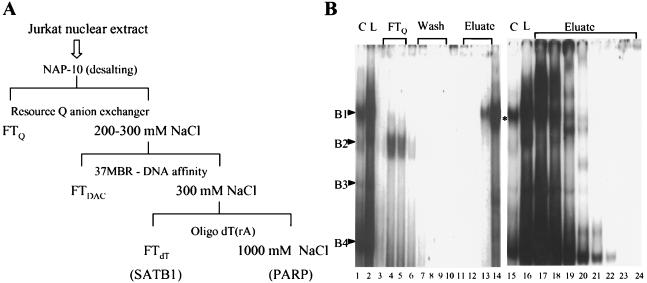
Because the Jurkat nuclear extract demonstrated the highest level of complex formation with MBR targets, we subjected it to a series of chromatographic steps to purify the protein constituents of several complexes. The purification scheme is shown in Fig. 3A. MSAs using 37MBR as the probe were used to monitor protein binding activity through all purification steps. Linear gradient elution from a Resource Q anion-exchange column using increasing concentrations of NaCl successfully resulted in a 100-fold purification of the B1 complex (Fig. 3B, fractions 13 and 14) and its separation from complexes B2, B3, and B4. B3 and B4 were also isolated independently of other complexes in fractions 34 and 35 (not shown). Active fractions containing complexes B1 and B3/B4 were then applied separately to a Sepharose 4B column to which concatenated 37MBR oligomers had been coupled; proteins were then eluted with an NaCl step gradient. Two bound proteins present in the B1-containing Resource Q fractions with apparent molecular masses of 103 and 118 kDa coeluted from the affinity column at 0.2 and 0.4 M NaCl (Fig. 4A). Three proteins of 70, 86, and >300 kDa were present in the high-salt eluates of the affinity column loaded with active Resource Q fractions containing the B3 and B4 complexes (Fig. 4D). These reproduced both the B3 and B4 complexes in MSAs (Fig. 4E).
FIG. 3.
(A) Schematic detail of chromatographic purification of the B1 complex. FT, flowthrough; DAC, DNA affinity chromatography. (B) MSA with the protein fractions obtained from the Resource Q column. The binding reaction was performed in the presence of 100 ng of poly(dI-dC) as nonspecific competitor with Jurkat nuclear extract (load [L]; lanes 2 and 16). Other lanes contain flowthrough (FT; lanes 3 to 5), column washes (lanes 6 to 10), and eluates obtained with a linear NaCl gradient (lanes 11 to 13 and 16 to 20). Purified B1 complex, corresponding to lanes 13 and 14, is indicated with an asterisk. C, standard MSA with Jurkat nuclear extract.
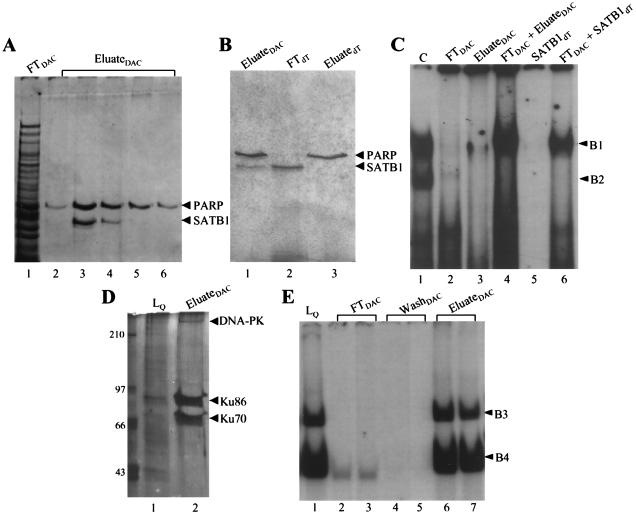
FIG. 4.
(A) Purification of SATB1 (complex B1) by DNA affinity chromatography (DAC). The silver-stained SDS–8% polyacrylamide gel shows protein fractions obtained from the 37-mer-Sepharose 4B column. Lane 1, flowthrough (FT); lanes 2 and 3, 0.2 M eluate from the NaCl gradient; lanes 3 and 4, 0.4 M eluate from the NaCl gradient. The positions of PARP and SATB1 are indicated. (B) Protein profile of fractions from a poly(rA)-oligo(dT)-Sepharose column for SATB1 (complex B1) purification. In this silver-stained gel (6%), lane 1 represents material loaded onto the poly(rA)-oligo(dT) column, lane 2 shows the unbound SATB1 fraction, and lane 3 contains the tightly bound PARP fraction eluted as described in Materials and Methods. (C) Reconstitution of Jurkat complex formation with fractions from affinity and poly(rA)-oligo(dT) columns for SATB1 (complex B1) purification. Binding reactions were performed with Jurkat nuclear extract (C; lane 1), flowthrough from the 37-mer affinity column (FT; lane 2), eluate from the affinity column (containing SATB1 and PARP; lane 3), FT and eluate together (lane 4), SATB1 alone obtained from the flowthrough of the poly(rA)-oligo(dT) column (lane 5), and flowthrough from the affinity column together with SATB1 from poly(rA)-oligo(dT) (lane 6). (D) Affinity column purification of proteins in the B4 complex; silver-stained SDS-polyacrylamide gel. Lane 1, material loaded onto the column from Sepharose Q; lane 2, proteins eluted from the affinity column as described in Materials and Methods. DNA-PK, DNA-dependent protein kinase. (E) Reconstitution of B3 and B4 complexes with high-salt eluates of affinity chromatography. MSAs were performed with B3 and B4 fractions from Sepharose Q (lane 1), flowthrough (lanes 2 and 3), and low-salt washes (lanes 4 and 5) of the affinity column and high-salt eluates (lanes 6 and 7). Positions of B3 and B4 are indicated.
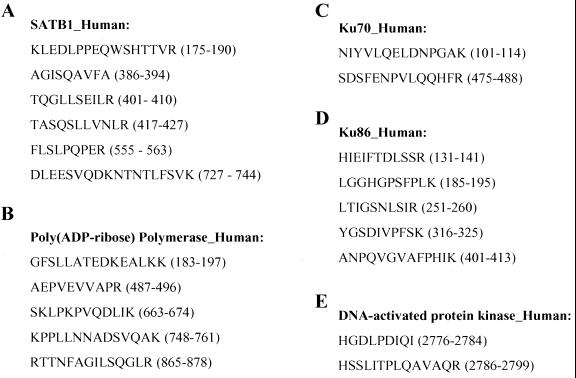
The proteins eluted from the affinity columns were then subjected to SDS-PAGE. The Coomassie blue-stained bands from B1- and B3/B4-specific gels were excised, and individual bands were digested with trypsin. The bands from the B1 complex shown in Fig. 4A were identified by using MS analysis as PARP (118 kDa) and SATB1 (103 kDa). SATB1 is a cell-type-restricted MAR-binding protein which is predominantly expressed in thymocytes (6). For SATB1, six oligopeptides, ranging in size from 9 to 18 amino acids, were identified as identical to the previously established SATB1 protein sequence (Fig. 5A). For PARP, five oligopeptides were identified (Fig. 5B). The three proteins from the B3 and B4 complexes were similarly identified as Ku70 (Fig. 5C), Ku86 (Fig. 5D), and the DNA-dependent protein kinase (Fig. 5E). All these identifications were consistent with our preliminary results from UV cross-linking.
FIG. 5.
MS identification of purified proteins from B1 and B4 complexes. Shown are the peptide fragments identified by MS analysis for SATB1 (A), PARP (B), Ku70 (C), Ku86 (D), and DNA-dependent protein kinase (E). Amino acid residue numbers of the peptides for each protein are listed in parentheses.
The final step of the purification scheme for the B1 complex was passage of the mixture containing SATB1 and PARP over a poly(rA)-oligo(dT)-cellulose column. We attempted this because of the reported affinity of PARP for a variety of DNA structures, including single strands and gaps, which SATB1 does not bind. In confirmation of these reported differences, SATB1 appeared in the flowthrough of this column, while PARP bound very tightly and was subsequently eluted with 1.0 M NaCl (Fig. 4B).
Reconstitution of 37MBR complexes with proteins purified from affinity chromatography and poly(rA)-oligo(dT) cellulose.
To determine if the complexes formed with whole nuclear extracts could be reproduced with purified protein preparations, we performed MSAs with eluate from the DNA affinity column, as well as the SATB1 and PARP components purified from poly(rA)-oligo(dT)-cellulose chromatography. When the SATB1-PARP mixture (Fig. 4C, lane 3) was incubated with 37MBR target, a complex of the appropriate size was observed. However, when flowthrough was added to the SATB1-PARP mixture, binding activity was substantially increased (Fig. 4C, lane 4), and the complex morphology widened considerably to reproduce the pattern observed with crude nuclear extracts. The same effect was observed with SATB1 purified from poly(rA)-oligo(dT)-cellulose. While purified SATB1 reproduced the B1 complex when present in high concentrations during MSA reactions, the binding activity was augmented in the presence of flowthrough from the DNA affinity column (Fig. 4C, compare lanes 5 and 6). For the MSAs shown in Fig. 4C, lanes 5 and 6, amounts of purified SATB1 and flowthrough from the DNA affinity column were chosen from the binding curves in Fig. 6 to illustrate the effect on binding more clearly. When we quantitated the dissociation constant with purified SATB1, the effect of flowthrough on binding was substantial: a fivefold decrease in the Kd, from 2.1 to 0.4 (Fig. 6). This effect was observed only on reconstitution of the complex with SATB1 and the flowthrough from the DNA affinity column of the B1 complex. Flowthrough from the B3/B4 affinity column, as well as several control variations of the binding reaction testing the effects of salt, buffer, and pH (9), had no influence on SATB1 affinity (not shown). The same effect of flowthrough on SATB1 binding was observed when we replaced SATB1 purified from Jurkat with SATB1 expressed in, and then purified from E. coli (not shown).
Presence of SATB1 in B1 complexes formed with Jurkat extracts.
To investigate further the roles of SATB1 and PARP in Jurkat complex formation, we repeated MSAs with Jurkat nuclear extracts in the presence of anti-SATB1 antibody (Fig. 7A). Available antibody preparations against SATB1 do not produce supershifts; rather, complex formation is inhibited (6a). As shown in lanes 2 and 3 of Fig. 7A, anti-SATB1 antibody, but not anti-PCNA antibody, had a marked inhibitory effect. Incubation of extracts with antibody before and after addition of target DNA gave the same result. As additional evidence supporting the presence of SATB1 in the Jurkat B1 complex, we scaled up an MSA with crude nuclear extract as well as with combined eluate and flowthrough of the DNA affinity column. These complexes were excised from MSA gels and then subjected to SDS-PAGE and Western blotting. SATB1 was clearly present in complexes formed with both crude extract (Fig. 7B, lane 2) and, as expected, with proteins purified from the affinity column (lane 3). In contrast, PARP was absent in complexes from both sources (Fig. 7C). No inhibition of complex formation or supershifting was observed when anti-PARP antibody was used in MSAs of B1 complex formation (data not shown).
Transient expression of SATB1 in MCF-7 cells.
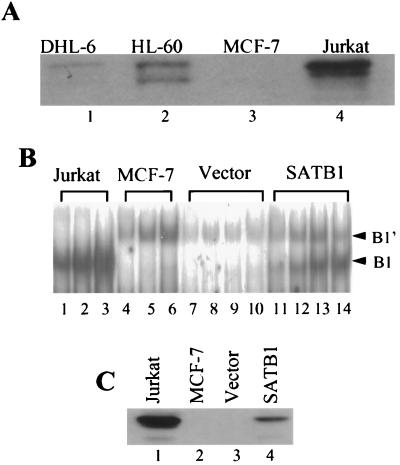
MSAs with nuclear extracts from cell lines such as MCF-7 did not demonstrate the B1 complex (Fig. 2A), suggesting the absence of Jurkat factors critical to complex formation. Given the previously described results concerning SATB1, complex formation in the cell lines that we have tested should therefore correlate with the presence or absence of this protein in nuclear extracts. We tested this hypothesis in several ways. First, Western blotting of nuclear extracts from three B1-complex-positive cell lines (Jurkat, SUDHL-6, and HL60) and one B1-complex-negative cell line (MCF-7) was performed; the level of SATB1 protein corresponded well with the level of complex formation (Fig. 8A). Levels of SATB1 were highest in Jurkat, where we observed the largest amount of B1 complex, and absent in MCF-7. Two cell lines with intermediate levels of complex formation (SUDHL-6 and HL60) demonstrated levels of SATB1 that fell between those of Jurkat and MCF-7. All positive cell lines shared a larger form of SATB1 equivalent to that purified from affinity chromatography. A second, smaller form appeared in HL60; an alternative form slightly larger than the HL60 second component was detected in Jurkat. Whether these forms represented specific degradation products, posttranslational modifications, or the products of alternative splicing has not yet been ascertained. Phosphorylation of SATB1 has been observed (T. Kohwi-Shigematsu, unpublished data).
FIG. 8.
(A) Western blot analysis of nuclear extracts from four cell lines. SDS-PAGE, followed by Western blotting and incubation with anti-SATB1 antibody, was performed with nuclear extracts from SUDHL-6 (DHL-6), HL60, MCF-7, and Jurkat cells. (B) SATB1 expression and B1 complex formation in the MCF-7 cell line. Transient expression of a SATB1 expression plasmid (SATB1) and a control plasmid (Vector) was performed as described in the text. MSAs were performed with increasing amounts of Jurkat nuclear extract (lanes 1 to 3), MCF-7 nuclear extract (lanes 4 to 6), and extracts from MCF-7 cells transiently transfected with either the vector alone (lanes 7 to 10) or the vector expressing SATB1 (lanes 11 to 14). Positions of the Jurkat (B1)- and MCF-7 (B1′)-specific complexes are indicated by arrowheads. (C) Western blot analysis of the same extracts used in the MSAs of panel B. Anti-SATB1 antibody was used.
Second, we transfected MCF-7 transiently with a SATB1 expression plasmid. As shown in Fig. 8B, expression of SATB1 resulted in the appearance of the B1 complex in MCF-7 nuclear extracts. This corresponded to the appearance of SATB1 protein by Western blotting (Fig. 8C). As expected, untransfected MCF-7 cells or cells transfected with the vector alone demonstrated neither B1 complex nor SATB1 protein (Fig. 8B and C).
SATB1 binding of the BCL2 MBR in vivo.
The high-affinity, cell cycle-regulated binding of SATB1 to the MBR strongly suggested that this region served as a MAR. Recently, a highly sensitive and specific assay, involving chromatin immunoprecipitation by SATB1 antibody followed by PCR amplification, has been used to demonstrate the association of SATB1 with target DNA sequences in vivo (5). Therefore, we tested Jurkat MBR DNA sequences for in vivo SATB1 binding in this assay. For comparison, we used the previously characterized, in vivo SATB1 binding sequence SBS-3 (Fig. 9B) and a region isolated from the HBB locus that does not have any SATB1 binding sequence as a negative control (Fig. 9C). DNA purified from chromatin immunoprecipitated with anti-SATB1 antiserum (Fig. 9A, lane S) but not preimmune serum (Fig. 9A, lane P) showed nearly as strong a PCR signal as the positive control (Fig. 9B), indicating the in vivo association of SATB1 with the MBR.
FIG. 9.
SATB1 binds to 279MBR in vivo. PCR amplification of immunoprecipitated DNA was performed as described in Materials and Methods. PCR products from DNA immunoprecipitated by preimmune serum (P) or anti-SATB1 antiserum (S) and genomic DNA purified from Jurkat cells (G) were analyzed by 2% agarose gels containing ethidium bromide. Three regions were examined from each preparation: BCL2 (A), SBS3 (positive control; B), and HBB (negative control [N]; C).
Cell cycle-dependent binding of the B1 complex.
We have previously noted cell cycle-dependent binding activity of the B1 complex in Jurkat cells fractionated by counterflow centrifugal elutriation. To determine if this variation corresponded to changes in SATB1 or PARP levels, we prepared Jurkat nuclear extracts from six different stages of the cell cycle (G1, G1/S, early S, middle S, late S, and G2/M) (Fig. 10A) fractionated on this occasion by FACS. As before, B1 complex formation was maximal at the G1/S boundary and early S, with a marked reduction in binding activity observed at mid-S (Fig. 10B). Quantitation by a PhosphorImager revealed a fourfold difference between G1/early S and mid-S. In contrast, the B4 complex showed no variation throughout the cycle; B3 showed a very modest elevation in late S and G2/M. We also monitored the level of an irrelevant binding factor, CCAAT binding factor (2); levels of complex formation corresponding to this factor also remained constant across the cell cycle (data not shown). The MCF-7 B1′ complex (Fig. 2A) also demonstrated no cell cycle-related changes in binding activity (data not shown). We attributed the rise of B1 complex at late S and G2/M, which was absent in our earlier studies, to a small contamination of our present Jurkat cells with a hyperdiploid component (Fig. 10A) in which some hyperdiploid G1 cells were included in the G2/M fraction of the diploid majority component.
FIG. 10.
(A) Cell cycle fractionation of Jurkat. Unsynchronized Jurkat cells were stained with Hoechst 33342 and separated by FACS into the six indicated fractions. Fraction 1, G1; fraction 2, G1/early S; fraction 3, early S (Se); fraction 4, mid-S (Sm); fraction 5, late S (Sl); fraction 6, G2/M. (B) Cell cycle-dependent activity of the Jurkat B1 complex. Nuclear extracts were prepared from Jurkat cells sorted as shown in panel A and used in MSAs with 37MBR as the probe. Positions of the four resulting complexes are marked. (C) Western blot analysis of nuclear extracts from different stages of the cell cycle. Extracts used in the MSAs (except G1/S) were subjected to Western blotting with anti-PARP antibody. (D) Western blot analysis of nuclear extracts from different stages of the cell cycle. Extracts used in the MSAs (except G1/S) were subjected to Western blotting with anti-SATB1 antibody.
Western blots of the nuclear extracts from different stages of the cell cycle with anti-PARP (Fig. 10C) and anti-SATB1 (Fig. 10D) antibodies did not reveal any changes in the levels or apparent modifications of these proteins that could explain the difference in binding activity.
DISCUSSION
We have demonstrated that the AT-rich region immediately flanking the BCL2 MBR is a binding site for the MAR protein SATB1. Following purification by multiple steps of column chromatography, including DNA affinity chromatography, this protein was identified unambiguously through MS analysis of tryptic peptides. We obtained additional evidence for this identity by a characteristic inhibition of complex formation on incubation of anti-SATB1 antibody with nuclear extracts during MSAs (Fig. 7A) and by Western blotting of gel-purified complex with the MBR AT-rich region as the target (Fig. 7B). Finally, introduction of a SATB1 expression plasmid into MCF-7, a cell line deficient in SATB1, was successful in inducing the formation of a complex identical to that of Jurkat, a cell line with very high levels of SATB1 expression (Fig. 8). While the SDS-PAGE migration of our SATB1 preparation produced a higher apparent molecular mass than predicted by the DNA sequence (103 versus 86 kDa), independent preparations of the protein from two different laboratories comigrated on our gels (data not shown; see also reference 18). Therefore, posttranslational modification is likely to have occurred.
It is probable that other proteins are required for MBR complex formation, although we have not yet determined whether these proteins are present in the final complex or simply enabling cofactors that do not remain assembled with SATB1. Our observations on the effects of proteins present in the flowthrough of the affinity column on MBR complex formation and SATB1 binding support this expectation. Purified SATB1, incubated alone with MBR target DNA, binds with relatively low affinity (2.1 nM) and produces tight B1 complexes on MSA gels. When flowthrough is added to the SATB1-DNA mixture, two changes occur. First, the SATB1 complex morphology widens considerably (Fig. 4C) to reproduce that obtained with Jurkat nuclear extracts. Second, the affinity increases fivefold (Fig. 6). While the affinity of DNA binding proteins may be artifactually increased following purification by buffer conditions (9), the increased affinity that we observed was not influenced by such factors as cation substitution and protein carrier but was destroyed by heat and proteinase digestion (data not shown). Effects of the flowthrough from B1 complex purification could not be substituted with the flowthrough from B4 complex purification. Therefore, additional proteins are required for high-affinity, morphologically identical binding of SATB1 to the MBR target DNA.
The identities of these additional and/or auxiliary proteins remain unknown. We viewed the copurification of SATB1 and PARP as an intriguing clue, since PARP is also a constituent protein of the nuclear matrix (14). Furthermore, copurification proceeded beyond the DNA affinity column; when the SATB1-PARP mixture was fractionated with an NAD affinity column, which should specifically retain PARP through binding of its NAD cofactor, both proteins were retained and coeluted. Only the poly(rA)-oligo(dT) column, which provided a number of DNA structures serving as substrates for PARP binding but not for SATB1 binding, allowed the separation of these two proteins.
However, a number of experiments have cast doubt on PARP as a cofactor in MBR complex formation. First of all, PARP was not detected in MBR complexes produced under a variety of experimental conditions (Fig. 7C). Second, immunoprecipitation of nuclear extracts with anti-SATB1 antibody did not result in the coprecipitation of PARP (data not shown). Finally, incubation of the two proteins in the presence of either MBR target or nonspecific DNA did not result in poly(ADP) ribosylation of SATB1 under conditions that produced self poly(ADP) ribosylation of PARP (data not shown). Given our observation (see above) that SATB1 was likely purified in a posttranslationally modified state, it is possible that poly(ADP) ribosylation was already complete. Therefore, while further biochemical studies on the nature of SATB1 modification will be required to exclude PARP as one of the auxiliary proteins of the MBR complex, current results are not encouraging in this regard.
Other candidates include the Ku proteins. We have previously demonstrated the presence of these proteins in complexes forming with the 279-bp MBR target (7). While it seems likely that the Ku proteins we purified in the B4 complex resulted from adventitious associations such as those observed by others during the purification of DNA binding proteins, our UV cross-linking studies suggested some shared bands among the four complexes that we produced with MBR target DNA. Thus, Ku proteins could have a precursor function in targeting successive binding factors to the AT-rich MBR site. In this regard, it is known that Ku can be targeted to specific DNA sequences through association with other DNA binding proteins, particularly transcriptional activators (3). Furthermore, there is recent evidence that Ku70/86 as well as PARP individually bind DNA targets from the MARs surrounding the IgH enhancer when these sequences are presented as closed circular DNA templates to eliminate the end-binding activity of Ku proteins. Ku70/86 and PARP make a protein complex in vivo and in vitro in the absence of DNA; as a result, their affinity to MARs is synergistically enhanced (9a). However, the Ku proteins that we purified from B3 and B4 complexes do not increase SATB1 binding to 37MBR (data not shown). Nonetheless, despite some of our current evidence to the contrary, the possibility of interactions among Ku70/86, PARP, and SATB1 requires further investigation.
Recently, the heat shock proteins Hsp70 and Hsp40 were observed to induce an affinity increase of magnitude similar to that for our SATB1 in an origin binding protein, E1, targeted to the human papillomavirus 11 origin of replication (17). The facilitated binding of E1 resulted in a stimulation of viral DNA synthesis (17). This is a provocative connection in several regards. First, MARs likely play a role in DNA replication, perhaps by organizing zones of replication initiation and/or serving as replicon boundaries. Second, BCL2 translocation at the MBR is associated with a change in the replication timing of the translocated BCL2 gene (Y. Sun and T. G. Krontiris, unpublished results). Therefore, it will be important to determine if proteins of the Hsp functional class can serve as auxiliary proteins for SATB1 binding.
The demonstration that SATB1 binding to the MBR occurs in vivo strongly suggests that the complex formation we have observed is functionally significant and that the MBR is a MAR. In fact, those genomic sequences, such as SBS-3, that bind to SATB1 in vivo were found to be associated with the nuclear matrix in vivo in both Jurkat T cells (5) and mouse thymocytes (S. Cai and T. Kohwi-Shigematsu, unpublished results). While further studies will expand these initial observations, the MBR acting as a MAR would have several interesting implications for the siting of BCL2 translocations. At the present time we can offer some additional evidence from DNA sequence comparisons and our other laboratory studies suggesting a SATB1-related functional role for the MBR. First, the AT-rich SATB1 binding site is evolutionarily conserved in the mouse bcl2 gene, although no such conservation exists for the remainder of the 3′ untranslated region of exon 3. Also, our recently completed in vivo footprinting studies within the MBR (F. Ye, K. Foldenauer, and T. G. Krontiris, unpublished results) demonstrate that the SATB1 binding site contains multiple, constitutive, DNase I-hypersensitive sites. In cells expressing SATB1, this region is also hypersensitive to KMnO4, an agent with specificity for single-stranded DNA. Such hypersensitivity is absent in cells lacking SATB1.
It will be interesting to determine whether cell cycle modulation of MBR complex formation, the existence of ancillary factors capable of modifying SATB1 affinity to the MBR, an AT-rich region apparently unwound in SATB1-expressing cells, and altered replication timing of the region following BCL2 translocation are all related through an MBR role in DNA replication as a MAR. In this regard it should be noted that all translocations in the MBR have the effect of removing the AT-rich region and replacing it with the MARs of the IgH enhancer region 1 to 2 kb downstream of each translocation breakpoint. If it also eventually transpires that MARs display intrinsic fragility, then the highly specific localization of BCL2 translocations may indeed result from both a propensity for DNA breakage and a subsequent, selectable change in functional properties.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health (CA51985) and funds from the Beckman Research Institute of the City of Hope to T. G. Krontiris and grants from the National Institutes of Health (CA 39681) and the Department of Energy (DE-AC03-76SF00098) to T. Kohwi-Shigematsu.
REFERENCES
- 1.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi W, Wu L, Coustry F, de Crombrugghe B, Maity S N. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- 3.Chung U, Igarashi T, Nishishita T, Iwanari H, Iwamatsu A, Suwa A, Mimori T, Hata K, Ebisu S, Ogata E, Fujita T, Okazaki T. The interaction between Ku antigen and REF1 protein mediates negative gene regulation by extracellular calcium. J Biol Chem. 1996;271:8593–8598. doi: 10.1074/jbc.271.15.8593. [DOI] [PubMed] [Google Scholar]
- 4.Cleary M L, Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci USA. 1985;82:7439–7443. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 6a.Dickinson L A, Kohwi-Shigematsu T. Nucleolin is a MAR/SAR DNA-binding protein specifically recognizing a region with high base-unpairing potential. Mol Cell Biol. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiCroce P A, Krontiris T G. The BCL2 major breakpoint region is a sequence- and cell-cycle-specific binding site of the Ku antigen. Proc Natl Acad Sci USA. 1995;92:10137–10141. doi: 10.1073/pnas.92.22.10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fried M G. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis. 1989;10:366–376. doi: 10.1002/elps.1150100515. [DOI] [PubMed] [Google Scholar]
- 9a.Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku antigen form a complex and synergistically bind to matrix attachment regions. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 10.Hellman U, Wernstedt C, Gonez J, Heldin C-H. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Biochemistry. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 11.Hockenberry D, Nunez G, Milliman C, Schreiber R D, Korsmeyer S J. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348:334–336. doi: 10.1038/348334a0. [DOI] [PubMed] [Google Scholar]
- 12.Hockenberry D M, Zutter M, Hickley W, Nahm M, Korsmeyer S A. BCL2 protein is topographically restricted in tissues characterized by apoptotic cell death. Proc Natl Acad Sci USA. 1991;88:6961–6965. doi: 10.1073/pnas.88.16.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadonaga J T, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufmann S H, Brunet G, Talbot B, Lamarr D, Dumas C, Shaper J H, Poirier G. Association of poly(ADP-ribose) polymerase with the nuclear matrix: the role of intermolecular disulfide bond formation, RNA retention, and cell type. Exp Cell Res. 1991;192:524–535. doi: 10.1016/0014-4827(91)90072-3. [DOI] [PubMed] [Google Scholar]
- 15.Kofler B, Wallraff E, Herzog H, Schneider R, Auer B, Schweiger M. Purification and characterization of NAD+:ADP-ribosyltransferase (polymerizing) from Dictyostelium discoideum. Biochem J. 1993;293:275–281. doi: 10.1042/bj2930275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Kohwi-Shigematsu T, deBelle I, Dickinson L A, Galande S, Kohwi Y. Identification of base-unpairing region-binding proteins and characterization of their in vivo binding sequences. Method Cell Biol. 1998;53:324–352. doi: 10.1016/s0091-679x(08)60885-7. [DOI] [PubMed] [Google Scholar]
- 16.Krowczynska A M, Rudders R A, Krontiris T G. The human minisatellite consensus at breakpoints of oncogene translocations. Nucleic Acids Res. 1990;18:1121–1127. doi: 10.1093/nar/18.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J-S, Kuo S-R, Makhov A M, Cyr D M, Griffith J D, Broker T R, Chow L T. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 18.McDonnell T J, Deane N, Platt F M, Nunez G, Jaeger U, McKearn J P, Korsmeyer S J. Bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 19.McGuire E A, Hockett R D, Pollock K M, Bartholdi M F, O'Brien S J, Korsmeyer S A. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989;9:2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagomi K, Kohwi Y, Dickinson L A, Kohwi-Shigematsu T. A novel DNA-binding motif in the nuclear matrix attachment DNA-binding protein SATB1. Mol Cell Biol. 1994;14:1852–1860. doi: 10.1128/mcb.14.3.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrovic A S, Young R L, Hilgarth B, Ambros P, Korsmeyer S J, Jaeger U. The Ig heavy chain 3′ end confers a posttranscriptional processing advantage to Bcl-2-IgH fusion RNA in t(14;18) lymphoma. Blood. 1998;91:3952–3961. [PubMed] [Google Scholar]
- 22.Seto M, Jaeger U, Hockett R D, Graniger W, Bennett S, Goldman P, Korsmeyer S J. Alternative promoters and exons, somatic mutation and deregulation of the bcl-2-lg fusion gene in lymphoma. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith G R. Chi hotspots of generalized recombination. Cell. 1983;34:709–710. doi: 10.1016/0092-8674(83)90525-1. [DOI] [PubMed] [Google Scholar]
- 24.Stahl D, Swiderek K M, Davis M T, Lee T D. Data-controlled automation of liquid chromatography/tandem mass spectrometry analysis of peptide mixtures. J Am Soc Mass Spectrom. 1996;7:532–540. doi: 10.1016/1044-0305(96)00057-8. [DOI] [PubMed] [Google Scholar]
- 25.Tsujimoto U, Cossman J, Jaffe E, Croce C M. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 26.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce C M. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229:1390–1393. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- 27.Wyatt R T, Rudders R A, Delellis R, Zelenetz A D, Krontiris T G. BCL2 oncogene translocation is mediated by a chi-like consensus. J Exp Med. 1992;175:1575–1588. doi: 10.1084/jem.175.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]