Abstract
The outbreak of COVID-19 pandemic and its consequences have inflicted a substantial damage on the world. In this study, it was attempted to review the recent coronaviruses appeared among the human being and their epidemic/pandemic spread throughout the world. Currently, there is an inevitable need for the establishment of a quick and easily available biosensor for tracing COVID-19 in all countries. It has been known that the incubation time of COVID-19 lasts about 14 days and 25% of the infected individuals are asymptomatic. To improve the ability to determine SARS-CoV-2 precisely and reduce the risk of eliciting false-negative results produced by mutating nature of coronaviruses, many researchers have established a real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assay using mismatch-tolerant molecular beacons as multiplex real-time RT-PCR to distinguish between pathogenic and non-pathogenic strains of coronaviruses. The possible mechanisms and pathways for the detection of coronaviruses by biosensors have been reviewed in this study.
Keywords: Coronavirus, COVID-19, Biosensor, Pandemic outbreak, SARS-CoV-1, SARS-CoV-2
Introduction
The outbreak of COVID-19 coronavirus pandemic has led to many challenges and unprecedented crises around the globe. The current coronavirus is the third highly pathogenic version of severe acute respiratory syndrome (SARS-CoV-1) coronavirus previously spread and recognized in China in 2002 [1]. SARS-CoV-1 rapidly spread across 29 countries and infected 8098 people, resulting in 774 deaths by the end of the epidemic in July 2003 [2–4]. On 4 April 2012, the second outbreak of acute respiratory disease was reported in Saudi Arabia. This novel coronavirus, called CoV 2c EMC/2012, was the first human coronavirus in lineage C [5, 6]. In September 2012, an international consensus changed its name to Middle East Respiratory Syndrome (MERS) [2, 6]. Most of the people infected with MERS-CoV and SARS-CoV-1 had similar symptoms such as high fever and acute lower respiratory infection symptoms [5–7]. Since September 2012, the World Health Organization (WHO) has reported 2494 laboratory-confirmed MERS-CoV cases leading to 858 deaths in 27 countries [7–9]. According to the WHO, the MERS-CoV infection still remains in some countries. For instance, in Saudi Arabia, around 19 new cases including 8 associated deaths were reported during December 2019 [10].
Coronaviruses belong to the large family of Coronaviridae [11]. Based on their genotype and serological characteristics, they were initially classified into three groups. The International Committee for Taxonomy of Viruses (ICTV) later changed this classification and categorized the coronaviruses into four genera named Alpha-coronavirus, Beta-coronavirus, Gama-coronavirus and Delta-coronavirus. All the four genera are found in mammals and can cause infection in humans [3, 6, 12]. The phylogenetic relationships among these coronaviruses reveal that Beta-coronaviruses are most important ones due to their animal–human and human–human transmission capabilities. As an evidence, three photogenic coronaviruses, namely SARS-CoV-1, MERS-CoV and SARS-CoV-2, are denoted as Beta-coronavirus [5, 13, 14]. Beta-coronaviruses are divided into four lineage subgroups (A, B, C and D). HCoV-HKV1 and HCoV-OC43 belong to lineage A, and lineage B includes SARS-CoV-1 and SARS-CoV-2. MERS-CoV is the first human coronavirus belonging to lineage C. Lineage D does not contain human transmittable coronaviruses [15, 16]. All the coronaviruses in B lineage are associated with severe pneumonia which is the same symptom in SARS-CoV-1 and SARS-CoV-2 (Figure 1).
Fig. 1.
Schematic diagram for the zoonotic origins and intermediate hosts of the most pathogenic coronaviruses: SARS-CoV-1, SARS-CoV-2 and MERS-CoV
SARS-CoV-2, probably originated from bat and/or pangolin, was spread in Wuhan, China early in 2020 [17, 18]. The genome of COVID-19 has already been sequenced and many outstanding research groups are now working hard to come up with the best treatment to abolish the coronavirus [18]. The immediate detection and management of COVID-19 depend on specific drugs or vaccines. However, the new coronaviruses have the potency to undergo a consistent mutation and recombination, leading to new serotypes and events. Hence, vaccine development cannot be considered as an ultimate solution. Although the molecular methods proposed for diagnosis of coronaviruses are standard and highly reliable and have high sensitivity and selectivity, they sometimes appear to be impractical as molecular tests require well equipped-laboratories which may not be available everywhere. Furthermore, the equipment required for PCR tests is expensive and the viral nucleic acids should be recognized in a limited period following the infection. Considering the time factor, the RT-PCR tests at optimal conditions take at least several hours and require an additional time for viral sample RNA preparation. In these tests, the viral RNA preparation steps are not flawless and may deal with some errors leading to incorrect negative or positive results [19, 20].
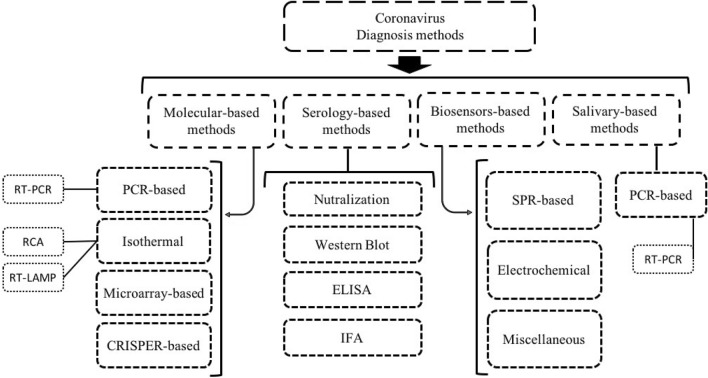
Knowing that the vaccine is not the only solution to overcome the current crisis, diagnosis of the infected individuals is of high importance in harnessing the coronavirus pandemic outbreak since a significant number of these individuals appear to be asymptomatic (confirmed by Center for Disease Control and Prevention (CDC), Atlanta, Georgia, USA). Ignoring the incubation time (up to 14 days), which has a pivotal role in prevalence of a pandemic, the appearance of asymptomatic patients has made the situations more complicated. There are several similarities in the genomes, proteins, and transmission pathways of coronaviruses. The aim of this study was to review, compare and evaluate the different methods proposed for detecting COVID-19. For this purpose, three highly pathogenic coronavirus strains, specifically COVID-19, were overviewed to compile comprehensive data about the detection of coronaviruses and their developed biosensors (Figure 2).
Fig. 2.
Overview of serological, molecular and biosensors methods for diagnosis of COVID-19
Molecular Methods for Coronavirus Diagnosis
PCR-Based Methods
Real-Time Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Real-time RT-PCR is currently the most favored method for discovery of any type of coronavirus owing to its dominant application in quantitative assessments [6, 10]. The PCR assessment of SARS-CoV-2 should thoroughly cover positive control, negative control, and internal control processes (Fig. 3).
Fig. 3.
Schematic illustration of the RT-PCR assay. Reprinted by permission from Ref. [21]
Various commercial RT-PCR kits are produced and employed for identification of SARS-CoV-2 in bio-fluid samples. Some of these kits are RT-PCR LAB-KIT™ Biomaxima, RT-PCR Kit for COVID-19 Coronavirus Biotec Biomedical, Std M nCoV Real-Time Kit SD Biosensor, Roche Cobas SARS-CoV-2 Test, Real-Time Multiplex RT-PCRLifeRiver, PowerChek™ Real-Time PCR Kogene, Novel Coronavirus Real-Time PCR Kit Getein Biotech, Perfect Lyo SARS-CoV-2 Real-Time PCR kit Jiangsu Bioperfectus Tech., RealStar 2019-nCoV Real-Time PCR Kit Altona Diagnostics, Real-Time Multiplex RT-PCR LifeRiver, Real-Time Multiplex PCR Daan Diagnostics, and SARS-CoV-2 Real-Time PCR LAB-KIT ™ Biomaximakit. Improvement of the real-time RT-PCR assay in terms of sensitivity, specificity and time of the test, has received much attention. TaqMan-based real-time RT-PCR is also used in the ordinary process of coronaviruses diagnosis [14]. To enhance the test sensitivity, dual TaqMan probes and/or more probes are used for coronavirus detection [7]. Corman et al. [10] used TaqMan-based real-time RT-PCR for SARS-CoV-2 detection targeting the RdRp gene, E gene and N gene. They reported the best result for sensitivity in E gene and RdRp gene assays (3.8 and 5.2 copies per reaction at 95% detection probability, respectively), and showed that the N gene was slightly less sensitive. In terms of specificity, no false-positive result was detected, and Q-PCR time was less than 40 min [5]. The commercial TaqMan probes, including 2019-nCoV TaqMan RT-PCR Kit (Catalog# TM67100) Norgen Biotech, TaqMan™ SARS-CoV-2 Assay Kit v2 (EUA-FDA) Thermo Fisher, and TaqPath™ COVID-19 Combo Kit Thermo Fisher, were applied for SARS-CoV-2 identification [6].
To achieve more reliable analytical results during the detection of SARS-CoV-2, the real-time RT-PCR assay was designed in a way to utilize the armored RNA as an external positive control (EPC). Application of the proper and stable controls in real-time RT-PCR reaction contributes to the reduction of false results [8]. Asuragen Company developed an unarmored RNA Quant® SARS-CoV-2 Control which could be used as an EPC in all the real-time RT-PCR-based tests for SARS-CoV-2 detection. Additionally, some researchers have established a real-time RT-PCR assay using the mismatch-tolerant molecular beacons as multiplex real-time RT-PCR in order to distinguish between pathogenic and nonpathogenic strains of coronaviruses, improve their ability to determine the coronaviruses precisely and mitigate the likelihood of obtaining the false-negative results due to the mutating nature of coronaviruses (Table 1) [9, 10].
Table 1.
Comparison of the properties of molecular assays, RT-PCR and serological assays (ELISA) used for the diagnosis of SARS-CoV-1
| Assay | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Refs |
|---|---|---|---|---|---|
| ELISA | 98.2% | 98.7 | 98.7 | 98.4 | [46] |
| RT-PCR | 52.2 | 78.7 | 74.5 | 58.1 | |
| ELISA | 33.3 | 80.4 | 33.3 | 80.4 | [70] |
| RT-PCR | 28.2 | 79.6 | 36.7 | 72.6 |
PPV positive predictive value, NPV negative predictive value
Chantal et al. [22] evaluated the primer–probe sets employed in 4 common diagnostic assays introduced by the WHO and developed by the Chinese CDC for N and nsp10 genes, the CDC of Atlanta, Georgia, USA for N gene, Charité for RdRp and E genes, and Hong Kong University (HKU) for N and nsp14 genes. They showed that the whole primer–probe sets, except for the set developed by Charité–Universitäts Medizin Berlin, could be employed to recognize SARS-CoV-2 at 500 coronavirus copies per reaction.
Isothermal Nucleic Acid Amplification-Based Methods
Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP)
Many laboratories have examined the RT-LAMP tests for the detection of coronavirus. RT-LAMP shows considerable specificity and sensitivity due to its exponential amplification characteristic and identifies six distinct target sequences by four different primers simultaneously. In RT-LAMP, the reaction occurs in less than 1 h at around 60–65 min. This technique can be easily performed and visualized for detection, does not require a thermocycler and has less background signal. However, the problems associated with optimization of primers and reaction conditions are considered as the disadvantages of RT-LAMP [23, 24]. In this method, amplification can be detected using fluorescence dye or magnesium pyrophosphate. In this way, pyro-phosphate or fluorescence can be monitored to tackle the shortcoming of the endpoint detection [23]. Pei Huang et al. [25] developed a nucleic acid visualization method by combining the RT-LAMP technique with the vertical flow visualization strip (RT-LAMP-VF) technique to diagnose MERS-CoV (Fig. 4). Shirato et al. [13] employed a quenching probe (QProbe) to screen the signals and improved the application of RT-LAMP for the detection of human MERS-CoV using fluorescent visualization. Cai et al. [11] showed that QProbe could prevent the probability of collecting the unpredicted signals from the primer-dimer or non-primer reactions during the diagnosis of MERS-CoV. Furthermore, this technique was successfully applied to identify HCoV-NL63 using the agarose gel electrophoresis with appropriate specificity and sensitivity [26]. RT-LAMP is temperature sensitive and shows the highest performance at a temperature range of 60–65 °C which usually confines its application. Cai et al. [11] established a modified type of LAMP that relied on phosphorothioated primer (PS-LAMP) to form more efficient hairpin and expansion at the termini of growing concatemers. This modification enabled the technique to operate at a much lower temperature of around 40 °C.
Fig. 4.

Schema of the RT-LAMP-VF technique. The RT-LAMP products are identified by a vertical flow visualization strip; the amplicons are labeled with biotin and FITC; after amplification, amplicons are labeled with biotin which can attach to gold nanoparticles linked with streptavidin to make a complex. The complexes, labeled with FITC, are captured by an anti-FITC antibody that is fixed on the text line of the strip, and the results are visualized as a colored band. Reprinted by permission from Ref. [25]
To date, several RT-LAMP assay-based protocols, with high specificity and sensitivity, have been developed for the identification of SARS-CoV-2 [12, 40]. Yan et al. [27] optimized the RT-LAMP assay for SARS-CoV-2 identification by the primers for orf1ab and S genes. They reported a specificity of 100% (95% CI 93.7%–100%) and sensitivity of 100% (95% CI 92.3%–100%). Using this assay, SARS-CoV-2 could be detected in the mean (± SD) time of 26.28 ± 4.48 min. The COVID-19 detection kits, which rely on commercially isothermal amplification techniques, have been produced by LAMP-COVID-19 SunStar Joint Stock Company and Colorimetric and Isothermal Detection COVID-19 (CE-IVD) Biotec Biomedical.
Rolling Circle Amplification (RCA)
Under isothermal conditions, the RCA DNA polymerase is applied to recurrently replicate the sequence and extend the circular primer. This technique can lead to signal amplification of 109 folds of each circle in 90 min. RCA proved to be able to set up an acceptable assay for sensitive detection of SARS-CoV-1 in both solid and liquid phases [24]. Advantageously, RCA can be carried out with a limited number of reagents under isothermal conditions. Moreover, it excludes the false-positive results which repeatedly occur in the PCR-based assays. Wang et al. [28] reported a detection sensitivity of close to the single template level detecting SARS-CoV-1 using RCA. To our knowledge, the commercial kits which rely on this technique for coronavirus detection are not available yet.
Microarray-Based Methods
Microarray is a high–throughput technique for simultaneous assessment of a large amount of DNA fragments on one chip (Table 2). Considering the high genetic diversity of coronaviruses, this assay can be simply used to detect and identify the strain. Long et al. [24] developed a global microarray system using a combination of a ligase detection reaction (LDR) and RT-PCR to assess six nucleotide positions (nt27827, nt22222, nt21721, nt19838, nt9479, and nt9404) for coronavirus detection. This tool can detect the coronaviruses including SARS-CoV-2 with high mutation rates. Chen et al. [29] established a hybrid microarray-based technique for diagnosis of a target coronavirus from 8 coronavirus strains. They demonstrated that the studied gene chip could detect the 8 coronavirus strains specifically. They also reported that 10–2 and 10–5 diluted cDNA could be detected by multi-PCR and the chip, respectively, demonstrating that the chip could be 1000× more sensitive than PCR.
Table 2.
The list of available molecular methods being used for coronavirus detection
| PCR-based methods |
Real-time reverse transcriptase-polymerase chain reaction (RT-PCR), TaqMan for different types of coronaviruses E, S, RdRp, N, nsp10 genes (SARS-CoV-2) QPCR time is up to 1 h, needs RNA extraction and cDNA synthesis before QPCR, which needs different times according to the in-use kits |
| Isothermal nucleic acid amplification-based methods |
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) Time of test is 30–40 min, needs RNA extraction and cDNA synthesis before QPCR, which have different time uses according to the in-use kits Rolling circle amplification (RCA) RCA needs 90 min, needs RNA extraction and cDNA synthesis before QPCR, which have different time uses according to the in-use kits |
| Microarray based methods |
Detect accurately single nucleotide polymorphism (SNP) mutations Spike (S) gene sequence of SARS-CoV-1, needs RNA extraction and cDNA synthesis before QPCR, which have different time uses according to the in-use kits. Subsequently needs amplification of the cDNA sample which takes about 1 h Mobile Analysis Platform (MAP) MERS detection Microarray using ligase detection reaction (LDR) and RT-PCR Detect single nucleotide polymorphism (SNP) mutations SARS-CoV-1 |
| Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-based methods |
SHERLOCK, CASLFA, CRISPR/Cas12-DETECTOR N and E genes (SARS-CoV-2) Time of test is < 40 min, needs RNA extraction and cDNA synthesis and LAMP before test |
Guo et al. [30] introduced a microarray to identify 24 single nucleotide polymorphism (SNP) mutations through the S gene sequence of SARS-CoV-1. The hybridization assessment showed that all the 19 samples enrolled were identified and genotyped with an accuracy of 100%. Luna et al. [31] programmed a non-fluorescent, cheap, low-density oligonucleotide array to identify the coronavirus genus with high sensitivity. Hardick et al. [32] evaluated the Mobile Analysis Platform (MAP) which was a microarray-chip-based diagnostic platform. They showed that MAP was an accurate, rapid, deployable technology for the identification of coronavirus including MERS.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas-Based Methods
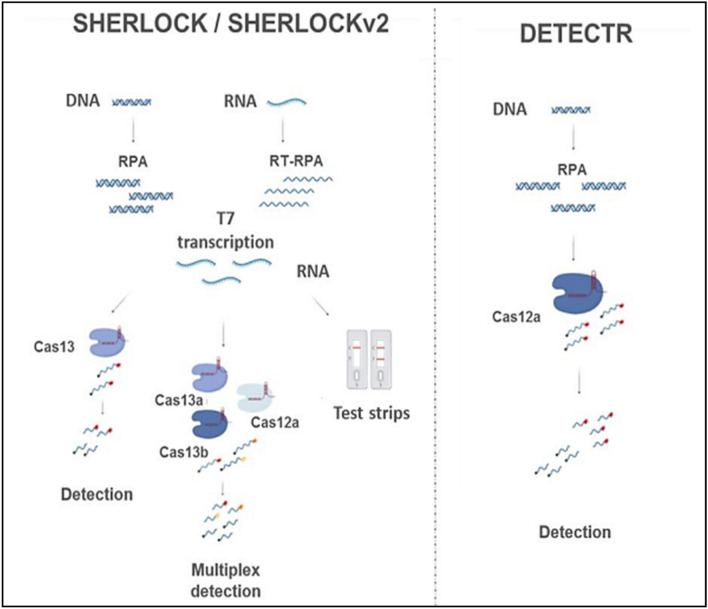
CRISPR/Cas systems have already been utilized for fast and mobile monitoring of nucleic acids, contributing to the detection of pathogens (Table 2). CRISPR/Cas9 mediated lateral flow nucleic assay (CASLFA) along with sgRNA anchoring-based hybridization (biotin-streptavidin) assay was applied to detect the genetically modified organisms (GMOs), Listeria monocytogenes and African swine fever virus (ASFV) [33]. Max J. Kellner et al. [34] combined isothermal pre-amplification and Cas13 and introduced a platform called SHERLOCK (specific high-sensitivity enzymatic reporter unlocking). They used this technique to specifically and sensitively detect the single molecules of DNA or RNA. This platform allowed for multiple, mobile, ultra-sensitive identification of DNA or RNA by fluorescence and was used to identify dengue and Zika viruses. Broughton et al. [35] attempted to develop a proper sub-40 min CRISPR/Cas12 based on lateral flow assay for the identification of SARS-CoV-2. The CRISPR-based DETECTR assay allowed for simple, visual, fast detection of SARS-CoV-2 with 100% and 95% negative predictive agreement and positive predictive agreement, respectively, in < 40 min. The DETECTR assay was carried out by RT–LAMP for viral pre-amplification or control of RNA target and by LbCas12a for trans-cleavage assay. Cas12 gRNAs are designed to specifically target the E gene and N gene regions of SARS-CoV-2 or broadly identify the related coronavirus strains. In the detection process, the undamaged FAM-biotinylated reporter molecule moves into the control capture line. After detection of the fitted target, the Cas–gRNA complex cleaves the reporter molecule that enters the target capture line [35, 36]. To our knowledge, the commercial kits using CRISPR/Cas systems to detect coronavirus are not available yet (Fig. 5).
Fig. 5.
Schematic diagram of CRISPR-Cas diagnostic platforms, SHERLOCK/v2 and DETECTR. Reprinted by permission from Ref. [37]
Serological Diagnostic Methods
Developing a sensitive, rapid and accurate method that directly determines the coronaviruses or their biological and molecular entities without sample preparation and amplification is a global concern. Hence, application of some methods such as serological assays with rapid, economic, sensitive and specific features in diagnosis of the infection caused by coronaviruses can be a good solution. The importance of serological tests in epidemiological investigations of coronaviruses has been highlighted [19, 20]. Rapid serological diagnostic tests can be vital in controlling the epidemic outbreak of coronavirus [20, 38]. Some of the most important serological assays for identification of SARS-CoV-1, SARS-CoV-2 and MERS-CoV are as follows:
Neutralization method This method determines the coronaviruses when the neutralizing antibodies in the serum of the patient inhibit the viral growth in the culture media.
Western Blot (WB) analysis In this method, the proteins existing in the serum of the patient are separated by gel electrophoresis and compared to the reference proteins of the coronavirus.
Enzyme-Linked Immunosorbent Assays (ELISA) It is a common and powerful method for the detection of proteins or protein–protein interactions by antibodies (Table 1).
Immunofluorescence Assays (IFA) It is a traditional laboratory technique used to detect the presence of antibodies by their abilities to react with specific antigens [39, 41].
Recombinant protein-based WB and ELISA methods are known to be highly sensitive for viral detection [42, 43]. Although IFA and ELISA exhibit high sensitivity (85–100%), they have very low specificity and selectivity [44, 45]. Figures 6 and 7 schematically illustrate the protein detection methods and neutralizing tests.
Fig. 6.
Overview of the principles, types and applications of protein detection
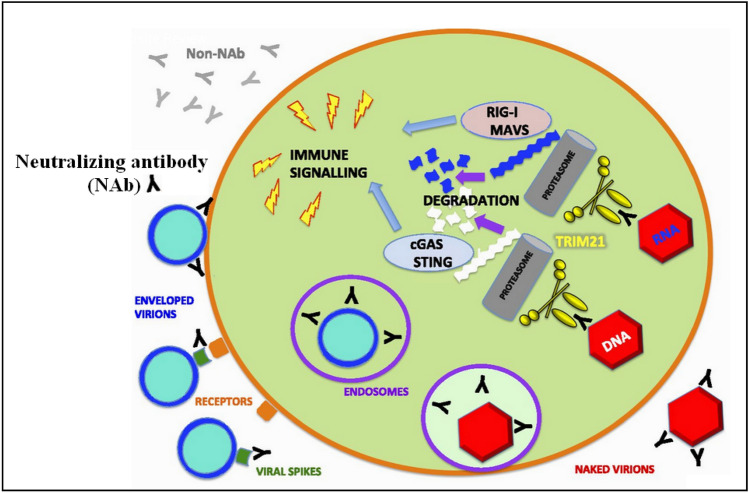
Fig. 7.
Neutralizing antibodies (black Y shapes) commonly bind to the surface proteins of the virus (dark green) and inhibit the interaction with receptors (orange) or subsequent events in the fusion process. Reprinted by permission from Ref. [46]
Since the spike protein of coronavirus for linking with and penetrating into the host cell is crucial, considerable efforts have been made to identify and detect the therapeutic antibodies that can target the spike protein [47, 48]. Therefore, it is of utmost urgency to study the cross-reactivity of the anti-SARS-CoV antibodies with the spike protein of SARS-CoV-2 in order to determine the therapeutic antibodies against SARS-CoV-2 [49]. The spike protein is capable of assembling a trimer structure on the viral surface and contains two different functional subunits: (1) S1 which connects the coronavirus to the cell surface, and (2) S2 that contributes to the internalization of the coronavirus in the host cell [50–52]. A part of S1 subunit is divided into receptor-binding domain (RBD) and N-terminal domain (NTD) [53]. RBD on the viral spike protein comprises a receptor-binding subdomain and a core. The receptor-binding subdomain reciprocally acts with the host cell receptor and establishes the connection between the coronavirus and the host cell [52]. MERS-CoV, SARS-CoV-1 and SARS-CoV-2 RBDs carry a highly similar structure in the core subdomains but significantly differ in the receptor-binding subdomains [49, 50]. In MERS-CoV, RBD interacts with the extracellular domain of human dipeptidyl peptidase 4 (DPP4). Therefore, RBD plays a key role in producing the MERS-CoV neutralizing antibodies and can be a good candidate for application in vaccine production. However, SARS-CoV-1 and SARS-CoV-2 RBDs reciprocally act with the host receptor, angiotensin-converting enzyme 2 (ACE2), to attach and enter the cells [38, 53–56]. First SARS-CoV-1 and SARS-CoV-2 use an identical receptor on the host cell surface to enter the cell and then the probable blocking factors or tested strategies for prevention of SARS-CoV-1 entrance inhibit SARS-CoV-2 [38]. Investigation of SARS-CoV-2 conformation with a series of neutralizing antibodies also indicated the probable interactions among SARS-CoV-2 RBD and specific antibodies of SARS-CoV-1 [49]. The similarity of RBD structures in SARS-CoV-1 and SARS-CoV-2 is about 73% [57]. Wang et al. [58] discovered a human monoclonal antibody which could neutralize both SARS-CoV-1 and SARS-CoV-2 in the cell culture. This antibody targeted an epitope that was in common between the two SARS-CoVs and thereby inhibited the coronaviruses in the cell culture [58].
Structural studies and modeling show that a conserved residue, called R395 in SARS-CoV-1 RBD and R408 in SARS-CoV-2 RBD, interacts with m396 neutralizing antibody. R395 and R408, in their structures, are able to form a salt bridge with D95 in m396 neutralizing antibody (m396-VL). The molecular analyses revealed that specific monoclonal antibodies of SARS-CoV-1 can be influential in neutralizing SARS-CoV-2 [57, 59]. Tian et al. [49] demonstrated that unlike some of the important SARS-CoV-specific neutralizing antibodies (e.g. m396, CR3014), a SARS-CoV-specific human monoclonal antibody (CR3022), despite the lack of overlap in the epitope with the ACE2 specific binding site in SARS-CoV-2 RBD, could link with SARS-CoV-2 RBD. Okba et al. [60] tested and validated various antigens (RBD, N, S1) of MERS-CoV using different ELISA methods. They found that, in identifying the SARS-CoV-2 antibodies, S1 was more specific than N and N was more sensitive than S1. The specific and sensitive serologic S1-based assay can be used to estimate the prevalence of MERS-CoV. The results of many studies confirm that S1 is a highly important antigen in MERS serology based on ELISA assays [43–45, 47–57, 59]. Amanat et al. [61] attempted to use the recombinant antigens extracted from the SARS-CoV-2 spike protein and develop an ELISA method according to their reactivity to the immunogenic spike protein. They finally generated two different types of spike protein: (1) a full-length spike protein, and (2) a smaller RBD fragment. A recent study showed that immunoassays could be performed only by fragments of the spike protein and expressing its full-length was difficult. However, N protein was easily expressed in a variety of expression vectors as it had a smaller size and no glycosylation sites [43]. Leung et al. [42] used a recombinant antigen of the N-terminal half of SARS-CoV-1 nucleocapsid protein in an IgG ELISA, and finally obtained 89% sensitivity and 94%–95% specificity. In another study, six fragments and full length of the nucleocapsid gene of SARS-CoV-1 were cloned and expressed to perform a rapid diagnosis and evaluate the response of human immune system to SARS-CoV-1 infection. Among them, a195 amino acid fragment from the C-terminal of a nucleocapsid protein (N195) could efficiently identify the antibodies against SARS-CoV-1. Using Western blotting assay, the specificity and sensitivity of this test were found to be 98.3 and 90.9%, respectively. This results demonstrated that N195 protein could be used to specifically identify the SARS-CoV-1 antibodies in humans [20]. Song et al. [62] developed a detection assay according to the identification of the hydrophilic regions of the MERS-CoV nucleocapsid gene. They selected five peptides, and showed that the synthesized, cloned hybridomas reacted to different epitopes and were not identified by the immunoblotting assay. Among the five monoclonal anti-MERS-CoV, P1 (amino acids 22 to 40) and P3 (amino acids 164 to 202) produced the best sensitivity and specificity results with the relative specificity and sensitivity of the assay being 100% and 93.90%, respectively. Figure 8 represents the visual immunochromatographic assay for MERS-CoV detection.
Fig. 8.
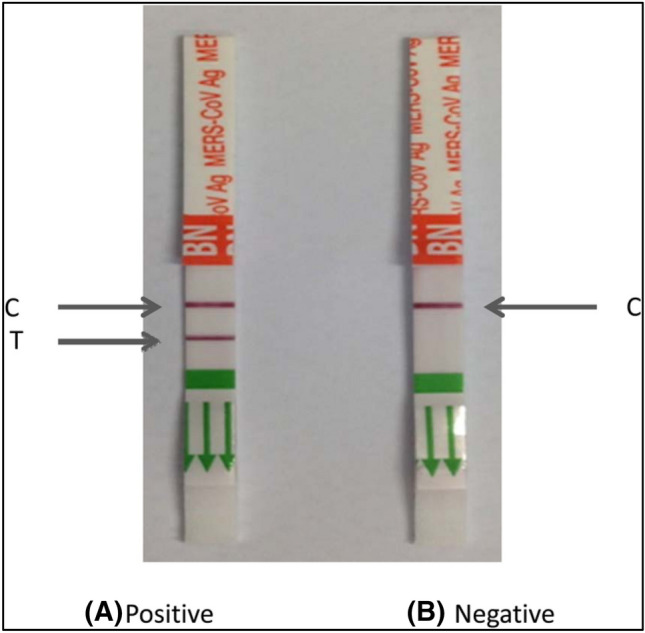
A visual immunochromatographic assay for detection of MERS-CoV. C and T represent control line and test line, respectively. Reprinted by permission from Ref. [62]
The NP-based studies indicated that the full-length nucleocapsid protein of MERS-CoV in a recombinant NP ELISA assay showed a high sensitivity with detection limits of 1 ng/ml and 100% specificity [63]. The antibody titers in the patients’ sera analyzed based on recombinant antigens were characterized as recombinant MERS-CoV spike (S or fragments of S) ELISA, inactivated MERS-CoV ELISA and nucleocapsid protein ELISA. A significant correlation was observed among RBD, S1, inactivated MERS-CoV ELISA and NP ELISAs. S and RBD ELISAs were highly correlated with the commercial S1 ELISA [64]. The reported results indicated the high correlation between molecular and serologic methods for evaluating coronaviruses (Table 1) [41, 65]. Taking advantage of the new technologies, various methods have been developed and examined in research laboratories based on immunological diagnostics and biological sensing.
Biosensors
Surface Plasmon Resonance (SPR) Based Biosensors
Qiu et al. [66] examined a dual-functional plasmonic biosensor for sensing SARS-CoV-2. They used a creative combination of surface plasmon resonance (SPR) and plasmonic photothermal (PPT) effect to detect the specific viral sequences in interaction with complementary DNA receptors immobilized on two-dimensional (2D) gold nano-islands (AuNIs) [66]. This fascinating biosensor exhibited a significant sensitivity to the SARS-CoV-2 sequences with a remarkable detection limit below the concentration of 0.22 pM. In addition, the reported biosensor was more selective and allowed for exact identification of the specific viral targets in a multigene complex sample. In another study, Shi et al. [67] developed a SPR biosensor for concurrent multiplex viral detection of SARS-CoV-1. This biosensor was fabricated by immobilizing nine respiratory-specific viral oligonucleotides in an SPR chip. The obtained results confirmed that the SPR biosensor could simultaneously detect the common respiratory coronaviruses. Yang et al. [68] developed another SPR method based on hybridizing of RNAs on the surface of a streptavidin-coated (SA) sensor chip to study RNA–protein interactions for SARS-CoV-1 viral analysis purposes. They revealed that the N protein of SARS-CoV-1 had a high binding affinity with the main sequence of the SARS-CoV-1 genome. Since this method shows the affinity of interactions with high sensitivity, it can be used to monitor the binding options for a given RNA target motif with one chip. Huang et al. [69] introduced a coupled method using a SPR-fluorescence fiber-optic biosensor for identification of SARS-CoV-1 coronavirus nucleocapsid protein (N protein) in human serum. The combination of sandwich immunoassay approach with local surface plasmon method provides a fascinating tool for determination of viral N-protein analysis at very low concentration of 0.1 pg/ml [69] (Fig. 9).
Fig. 9.
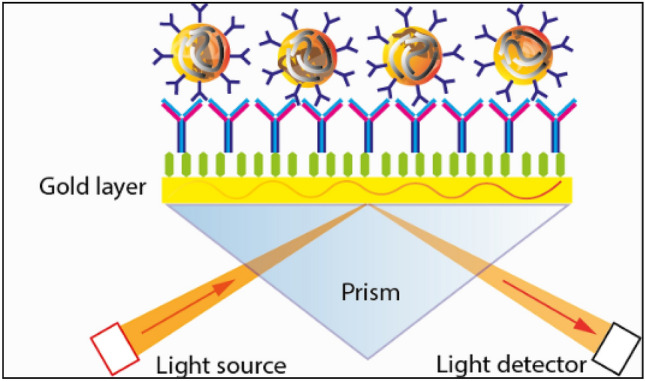
Schematic diagram of SPR Kretschmann geometry. https://dqmp.unige.ch/the-lta-contributes-to-the-research-on-the-covid-19/
Electrochemical Biosensors
Abad-Valle et al. [70] designed an electrochemical enzymatic biosensor base on square wave voltammetry to determine the SARS-CoV-1 sequence. They employed 100 nm sputtered gold film labeled with a complementary strand of 30-mer sequence viral sequence. Through blocking the film surface and subsequent hybridization with the biotin-conjugated SARS-CoV-1 strand, the interaction with alkaline phosphatase-labeled streptavidin took place and facilitated the indirect electrochemical viral detection. In another study, Chang et al. [71] fabricated a flexural plate wave (FPW) phase shift sensor by employing microelectromechanical systems (MEMS) to develop a portable and non-portable SARS-CoV-1 biosensing detector. The phase delay occurrence in combining viral spike protein to the human cell receptors of angiotensin-converting enzyme-2 (hACE2) was the conceptual mechanism of viral measurement in the presented FPW sensor. Zari et al. [72] employed an electrochemical biosensor method for label-free detection of specific DNA sequences with the potential to be applied as a coronavirus detector. They utilized Au electrode, instead of common carbon and glassy carbon electrodes, in square wave voltammetry procedure for oxidation of specific DNA fragments. The proposed method was capable of measuring adenine and guanine contents in hydrolyzed DNA. The authors claimed that their electrochemical biosensor could specifically identify the sequences from single strand avian coronavirus (a coronavirus found in birds) followed by hybridization and hydrolysis reactions on the biosensor surface. Ishikawa et al. [73] targeted the same viral biomarker of N-protein using indium oxide (In2O3) nanowire biosensor. They utilized antibody mimic proteins (AMPs) to interact with target biomarkers with high affinity and specificity. Implementing this label-free electrical biosensor, the SARS-CoV N protein was determined in bovine serum albumin medium at subnanomolar (< 1 nM) concentrations. In another systematic study, Ishikawa [74], developed carbon nanotube (CNT) FET biosensors to investigate the effect of density on detection performance [74]. The reported CNT FET biosensor was employed in identification of N protein as a SARS-CoV-1 biomarker. The results indicated that the mentioned biosensor had a high level of performance both in detection limit (1 pM) and selectivity (possibility of detecting SARS N protein in the complex matrix). Using a carbon electrode modified by gold nanoparticles array, Laygah et al. [75] fabricated an electrochemical immunosensor for diagnosis of MERS coronavirus. This biosensor targeted the spike protein as a MERS-CoV biomarker. In the mentioned study, the sensing mechanism was dependent on indirect competition of free coronaviruses in the sample and immobilized spike protein on the electrode for attachment to the constant number of antibodies added to the analyte vial. Moreover, the voltammetric responses were collected through square wave voltammetry, and the detection time of assay was less than 30 min and the obtained viral detection limit was 1.0 pg/ml [76]. Koo et al. [76] introduced a rapid label-free detection assay for diagnosis of viral RNA of MERS-CoV. In this study, the bio-optical sensor termed iROAD provided a one-step viral RNA amplification in less than 20 min, and the detection limit of the iROAD assay was reported to be 10-times more sensitive than that of the RT-PCR method.
Miscellaneous Methods
In an indirect approach, Kilianski et al. [77] investigated 3CLpro activity in MERS-CoV using luciferase-based biosensors. The biosensor assay developed with protease expression facilitated the rapid assessment of protease activity in coronaviruses and the determination of protease inhibitors. Lin et al. [78] developed a DNA-tile-based biosensor in a self-assembled sensing platform which could successfully detect SARS-CoV-1 viral DNA. They applied hybridization chain reaction (HCR) to the DNA nanoarray probes and obtained an amplified fluorescent signal for the identification of SARS-CoV-1 viral DNA and ATP. Zhou et al. [79] investigated the proteolytically inactive mutations in coronavirus 3C-like protease. They established an efficient, rapid, sensitive luciferase-based biosensor for in vivo monitoring of the PDEV 3CLpro gene activity. 3C-like (3CLpro) proteases play an important role in viral replications which make them attractive targets for anti-viral therapeutics and diagnostics. Park et al. [80] investigated the roles of protein nanopatterns and biosensor in the identification of SARS-CoV-2 using gold binding polypeptide. Using a gold binding polypeptide as a linking partner, they introduced an efficient strategy to immobilize proteins on a gold surface. In their study, SARS-CoV-2 envelope (E) protein, as a model protein, was immobilized on the surface of gold nanoparticles via linking with polypeptide partners. Accordingly, E protein interacted with its antibody and was changed in terms of absorbance, fluorescent emission color and intensity, which allowed for the diagnosis of SARS-CoV-2 [80]. Park et al. [81] also attempted to determine SARS-CoV-1 utilizing an optical biosensor based on functionalized photonic nanocrystals. They detected SARS-CoV-1 using the antigen–antibody interaction determined by the functional photonic nanocrystals with a low-cost robust optical-readout of resonance peak shift. In another study, Seo et al. [82] introduced a field emission transistor (FET)-based biosensor for fast analysis of SARS-CoV-2 in human nasopharyngeal swab specimens. This FET biosensor was fabricated using 2D graphene and specifically targeted the spike protein of SARS-CoV-2. They subjected their FET biosensor to viral S1 protein concentration and obtained ultra-trace detection limits of 1 fg/ml and 100 fg/ml for (PBS) and clinical samples, respectively. Qi et al. [83] applied the biosensor technique based on imaging ellipsometry to obtain serial serum samples and neutralizing monoclonal antibodies from the patients infected with 10 SARS-CoV-2 and 12 volunteers without SARS-CoV-2. They evaluated the kinetic process of the interaction between SARS-CoV-2 and antibodies using the real-time function of the biosensor. The obtained results complied with the ELISA tests which revealed that h12 antibody had a higher affinity compared to b1 antibody. They reported the affinities of h12 and b1 antibodies as 1.36 × 107 M−1 and 9.5 × 106 M−1, respectively. Application of the biosensor based on imaging ellipsometry, as a label-free method, proved to be an efficient mechanism for assessing the serum samples taken from the patients with SARS-CoV-2 and the affinity of their antibodies with SARS coronavirus.
Roh et al. [84] presented a lab-on-chip (LOC) toll developed by the arrangement of quantum dots-conjugated RNA aptamer on the chip for sensitive and quantitative targeting of SARS-CoV-1N proteins. The results indicated that the QDs-conjugated RNA aptamer could specifically hybridize the immobilized SARS-CoV-1N protein on the glass chip surface. They performed the identification process relying on the optical signal variation of a QDs-supported RNA aptamer interacting on the immobilized protein chip. Application of an optical QDs-based RNA aptamer chip led to the SARS-CoV-1N protein concentration of 0.1 pg/ml. Weng et al. [85] employed molybdenum disulfide (MoS2) nanosheets (which had a high fluorescence-quenching ability along with a dye-labeled antibody) to explore another sensitive, cheap, rapid biosensor for viral detection. They developed a functionalized MoS2 with the antibody to fabricate a fluorescent immunosensor which was responsible for establishing the fluorescence resonance energy transfer (FRET) between the fluorescence dye and MoS2 during the interaction of antibody and antigen. The used assay setup was achieved on a cheap cotton thread-based microfluidic platform which had acceptable flexibility and wicking property.
Salivary Detection Methods
Salivary diagnosis of viral infection has received much attention due to being non-invasive, easiness to collect saliva and low cost [86]. Different approaches, such as collection of saliva by cough, simple swab or the whole saliva collection and passive collection from posterior oro-pharynx, have been used to collect saliva. Besides, saliva has been reported as a positive detection method for SARS-CoV-2 [87]. The merits of using saliva samples to detect SARS-CoV-2, including self-collection and sample collection outside the hospital, can lead to the mitigation of nosocomial transmission risks and reduction of test waiting time and storage costs [88]. It has been approved that the salivary-based diagnostic method for SARS-CoV-2 using the RT-PCR has high detection accuracy for positive cases but may lack accuracy for the diagnosis of false-positive cases [87]. In addition to RT-PCR tests for the diagnosis of SARS-CoV-2, other cheap and fast diagnostic methods for mass screening studies have been approved by the FDA under the EUA (Emergency Use Authorization) [89]. The diagnostic sensitivity and specificity of saliva and sputum in the controlled treatment studies with a smaller size of samples are of high importance. Although most of the controlled treatment studies have validated the use of salivary detection in smaller populations, it may be too early to rely on salivary detection as the only method for the diagnosis of SARS-CoV-2 [87].
Conclusions
Detection of COVID-19 and application of vaccines are considered as the vital actions that should be taken throughout the world. Therefore, many researches should focus on rapid and highly accurate COVID-19 detection methods with low price in order to allow for all countries to sufficiently use the related kits and devices. Some detection methods, such as visual and electrochemical biosensors which do not require laboratory analysis, may be more rapid and cheaper. Such methods can enable everyone to find out his/her infection through a simple test at home. However, PCR-based methods are excellent in definite confirmation of the infection after the initial positive contraction of the patient. More importantly, some infected cases may be exposed to the risk of COVID-19 reinfection due to the lack of antibodies. This can lead to lack of trust in detection of coronavirus through the identification of antibodies in the patient’s blood. Furthermore, the antigen-based detection methods for COVID-19 may overlap with other types of coronavirus detection methods since there is a wide range of similarity among the coronaviruses (for example, in spike protein and/or any other protein in the structure of COVID-19). Therefore, it is suggested to take into account the properties of each detection method that is being used for the identification of coronavirus (COVID-19).
Acknowledgements
The authors would like to acknowledge all who assisted in preparation and conduction of this work, especially Saeed Shaverdy and Esmail Dostkhah for their collaboration in designing the graphics of the paper.
Declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mehrnaz Entesari, Email: m.entesaei@bondagroup.com.
Mina Zamani, Email: mina.zamani1367@gmail.com.
Mohammad Heidarizadeh, Email: hz.mohamad@yahoo.com.
Rasoul Moradi, Email: rmoradi@khazar.org.
Fatemeh Khakdan, Email: f.khakdan@semnan.ac.ir.
Fariba Rafiei, Email: fariba.rafiee@ut.ac.ir.
References
- 1.Holmes KV, Enjuanes L. The SARS coronavirus: A postgenomic era. Science. 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biology. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishiura H, Kobayashi T, Miyama T, Suzuki A, Jung S, Hayashi K, Kinoshita R, Yang Y, Yuan B, Akhmetzhanov AR, Linton NM. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) International Journal of Infectious Diseases. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, Boheemen S, Gopal R, Ballhause M, Bestebroer T, Muth T, Müller M, Drexler JF, Zambon M, Osterhaus A, Fouchier R, Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 6.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, van der Haagmans BL, van den Veer B, Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yip SP, To SST, Leung PH, Cheung TS, Cheng PK, Lim WW. Use of dual TaqMan probes to increase the sensitivity of 1-step quantitative reverse transcription-PCR: Application to the detection of SARS coronavirus. Clinical Chemistry. 2005;51:1885–1888. doi: 10.1373/clinchem.2005.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu XF, Pan JC, Ye R, Xiang HQ, Kou Y, Huang ZC. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. Journal of Clinical Microbiology. 2008;46:837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadjinicolaou AV, Farcas GA, Demetriou VL, Mazzulli T, Poutanen SM, Willey BM, Low DE, Butany J, Asa SL, Kain KC, Kostrikis LG. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): A general methodology for detecting rapidly mutating viruses. Archives of Virology. 2011;156:671–680. doi: 10.1007/s00705-010-0906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciftci S, Neumann F, Hernández-Neuta I, Hakhverdyan M, Bálint Á, Herthnek D, Madaboosi N, Nilsson M. A novel mutation tolerant padlock probe design for multiplexed detection of hypervariable RNA viruses. Science and Reports. 2019;9:2872. doi: 10.1038/s41598-019-39854-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai S, Jung C, Bhadra S, Ellington AD. Phosphorothioated primers lead to loop-mediated isothermal amplification at low temperatures. Analytical Chemistry. 2018;90:8290–8294. doi: 10.1021/acs.analchem.8b02062. [DOI] [PubMed] [Google Scholar]
- 12.Yan C, Cui J, Huang L, Du B, Chen L, Xue G, Li S, Zhang W, Zhao L, Sun Y, Yao H, Li N, Zhao H, Feng Y, Liu S, Zhang Q, Liu D, Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clinical Microbiology & Infection. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirato K, Semba S, El-Kafrawy SA, Hassan AM, Tolah AM, Takayama I, Kageyama T, Notomi T, Kamitani W, Matsuyama S, Azhar EI. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. Journal of Virological Methods. 2018;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Wu X, Wan Z, Li Y, Zuo L, Qin J, Jin X, Zhang C. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virologica Sinica. 2020;35:344–347. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baek YH, Um J, Antigua KJC, Park JH, Kim Y, Oh S, Kim Y, Choi WS, Kim SG, Jeong JH, Chin BS, Nicolas HDG, Ahn JY, Shin KS, Choi YK, Park JS, Song MS. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerging Infectious Diseases. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bath C, Scott M, Sharma PM, Gurung RB, Phuentshok Y, Pefanis S, Colling AS, Balasubramanian N, Firestone SM, Ungvanijban S, Ratthanophart J, Allen J, Rawlin G, Fegan M, Rodoni B. Further development of a reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of foot-and-mouth disease virus and validation in the field with use of an internal positive control. Transboundary and Emerging Diseases. 2020;16:331–334. doi: 10.1111/tbed.13589. [DOI] [PubMed] [Google Scholar]
- 17.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nature Medicine. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Zhang Z, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Chen W, Xiao L, Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 19.Yong SEF, Anderson DE, Wei WE, Pang J, Chia WN, Tan CW, Teoh YL, Rajendram P, Toh MPHS, Poh C, Koh VTJ, Lum J, Suhaimi NAM, Chia PY, Chen MIC, Vasoo S, Ong B, Leo YS, Wang L, Lee VJM. Connecting clusters of COVID-19: An epidemiological and serological investigation. The Lancet Infectious Diseases. 2020 doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Q, Chong KH, Chng HH, Leung B, Ling AE, Wei T, Chan SW, Ooi EE, Kwang J. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clinical and Diagnostic Laboratory Immunology. 2004;11:417–422. doi: 10.1128/CDLI.11.2.417-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu R, Fu A, Deng Z, Li Y, Liu T. Promising methods for detection of novel coronavirus SARS-CoV-2. View. 2020;1:e4. doi: 10.1002/viw2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogels CB, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Landry ML, Foxman EF, Grubaugh ND. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv. 2020 doi: 10.1101/2020.03.30.20048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochemical and Biophysical Research Communications. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 24.Long WH, Xiao HS, Gu XM, Zhang QH, Yang HJ, Zhao GP, Liu JH. A universal microarray for detection of SARS coronavirus. Journal of Virological Methods. 2004;121:57–63. doi: 10.1016/j.jviromet.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang P, Wang H, Cao Z, Jin H, Chi H, Zhao J, Yu B, Yan F, Hu X, Wu F, Jiao C, Hou P, Xu S, Zhao Y, Feng N, Wang J, Sun W, Wang T, Gao Y, Yang S, Xia X. A rapid and specific assay for the detection of MERS-CoV. Frontiers in Microbiology. 2018 doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyrc K, Milewska A, Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. Journal of Virological Methods. 2011;175:133–136. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. Journal of Autoimmunity. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, Potter SJ, Lin Y, Cunningham AL, Dwyer DE, Su Y, Ma X, HouSaksena YNK. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. Journal of Clinical Microbiology. 2005;43:2339–2344. doi: 10.1128/JCM.43.5.2339-2344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Li J, Deng Z, Xiong W, Wang Q, Hu Y. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo X, Geng P, Wang Q, Cao B, Liu B. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. Journal of Microbiology and Biotechnology. 2014;24:1445–1454. doi: 10.4014/jmb.1404.04024. [DOI] [PubMed] [Google Scholar]
- 31.de Souza Luna LK, Heiser V, Regamey N, Panning M, Drexler JF, Mulangu S, Poon L, Baumgarte S, Haijema BJ, Kaiser L, Drosten C. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. Journal of Clinical Microbiology. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardick J, Metzgar D, Risen L, Myers C, Balansay M, Malcom T, Rothman R, Gaydos C. Initial performance evaluation of a spotted array Mobile Analysis Platform (MAP) for the detection of influenza A/B, RSV, and MERS coronavirus. Diagnostic Microbiology and Infectious Disease. 2018;91:245–247. doi: 10.1016/j.diagmicrobio.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Xiong E, Tian T, Cheng M, Lin W, Wang H, Zhang G, Sun J, Zhou X. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14:2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 34.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nature Protocols. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan CY, Guevara H, Wadford DA, Chen JS, Chiu CY. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J. Pathogenicity and transmissibility of 2019-nCoV—A quick overview and comparison with other emerging viruses. Microbes and Infection. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kostyusheva, A. Brezgin, S. Babin, Y. Vasil'eva, I. Kostyushev, D. Chulanov, V. (2020) CRISPR-Cas systems for diagnosing infectious diseases. Preprints. 10.20944/preprints202002.0007.v1 [DOI] [PMC free article] [PubMed]
- 38.Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pacific Journal of Allergy and Immunology. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 39.Chan KH, Poon LL, Cheng V, Guan Y, Hung I, Kong J, Yam LY, Seto WH, Yuen KY, Peiris JSM. Detection of SARS coronavirus in patients with suspected SARS. Emerging Infectious Diseases. 2004;10:294. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park GSKu, Baek K, Kim SH, Kim SJ, Kim SI, Maeng BT, J. S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2. The Journal of Molecular Diagnostics. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu HS, Chiu SC, Tseng TC, Lin SF, Lin JH, Hsu YF, Wang MC, Lin TL, Yang WZ, Ferng TL. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerging Infectious Diseases. 2004;10:305. doi: 10.3201/eid1002.030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung DT, Chi M, Hang T, Chun F, Hung MS, Chan PK, Cheung JLK, Niu H, Tam JSL, Lim PL. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. Journal of Infectious Diseases. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Infantino M, Damiani A, Gobbi FL, Grossi V, Lari B, Macchia D, Casprini P, Veneziani F, Villalta D, Bizzaro N. Serological assays for SARS-CoV-2 infectious disease: Benefits, limitations and perspectives. Israel Medical Association. 2020;22:203–210. [PubMed] [Google Scholar]
- 44.Che XY, Qiu LW, Liao ZY, Wang YD, Wen K, Pan YX, Hao W, Mei YB, Cheng VC, Yuen KY. Antigenic cross-reactivity between severe acute respiratory syndrome: Associated coronavirus and human coronaviruses 229E and OC43. Journal of Infectious Diseases. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dijkman R, Jebbink MF, Gaunt E, Rossen JW, Templeton KE, van der Kuijpers TW, Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. Journal of Clinical Virology. 2012;53:135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klasse P. Collusion between neutralizing antibodies and other immune factions in the destruction of adenoviral vectors. Proceedings of the National Academy of Sciences. 2018;115:10201–10203. doi: 10.1073/pnas.1814275115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mupapa K, Massamba M, Kibadi K, Kuvula K, Bwaka A, Kipasa M, Colebunders R, Muyembe-Tamfum JJ, Committee T. Treatment of Ebola hemorrhagic fever with blood transfusions from convalescent patients. Journal of Infectious Diseases. 1999;179:S18–S23. doi: 10.1086/514298. [DOI] [PubMed] [Google Scholar]
- 48.Arabi Y, Balkhy H, Hajeer AH, Bouchama A, Hayden FG, Al-Omari A, Al-Hameed FM, Taha Y, Shindo N, Whitehead J, Merson L, AlJohani S, Al-Khairy K, Carson G, Luke TC, Hensley L, Al-Dawood A, Al-Qahtani S, Modjarrad K, Sadat M, Rohde G, Leport C, Fowler R. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: A study protocol. Springerplus. 2015;4:709. doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging Infectious Diseases. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Research. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proceedings of the National Academy of Sciences. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends in Immunology. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV: A target for vaccine and therapeutic development. Nature Reviews Microbiology. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: A key target for antivirals. Expert Opinion on Therapeutic Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Shi W, Joyce MG, Modjarrad K, Zhang Y, Leung K, Lees CR, Zhou T, Yassine HM, Kanekiyo M, Yang Z, Chen X, Becker MM, Freeman M, Vogel L, Johnson JC, Olinger G, Todd JP, Bagci U, Solomon J, Mollura DJ, Hensley L, Jahrling P, Denison MR, Rao SS, Subbarao K, Kwong PD, Mascola JR, Kong WP, Graham BS. Evaluation of candidate vaccine approaches for MERS-CoV. Nature Communications. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prabakaran P, Gan J, Feng Y, Zhu Z, Choudhry V, Xiao X, Ji X, Dimitrov DS. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. Journal of Biological Chemistry. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C, Li W, Drabek D, van Okba NMA, Haperen R, van Osterhaus ADME, Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch BJ. A human monoclonal antibody blocking SARS-CoV-2 infection. Nature Communications. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pak JE, Sharon C, Satkunarajah M, Auperin TC, Cameron CM, Kelvin DJ, Seetharaman J, Cochrane A, Plummer FA, Berry JD. Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain. Journal of Molecular Biology. 2009;388:815–823. doi: 10.1016/j.jmb.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okba NM, Raj VS, Widjaja I, de GeurtsvanKessel CH, Bruin E, Chandler FD, Park WB, Kim NJ, Farag EA, Al-Hajri M. Sensitive and specific detection of Low-level antibody responses in Mild Middle East respiratory syndrome coronavirus infections. Emerging Infectious Diseases. 2019;25:1868. doi: 10.3201/eid2510.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amanat, F. Nguyen, T. Chromikova, V. Strohmeier, S. Stadlbauer, D. Javier, A. Jiang, K. Asthagiri-Arunkumar, G. Polanco, J. Bermudez-Gonzalez, M.(2020) A serological assay to detect SARS-CoV-2 seroconversion in humans, MedRxiv.10.1101/2020.03.17.20037713 [DOI] [PMC free article] [PubMed]
- 62.Song D, Ha G, Serhan W, Eltahir Y, Yusof M, Hashem F, Elsayed E, Marzoug B, Abdelazim AA, Muhairi S. Development and validation of a rapid immunochromatographic assay for detection of Middle East respiratory syndrome coronavirus antigen in dromedary camels. Journal of Clinical Microbiology. 2015;53:1178–1182. doi: 10.1128/JCM.03096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, Chan KH, Kang Y, Chen H, Luk HK, Poon RW, Chan JF, Yuen KY, Xia N, Lau SK. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerging Infectious Diseases. 2015;4:1–5. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, Wang H, Deng Y, Song T, Lan J, Wu G, Ke C, Tan W. Characterization of anti-MERS-CoV antibodies against various recombinant structural antigens of MERS-CoV in an imported case in China. Emerging Infectious Diseases. 2016;5:1–12. doi: 10.1038/emi.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izzo M, Kirkland P, Gu X, Lele Y, Gunn A, House J. Comparison of three diagnostic techniques for detection of rotavirus and coronavirus in calf faeces in Australia. Australian Veterinary Journal. 2012;90:122–129. doi: 10.1111/j.1751-0813.2011.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qiu GZ, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;5:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 67.Shi L, Sun Q, He J, Xu H, Liu C, Zhao C, Xu Y, Wu C, Xiang J, Gu D, Long J, Lan H. Development of SPR biosensor for simultaneous detection of multiplex respiratory viruses. Bio-Medical Materials and Engineering. 2015;26:S2207–S2216. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- 68.Yang Y, Wang Q, Guo D. A novel strategy for analyzing rna-protein interactions by surface plasmon resonance biosensor. Molecular Biotechnology. 2008;40:87–93. doi: 10.1007/s12033-008-9066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang JC, Chang YF, Chen KHSu, Lee LC, Chen CW, Chen CC, Chou YMA. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosensors & Bioelectronics. 2009;25:320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abad-Valle P, Fernández-Abedul MT, Costa-García A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosensors & Bioelectronics. 2005;20:2251–2260. doi: 10.1016/j.bios.2004.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang WY, Sung PH, Chu CH, Shih CJ, Lin YC. Phase detection of the two-port FPW sensor for biosensing. IEEE Sensors Journal. 2008;8:501–507. [Google Scholar]
- 72.Zari N, Mohammedi H, Amine A, Ennaji MM. DNA hydrolysis and voltammetric determination of guanine and adenine using different electrodes. Analytical Letters. 2007;40:1698–1713. [Google Scholar]
- 73.Ishikawa FN, Chang HK, Curreli M, Liao HI, Olson CA, Chen PC, Zhang R, Roberts RW, Sun R, Cote RJ, Thompson M, Zhou E. Label-free, electrical detection of the SARS virus N-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano. 2009;3:1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishikawa FN, Curreli M, Olson CA, Liao HI, Sun R, Roberts RW, Cote RJ, Thompson ME, Zhou C. Importance of controlling nanotube density for highly sensitive and reliable biosensors functional in physiological conditions. ACS Nano. 2010;4:6914–6922. doi: 10.1021/nn101198u. [DOI] [PubMed] [Google Scholar]
- 75.Layqah LA, Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchimica Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koo B, Jin CE, Lee TY, Lee JH, Park MK, Sung H, Park SY, Lee HJ, Kim SM, Kim JY, Kim SH, Shin Y. An isothermal, label-free, and rapid one-step RNA amplification/detection assay for diagnosis of respiratory viral infections. Biosensors & Bioelectronics. 2017;90:187–194. doi: 10.1016/j.bios.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kilianski A, Mielech AM, Deng X, Baker SC. Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. Journal of Virology. 2013;87:11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin C, Nangreave JK, Li Z, Liu Y, Yan H. Signal amplification on a DNA-tile-based biosensor with enhanced sensitivity. Nanomedicine. 2008;3:521–528. doi: 10.2217/17435889.3.4.521. [DOI] [PubMed] [Google Scholar]
- 79.Zhou J, Fang L, Yang Z, Xu S, Lv M, Sun Z, Chen J, Wang D, Gao J, Xiao S. Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. The FASEB Journal. 2019;33:14575–14587. doi: 10.1096/fj.201901624RR. [DOI] [PubMed] [Google Scholar]
- 80.Park TJ, Lee SY, Lee SJ, Park JP, Yang KS, Lee KB, Ko S, Park JB, Kim T, Kim SK, Shin YB, Chung BH, Ku SJ, Kim DH, Choi IS. Protein nanopatterns and biosensors using gold binding polypeptide as a fusion partner. Analytical Chemistry. 2006;78:7197–7205. doi: 10.1021/ac060976f. [DOI] [PubMed] [Google Scholar]
- 81.Park TJ, Lee SK, Yoo SM, Yang SM, Lee SY. Development of reflective biosensor using fabrication of functionalized photonic nanocrystals. Journal of Nanoscience and Nanotechnology. 2011;11:632–637. doi: 10.1166/jnn.2011.3269. [DOI] [PubMed] [Google Scholar]
- 82.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 83.Qi C, Duan JZ, Wang ZH, Chen YY, Zhang PH, Zhan L, Yan XY, Cao WC, Jin G. Investigation of interaction between two neutralizing monoclonal antibodies and SARS virus using biosensor based on imaging ellipsometry. Biomedical Microdevices. 2006;8:247–253. doi: 10.1007/s10544-006-8305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roh C, Jo SK. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. Journal of Chemical Technology and Biotechnology. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weng X, Neethirajan S. Immunosensor based on antibody-functionalized MoS2 for rapid detection of avian coronavirus on cotton thread. IEEE Sensors Journal. 2018;18:4358–4363. doi: 10.1109/JSEN.2018.2829084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Corstjens PL, Abrams WR, Malamud D. Detecting viruses by using salivary diagnostics. The Journal of the American Dental Association. 2012;143:12S–18S. doi: 10.14219/jada.archive.2012.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warsi I, Khurshid Z, Shazam H, Umer MF, Imran E, Khan E, Goodson J. Saliva exhibits high sensitivity and specificity for the detection of SARS-COV- Diseases. 2021;9:38. doi: 10.3390/diseases9020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, Leung WS, Chik TS, Choi CY, Kandamby DH, Lung DC. Consistent detection of 2019 novel coronavirus in saliva. Clinical Infectious Diseases. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.FDA. U.S. Food and Drug Administration Emergency Use Authorizations; FDA: Muntinlupa, PH, USA, 2021. Available online: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergencyuse-authorization. Accessed on 26 March 2021.