Abstract
The mouse tubby phenotype is characterized by maturity-onset obesity accompanied by retinal and cochlear degeneration. A positional cloning effort to find the gene responsible for this phenotype led to the identification of tub, a member of a novel gene family of unknown function. A splice defect mutation in the 3′ end of the tub gene, predicted to disrupt the C terminus of the Tub protein, has been implicated in the genesis of the tubby phenotype. It is not clear, however, whether the Tub mutant protein retains any biological activity, or perhaps has some dominant function, nor is it established that the tubby mutation is itself responsible for all of the observed tubby phenotypes. To address these questions, we generated tub-deficient mice and compared their phenotype to that of tubby mice. Our results demonstrate that tubby is a loss-of-function mutation of the tub gene and that loss of the tub gene is sufficient to give rise to the full spectrum of tubby phenotypes. We also demonstrate that loss of photoreceptors in the retina of tubby and tub-deficient mice occurs by apoptosis. In addition, we show that Tub protein expression is not significantly altered in the ob, db, or melanocortin 4 receptor-deficient mouse model of obesity.
The tubby strain of obese mice arose spontaneously in a mouse colony at the Jackson Laboratory (5). The tubby phenotype is inherited in an autosomal recessive manner and is characterized by late-onset weight gain accompanied by progressive retinal and cochlear degeneration (16, 17). The combination of these phenotypes resembles human syndromes, such as Usher's (retinal and cochlear degeneration), Bardet-Biedl, and Alstrom's (obesity and sensory deficits).
The obesity of tubby mice is relatively mild and late in onset, resembling the weight gain in human populations more closely than that observed in other murine models of obesity, such as obese (ob) and diabetes (db) (4). Weight gain in tubby mice occurs relatively slowly, and the mice reach about twice the weight of their unaffected siblings. Tubby mice are not sterile, but they become infertile after significant obesity develops. The retinal and cochlear degeneration has been proposed to be due to apoptosis of retinal and cochlear neurosensory cells.
The identity of the tub gene was determined by positional cloning (11, 14). The expression pattern of tub mRNA appears to be specific for the nervous system (10). tub is a member of a novel gene family of unknown function (7). Tub family proteins are quite hydrophilic but otherwise lack any distinguishing features that might provide insight into their function. There are no recognizable structural motifs or obvious homology to any known proteins.
The tubby mutation is a G-to-T transversion that abolishes the donor splice site in the penultimate exon (exon 11), resulting in an aberrant transcript (11). Translation of intron sequence results in the substitution of the Tub C-terminal 44 amino acids with 24 different amino acids encoded by the intron. The aberrant transcript is expressed at elevated levels in tubby mice (5), but Tub expression at the protein level has not been investigated.
The C terminus is highly conserved among Tub family members (15), suggesting that the tubby mutation may have disrupted a domain important for the biological function of tub. Strikingly, mutation of the tub family member TULP1, at a position exactly corresponding to that mutated in the murine tub allele, was recently found to be the cause of a form of retinitis pigmentosa (RP 14) in humans (1, 7). Three other pathogenic missense mutations of TULP1 have been characterized (7), and they all fall within the highly conserved carboxy-terminal 250 amino acids of Tub.
Despite elucidation of the molecular basis of the tubby mutation, it remains unclear whether the tubby phenotype represents a complete or partial loss of function or possibly even a gain of function. Furthermore, it has not been established whether the suite of tubby phenotypes (obesity, insulin resistance, and retinal and cochlear degeneration) are necessarily all attributable to mutation of the tub gene. It has not been ruled out that some features of the phenotype may be caused by a tightly linked, as yet unidentified gene.
MATERIALS AND METHODS
Targeting of tub.
Murine tub gene sequences were isolated from a 129/Sv genomic bacterial artificial chromosome library (Research Genetics). To generate a targeting construct, an approximately 4.5-kb HpaI-XbaI fragment from the 3′ end of the tub gene, spanning exons 9 to 12, was subcloned into the XbaI site of pNEB 193 (New England Biolabs). An approximately 2.6-kb HincII-EagI fragment derived from just upstream of exon 1 was then subcloned into the BssHII site of the pNEB193 polylinker, situating it upstream of the 3′ tub genomic fragment, in the same transcriptional orientation. A PGK-neo expression cassette, consisting of the neo gene under the transcriptional control of the mouse phosphoglycerate kinase (PGK-1) promoter and the PGK-1 poly(A) addition site, was obtained from the plasmid pKJ1 (13). The cassette was subcloned into the PacI site of the pNEB193 polylinker, between the 5′ and 3′ tub homologous sequences. The vector was linearized with XhoI digestion prior to electroporation.
MPI-2-71.6 embryonic stem (ES) cells, derived from C57BL/6J-Hprtb-m3 mice, were cultured on mitotically inactive G418-resistant mouse embryo fibroblast feeder cells as described elsewhere (18). Cells were electroporated with the linearized targeting vector as described previously (8) and screened for homologous recombination with the vector by Southern blot hybridization using the 5′ flanking probe shown in Fig. 1A. One targeted clone was detected out of 190 clones screened.
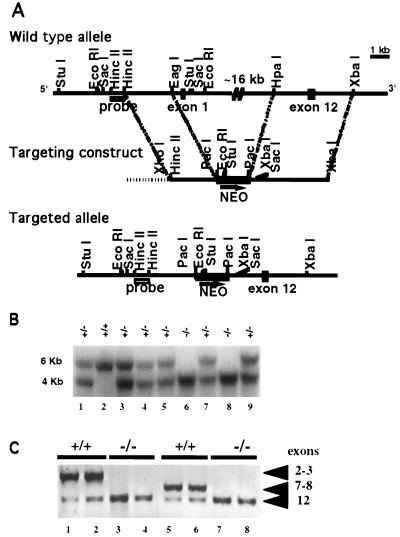
FIG. 1.
Disruption of the mouse tub locus. (A) Schematic representation and partial restriction maps of the tub wild-type locus, targeting vector, and targeted allele. Closed boxes represent tub exons 1 and 12, the hatched box indicates the 5′ flanking probe, the shaded box is the PGK-neo expression cassette, and the horizontal dotted line is plasmid DNA. The arrow below neo indicates the direction of transcription. (B) Southern blot analysis of EcoRI-digested tail DNA from F2 progeny hybridized with the radiolabeled probe shown in panel A. The wild-type (+/+) hybridizing band is 6 kb; the band from homozygous mutant mice (−/−) is 4 kb; mice heterozygous for the mutation (+/−) show both 6- and 4-kb bands. (C) PCR analysis of tail DNA of F2 +/+ and −/− mice. Each PCR was done in duplicate. The primers are from exons 2 and 3 (lanes 1 to 4) or 7 and 8 (lanes 5 to 8) and 12 (all lanes). Exons 2, 3, 7, and 8 are absent in −/− mice. Exon 12 is present in both +/+ and −/− mice.
Generation of mutant mice.
The targeted ES clone was injected into BALB/cByJ blastocysts as described previously (2) to generate chimeras. Male chimeras were bred with C57BL/6 females. Black offspring heterozygous for the mutation were interbred to generate F2 progeny for analysis. Mice were maintained on a 12-h light/12-h dark cycle, fed PMI 5021 chow containing 9% fat, and provided water ad libitum.
Southern blot hybridization and PCR analysis.
For Southern blot analysis, genomic DNAs from ES cells and tail biopsies were prepared as described by Laird et al. (12). EcoRI-digested genomic DNA (approximately 20 μg) was electrophoresed through a 1% agarose gel, transferred to a Hybond N+ membrane (Amersham), and hybridized with the 32P-labeled 5′ flanking probe depicted in Fig. 1A.
Genomic DNA derived from tail biopsies of tub+/+ and tub−/− mice was used as the template for PCR analysis. The primers used were derived from sequences contained in exons 2 (forward, TGAGGCAGCAGAAGC), 3 (reverse, CACTGCTGCTGAGGTAGGACTC), 7 (forward, GGACAAGAAGGGGATGGAC), 8 (reverse, GTTGGGTCCACAGAGATGATGGA), and 12 (forward, GGGTAGCAGAAGATGTGT; reverse, GCAGCAGAGGCAGAGC) of tub. The reactions were carried out as described in the legend to Fig. 1C.
Immunoprecipitation and Western blots.
Chinese hamster ovary (CHO) cells were transfected with either an empty expression vector (pN8ɛ) or an expression vector containing hemagglutinin epitope (HA)-tagged wild-type or mutant Tub (8 μg/100-mm-diameter plate) by the Lipofectamine method (Gibco-BRL, Gaithersburg, Md.). At 48 h posttransfection, cells were lysed in 400 μl of lysis buffer (20 mM HEPES [pH 7.5], 0.15 M NaCl, 0.2 mM EDTA, 1.5 mM MgCl2, 1 mM dithiothreitol, 1 mM sodium vanadate, 10 mM NaF, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1 μg each of leupeptin, pepstatin, and aprotinin per ml). An aliquot was taken from the cleared whole cell lysate and boiled in sodium dodecyl sulfate (SDS)-containing sample buffer. Alternatively, the lysates were incubated with either anti-HA (10 μl; 12CA5; Boehringer Mannheim) or anti-Tub (5 μl; raised against full-length His6-tagged wild-type Tub [murine] and kindly provided by Rene Devos, Roche-Gent Research Institute) antibody in the presence of protein A-Sepharose beads (Pharmacia Biotech, Piscataway, N.J.). The precipitates were then washed three times with high-salt wash buffer (20 mM HEPES [pH 7.5], 0.3 M NaCl, 2.5 mM MgCl2, 0.5% Triton X-100) and twice with low-salt wash buffer (20 mM HEPES [pH 7.5], 0.05 M NaCl, 2.5 mM MgCl2, 0.5% Triton X-100) and boiled in SDS-containing sample buffer. Lysates from brain and spleen were generated by mincing the organs with a razor blade and then douncing them with a tight-fitting pestle in EBC buffer (50 mM Tris-HCl [pH 8], 120 mM NaCl, 0.5% NP-40) supplemented with Complete Mini protease inhibitor cocktail tablets (Boehringer Mannheim). Dounced lysates were pelleted in a microcentrifuge at 12,000 rpm for 15 min at 4°C. Supernatants were boiled in SDS-containing sample buffer.
Lysates and immunoprecipitates were resolved by polyacrylamide gel electrophoresis (PAGE) in 8 or 10% polyacrylamide gels (Novex), transferred to nitrocellulose, and probed with the anti-Tub antiserum (1:1,000). Following incubation with the primary antibody, the membranes were washed and incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody (Amersham). Blots were developed by enhanced chemiluminescence (Amersham) according to the manufacturer's directions.
Weight measurements.
Ten animals of each sex and genotype (+/+, tub/tub, and −/−), were maintained on a chow diet ad libitum. Their weight was monitored once a week for a period of 15 weeks, starting at 5 weeks of age, using a Sartorius model 14800 P balance.
Histological analysis of the retina and TUNEL apoptosis assay for fragmented DNA.
Eight-week-old male mice of each genotype (+/+, tub/tub, and −/−) were anesthetized and perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA)–PBS. Eyes were removed for processing and embedding in paraffin, using a TissueTek tissue processor. For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay, 5-μm cross sections were cut, deparaffinized, and postfixed with 4% PFA–PBS for 15 min. Following a 5-min wash with PBS, sections were digested with proteinase K (2 μg/ml) at 37°C for 15 min, fixed again with 4% PFA–PBS for 15 min, washed twice with Tris-buffered saline for 1 min, and washed twice with H2O for 1 min. Terminal transferase labeling of fragmented DNA was performed with a Boehringer Mannheim fluorescein in situ death detection kit (product no. 1-684-795) essentially as described. Briefly, sections were incubated in reaction mixture (100-μl volume/slide) for 90 min at 37°C and washed three times in Tris-buffered saline–0.1% Tween. Slides were visualized and photographed with a Zeiss BX100 microscope equipped with a fluorescein isothiocyanate filter (magnification, ×40). For comparison, adjacent retina sections were stained with hematoxylin and eosin (magnification, ×400).
RESULTS
Generation of tub-deficient mice.
The tub gene was inactivated by gene targeting in C57BL/6 ES cells. Approximately 16 kb of tub genomic sequence, spanning exons 1 through 8, were deleted from the tub locus by homologous recombination, using the targeting vector shown in Fig. 1A. This represents deletion of over 60% of the coding sequence. Interbreeding of mice heterozygous for the mutation generated F2 progeny that were wild type, heterozygous, and homozygous for the targeted allele (Fig. 1B), with the expected 1:2:1 distribution. PCR analysis verified the predicted absence of tub exons 2, 3, 7, and 8, and the presence of exon 12, in homozygous mutant mice (Fig. 1C). Together with the Southern analyses (Fig. 1B), these data confirm the generation of mice carrying a large deletion of the tub coding sequence. tub−/− mice were viable and had no obvious defects at birth or in the neonatal period.
Expression of the Tub protein in wild-type and tub−/− mice.
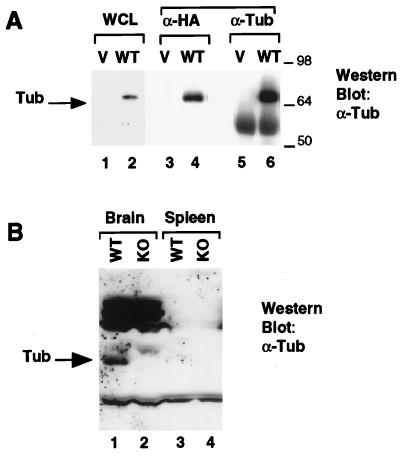
We used an anti-Tub antiserum raised against His6-tagged full-length wild-type murine Tub to assess expression of Tub at the protein level. As shown in Fig. 2A, this antiserum recognizes HA-tagged wild-type Tub protein expressed in CHO cells. Lanes 1 and 2 show a Western blot of lysate from CHO cells transfected with either the empty vector (lane 1) or a vector expressing HA-Tub (lane 2). The antibody recognizes a specific band of the correct size only in the cells transfected with HA-Tub. Furthermore, when the lysates are immunoprecipitated with an antibody against the HA tag, the anti-Tub antiserum recognizes the HA-tagged tub protein (lane 4). Finally, transfected Tub protein is specifically immunoprecipitated by the anti-Tub antiserum (lane 6). The band below Tub in lanes 5 and 6 is the immunoglobulin heavy chain.
FIG. 2.
Expression of the Tub protein in wild-type (WT) and tub−/− mice. (A) Anti-Tub immunoblot of HA-Tub expressed in CHO cells. Lanes 1, 3, and 5, transfection with the empty vector (V); lanes 2, 4, and 6, transfection with HA-Tub. Samples were resolved by SDS-PAGE in a 10% gel, transferred to nitrocellulose, and probed with the anti-Tub antibody. Lanes 1 and 2, blot of whole cell lysate (WCL); lanes 3 and 4, blot of anti-HA immunoprecipitates; lanes 5 and 6, blot of anti-Tub immunoprecipitates. Sizes are indicated in kilodaltons. (B) Brain and spleen lysates from wild-type and tub knockout mice were resolved by SDS-PAGE in an 8% gel and transferred to nitrocellulose. The membrane was probed with anti-Tub antibody. The Tub protein, indicated by the arrow, is detectable in brain lysates from wild-type (lane 1) but not tub knockout (KO; lane 2) mice. No Tub expression is detected in spleen lysates from either wild-type (lane 3) or tub knockout (lane 4) mice.
To confirm that tub is not expressed in the gene-targeted mice, we compared the protein levels of Tub in brain lysates of wild-type and tub−/− mice by Western blotting (Fig. 2B). We chose the brain because tub mRNA is extensively expressed there (5, 10). As an additional negative control, lysates were also prepared from the spleen, which was not expected to express tub (19). Lysates containing equivalent amounts of protein were resolved by SDS-PAGE and probed with anti-Tub antiserum. As shown in the Western blot in Fig. 2B, a specific band was detected in the lane with brain lysate from wild-type (lane 1) but not tub−/− (lane 2) mice, verifying the null mutation of tub in the knockout mice. No tub expression was detected in spleens of either wild-type or tub−/− mice (Fig. 2B, lanes 3 and 4).
Weight gain of tubby and tub−/− mice.
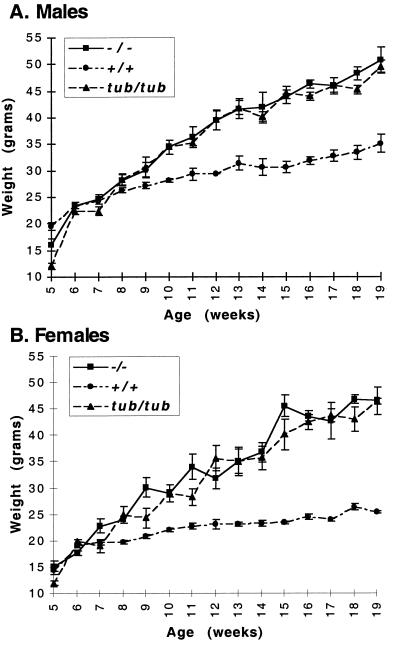
The weight gains of tubby (tub/tub) mice, homozygous mutant (tub−/−) targeted mice, and their wild-type (+/+) littermate controls were monitored regularly over a 19-week period. All mice were on the C57BL/6 strain background. As shown in Fig. 3, the weights of all genotypes were comparable for the first 8 weeks of life. At this time, the weight of both male (Fig. 3A) and female (Fig. 3B) tub/tub mice begins to diverge from that of the wild-type mice, and at 19 weeks of age, the weight of the tub/tub mice is almost twice that of their wild-type littermates. This finding is consistent with a previous characterization of tubby weight gain (5), although in our study, the weight of the tubby mice diverges from that of the wild-type animals slightly earlier (8 weeks rather than 12 weeks). As shown, the weight gain of the tub−/− mice was indistinguishable from that of the tub/tub mice (Fig. 3). Both male and female heterozygous mice (+/tub or +/−) exhibited weight curves indistinguishable from that of wild-type mice (data not shown).
FIG. 3.
Weight gain of +/+, tub/tub, and −/− mice: (A) Weight gain of male wild-type (+/+), tubby (tub/tub), and tub-deficient (−/−) mice. The weights of 10 mice of each genotype were measured at the indicated times. (B) Weight gain of female wild-type, tubby, and tub-deficient mice. The weights of 10 mice of each genotype were measured at the indicated times.
Retinal degeneration in tubby and tub knockout mice.
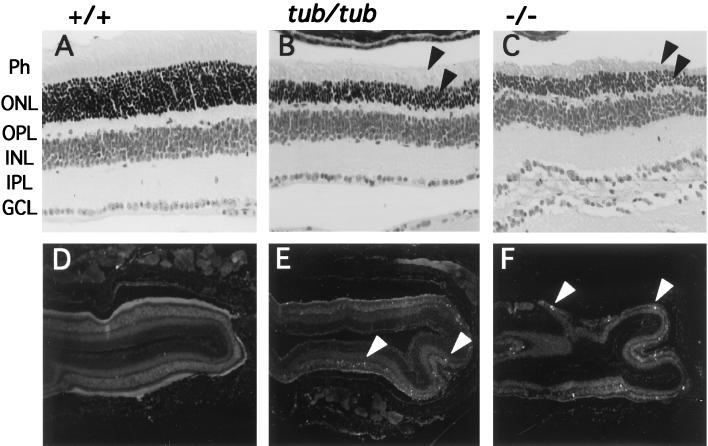
Earlier studies have shown that tubby mice exhibit early progressive retinal degeneration, possibly due to apoptosis (16, 17). Photoreceptor cells, the main cell type lost, have been shown to express Tub (19). We examined the retinas of 8-week-old wild-type (+/+), tub/tub, and knockout (tub−/−) mice. It is evident from the histological sections shown in Fig. 4B and C that tubby and tub−/− mice exhibit severe and comparable retinal degeneration at this age, with significant loss of the photoreceptor and outer nuclear layers (arrows) relative to their wild-type littermates (Fig. 4A). The remaining layers of the retina appear normal. The retinas of the tub+/− mice are unaffected and appear indistinguishable from those of wild-type mice (not shown).
FIG. 4.
Histological section and TUNEL assay of wild-type, tub/tub, and tub−/− mouse retinas. (A to C) Histological sections of retinas from wild-type (A), tub/tub (B), and tub−/− (C) mice at 8 weeks of age. Photoreceptors (Ph) and outer nuclear layers (ONL) are reduced in tub/tub and tub−/− mice (arrows) compared to wild-type mice. All other layers appear normal. Magnification, ×400. OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. (D to F) In situ TUNEL assay of retinas from wild-type (D), tub/tub (E), and tub−/− (F) mice at 8 weeks of age. Apoptotic figures (indicated by arrowheads) are present in retinas derived from tub/tub and tub−/− but not wild-type mice. Magnification, ×40.
Figure 4D to F shows TUNEL staining of the same retinas at lower magnification. This assay reveals the presence of cells undergoing apoptosis in retinas from tub/tub and tub−/− mice (Fig. 4E and F, arrows) but not in retinas from wild-type littermates (Fig. 4D). This finding is consistent with a recent report (9) and demonstrates that the retinal degeneration observed in tubby and tub knockout mice is in fact due to apoptosis of the photoreceptor cells.
Expression of the Tub protein in tubby mice and other mouse models of obesity.
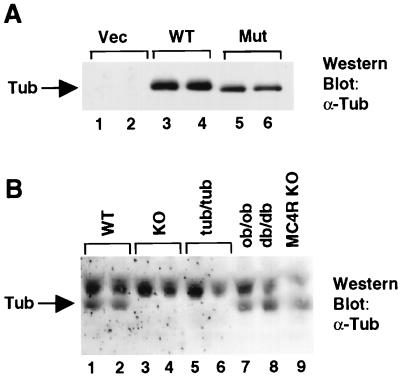
The tubby mutant protein is 20 residues shorter than wild-type Tub. As shown by Western blotting, using the anti-Tub antiserum (Fig. 5A), HA-tagged mutant tubby protein, overexpressed in CHO cells, migrates slightly but detectably faster (lanes 5 and 6) than HA-Tub (lanes 3 and 4) on an SDS-polyacrylamide gel. We used the anti-Tub antibody to investigate the expression of this protein in tub/tub mice.
FIG. 5.
(A) Anti-Tub immunoblot of whole cell lysates from CHO cells transfected with the empty vector (Vec; lanes 1 and 2), HA-Tub (WT; lanes 3 and 4), or HA-mutant Tub (Mut; lanes 5 and 6). (B) Brain lysates were resolved by SDS-PAGE and probed with the anti-Tub antiserum as for Fig. 3. Each lane represents an individual mouse of the indicated genotype. KO, knockout; MC4R, melanocortin 4 receptor.
By Northern analysis and in situ hybridization, tub mRNA levels were shown to be higher in tub/tub mice than in wild-type littermates (11). To examine Tub expression at the protein level in tubby mice and compare it to expression in wild-type and tub knockout mice, and other models of obesity, brain lysates were resolved by SDS-PAGE and probed with the anti-Tub antiserum. Each lane in Fig. 5B represents an individual mouse. As in Fig. 3, the Tub protein is detected in brain lysates from wild-type mice (Fig. 6, lanes 1 and 2), but undetectable in brain lysates from tub knockout mice (lanes 3 and 4). Unexpectedly, Tub protein was also not detected in lysates from tubby (tub/tub) mice (lanes 5 and 6), indicating that the Tub mutant protein is either absent or present at levels below the limit of detection in tubby mice.
Expression levels of Tub in brain lysates of ob, db, and melanocortin 4 receptor-deficient mice (lanes 7 to 9) was also assessed to determine whether alteration of Tub protein expression occurs in these models of murine obesity. No significant change in Tub protein level was observed, suggesting that alteration of Tub expression may not be a common feature of mouse models of obesity.
DISCUSSION
In this study we have generated tub-deficient mice by targeted deletion of tub exons 1 to 8 in C57BL6 ES cells. Mice homozygous for the targeted allele (tub−/−) are phenotypically indistinguishable from tubby mice with regard to weight gain and retinal degeneration. These observations demonstrate that these two very different tubby phenotypes are caused by disruption of a single gene, not two closely linked genes.
If the mutated allele of tub expressed by the tubby (tub/tub) mice retained some residual function, one might expect the phenotype of the tub knockout mice to be more severe than that of tubby mice. Conversely, if the tubby allele was nonfunctional, one might expect the phenotypes of tubby and tub knockout mice to be identical. Our observation that tub−/− and tubby mice have indistinguishable phenotypes supports the hypothesis that tubby is a null allele. In addition, we unexpectedly found that the expression levels of the mutant protein in tubby mice are so low as to be undetectable. Since Northern blot and in situ analyses (11) indicate that tub mRNA levels are elevated in tubby mice, the defect must be at the posttranscriptional level. The elevated mRNA levels in tubby mice may represent an attempt to upregulate expression of the Tub protein. The inability to detect tubby mutant protein in spite of the elevated mRNA levels may be due to protein instability, perhaps resulting from disruption of the highly conserved carboxy terminus of Tub. It is possible that this region forms an as yet unrecognized structural motif required for stability of the tubby protein.
We also show that the retinal degeneration observed in tub/tub and tub knockout mice is due to apoptosis. Programmed cell death has been proposed as the cause of both retinal and cochlear degeneration in tubby mice (16, 17), and this has now been formally demonstrated for retinal degeneration by our data as well as a very recent report (9). This result suggests that Tub function is important for the survival of retinal photoreceptor cells in the adult animal.
It is possible that the late-onset obesity observed in tubby mice is similarly due to the apoptotic loss of cells in hypothalamic nuclei involved in body weight regulation. Tubby obesity and hypersinulinemia are reminiscent of the effect of surgical ablation of the ventral-medial hypothalamus (3). We searched for, but did not find, any cellular alterations in the hypothalamus of tubby and tub−/− mice (data not shown). However, we cannot rule out the possibility of subtle changes that we may not be able to detect, such as the loss of a small subpopulation of neurons.
It is possible that the defect leading to obesity in tubby mice lies in the peripheral rather than the central nervous system. It is interesting that unilateral denervation of fat pads results in increased size of the denervated pad but not of the contralateral pad (6), suggesting that fat innervation is important for the control of body weight.
If tub function is important for neuronal survival, why does absence of tub not result in a more severe phenotype, given the extensive expression of Tub in the brain and other neuron-derived tissues (19)? One possibility is that the Tub protein does not play a critical role in most of the tissues where it is expressed. Alternatively, there may be redundancy between Tub family members. Tub is a member of a newly discovered class of proteins, only a few of which have been cloned. According to one estimate, the family consists of approximately 6 to 10 members (15). Mouse tub may be functional in many neuronal tissues, but a phenotype may be seen only in the few tissues where other family members are not expressed or cannot compensate for tub loss.
While we now have a wealth of genetic data relating to Tub, the biochemical function of the protein remains unclear. Although Tub lacks known protein motifs, Tub family members share a remarkably conserved carboxy terminus, suggesting that this domain is very important for Tub function. Furthermore, Tub-like proteins are found in a variety of organisms, including plants, worms, flies, and mammals, suggesting that Tub may have an evolutionarily conserved biological function.
Tub's hydrophilic character and lack of signal sequence or transmembrane domains suggest that Tub is an intracellular protein. Our data showing that the retinal degeneration resulting from loss of Tub function is due to apoptosis suggest that Tub may play a role in the intracellular pathways involved in the survival of neurosensory cells. We have previously demonstrated that the Tub protein can be phosphorylated on tyrosine in response to insulin (10). A better understanding of Tub function awaits further elucidation of the biochemical properties of the protein and the pathways in which it acts.
ACKNOWLEDGMENTS
We thank Rene Devos and colleagues at Roche-Gent Research Institute for the anti-Tub sera. We thank Lou Tartaglia, Jim Lillie, Frank Lee, and Bob Tepper for helpful discussions and advice, Suzy Dembsky for help with tissue dissection, Pei Ge and Michael Donovan for pathology, and Tanya Macek for genotyping. We are also grateful to all the members of Millennium's Metabolic Diseases Biology team for helpful and stimulating discussions.
This work was partly supported by Hoffmann-La Roche, Inc.
REFERENCES
- 1.Banerjee P, Kleyn P W, Knowles J A, Lewis C A, Ross B M, Parano E, Kovats S G, Lee J J, Penchaszadeh G K, Ott J, Jacobson S G, Gilliam T C. TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nat Genet. 1998;18:177–179. doi: 10.1038/ng0298-177. [DOI] [PubMed] [Google Scholar]
- 2.Bradley A. Production and analysis of chimaeric mice. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, England: IRL; 1987. pp. 113–115. [Google Scholar]
- 3.Bray G A, Inoue S, Nishizawa Y. Hypothalamic obesity. The autonomic hypothesis and the lateral hypothalamus. Diabetologia. 1981;20:366–377. doi: 10.1007/BF00254505. [DOI] [PubMed] [Google Scholar]
- 4.Coleman D L. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia. 1978;14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- 5.Coleman D L, Eicher E M. Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered. 1990;81:424–427. doi: 10.1093/oxfordjournals.jhered.a111019. [DOI] [PubMed] [Google Scholar]
- 6.Cousin B, Casteilla L, Lafontan M, Ambid L, Langin D, Berthault M F, Penicaud L. Local sympathetic denervation of white adipose tissue in rats induces preadipocyte proliferation without noticeable changes in metabolism. Endocrinology. 1993;133:2255–2262. doi: 10.1210/endo.133.5.8404678. [DOI] [PubMed] [Google Scholar]
- 7.Hagstrom S A, North M A, Nishina P L, Berson E L, Dryja T P. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18:174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 8.Huszar D, Lynch C A, Fairchild-Huntress V, Dunmore J H, Fang Q, Berkemeier L R, Gu W, Kesterson R A, Boston B A, Cone R D, Smith F J, Campfield L A, Burn P, Lee F. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda S, He W, Ikeda A, Naggert J K, North M A, Nishina P M. Cell-specific expression of tubby gene family members (tub, Tulp1,2, and 3) in the retina. Investig Ophthalmol Vis Sci. 1999;40:2706–2712. [PubMed] [Google Scholar]
- 10.Kapeller R, Moriarty A, Strauss A, Stubdal H, Theriault K, Siebert E, Chickering T, Morgenstern J P, Tartaglia L A, Lillie J. Tyrosine phosphorylation of Tub and its association with Src homology 2 domain-containing proteins implicate Tub in intracellular signaling by insulin. J Biol Chem. 1999;275:24980–24986. doi: 10.1074/jbc.274.35.24980. [DOI] [PubMed] [Google Scholar]
- 11.Kleyn P W, Fan W, Kovats S G, Lee J J, Pulido J C, Wu Y, Berkemeier L R, Misumi D J, Holmgren L, Charlat O, Woolf E A, Tayber O, Brody T, Shu P, Hawkins F, Kennedy B, Baldini L, Ebeling C, Alperin G D, Deeds J, Lakey N D, Culpepper J, Chen H, Glucksmann-Kuis M A, Moore K J, et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 12.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBurney M W, Sutherland L C, Adra C N, Leclair B, Rudnicki M A, Jardine K. The mouse Pgk-1 gene promoter contains an upstream activator sequence. Nucleic Acids Res. 1991;19:5755–5761. doi: 10.1093/nar/19.20.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noben-Trauth K, Naggert J K, North M A, Nishina P M. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- 15.North M A, Naggert J K, Yan Y, Noben-Trauth K, Nishina P M. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci USA. 1997;94:3128–3133. doi: 10.1073/pnas.94.7.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohlemiller K K, Hughes R M, Lett J M, Ogilvie J M, Speck J D, Wright J S, Faddis B T. Progression of cochlear and retinal degeneration in the tubby (rd5) mouse. Audiol Neurootol. 1997;2:175–185. doi: 10.1159/000259242. [DOI] [PubMed] [Google Scholar]
- 17.Ohlemiller K K, Hughes R M, Mosinger-Ogilvie J, Speck J D, Grosof D H, Silverman M S. Cochlear and retinal degeneration in the tubby mouse. Neuroreport. 1995;6:845–849. doi: 10.1097/00001756-199504190-00005. [DOI] [PubMed] [Google Scholar]
- 18.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, England: IRL; 1987. pp. 71–112. [Google Scholar]
- 19.Sahly I, Gogat K, Kobetz A, Marchant D, Menasche M, Castel M, Revah F, Dufier J, Guerre-Millo M, Abitbol M M. Prominent neuronal-specific tub gene expression in cellular targets of tubby mice mutation. Hum Mol Genet. 1998;7:1437–1447. doi: 10.1093/hmg/7.9.1437. [DOI] [PubMed] [Google Scholar]