Abstract
Background
It was reported that long-noncoding RNAs (lncRNAs) had been identified as a novel class of regulators related to various cancers. RPARP-AS1, a differentially-expressed gene, was found in analysis of the gene expression profile of CRC from GEO database. However, its function has not been clear.
Methods
RPARP-AS1 expression was determined by qPCR and Startbase3 analysis. Knockdown of RPARP-AS1 in CRC cell lines was performed by RNAi technology, named si-RPARP-AS1 HCT116 and si-RPARP-AS1 LoVo. Cell proliferation was examined by CCK8 and colony formation assay. RNA pull-down and Luciferase reporter assay were performed to confirm the interaction between RPARP-AS1 and miR-125a-5p.
Results
In the study, we found that the expression of RPARP-AS1 was significantly up-regulated in CRC tissues and multiple CRC cell lines, which was closely related to poor prognosis of CRC patients. Loss-of-function studies indicated that knockdown of RPARP-AS1 inhibited CRC cell proliferation, migration and invasion in HCT116 and LoVo cell lines. Results of research on the mechanisms showed that RPARP-AS1 functioned as a competitive endogenous RNA (ceRNA) to sponge miR-125a-5p, therefore promoting CRC procession.
Conclusion
In summary, these results indicated that RPARP-AS1/miR-125a-5p axis played a positive role in promoting cell proliferation, migration and invasion in CC. It may be as a biomarker used to evaluate CRC prognosis.
Keywords: RPARP-AS1, miR-125a-5p, colon cancer, lncRNA, proliferation, migration, invasion
Introduction
Colon cancer is the fourth most commonly diagnosed cancer worldwide, with more than 600,000 deaths estimated each year.1 The incidence rate of CRC in city area is far higher than that in rural.2 At the time of diagnosis, most of the patients were in advanced stage.3 According to the latest statistics in 2015, the incidence rate of CRC in China accounted for fourth of all cancers, and the mortality rate was fifth, which showed an increasing trend.4 The incidence rate of CRC accounts for fourth of all cancers in the United States, while the mortality rate is second. From 2008 to 2011, incidence rate of CRC dropped by at 4% or even higher.5 The treatment of CRC mainly includes surgery, chemotherapy, radiotherapy, targeted therapy and immunotherapy.6 Targeted therapy is known for its safety and efficacy, which can improve the survival time and quality of life of patients.6 However, due to the existence of KRAS, NRAS, BRAF or PIK3CA mutations resulting in persistent activation, some patients cannot benefit from targeted therapy.7 Therefore, it is very important to study the underlying molecular mechanism of CRC and develop new targeting sites and drugs.
Long noncoding RNA (lncRNA) transcripts are between 200 nt and 100 kb in length.8 They do not encode or rarely encode proteins, and have become the focus of scientific research in recent years. The analysis of human genome revealed that lncRNA content was huge, accounting for about 4–9%.9 They play various biological roles by regulating transcription, epigenetics and post-transcriptional translation. The role of lncRNA in the development of disease has been gradually revealed, including cancer, diabetes and nervous system disease. For example, lncRNAs, such as TNXA, CTA-134P22.2, CTC-276P9.1, KRT19p3 and H19 were identified in bladder cancer and adjacent tissues.10 H19 could activate Wnt/β-Catenin and down-regulate E-cadherin by binding with EZH2, thus promoting the metastasis of bladder cancer cells.11 lncRNA plays a more significant role in the prevention, diagnosis and treatment of CC. Related studies have classified lncRNAs in the course of CC, including PVT-1, MALAT-1, ncRNA, CCAT1-l and PRNCR1. lncRNA HNF1A-AS1 was highly expressed in CC, and promoted cell proliferation through miR-34a/SIRT1/TP53 and Wnt signal transduction pathways. In addition, HNF1A-AS1 was associated with lymph node metastasis.12
In a variety of tumors, lncRNA, miRNA and functional gene jointly construct a complex regulatory network to regulate the proliferation, migration, invasion, apoptosis and drug response of tumor cells. In the last few decades, a lot of research had proved that lncRNAs were involved in gene expression network by sponging miRNAs, regulating translation, modifying histone and chromatin, and so on.13,14 In the study, we found a differentially-expressed gene, RPARP-AS1, in analysis of the gene expression profile of CRC from GEO database which was positively correlated to poor prognosis of CC. RPARP-AS1 could sponge miR-125a-5p to decrease its abundance in cytoplasm which promoted tumor cell proliferation, migration and invasion.
Methods
Patients and Specimens
Thirty patients with CRC in the Affiliated Hospital of North Sichuan Medical College were selected to obtain CRC tissues and corresponding paracancerous tissues. All of the patients had not received any preoperative treatment, including chemotherapy, radiotherapy or targeted therapy which signed informed consent in advance. The obtained specimens were frozen in liquid nitrogen immediately and then transferred to a refrigerator at −80°C for further study. The study was performed under a project license (KE-YT-2018-00) by the Ethics Committee of the Affiliated Hospital of North Sichuan Medical College, which accorded with the ethical standards formulated in the Helsinki Declaration.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted by using TRIzol reagent (Thermo Fisher Scientific, United States). The RNA was quantified by using NanoDrop ND2000 microspec-trophotometer (Thermo Fisher Scientific, United States). Four microgram of total RNA was then reversely transcribed into cDNA according to the protocol of the Reverse Transcription System (Promega, United States). Quantitative RT-PCR analysis was carried out by using the UltraSYBR Mixture kit (CWBIO, China). In this study, the primers used were synthesized by Sangon Biotech (China). The expression levels of relevant genes were determined by the ΔΔCT method.
Luciferase Reporter Assay
For analysis of miR-125a-5p regulated by RPAPP-AS1, the reporter plasmid was constructed by fusing RPAPP-AS1 cDNA into the 3’UTR region of Luciferase gene. Then the synthesized miR-125a-5p with the reporter plasmid were co-transfected into HCT116 and LoVo cells. The LUC activities were quantified using the Luciferase Assay System (Promega, United States) which represent the regulation of RPAPP-AS1 on miR-125a-5p.
Cell Transfection
In order to knock down RPARP-AS1 expression, three siRNA targeting RPARP-AS1 were designed, and three Lentivirus vector containing a siRNA separately were synthesized by BioMiao Biological Technology (Beijing, China), as well as the plasmid for RPARP-AS1 to enhance its expression. HCT116 and LoVo cells were transiently transfected with si-RPARP-AS1, si-NC and OE-RPARP-AS1 by using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
CCK8
One hundred microlitre cell suspension was inoculated in 96-well plate which was subsequently cultured at 37°C, 5% CO2. Add 10 µL CCK8 solution. The plate continued to be cultured for 1–4 h. Cell proliferation analysis of HCT118 and LoVo cells was performed using a Microplate reader (BioTek, United States).
Colony Formation Assay
The logarithmic cells were suspended and inoculated in dishes containing culture medium. The dishes were cultured at 37°C for 2–3 weeks until clones being observed. The clones were fixed with 4% paraformaldehyde and subsequently stained with GIMSA solution for 10–30 min. The clone formation rate was evaluated by formula “Clone formation rate=Number of Clones/Number of Cells×100%”.
Transwell
Transwell chamber was put into the culture plate. The upper and lower chambers were filled with liquid medium and solid medium separately. The upper and lower layers of culture medium were separated by a polycarbonate membrane. For the study of cell invasion experiment, EMC was added into the lower medium. The cells were suspended in serum-free medium containing BSA reaching to 5×105/mL. Then 200 μL of suspension cells were added into the upper chamber and then cultured in an incubator at 37°C, 5% CO2 for 12–48 h.
RNA Pull-Down
Biotin-labeled miR-125a-5p sense and antisense hybridization probes were obtained by transcription in vitro (Roche, Switzerland). The probes were incubated with cytoplasmic extract from HCT118 and LoVo cells to form miRNA-lncRNA compounds which were separated using magnetic beads labeled with avidin. Finally, qRT-PCR analysis determined the enrichment of RPAPP-AS1 in the immunoprecipitated RNA.
Statistical Analysis
GraphPad Prism software was used to perform Statistical analysis and generate column diagram. The statistically significant differences were evaluated by Student’s t-test (*P-value < 0.05, **P-value < 0.01, ***P-value < 0.001).
Result
RPARP-AS1 Expression Increased in Colon Cancer with Poor Prognosis
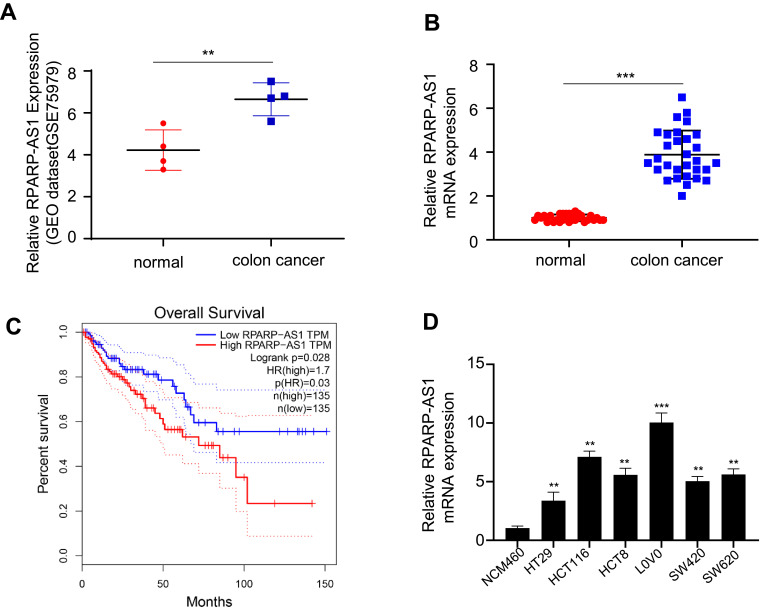
A differentially-expressed gene, RPARP-AS1, was found in analysis of the gene expression profile of CRC from GEO database. RPARP-AS1 expression was significantly increased in CRC tissues than that in the adjacent tissues (Figure 1A). To confirm this conclusion, 30 pairs of CRC and adjacent tissues were collected in which RPARP-AS1 expression was detected by qRT-PCR. As shown in Figure 1B, RPARP-AS1 expression was significantly higher in CRC tissues than that in the adjacent tissues which was consistent with Figure 1A. Further, we analyzed the relationship between RPARP-AS1 expression and CRC using data from TCGA database by GEPIA. The results demonstrated that highly-expressed RPARP-AS1 was greatly correlated with poor prognosis of CRC (Figure 1C). It aroused our interest to study the biological function of RPARP-AS1. Among all cell lines, the expression of RPARP-AS1 was highest in HCT116 and LoVo (Figure 1D), which were selected for further study.
Figure 1.
RPARP-AS1 was highly expressed in colon cancer with poor prognosis. (A) RPARP-AS1 expression in colon cancer was significantly higher than that in the adjacent tissues from GSE75970 (P-value < 0.01). (B) RPARP-AS1 expression in colon cancer than that in the adjacent tissues from clinical samples (P-value < 0.001). (C) GEPIA analysis of TCGA database found that patients with high expression of RPARP-AS1 had poor prognosis (P-value < 0.05). (D) RPARP-AS1 expression in all colon cancer cell lines (HT29, HCT116, hct8, LoVo, sw420, SW620) was higher than that in control group cell line (NCM460) (**P-value < 0.01; ***P-value < 0.001).
RPARP-AS1 Gene Silencing Inhibited Cell Proliferation
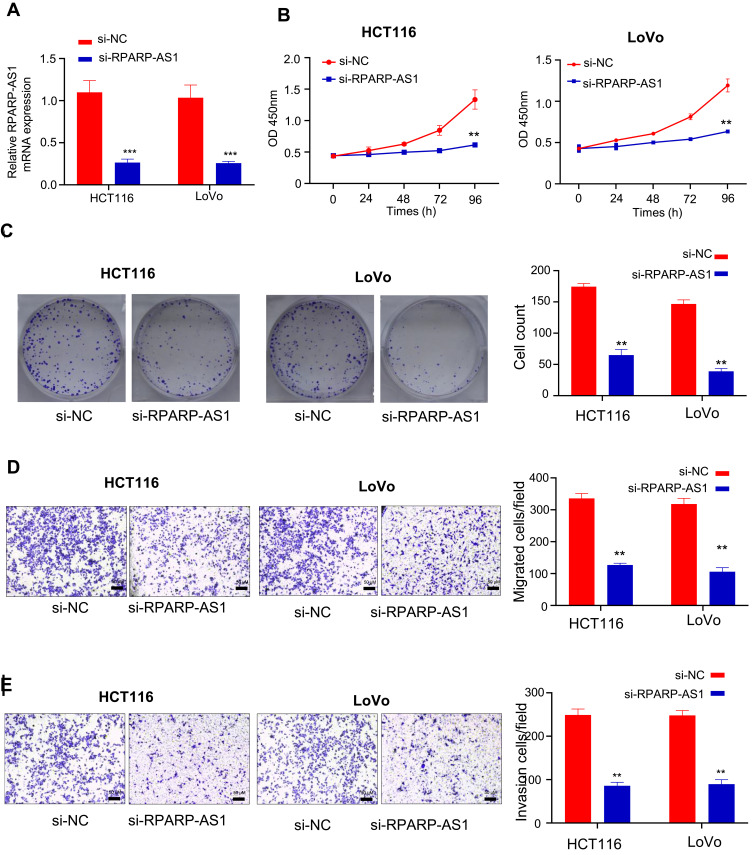
The lentivirus vectors silencing RPARP-AS1 gene expression were introduced into HCT116 and LoVo cell lines separately. As a result, RPARP-AS1 expression significantly decreased (Figure 2A), named si-RPARP-AS1 HCT116 and si-RPARP-AS1 LoVo cell lines. In the course of experiment, we designed three siRNA, in which si#1 and si#2 effectively knocked down the expression of RPARP-AS1 (Supplementary Figure 1E), the sequences as follows: si#1, 5'-AAGAAAAGUAAAUAGGCC-3'; si#2, 5'-UAAGAAAAGUAAAUAGGC-3'. We selected the vector si#1 for further study of gene function. Considering that RPARP-AS1 was associated with poor prognosis of CRC (Figure 1C), we guessed that it may be involved in promoting tumor cell proliferation, migration or invasion. Therefore, we performed CCK8, colony formation assay and TransWell to confirm the role of RPARP-AS1 in CC. After 24 h of culture, the proliferation of si-RPARP-AS1 HCT116 and LoVo cells was lower than that of normal control (si-NC) cells by CCK8. With the extension of culture time, gap was becoming greater and greater until culture for 96 h (Figure 2B). Consistent with the results of CCK8, si-RPARP-AS1 HCT116 and si-RPARP-AS1 LoVo cells formed fewer clones (Figure 2C). In addition, Transwell results showed that the migration of si-RPARP-AS1 HCT116 and LoVo cells significantly reduced than that of si-NC groups (Figure 2D), as well as the invasion of si-RPARP-AS1 cells (Figure 2E). In contrast, over-expression of RPARP-AS1 promoted the proliferation and invasion in HCT116 cells (Supplementary Figure 1A–D).
Figure 2.
RPARP-AS1 knockdown in HCT116 and LoVo cell lines inhibited cell proliferation. (A) RPARA-AS1 expression significantly decreased in both HCT116 and LoVo cell lines mediated with lentiviral vectors (P-value < 0.001). (B) CCK-8 was used to detect cell proliferation of HT29 and HCT116. Knockdown of RPARP-AS1 decreased the light absorption value of cells at 450 nm (P-value < 0.01). (C) Colony formation assay confirmed that knockdown of RPARP-AS1 decreased the colony-forming ability of HCT116 and LoVo cells (P-value < 0.01). (D) TransWell (without Matrigel) showed that knockdown of RPARP-AS1 decreased the migration ability of HCT116 and LoVo cells (P-value < 0.01). (E) TransWell (with Matrigel) showed that knockdown of RPARP-AS1 decreased the invasion ability of HCT116 and LoVo cells (P-value < 0.01). (**P-value < 0.01; ***P-value < 0.001).
RPARP-AS1 Sponged miR-125a-5p
Furthermore, we studied the mechanism of RPARP-AS1 in promoting tumor cell proliferation, migration and invasion in CC. The sub-cellular localization of RPARP-AS1 was explored by Nucleocytoplasmic separation. The results showed that RPARP-AS1 was mainly expressed in cytoplasm (Figure 3A). Combined with previous research, RPARP-AS1 may function by sponging miRNA. The potential miRNA was analyzed by Startbase3 and the binding site of miR-125a-5p was found in RPARP-AS1 (Figure 3B). RNA pull-down and Luciferase reporter assay were performed to analyze the interaction between RPARP-AS1 and miR-125a-5p. As shown in Figure 3C, miR-125a-5p probe could effectively enrich RPARP-AS1. Also, over-expression of miR-125a-5p could bind to RPARP-AS1 and inhibit the activity of Luciferase in both HCT116 and LoVo cells. Corresponding to it, miR-125a-5p expression significantly increased in si-RPARP-AS1 cells than that in si-NC groups (Figure 3D). Therefore, RPARP-AS1 sponged miR-125a-5p in CC. In line with expectations, miR-125a-5p expression was lower in CRC using 30 pairs of clinical samples (Figure 3E) and data from TCGA (Figure 3G), which was negatively correlated with RPARP-AS1 expression (Figure 3F and H).
Figure 3.
RPARP-AS1 sponged miR-125a-5p. (A) RPARP-AS1 was mainly expressed in nucleus, but not in cytoplasm (U6 and GAPDH were used as internal parameters of nucleus and cytoplasm respectively). (B) The binding site of miR-125a-5p was found in RPARP-AS1 by analyzing the startbase3 database. (C) RNA pull-down assay was performed in HT29 and HCT116 cells. miR-125a-5p probe enriched more RPARP-AS1 than NC probe (Left). Luciferase reporter experiment showed that miR-125a-5p could inhibit the activity of luciferase coupling with RPARP-AS1, but miR-NC could not in both HCT116 (Middle) and LoVo (Right) cell lines. (D) qRT-PCR was used to detect the expression of mir-125a-5p after knockdown of RPARP-AS1 in HT29 and HCT116 cells. Compared with si-NC groups, miR-125a-5p expression increased in si-RPARP-AS1 cell lines. (E) miR-125a-5p expression decreased significantly in CRC than that in paracarcinomatic tissue by qRT-PCR (n=30, P-value<0.001). (F) Consistent with those in Figure E, miR-125a-5p reduced in CRC significantly using data from TCGA by Startbase3 analysis. (G) The correlation between the expression of RPARP-AS1 and miR-125a-5p in 30 cases of CRC which was negatively correlated (P-value<0.001). (H) The expression of RPARP-AS1 and miR-125a-5p in CRC was also negatively correlated using data from TCGA by Startbase3 analysis. (**P-value < 0.01; ***P-value < 0.001).
RPARP-AS1 Regulated CRC Progression by Sponging miR-125a-5p
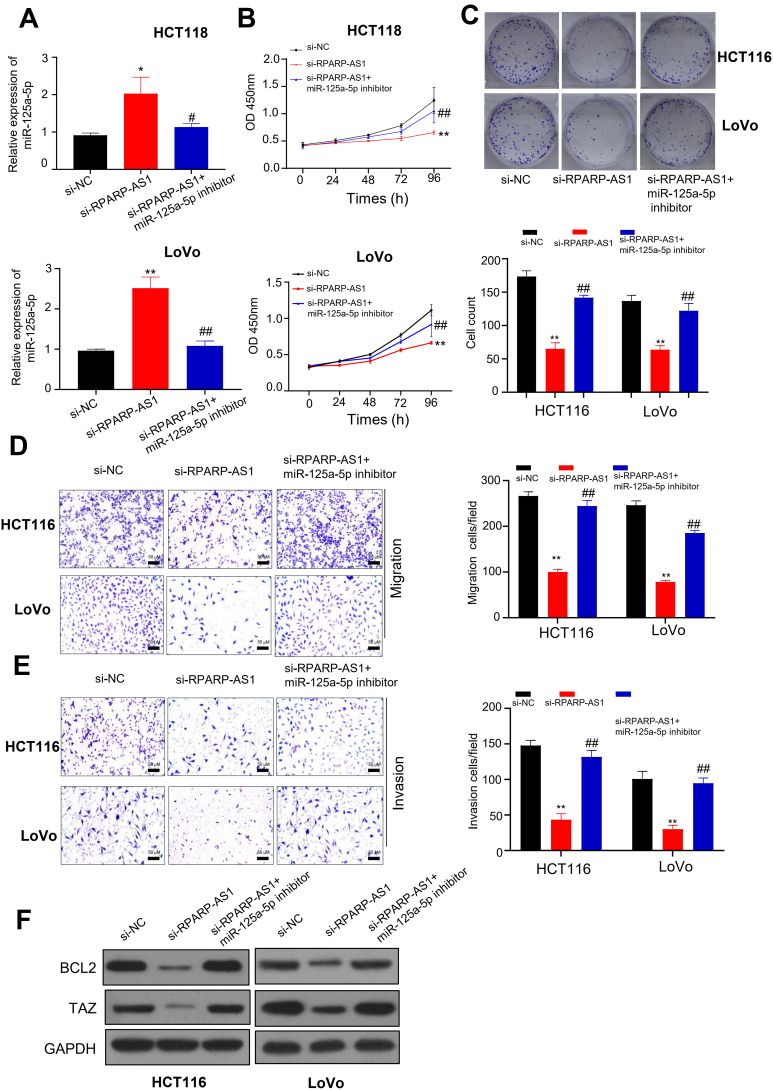
Above results proved that RPARP-AS1 could sponge miR-125a-5p directly. Next, we studied the role of RPARP-AS1/miR-125a-5p axis in CRC procession. As described in Result 2, knockdown of RPARP-AS1 decreased the proliferation, migration and invasion in HCT116 and LoVo cell lines (Figure 2). In this result, miR-125a-5p expression increased in si-RPARP-AS1 cell lines than that in si-NC groups. miR-125a-5p inhibitor was then introduced in the study which could inhibit miR-125a-5p expression (Figure 4A). In addition, miR-125a-5p inhibitor could reverse decreased tumor cell proliferation by CCK8 (Figure 4B), clone formation by colony formation assay (Figure 4C), migration (Figure 4D) and invasion (Figure 4E) by TransWell. BCL2 and TAZ were targets of miR-125a-5p in CRC. Thus, we detected the expression of BCL2 and TAZ through Western blot. As shown in Figure 4F, silencing RPARP-AS1 inhibits BCL2 and TAZ expression. However, when co-transfected with miR-125a-5p, BCL2 and TAZ expression is increased. These results indicated that RPARP-AS1 expression increased which promoting tumor cell proliferation, migration and invasion in CRC by sponging miR-125a-5p.
Figure 4.
RPARP-AS1 sponged miR-125a-5p to regulate tumor cell proliferation, migration or invasion. (A) miR-125a-5p expression increased in si-RPARP-AS1 cells which partially decreased by miR-125a-5p inhibitor in HCT116 and LoVo cell lines. (B) The proliferation of si-RPARP-AS1 cells decreased than that of si-NC groups which partially increased by miR-125a-5p inhibitor in HCT116 and LoVo cell lines. (C) Colony formation assay showed that siRPAPR-AS1 cells formed fewer clones than si-NC cells which partially increased by miR-125a-5p inhibitor in HCT116 and LoVo cell lines. (D) TransWell (without Matrigel) showed that knockdown of RPARP-AS1 decreased the migration ability of HCT116 and LoVo cells which was partially recovered by miR-125a-5p inhibitor. (E) TransWell (with Matrigel) showed that knockdown of RPARP-AS1 decreased the invasion ability of HCT116 and LoVo cells which partially increased by miR-125a-5p inhibitor. (F) Western blot showed that knockdown of RPARP-AS1 decreased the expression of BCL2 and TAZ which partially increased by miR-125a-5p. (*P-value < 0.05, **P-value < 0.01, compared with si-NC; #P-value < 0.05, ##P-value < 0.01, compared with si-RPARP-AS1).
Discussion
lncRNAs are a class of RNA molecules with a length of more than 200 nucleotides.8 They do not participate in protein coding, but directly regulate various epigenetics in the form of RNA and participate in transcription and post-transcriptional protein coding. In CRC, lncRNA expression is closely related to the occurrence and development of CRC. In 2011, Graham et al15 found that CRNDE-h was up-regulated in plasma and tissues of CRC patients. Ellis et al16 found that CRNDE affected insulin/insulin-like growth factor (IGF) on regulation of glucose metabolism and lactate secretion through PI3K/Akt/mTOR and Raf/MAPK pathways, whose expression was up-regulated in CRC cells. Dai et al17 further confirmed that CRNDE was highly expressed in tissues, serum and cell lines of CRC patients. It was suggested that CRNDE could activate or inhibit Ras/MAPK and Wnt/β-catenin signaling pathways through competitive binding of miRNA, thus regulating the proliferation, invasion and apoptosis of CRC cells. Therefore, CRNDE might be used as one of the markers for early diagnosis of CRC.18 In 2010, Tsang et al10,19 found that over-expression of miR-675 promoted the growth of CRC cells, indicating that H19 might be related to the occurrence of CRC. Liang et al20 found that H19 was highly expressed in mesenchymal-like carcinoma cells and primary CRC, and the stable expression of H19 could promote EMT and accelerate tumor growth in vivo and in vitro. Yang et al21 found that H19, as a competitive endogenous RNA (ceRNA) of miR-138 and miR-200a, regulated the expression of multiple genes involved in EMT and accelerates the growth of CRC cells. Schwarzenbach22 found the effect of H19 on RB1-E2F pathway and Cdk8-β-Catenin signal transduction. It is speculated that H19, which is highly expressed in primary CRC, can be detected in blood and may be a potential biomarker for diagnosis or prognosis of CRC. Therefore, lncRNA plays an important role in the diagnosis, prognosis, progress and treatment of CRC. In this study, we found a differentially expressed gene, RPARP-AS1, by analyzing the CRC gene expression profile. Consistent with the previous results, RPARP-AS1 was highly expressed in CRC (Figure 1).
In the study, we demonstrated that RPARP-AS1 could promote the proliferation, migration and invasion of tumor cells through sponging mir-125a-5p (Figure 3). Previous studies have shown that mir-125a-5p is downregulated and played a negative role in tumorigenesis and development including colorectal cancer.23–26 miR-125a-5p could inhibit the growth, invasion and metastasis of gastric cancer, breast cancer, liver cancer, lung cancer and CRC.27 Consistent with these reports, we also found downregulation of miR-125a-5p in CC. As a tumor suppressor, over-expression of miR-125a-5p inhibited proliferation and induces apoptosis in CC through targeting BCL2, BCL2L12, MCL-128 and TAZ,29 which are known as tumor suppressors in CC. In our study, silencing RPARP-AS1 could inhibits BCL2 and TAZ expression, which could be reversed by co-transfected with miR-125a-5p inhibitor. Therefore, RPARP-AS1/miR-125a-5p may be involved in promoting CRC by regulating the expression of BCL2, BCL2L12, MCL-1 and TAZ genes.
In conclusion, our results demonstrate that RPARP-AS1 is highly expressed in CC, which can promote the proliferation, migration and invasion of CRC cells through sponging tumor suppressor miR-125a-5p.
Acknowledgments
Yongjun Ren and Caixia Zhao are co-first authors for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Argiles G, Tabernero J, Labianca R, et al. Localised colon cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31(10):1291–1305. doi: 10.1016/j.annonc.2020.06.022 [DOI] [PubMed] [Google Scholar]
- 2.Jahanafrooz Z, Mosafer J, Akbari M, et al. Colon cancer therapy by focusing on colon cancer stem cells and their tumor microenvironment. J Cell Physiol. 2020;235(5):4153–4166. doi: 10.1002/jcp.29337 [DOI] [PubMed] [Google Scholar]
- 3.Auclin E, Zaanan A, Vernerey D, et al. Subgroups and prognostication in stage III colon cancer: future perspectives for adjuvant therapy. Ann Oncol. 2017;28(5):958–968. doi: 10.1093/annonc/mdx030 [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 5.Benson AB 3rd, Venook AP, Cederquist L, et al. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(3):370–398. doi: 10.6004/jnccn.2017.0036 [DOI] [PubMed] [Google Scholar]
- 6.Ruan H, Leibowitz BJ, Zhang L, et al. Immunogenic cell death in colon cancer prevention and therapy. Mol Carcinog. 2020;59(7):783–793. doi: 10.1002/mc.23183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu H-C, Thiam TK, Lu Y-J, et al. Mutations of KRAS/NRAS/BRAF predict cetuximab resistance in metastatic colorectal cancer patients. Oncotarget. 2016;7(16):22257–22270. doi: 10.18632/oncotarget.8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Yu B, Jin Q, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA RP11-159K7.2/miR-206/DNMT3A axis. J Cell Mol Med. 2020;24(12):6781–6795. doi: 10.1111/jcmm.15331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranzani V, Rossetti G, Panzeri I, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16(3):318–325. doi: 10.1038/ni.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y-P, Bian X-J, Ye D-W, et al. Long noncoding RNA expression signatures of bladder cancer revealed by microarray. Oncol Lett. 2014;7(4):1197–1202. doi: 10.3892/ol.2014.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo M, Li Z, Wang W, et al. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033 [DOI] [PubMed] [Google Scholar]
- 12.Fang C, Qiu S, Sun F, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012 [DOI] [PubMed] [Google Scholar]
- 13.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–565. doi: 10.1101/gr.6036807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham LD, Pedersen SK, Brown GS, et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2(8):829–840. doi: 10.1177/1947601911431081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843(2):50–62. doi: 10.1016/j.bbamcr.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 17.Dai M, Li S, Qin X. Colorectal neoplasia differentially expressed: a long noncoding RNA with an imperative role in cancer. Onco Targets Ther. 2018;11:3755–3763. doi: 10.2147/OTT.S162754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hongzhen Z, Yanyu L, Xuexiang L, et al. The diagnostic and prognostic significance of long non-coding RNA CRNDE in pan-cancer based on TCGA, GEO and comprehensive meta–analysis. Pathol Res Pract. 2019;215(2):256–264. doi: 10.1016/j.prp.2018.11.008 [DOI] [PubMed] [Google Scholar]
- 19.Tsang WP, Ng EKO, Ng SSM, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. doi: 10.1093/carcin/bgp181 [DOI] [PubMed] [Google Scholar]
- 20.Liang W-C, Fu W-M, Wong C-W, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi: 10.18632/oncotarget.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Ning N, Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing beta-catenin expression in colorectal cancer. Biomed Res Int. 2017;2017:2767484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarzenbach H. Biological and clinical relevance of H19 in colorectal cancer patients. EBioMedicine. 2016;13:9–10. doi: 10.1016/j.ebiom.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Ma Y, Liu C, et al. Reduced miR-125a-5p level in non-small-cell lung cancer is associated with tumour progression. Open Biol. 2018;8(10):180118. doi: 10.1098/rsob.180118 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Cai M, Chen Q, Shen J, et al. Retracted: epigenetic silenced miR-125a-5p could be self-activated through targeting Suv39H1 in gastric cancer. J Cell Mol Med. 2018;22(10):4721–4731. doi: 10.1111/jcmm.13716 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Tang L, Zhou L, Wu S, et al. MiR-125a-5p inhibits colorectal cancer cell epithelial–mesenchymal transition, invasion and migration by targeting TAZ. Onco Targets Ther. 2019;12:3481–3489. doi: 10.2147/OTT.S191247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L-B, Feng L, He J, et al. MiR-125a-5p inhibits the proliferation and invasion of breast cancer cells and induces apoptosis by targeting GAB2. Math Biosci Eng. 2019;16(6):6923–6933. doi: 10.3934/mbe.2019347 [DOI] [PubMed] [Google Scholar]
- 27.Naidu S, Shi L, Magee P, et al. PDGFR-modulated miR-23b cluster and miR-125a-5p suppress lung tumorigenesis by targeting multiple components of KRAS and NF-kB pathways. Sci Rep. 2017;7(1):15441. doi: 10.1038/s41598-017-14843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong Z, Liu N, Lin L, et al. MiR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed Pharmacother. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036 [DOI] [PubMed] [Google Scholar]
- 29.Yang M, Tang X, Wang Z, et al. miR-125 inhibits colorectal cancer proliferation and invasion by targeting TAZ. Biosci Rep. 2019;39(12):BSR20190193. [DOI] [PMC free article] [PubMed] [Google Scholar]