Abstract
Guide RNAs (gRNAs) are small RNAs that provide specificity for uridine addition and deletion during mRNA editing in trypanosomes. Terminal uridylyl transferase (TUTase) adds uridines to pre-mRNAs during RNA editing and adds a poly(U) tail to the 3′ end of gRNAs. The poly(U) tail may stabilize the association of gRNAs with cognate mRNA during editing. Both TUTase and gRNAs associate with two ribonucleoprotein complexes, I (19S) and II (35S to 40S). Complex II is believed to be the fully assembled active editing complex, since it contains pre-edited mRNA and enzymes thought necessary for editing. Purification of TUTase from mitochondrial extracts resulted in the identification of two chromatographically distinct TUTase activities. Stable single-uridine addition to different substrate RNAs is performed by the 19S complex, despite the presence of a uridine-specific 3′ exonuclease within this complex. Multiple uridines are added to substrate RNAs by a 10S particle that may be an unstable subunit of complex I lacking the uridine-specific 3′ exonuclease. Multiple uridines could be stably added onto gRNAs by complex I when the cognate mRNA is present. We propose a model in which the purine-rich region of the cognate mRNA protects the uridine tail from a uridine exonuclease activity that is present within the complex. To test this model, we have mutated the purine-rich region of the pre-mRNA to abolish base-pairing interaction with the poly(U) tail of the gRNA. This RNA fails to protect the uridine tail of the gRNA from exoribonucleolytic trimming and is consistent with a role for the purine-rich region of the mRNA in gRNA maturation.
During kinetoplastid RNA editing, uridine residues are posttranscriptionally added to or removed from mitochondrial pre-mRNAs. Editing occurs at specific sites in the mRNA to produce mature mRNA coding sequences (for recent reviews, see references 3, 15, and 26). Current in vitro evidence for kinetoplastid RNA insertional editing supports the multistep enzymatic process first proposed by Blum et al. (4, 7, 9, 11, 16, 25). First, an editing site-specific endonuclease recognizes and cleaves the pre-mRNA. Second, a terminal uridylyl transferase (TUTase) adds uridine (U) residues to the 3′ terminus of the 5′ cleavage fragment, and then an RNA ligase joins the cleaved halves of the mRNA together, completing one round of insertional editing. In kinetoplastid RNA deletional editing, the pre-mRNA contains U residues that must be removed by a U-specific exonuclease to form a mature mRNA coding sequence (9). In this model of RNA editing, both the TUTase and the U-specific exonuclease operate at the same step to help ensure that the proper number of U residues are added or deleted.
The editing site-specific endonuclease, TUTase, and RNA ligase activities have been shown to cosediment in glycerol gradients, suggesting that these activities may be part of high-molecular-weight complexes (1, 8, 16, 18, 20, 22, 25). In Trypanosoma brucei, two complexes are proposed to be involved in RNA editing (20). Complex I is a ribonucleoprotein (RNP) that sediments at 19S and consists of guide RNA (gRNA), TUTase, RNA ligase, and an editing site-specific endonuclease. Complex II is also an RNP but sediments at 35S to 40S and contains pre-mRNA in addition to gRNA, TUTase, RNA ligase, and an editing site-specific endonuclease. Because complex II contains pre-edited mRNA, it is likely to be the fully assembled active editing complex.
gRNAs are key molecules in the editing process. The specificity of U addition and/or deletion is directed by these small transcripts, which can fully base pair with the edited mRNA. Although there are hundreds of different gRNA sequences, gRNAs have some conserved features. For example, the 5′ ends of all gRNAs contain a 4- to 14-nucleotide anchor sequence, which is proposed to initiate the RNA editing reaction by base pairing with the pre-edited mRNA immediately 3′ of the editing site (5, 25). Also, gRNAs contain an internal 30- to 40-nucleotide sequence that is complementary to the edited mRNA and provides the information for correct editing of the mRNA (4). gRNAs undergo posttranscriptional addition of U residues at the 3′ terminus (6). The poly(U) tail length varies between about 5 and 24 U residues and is critical for efficient in vitro editing reactions to occur (16, 25). Although the precise function of the U tail is unknown, it has been proposed to assist in RNA editing by forming a duplex with the purine-rich regions of the mRNA commonly found at the pre-edited sites (6). In this way, the U tail could act as a tether and stabilize the 5′ pre-mRNA fragment after endonuclease cleavage. Recently, RNA-RNA cross-linking studies have shown that the U tail may interact with the pre-mRNA, preferring the purine-rich sites close to the first few editing sites (17). This idea is attractive, since this base pairing would increase the stability of what is initially a weak interaction between the anchor duplex of the gRNA and the pre-mRNA.
MATERIALS AND METHODS
Materials.
Nucleotide triphosphates (NTPs) were purchased from Sigma, and radioactively labeled nucleotides were from DuPont New England Nuclear. Homopolymers and full-length gRNAs were synthesized on an Applied Biosystems 392 DNA/RNA synthesizer. Bacterial alkaline phosphatase, T7 polymerase, Escherichia coli poly(A) polymerase, and T4 polynucleotide kinase were purchased from Bethesda Research Laboratories.
Isolation of mitochondria and glycerol gradient sedimentation.
Procyclic T. brucei TREU 667 was grown at 26°C in Cunningham medium supplemented with 10% heat-inactivated fetal bovine serum and gentamicin sulfate (25 μg/ml) (10). Cells were harvested when they reached a density of 1 × 107 to 1.5 × 107 cells/ml. Mitochondria were isolated as described by Rohrer et al. (21). Mitochondrial extract was prepared and sedimented on a 10 to 30% glycerol gradient as described by Pollard et al. (20).
Preparation of RNAs.
RNA primers (50 pmol) without a 5′ phosphate were 5′ 32P labeled with T4 polynucleotide kinase in accordance with manufacturer recommendations. For 3′-end labeling, RNA primers (50 pmol) were extended by a single nucleotide in a 10-μl reaction mixture containing 1.74 U of poly(A) polymerase, 60 mM KCl, 20 mM Tris (pH 7.8), 5 mM magnesium chloride, 10 mM manganese acetate, and 66 pmol of [α-32P]NTP. All RNAs were gel purified on preparative denaturing 8% polyacrylamide sequencing gels. The sequences of the gRNAs used are in the gRNA sequence database (27). For gA6-[14] and gA6-[48], the 3′-most terminal U was not included, unless otherwise designated. A6U2 and CybΔ5′ mRNA sequences were transcribed in vitro as previously described (12, 16).
Enzyme assays.
The standard TUTase assay was performed in an 18-μl reaction mixture which consisted of 6 μl of a partially purified 19S glycerol gradient fraction or 1 μl of the purified complex, 50 mM KCl, 20 mM Tris (pH 7.9), 5 mM magnesium chloride, 1 mM UTP, and 1 pmol of RNA substrate (approximately 50,000 cpm). This reaction mixture was incubated at room temperature for 90 min. Acid precipitation assays for TUTase were performed as previously described (20). U-specific exonuclease assays were performed essentially the same way as the standard TUTase assay, but UTP was omitted. Ligase adenylation was performed as previously described (23).
gRNA U-tailing experiments used 1 pmol of a synthetic gRNA for ATPase subunit 6 mRNA editing (gA6-[14] with no U tail) with or without 5 pmol of cognate ATPase subunit 6 pre-mRNA (A6U2) (16) or control cytochrome b pre-mRNA (CybΔ5′) (12) per assay. When nucleotides were added, the reaction mixtures contained the designated NTP at 1 mM. RNAs were recovered by ethanol precipitation as described above. Products were separated on a denaturing 8% polyacrylamide sequencing gel containing 7 M urea and visualized by autoradiography. Quantitation was performed on an Applied Biosystems Storm phosphorimager.
Purification.
Purifications were performed with 20 liters of T. brucei (approximately 1.5 × 107 cells/liter). Mitochondrial extract was prepared as described previously (20). Extract was cleared by sedimentation (12,000 × g, 10 min, 4°C) prior to running on a heparin-Sepharose column (5-ml packed volume; Pharmacia). The column was washed with buffer A (20 mM Tris [pH 7.9], 100 mM KCl, 1 mM EDTA). Fractions were eluted with a salt gradient (100 to 600 mM KCl). Fractions active for single-U addition and ligase (eluting at approximately 300 mM KCl) were then pooled and diluted with buffer A to reduce the salt concentration to below 100 mM KCl. This material was then loaded onto a Q-Sepharose column (1-ml packed volume; Pharmacia). After washing with 150 mM KCl, 1-ml fractions were collected from a 150 to 600 mM KCl 16-ml gradient. Fractions were assayed, and activities were found to elute at approximately 200 mM KCl. Active fractions were pooled (2 ml), and proteins were separated on a 10 to 30% glycerol gradient as previously described (20) except that MgCl was omitted.
Metabolic labeling and isolation of mitochondria.
Metabolic pulse-labeling of mitochondrial vesicles was performed as previously described (13). Metabolically labeled mitochondria were solubilized with Triton X-100, and radiolabeled complexes were separated on a glycerol gradient (20). Further separation of labeled complexes was performed on native 3 to 20% acrylamide gels. Labeled RNAs from glycerol gradient fractions were isolated by phenol-chloroform extraction and then analyzed on a denaturing 6% polyacrylamide–7 M urea gel. Pulse-chase experiments were conducted as for the metabolic labeling experiments, except that excess unlabeled UTP (5 mM) was added at the designated time points.
RESULTS
TUTase activity sediments with 19S and 35S-to-40S complexes.
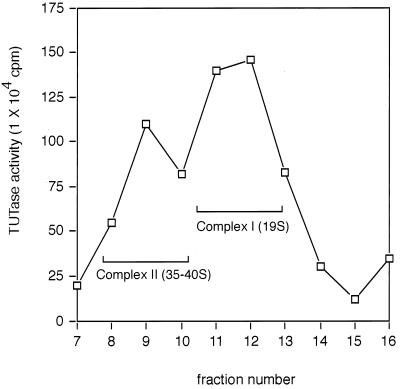
In order to examine TUTase activity associated with editing complexes, mitochondrial lysates were fractionated on linear 10 to 30% glycerol gradients (20). TUTase activity within each fraction was assayed by measuring the incorporation of [α32P]UTP into trichloroacetic acid-precipitable RNA (Fig. 1). The major peak of TUTase activity sedimented at 19S, consistent with its being part of previously defined editing complex I (19S) (20). A second, smaller peak consistently sedimented at 35S to 40S, suggesting that TUTase activity is also present in complex II. Gradient fractions containing particles larger than 40S had no significant TUTase activity.
FIG. 1.
TUTase activity sediments with RNP complexes. Mitochondrial extract was sedimented on a 10 to 30% glycerol gradient and fractionated into 16 fractions, and TUTase activity was measured by monitoring the incorporation of [α32P]UTP into yeast total tRNA by using the trichloroacetic acid assay as described in Materials and Methods. Gradient fractions containing complexes I (19S) and II (35S to 40S) are indicated.
gRNA poly(U) tail length in complex I and II RNPs.
The next experiments examined the posttranscriptional addition of UTP into endogenous gRNAs associated with complexes I and II. Isolated mitochondria were incubated in the presence of [α-32P]UTP under conditions that suppress transcription. Previous work has shown that such conditions produce radiolabeled editing complexes due to incorporation of [α-32P]UTP into RNAs by TUTase (19, 22). The identities of various labeled RNAs have previously been shown by hybridization experiments (13). Mitochondrial mRNAs incorporate UTP because of RNA editing, while 9S and 12S rRNAs receive long U tails (2), as do gRNAs (6).
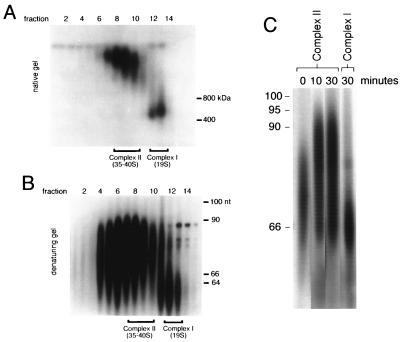
Pulse-labeled mitochondria were detergent solubilized, and editing complexes were separated by centrifugation on glycerol gradients. Aliquots from each fraction were run on nondenaturing polyacrylamide gradient gels, and native RNPs were identified by autoradiography (Fig. 2A). Two major RNA-containing complexes were identified in fractions corresponding to 19S complex I and 35S to 40S complex II. To examine the sizes of the gRNAs in the mitochondrial RNPs, RNA from each fraction was isolated and resolved on denaturing gels (Fig. 2B). The average sizes of the gRNAs present in these two editing complexes differed by 15 nucleotides.
FIG. 2.
Posttranscriptional labeling of isolated mitochondria. Isolated mitochondria were pulse-labeled with [α32P]UTP under conditions that arrest transcription but support polyuridylation of gRNAs. Even numbers at the top of both panels A and B represent glycerol gradient fractions. (A) Native-gel separation of pulse-labeled mitochondrial RNPs fractionated on a 10 to 30% glycerol gradient. Fractions containing complexes I and II are indicated. Protein molecular size markers are indicated in kilodaltons. (B) Analysis of labeled gRNAs present in complexes I and II from the posttranscriptional pulse-labeling experiments on denaturing sequencing gel. Fractions containing complexes I and II are indicated. nt, nucleotides. (C) Time course of pulse-chase-labeled complex II gRNAs shown in panel B. Isolated mitochondria were pulsed with [α32P]UTP for 3 min and then incubated with excess unlabeled UTP for the times shown (for details, see Materials and Methods). DNA size standards are indicated in nucleotides.
Pulse-chase experiments also determined the overall kinetics of UTP incorporation into gRNAs. For these studies, an excess of unlabeled UTP was added after a 3-min [α-32P]UTP pulse. Mitochondria were incubated for another 10 or 30 min (Fig. 2C), and then the RNAs were recovered and analyzed on a denaturing gel. The length of complex II-associated gRNAs increased by approximately 15 nucleotides between the 3-min pulse and the 10-min chase time. Longer chase times of up to 30 min did not radically increase the size (Fig. 2C, 30 min). The length of the complex I-associated gRNAs remained short during the course of the pulse-chase experiments (Fig. 2C, compare complexes II and I at 30 min). The size distribution of the radiolabeled gRNAs in complex I is consistent with primary gRNA transcripts lacking a 3′ poly(U) tail. Larger gRNA transcripts, which likely have posttranscriptionally added poly(U) tails of ∼15 to 20 nucleotides, are enriched within editing complex II RNPs. These results suggest that factors within complex II can influence the formation or stability of the 3′ poly(U) tail on gRNAs.
TUTase reactivity on different RNA substrates.
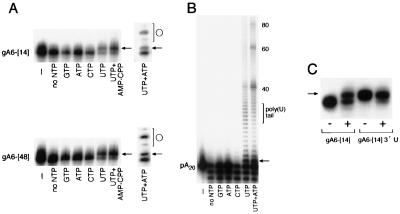
We have examined nucleotide addition onto different RNAs by glycerol gradient-purified complex I TUTase. Complex I-associated TUTase activity was assayed with 5′- or 3′-end labeled gRNAs and poly(A) RNA in the presence of various nucleotides. This allowed us to evaluate both nucleotide specificity and RNA preferences. The ability of TUTase to add U residues to three full-length gRNAs (gA6-[14], gA6-[48], and gCyb-[558]) was examined. These gRNAs were 5′ end labeled and reacted with TUTase in the presence of GTP, ATP, CTP, UTP, or UTP plus AMP-CPP. AMP-CPP is an ATP analogue that contains a nonhydrolyzable α-β phosphate bond and is an inhibitor of RNA ligase (23). gRNAs gA6-[14] and gA6-[48] were excellent substrates for the addition of a single nucleotide (n+1) but not for U tail formation (Fig. 3A). Single-U addition to gCyb[558] was also observed (data not shown). A long exposure of the gel shown in Fig. 3A revealed a small amount of product which resulted from the addition of multiple U residues; however, the major product consisted of gRNAs containing one added U. This result was consistent among different preparations, and often over 60% of the gRNA could be converted to the n+1 product. The presence of AMP-CPP did not affect the formation of this n+1 product, and the addition of 1 mM ATP to UTP-containing reaction mixtures did not promote any further gRNA elongation. However, the addition of both UTP and ATP promoted the self-ligation of the gRNA to form circular RNAs (Fig. 3A, ATP+UTP lane). Consistent with its being a circular RNA, this product migrated at different positions on different-percentage polyacrylamide gels (data not shown). The circularization of T. brucei RNAs, including gRNAs, by RNA ligase has previously been described by other groups (22, 28).
FIG. 3.
U addition to different RNA substrates using glycerol gradient-purified complex I. (A) Predominately one stable U is added to gRNAs. 5′-end-labeled gA6-[14] and gA6-[48] were incubated with the designated NTP at 1 mM. Arrows point to the single-U addition product, and circles designate circularization of the gRNA by RNA ligase present in the extract. A minus sign indicates buffer without enzyme. (B) A poly(A) RNA is a substrate for both a single U addition and a poly(U) tail. The nucleotides added are indicated at the bottom. The arrow points to the single-U addition product, and the bracket shows the 10- to 15-nucleotide poly(U) tail. A minus sign represents buffer without enzyme. The values on the right are numbers of nucleotides. (C) A 5′-end-labeled gA6-[14] gRNA that already contains a single 3′ U is not a substrate for another U. The plus and minus symbols refer to with- and without-enzyme conditions, respectively. All products were analyzed by 8% denaturing PAGE and visualized by autoradiography.
To further examine the substrate preferences of TUTase, we tested ribohomopolymers poly(G), poly(A), poly(C), and poly(U) for the ability to serve as substrates for TUTase addition. Both poly(A) (Fig. 3B) and poly(U) (data not shown) are substrates for the complex I-associated TUTase, while neither poly(G) nor poly(C) can serve as an efficient substrate (data not shown). Poly(A) RNA, radiolabeled at the 5′ end, is also very efficient at receiving a single added U (Fig. 3B, arrow). However, this RNA was unlike any other RNA we had examined in that it was a substrate for the addition of a distinct 10- to 15-U tail product. The addition of 10 to 15 U residues was independent of poly(A) substrate RNA length (data not shown). The poly(A) substrate was only extended in the presence of UTP (Fig. 3B). Neither UMP nor UDP could substitute for UTP in the extension of the poly(A) substrate (data not shown). Longer exposures of the autoradiograph revealed that only a very small amount of other nucleotides could be added onto either poly(A) or gRNAs (Fig. 3A and B and data not shown). The presence of ATP along with UTP in these reaction mixtures slightly skewed the distribution of the U added and considerably increased the amount of the poly(A) substrate ligation products migrating by about 40 and 60 nucleotides (Fig. 3B, ATP+UTP lane). Interestingly, these ligation products depended upon the presence of UTP, as ATP alone produced no ligated poly(A) RNA (Fig. 3B, ATP lane). These results show that the TUTase activity associated with the 19S complex is specific for UTP and does not require ATP. To summarize, both gRNAs and poly(A) are substrates for a major single-U addition, yet only the poly(A) RNA produced a significant amount of a 10- to 15-nucleotide poly(U) addition.
A polymerase is said to be distributive when only a single nucleotide is added per binding event. It is possible that the n+1 addition to the gRNAs is simply the result of U addition by a distributive enzyme and that the poly(U) tail addition to the poly(A) RNA is the product of a different enzyme that is not active on the gRNA. In principle, the major n+1 product could simply be the consequence of limiting amounts of a distributive enzyme. To test this, we prepared a gRNA substrate that already contained a 3′ added U. When we tested this gRNA in the TUTase assay, we found that it was not a substrate for U addition (Fig. 3C, compare A6-[14]3′U to control gA6-[14]). Thus, our initial interpretation was that the n+1 product was not simply a result of having limiting amounts of a distributive enzyme present.
We also synthesized 5′-end-labeled gRNAs with poly(U) tails varying in length from 2 to 20 U residues. When complex I TUTase was added to these reaction mixtures, we observed rapid exonuclease trimming of the gRNA to a length corresponding to the native gRNA sequence and n+1 product lengths (data not shown). This result is not altogether surprising, since a 3′ U-specific exoribonuclease activity has been shown to copurify with a functional 19S editing complex (22). Thus, competing activities of TUTase and 3′ U-specific exoribonuclease are present within the glycerol gradient 19S complex I fractions.
Single-U addition copurifies with RNA ligase.
The above-described experiments did not address the issue of whether the n+1 addition and the poly(U) tail are the result of a single enzyme or multiple enzymes. To address this, we purified the TUTase activities starting from isolated mitochondria. In this purification, we monitored RNA ligase activity, since RNA ligase can be radiolabeled with ATP and is therefore a convenient marker for editing complexes (23). Heparin-Sepharose chromatography was chosen as our initial step in TUTase purification, since it bound most of the RNA ligase and TUTase activities and the majority of mitochondrial proteins did not bind. However, a significant amount of nuclease activity for gRNAs was present in the fractionation. The poly(A) substrate was more resistant to nuclease degradation in these assays than gRNAs. For this reason, poly(A) RNA and gRNA substrates were sometimes substituted in the TUTase assays. Monitoring of U addition to the poly(A) RNA also allowed us to assay for both single-U and poly(U) additions.
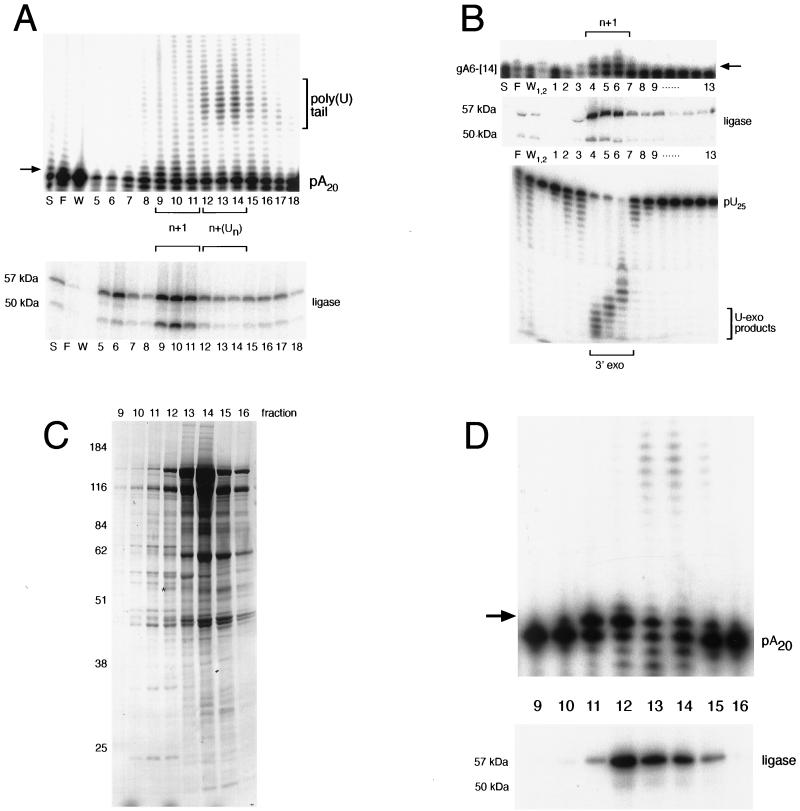
When we assayed fractions from the heparin-Sepharose separation, we found that n+1 addition activity was chromatographically distinct from the multiple-U addition activity on a poly(A) substrate (Fig. 4A). Fractions that contained major 10- to 15-U addition products (lanes 12 to 14) also contained single-U addition products. The peak of RNA ligase coincided with the single-U addition activity and not the 10- to 15-nucleotide U addition activity (Fig. 4A, ligase). We reasoned that fractions containing both TUTase and RNA ligase activities would be enriched in editing complexes. Thus, we pooled fractions 9 to 11 for further purification by Q-Sepharose chromatography.
FIG. 4.
Single-U addition activity copurifies with RNA ligase and exonuclease. (A) RNA ligase and multiple-U addition activities are chromatographically distinct on heparin-Sepharose. A 1-pmol sample of 5′-labeled poly(A) RNA (designated pA20) was incubated with 1 μl of each fraction in the presence of 1 mM UTP. RNA ligase in each of the corresponding fractions was adenylated and is shown at the bottom. (B) Single-U addition activity cofractionates with U exonuclease (U-exo) activity. Fractions 9 to 11 from the heparin-Sepharose column (A) were pooled and further purified on Q-Sepharose. A 1-pmol sample of 3′-labeled A6-[14] was incubated with 1 μl of each fraction in the presence of 1 mM UTP. RNA ligase in each of the fractions was adenylated and is shown at the bottom. For the exonuclease assay, 1 pmol of 5′-end-labeled poly(U) RNA (designated pU25) was incubated with 1 μl of each fraction. For the TUTase assays, single-U addition is shown by an arrow. S, F, and W, respectively, indicate starting material, flowthrough, and wash. (C) Silver staining of proteins present in the glycerol gradient purification step. Fractions 4 and 5 from the Q-Sepharose column (B) were pooled, and the complexes were separated on a 10-to-30% glycerol gradient. Following adenylation, protein was isolated and analyzed by SDS–10% PAGE. Size markers are shown at the left, and sizes are given in kilodaltons. Gradient fractions are indicated above. An asterisk indicates the 57-kDa radioactively labeled ligase. (D) Single-U addition products coincide with a stable 19S complex. A 1-pmol sample of 5′-labeled poly(A) RNA was incubated with 1 μl of each glycerol gradient fraction in the presence of 1 mM UTP. RNAs were recovered and analyzed by 8% urea–PAGE. Adenylated ligase that corresponds to these fractions is shown at the bottom.
On Q-Sepharose, most of the RNA ligase and TUTase activity bound while the majority of proteins flowed through the column during loading. We were able to analyze single-U addition to a 5′-end-labeled gA6-[14] RNA across these fractions (Fig. 4B, top). Again, single-U addition coincided with the peak of RNA ligase (Fig. 4B, middle, lanes 4 to 6). We also analyzed the fractions for 3′ U-specific exoribonuclease across these fractions (Fig. 4B, bottom), which has been previously shown to copurify with an editing complex (22). This activity also peaked in fractions containing a single-U addition and RNA ligase, suggesting that these activities can reside within the same complex.
In order to determine whether the TUTase activity purified on the Q-Sepharose column was associated with a stable editing complex, pooled fractions (4 and 5) were sedimented on a 10 to 30% glycerol gradient. Proteins in each glycerol gradient fraction were then analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (Fig. 4C). Most of the protein from the Q-Sepharose pool sedimented in fraction 14, where free protein and small complexes sediment. Bradford protein assays revealed that combined fractions 11 and 12 contained approximately 15% of the starting total protein (Table 1). Only ∼60% of the adenylated RNA ligase present in the glycerol gradient sedimented at 19S, suggesting that some dissociation of the complexes occurred during purification (Fig. 4D, bottom, ligase). When TUTase was assayed across this gradient, we found that U addition activity predominately cosedimented with the 19S complex (Fig. 4D, top, lanes 11 and 12). Interestingly, the 10- to 15-nucleotide U addition activity that was not previously detected in the Q-Sepharose pool was present in the ∼10S fractions that contained free protein (Fig. 4C, lanes 13 and 14).
TABLE 1.
Purification of single-U addition activity
| Fractiona | Total protein (μg) | n+1 Ub
|
Ligasec
|
||||
|---|---|---|---|---|---|---|---|
| Total activity (counts)d | Sp act (counts/μg) | Purification (fold) | Total activity (counts) | Sp act (counts/μg) | Purification (fold) | ||
| Mitochondria | 90,644 | 213,742 | 2.36 | NAe | 1,054,606 | 11.63 | NA |
| Heparin-Sepharose | 9,743 | 171,139 | 17.56 | 7.45 | 3,400,831 | 349.04 | 30.0 |
| Q-Sepharose | 960 | 209,049 | 217.67 | 92.31 | 3,509,089 | 3,653.82 | 314.05 |
| Glycerol gradient | 140 | 374,897 | 2,677.84 | 1,135.62 | 1,502,781 | 10,734.15 | 922.61 |
The mitochondrial fraction is the clarified (20,000 × g for 15 min) supernatant of the 0.5% Triton X-100-solubilized mitochondrial lysate described in Materials and Methods. The heparin-Sepharose, Q-Sepharose, and glycerol gradient fractions correspond to the pooled peaks of single-U addition and ligase activities from each chromatography step.
n+1 U refers to single-U addition, as assayed by single-U extension on a poly(A)20 RNA substrate.
ligase refers to the quantitation of adenylated 57-kDa ligase.
Counts are arbitrary units quantitated from an Applied Biosystems phosphorimager with the background subtracted.
NA, not applicable.
The results of our purification are outlined in Table 1. The single-U TUTase activity was purified over 1,000-fold. This represents significant purification, since the starting mitochondrial extract represents approximately 10% of the total T. brucei protein. Likewise, ligase was purified over 900-fold, further supporting the idea that these two enzymes are associated within a single complex.
In a separate purification scheme, we pursued isolation of the 10- to 15-nucleotide U addition activity. This TUTase activity sedimented at ∼10S and contained very little, if any, RNA ligase and 3′ U-specific exonuclease (data not shown). Furthermore, preliminary evidence suggests that this ∼10S particle is a subunit of the 19S complex and can be dissociated by treatment with a high salt concentration.
Role of mRNA in formation of the gRNA poly(U) tail.
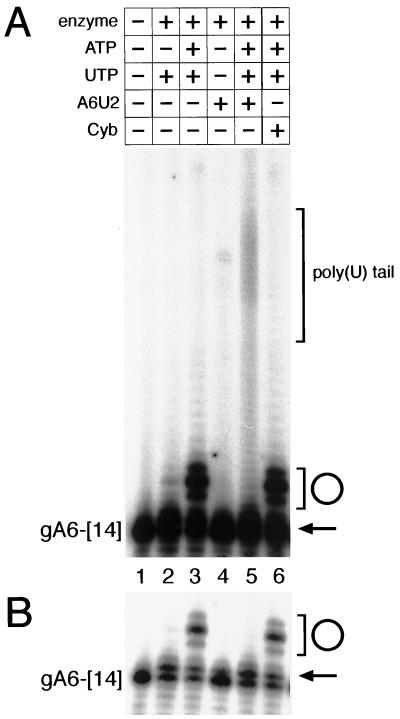
Given that the 19S complex adds only a single U to gRNAs, we wanted to understand how the 35S-to-40S complex was able to generate gRNAs containing poly(U) tails. A major difference between editing complexes I and II is the presence of pre-mRNA (20). To determine whether the formation of the gRNA poly(U) tail might be influenced by the presence of its cognate mRNA, we added synthetic pre-mRNAs to in vitro reaction mixtures containing purified editing complex and 5′-labeled gRNAs (Fig. 5). The addition of A6U2 pre-mRNA to the TUTase reactions containing the A6-[14] gRNA resulted in the addition of approximately 30 to 40 U residues to the gRNA [Fig. 5, lane 5, bracket labeled poly(U) tail]. A shorter exposure of this autoradiogram revealed a prominent n+1 product (arrow) that was present in reaction mixtures with and without added mRNA (Fig. 5B, lanes 2, 3, 5, and 6). The addition of noncognate pre-mRNA (Cyb) did not promote gRNA elongation (lane 6). As expected, a major product present in reaction mixtures that contained ATP was circular gRNA (Fig. 5A and B, bracket and circle). Cognate mRNA, but not noncognate mRNA, was sufficient to abolish gRNA circularization (compare lane 5 to lane 6). Addition of ATP (compared to UTP alone) to these reaction mixtures increased the amount, but not the size distribution, of tailed products (data not shown). These results were also reproduced by using gCyb[558] and 5′ΔCyb mRNA substrates (data not shown). The reason for the ATP stimulation of the poly(U) extension reaction is unclear but may be ATP-dependent conformational changes in the editing complex. This possibility remains untested. The ATP stimulation of gRNA circularization is not unexpected. At low RNA concentrations, intramolecular ligation and circularization are favored over intermolecular ligation and dimer formation. Also favoring circularization is the close proximity of the 5′ and 3′ ends of gRNAs (24). For these reasons, it is likely that these gRNAs would be excellent substrates for self-ligation and form circular RNAs. Whether circular gRNAs have any physiological relevance or whether they are simply in vitro artifacts is unknown. Circularization of the gRNAs could certainly interfere with poly(U) tail formation, since the 3′ OH would be unavailable for poly(U) addition.
FIG. 5.
Addition of cognate mRNA promotes addition of a stable poly(U) tail to gRNA. (A) Samples (200 nmol) of synthetic gRNAs with (+) or without (−) 1 pmol of cognate mRNA per assay (A6U2). Cognate mRNA or 1 pmol of nonspecific control RNA (CybΔ5′ mRNA) was added with the gRNA prior to the addition of buffer, nucleotides, and extract. The contents of each reaction mixture are indicated above the respective lane. The starting substrate gRNA A6-[14] is labeled at the left, and gRNA gA6-[14] with a poly(U) tail is shown by the bracket at the right [poly(U) tail]. (B) Lower exposure of the autoradiogram in panel A. The circle and bracket depict circular gRNA products, and the arrow depicts single-U addition. Products of the reactions were resolved by denaturing 8% PAGE and visualized by autoradiography.
The efficiency of these reactions is comparable to that of the in vitro editing reactions (16, 25), with only a small percentage being converted to poly(U)-tailed gRNA. Phosphorimager analysis quantitated a 2.5-fold increase in poly(U)-tailed gRNA in reaction mixtures that contained cognate pre-edited mRNA compared to control reaction mixtures that contained noncognate pre-mRNA. There was no significant increase in poly(U) tailing between the reaction mixtures with no added mRNA and control reaction mixtures with added noncognate pre-mRNA.
Polypurine-rich region of the pre-mRNA stabilizes the gRNA poly(U) tail.
In our initial studies with crude editing complexes, we found that poly(A) ribohomopolymer is a substrate for the addition of multiple U residues (Fig. 3B). One possible mechanism by which the poly(A) substrate can receive a strong 10- to 15-nucleotide U tail is by snap-back hybridization between the newly synthesized poly(U) tail and the poly(A) substrate sequence. In this model, approximately 10 to 15 U residues would be added by the TUTase, which would then fold back to hybridize with the poly(A) sequence. The added U residues would be base paired with the poly(A) substrate and thus be inaccessible to both TUTase and 3′ U-specific exonuclease. In a similar manner, the gRNA poly(U) tail might interact with purine residues in the pre-mRNA and thus protect newly added U residues on the gRNA from U-specific exoribonucleolytic trimming. To test this idea, we created a mutant RNA (RΔY) in which the purine-rich region of the pre-edited site was modified so that it contained predominately pyrimidine residues. This mutation should abolish base-pairing interactions between the gRNA poly(U) tail and the mRNA.
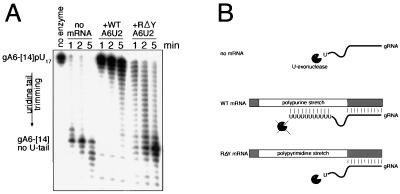
In the absence of cognate wild-type mRNA, the poly(U) tail of the gRNA was trimmed back to the native gRNA sequence by the U-specific exonuclease present in purified 19S complex (Fig. 6A, no-mRNA lanes). Within 1 min, the gRNA poly(U) tail was completely cleaved back to the native gRNA sequence (Fig. 6A, gA6-[14] no U-tail). Addition of cognate wild-type sequence A6U2 mRNA essentially prevented trimming, and most of the poly(U) tail remained intact even at the later time points (Fig. 6A, +WT A6U2 lanes). However, the purine-to-pyrimidine substitution mutant pre-mRNA could not reproduce this protection (Fig. 6A, +RΔY lanes), and at the 5-min time point, most of the U tail was effectively removed. Thus, the presence of a polypurine region upstream of the editing site protects the poly(U) tail from degradation, possibly by A · U and G · U base pairing. The pre-edited region of the A6U2 mRNA contains a purine-rich region approximately 35 nucleotides long, and can base pair with the poly(U) tail, making the 3′ nucleotide inaccessible to exoribonucleolytic trimming. The mutant RΔY pre-mRNA, having no purine-rich region to base pair with the poly(U) tail, is not able to protect from U-specific 3′ trimming (Fig. 6B).
FIG. 6.
Polypurine-rich region of the pre-edited mRNA stabilizes the gRNA poly(U) tail. (A) The starting RNA substrate is gA6-[14] with a 17-nucleotide poly(U) tail (gA6-[14]pU17). The substrate RNA and 19S complex from the Q-Sepharose pool was incubated in the absence of mRNA (no mRNA), in the presence of cognate wild-type A6U2 mRNA (+WT A6U2), or in the presence of cognate purine-rich region substitution mutant mRNA (+RΔY A6U2). Products of U-specific 3′ exoribonucleolytic trimming are indicated at the left (gA6-[14] no U-tail). Incubation times are given above the lanes. The arrow labeled uridine tail trimming indicates the direction of product formation. (B) Cartoon depicting reactions shown in panel A. In the absence of cognate wild-type mRNA, the gRNA poly(U) tail is subject to U-specific 3′ exoribonucleolytic trimming. In the presence of cognate wild-type mRNA, the poly(U) tail can hybridize to the polypurine-rich region on the mRNA and thus hinder U trimming. In the polypyrimidine-rich mutant, the U tail cannot base pair with the mRNA and thus is susceptible to U trimming.
DISCUSSION
The results presented here provide several new insights into the TUTase activity present in T. brucei mitochondria. We found that the 19S editing complex stably added a single U to RNAs. This is likely due to competing activities between the complex-associated U-specific 3′ exonuclease and TUTase. However, gRNAs received multiple U residues in the presence of cognate mRNA, suggesting that complex II contains a pool of gRNAs with stable poly(U) tails. We also show that the polypurine-rich regions within the pre-edited mRNA confer stability on the gRNA poly(U) tail.
Glycerol gradient sedimentation of crude mitochondrial extracts revealed that TUTase activity associates with both 19S complex I and 35S-to-40S complex II (Fig. 1). By using an assay, we found that both complexes contained single- and multiple-U addition activities when poly(A) RNA was used as a substrate. However, we found that both of these complexes added predominately a single U to gRNA substrates (Fig. 3A and data not shown). TUTase activity may be present in the upper gradient fractions that correspond to ∼10S (fractions 13 to 15), but nucleases within these same fractions would mask such activity (Fig. 1).
When isolated mitochondria are incubated in the presence of [α-32P]UTP, editing complexes are labeled (18–20). This labeling is the result of TUTase activity and not mitochondrial transcription (13). We found that two major RNP complexes became labeled during [α-32P]UTP incubation (Fig. 2). Based on prior studies, it is likely that these two UTP-labeled complexes represent editing complexes I and II (20). Those studies suggested that complex I functions as a gRNA maturation complex that mediates the formation of the poly(U) tail. This proposal was based on the colocalization of TUTase and gRNAs, but not mRNAs, within complex I. However, the ability of complex I to polyuridylate gRNAs was not examined. In our initial studies, we found that the TUTase present in complex I could add only a single U to three different gRNAs.
It has been shown that [α-32P]UTP incorporates into both mRNA and gRNA in isolated mitochondria (13, 18). In the experiments described here, we evaluated the lengths of the labeled gRNAs. We found evidence that complex II gRNAs are able to receive approximately 15 U residues, the average length of a gRNA poly(U) tail. Pulse-chase analysis of complex II gRNAs showed that the U tails were added rapidly (within 5 min) and appeared to be stable throughout the experiment. In contrast, gRNAs associated with complex I did not increase significantly in size during the time course. These experiments suggested that the addition of a 3′ poly(U) tail to gRNAs occurs within complex II.
Complex I TUTase from the glycerol gradient seems to have very stringent nucleotide specificity for U. This result reinforced the evidence that we were studying a TUTase and not a poly(A) polymerase or another contaminating polymerase. An interesting product in these assays was ligated RNA, which became readily detectable when ATP was added. Self-ligated circular gRNAs were the prominent ligation product (Fig. 1), since the 5′ and 3′ ends are in close proximity to each other in the secondary structure of the gRNA (24). Circular gRNAs were formed when we used 5′-end-labeled gRNA, due to the presence of a 5′ monophosphate on this RNA. Neither uniformly labeled gRNA from T7 runoff transcriptions (which contains a 5′ triphosphate) nor 3′-labeled gRNA [which contains a 5′ OH from poly(A) polymerase addition] would produce these circles (data not shown).
A gRNA substrate that a priori contains a single 3′ U nucleotide is not a substrate for the addition of a second U (Fig. 3C). This result eliminated the possibility that the single U addition was the result of limiting amounts of a distributive TUTase. It is possible that the 19S TUTase prefers to add a single U to an RNA that does not already contain a 3′ U. A TUTase that prefers to add a single U has been described, although the activity on an RNA that a priori contains a single 3′ U nucleotide was not examined (29). Alternatively, what appears to be a single U addition may be the result of competing multiple-U-adding TUTase and 3′ U-specific exonuclease activities. In this scenario, an RNA that contains a single U would be a poor substrate for the exonuclease. Recently, it was found that RNase E from E. coli would trim back 3′ poly(U) tails to leave a single nucleotide uridylate remnant (14).
On glycerol gradients, we found that the single-U addition activity cosedimented with the purified 19S complex. This finding suggests that this TUTase activity is part of complex I (Fig. 1 to 4). However, we have not analyzed whether our purified complex contained either gRNAs or mRNAs, and it is possible that other factors were stripped away during the purification. Multiple-U extension activity appears to dissociate from the 19S complex, and treatment of the purified 19S complex with a high salt concentration increases the amount of multiple-U addition products (data not shown). At this stage, however, we cannot exclude the possibility that the multiple-U addition TUTase activity is a different TUTase also present in the 19S fraction which is masked by the U-specific exonuclease during purification. Clearly, these results warrant further investigations into how TUTase assembles into 19S editing complexes.
In an effort to reconstitute gRNA polyuridylation, we sought conditions that would shift the gRNA n+1 product to gRNAs containing a poly(U) tail. The addition of ATP along with UTP did not promote gRNA elongation, nor did the presence of a nonhydrolyzable ATP analogue (AMP-CPP) inhibit gRNA n+1 TUTase activity. Since gRNAs interact with mRNAs and our mitochondrial metabolic labeling results showed gRNA poly(U) extension to occur in complex II, where mRNA is present, we examined whether gRNAs could be polyuridylated when complexed with cognate pre-mRNA (Fig. 5). Cognate pre-mRNA, but not noncognate pre-mRNA, was sufficient to promote multiple-U tail formation. Presumably, the 5′ anchor of the gRNA would act to specify the interaction with its cognate mRNA. However, we have not excluded the possibility that the purine-rich regions of other noncognate pre-mRNAs can stimulate gRNA poly(U) tail addition. One might expect that higher concentrations of noncognate mRNA are required to produce the same effect, although we have not tested this possibility.
Previous studies suggested that the poly(U) tail of the gRNA interacts with the pre-edited region of the mRNA (6, 17). Our data suggest that interaction of the newly synthesized poly(U) tail of the gRNA with the purine-rich region of the mRNA protects the gRNA 3′ tail from editing complex-associated 3′ U-specific exonuclease activity. Alternatively, it is possible that the purine-rich region is required for association of the gRNA with the editing complex and that the association of gRNA with the editing complex, and not pre-mRNA, protects the poly(U) tail from riboexonucleolytic trimming. It would be very interesting if polypurine-rich RNAs were preferred RNAs for association with editing complexes, since the polypurine-rich regions are common at editing sites. However, the exact RNA preferences (if there are any) that govern association with either the 19S or 35S-to-40S editing complex are not known. By changing the purine-rich region to a pyrimidine-rich region, we abolished its ability to base pair with the poly(U) tail. Interactions between the gRNA 5′ anchor and the mRNA do not seem to be critical for protection of the poly(U) tail, since both RΔY and wild-type mRNAs contain the same 5′ anchor. It is more likely that the 3′ U-specific exonuclease prefers a non-base-paired nucleotide and that duplexing of the poly(U) tail protects from exoribonucleolytic trimming.
How does the single-U addition activity present in the 19S complex relate to mRNA editing? One possibility is that editing occurs through multiple rounds of single-U addition and deletion, even at sites where many U residues must be added or deleted (4). We have found that the purified 19S complex is capable of performing U addition editing and that predominately a single U is added to exogenously supplied mRNA 5′ cleavage fragments (data not shown). A second possibility is that U addition to mRNA 5′ cleavage fragments is similar to the mRNA-dependent gRNA U addition described in this report. In this possibility, multiple U residues would be added, which would then base pair with the guiding nucleotides of the gRNA. Base-pairing interactions would then protect the newly added U residues from U-specific exonuclease.
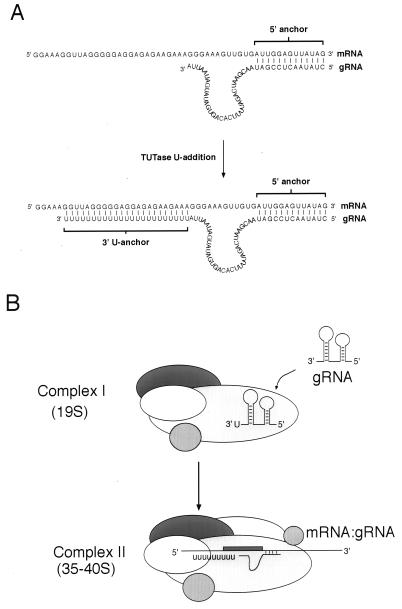
These results support a model in which gRNA maturation occurs in complex II (Fig. 7). gRNAs that are associated with complex I are subject to both U addition and riboexonucleolytic trimming. These gRNAs would contain a stable 3′ U residue. Upon assembly of complex II and prior to editing, these gRNAs would receive multiple added U residues which would be stabilized by the purine-rich regions of the pre-mRNA.
FIG. 7.
Proposed model of gRNA maturation. (A) gA6-[14] gRNA with and without a poly(U) tail shown hybridized to the cognate pre-mRNA substrate A6U2. The brackets represent gRNA anchor sequences. (B) Model of complex assembly and gRNA maturation. gRNA is thought to associate with complex I, where one U may be added at the 3′ end. The cognate mRNA may then associate via the gRNA anchor sequence, disrupting the secondary structure of the gRNA. U residues are then added to the gRNA, which form base pairs with the purine-rich region of the pre-edited region. The pre-edited region of the mRNA is represented as a box.
ACKNOWLEDGMENTS
We thank Robert Sabatini, Susan Madison-Antenucci, Mike Miller, Karen Bertrand, and Jayleen Grams for helpful discussions and insightful advice.
This work was supported by PSA grant CA 60151 to B.K.A. and PHS grant AI 21401 to S.L.H.
REFERENCES
- 1.Adler B K, Hajduk S L. Guide RNA requirement for editing-site-specific endonucleolytic cleavage of pre-edited mRNA by mitochondrial ribonucleoprotein particles in Trypanosoma brucei. Mol Cell Biol. 1997;17:5377–5385. doi: 10.1128/mcb.17.9.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler B K, Harris M E, Bertrand K I, Hajduk S L. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol Cell Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfonzo J D, Thiemann O, Simpson L. The mechanism of U insertion/deletion RNA editing in kinetoplastid. Nucleic Acids Res. 1997;25:3751–3759. doi: 10.1093/nar/25.19.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 5.Blum B, Simpson L. Formation of gRNA/mRNA chimeric molecules in vitro, the initial step of RNA editing, is dependent on an anchor sequence. Proc Natl Acad Sci USA. 1992;89:11944–11948. doi: 10.1073/pnas.89.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 7.Byrne E M, Connell G J, Simpson L. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 1996;15:6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 8.Corell R A, Read L K, Riley G R, Nellissery J K, Allen T E, Kable M L, Wachal M D, Seiwert S D, Myler P J, Stuart K D. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol Cell Biol. 1996;16:1410–1418. doi: 10.1128/mcb.16.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Reyes J, Sollner-Webb B. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc Natl Acad Sci USA. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- 11.Frech G C, Simpson L. Uridine insertion into preedited mRNA by a mitochondrial extract from Leishmania tarentolae: stereochemical evidence for the enzyme cascade model. Mol Cell Biol. 1996;16:4584–4589. doi: 10.1128/mcb.16.8.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris M, Decker C, Sollner-Webb B, Hajduk S. Specific cleavage of pre-edited mRNAs in trypanosome mitochondrial extracts. Mol Cell Biol. 1992;12:2591–2598. doi: 10.1128/mcb.12.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris M E, Moore D R, Hajduk S L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- 14.Huang H, Liao J, Cohen S N. Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E. coli ribonuclease E. Nature. 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- 15.Kable M L, Heidmann S, Stuart K D. RNA editing: getting U into RNA. Trends Biochem Sci. 1997;22:162–166. doi: 10.1016/s0968-0004(97)01041-4. [DOI] [PubMed] [Google Scholar]
- 16.Kable M L, Seiwert S D, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA [see comments] Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 17.Leung S S, Koslowsky D J. Mapping contacts between gRNA and mRNA in trypanosome RNA editing. Nucleic Acids Res. 1999;27:778–787. doi: 10.1093/nar/27.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peris M, Frech G C, Simpson A M, Bringaud F, Byrne E, Bakker A, Simpson L. Characterization of two classes of ribonucleoprotein complexes possibly involved in RNA editing from Leishmania tarentolae mitochondria. EMBO J. 1994;13:1664–1672. doi: 10.1002/j.1460-2075.1994.tb06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peris M, Simpson A M, Grunstein J, Liliental J E, Frech G C, Simpson L. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol Biochem Parasitol. 1997;85:9–24. doi: 10.1016/s0166-6851(96)02795-8. [DOI] [PubMed] [Google Scholar]
- 20.Pollard V W, Harris M E, Hajduk S L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohrer S P, Michelotti E F, Torri A F, Hajduk S L. Transcription of kinetoplast DNA minicircles. Cell. 1987;49:625–632. doi: 10.1016/0092-8674(87)90538-1. [DOI] [PubMed] [Google Scholar]
- 22.Rusche L N, Cruz-Reyes J, Piller K J, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabatini R, Hajduk S L. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J Biol Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 24.Schmid B, Riley G R, Stuart K, Goringer H U. The secondary structure of guide RNA molecules from Trypanosoma brucei. Nucleic Acids Res. 1995;23:3093–3102. doi: 10.1093/nar/23.16.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seiwert S D, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 26.Sloof P, Benne R. RNA editing in kinetoplastid parasites: what to do with U. Trends Microbiol. 1997;5:189–195. doi: 10.1016/S0966-842X(97)01034-2. [DOI] [PubMed] [Google Scholar]
- 27.Souza A E, Hermann T, Göringer H U. The guide RNA database. Nucleic Acids Res. 1997;25:104–106. doi: 10.1093/nar/25.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White T C, Borst P. RNA end-labeling and RNA ligase activities can produce a circular rRNA in whole cell extracts from trypanosomes. Nucleic Acids Res. 1987;15:3275–3290. doi: 10.1093/nar/15.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zabel P, Dorssers L, Wernars K, Van Kammen A. Terminal uridylyl transferase of Vigna unguiculata: purification and characterization of an enzyme catalyzing the addition of a single UMP residue to the 3′-end of an RNA primer. Nucleic Acids Res. 1981;9:2433–2453. doi: 10.1093/nar/9.11.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]