Abstract
Coronavirus disease 2019 (COVID-19) has a broad range of clinical manifestations, highlighting the need for specific diagnostic tools to predict disease severity and improve patient prognosis. Recently, calprotectin (S100A8/A9) has been proposed as a potential biomarker for COVID-19, as elevated serum S100A8/A9 levels are associated with critical COVID-19 cases and can distinguish between mild and severe disease states. S100A8/A9 is an alarmin that mediates host proinflammatory responses during infection and it has been postulated that S100A8/A9 modulates the cytokine storm; the hallmark of fatal COVID-19 cases. However, it has yet to be determined if S100A8/A9 is a bona-fide biomarker for COVID-19. S100A8/A9 is widely implicated in a variety of inflammatory conditions, such as cystic fibrosis (CF) and chronic obstructive pulmonary disorder (COPD), as well as pulmonary infectious diseases, including tuberculosis and influenza. Therefore, understanding how S100A8/A9 levels correlate with immune responses during inflammatory diseases is necessary to evaluate its candidacy as a potential COVID-19 biomarker. This review will outline the protective and detrimental roles of S100A8/A9 during infection, summarize the recent findings detailing the contributions of S100A8/A9 to COVID-19 pathogenesis, and highlight its potential as diagnostic biomarker and a therapeutic target for pulmonary infectious diseases, including COVID-19.
Keywords: S100A8/A9, COVID-19, pulmonary pathogens
Graphical Abstract
1. Introduction
Coronavirus disease 2019 (COVID-19), an infectious disease that affects the lungs and the immune system, has spread to hundreds of countries and territories since its outbreak in December 2019 [1]. Since then, the global burden of the disease has risen to nearly 145 million cases and it is estimated that over 3 million deaths, globally, can be attributed to COVID-19 [2]. In 2020, COVID-19 was the third leading cause of death in the United States and fourth leading cause of death globally [3], [4].
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, results in heterogenous clinical manifestations ranging from asymptomatic, mild disease (including fever, cough, fatigue, sore throat, muscle ache, diarrhea, dyspnea, and headache) to severe illness (such as disseminated intravascular coagulation, respiratory failure, septic shock, multiorgan failure (MOF), and death) [5], [6], [7]. Patients that present with mild disease typically develop an efficient immune response. However, approximately 15% of patients progress to severe pneumonia while about 5% of patients must be admitted to the intensive care unit (ICU) following the development of critical complications, such as acute respiratory distress syndrome (ARDS) [5]. Determining the factors that influence patient risk for disease progression is vital for developing a strategy to monitor the treatment of COVID-19 patients and decrease mortality rates.
A hallmark of poor COVID-19 prognosis includes hypercytokinemia, often termed as cytokine storm, which results in the sudden worsening of COVID-19 patients due to a dysregulated immune response can result in severe COVID-19 disease, sepsis, MOF, and death [8], [9], [10]. Cytokine storm is characterized by the overproduction and release of over 150 inflammatory cytokines due to overactivation and hyperproliferation of T cells, macrophages, and NK cells. Accumulating evidence suggests that interleukin-6 (IL-6) may be a potential therapeutic target to mediate the cytokine storm in patients with severe COVID-19, as IL-6 serum levels correlate with disease mortality in patients with fatal COVID-19 cases [11], [12]. However, while IL-6 has previously been implicated as a notable marker of the cytokine storm, other recent studies have also described the cytokine storm to be associated with expression of an alarmin called calprotectin (S100A8/A9) [13], [14].
2. Structure and biological functions of alarmin S100A8/A9
Alarmins are endogenous, immune-activating proteins that are released to initiate and amplify inflammatory immune responses [15], [16]. Following microbial infection, alarmins relay intercellular defense signals by interacting with pattern recognition receptors (PRRs) to activate immune responses, which involves the recruitment of neutrophils and monocytes to eliminate foreign microorganisms, the activation of resident leukocytes, and the production of inflammatory mediators [15], [16], [17], [18], [19]. However, uncontrolled production or sustained expression of alarmins can be harmful or even fatal to the host by inducing cytokine storm or excessive inflammation, as is observed in severe COVID-19 cases [8], [20], [21], [22], [23], [24], [25].
S100 proteins are recognized as a major category of alarmins, among defensins, interleukins, heat shock proteins, high-mobility group proteins, cathelin-related anti-microbial peptides, and eosinophil derived neurotoxins [16], [26]. S100A8 and S100A9, also known as myeloid-related protein 8 (MRP8) and MRP14, are members of the S100 protein family [1]. Similar to other S100 proteins, both S100A8 and S100A9 have a helix-loop motif containing charged amino-acid residues that result in high affinity for divalent ions, such as Ca2+ and Zn2+ [1]. Together, S100A8 and S100A9 form the calcium-binding heterodimer known as calprotectin (S100A8/A9), which makes up approximately 45% of the cytoplasmic proteins present in human neutrophils [11]. While both S100A8 and S100A9 form non-covalent homodimers, the heterodimer formation is more stable and prominent at higher calcium concentrations as compared to either homodimer configuration [27]. It’s small size (24 kDa) allows it to easily diffuse between the tissue and blood and its resistance to degradation emphasizes its usefulness as a sensitive and dynamic marker for neutrophil activation throughout the body [1], [28], [29]. Upon binding divalent ions, S100A8/A9 undergoes a conformational change that allows it to exert many of its functions, such as regulating leukocyte migration and trafficking, amplifying inflammatory responses, and modulating cell chemotaxis [1], [28], [30]. Intracellularly, high levels of S100A8/A9 are stored in neutrophils and myeloid-derived dendritic cells, where it is involved in cytoskeleton rearrangement and arachidonic acid metabolism [1].
Following neutrophil activation, S100A8/A9 is released extracellularly during neutrophil extracellular trap (NET) formation and can also be passively released from neutrophils dying of necrosis. However, S100A8 and S100A9, like other S100 proteins, lack a secretion signal sequence for the classic endoplasmic reticulum/golgi transport. Instead, S100A8/A9 release relies on an alternative energy-dependent pathway whereby activated monocytes/macrophages and neutrophils require an intact cytoskeleton, activation of protein kinase C (PKC), and microtubule formation for S100A8 and S100A9 secretion [27]. Furthermore, the increased release of S100A8 and S100A9 are inversely related to their de novo synthesis, which suggests that S100A8/A9 extracellular signaling is restricted to distinct differentiation stages of monocytes [31].
While S100A8/A9 functions in a protective capacity to mediate the host response against pathogens, its detrimental role in promoting enhanced disease states has also been documented. S100A8/A9 is an endogenous ligand of Toll-like receptor 4 (TLR4) and the receptor for advanced glycation end products (RAGE). It binds upstream of TNF and CXCL8 secretion to promote NF-B signaling and the secretion of inflammatory proteins, such as IL-6 [1], [22], [29], [32], [33], [34]. However, tissue damage can sustain amplified S100A8/A9 production, which promotes autocrine production, resulting in a hyperinflammation loop that prevents the protein from exerting its protective function [29], [35], [36], [37], [38]. This sustained over-expression of S100A8/A9 can contribute to uncontrolled inflammation and cytokine storm, as has been observed in COVID-19 cases.
3. S100A8/A9 modulates pro-inflammatory responses during host infection
Due to its role in inflammation, S100A8/A9 has been investigated and associated with a number of inflammatory conditions such as arthritis, chronic obstructive pulmonary disorder (COPD), and inflammatory bowel disease (IBD) [39], [40], [41]. While S100A8/A9 secretion can occur due to inflammation caused by metabolic inflammatory conditions, autoimmune diseases, and degenerative diseases, S100A8/A9 is also notably upregulated following infection. In response to microbial infection, neutrophils, macrophages, and monocytes induce the expression and secretion of S100A8/A9 to mediate pro-inflammatory responses by stimulating leukocyte recruitment and inducing cytokine release [29]. This section will detail a comprehensive overview of S100A8/A9 in response to both viral and bacterial infections.
4. Viral infections
Infection with SARS-CoV-2 results in the activation of innate and adaptive immune responses that, when dysregulated, can cause local and systemic tissue damage [42], [43]. Neutrophils are typically the first responders to acute infection and amplify the host inflammatory response by releasing pro-inflammatory cytokines. However, sustained increase in pro-inflammatory signaling can lead to cytokine storm, which can result in MOF and death, as is observed during fatal COVID-19 infection [8], [9]. Patients with severe or critical COVID-19 have increased neutrophil accumulation due to recruitment by inflammatory cytokines, with 100% of patients (n = 82) experiencing neutrophilia within 24 h of their death [44], [45].
Recent studies have investigated the mechanisms of uncontrolled immune activation associated with aberrant cytokine responses to better understand COVID-19 pathogenesis. SARS-CoV-2 infection has been shown to significantly decrease B-cell counts and induce genes that are involved in neutrophil chemotaxis and anti-bacterial humoral responses, rather than inducing canonical type I IFNs [46]. Additionally, SARS-CoV-2 infection has been shown to induce the formation of a group of immature, non-canonical, aberrant neutrophils [46], [47], [48].
Patients with severe COVID-19 cases exhibit significantly increased neutrophil counts and are depleted of B cells, NK cells, and CD4+ and CD8+ T cells [23], [48], [49], [50], [51], [52]. Because neutrophils are typically activated during bacterial infection to phagocytose, kill, and digest invading pathogens, it was originally postulated that the observed increase in neutrophils in severe COVID-19 patients was due to coinfection of bacteria, such as Streptococcus pneumoniae. However, accumulating evidence suggests that the increase in neutrophil counts may be attributed to the presence of an aberrant, immature neutrophil subset induced by SARS-CoV-2 infection [46], [47], [48], [53].
CD10 and CD101 are neutrophil markers of maturation, where CD10lowCD101- neutrophils represent an immature cell subsets, and patients with varying COVID-19 severity displayed distinct phenotypes of these circulating innate immune cells [48], [54]. Severe COVID-19 patients had an expansion of circulating neutrophils in the peripheral blood coupled with a higher amount of circulating CD10lowCD101- neutrophils as compared with controls or patients with mild disease ( Fig. 1) [48]. Similarly, patients with severe COVID-19 who progressed to ARDS had elevated levels of immature neutrophils, which were termed “developing neutrophils” as they lacked the canonical neutrophil markers CXCR2 and FCG3RB [46], [47]. While the function and derivation of this aberrant neutrophil subset is still under investigation, it has been determined that the formation of this abnormal neutrophil group is induced specifically by coronavirus infection.
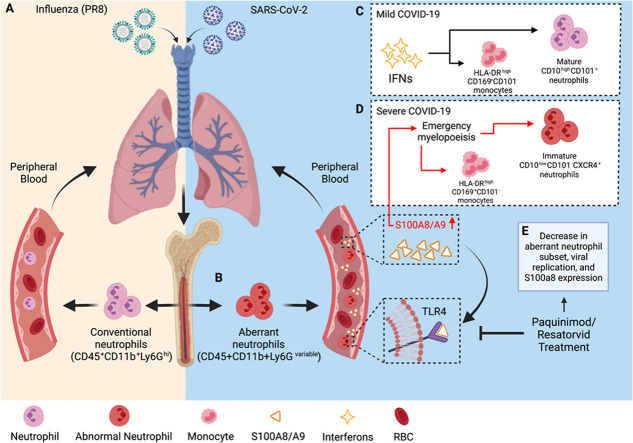
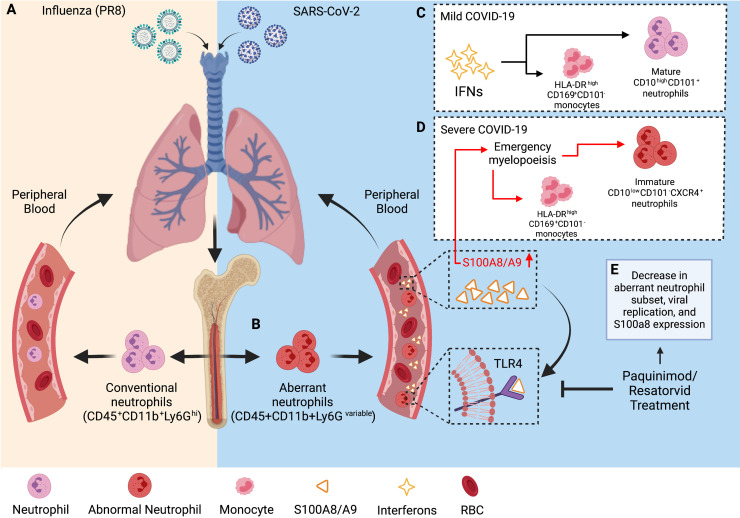
Fig. 1.
SARS-CoV-2 infection induces a dysregulated antiviral innate immune response. (A) Typical antiviral response following exposure to influenza. (B) Aberrant CD45+CD11b+Ly6Gvariable neutrophil subset induced following exposure to SARS-CoV-2. Immune response in (C) mild and (D) severe COVID-19 cases where patients with severe COVID-19 had increased circulating levels of S100A8/A9 that drives emergency myelopoiesis and the formation of an aberrant immature neutrophil subset. (E) Treatment with S100A9 inhibitors blocks S100A8/A9-mediated TLR4 signaling and decreases viral replication, S100a8 expression, and diminish the number of aberrant CD45+CD11b+Ly6Gvariable neutrophils in mice. Created with Biorender.com
Ly6G is a widely used marker for neutrophils and low Ly6G levels suggest neutrophil cell immaturity. Mice infected with either murine coronavirus (mouse hepatitis virus, MHV) or SARS-CoV-2 accumulated a subset of CD45+CD11b+Ly6Gvariable neutrophils in comparison to mice in the control group, which contained the typical CD45+CD11b+Ly6Ghigh neutrophils. Furthermore, when challenged with other infectious stimuli (including influenza A virus (IAV), encephalomyocarditis virus (EMCV), herpes simplex virus (HSV), and lipopolysaccharide (LPS)), the neutrophil population remained CD45+Cd11b+Ly6Ghigh despite the fluctuation in neutrophil count between groups. These observations supported the notion that invasion and formation of the aberrant neutrophil group was specifically induced by SARS-CoV-2 infection (Fig. 1) [46].
In addition to the presence of the abnormal immature neutrophil subset, S100A8 and S100A9 expression is elevated in peripheral blood nucleated cells from patients with severe COVID-19 as compared with controls and patients with mild disease [48]. The same trends are observed in non-human-primate (NHP) studies as well, where SARS-CoV-2-infected macaques exhibited robust expression of S100A8 between 3 and 5 days post infection (dpi) [46]. In fact, S100A8 was the most significantly induced gene among all known alarmins and its upregulation corresponded with an increase in the viral lung burden [46]. Consistent with these findings, the lung samples from post-mortem COVID-19 patients also had elevated expression of S100a8 and neutrophil marker genes [46]. In COVID-19 positive patients, S100A8 expression was markedly higher compared to healthy subjects, supporting previous findings that indicate the potential of S100A8/A9 as a biomarker for COVID-19 disease severity and clinical outcomes. Consistent with the observations from the NHP experiment, SARS-CoV-2-infected human ACE2 transgenic mice exhibited robust anti-bacterial humoral response, neutrophil chemotaxis, and upregulated transcription of S100a8 and neutrophil marker genes (Ly6G, Mmp8 etc.) at 5 dpi, corresponding to when mice became sicker.
These increases in expression and atypical anti-viral responses are specific to SARS-CoV-2 infection. Exposure to influenza A virus (IAV), a single-stranded RNA virus, can also result in severe respiratory tract infections. In high-risk individuals, including the elderly and immunocompromised patients, IAV infection causes exacerbated airway inflammation that can develop into severe cases of pneumonia [41]. During IAV infection, toll-like receptors (TLRs) and cytosolic PRRs are required for an effective innate immune response [41]. While these receptors are necessary to surmount a strong immune response against pathogens, PRRs can also be detrimental to the host [41]. TLR4 is activated during IAV infection but has previously been shown to contribute to exacerbated lung disease and mortality in animal models [41]. S100A8/A9 interacts with TLR4 to mediate proinflammatory responses, and it has been demonstrated that DDX21/TRIF and TLR4/MyD88 pathways are required for S100A9 expression and activity [41]. However, IAV infection induced canonical anti-viral responses by activating type I IFN signaling and did not result in elevated expression of S100a8 expression nor neutrophil marker genes as had been observed following SARS-CoV-2 expression (Fig. 1) [46].
Similarly, S100a8 expression was measured in the blood and lungs of C57BL/6 mice infected with EMCV or HSV-1, but neither viral infection resulted in the induction of S100A8 or neutrophil chemotaxis transcription. However, when interferon-gamma alpha receptor (IFNAR)-deficient mice were infected with MHV-59, S100a8 and Ly6G expression were sharply upregulated and genes induced by MHV were enriched in neutrophil chemotaxis and anti-bacterial pathways, suggesting the upregulation of S100a8 and observed chemotaxis is attributed specifically to infection with coronaviruses and is involved in the formation of fatal SARS-CoV-2 infections [46].
However, it is possible that mature neutrophils harboring large amounts of S100A8/A9 migrate to the lungs where S100A8/A9 is released, which amplifies the aberrant innate anti-viral response in a positive feedback loop. The uncontrolled, excessive production of S100A8/A9 may strongly stimulate the TLR4 signal, which then results in the induction of an aberrant neutrophil subset and imbalance of immune response as observed during SARS-CoV-2 infection [46]. S100A8/A9 binds to TLR4 and RAGE to mediate pro-inflammatory responses and mice lacking MyD88, an essential adaptor protein which acts as a signal transducer in the IL-1 and TLR signaling pathways, exhibit suppression of TLR4 signaling. However, when COVID-infected mice were treated with recombinant S100A8/A9, expression of S100a8 and neutrophil chemokine Cxcl2 are induced in a TLR-4/Myd88-dependent manner, reflecting the ability of S100A8/A9 to induce expression of itself. Taken together, these results support the notion that a positive loop could be the source of observed aberrant immune responses [46].
Alternatively, it has been postulated that the burst of S100A8/A9 detected in COVID-19 patients may trigger downstream NF-KB-driven emergency myelopoiesis, which could also account for the generation of immature cells (Fig. 1) [55], [56]. In contrast, it has also been proposed that SARS-CoV-2 directly stimulates the expression of S100A8/A9 by TLR4 signaling. It has been predicted that the binding of antigenic epitopes within the full-length S protein of SARS-CoV-2 could bind to TLR4 to activate an immune response [57]. While the function and derivation of this aberrant neutrophil subset is still under investigation, understanding how S100A8/A9 mechanistically contributes to the emergence of this population will provide valuable insight into how the targeted inhibition of S100A8/A9 could improve COVID-19 outcomes.
5. Bacterial infections
Tuberculosis (TB) is a global health threat, with nearly a quarter of the world’s population estimated to be infected with Mycobacterium tuberculosis (Mtb), the causative agent of TB. Mtb proliferates in the lungs and individuals with active TB (ATB) disease can transmit the bacteria to other individuals. Understanding what drives TB pathogenesis is crucial for decreasing mortality and transmission rates, especially in low-resource and high TB-incidence areas. Elevated serum S100A8/A9 levels have been implicated in enhanced TB disease, along with increased neutrophil accumulation [58]. Not only are S100A8/A9 levels correlated to neutrophil accumulation, but it has also been determined that the alarmin directly mediates neutrophil accumulation by regulating CD11b expression on myeloid cells [59]. When neutrophils from WT Mtb-infected mice were extracted and adoptively transferred to Mtb-infected mice that lacked a functional copy of S100A9, CD11b expression increased significantly, suggesting that S100A8/A9-deficiency is directly involved in regulating CD11b expression on neutrophils. In addition, S100A8/A9-deficiency depleted neutrophil accumulation to the lung as well as in granulomas and was associated with decreased lung bacterial burden and increased host-control over Mtb infection.
Cystic fibrosis (CF) is a genetic condition which can affect the lungs, pancreas, liver, kidneys, and intestine. Cells involved in mucous production are primarily affected by the disorder, which can result in persistent lung infections. CF is characterized by chronic, neutrophil-dominated inflammation, and S100A8/A9 has been implicated as a CF antigen where elevated sputum and serum S100A8/A9 levels have been reported in exacerbated CF patients [60]. However, S100A8/A9 levels from the sputum of CF patients are comparable to those analyzed from COPD patients and may not be ideal for CF diagnostics. Fecal S100A8/A9 levels have also been implicated in CF enteropathy, which occurs due to pancreatic insufficiency (PI) and resulting maldigestion and malabsorption of nutrients [61]. In CF patients, antibiotic treatment for respiratory exacerbation reduced fecal S100A8/A9 levels as compared to pre-treatment values, suggesting that treating inflammation in the respiratory system induced changes in the gastrointestinal tract [62]. These findings are intriguing, as they indicate that S100A8/A9 levels can be affected by external treatment of inflammatory conditions in other systems.
6. S100A8/A9 as a potential clinical biomarker
To improve the treatment of COVID-19 patients, the determinants of fatal outcomes of severe COVID-19 patients must be identified [14]. Identifying and utilizing a predictive biomarker that is indicative of disease severity can allow physicians to treat patients effectively for better prognosis. While recent studies have identified several biomarkers that are significantly associated with poor prognosis in COVID-19 patients (such as lymphopenia, C-reactive protein (CRP), calcitonin (CT), D-dimer, creatine kinase, aspartate aminotransferase, alanine transaminase, and creatinine and serum amyloid A) accumulating evidence suggests that S100A8/A9 may be a better biomarker than those previously identified [1], [14], [48], [63], [64], [65], [66].
S100A8/A9 is more appealing as biomarker over commonly used inflammatory biomarkers, such as CRP and CT, which require de novo protein biosynthesis. However, S100A8/A9 is synthesized and secreted locally at the inflammatory site before being released into the bloodstream, giving it a decisive kinetic advantage as it may be the first host response to inflammatory conditions [1], [6]. S100A8/A9 has a documented half life of 5 h and its stability at room temperature contributes favorably to its potential as a clinical biomarker in inflammatory diseases, including COVID-19 [1], [48], [51]. The local production and secretion is beneficial, as S100A8/A9 can be detected in several bodily fluids (such as stool, serum, synovial, and salivary media) and has already been investigated as a potential biomarker for a number of inflammatory conditions, such as CF, IBS, and rheumatoid arthritis [1], [30], [67]. Due to its elevated levels in other bodily samples and its use as a biomarker in other diseases, the specificity of S100A8/A9 must be determined to indicate if poor prognosis in patients with COVID-19 can be accurately predicted in the presence of other inflammatory comorbidities.
Plasma S100A8/A9 levels are significantly higher in control, mild, and severe COVID-19 patients with comorbidities as compared to those without comorbidities [48]. However, the observed increase in S100A8/A9 in patients with severe COVID-19 far exceeded correlations associated with comorbidities, which included obesity, diabetes mellitus, cardiovascular and respiratory disease, and cancer [48]. Furthermore, when S100A8/A9 plasma levels were modeled using multivariable linear regression to take into account potential confounding factors (such as age, sex, and comorbidities), the previously observed correlations with neutrophil count, fibrinogen, and D dimers, remained statistically significant [48]. Therefore, further studies are needed to determine potential confounding factors should S100A8/A9 be used as a clinical biomarker.
As previously mentioned, it was originally thought that co-infection with bacteria caused the increase in neutrophil count in patients with severe COVID-19. During infection with Mtb, S100A8/A9 is implicated in mediating neutrophil accumulation by regulating CD11b expression, resulting in enhanced TB disease. Furthermore, serum S100A8/A9 levels are indicative of TB disease state [58], [59]. Therefore, it is not unreasonable to assume that bacterial co-infections may decrease the capacity for S100A8/A9 to specifically predict COVID-19 progression. Although some patients with severe COVID-19 had co-infections with bacteria, it did not significantly modify S100A8/A9 levels [48]. Together, these results indicate that plasma levels of S100A8/A9 can discern between severe COVID-19 patients and patients with mild COVID-19. Importantly, the increase observed in severe cases is independent of confounding factors for prognosis, including advanced age, comorbidities, or concurrent bacterial infection, which have only minor effects on plasma S100A8/A9 levels [48].
It has been previously demonstrated that S100A8/A9 levels are indicative of COVID-19 disease severity, where mild, moderate, and severe cases of COVID-19 correspond to low, mid-range, and high S100A8/A9 levels, respectively [48]. While serum levels in healthy individuals are typically below 1 ug/mL, these levels are reported to rise by nearly 100-fold following inflammation [1], [30]. Though multiple studies have found that serum samples from clinical COVID-19 patients contain elevated S100A8/A9 levels that correspond to disease severity, Bauer et al. investigated S100A8/A9 levels in regard to prediction of prognosis in COVID-19 positive patients admitted to the emergency department (ED) [6], [46], [48], [68]. The authors demonstrated that patients that suffered from MOF within 72 h of sample collection had increased S100A8/A9 levels compared to patients that did not suffer from MOF [6]. The area under the receiver operating characteristic curve (AUROC) was calculated for S100A8/A9, which was compared to biomarkers that are routinely used for clinical evaluation after admission to the ED. Compared to lactate, CRP, and CT, S100A8/A9 had the highest AUROC value (0.87) for capacity to predict MOF in patients within 72 h of sample collection (lactate, 0.79; CRP, 0.70; CT, 0.75). In patients who experienced MOF at any point after ICU admission, the calculated AUROC of S100A8/A9 increased to 0.91, in comparison to lactate (0.82), CRP (0.83), and CT (0.76) [6] ( Table 1).
Table 1.
Biomarker prediction of COVID-19 patient outcomes.
| Prediction | Biomarker | AUC value | 95% CI | p-value | Author |
|---|---|---|---|---|---|
| MOF in patients (within 72 h) | |||||
| S100A8/A9 | 0.87 | 0.63–1.00 | 0.03 | Bauer et al.[6] | |
| Lactate | 0.79 | 0.38–1.00 | 0.09 | ||
| CRP | 0.70 | 0.34–1.00 | 0.26 | ||
| CT | 0.75 | 0.37–1.00 | 0.15 | ||
| MOF in patients | |||||
| S100A8/A9 | 0.91 | 0.77–1.00 | < 0.01 | Bauer et al.[6] | |
| 0.64 | 0.03 | Kaya et al.[70] | |||
| Lactate | 0.82 | 0.56–1.00 | 0.02 | Bauer et al.[6] | |
| CRP | 0.83 | 0.58–1.00 | 0.08 | ||
| CT | 0.76 | 0.47–1.00 | |||
| ICU Treatment | |||||
| S100A8/A9 | 0.70 | 0.42–0.99 | 0.15 | Bauer et al.[6] | |
| Lactate | 0.80 | 0.58–1.00 | 0.03 | ||
| CRP | 0.66 | 0.36–0.96 | 0.27 | ||
| CT | 0.60 | 0.29–0.90 | 0.51 | ||
| 90 Day Mortality | |||||
| S100A8/A9 | 0.85 | 0.54–1.00 | 0.14 | Bauer et al.[6] | |
| Lactate | 0.88 | 0.71–1.00 | 0.10 | ||
| CRP | 1 | 1 | 0.01 | ||
| CT | 1 | 1 | 0.03 | ||
| Survivors v. Fatal | |||||
| S100A8/A9 | 0.80 | 0.691–0.894 | 0.001 | De Guadiana Romualdo et al.[69] | |
| Ferritin | 0.69 | 0.566–0.800 | 0.080 | ||
| CRP | 0.791 | 0.673–0.881 | 0.003 | ||
| D-dimer | 0.869 | 0.763–0.939 | < 0.001 | ||
| GDF-15 | 0.892 | 0.792–0.955 | < 0.001 | ||
| Ventilation | |||||
| S100A8/A9 | 0.794 | N/R | N/R | Shi et al.[68] | |
| LDH | 0.699 | N/R | N/R | ||
| CRP | 0.614 | N/R | N/R | ||
| Ferritin | 0.562 | N/R | N/R |
Similar studies have investigated the predictive capacity of S100A8/A9. Serum S100A8/A9 levels, while increased in fatal COVID-19 cases, had an AUROC value of 0.80 in comparison to ferritin (0.69), CRP (0.791), D-dimer (0.869) and GDF-15 (0.892) [69]. Similarly, serum S100A8/A9 levels were significantly higher in patients admitted to the ICU, with a predictive AUROC value of 0.64. However, it should be noted that its predictive power was not compared to other clinically relevant biomarkers [70]. Lastly, S100A8/A9 levels were increased in patients who required ventilation as compared to those who did not. The capacity of S100A8/A9 to predict ventilation needs had a calculated AUROC value of 0.794 in contrast to LDH (0.699), CRP (0.614), and ferritin (0.562) [68] (Table 1). Together, these initial findings provide promising evidence that S100A8/A9, as compared to other biomarkers, may have better predictive capacity for multiple aspects of COVID-19 prognosis.
Clinically, S100A8/A9 is currently used to distinguish between inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS). Individuals with IBD have inflammation or destruction of the bowel wall, which can lead to lesions or narrowing of the intestines. IBD is a broad term which encompasses two major forms of chronic intestinal inflammation: Crohn’s disease and ulcerative colitis (UC) [71]. In contrast, IBS does not cause ulcers nor lesions within the bowel and is mainly associated with the colon. Both IBD and IBS have similar symptoms, such as abdominal pain and diarrhea, and can be difficult to distinguish from one another without endoscopy. Because S100A8/A9 is heavily implicated in inflammation, relatively stable for days at room temperature, and non-invasive, it was investigated as a potential biomarker where samples could be collected at home and analyzed at the lab. Recently, technological advances have made it possible to rapidly quantify fecal S100A8/A9 by benchtop machinery following sample analysis via lateral flow assay. While these machines can rapidly detect and analyze S100A8/A9 levels, they are only calibrated for fecal samples and cannot be used to accurately detect S100A8/A9 in other human samples. However, the existence of rapid S100A8/A9 quantification technology is promising, as perhaps the technology can be modified for use for other inflammatory conditions and human samples to improve patient outcomes.
Infection with Mtb, the causative agent of tuberculosis, can result in either latent infection (LTB) or ATB disease. While latently infected individuals are likely not able to transmit the disease to others, there remains a 10% lifetime risk that LTB patients may progress to ATB disease. Currently, there is an unmet clinical need for rapid, point-of-care TB diagnostic tests to decrease mortality and transmission rates, as available TB diagnostics are limited in their capacity to rapidly distinguish between TB disease states and may also require lab settings to analyze samples, which can be challenging in high TB-incidence, low-resource settings.
S100A8/A9 has been implicated as a driver of TB pathogenesis in both human and animal models, whereby exacerbated lung inflammation can be attributed to S100A8/A9-mediated neutrophil accumulation [58], [59]. Furthermore, S100A9-deficient mice had lower rates of LTB reactivation and bacterial lung burden when compared with B6 mice, indicating that S100A8/A9 protein expression is also associated with LTB progression S100A8/A9 [59]. In addition, serum S100A8/A9 levels increase with TB disease severity and can distinguish between ATB and HC (AUC = 0.9083) and, to a lesser extent, ATB and LTB (AUC = 0.8575) [59]. While there are currently no tests that can distinguish between LTB and ATB disease, the knowledge that serum S100A8/A9 levels can distinguish between these disease states is promising for the development of a new diagnostic test. Excitingly, serum mRNA levels of S100A8 and S100A9 have been shown to increase in LTB progressors six months prior to ATB diagnosis as compared to LTB non-progressors, who did not significantly increase S100A8 or S100A9 expression [59]. These results are encouraging for the potential future development of a new TB diagnostic test that may be able to distinguish between LTB progressors and non-progressors, allowing patients to receive treatment before the onset of ATB disease to improve their prognosis and reduce TB transmission.
7. S100A8/A9 as a potential therapeutic target
As previously discussed, aberrant neutrophil groups account for the emergence of COVID-19 symptoms in mice and may be responsible for the formation of fatal infections by SARS-CoV-2. Both S100A8 and S100A9 have been shown to influence neutrophil accumulation in inflammatory infectious diseases, such as tuberculosis [59]. Therefore, S100A8/A9 has recently been investigated as a potential target for small molecule therapeutics to suppress the uncontrolled immune response associated with severe COVID-19 cases [46].
Paquinimod is an S100A9-specific small molecule inhibitor that prevents the binding of S100A9 to TLR4, suggesting that it can be used to block the function of S100A8/A9 [72], [73]. Mice treated with paquinimod had lower amounts of pulmonary interstitial damage, inhibited viral replication, and a reduced accumulation of aberrant neutrophil subsets. Specifically, the aberrant neutrophil subset returned to normal CD45+ CD11b+Ly6Ghigh neutrophils following treatment with paquinimod, and both S100a8 and Ly6g expression were reduced in mice. Mice infected with IAV that lacked excessive S100a8 expression and the abnormal myeloid cell subset did not have improved outcomes following paquinimod treatment. In short, it is postulated that the inhibitor blocked the function of S100A8/A9 specifically and prevented the accumulation of aberrant neutrophils and fatal outcome [46].
Because paquinimod treatment rescued mice from severe COVID-19, likely by preventing the interaction between S100A8/A9 and TLR4 from occurring, it was suggested that the TLR4 signaling pathway could play an important role in critical SARS-CoV-2 infections. Administration of resatorvid, a selective TLR4 inhibitor that downregulates the expression of downstream TLR4 signaling molecules, to control mice and mice infected with SARS-CoV-2, resulted in a decrease to the aberrant neutrophil subset, the viral replication in the lungs, and both S100a8 and Ly6g expression (Fig. 1) [46]. However, S100A8/A9 is also known to interact with RAGE. To determine the importance of S100A8/A9-RAGE interactions in severe COVID-19 cases, mice were treated with azeliragon, a RAGE inhibitor. Azeliragon treatment did not significantly prevent the production of the aberrant neutrophil subset or viral replication, indicating that the TLR4 pathway may play a critical role in fatal COVID-19 cases [46]. Together, these results highlight the therapeutic target potential of S100A8/A9 in improving host control over the infection by suppressing the abnormal immune response caused by SARS-CoV-2 infection.
Because S100A8, S100A9, and S100A8/A9 are implicated in several inflammatory conditions, a variety of S100-specific inhibitors have been developed for therapeutic treatment. Notably, quinoline-3-carboxamide derivatives, such as tasquinimod, have been shown to bind to S100A9 and the S100A8/A9 complex to block the interaction of S100A9 with TLR4 and RAGE [1], [28]. Aside from treating inflammatory infectious diseases, inhibiting S100A8/A9 has been shown to decrease lung inflammation during IAV infection, prevent liver injury and bacterial dissemination during human sepsis and endotoxemia, and improve the clinical score of rheumatoid arthritis patients by nearly 50% [41], [74]. Additionally, S100A8/A9 inhibition has been shown to alleviate worsening of disease in other inflammatory conditions, such as tuberculosis, where Mtb-infected B6 mice treated with FPS-ZM1, another RAGE inhibitor, decreased the bacterial lung burden to levels comparable to Mtb-infected S100A9-deficient mice [59].
8. Conclusions
Although S100A8/A9 has been implicated in inflammatory conditions, such as IBS, CF, and rheumatoid arthritis, it acts distinctively during SARS-CoV-2 infection to drive the formation and chemotaxis of a group of aberrant neutrophils, which are thought to drive fatal coronavirus infections and cytokine storm. While its role in chemotaxis has previously been identified in TB disease pathogenesis, S100A8/A9 notably affects the formation of immature neutrophil subsets during SARS-CoV-2 infection, possibly through its interactions with TLR4. Furthermore, while elevated serum and plasma levels of S100A8/A9 are indicative of worsening COVID-19 prognosis, it is possible that preventing S100A8/A9-TLR4 interactions through paquinimod or resatorvid treatment may improve disease outcome by limiting excessive S100a8 and Ly6g expression, thus reducing viral lung burden and diminishing the aberrant neutrophil population as has been observed in mice. However, additional studies are needed to confirm the changes in disease phenotype in other models following treatment of paquinimod or resatorvid. Future research should be aimed at distinguishing the function and mechanism of induction of the aberrant neutrophil subset, as it will provide valuable information on potential contributions to deteriorating COVID-19 cases and how to prevent the subset from worsening disease outcomes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Washington University in St. Louis, National Institutes of Health (USA) Grant #AI134236-05S1 to SAK, and the Washington University (USA) T32 #HL007317-42 Pulmonary Training Grant to LM.
Biographies

Leah Mellett is a pre-doctoral student at Washington University in Saint Louis. She received her Bachelor of Science in May 2018 from Beloit College, majoring in Biochemistry. She is pursuing her doctorate in Biology and Biomedical Sciences at Washington University in Saint Louis and plans to graduate in 2024. Since September 2019, she has been involved in Dr. Khader’s research lab and is currently studying S100A8/A9 in the context of tuberculosis.

Shabaana Khader received her PhD in Biotechnology from Madurai Kamaraj University, India where she studied host-pathogen interactions during the mycobacterial disease, leprosy. Dr. Khader then carried out her Post-doctoral training at the Trudeau Institute, NY, where she continued studying host immune responses to another globally relevant mycobacterial disease, tuberculosis. During her stay at the Trudeau Institute, Dr. Khader demonstrated a critical role for the cytokine Interleukin-17 in vaccine-induced immunity to tuberculosis, as well as described seminal roles for IL-12 cytokines in tuberculosis. Dr. Khader then joined the University of Pittsburgh in 2007 as Assistant Professor in the Department of Pediatrics where her lab continued to study the role of cytokines in immunity to intracellular pathogens such as Mycobacterium tuberculosis and Francisella tularensis. In 2013, Dr. Khader and her research team moved to the Department of Molecular Microbiology at the Washington University in St. Louis, where she is now a tenured Professor, in the Department of Molecular Microbiology. Her lab is focused on the immunopathogenesis of tuberculosis including aspects of both innate and adaptive immunity as well as vaccine-induced immunity to tuberculosis.
References
- 1.S. Wang, R. Song, Z. Wang, Z. Jing, S. Wang, J. Ma, S100A8/A9 in Inflammation. Front Immunol [Internet]. 2018 Jun 11 [cited 2021 Jan 20];9. Available from: 〈https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6004386/〉.
- 2.COVID-19 Map [Internet], Johns Hopkins Coronavirus Resource Center. [cited 2021 Apr 22]. Available from: 〈https://coronavirus.jhu.edu/map.html〉.
- 3.C. Troeger, Just How Do Deaths Due to COVID-19 Stack Up? Think Global Health [Internet]. Available from: 〈https://www.thinkglobalhealth.org/article/just-how-do-deaths-due-covid-19-stack〉.
- 4.F.B. Ahmad, Provisional Mortality Data — United States, 2020. MMWR Morb Mortal Wkly Rep [Internet]. 2021, 70 [cited 2021 Sep 2];70–522. Available from: https://www.cdc.gov/mmwr/volumes/70/wr/mm7014e1.htm. [DOI] [PMC free article] [PubMed]
- 5.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:10–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer W., Diehl-Wiesenecker E., Ulke J., Galtung N., Havelka A., Hegel J.K., Tauber R., Somasundaram R., Kappert K. Outcome prediction by serum calprotectin in patients with COVID-19 in the emergency department. J. Infect. 2021;82(4):84–123. doi: 10.1016/j.jinf.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y., Tu L., Zhu P., Mu M., Wang R., Yang P., Wang X., Hu C., Ping R., Hu P., Li T., Cao F., Chang C., Hu Q., Jin Y., Xu G. Clinical features of 85 fatal cases of COVID-19 from Wuhan. A retrospective observational study. Am. J. Respir. Crit. Care Med. 2020;201(11):1372–1379. doi: 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.K., Roth J., Oppenheim J.J., Tracey K.J., Vogl T., Feldmann M., Horwood N., Nanchahal J. Alarmins: awaiting a clinical response. J. Clin. Invest. 2012;122(8):2711–2719. doi: 10.1172/JCI62423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Pia L., Risson E., Saffern M., Salomé B., Esai Selvan M., Spindler M.P., Tan J., van der Heide V., Gregory J.K., Alexandropoulos K., Bhardwaj N., Brown B.D., Greenbaum B., Gümüş Z.H., Homann D., Horowitz A., Kamphorst A.O., Curotto de Lafaille M.A., Mehandru S., Merad M., Samstein R.M., Agrawal M., Aleynick M., Belabed M., Brown M., Casanova-Acebes M., Catalan J., Centa M., Charap A., Chan A., Chen S.T., Chung J., Bozkus C.C., Cody E., Cossarini F., Dalla E., Fernandez N., Grout J., Ruan D.F., Hamon P., Humblin E., Jha D., Kodysh J., Leader A., Lin M., Lindblad K., Lozano-Ojalvo D., Lubitz G., Magen A., Mahmood Z., Martinez-Delgado G., Mateus-Tique J., Meritt E., Moon C., Noel J., O’Donnell T., Ota M., Plitt T., Pothula V., Redes J., Reyes Torres I., Roberto M., Sanchez-Paulete A.R., Shang J., Schanoski A.S., Suprun M., Tran M., Vaninov N., Wilk C.M., Aguirre-Ghiso J., Bogunovic D., Cho J., Faith J., Grasset E., Heeger P., Kenigsberg E., Krammer F., Laserson U. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong J.P., Viswanathan S., Wang M., Sun L.-Q., Clark G.C., D’Elia R.V. Current and future developments in the treatment of virus-induced hypercytokinemia. Future Med. Chem. 2017;9(2):169–178. doi: 10.4155/fmc-2016-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368 doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., Hirano T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., Wei J., Luo H., Zhu H., Huang L., Meng F., Huang L., Wang N., Zhou X., Zhao L., Chen X., Mao Z., Chen C., Li Z., Sun Z., Zhao J., Wang D., Huang G., Wang W., Zhou J. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:1–3. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oppenheim J.J., Yang D. Alarmins: chemotactic activators of immune responses. Curr. Opin. Immunol. 2005;17(4):359–365. doi: 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Yang D., Han Z., Oppenheim J.J. Alarmins and immunity. Immunol. Rev. 2017;280(1):41–56. doi: 10.1111/imr.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianchi M.E. DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukocyte Biol. 2007;81(1):1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 18.Chen G.Y., Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 2010;10(12):826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathan C. Points of control in inflammation. Nature. 2002;420(6917):846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 20.Cher J.Z.B., Akbar M., Kitson S., Crowe L.A.N., Garcia-Melchor E., Hannah S.C., McLean M., Fazzi U.G., Kerr S.C., Murrell G., Millar N.L. Alarmins in frozen shoulder: a molecular association between inflammation and pain. Am. J. Sports Med. 2018;46(3):671–678. doi: 10.1177/0363546517741127. [DOI] [PubMed] [Google Scholar]
- 21.R. Kang, HMGB1 in health and disease, 2014;113. [DOI] [PMC free article] [PubMed]
- 22.Patel S. Danger-Associated Molecular Patterns (DAMPs): the Derivatives and Triggers of Inflammation. Curr Allergy Asthma Rep. 2018;18(11):63. doi: 10.1007/s11882-018-0817-3. [DOI] [PubMed] [Google Scholar]
- 23.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giri K., Pabelick C.M., Mukherjee P., Prakash Y.S. Hepatoma derived growth factor (HDGF) dynamics in ovarian cancer cells. Apoptosis. 2016;21(3):329–339. doi: 10.1007/s10495-015-1200-7. [DOI] [PubMed] [Google Scholar]
- 27.Gazzar M.E. Immunobiology of S100A8 and S100A9 proteins and their role in acute inflammation and sepsis. Int. J. Immunol. Immunother. [Internet] 2015;2(2) 〈https://clinmedjournals.org/articles/ijii/international-journal-of-immunology-and-immunotherapy-ijii-2-013.php?jid=ijii〉 (Available from) [Google Scholar]
- 28.Bresnick A.R. S100 proteins as therapeutic targets. Biophys. Rev. 2018;10(6):1617–1629. doi: 10.1007/s12551-018-0471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogl T., Tenbrock K., Ludwig S., Leukert N., Ehrhardt C., van Zoelen M.A.D., Nacken W., Foell D., van der Poll T., Sorg C., Roth J. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007;13(9):1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 30.Ometto F., Friso L., Astorri D., Botsios C., Raffeiner B., Punzi L., Doria A. Calprotectin in rheumatic diseases. Exp. Biol. Med. ((Maywood)) 2017;242(8):859–873. doi: 10.1177/1535370216681551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rammes A., Roth J., Goebeler M., Klempt M., Hartmann M., Sorg C. Myeloid-related protein (MRP) 8 and MRP14, calcium-binding proteins of the s100 family, are secreted by activated monocytes via a novel, tubulin-dependent pathway*. J. Biol. Chem. 1997;272(14):9496–9502. doi: 10.1074/jbc.272.14.9496. [DOI] [PubMed] [Google Scholar]
- 32.Simard J.-C., Noël C., Tessier P.A., Girard D. Human S100A9 potentiates IL-8 production in response to GM-CSF or fMLP via activation of a different set of transcription factors in neutrophils. FEBS Lett. 2014;588(13):2141–2146. doi: 10.1016/j.febslet.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 33.Riva M., Källberg E., Björk P., Hancz D., Vogl T., Roth J., Ivars F., Leanderson T. Induction of nuclear factor-κB responses by the S100A9 protein is Toll-like receptor-4-dependent. Immunology. 2012;137(2):172–182. doi: 10.1111/j.1365-2567.2012.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narumi K., Miyakawa R., Ueda R., Hashimoto H., Yamamoto Y., Yoshida T., Aoki K. Proinflammatory proteins S100A8/S100A9 activate NK cells via interaction with RAGE. JI. 2015;194(11):5539–5548. doi: 10.4049/jimmunol.1402301. [DOI] [PubMed] [Google Scholar]
- 35.Austermann J., Friesenhagen J., Fassl S.K., Ortkras T., Burgmann J., Barczyk-Kahlert K., Faist E., Zedler S., Pirr S., Rohde C., Müller-Tidow C., von Köckritz-Blickwede M., von Kaisenberg C.S., Flohé S.B., Ulas T., Schultze J.L., Roth J., Vogl T., Viemann D. Alarmins MRP8 and MRP14 induce stress tolerance in phagocytes under sterile inflammatory conditions. Cell Rep. 2014;9(6):2112–2123. doi: 10.1016/j.celrep.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Freise N., Burghard A., Ortkras T., Daber N., Imam Chasan A., Jauch S.-L., Fehler O., Hillebrand J., Schakaki M., Rojas J., Grimbacher B., Vogl T., Hoffmeier A., Martens S., Roth J., Austermann J. Signaling mechanisms inducing hyporesponsiveness of phagocytes during systemic inflammation. Blood. 2019;134(2):134–146. doi: 10.1182/blood.2019000320. [DOI] [PubMed] [Google Scholar]
- 37.Ulas T., Pirr S., Fehlhaber B., Bickes M.S., Loof T.G., Vogl T., Mellinger L., Heinemann A.S., Burgmann J., Schöning J., Schreek S., Pfeifer S., Reuner F., Völlger L., Stanulla M., von Köckritz-Blickwede M., Glander S., Barczyk-Kahlert K., von Kaisenberg C.S., Friesenhagen J., Fischer-Riepe L., Zenker S., Schultze J.L., Roth J., Viemann D. S100-alarmin-induced innate immune programming protects newborn infants from sepsis. Nat. Immunol. 2017;18(6):622–632. doi: 10.1038/ni.3745. [DOI] [PubMed] [Google Scholar]
- 38.Kuipers M.T., Vogl T., Aslami H., Jongsma G., van den Berg E., Vlaar A.P.J., Roelofs J.J.T.H., Juffermans N.P., Schultz M.J., van der Poll T., Roth J., Wieland C.W. High levels of S100A8/A9 proteins aggravate ventilator-induced lung injury via TLR4 signaling. PLOS One. 2013;8(7) doi: 10.1371/journal.pone.0068694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goyette J., Geczy C.L. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids. 2011;41(4):821–842. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- 40.Lorenz E., Muhlebach M.S., Tessier P.A., Alexis N.E., Duncan Hite R., Seeds M.C., Peden D.B., Meredith W. Different expression ratio of S100A8/A9 and S100A12 in acute and chronic lung diseases. Respir. Med. 2008;102(4):567–573. doi: 10.1016/j.rmed.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai S.-Y., Segovia J.A., Chang T.-H., Morris I.R., Berton M.T., Tessier P.A., et al. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. [Internet] 2014;10(1) doi: 10.1371/journal.ppat.1003848. 〈https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3879357/〉 (Available from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y., Liu X., Xiong L., Cai K. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J. Med. Virol. 2020;92(9):1449–1459. doi: 10.1002/jmv.25822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Du X., Chen J., Jin Y., Peng L., Wang H.H.X., Luo M., Chen L., Zhao Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020;81(1):e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B., Zhou X., Qiu Y., Song Y., Feng F., Feng J., Song Q., Jia Q., Wang J. Clinical characteristics of 82 cases of death from COVID-19. PLOS One. 2020;15(7) doi: 10.1371/journal.pone.0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Q., Zhao Y., Li J., Liu J., Yang X., Guo X., Kuang M., Xia H., Zhang Z., Cao L., Luo Y., Bao L., Wang X., Wei X., Deng W., Wang N., Chen L., Chen J., Zhu H., Gao R., Qin C., Wang X., You F. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021;29(2):222–235.e4. doi: 10.1016/j.chom.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., Rameau P., Catelain C., Alfaro A., Dussiau C., Friedrich C., Sourdeau E., Marin N., Szwebel T.A., Cantin D., Mouthon L., Borderie D., Deloger M., Bredel D., Mouraud S., Drubay D., Andrieu M., Lhonneur A.S., Saada V., Stoclin A., Willekens C., Pommeret F., Griscelli F., Ng L.G., Zhang Z., Bost P., Amit I., Barlesi F., Marabelle A., Pène F., Gachot B., André F., Zitvogel L., Ginhoux F., Fontenay M., Solary E. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal. Transduct Target Ther. [Internet] 2020:5. doi: 10.1038/s41392-020-0148-4. 〈https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7100419/〉 (Available from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., Kuthuru O., Apostolidis S.A., Bershaw L., Dougherty J., Greenplate A.R., Pattekar A., Kim J., Han N., Gouma S., Weirick M.E., Arevalo C.P., Bolton M.J., Goodwin E.C., Anderson E.M., Hensley S.E., Jones T.K., Mangalmurti N.S., Luning Prak E.T., Wherry E.J., Meyer N.J., Betts M.R. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 52.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., De Domenico E., Wendisch D., Grasshoff M., Kapellos T.S., Beckstette M., Pecht T., Saglam A., Dietrich O., Mei H.E., Schulz A.R., Conrad C., Kunkel D., Vafadarnejad E., Xu C.J., Horne A., Herbert M., Drews A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., Müller-Redetzky H., Heim K.M., Machleidt F., Uhrig A., Bosquillon de Jarcy L., Jürgens L., Stegemann M., Glösenkamp C.R., Volk H.D., Goffinet C., Landthaler M., Wyler E., Georg P., Schneider M., Dang-Heine C., Neuwinger N., Kappert K., Tauber R., Corman V., Raabe J., Kaiser K.M., Vinh M.T., Rieke G., Meisel C., Ulas T., Becker M., Geffers R., Witzenrath M., Drosten C., Suttorp N., von Kalle C., Kurth F., Händler K., Schultze J.L., Aschenbrenner A.C., Li Y., Nattermann J., Sawitzki B., Saliba A.E., Sander L.E., Deutsche COVID- OMICS Initiative ( Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng L.G., Ostuni R., Hidalgo A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 2019;19(4):255–265. doi: 10.1038/s41577-019-0141-8. [DOI] [PubMed] [Google Scholar]
- 55.Basiorka A.A., McGraw K.L., Eksioglu E.A., Chen X., Johnson J., Zhang L., Zhang Q., Irvine B.A., Cluzeau T., Sallman D.A., Padron E., Komrokji R., Sokol L., Coll R.C., Robertson A.A.B., Cooper M.A., Cleveland J.L., O’Neill L.A., Wei S., List A.F. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood. 2016;128(25):2960–2975. doi: 10.1182/blood-2016-07-730556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X., Eksioglu E.A., Zhou J., Zhang L., Djeu J., Fortenbery N., Epling-Burnette P., Van Bijnen S., Dolstra H., Cannon J., Youn J.I., Donatelli S.S., Qin D., De Witte T., Tao J., Wang H., Cheng P., Gabrilovich D.I., List A., Wei S. Induction of myelodysplasia by myeloid-derived suppressor cells. J. Clin. Invest. 2013;123(11):4595–4611. doi: 10.1172/JCI67580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharya M., Sharma A.R., Mallick B., Sharma G., Lee S.S., Chakraborty C. Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104587. 104587–104587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gopal R., Monin L., Torres D., Slight S., Mehra S., McKenna K.C., Fallert Junecko B.A., Reinhart T.A., Kolls J., Báez-Saldaña R., Cruz-Lagunas A., Rodríguez-Reyna T.S., Kumar N.P., Tessier P., Roth J., Selman M., Becerril-Villanueva E., Baquera-Heredia J., Cumming B., Kasprowicz V.O., Steyn A.J., Babu S., Kaushal D., Zúñiga J., Vogl T., Rangel-Moreno J., Khader S.A. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am. J. Respir. Crit. Care Med. 2013;188(9):1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott N.R., Swanson R.V., Al-Hammadi N., Domingo-Gonzalez R., Rangel-Moreno J., Kriel B.A., Bucsan A.N., Das S., Ahmed M., Mehra S., Treerat P., Cruz-Lagunas A., Jimenez-Alvarez L., Muñoz-Torrico M., Bobadilla-Lozoya K., Vogl T., Walzl G., du Plessis N., Kaushal D., Scriba T.J., Zúñiga J., Khader S.A. S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J. Clin. Invest. 2020;130(6):3098–3112. doi: 10.1172/JCI130546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gray R.D., Imrie M., Boyd A.C., Porteous D., Innes J.A., Greening A.P. Sputum and serum calprotectin are useful biomarkers during CF exacerbation. J. Cyst. Fibros. 2010;9(3):193–198. doi: 10.1016/j.jcf.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Parisi G.F., Papale M., Rotolo N., Aloisio D., Tardino L., Scuderi M.G., Di Benedetto V., Nenna R., Midulla F., Leonardi S. Severe disease in Cystic Fibrosis and fecal calprotectin levels. Immunobiology. 2017;222(3):582–586. doi: 10.1016/j.imbio.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Schnapp Z., Hartman C., Livnat G., Shteinberg M., Elenberg Y. Decreased fecal calprotectin levels in cystic fibrosis patients after antibiotic treatment for respiratory exacerbation. J. Pediatr. Gastroenterol. Nutr. 2019;68(2):282–284. doi: 10.1097/MPG.0000000000002197. [DOI] [PubMed] [Google Scholar]
- 63.Malik P., Patel U., Mehta D., Patel N., Kelkar R., Akrmah M., et al. Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ EBM. 2020 doi: 10.1136/bmjebm-2020-111536. bmjebm-2020-111536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57:1–11. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. (CCLM) 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 66.Effenberger M., Grabherr F., Mayr L., Schwaerzler J., Nairz M., Seifert M., Hilbe R., Seiwald S., Scholl-Buergi S., Fritsche G., Bellmann-Weiler R., Weiss G., Müller T., Adolph T.E., Tilg H. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69(8):1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haga H.J., Brun J.G., Berntzen H.B., Cervera R., Khamashta M., Hughes G.R. Calprotectin in patients with systemic lupus erythematosus: relation to clinical and laboratory parameters of disease activity. Lupus. 1993;2(1):47–50. doi: 10.1177/096120339300200108. [DOI] [PubMed] [Google Scholar]
- 68.Shi H., Zuo Y., Yalavarthi S., Gockman K., Zuo M., Madison J.A., et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. medRxiv [Internet] 2020 doi: 10.1002/JLB.3COVCRA0720-359R. 〈https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7274221/〉 (Available from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luis García de Guadiana Romualdo R., Mulero M.D.R., Olivo M.H., Rojas C.R., Arenas V.R., Morales M.G., Abellán A.B., Conesa-Zamora P., García-García J., Hernández A.C., Morell-García D., Dolores Albaladejo-Otón M., Consuegra-Sánchez L. Circulating levels of GDF-15 and calprotectin for prediction of in-hospital mortality in COVID-19 patients: a case series. J. Infect. 2021;82(2):e40–e42. doi: 10.1016/j.jinf.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaya T., Yaylacı S., Nalbant A., Yıldırım İ., Kocayiğit H., Çokluk E., Şekeroğlu M.R., Köroğlu M., Güçlü E. Serum calprotectin as a novel biomarker for severity of COVID-19 disease. Ir. J. Med. Sci. [Internet] 2021 doi: 10.1007/s11845-021-02565-8. Available from: http://link.springer.com/10.1007/s11845-021-02565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Rheenen P.F., Van de Vijver E., Fidler V. Faecal calprotectin for screening of patients with suspected inflammatory bowel disease: diagnostic meta-analysis. BMJ. 2010;341(jul15 1):c3369. doi: 10.1136/bmj.c3369. c3369–c3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schelbergen R.F., Geven E.J., van den Bosch M.H.J., Eriksson H., Leanderson T., Vogl T., Roth J., van de Loo F.A.J., Koenders M.I., van der Kraan P.M., van den Berg W.B., Blom A.B., van Lent P.L.E.M. Prophylactic treatment with S100A9 inhibitor paquinimod reduces pathology in experimental collagenase-induced osteoarthritis. Ann. Rheum Dis. 2015;74(12):2254–2258. doi: 10.1136/annrheumdis-2014-206517. [DOI] [PubMed] [Google Scholar]
- 73.Björk P., Björk A., Vogl T., Stenström M., Liberg D., Olsson A., Roth J., Ivars F., Leanderson T. Identification of human S100A9 as a novel target for treatment of autoimmune disease via binding to quinoline-3-carboxamides. Akira S, editor. PLoS Biol. 2009;7(4) doi: 10.1371/journal.pbio.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cesaro A., Anceriz N., Plante A., Pagé N., Tardif M.R., Tessier P.A. An inflammation loop orchestrated by S100A9 and calprotectin is critical for development of arthritis. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045478. [DOI] [PMC free article] [PubMed] [Google Scholar]