Abstract
The proline utilization pathway in Saccharomyces cerevisiae is regulated by the Put3p transcriptional activator in response to the presence of the inducer proline and the quality of the nitrogen source in the growth medium. Put3p is constitutively bound to the promoters of its target genes, PUT1 and PUT2, under all conditions studied but activates transcription to the maximum extent only in the absence of rich nitrogen sources and in the presence of proline (i.e., when proline serves as the sole source of nitrogen). Changes in target gene expression therefore occur through changes in the activity of the DNA-bound regulator. In this report, we demonstrate by phosphatase treatment of immunoprecipitates of extracts metabolically labeled with 32P or 35S that Put3p is a phosphoprotein. Examination of Put3p isolated from cells grown on a variety of nitrogen sources showed that it was differentially phosphorylated as a function of the quality of the nitrogen source: the poorer the nitrogen source, the slower the gel migration of the phosphoforms. The presence of the inducer does not detectably alter the phosphorylation profile. Activator-defective and activator-constitutive Put3p mutants have been analyzed. One activator-defective mutant appears to be phosphorylated in a pattern similar to that of the wild type, thus separating its ability to be phosphorylated from its ability to activate transcription. Three activator-constitutive mutant proteins from cells grown on an ammonia-containing medium have a phosphorylation profile similar to that of the wild-type protein in cells grown on proline. These results demonstrate a correlation between the phosphorylation status of Put3p and its ability to activate its target genes and suggest that there are two signals, proline induction and quality of nitrogen source, impinging on Put3p that act synergistically for maximum expression of the proline utilization pathway.
Saccharomyces cerevisiae cells can sense the quality of the nitrogen source in their environment, enabling them to utilize preferred nitrogen-containing compounds over nonpreferred ones or to express pathways for the utilization of alternative nitrogen sources when the preferred ones have been consumed. Although very little is known about the sensing mechanism itself, work over the last decade has led to the discovery of a set of regulatory proteins, the GATA factors, whose role is to regulate, in both positive and negative directions, the expression of pathways of nitrogen assimilation in yeast. These proteins, Gln3p (26), Nil1p/Gat1p (10, 44), Dal80p/Uga43p (12, 13), and Nil2p/Gzf3p/Deh1p (11, 34, 42), are involved in a complex set of regulatory loops, competition for GATA binding sites, and possibly even some autoregulation. Recently, the coactivator Ada1p, isolated as Gan1p, was identified as a link between the GATA binding proteins and the basal transcriptional machinery (41). Global nitrogen repressor Ure2p is believed to interact with Gln3p to obtain appropriate expression of a variety of nitrogen assimilatory pathways (3; reviewed by Magasanik [23]).
In their natural habitat, S. cerevisiae cells are found on grapes and in grape must, a nitrogen-poor environment where the most abundant nitrogen source is proline (2). Although proline is the least-preferred nitrogen source for many laboratory yeast strains and although its utilization results in the slowest growth rates, yeast cells have evolved a regulatory circuit that enables them to use the proline in the environment when preferred nitrogen sources are no longer available. The flux of proline into yeast cells is controlled by the activities of the general amino acid permease Gap1p and the proline-specific permease, Put4p (21). These permeases are regulated by nitrogen repression and do not respond to proline induction (17, 21, 43). The enzymes of the proline utilization pathway are induced by the presence of proline (6), and their levels reflect internal proline levels. The PUT1 and PUT2 genes encoding the enzymes of the pathway are regulated by Put3p, a member of the Zn(II)2Cys6 binuclear cluster protein family (4, 6, 7, 15, 24, 25, 40, 45, 49) and a close relative of Gal4p, the activator of the galactose utilization pathway. In vivo, Put3p binds the promoters of PUT1 and PUT2 in the presence or absence of proline and without regard to the quality of the nitrogen sources present in the growth medium (1) but activates transcription to a maximum level when proline is the sole source of nitrogen. PUT1 and PUT2 are repressed by Ure2p and in some, but not all, strain backgrounds are regulated by some of the GATA factors (9, 14, 50).
This report presents the results of studies on wild-type and regulation-defective mutant Put3 proteins in cells grown in media containing different nitrogen sources. We show that Put3p is differentially phosphorylated as a function of the quality of the nitrogen source and that the slowest-migrating species of Put3p are correlated with elevated transcriptional activity. Analysis of the Put3p phosphoforms of activator-defective and activator-constitutive mutants leads to the suggestion that altered phosphorylation status may be one of two signals (proline induction being the other) that is required for maximum transcriptional activity by Put3p.
MATERIALS AND METHODS
Strains and plasmids.
The protease-deficient strain BJ2168 (MATa prb1-1122 pep4-3 prc1-451 leu2 trp1 ura3-52 gal2 [19]) was a gift from J. Thorner. Strain DB1000 was derived from BJ2168 by transformation with a put3::LEU2 DNA fragment from plasmid pDNB118 (see below). This deletion allele removes PUT3 from −190 to +2896 bp (where +1 is the start of the opening reading frame) and is missing codons 1 to 966. Strain DB8-5C (MATα put3-316 ura3-52 TRP1::PUT2-lacZ) carries a mutation in the central domain of the PUT3 gene that converts a glycine at position 532 to arginine (15). Strains BJ2168 and DB1000 are from the S288C background, and strain DB8-5C is from the Σ1278b background.
The plasmids used in this study are listed in Table 1. Plasmid pDB37 (25) carries about 11 kb of genomic DNA of the PUT3 region on chromosome XI inserted into high-copy-number plasmid YEp24; the PUT3 gene is driven by its natural promoter in this construct. Plasmid pYEX4T-1 (Amrad Biotech) is a high-copy-number yeast shuttle vector that contains the S. cerevisiae CUP1 promoter fused to a glutathione S-transferase gene (GST) with a thrombin cleavage site followed by a polylinker region. Plasmid pHB3, containing the CUP1 promoter fused to GST, a thrombin cleavage site, and the PUT3 open reading frame, was constructed in the following way. Plasmid pHB7 contains the entire PUT3 opening reading frame located between an engineered SmaI site immediately upstream of the ATG of PUT3 and a NotI site downstream of the stop codon and polyadenylation sequences in baculovirus expression vector pVL1393 (Pharmingen). A SmaI-NotI fragment containing PUT3 from plasmid pHB7 was inserted into the polylinker of pYEX4T-1 to form plasmid pHB3. This plasmid produces copper-inducible GST-Put3p, which can be cleaved by thrombin to form full-length Put3p.
TABLE 1.
Plasmids used in this study
| Name | Description | Source or reference |
|---|---|---|
| YEp24 | URA3 2μm (high-copy-number plasmid) | D. Botstein |
| pDB37 | PUT3 in YEp24 | 25 |
| pDB193 | put3-75 in YEp24 | 24 |
| pHB6 | put3-316 in YEp24 | This work |
| pHB7 | SmaI-NotI fragment of PUT3 ORF in pLV1393 | This work |
| pYEX4T-1 | GST URA3 leu2d 2μm (high-copy-number plasmid) | Amrad Biotech |
| pHB3 | GST-PUT3 in pYEX4T-1 | This work |
| YCp50 | URA3 CEN-ARS (low-copy-number plasmid) | 18 |
| pDNB109 | 7.7-kb KpnI fragment of PUT3 in YCp50 | D. Barber |
| pDNB118 | put3::LEU2 in YCp50 | D. Barber |
| pDB120 | PUT3c-903 URA3 (integrating plasmid) | 24 |
| pMB3 | PUT3c-903 URA3 in YEp24 | This work |
| pDB191 | PUT3c-914 URA3 (integrating plasmid) | 24 |
| pMB4 | PUT3c-914 URA3 in YEp24 | This work |
| pDB130 | PUT3c-683 URA3 (integrating plasmid) | 24 |
| pMB5 | PUT3c-683 URA3 in YEp24 | This work |
| pMB6 | PUT2-lacZ CEN ARS | This work |
| pMB7 | PUT2-lacZ TRP1 CEN ARS | This work |
Plasmid pHB6, carrying the put3-316 mutation in plasmid YEp24, was constructed as follows. Plasmid pHB4 is a YCp50 (18) derivative which carries a 4-kb KpnI fragment corresponding to DNA 5′ and 3′ of PUT3 but from which the 3.7-kb SnaBI fragment of PUT3 is completely deleted. Plasmid pHB4 was linearized at its unique SnaBI site and used to rescue the put3-316 allele by gap repair (30) from strain DB8-5C. DNA was isolated from Ura+ yeast transformants and screened for put3-containing DNA. The appropriate plasmid, pHB5, was amplified in Escherichia coli, and the 3.7-kb put3-316 DNA fragment was ligated to plasmid YEp24 cut with SmaI to form high-copy-number plasmid pHB6 carrying the mutant allele. As expected, put3Δ strains carrying plasmid pHB6 failed to grow on proline-containing medium but did make Put3p that was detectable by immunoblotting.
Plasmid pDNB118 is a YCp50 derivative that carries a 7.7-kb KpnI fragment of the PUT3 gene, in which a SacII-BstUI fragment of the LEU2 gene was inserted between the SacII site at position −190 bp and the PvuII site at position +2896 of PUT3. This put3::LEU2 allele removes codons 1 to 966 of PUT3.
High-copy-number plasmids carrying each of the constitutive PUT3c alleles PUT3c-903 (L903R), PUT3c-914 (N914I), and PUT3c-683 (S683F) were constructed as follows. The 3.7-kb SnaBI fragments carrying PUT3c were isolated from each of the integrating vectors pDB120, pDB191, and pDB130 (24) and ligated to plasmid YEp24, digested with SmaI. The resulting plasmids were called pMB3, pMB4, and pMB5, respectively. Each encoded a mutant Put3p that could activate transcription of PUT2-lacZ in the absence of proline.
Plasmid pMB6 is a derivative of plasmid YCp50 containing CEN, ARS, and PUT2-lacZ but lacking a yeast selectable marker. It was constructed by ligating the 4.1-kb EcoRI-NsiI, PUT2-lacZ fragment from plasmid pABC4 (39) to a 5.4-kb EcoRI-NsiI fragment of plasmid YCp50. A 1.4-kb EcoRI fragment containing the TRP1 gene from plasmid pJH-W1 (46) was then inserted into the unique EcoRI site of plasmid pMB6 to form plasmid pMB7, a low-copy-number plasmid with TRP1 and PUT2-lacZ.
Media.
The minimal medium used in this study has been previously described (5). Glucose (2%) was the carbon source. Nitrogen sources were ammonium sulfate (0.2%), γ-aminobutyric acid (GABA; 0.1%) without or with proline (0.1%), urea (0.1%) without or with proline (0.1%), and proline (0.1%) alone. Supplements of tryptophan, uracil, or leucine were added when required. For copper induction of GST-Put3p encoded by leu2d-bearing plasmid pHB3, 50 μM copper sulfate was added for 5 h in the absence of leucine. In standard yeast nitrogen base medium without additional copper, the amount of GST-Put3p was induced to about half the amount observed with copper addition.
For metabolic labeling experiments, low-phosphate and -sulfate medium (LPSM) was used. LPSM is identical to standard yeast nitrogen base except for the concentrations of KH2PO4 (50 μM), (NH4)2SO4 (20 μM), KCl (500 mg/liter), and MgCl2 (600 mg/liter). Required auxotrophic supplements, glucose (2%), and a nitrogen source were added. For 32P labeling, the (NH4)2SO4 concentration was increased to 2 mM. For 35S labeling, the KH2PO4 concentration was increased to 5 mM and asparagine (0.1%) was used as a rich nitrogen source to replace ammonium sulfate.
Metabolic labeling.
Precultures (2.5 ml) were grown in standard minimal medium with ammonium sulfate or proline as the sole nitrogen source to an optical density at 600 nm (OD600) of 0.8 to 1.2 (exponential phase). The cultures were diluted twofold with LPSM and incubated for 5 h at 30°C with aeration. They were used to inoculate 5 ml of fresh LPSM medium at an OD600 of 0.02 to 0.1 and incubated overnight for phosphate depletion. Cultures (OD600 = 0.8 to 1.2) were harvested by centrifugation at 2,000 × g for 5 min at room temperature. Cells were resuspended in prewarmed fresh LPSM and allowed to recover for 1 h. Cu2SO4 (50 μM) was added at this step when induction of GST-Put3p expression was required. 32P-labeled orthophosphate (1 mCi; carrier and HCl free; Amersham) was added, and incubation continued for 2 to 4 h. The procedure for 35S-labeling experiments was similar to that described above, except 35S-labeled methionine and cysteine (1 mCi; Pro-mix L 35S in vitro cell labeling mixture; Amersham) was used and the labeling time was 1 to 2 h, followed by a 30-min chase with unlabeled methionine and cysteine.
Preparation of whole-cell extracts for immunoblotting and IP.
To inhibit the activity of phosphatases, cells were treated with phosphatase inhibitors (sodium pyrophosphate, sodium azide, and sodium fluoride, each at 10 mM, and sodium metavanadate and sodium orthovanadate, each at 0.4 mM) before harvesting. Cells were washed twice with phosphate-buffered saline (PBS; pH 7.4) containing the phosphatase inhibitors. For immunoblotting analysis, cells were broken in 1× Laemmli (20) sample buffer (LSB; 40 μl per total OD600 unit) by being vortexed with glass beads (30 μl per total OD600 unit) for 3 min (alternating 1 min of vortexing and 1 min of chilling on ice). The extracts were clarified by centrifugation at 16,000 × g in a microcentrifuge for 5 min at 4°C. Supernatants were transferred to clean tubes and boiled for 5 min.
For immunoprecipitation (IP) analysis, cell pellets were resuspended in 300 μl of IP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 10 mM dithiothreitol, 0.1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) with a protease inhibitor cocktail (0.5 mM phenylmethylsulfonyl fluoride and 10 μg of benzamidine, 2 μg of leupeptin, 1 μg of pepstatin A, and 2 μg of aprotinin per ml). Acid-rinsed, cold glass beads (100 μl) were added to each tube, and the cells were broken by vortexing (6-min cycles of 1 min of vortexing alternating with 1 min of chilling on ice). Whole-cell extracts were collected and clarified by centrifugation in a microcentrifuge at 16,000 × g for 20 min at 4°C. Monoclonal anti-GST (1 μl; Santa Cruz Biotechnology Inc.) or polyclonal anti-Put3 (0.5 μl) was added to each sample, and the samples were incubated with rotation for 2 to 4 h at 4°C. Protein A-Sepharose (50 μl; 50/50 slurry in IP buffer; Sigma Chemical Co.) was added to each of the mixtures, and the incubation was continued overnight. The beads were allowed to settle for 5 min before centrifugation at low speed (500 × g) and were subsequently washed twice with the same buffer and four times with Tris-buffered saline (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). For additional treatments, the suspensions were divided into equal aliquots before the final centrifugation.
For thrombin cleavage of GST-Put3p, equal amounts of anti-GST immunoprecipitates were placed in tubes with 30 μl of thrombin buffer (20 mM Tris-HCl [pH 8.4], 150 mM NaCl, 2.5 mM CaCl2). Thrombin (Sigma; 0.1 U) was added to half the tubes, and all samples were incubated at 17°C for 16 h. The reactions were stopped by addition of 30 μl of 2× LSB, and samples were boiled for 5 min before being loaded onto SDS gels.
For dephosphorylation with calf intestinal phosphatase (CIP), IP pellets were resuspended in 30 μl of CIP buffer (50 mM Tris-HCl [pH 7.9], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol) and 10 U of CIP (New England Biolabs) with or without phosphatase inhibitors. The reaction mixture was incubated at 37°C for 1 h, and the reaction was terminated by addition of 30 μl of 2× LSB and boiling for 5 min.
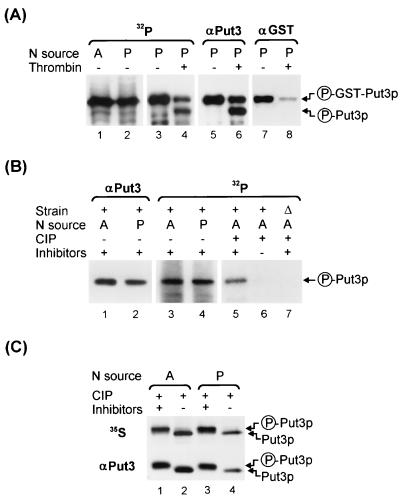
The SDS polyacrylamide gel electrophoresis (PAGE) gels shown in Fig. 1 were run under conditions that do not resolve the Put3p phosphoforms.
FIG. 1.
GST-Put3p and Put3p are phosphoproteins. (A) 32P labeling and IP of GST-Put3p. Extracts were made from cells of strain DB1000 carrying plasmid pHB3 (GST-PUT3) that were grown in a low-phosphate medium with ammonia (A; lane 1) or proline (P; lane 2) as the sole nitrogen source, labeled with 32P, and immunoprecipitated with mouse monoclonal anti-GST, as described in Materials and Methods. The immunoprecipitated GST-Put3p from the proline culture (lane 3) was incompletely digested with thrombin to release full-length Put3p (lane 4). After the signal decayed, the membrane was probed with anti-Put3p (lanes 5 and 6) and then reprobed with anti-GST antibodies (lanes 7 and 8) for detection of GST-Put3p and the cleaved Put3p. The proteins were resolved on a 12.5% (12.57% total acrylamide concentration [T], 0.5% cross-linker concentration [C]) polyacrylamide gel and transferred to a PVDF membrane. Circled P, phosphorylated form. (B) 32P labeling and IP of Put3p. Extracts were made from cells of strain DB1000 carrying plasmid pDB37 (PUT3) grown in a low-phosphate medium with ammonia (lane 3) or proline (lane 4) as the sole nitrogen source or carrying YEp24 grown in a low-phosphate medium with ammonia (lane 7), labeled with 32P, and immunoprecipitated with polyclonal anti-Put3p, as described in Materials and Methods. The proteins were resolved on a 7.5% (7.65% T, 2% C) polyacrylamide gel and transferred to a PVDF membrane. After the signal decayed, the membrane was probed with anti-Put3p antibody (lanes 1 and 2). The 32P-labeled immunoprecipitates from the ammonia cultures were treated with CIP in the presence (+) or absence (−) of phosphatase inhibitors (lanes 5 and 6, wild type; lane 7, put3Δ). (C) 35S labeling of Put3p. Extracts of 35S-labeled DB1000 carrying plasmid pDB37 grown on ammonia- or proline-containing medium were immunoprecipitated with anti-Put3p antibody (lanes 1 to 4, upper section) and treated with CIP in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of phosphatase inhibitors. The proteins were resolved on a 7.5% (7.65% T, 2% C) polyacrylamide gel and transferred to a PVDF membrane. After the signal decayed, the membrane was probed with anti-Put3p antibody (lanes 1 to 4, lower section). These gels were run under conditions that do not resolve isoforms of Put3p.
Analysis of Put3p isoforms by SDS-PAGE and immunodetection.
For high resolution of Put3p isoforms on the denaturing gels shown in Fig. 2 and 3, Tris concentrations in the gel (0.75 M) and running buffer (0.05 M) were increased as described by Okajima et al. (29). The separated polypeptides were transferred to polyvinylidene difluoride (PVDF) membranes (Polyscreen; NEN) in 1× Towbin buffer (47). Membranes with radioisotope-labeled samples were exposed to Kodak X-OMAT film. After decay of radioactivity, the membranes were subjected to immunodetection. Briefly, the nonspecific sites on the membrane were blocked by a 30-min incubation in blocking reagent (3 to 5% nonfat dry milk dissolved in PBST [PBS, pH 7.4, plus 0.04% Tween-20]) at room temperature. Primary antibody was added directly to the blocking reagent at a 1:1,000 dilution for anti-Put3p antibody (50) or at a final concentration of 0.1 μg/ml for anti-GST antibody. The incubation was continued for 1 h and was followed by washes with PBST. After a 10-min incubation in blocking reagent, secondary antibody was added directly to the blocking reagent (1:5,000 dilution of horseradish peroxidase [HRP]-conjugated anti-rabbit immunoglobulin G [IgG] or 1:1,000 dilution of HRP-conjugated anti-mouse IgG). After a 1-h incubation, the membranes were washed with PBST and the ECL chemiluminescence protocol (Amersham) was used to detect the proteins, following the instructions of the manufacturer.
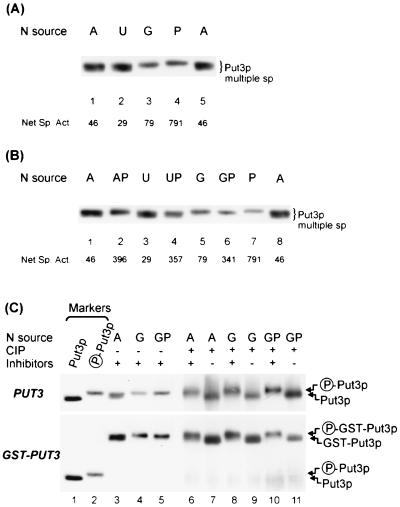
FIG. 2.
Put3p is differentially phosphorylated as a function of the quality of the nitrogen source. (A) Isoforms of Put3p. Extracts of strain DB1000 carrying plasmid pDB37 and grown on minimal media with different nitrogen sources were subjected to SDS–6% PAGE (6.12% T, 2% C) and immunoblotting with anti-Put3p antiserum. Lanes: 1 and 5, ammonia; 2, urea; 3, GABA; 4, proline. Below each lane is the net specific activity (Net Sp. Act.) of β-galactosidase from strain DB1000 (put3Δ) carrying plasmids pDB37 (PUT3) and pMB7 (PUT2-lacZ) and grown under the same conditions. (B) Addition of the inducer has little effect on the migration of Put3p isoforms. Extracts of DB1000 carrying plasmid pDB37 were treated as described for panel A. Lanes: 1 and 8, ammonia; 2, ammonia plus proline; 3, urea; 4, urea plus proline; 5, GABA; 6, GABA plus proline; 7, proline. Below each lane is the net specific activity of β-galactosidase from strain DB1000 (put3Δ) carrying plasmids pDB37 (PUT3) and pMB7 (PUT2-lacZ) and grown under the same conditions. (C) Isoforms of Put3p and GST-Put3p are due to differential phosphorylation. Extracts of DB1000 carrying plasmid pDB37 (PUT3) or pHB3 (GST-PUT3) were made from cells grown in minimal media with ammonia (lane 3), GABA (lane 4), or GABA plus proline (lane 5) as the sole nitrogen sources and analyzed as described for panel A. Markers: lane 1, extract from strain DB1000 carrying plasmid pHB6 (put3-75) grown on minimal GABA medium; lane 2, extract from strain DB1000 carrying plasmid pDB37 (PUT3) grown on minimal proline medium. Immunoprecipitates from the same extracts were treated with CIP in the presence (lanes 6, 8, and 10) or absence (lanes 7, 9, and 11) of phosphatase inhibitors.
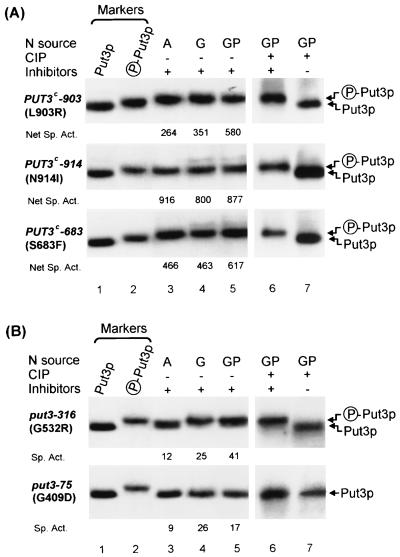
FIG. 3.
Phosphorylation is altered in regulatory Put3p mutants. (A) Activator-constitutive mutants have unique phosphorylation profiles of Put3p. Extracts of strain DB1000 carrying plasmids pMB3 (PUT3c-903, upper section), pMB4 (PUT3c-914, middle section), or pMB5 (PUT3c-683, lower section) grown on minimal medium with ammonia (lane 3), GABA (lane 4), or GABA plus proline (lane 5) were subjected to SDS–6% PAGE (6.12% T, 2% C) and immunoblotting with anti-Put3p antiserum. Extracts from GABA-plus-proline-grown cultures were immunoprecipitated with anti-Put3p antiserum and treated with CIP in the presence (lane 6) or absence (lane 7) of phosphatase inhibitors. Markers: lane 1, extract from strain DB1000 carrying plasmid pHB6 (put3-75) grown on minimal GABA medium; lane 2, extract from strain DB1000 carrying plasmid pDB37 (PUT3) grown on minimal proline medium. The net specific activities (Net Sp. Act.) of β-galactosidase from a PUT2-lacZ gene in each strain are given below lanes 3 to 5. (B) Two activator-defective Put3p mutants differ from each other in their phosphorylation profiles. Extracts of strain DB1000 carrying plasmids pHB6 (put3-316; upper section) or pDB193 (put3-75; lower section) grown on minimal medium with ammonia (lane 3), GABA (lane 4), or GABA plus proline (lane 5) were subjected to SDS-PAGE and immunoblotting with anti-Put3p antiserum. Extracts from GABA-plus-proline-grown cultures were immunoprecipitated with anti-Put3p antiserum and treated with CIP in the presence (lane 6) or absence (lane 7) of phosphatase inhibitors. Markers are as described for panel A. Full specific activities of β-galactosidase from a PUT2-lacZ gene in each strain are given below lanes 3 to 5.
Growth of yeast strains, extract preparation, and β-galactosidase assays.
These methods have been described previously (24). The units of specific activity are nanomoles of o-nitrophenol formed per minute per milligram of protein. The numbers represent the average of two determinations; variation was <20%. Net specific activity indicates the specific activity due to PUT3 activation of PUT2-lacZ; the background value of a put3Δ strain carrying plasmid YEp24 and grown on the same medium was subtracted in each case. The nature of this background activity remains unknown.
RESULTS
Effect of Put3p gene dosage and genetic background on the regulation of proline utilization.
Like many fungal regulatory proteins, Put3p is present in extremely low levels in S. cerevisiae cells and is highly sensitive to proteolysis. To facilitate the study of posttranslational modifications of Put3p, it was necessary to work with a protease-deficient strain (derived from the S288C background) and to increase the expression of wild-type or tagged PUT3 genes. The genes encoding wild-type and GST-tagged Put3p were first placed on high-copy-number plasmids, and the effect of higher gene dosage in a put3Δ strain was examined. GST-Put3p complements a put3Δ strain for growth on proline and activates PUT1 and PUT2 in a manner indistinguishable from that of the untagged Put3p under all conditions tested (data not shown). Increased dosage of the wild-type PUT3 gene led to overproduction of the Put3 protein by 30-fold compared to that made from the genomic copy in a wild-type strain (data not shown). Increased gene dosage and copper induction led to overproduction of GST-Put3p by at least twice that observed for overexpressed Put3p from plasmid pDB37 (data not shown). However, the increase in the level of Put3p or GST-Put3p did not affect the normal regulation of Put3p target genes in an otherwise wild-type strain (data not shown).
The absence of the major vacuolar proteases in the S288C-derived strain used in this work also had no effect on the normal expression and proline inducibility of the PUT genes, although different genetic backgrounds differed in absolute levels of target gene expression and induction ratios (Table 2; Fig. 2A and B; data not shown). In general, the laboratory yeast strains used in European laboratories (e.g., Σ1278b) are more sensitive to nitrogen derepression and induction by specific nitrogen sources than strains used in North American laboratories (e.g., S288C). There are also significant differences in growth rates on specific sources of nitrogen. For example, Σ1278b-derived strains grow on a minimal medium containing glucose and proline with a 3-h doubling time, while many S288C-derived strains have >8-h doubling times on this medium and some cannot maintain balanced growth (M. C. Brandriss, unpublished results). Σ1278b-derived strains grow more slowly on a medium containing urea than on one containing ammonia. In contrast, S288C strains grow with comparable doubling times on media containing either of these nitrogen sources.
TABLE 2.
Regulation by wild-type and mutant forms of Put3p in a protease-deficient strain
| Allele | Mutation | Sp. act. of β-galactosidasea with medium containing:
|
|||
|---|---|---|---|---|---|
| Amm | GABA | GABA + Pro | Pro | ||
| Vector | None | 20 | 55 | 44 | NG |
| PUT3 | Wild type | 66 | 134 | 385 | 791 |
| PUT3c-903 | L903R | 284 | 406 | 624 | 938 |
| PUT3c-914 | N914I | 936 | 855 | 921 | 945 |
| PUT3c-683 | S683F | 486 | 518 | 661 | 908 |
| put3-75 | G409D | 9 | 26 | 17 | NG |
| put3-316 | G532R | 12 | 25 | 41 | NG |
Strain DB1000 (pep4-3 prb1-1122 prc1-451 put3::LEU2 ura3-52 leu2 trp1) carried two plasmids in each experiment: the low-copy-number plasmid pMB7 (TRP1 PUT2-lacZ) and a high-copy-number (URA3) plasmid with the indicated allele of PUT3. The strain was grown on minimal media containing glucose (2%) and ammonium sulfate (Amm; 0.2%), GABA (0.1%) without or with proline (Pro; 0.1%), or proline alone (0.1%). Each PUT3 gene was driven by its natural promoter. Specific activities are the averages of measurements on two independent transformants; variation was <20%. Units are nanomoles of o-nitrophenol formed per minute per milligram of protein. The vector is plasmid YEp24, and the enzyme levels measured represent Put3p-independent (background) expression of PUT2-lacZ. NG, no growth.
Put3p is a phosphoprotein.
To determine if phosphorylation played a role in the regulation of Put3p activity, we examined both GST-Put3p and Put3p in metabolic labeling experiments. The protease-deficient put3Δ strain DB1000 carrying a plasmid-borne GST-PUT3 gene was metabolically labeled with 32P in a low-phosphate minimal medium containing either ammonium sulfate or proline as the sole source of nitrogen, as described in Materials and Methods. IP of GST-Put3p with a monoclonal anti-GST antibody yielded a labeled species of the predicted molecular mass (136 kDa) from both cultures (Fig. 1A, lanes 1 and 2). Partial thrombin digestion of the immunoprecipitate from the proline culture resulted in the production of two new labeled species with the expected molecular masses for full-length Put3p (111 kDa; Fig. 1A, lanes 3 and 4) and GST (23 kDa; data not shown). These assignments were confirmed by probing the same membrane, after the 32P decayed, with anti-Put3p and anti-GST antibodies (Fig. 1A, lanes 5 to 8, and data not shown). Identical results were obtained with the immunoprecipitate from the ammonia culture (data not shown).
When Put3p was immunoprecipitated from 32P-labeled cells of strain DB1000 carrying plasmid pDB37 with polyclonal anti-Put3p antiserum in experiments similar to those described above, a radioactive species the size of full-length Put3p was detected (Fig. 1B, lanes 3 and 4) and confirmed by immunoblotting (Fig. 1B, lanes 1 and 2). The specificity of the IP was demonstrated with a metabolically labeled put3Δ strain, where no labeled species were detected (Fig. 1B, lane 7). Subsequent treatment with CIP caused the Put3p band (Fig. 1B, lane 5) to disappear (Fig. 1B, lane 6). In a similar experiment, phosphatase treatment of Put3p immunoprecipitates from cultures metabolically labeled with 35S revealed a faster-migrating species on SDS gels, confirmed by immunoblotting to be Put3p (Fig. 1C, lanes 1 to 4). Based on these results, we conclude that both GST-Put3p and Put3p exist as phosphorylated proteins in vivo when cells are grown on ammonia- or proline-containing media and that phosphorylated and nonphosphorylated isoforms can be distinguished by their different mobilities by SDS-PAGE.
Changes in Put3p phosphorylation status are correlated with the quality of the nitrogen source.
The phosphorylated forms of Put3p were examined in cells grown in media containing different nitrogen sources and in the presence or absence of the inducer, proline. Ammonia is a rich nitrogen source that fully represses the expression of genes of many alternative nitrogen assimilatory pathways. GABA and proline are much poorer nitrogen sources in which nitrogen repression of alternative pathways is relieved. In the DB1000 strain background, urea is as good a source of nitrogen as ammonia in terms of growth rate and nitrogen repression. On the basis of growth rate measurements for this strain, the nitrogen sources can be ranked in terms of quality as follows: ammonia = urea > GABA > proline. The effect of induction by proline can be observed when proline is added to media containing another nitrogen source. Maximum expression of the proline utilization pathway occurs when proline is the sole source of nitrogen; nitrogen repression is minimal, and proline induction is maximal.
Extracts from cultures of cells grown on different nitrogen sources were examined by SDS-PAGE and immunoblotting with anti-Put3p antiserum. Put3p (and GST-Put3p [not shown]) from cells grown in an ammonia- or urea-containing medium appeared as a broad band, suggestive of multiple species (Fig. 2A, lanes 1, 2, and 5), whereas Put3p (and GST-Put3p [not shown]) from cells grown on GABA or proline migrated as a sharper band with slower mobility (Fig. 2A, lanes 3 and 4). Put3p from extracts of proline cultures migrated more slowly than that from extracts of GABA cultures. The addition of proline to ammonia-, urea-, or GABA-containing cultures had little observable effect on the migration of the Put3p band (Fig. 2B; compare lanes 3 and 4 and 5 and 6). The slight shift in Put3p from cultures grown on ammonia plus proline in lane 2 of Fig. 2B was not reproducible in other experiments, and the migration and size of the Put3p band were usually identical to those observed in lane 1. Put3p isolated from cells grown on GABA plus proline medium migrates faster than Put3p isolated from cells grown on proline alone (Fig. 2B; compare lanes 6 and 7).
Phosphatase treatment of immunoprecipitates of Put3 or GST-Put3 proteins from cells grown in each condition demonstrated that the difference in band migration is due to differential phosphorylation. The Put3p and GST-Put3p bands each collapse to one tight, fast-migrating band (Fig. 2C, lanes 6 to 11). The differences in the band patterns in immunoblots can be more easily observed when the migration of the bands is compared to that of the fastest-migrating, nonphosphorylated species (a mutant form of Put3p that is unable to bind DNA; see below) and to the slowest-migrating species found in proline-grown cells (Fig. 2C, lanes 1 and 2). It appears that the Put3p species from ammonia-grown cultures is a more heterogeneous mixture than the species observed in GABA- or proline-grown cells. The phosphoforms of Put3p in GABA and GABA-plus-proline extracts (Fig. 2C, lanes 4 and 5) migrate slightly faster than the wild-type Put3p from a proline culture (Fig. 2C, lane 2).
Although Put3p migrates on gels more slowly as the quality of the nitrogen source diminishes, we cannot state at this time that the amount of total phosphorylation of Put3p increases. Until additional biochemical analyses on the phosphoforms are carried out, we will refer to changes in phosphorylation profiles or status rather than hypophosphorylation or hyperphosphorylation. Based on these observations, we conclude that Put3p undergoes a change in phosphorylation status as a function of the quality of the nitrogen source. The presence of proline itself does not trigger a detectable change in the phosphorylation profile; in fact, when proline is added to a medium containing GABA, the growth rate increases as the quality of the nitrogen source improves and the band appears to migrate slightly faster (visible for GST-Put3p; Fig. 2C, lower section; compare lanes 4 and 5.)
Transcriptional activation by Put3p in response to nitrogen derepression correlates with altered phosphorylation profiles.
Expression of the reporter PUT2-lacZ gene present in strain DB1000 was measured in extracts prepared under the same conditions as those used in the immunoblotting experiments and is shown below each lane in Fig. 2A and B. The activation of PUT2 in response to nitrogen derepression parallels the appearance of slower-migrating Put3p species (Fig. 2A). Ammonia and urea are repressing sources of nitrogen, and PUT2 was expressed at a low level under these conditions. Nitrogen derepression on a GABA-containing medium caused expression to increase almost threefold. However, whenever proline was added to each medium, the expression of PUT2 increased without detectably altering the migration pattern of the phosphoforms. For example, the phosphorylation profiles for the urea and urea-plus-proline cultures (lanes 3 and 4) were similar to each other, as were those for the GABA and GABA-plus-proline cultures (lanes 5 and 6), but the activation of PUT2 increased 12-fold when proline was added to urea-containing medium or 4-fold when proline was added to GABA-containing medium (Fig. 2B). Thus, two different signals, nitrogen derepression and proline induction, affect Put3p activity, resulting in increased PUT2 gene expression. When proline is the sole source of nitrogen, both induction and nitrogen derepression occur, resulting in the slowest-migrating Put3p species and maximum target gene expression.
Activator-constitutive and activator-defective Put3p mutants are altered in their phosphorylation profiles.
Mutations in the PUT3 gene that led to either constitutive (proline-independent) or noninducible expression of its target genes have been previously characterized (4, 7, 15, 24, 25). To examine the regulatory behavior of each mutant Put3 protein, strain DB1000 carrying each mutant gene on a high-copy-number plasmid was grown in media containing different nitrogen sources. The phosphorylation status of these mutant proteins was examined by SDS-PAGE and immunoblotting, and their abilities to activate the transcription of a PUT2-lacZ reporter gene were measured.
In contrast to the behavior of wild-type Put3p (broad, faster-migrating band in ammonia extracts; narrow, slower-migrating band in proline extracts) (Fig. 2A, lanes 4 and 5), each of the activator-constitutive proteins appeared to have a phosphorylation profile on an ammonia-containing medium similar to that of the wild-type protein derived from proline-grown cultures (Fig. 3A; compare lane 3 to lane 2 in each section). The mobilities of these mutant proteins did not decrease further when a poorer nitrogen source was substituted (Fig. 3A, lanes 4 and 5). There were subtle differences in the phosphorylation profiles, as judged by the difference in migration of the phosphatase-treated and untreated proteins (Fig. 3A, lanes 6 and 7). The migration of the phosphatase-treated mutant Put3 proteins appeared similar to that of the phosphatase-treated wild-type protein, indicating that the single amino acid changes did not cause the observed differences in migration (data not shown). Unlike the situation with the wild-type Put3p, in which changes in phosphorylation appear to be a response to a decrease in the quality of the nitrogen source, these activator-constitutive mutants behave as if the environment is always nitrogen poor, even in the presence of ammonia. They are defective in their ability to sense the quality of the nitrogen source.
PUT2 expression from the activator-constitutive alleles PUT3c-683, PUT3c-903, and PUT3c-914 on high-copy-number plasmids was similar to that of the mutant genomic copies previously described in a different strain background (15, 24) (Table 2). High levels of PUT2 expression, as indicated below each lane in Fig. 3A, correlate with the phosphorylation profile observed. The PUT3c-903 and PUT3c-683 alleles are still somewhat proline responsive, as indicated by the significant increase in expression when proline was added to GABA medium and on proline-only medium (Table 2). In contrast, the PUT3c-914 allele is fully constitutive, showing the same high level of expression on each medium tested, including proline-only medium.
A comparison of PUT2 expression with the Put3p band pattern on SDS gels (Fig. 3A, lanes 3 to 5 [net specific activities], and Table 2) supports the conclusion that there is a correlation between phosphorylation status and activation by Put3p.
Strains expressing the activator-defective proteins are unable to use proline as the sole nitrogen source because they cannot induce target gene expression (Table 2 and Fig. 3, lanes 3 to 5). The level of expression of PUT2 in these strains is very low, below what is observed in a put3Δ strain. At present, we cannot explain this result, but it may relate to the presence of other zinc cluster proteins that can bind UASPUT and activate transcription of the PUT genes to a low level in the absence of Put3p but that have no effect when a mutant Put3p is present (M. D'Alessio and M. C. Brandriss, unpublished results). Previous work demonstrated that the Put3-316 mutant protein was able to bind DNA in vitro and had steady-state levels similar to those of the wild-type protein. We were unable to detect DNA binding in extracts containing the Put3-75 mutant protein, and it was present at lower steady-state levels than the wild type protein (15).
Mutant Put3 proteins in extracts from these strains differed from each other in the migration patterns observed on SDS-PAGE (Fig. 3B, lanes 3 to 5). The phosphoforms of the G532R protein (encoded by the put3-316 allele) resembled those of the wild type under all conditions examined. Although it can respond to the quality of the nitrogen source, the Put3-316 protein may be defective in its response to the presence of proline and therefore lacks one of the two signals for maximal activity, leading to its Put− phenotype. In contrast, the migration of the G409D protein (encoded by the put3-75 allele) did not shift after phosphatase treatment when isolated from cells grown under any conditions tested, and the protein is apparently not detectably phosphorylated. Since this protein failed to bind DNA in our in vitro assays, it was not surprising to find that it was not phosphorylated. As observed previously, neither of these mutant proteins could activate PUT2 appreciably (Table 2 and Fig. 3B).
Because the Put3p-316 mutant protein can bind DNA and has a phosphorylation profile resembling that of the wild-type strain in response to changes in the nitrogen source but cannot activate its target genes, we conclude that the change in Put3p phosphorylation is a cause, rather than a consequence, of transcriptional activation in this system, and is, along with proline, required for maximal activity of Put3p.
DISCUSSION
We have provided evidence that the Put3p transcriptional activator is a phosphoprotein whose phosphorylation status varies as a function of the quality of the nitrogen source present in the medium. Rapidly migrating forms of Put3p are correlated with low levels of target gene expression, while slower-migrating forms are correlated with high levels of target gene expression. We suggest that the change in phosphorylation status is not merely a consequence of transcriptional activation but is required for high levels of PUT gene expression. This working model is based on the behavior of a Put3p mutant that cannot activate transcription of its target genes but that shows the phosphorylation profiles of the wild type and is supported by the profiles of the constitutively active Put3p mutants. Furthermore, because both proline and nitrogen derepression affect the transcription of the PUT genes, we believe that these two inputs act synergistically. Maximum expression of the PUT genes is achieved when both conditions are met, i.e., when proline serves as the sole nitrogen source.
These findings force us to modify our previous conclusions concerning nitrogen repression and the action of Put3p. In a previous report (15), we observed that the PUT genes continued to respond to nitrogen derepression even in a put3Δ strain and concluded that Put3p was not responsible for nitrogen derepression and therefore did not respond to nitrogen excess or limitation. The data presented here indicate that there are both Put3p-dependent and Put3p-independent aspects to nitrogen regulation of this pathway and that the protein does indeed respond to changes in nitrogen source by changes in its phosphorylation status.
Put3p may regulate its target genes by cycling between active and inactive states through changes in conformation due to posttranslational modifications, inducer binding, or both. The data presented here suggest that changes in either the nitrogen environment or proline induction can increase target gene expression to a small extent. This hypothesis is consistent with our previous observation that target gene expression increased two- to threefold as the quality of the nitrogen source diminished even in the absence of the inducer (15, 50) and an early observation that addition of proline to ammonia-grown cells also resulted in increased PUT gene expression in spite of the presence of a rich nitrogen source (6). At this time, we do not know whether these inputs are dependent on, or independent of, each other. Based on the data presented in this report, we hypothesize that the activator-constitutive mutants are insensitive to the nitrogen repression signal and are in an “on” conformation inappropriately and that some can be further stimulated by the proline signal. Conversely, the activator-defective mutant protein receives the nitrogen derepression signal appropriately but can no longer respond to proline, resulting in a failure to convert to a fully “on” state and an inability to produce adequate levels of the PUT gene products.
A comparison of Put3p with Gal4p, one of the best-characterized regulators and the prototype of the Zn(II)2Cys6 binuclear cluster class of proteins, shows intriguing similarities and differences in the way the two proteins appear to be regulated. Put3p is phosphorylated under all conditions examined. In contrast, Gal4p exists in three relatively discrete isoforms (referred to as non-, hypo-, and hyperphosphorylated) whose levels correspond to the presence of galactose as well as glucose repression (27, 28). These authors also showed that gal80 mutants that constitutively express the galactose pathway contained the hyperphosphorylated form in the absence of added galactose. They concluded that the presence of the hyperphosphorylated species was correlated with activation of the GAL genes and hypothesized that changes in the phosphorylation status of Gal4p are responsible for differences in its activity. Subsequently, Parthun and Jaehning (31) demonstrated an in vitro correlation between Gal4p phosphorylation and galactose induction and showed that the unphosphorylated form could bind DNA as well as the phosphorylated form.
Three activator-defective gal4 alleles encoding mutations of amino acids located in the central domain of Gal4p (S322F, L331P, S352F) produced proteins that could bind DNA but that were not phosphorylated (28). Activator-competent pseudorevertants of several of these mutants regained the ability to become hyperphosphorylated. In contrast, the put3-316 mutation in the Put3p central domain produced a DNA-binding-competent activator-defective protein whose phosphorylation profile responded like the wild-type protein to changes in nitrogen source. The amino acid (arginine) at position 532 in this mutant replaced a highly conserved glycine (15) found in all the central domains of members of the binuclear cluster protein class (8, 37).
Mylin et al. (28) demonstrated that a DNA binding-defective gal4 mutant contained the hypo- but not the hyperphosphorylated form of Gal4p. In contrast, the activator-defective mutant protein (Put3-75p) that is less stable than the wild type and that failed to bind DNA (15) was not detectably phosphorylated under any condition. We doubt that its lack of phosphorylation is responsible for its inability to bind DNA because unphosphorylated amino-terminal fragments of Put3p produced in E. coli can bind DNA (15, 33); Gal4p, whose DNA-binding domain has the same structure as that of Put3p (45, 49), can also bind DNA when unphosphorylated (31), as can amino-terminal fragments produced from E. coli (33). Ammonia repression itself does not interfere with DNA binding by Put3p because in vivo footprinting experiments demonstrated that Put3p binds DNA even when ammonia is the sole nitrogen source (1). The put3-75 mutation causes a glycine-to-arginine substitution at position 409 in a region of unknown function. It may cause a change in conformation that is not compatible with DNA binding or results in a protein that cannot be a substrate for kinase activity.
Sadowski et al. (36) identified Ser837 as a major site of Gal4p phosphorylation, which they found was not required for transcriptional activation, and concluded from this and other observations that the phosphorylation of Gal4p is a consequence, rather than a cause, of transcriptional activation. More recently, this laboratory identified Ser699 as a site required for galactose-inducible transcription (35). To date, it remains unresolved in the published literature whether phosphorylation of Gal4p is a requirement for, or a consequence of, transcriptional activation.
The differential phosphorylation observed for Put3p in response to nitrogen repression and derepression is the first example reported for a regulator of a nitrogen assimilatory pathway. However, these findings resemble those previously described for two Saccharomyces transcriptional activators, Cat8p (16) and Sip4p (22), that also belong to the Zn(II)2Cys6 binuclear cluster family and that respond to changes in carbon repression and derepression by differential phosphorylation. Cat8p activates expression of the gluconeogenic genes whose products are required for the utilization of nonfermentable carbon sources such as ethanol. Under repressing conditions, Cat8p migrates as two species, a nonphosphorylated form, Cat8pI, and a phosphorylated form, Cat8pII. Under derepressing conditions, a hyperphosphorylated species, Cat8pIII, that depends (directly or indirectly) on the Snf1 kinase is formed (32). As the quality of the carbon source diminished (glucose > maltose > raffinose > galactose > ethanol), increasing amounts of Cat8pIII were found. Sip4p is also phosphorylated in a Snf1-dependent manner in response to low glucose (22). These workers suggest that the phosphorylation of Sip4p may increase its ability to turn on its target genes.
To date, no kinases that play a role in nitrogen metabolism analogous to the one played by Snf1p in carbon metabolism have been identified in S. cerevisiae. Phosphorylation is known to be important in the activity and stability of specific and general amino acid permeases. The protein kinase homolog Npr1p is believed to be involved in activating the general amino acid permease (Gap1p) under nitrogen derepressing conditions (48). Tor1p and Tor2p are phosphatidylinositol kinase homologs that may play a role in stabilizing amino acid permeases (e.g., the tryptophan transporter Tat2p) in nutrient-rich conditions and appear to control the phosphorylation and activity of Npr1p (38). The development of genetic and molecular approaches to isolate the kinases, phosphatases, and other proteins responsible for relaying information on the quality of the nitrogen environment to regulators such as Put3p will be essential for a fuller understanding of this signaling process.
ACKNOWLEDGMENTS
We thank C. Michels and J. Thorner for gifts of strains and plasmids and D. Barber for the construction of plasmids pDNB109 and pDNB118 and strains DB1000 and DB8-5C. We are grateful to members of the laboratory for helpful discussions and to S. Garrett and S. A. des Etages for critical reading of the manuscript.
This work was supported by Public Health Service grant 5 R01 GM40751 from the National Institutes of Health and grant 21-98 from the Foundation of UMDNJ.
REFERENCES
- 1.Axelrod J D, Majors J, Brandriss M C. Proline-independent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol Cell Biol. 1991;11:564–567. doi: 10.1128/mcb.11.1.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisson L F. Proceedings of the International Symposium on Nitrogen in Grapes and Wine. Davis, Calif: American Society for Enology and Viticulture; 1991. Influence of nitrogen on yeast and fermentation of grapes; pp. 78–89. [Google Scholar]
- 3.Blinder D, Coschigano P E, Magasanik B. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandriss M C. Evidence for positive regulation of proline utilization in Saccharomyces cerevisiae. Genetics. 1987;117:429–435. doi: 10.1093/genetics/117.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandriss M C. Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J Bacteriol. 1979;138:816–822. doi: 10.1128/jb.138.3.816-822.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandriss M C, Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979;140:498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandriss M C, Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: mutation causing constitutive enzyme expression. J Bacteriol. 1979;140:504–507. doi: 10.1128/jb.140.2.504-507.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chasman D I, Kornberg R D. Gal4 protein: purification, association with Gal80 protein, and conserved domain. Mol Cell Biol. 1990;10:2916–2923. doi: 10.1128/mcb.10.6.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffman J A, Rai R, Cooper T G. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:6910–6918. doi: 10.1128/jb.177.23.6910-6918.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman J A, Rai R, Cunningham T, Svetlov V, Cooper T G. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffman J A, Rai R, Loprete D M, Cunningham T, Svetlov V, Cooper T G. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3416–3429. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coornaert D, Vissers S, Andre B, Grenson M. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr Genet. 1992;21:301–307. doi: 10.1007/BF00351687. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham T S, Cooper T G. Expression of DAL80, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol Cell Biol. 1991;11:6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daugherty J R, Rai R, El Berry H, Cooper T G. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1993;175:64–73. doi: 10.1128/jb.175.1.64-73.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.des Etages S G, Falvey D A, Reece R J, Brandriss M C. Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics. 1996;142:1069–1082. doi: 10.1093/genetics/142.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedges D, Proft M, Entian K-D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jauniaux J-C, Vandenbol M, Vissers S, Broman K, Grenson M. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. Cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur J Biochem. 1987;164:601–606. doi: 10.1111/j.1432-1033.1987.tb11169.x. [DOI] [PubMed] [Google Scholar]
- 18.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones E W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lasko P, Brandriss M C. Proline transport in Saccharomyces cerevisiae. J Bacteriol. 1981;148:241–247. doi: 10.1128/jb.148.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 283–317. [Google Scholar]
- 24.Marczak J E, Brandriss M C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991;11:2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marczak J E, Brandriss M C. Isolation of constitutive mutations affecting the proline utilization pathway in Saccharomyces cerevisiae and a molecular analysis of the PUT3 transcriptional activator. Mol Cell Biol. 1989;9:4696–4705. doi: 10.1128/mcb.9.11.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell A P, Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mylin L M, Bhat P J, Hopper J E. Regulated phosphorylation and dephosphorylation of GAL4, a transcriptional activator. Genes Dev. 1989;3:1157–1165. doi: 10.1101/gad.3.8.1157. [DOI] [PubMed] [Google Scholar]
- 28.Mylin L M, Johnston M, Hopper J E. Phosphorylated forms of GAL4 are correlated with ability to activate transcription. Mol Cell Biol. 1990;10:4623–4629. doi: 10.1128/mcb.10.9.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okajima T, Tanabe T, Yasuda T. Nonurea sodium dodecyl sulfate-polyacrylamide gel electrophoresis with high-molarity buffers for the separation of proteins and peptides. Anal Biochem. 1993;211:293–300. doi: 10.1006/abio.1993.1272. [DOI] [PubMed] [Google Scholar]
- 30.Orr-Weaver T L, Szostak J, Rothstein R. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 31.Parthun M R, Jaehning J A. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol Cell Biol. 1992;12:4981–4987. doi: 10.1128/mcb.12.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randez-Gil F, Bojunga N, Proft M, Entian K-D. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol. 1997;17:2502–2510. doi: 10.1128/mcb.17.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reece R J, Ptashne M. Determinants of binding site-specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 34.Rowen D W, Esiobu N, Magasanik B. Role of GATA factor Nil2p in nitrogen regulation of gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3761–3766. doi: 10.1128/jb.179.11.3761-3766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadowski I, Costa C, Dhanawansa R. Phosphorylation of Gal4p at a single C-terminal residue is necessary for galactose-inducible transcription. Mol Cell Biol. 1996;16:4879–4887. doi: 10.1128/mcb.16.9.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadowski I, Neidbala D, Wood K, Ptashne M. GAL4 is phosphorylated as a consequence of transcriptional activation. Proc Natl Acad Sci USA. 1991;88:10510–10514. doi: 10.1073/pnas.88.23.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schjerling P, Holmberg S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddiqui A H, Brandriss M C. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol Cell Biol. 1988;8:4634–4641. doi: 10.1128/mcb.8.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddiqui A H, Brandriss M C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989;9:4706–4712. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soussi-Boudekou S, Andre B. A co-activator of nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:753–762. doi: 10.1046/j.1365-2958.1999.01187.x. [DOI] [PubMed] [Google Scholar]
- 42.Soussi-Boudekou S, Vissers S, Urrestarazu A, Jauniaux J-C, Andre B. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol. 1997;23:1157–1168. doi: 10.1046/j.1365-2958.1997.3021665.x. [DOI] [PubMed] [Google Scholar]
- 43.Stanbrough M, Magasanik B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanbrough M, Rowen D W, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaminathan K, Flynn P, Reece R J, Marmorstein R. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat Struct Biol. 1997;4:751–759. doi: 10.1038/nsb0997-751. [DOI] [PubMed] [Google Scholar]
- 46.Tomenchok D M, Brandriss M C. Gene-enzyme relationships in the proline biosynthesis pathway of Saccharomyces cerevisiae. J Bacteriol. 1987;169:5346–5372. doi: 10.1128/jb.169.12.5364-5372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towbin H, Staehelin J, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandenbol M, Jauniaux J-C, Grenson M. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encodes a protein kinase homologue. Mol Gen Genet. 1990;222:393–399. doi: 10.1007/BF00633845. [DOI] [PubMed] [Google Scholar]
- 49.Walters K J, Dayie K T, Reece R J, Ptashne M, Wagner G. Structure and mobility of the PUT3 dimer. Nat Struct Biol. 1997;4:744–750. doi: 10.1038/nsb0997-744. [DOI] [PubMed] [Google Scholar]
- 50.Xu S, Falvey D A, Brandriss M C. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2321–2330. doi: 10.1128/mcb.15.4.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]