Abstract
Activation of insulin gene transcription specifically in the pancreatic β cells depends on multiple nuclear proteins that interact with each other and with sequences on the insulin gene promoter to build a transcriptional activation complex. The homeodomain protein PDX-1 exemplifies such interactions by binding to the A3/4 region of the rat insulin I promoter and activating insulin gene transcription by cooperating with the basic-helix-loop-helix (bHLH) protein E47/Pan1, which binds to the adjacent E2 site. The present study provides evidence that the homeodomain of PDX-1 acts as a protein-protein interaction domain to recruit multiple proteins, including E47/Pan1, BETA2/NeuroD1, and high-mobility group protein I(Y), to an activation complex on the E2A3/4 minienhancer. The transcriptional activity of this complex results from the clustering of multiple activation domains capable of interacting with coactivators and the basal transcriptional machinery. These interactions are not common to all homeodomain proteins: the LIM homeodomain protein Lmx1.1 can also activate the E2A3/4 minienhancer in cooperation with E47/Pan1 but does so through different interactions. Cooperation between Lmx1.1 and E47/Pan1 results not only in the aggregation of multiple activation domains but also in the unmasking of a potent activation domain on E47/Pan1 that is normally silent in non-β cells. While more than one activation complex may be capable of activating insulin gene transcription through the E2A3/4 minienhancer, each is dependent on multiple specific interactions among a unique set of nuclear proteins.
Like expression of other cell-type-specific genes, expression of the insulin gene depends on the actions of a unique set of nuclear activators. These activators cooperate synergistically in building a transcriptional activation complex that binds to the regulatory domains of the gene and activates the basal RNA polymerase machinery (reviewed in reference 7). The complexity and specificity of the interactions among these activators limit the cell types capable of building a functional activation complex. Dissection of these interactions provides insight into the mechanism by which insulin expression is limited to the correct cell type.
In adult mammals, activation of the insulin gene is tightly restricted to the β cells in the pancreatic islets of Langerhans, where it is expressed at high levels. This specificity is reflected in the restricted function of the insulin promoter, the proximal few hundred base pairs of which can replicate the specificity of the intact gene (19, 50). Because of the complexity of the intact promoter (9, 13, 24), a short portion of the rat insulin I promoter between bp −247 and −197 upstream from the transcription initiation site has been used as a model of the types of synergistic interactions that combine to give the characteristic activity of the full promoter (15). This 50-bp fragment contains at least three distinct DNA-binding sites named E2, A3, and A4 (13). The E and A elements synergize: neither has significant activity on its own, but in combination E and A elements produce β-cell specific transcriptional activation (15, 23).
The E2 element functions as a recognition site for dimers of basic helix-loop-helix (bHLH) proteins, including a heterodimer of the ubiquitous bHLH protein E47/Pan1 and the neuroendocrine specific bHLH protein BETA2/NeuroD1 (35). The A elements each contain the sequence TAAT and have been shown to bind to several homeodomain proteins found in β-cell nuclei (12, 16, 22, 32, 37). Two of these homeodomain proteins, PDX-1 (also known as IPF-1 [37], STF-1 [28], IDX-1 [32], IUF-1 [29], and GSF [30]) and Lmx1.1 can bind to the A3 and A4 (collectively referred to as A3/4) sites and activate the E2A3/4 minienhancer by synergizing with E47/Pan1 bound to the E2 site (16, 38, 40).
The LIM homeodomain protein Lmx1.1 contains two LIM domains that form zinc-binding structures in the amino end of the molecule. The second of these two LIM domains (LIM2) directly binds to the bHLH domain of E47/Pan1 and mediates the synergy between Lmx1.1 and E47/Pan1. This interaction is specific, since analogous domains from other LIM proteins and bHLH proteins cannot substitute for the LIM2 domain of Lmx1.1 or the bHLH domain of E47/Pan1, respectively (20).
PDX-1 plays an important role both in the development of the pancreas and in maintaining β-cell function. Mice with a targeted disruption of the pdx1 gene selectively lack a pancreas (2, 21, 36). Similar pancreatic agenesis has been found in a human patient with a single nucleotide deletion in the pdx1 gene (46). If the pancreas is allowed to develop with an intact pdx1 gene, and the pdx1 gene is disrupted only in mature β cells, diabetes ensues due to impaired β-cell function (3). This impairment presumably results from the loss of PDX-1 activation of β-cell genes, since PDX-1 has been shown to activate a variety of pancreatic islet-specific genes, including those encoding insulin, somatostatin, glucokinase, islet amyloid polypeptide, and glucose transporter type 2 (28, 32, 37, 44, 49, 51, 52). It should be noted, however, that the β cells lacking PDX-1 continue to express insulin (3).
The molecular basis of the PDX-1–E47/Pan interaction is unknown. The DNA-binding domains of both proteins, as well as the activation domains, are required. The transcription activation domain of PDX-1 is located in the amino end, upstream of the homeodomain (38), and is composed of five subdomains which are conserved between PDX-1 and the related Xenopus homeoprotein XIHbox8 (40). Interestingly, when the unrelated activation domain of herpesvirus VP16 is substituted for the PDX-1 activation domain, the chimeric VP16–PDX-1 protein can still synergize with E47/Pan1, even though the only portion of PDX-1 that is retained in the chimeric protein is the DNA-binding domain, the homeodomain (38). This result suggests that the synergy between PDX-1 and E47/Pan1 could result from cooperative DNA binding and the formation of a stable DNA-binding complex with both proteins. Previous studies, however, have been unable to demonstrate cooperative DNA binding in vitro (38).
In this study, we directly compare the transcriptional activation properties of PDX-1 and Lmx1.1. Even though both homeoproteins interact with a common factor, E47/Pan1, and activate insulin gene transcription through the same promoter element, the two proteins interact with E47/Pan1 by distinct mechanisms. We also demonstrate that the β-cell-specific bHLH factor BETA2/NeuroD1 and the nuclear high-mobility-group protein I(Y) [HMG I(Y)] contribute to PDX-1–E47/Pan1 synergy through direct interactions with the homeodomain of PDX-1. These results demonstrate that PDX-1 acts multifunctionally as a center of protein-protein interactions in an intricate complex controlling β-cell-specific gene expression in vivo.
MATERIALS AND METHODS
Cell culture and transfections.
The Syrian hamster kidney fibroblast line BHK-21 was maintained as described previously (16). The COS7 line was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The mouse islet cell lines αTC1.6 and βTC3 were maintained in Dulbecco's modified Eagle's medium supplemented with 15% horse serum and 2.5% fetal bovine serum. High Five insect cells (Invitrogen) from Trichoplusia ni were grown in Grace's insect medium supplemented with 10% heat-inactivated fetal bovine serum at 27°C without CO2.
For mammalian cell transfections, cells were plated on six-well plates at a density of 5 × 104 (BHK-21) or 5 × 105 (COS7, αTC1.6, and βTC3) cells per well on the day before transfection. A total of 3 μg of plasmid DNA was mixed with 6 μl of Superfect reagent (Qiagen), and transfections were performed according to the manufacturer's protocol. Cells were harvested 40 to 48 h after transfection. Luciferase activities, measured with the Promega luciferase assay system, were normalized to β-galactosidase activities derived from a cotransfected thymidine kinase promoter-driven β-galactosidase control plasmid (0.8 μg of plasmid DNA per transfection) assayed with a Luminescent β-Galactosidase Detection Kit II (Clontech). The total amount of cDNA expression plasmid DNA was kept constant for individual transfections (a total of 1.2 μg of plasmid DNA per transfection) by adding the pBAT12 vector without insert cDNA. Each data point represents the average of at least three independent transfections ± standard error of the mean.
For insect cell transfections, High Five cells were plated on six-well plates at a density of 105 per well on the day before transfection. A total of 1.7 μg of plasmid DNA was mixed with 5.1 μl of 1 mM TransFast transfection reagent (Promega) and 600 μl of serum-free medium, and transfections were performed according to the manufacturer's protocol. Cells were harvested 40 to 48 h after transfection. Luciferase activities were measured with the Promega luciferase assay system and normalized to the concentration of the cell extract. The total amount of cDNA expression plasmid DNA was kept constant for individual transfections (a total of 0.7 μg of plasmid DNA per transfection) by adding the pBAT12 vector without insert cDNA. Each data point represents the average of at least three independent transfections ± standard error of the mean.
The reporter construct used for all transfection experiments (1 μg of plasmid DNA per transfection) was the luciferase gene under the control of a minimal prolactin promoter and five copies of the rat insulin I E2A3/4 minienhancer (pFOXluc.prl.5FF1) or a mutant minienhancer (pFOXluc.prl.5mC1 or pFOXluc.prl.5mEF1) as previously described (16). The expression plasmids for PDX-1, Lmx1.1, E47/Pan1, BETA2/NeuroD1, and HMG I(Y) were constructed by subcloning the coding region of each individual cDNA into the polylinker downstream of the cytomegalovirus (CMV) promoter of the pBAT expression vectors (16). Procedures for cloning of the Syrian hamster cDNAs of PDX-1, Lmx1.1, and E47/Pan1 have been described previously (14, 16, 42). The full-length mouse BETA2/NeuroD1 cDNA was obtained by ligating a 462-bp fragment of the 5′ coding region sequences including the transcription initiation site (generated by PCR from a βTC3 cDNA library) with a 1.5-kb 3′ fragment containing the first stop codon (kindly provided by Jacqueline E. Lee, University of Colorado at Boulder). The coding sequences of the Syrian hamster HMG I(Y) cDNA, in the HMG Y splice form, was originally isolated from a HIT T15 M2.2.2. cDNA library as described previously (16). Since the clone contained 5′ untranslated sequences, the coding sequence was generated by PCR from the original. The deletion mutants of E47/Pan1 were generated by PCR.
In vitro protein-protein interaction assays.
Glutathione S-transferase (GST) fusion proteins were produced in Escherichia coli BL21 competent cells via the pPIG plasmid system (20). In vitro-translated and [35S]methionine-labeled proteins were prepared using the TNT coupled reticulocyte lysate system (Promega) according to the manufacturer's protocol. Fifteen microliters of labeled protein was mixed with 1 μg of each GST fusion protein bound to 20 μl of glutathione-agarose beads in a total volume of 600 μl of interaction buffer (40 mM HEPES [pH 7.5], 50 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 0.5% [vol/vol] Nonidet P-40) (45). Samples were then incubated for 1 h at 4°C with a gentle rocking, and the beads were washed three times with interaction buffer. The bound proteins were eluted with 25 μl of Laemmli buffer, and 15-μl aliquots of the eluted proteins were fractionated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and visualized by autoradiography.
The PDX-1, E47/Pan1, and BETA2/NeuroD1 deletion mutants were generated by PCR from the pBAT expression plasmids and subcloned into either the pBAT11 in vitro transcription vector driven by the T7 promoter (16) or the pPIG vector. The mouse Mash1 and MyoD cDNAs were the gracious gifts of David Anderson (California Institute of Technology) and Eric Olson (University of Texas Southwestern Medical Center at Dallas), respectively. The Id1 cDNA was generated by PCR from rat islet cDNA.
Electrophoretic mobility shift assays (EMSAs).
The PDX-1 homeodomain (amino acids 138 to 213), BETA2/NeuroD1 bHLH domain (amino acids 94 to 162), and full-length HMG I(Y) proteins were prepared as His6-tagged proteins produced in E. coli BL21(DE3)/pLysS by using the pET15b plasmid system (Novagen) and were purified with a Ni-nitrilotriacetic acid spin kit (Qiagen) according to the manufacturer's protocol. The E47/Pan1 bHLH domain (amino acids 537 to 602) and HMG I(Y) were produced in E. coli BL21 cells as a GST fusion protein in the pPIG plasmid system. The GST-E47/Pan1 bHLH was bound to glutathione-Sepharose beads and cleaved with thrombin (1 U per mg of immobilized recombinant protein) for 2 h at 20°C in cleavage buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 2.5 mM CaCl2). Ten-microliter aliquots of the recombinant proteins were mixed with Laemmli buffer and quantified by staining SDS-polyacrylamide gels with Coomassie blue, with various known amounts of bovine serum albumin as standards. Rat insulin I E2A3/4 probes were labeled with [γ-32P]ATP and T4 polynucleotide kinase. The sequence for the top strand of the double-stranded E2A3/4 probe (EA probe) is GATCCTTCATCAGGCCATCTGGCCCCTTGTTAATAATCTAATTACCCTAGGTCTAA (E2 and A3/4 elements are underlined). The sequence for the top strand of the double-stranded E2A3/4 probe with a 15-bp insertion between the E2 and A3/4 elements (EA-I probe) is GATCCCTTCATCAGGCCATCTGGCCCGAGTCCAGCCTCGAGCTTGTTAATAATCTAATTACCCTAGGTCTAA (inserted bases are italicized). The proteins were mixed with the binding buffer [10 mM HEPES, 75 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 3% (vol/vol) Ficoll, 100 ng of poly(dI-dC), 1 μg of bovine serum albumin] and 20,000 cpm of labeled probe with or without specific competitors in a total volume of 20 μl. Ten microliters of the binding reaction mixture was electrophoresed on 7% nondenaturing polyacrylamide gels and visualized by autoradiography.
RESULTS
Comparison of PDX-1 and Lmx1.1 synergy with E47/Pan1.
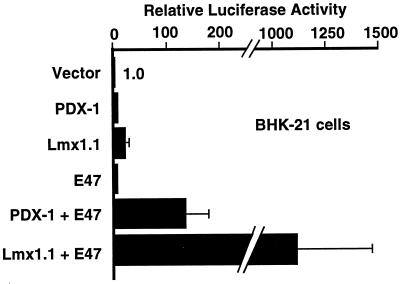
Although both PDX-1 and Lmx1.1 homeoproteins have been reported to synergize with E47/Pan1 in activating the rat insulin I E2A3/4 minienhancer (previously called the FF minienhancer) (16, 38), the two effects have not been directly compared. Therefore, we first compared the magnitude of PDX-1–E47/Pan1 synergy with that of Lmx1.1–E47/Pan1 synergy by cotransfection reporter gene analyses using several different cell lines (Fig. 1 and Table 1). The cDNA encoding each protein was inserted into a separate mammalian expression plasmid downstream of the CMV promoter. The reporter construct is plasmid pFOXluc.prl.5FF1 with the firefly luciferase gene inserted downstream of the rat prolactin minimal promoter and five copies of the rat insulin I E2A3/4 minienhancer. The reporter construct was then transfected with the cDNA expression plasmids into cultured cells, and luciferase activity was used as a gauge of transcriptional activation through the minienhancer.
FIG. 1.
Lmx1.1 and E47/Pan1 give greater transcriptional synergy than PDX-1 and E47/Pan1. BHK-21 cells were transfected with a reporter plasmid (pFOXluc.prl.5FF1) containing five copies of the insulin E2A3/4 minienhancer linked to the minimal prolactin promoter driving luciferase gene expression and with expression plasmids containing the CMV promoter driving expression of the cDNAs shown. Luciferase activity in the cells transfected with the reporter plasmid and the parent expression plasmid without insert is set to 1.0.
TABLE 1.
Comparison of minienhancer activation by Lmx1.1 and PDX-1
| Cell line | Lmx1.1 activation relative to PDX-1 activationa
|
||
|---|---|---|---|
| Basalb | Synergisticc | Ratio, synergistic/basald | |
| BHK-21 | 6.3 ± 0.8 | 10.0 ± 1.9 | 1.6 ± 0.2 |
| COS7 | 0.5 ± 0.09 | 13.4 ± 1.7 | 30.6 ± 9.3 |
| αTC1.6 | 8.4 ± 1.0 | 9.4 ± 1.2 | 1.1 ± 0.09 |
| βTC3 | 1.4 ± 0.07 | 4.1 ± 0.3 | 3.0 ± 0.2 |
BHK-21, COS7, αTC1.6, and βTC3 cells were transfected with a reporter plasmid (pFOXluc.prl.5FF1) containing five copies of the insulin E2A3/4 minienhancer linked to the minimal prolactin promoter driving luciferase gene expression and with expression plasmids containing the CMV promoter driving expression of the cDNAs shown. The data show luciferase activity in the cells transfected with Lmx1.1 relative to activity in the PDX-1-transfected cells.
Ratio of activity in the cells transfected with Lmx1.1 relative to the activity from the PDX-1-transfected cells in the absence of E47. The mean ± standard error of the mean for at least three independent transfections is shown.
Ratio of activity in the cells transfected with Lmx1.1 relative to the activity from the PDX-1-transfected cells in the presence of E47. The mean ± standard error of the mean for at least three independent transfections is shown.
Mean ± standard error of the mean of the ratio of synergistic to basal activity for Lmx1.1 relative to PDX-1.
In all cell types tested, for both the PDX-1–E47/Pan1 and Lmx1.1–E47/Pan1 combinations, synergy is observed: cotransfection of DNA plasmids expressing the two proteins gives a level of transcriptional activity that is significantly greater than the sum of the individual activities of the two proteins expressed separately (shown for BHK-21 cells in Fig. 1; data for other cell lines not shown). The luciferase activities of PDX-1–E47/Pan1 cotransfection relative to the basal activity of PDX-1 by itself are 16.6 ± 8.4, 341.7 ± 53.5, 5.6 ± 0.9, and 3.0 ± 0.6 in BHK-21, COS7, αTC1.6, and βTC3 cells, respectively. It should be noted that both basal activation and synergistic activation is low in βTC3 cells due to the high level of activity of the E2A3/4 minienhancer in these cells in the absence of any additional transcription factors.
Despite clear evidence of synergy, the PDX-1–E47/Pan1 combination gives a lower level of transcriptional activation than the Lmx1.1–E47/Pan1 combination in all four cell lines (Table 1). However, the basal level of transcriptional activation by Lmx1.1 alone is not always greater than that of PDX-1 (Table 1), indicating that the greater synergy of the Lmx1.1–E47/Pan1 combination may not simply result from a higher level of Lmx1.1 protein expression or Lmx1.1 DNA binding.
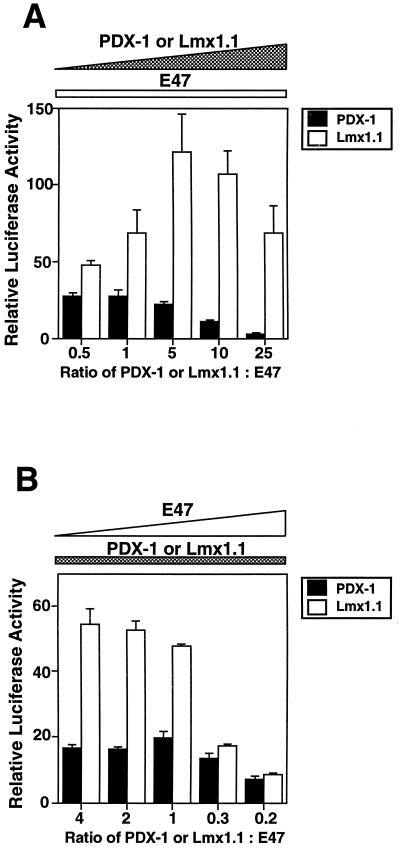
Although the level of transcriptional activation from the Lmx1.1–E47/Pan1 combination is consistently greater than that from the PDX-1–E47/Pan1 combination, the magnitude of synergy could depend on the absolute concentrations of the two homeodomain proteins or their concentrations relative to E47/Pan1. We therefore performed the same reporter gene analyses with increasing amounts of the PDX-1 or Lmx1.1 expression plasmid relative to the amount of E47/Pan1 expression plasmid (Fig. 2A) or vice versa (Fig. 2B). The activity of the Lmx1.1–E47/Pan1 combination is greater than that of the PDX-1–E47/Pan1 combination at all ratios of the expression plasmids tested. Interestingly, PDX-1–E47/Pan1 synergy is markedly decreased by increasing the amount of PDX-1 (Fig. 2A), whereas Lmx1.1–E47/Pan1 synergy is markedly decreased by excess E47/Pan1 (Fig. 2B). Transcription activity was not decreased by increasing the amount of any of the cDNAs when transfected alone (data not shown). These results suggest that both PDX-1 and Lmx1.1 may be able to interact with E47/Pan1 in solution, and the inhibitory effect from an excess of one protein could be due to excess free protein competing with and destabilizing DNA-bound protein.
FIG. 2.
Excess PDX-1 decreases synergy with E47/Pan1, and excess E47/Pan1 decreases synergy with Lmx1.1. (A) The pFOXluc.prl.5FF1 reporter plasmid was cotransfected with 0.02 μg of the E47/Pan1 expression plasmids and increasing amounts (0.01, 0.02, 0.1, 0.2, and 0.5 μg) of the PDX-1 or Lmx1.1 expression plasmid into BHK-21 cells. Luciferase activity in the cells transfected with the reporter plasmid and the E47/Pan1 expression plasmid alone is set at 1.0. (B) The reporter plasmid was cotransfected with 0.2 μg of the PDX-1 or Lmx1.1 expression plasmid and increasing amounts (0.05, 0.1, 0.2, 0.6, and 1.0 μg) of the E47/Pan1 expression plasmid into BHK-21 cells. The luciferase activity in the cells transfected with the reporter and the PDX-1 or Lmx1.1 expression plasmid alone is set at 1.0.
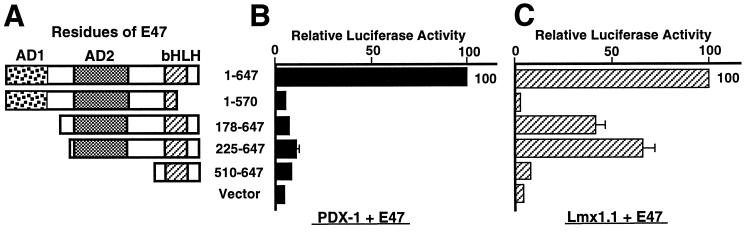
E47/Pan1 contains two distinct transcription activation domains. The first domain (AD1; amino acids 1 to 153) is consistently active in all cell types, whereas the second domain (AD2; amino acids 321 to 476), which includes a characteristic leucine zipper structure, functions selectively in pancreatic β-cell lines (5). Truncated E47/Pan1 constructs demonstrate the importance of these domains in synergistic activation (Fig. 3). Both PDX-1–E47/Pan1 synergy and Lmx1.1–E47/Pan1 synergy are lost when the C terminus of the bHLH domain of E47/Pan1 is partially deleted (E47 1-570 [Fig. 3A]), demonstrating that this domain is critical for synergy. Similarly, the truncation of both transcription activation domains of E47/Pan1 abolishes the synergy with PDX-1 or Lmx1.1, even in the presence of the intact bHLH domain (E47 510-647). Interestingly, Lmx1.1 can synergize with an N-terminally truncated E47/Pan1 in which AD2 is retained, whereas PDX-1 fails to synergize with any E47/Pan1 construct lacking AD1 (E47 178-647 and 225-647). These results indicate that the β-cell-specific AD2 plays an essential role in Lmx1.1–E47/Pan1 synergy whereas the nonspecific AD1 is required for PDX-1–E47/Pan1 synergy.
FIG. 3.
PDX-1 and Lmx1.1 cooperate with different activation domains of E47/Pan1. The pFOXluc.prl.5FF1 reporter plasmid was cotransfected with a plasmid expressing the wild-type or truncated E47/Pan1 cDNA and a plasmid expressing either the PDX-1 or the Lmx1.1 cDNA into BHK-21 cells. Luciferase activity in the cells transfected with the reporter, the homeobox cDNA expression plasmid, and the wild-type E47/Pan1 (amino acids 1 to 647) expression plasmid is set at 100. (A) Diagram showing the E47/Pan1 deletions used. (B) Results for cells transfected with the E47/Pan1 and PDX-1 expression plasmids. (C) Results for cells transfected with the E47/Pan1 and Lmx1.1 expression plasmids.
PDX-1 and E47/Pan1 physically interact in vitro.
The above results suggest that PDX-1 and Lmx1.1 interact with E47/Pan1 by distinct mechanisms. We have previously shown that Lmx1.1 physically interacts with E47/Pan1 in the absence of DNA (20). Other investigators have not succeeded in detecting a direct physical interaction between PDX-1 and E47/Pan1 by coimmunoprecipitation (38), but the antibodies used for coimmunoprecipitation may destabilize the PDX-1–E47/Pan1 complex.
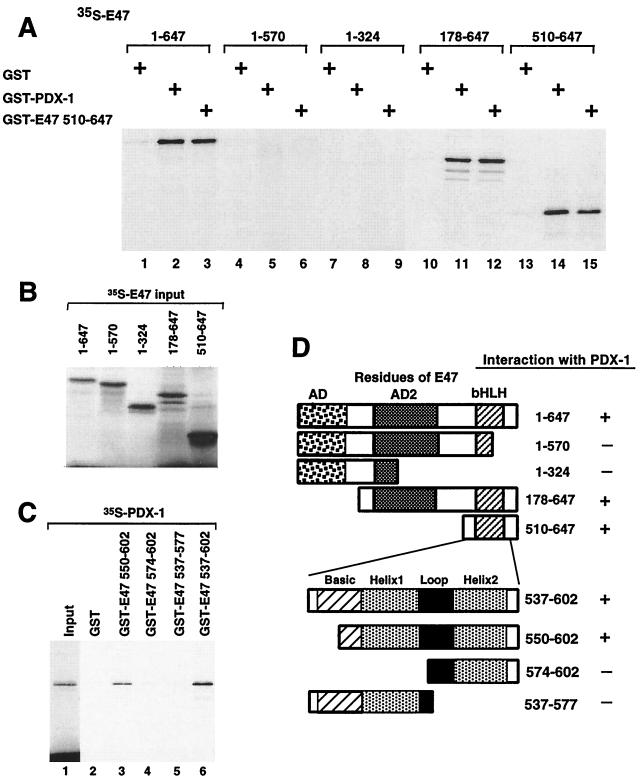
We performed in vitro protein binding assays using 35S-labeled full-length E47/Pan1 and various deletion mutants to test the interactions with a PDX-1–GST fusion protein (Fig. 4A). The labeled protein was mixed with the GST fusion protein in solution, and the mixture was added to glutathione-Sepharose beads and washed. The retained protein was separated by SDS-polyacrylamide gel electrophoresis (PAGE). The wild-type 35S-E47/Pan1 binds to GST–PDX-1, indicating that PDX-1 physically interacts with E47/Pan1 in vitro (Fig. 4A, lane 2). The binding is apparently as efficient as the homodimerization of E47/Pan1 (lane 3). The N-terminally truncated mutants of E47/Pan1 that contain the intact bHLH domain can interact with PDX-1 (lanes 11 and 14), whereas binding is lost when the C-terminal bHLH domain is deleted (lanes 5 and 8), demonstrating that the bHLH domain of E47/Pan1 is crucial for this interaction.
FIG. 4.
PDX-1 physically interacts with the HLH domain of E47/Pan1 in vitro. (A) In vitro-translated, 35S-labeled E47/Pan1 (wild type and deletion mutants) was incubated with GST–PDX-1 fusion protein immobilized on glutathione-Sepharose beads, and bound proteins were resolved on SDS-PAGE followed by autoradiography. GST alone and GST fused to E47/Pan1 amino acids 510 to 647 (including the bHLH dimerization domain) were used as controls. (B) Ten percent of the 35S-labeled E47/Pan1 (wild-type and deletion mutant) proteins used in the binding assay were resolved by SDS-PAGE. (C) In vitro-translated, 35S-labeled PDX-1 protein was incubated with GST alone or with GST fused to the truncated E47/Pan1 proteins shown. Bound proteins were immobilized on glutathione-Sepharose beads and resolved by SDS-PAGE followed by autoradiography. Ten percent of the 35S-labeled PDX-1 protein used in the binding assay was loaded in lane 1. (D) Structures of the E47/Pan1 deletion mutants and summary of the results.
Further truncations of the bHLH domain of E47/Pan1 demonstrate that an intact HLH domain is required for the interaction with PDX-1 (Fig. 4C). In this experiment, PDX-1 was labeled with 35S and E47/Pan1 was fused to GST because the truncated E47/Pan1 proteins were too small to fractionate by SDS-PAGE. Deletion of the first seven amino acids of the basic domain of E47/Pan1 (Pan1 550-602) abolishes DNA binding (data not shown) but does not affect the interaction with PDX-1 (Fig. 4C, lane 3), whereas deletion of either helix abolishes the interaction (Pan1 574-602 and 537-577 [lanes 4 and 5]). Similar results were observed for the interaction with Lmx1.1 (data not shown).
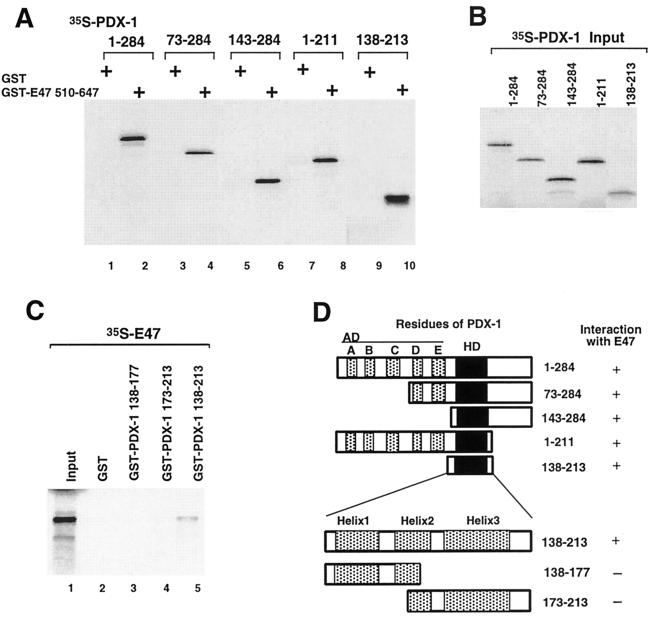
We next tested wild-type and truncated PDX-1 for interaction with the bHLH domain of E47/Pan1, and the interaction domain of PDX-1 maps to the homeodomain (Fig. 5A). We then prepared GST fusion proteins of intact and truncated PDX-1 homeodomains to test the interactions with in vitro-translated E47/Pan1 (Fig. 5C). Removal of either the N-terminal or C-terminal helix of the PDX-1 homeodomain eliminates the interaction with E47/Pan1 (Fig. 5C, lanes 3 and 4), indicating that the intact homeodomain of PDX-1 is required for interaction with E47/Pan1. These results are summarized in Fig. 5D.
FIG. 5.
The intact PDX-1 homeodomain interacts physically with E47/Pan1. (A) In vitro-translated, 35S-labeled PDX-1 (wild type and deletion mutants) was incubated with GST-E47/Pan1 fusion protein immobilized on glutathione-Sepharose beads, and bound proteins were resolved by SDS-PAGE followed by autoradiography. GST alone was used as a control. (B) Ten percent of the 35S-labeled PDX-1 (wild-type and deletion mutant) proteins used in the binding assay were resolved by SDS-PAGE. (C) In vitro-translated, 35S-labeled E47/Pan1 protein was incubated with GST alone or with GST fused to the truncated PDX-1 proteins shown. Bound proteins were immobilized on glutathione-Sepharose beads and resolved by SDS-PAGE followed by autoradiography. Ten percent of the 35S-labeled E47/Pan1 protein used in the binding assay was loaded in lane 1. (D) Structures of the PDX-1 deletion mutants and summary of the results. AD, transcription activation domain; HD, homeodomain; A to E, conserved segments of the PDX-1 activation domain as described by Peshavaria et al. (40).
PDX-1 physically interacts with class B bHLH proteins.
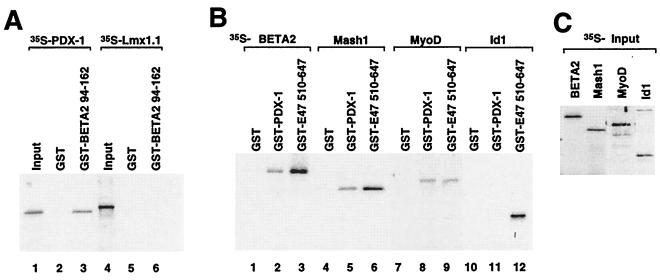
Since the entire HLH domain of E47/Pan1, the structure of which is conserved among the bHLH protein families (reviewed in reference 33), is required for the interaction with PDX-1 and Lmx1.1, we tested whether the bHLH domain of BETA2/NeuroD1, a member of the B class bHLH proteins that is expressed selectively in pancreatic islet cells (35), can physically associate with PDX-1 or Lmx1.1 (Fig. 6A). Interestingly, PDX-1, but not Lmx1.1, can interact with the bHLH domain of BETA2/NeuroD1 (BETA2 94-162 [Fig. 6A, lanes 3 and 6). In addition, PDX-1 interacts with other class B bHLH proteins, including Mash1 and MyoD, and the affinities are similar to the heterodimerization of these proteins with E47/Pan1 (Fig. 6B, lanes 5, 6, 8, and 9). Similar results were observed with the isolated PDX-1 homeodomain, indicating that these interactions are mediated via the homeodomain of PDX-1 (data not shown). However, the Id class HLH protein Id1 does not interact with PDX-1 (lane 11). The hematopoietic cell-specific bHLH protein Tal1 also does not interact with PDX-1 (data not shown), suggesting that PDX-1 recognizes more than the HLH structure alone, and certain conserved amino acids may be required. In contrast, Lmx1.1 does not interact with these class B bHLH proteins or Id1 in similar assays (data not shown). Thus, unlike Lmx1.1, PDX-1 can interact with a diverse group of bHLH proteins, including the islet cell-specific bHLH protein BETA2/NeuroD1.
FIG. 6.
PDX-1 physically interacts with tissue-specific bHLH proteins. (A) In vitro-translated, 35S-labeled PDX-1 or Lmx1.1 protein was incubated with GST alone or with the GST-BETA2 bHLH domain (containing amino acids 94 to 162 of BETA2/NeuroD). Bound proteins were immobilized on glutathione-Sepharose beads and resolved by SDS-PAGE followed by autoradiography. Ten percent of the 35S-labeled PDX-1 and Lmx1.1 proteins used in the binding assay were loaded in lanes 1 and 4, respectively. (B) 35S-labeled full-length HLH proteins BETA2/NeuroD, Mash1, MyoD, and Id1 were tested as for panel A for interaction with GST–PDX-1. GST alone and GST fused to E47/Pan1 amino acids 510 to 647 (including the bHLH dimerization domain) were used as controls. (C) Ten percent of the 35S-labeled HLH proteins used for the assay were resolved by SDS-PAGE.
BETA2/NeuroD1, E47/Pan1, and PDX-1 form a ternary complex on the insulin promoter.
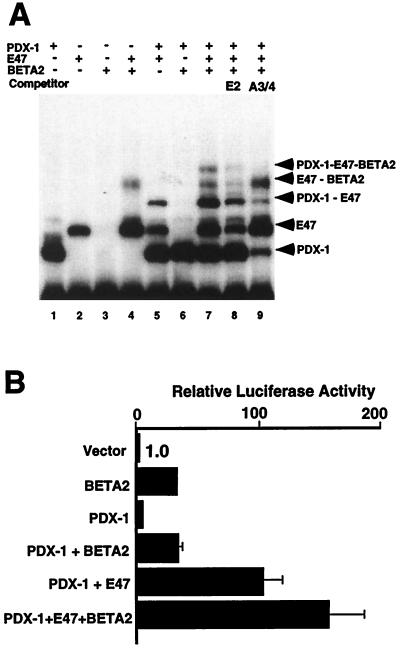
While the preceding results demonstrate that the DNA-binding domains of PDX-1, E47/Pan1, and BETA2/NeuroD1 can interact in solution, for these interactions to affect transcription of the insulin gene they need to form a stable ternary complex on the insulin promoter. Therefore, we tested whether the three proteins can form a stable ternary complex on the insulin E2A3/4 minienhancer by EMSA (Fig. 7A). Consistent with a previous study of the DNA-binding properties of BETA2/NeuroD1 (35), the bHLH domain of BETA2/NeuroD1 does not bind DNA by itself (lane 3) but does bind DNA as a BETA2/NeuroD1–E47/Pan1 heterodimer complex (lane 4). When the homeodomain of PDX-1 and the bHLH domain of E47/Pan1 are coincubated with the insulin E2A3/4 minienhancer probe, an additional DNA-bound complex is observed, indicating that the homeodomain of PDX-1 and the bHLH domain of E47/Pan1 can form a ternary complex on the probe (lane 5). When the homeodomain of PDX-1 and bHLH domains of E47/Pan1 and BETA2/NeuroD1 are incubated together with the E2A3/4 probe, a slower-migrating complex is observed in addition to the PDX-1–E47/Pan1 complex and the BETA2/NeuroD1–E47/Pan1 heterodimer complex (lane 7). This slower-migrating complex is reduced by coincubation with a 100-fold molar excess of nonradiolabeled E2 element (lane 8) or A3/4 element (lane 9).
FIG. 7.
BETA2/NeuroD can form a ternary complex with the bHLH domain of E47/Pan1 and the homeodomain of PDX-1 on the E2A3/4 element. (A) The PDX-1 homeodomain (320 pg), E47/Pan1 bHLH domain (30 ng), and BETA2/NeuroD bHLH domain (3 ng) recombinant proteins were tested by EMSA for the ability to bind the 32P-labeled rat insulin E2A3/4 probe. One hundred-fold molar excess of nonlabeled E2 (lane 8) or A3/4 (lane 9) oligonucleotide was used as a specific competitor. (B) BHK-21 cells were transfected with 1 μg of the reporter as described for Fig. 1 and 0.2 μg of the expression plasmid DNAs of PDX-1, E47/Pan1, and BETA2/NeuroD as shown. Luciferase activity in the cells transfected with the reporter plasmid and the pBAT12 plasmid with no insert is set to 1.0.
To test whether BETA2/NeuroD1 affects the synergistic activation of the insulin minienhancer by the PDX-1–E47/Pan1 combination, we expressed all three proteins in transfected BHK-21 cells (Fig. 7B). BETA2/NeuroD1 does not synergize with PDX-1 in the absence of E47/Pan1; it does, however, increase the PDX-1–E47/Pan1 synergy, although only by an amount approximately equal to the basal activation of BETA2/NeuroD1 by itself.
Cooperative DNA binding.
Although PDX-1 and E47/Pan1 can form a stable ternary complex on the insulin promoter, these experiments provide no visually obvious evidence of cooperative DNA binding by the two proteins, consistent with a previous report (38). We also detected no change in the preferences of PDX-1 for the different binding sites within the A3/4 region in the presence of E47/Pan1 (data not shown).
To detect more subtle evidence of cooperative DNA binding by the two proteins, however, we used a quantitative analysis. If PDX-1 and E47/Pan1 bind to DNA independently and bind to the same DNA molecule only by random chance, then cobinding should occur only at a frequency equal to the product of each independent binding event. To test if the two proteins truly bind to DNA independently, we performed an EMSA with the PDX-1 and E47/Pan1 binding domains and used a phosphorimager to measure the fraction of E2A3/4 minienhancer probe bound by each protein separately and by the two proteins together. The ratio of actual to predicted cobinding of the two proteins is the cooperativity index shown in Table 2 and Fig. 8B.
TABLE 2.
Cooperative DNA binding by the PDX-1 homeodomain and E47 bHLH domain
| % of probe bounda
|
Cooperativity indexc | P valued | |||
|---|---|---|---|---|---|
| PDX-1 | E47 | PDX-1 + E47
|
PDX-1 + E47
|
||
| Actual | Predictedb | ||||
| 3.4 ± 0.2 | 5.1 ± 1.2 | 1.1 ± 0.2 | 0.3 ± 0.1 | 3.5 ± 0.2 | <0.01 |
The homeodomain of PDX-1 (48 pg) and bHLH domain of E47/Pan1 (700 pg) were incubated with the 32P-labeled rat insulin E2A3/4 probe, and the percentage of probe bound separately by each protein and cobound by both proteins was measured with a phosphorimager. The mean ± standard error of the mean for four independent binding reactions is shown.
Defined as total PDX-1 binding × total E47 binding, where the total binding for each protein is the fraction of all probe bound by that protein.
Ratio of actual to predicted cobinding of PDX-1 and E47.
A comparison of actual and predicted cobinding of PDX-1 and E47 was used to calculate P values by the paired t test.
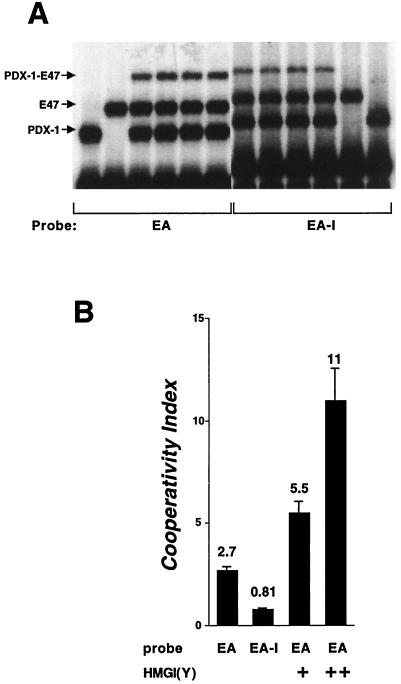
FIG. 8.
Modest cooperative DNA binding by E47 and PDX1. (A) The homeodomain of PDX-1 (160 pg; lanes 1, 3 to 10, and 12) and the bHLH domain of E47/Pan1 (6 ng; lanes 2 to 11) were incubated with the 32P-labeled rat insulin E2A3/4 probe (EA; lanes 1 to 6) or with a similarly labeled probe containing a 15-bp insertion between the E and A binding sites (EA-I; lanes 7 to 12) and analyzed by EMSA. (B) The complexes from at least five independent EMSAs were quantified with a phosphorimager, and the cooperativity index was calculated as defined in Table 2. HMG I(Y) experiments were performed as for Fig. 9, with 37 ng (+) and 110 ng (++) of HMG I(Y). Mean ± standard error of the mean is shown.
As can be seen in Table 2, significantly more probe is bound by the two proteins together than can be explained by independent binding. To determine whether cooperative binding requires that the two proteins bind to adjacent sites on the probe, cobinding was assayed with the EA-I probe containing a 15-bp insertion (one and a half helical turns) between the E and A binding sites. Cobinding is reduced, and there is no evidence for cooperativity (cooperativity index of 0.81) when the two sites are separated (Fig. 8). These data demonstrate that the two binding domains, which can physically interact in solution, can also bind to adjacent sites on DNA in a cooperative fashion.
HMG I(Y) interacts with the PDX-1–E47/Pan1 complex.
While these data demonstrate cooperative DNA binding, it seems unlikely that this modest degree of cooperativity can explain the much larger functional synergy between PDX-1 and E47/Pan1 as detected by transcription assays. In the nucleus of an intact cell, however, additional proteins may modify these DNA-binding properties. The presence of proteins that stabilize the PDX-1–E47/Pan1 complex on DNA or that modify DNA structure could favor cooperative DNA binding by the two proteins in vivo.
When we isolated the cDNA encoding Lmx1.1 (16), we also isolated a hamster β-cell cDNA encoding HMG I(Y) due to the affinity of the encoded protein for the rat insulin I A3/4 DNA probe. HMG I(Y) is a nonhistone, chromatin-associated nuclear protein that binds to the minor groove of AT-rich stretches of DNA (reviewed in references 6, 17, and 18). It can alter the binding of various transcription factors through its effects on DNA structure as well as through direct protein-protein interactions (1). HMG I(Y) binds to the A3/4 region of the insulin promoter (data not shown).
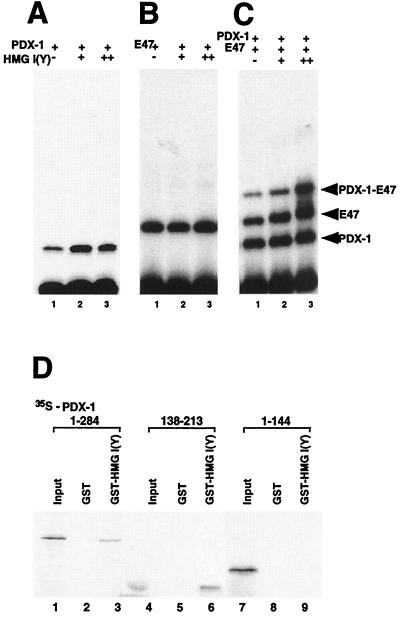
DNA binding of PDX-1 to the E2–A3/4 probe is increased in the presence of HMG I(Y) (Fig. 9A), and cobinding by PDX-1 and E47/Pan1 is even more dramatically increased (Fig. 9C). This increase in binding was quantified in a parallel experiment using a phosphorimager. The addition of 37 ng of His6-tagged HMG I(Y) per 10-μl binding reaction mixture resulted in a 2.4- ± 0.25-fold increase in the cooperativity index (as defined in Table 2), and the addition of 110 ng caused a 4.8- ± 0.7-fold increase (Fig. 8B). It should be noted that a new, slower-migrating band that would indicate the addition of HMG I(Y) to the complex is not always observed in these experiments. Similar observations have been reported in earlier studies (25) and may result from HMG I(Y) contributing to formation of the complex without remaining in the complex through electrophoresis.
FIG. 9.
HMG I(Y) interacts with the activation complex on the E2A3/4 minienhancer. (A) The homeodomain of PDX-1 (160 pg) was incubated with the 32P-labeled rat insulin E2A3/4 probe with GST-HMG I(Y) (160 ng in lane 2; 320 ng in lane 3) or GST alone (320 ng in lane 1; 160 ng in lane 2) and analyzed by EMSA. (B) The bHLH domain of E47/Pan1 (6 ng) was incubated with the 32P-labeled rat insulin E2A3/4 probe with GST-HMG I(Y) (160 ng in lane 2; 320 ng in lane 3) or GST alone (320 ng in lane 1; 160 ng in lane 2) and analyzed by EMSA. (C) The homeodomain of PDX-1 (160 pg) and bHLH domain of E47/Pan1 (6 ng) were incubated with the 32P-labeled rat insulin E2A3/4 probe with GST-HMG I(Y) (160 ng in lane 2; 320 ng in lane 3) or GST alone (320 ng in lane 1; 160 ng in lane 2) and analyzed by EMSA. (E) In vitro-translated, 35S-labeled PDX-1 protein (wild type and deletion mutants) was incubated with GST alone or with GST-HMG I(Y). Bound proteins were immobilized on glutathione-Sepharose beads and resolved by SDS-PAGE followed by autoradiography. Ten percent of the 35S-labeled PDX-1 proteins were loaded in lanes 1, 4, and 7.
HMG I proteins can bind to transcription factors as well as DNA, as exemplified by the interaction between HMG I(Y) and the Pou domain of the POU homeodomain protein Oct-2A (1). In an in vitro interaction assay, both wild-type PDX-1 and the homeodomain of PDX-1 associate with HMG I(Y) (Fig. 9D, lanes 3 and 6) but the C-terminally truncated mutant of PDX-1 that lacks the homeodomain (PDX-1 1-144) does not (lane 9), indicating that HMG I(Y) also interacts with PDX-1 via its homeodomain.
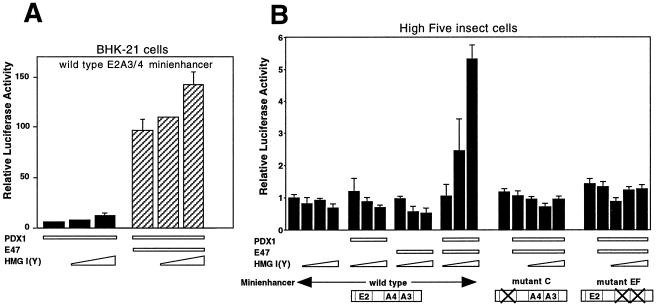
In cotransfection assays using BHK-21 cells, HMG I(Y) enhances both basal activation by PDX-1 and the synergistic activation of PDX-1 and E47/Pan1 on the E2A3/4 minienhancer in a dose-dependent manner (Fig. 10A) but does not affect transcription from the E2A3/4 minienhancer by itself (data not shown). This effect of HMG I(Y) in BHK-21 cells is modest, probably because mammalian cells already express high levels of the HMG I proteins.
FIG. 10.
HMG I(Y) contributes to the synergy of PDX-1 and E47/Pan1. (A) Transfections were performed in BHK-21 cells as for Fig. 1A, except for the addition of increasing amounts (0.2 and 0.4 μg) of the HMG I(Y) expression plasmid. Luciferase activity in the cells transfected with the reporter plasmid and the pBAT12 plasmid with no insert is set to 1.0. (B) High Five insect cells were transfected with the pFOXluc.prl.5FF1 reporter construct (1.0 μg) as for panel or with reporter constructs with mutations of the E2 (pFOXluc.prl.5mC1) or A3/4 (pFOXluc.prl.5mEF1) sites. Expression plasmids containing the CMV promoter driving expression of the cDNAs for PDX-1 (100 ng), E47/Pan1 (100 ng), or HMG I(Y) (20, 100, or 500 ng) were cotransfected. Luciferase activity in the cells transfected with the reporter plasmid and the parent expression plasmid without insert is set to 1.0.
Insect cells have been reported to have lower endogenous levels of HMG I(Y) and are more responsive to exogenous HMG I(Y) (25). In the insect cell line High Five, PDX-1 and E47/Pan1 do not activate the E2A3/4 minienhancer either individually or together. The addition of HMG I(Y), however, allows for strong cooperative activation of the minienhancer by the three proteins together, but only if both the E and the A binding sites are intact in the minienhancer (Fig. 10B).
DISCUSSION
These studies demonstrate a physical interaction between PDX-1 and E47/Pan1 in solution. The interaction involves the homeodomain of PDX-1 and the HLH domain of E47/Pan1 and apparently requires the structural integrity of these domains since smaller subdomains do not interact. We provide evidence that this interaction, along with interactions with HMG I(Y), contributes to transcriptional synergy by increasing binding of the protein complex to DNA, but other cooperative interactions may contribute to synergy as well.
The organization of the insulin promoter with tightly juxtaposed E and A sites strongly suggests a model of cooperative DNA binding by E47/Pan1 and PDX-1 leading to the assembly of an activation complex on the EA minienhancers. Synergy would then result from the lower free energy of cooperative versus independent DNA binding by the two proteins. This model is supported by the capacity of the two proteins to form a ternary complex on the E2A3/4 minienhancer and the high affinity between the DNA-binding domains of the two proteins. Further supportive evidence comes from a prior study showing that the isolated PDX-1 homeodomain linked to the unrelated activation domain from the VP16 transcriptional activator is sufficient to synergize with E47/Pan1 on the insulin promoter, although the degree of synergy is less than that observed with the intact PDX-1 protein (38). These data suggest that simple recruitment of multiple activators to the promoter through cooperative DNA binding is alone sufficient for synergistic activation of the insulin promoter.
But the degree of cooperative DNA binding with purified E47/Pan1 and PDX-1 in vitro is small. While we were able to demonstrate cooperative DNA binding by the same two domains that interact in solution, this effect is not as dramatic as the transcriptional synergy observed in vivo, consistent with what other investigators have seen (38). With the addition of BETA2/NeuroD1, an increased fraction of the E47/Pan1– BETA2/NeuroD1 heterodimer shifts up to the larger complex that includes PDX-1, but the overall effect is still modest.
An EMSA with purified proteins, however, does not necessarily reflect the normal environment of a cell nucleus in vivo. Unlike the EMSA, in the nucleus the presence of many other proteins may influence the DNA binding of the two proteins and their degree of cooperativity. These nuclear proteins could alter DNA binding by either protein, stabilize the protein-protein interaction, or modify the DNA structure in the E2A3/4 region. Any of these effects could improve the energetic advantage of the protein-protein interaction on DNA and thereby increase cooperative DNA binding.
In this regard, it is interesting that the HMG I(Y) protein binds to the A3/4 region. HMG I(Y) is a highly conserved, ubiquitously and abundantly expressed nuclear protein containing three short basic amino acid repeats that contact the minor groove of DNA along A/T-rich stretches and reduce the flexibility of the DNA helix. By reducing the energetic costs of DNA distortion, HMG I(Y) and similar architectural proteins play an essential role in the assembly of higher-order nucleoprotein complexes (7, 17, 47). In addition to binding to DNA, HMG I(Y) physically associates in vitro with transcription factors including the POU homeodomain proteins Oct-2A (1) and Tst-1/Oct-6 (27), and we have now shown that it also associates with the PDX-1 homeodomain. Through DNA binding, protein-protein interaction, or both, HMG I(Y) enhances both the DNA-binding and activation potential of PDX-1 alone and in combination with E47/Pan1.
Cooperative interactions with HMG I(Y) are not a common feature of all homeodomain proteins. HMG I proteins inhibit DNA binding by the engrailed homeodomain protein and inhibit both DNA binding and transcriptional activation by HOXD9 (4). In contrast, another chromosomal binding protein, HMG1, enhances DNA binding and transcriptional activation by homeodomain protein HOXD9 but not HOXD8 (53). These complex interactions increase the DNA-binding specificity of individual homeodomain proteins and help define the scope of their functional gene targets in vivo.
Taken together, these studies demonstrate that the PDX-1 homeodomain should be viewed not only as a DNA-binding domain but also as a protein-protein interaction domain capable of multiple, complex interactions. Here we have shown that the homeodomain can interact with the E47/Pan1 and BETA2/NeuroD1 HLH domains and the HMG I(Y) DNA architectural protein. In addition, the PDX-1 homeodomain, along with the adjacent FPWMK peptide, also interacts with PBX1 (39), a homeodomain protein that interacts with a variety of other homeodomain transcription factors including members of the HOX family (8, 48). Association with PBX1 alters the binding specificity of PDX-1. PDX-1 and PBX1 bind cooperatively to sites in the somatostatin promoter, but not to the A3/4 region of the insulin promoter (39), and high levels of PBX1 decrease the affinity of PDX-1 for the A3/4 element in vitro (data not shown). It seems possible that in the presence of E47/Pan1, this inhibitory interaction is destabilized, further enhancing the cooperativity of DNA binding by PDX-1 and E47/Pan1 to the A3/4 sites in vivo. In sum, these various negative and positive interactions combine to build a functional transcription activation complex on the insulin promoter with the PDX-1 homeodomain at the center.
Simple cooperative recruitment of transcriptional activators to the E2A3/4 minienhancer may not fully explain all of the transcriptional synergy between E47/Pan1 and PDX-1. Once recruited to the minienhancer, the clustering of multiple activation domains appears to have a synergistic effect on transcription, since removal of either the amino-terminal activation domain from PDX-1 (data not shown) (38, 40) or the nonspecific AD1 activator from E47/Pan1 causes near complete loss of transcriptional synergy. This effect is apparently not due to the interruption of additional specific interactions between the two proteins, since the function of AD1 is not context dependent (5, 31) and the activation domain of PDX-1 can be replaced by the viral VP16 activation domain (38). As has been demonstrated for enhancers from other genes, such as the beta interferon and T-cell receptor α gene enhancers, grouping of activators on DNA produces clusters of protein interaction sites that cooperatively recruit or stabilize binding of the RNA polymerase II transcription initiation complex (reviewed in reference 7). Non-DNA-binding coactivators may assist in this process by providing additional stabilizing interactions linking the DNA-bound transcriptional activators with the basal transcription machinery. In this regard, it is interesting that the p300 coactivator has been found to interact both physically and functionally with the activation domains of BETA2/NeuroD1 and E47/Pan1 (11, 34, 41).
If cooperative DNA binding and clustering of activation domains explains the transcriptional synergy between PDX-1 and E47/Pan1, then the greater synergy between Lmx1.1 and E47/Pan1 could result from stronger cooperative binding or a stronger activation domain on Lmx1.1. But the evidence indicates that the interaction between Lmx1.1 and E47/Pan1 is fundamentally different, since synergy persists when either the Lmx1.1 activation domain (16) or E47/Pan1 AD1 is removed. Additional intra- and intermolecular interactions amplify the transcriptional cooperativity of Lmx1.1 and E47/Pan1. First, the LIM domains of Lmx1.1 function as inhibitory domains in the absence of E47/Pan1, suppressing the inherent activity of the Lmx1.1 activation domain (16, 20). By interacting specifically with the second LIM domain, the E47/Pan1 bHLH domain may interrupt the intramolecular interaction and relieve the inhibition (20). This relief of an intramolecular repression may be very similar to the mechanism by which the zinc finger transcription factor GATA-4 unmasks the activation domain of the homeodomain factor Nkx2.5 when the two proteins bind to adjacent sites in cardiac gene promoters (10, 26, 43). Second, AD2 of E47/Pan1, which normally functions only in β cells (5), is activated in non-β-cells by interaction with Lmx1.1. Activation of AD2 could be due to conformational changes in E47/Pan1 that permit AD2 to function as an activator, or it could be due to specific cooperation between AD2 and Lmx1.1 in recruiting coactivators and the RNA polymerase II transcription initiation complex. We presume that these interactions generate the additional transcriptional potency of the Lmx1.1–E47/Pan1 combination.
To produce an activation complex that is unique to β-cell nuclei, the protein-protein interactions that secure the complex must be specific, not easily replaced by other protein interactions in the nuclei of other cells. The capacity to interact with other proteins may be a common feature of many homeodomains (8, 10, 26, 43, 48), but the types of interactions and precise interacting partners vary. For example, other homeodomains, including homeodomains from the β-cell factors Lmx1.1, Isl-1, and Cdx2/3, cannot substitute for the PDX-1 homeodomain in the interaction with the E47/Pan1 HLH domain (reference 20 and unpublished data).
It seems likely, however, that other factors in the β cell can substitute for the overall function of PDX-1 in the activation complex on the EA minienhancers. When PDX-1 is removed from the β cell by Cre-mediated deletion of the PDX-1 gene, β cells continue to express insulin (3). This result leaves open the question of which A-element-binding protein actually forms part of the activation complex on the intact insulin gene in vivo.
ACKNOWLEDGMENTS
We thank H. Ee, S. Smith, and H. Watada for critical reading of the manuscript, and we thank G. M. Grodsky and members of our laboratory for helpful discussions. We also thank Joey Leung and Yi Zhang for technical assistance and Leslie Spector for helping in preparation of the manuscript.
This work is supported by NIH grant DK-21344, a Juvenile Diabetes Foundation fellowship (to K.O.), and a Howard Hughes postdoctoral fellowship (to R.G.M.).
REFERENCES
- 1.Abdulkadir S A, Casolaro V, Tai A K, Thanos D, Ono S J. High mobility group I/Y protein functions as a specific cofactor for Oct-2A: mapping of interaction domains. J Leukoc Biol. 1998;64:681–691. doi: 10.1002/jlb.64.5.681. [DOI] [PubMed] [Google Scholar]
- 2.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 3.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. β-Cell-specific inactivation of the mouse ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlotta P, Rustighi A, Mantovani F, Manfioletti G, Giancotti V, Tell G, Damante G. High mobility group I proteins interfere with the homeodomains binding to DNA. J Biol Chem. 1997;272:29904–29910. doi: 10.1074/jbc.272.47.29904. [DOI] [PubMed] [Google Scholar]
- 5.Aronheim A, Shiran R, Rosen A, Walker M D. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci USA. 1993;90:8063–8067. doi: 10.1073/pnas.90.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin M, Lehn D A, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 7.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 8.Chan S K, Jaffe L, Capovilla M, Botas J, Mann R S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 9.Dumonteil E, Philippe J. Insulin gene: organisation, expression and regulation. Diabetes Metab. 1996;22:164–173. [PubMed] [Google Scholar]
- 10.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutural cofactors. EMBO J. 1997;18:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 12.Emens L A, Landers D W, Moss L G. Hepatocyte nuclear factor 1α is expressed in a hamster insulinoma line and transactivates the rat insulin I gene. Proc Natl Acad Sci USA. 1992;89:7300–7304. doi: 10.1073/pnas.89.16.7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, Imura H, Kennedy G, Madsen O, Melloul D, et al. The insulin gene promoter. A simplified nomenclature. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 14.German M S, Blanar M A, Nelson C, Moss L G, Rutter W J. Two related helix-loop-helix proteins participate in separate cell-specific complexes that bind the insulin enhancer. Mol Endocrinol. 1991;5:292–299. doi: 10.1210/mend-5-2-292. [DOI] [PubMed] [Google Scholar]
- 15.German M S, Moss L G, Wang J, Rutter W J. The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical beta-cell nuclear complexes. Mol Cell Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.German M S, Wang J, Chadwick R B, Rutter W J. Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- 17.Grosschedl R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 18.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Heritable formation of pancreatic β-cell tumors in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson J D, Zhang W, Rudnick A, Rutter W J, German M S. Transcriptional synergy between LIM-homeodomain proteins and basic helix-loop-helix proteins: the LIM2 domain determines specificity. Mol Cell Biol. 1997;17:3488–3496. doi: 10.1128/mcb.17.7.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson O, Walker M D, Rutter W J, Edlund T. Individual protein-binding domains of the insulin gene enhancer positively activate β-cell-specific transcription. Mol Cell Biol. 1989;9:823–827. doi: 10.1128/mcb.9.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy G, German M. Insulin gene regulation. In: LeRoith D, Olefsky J, Taylor S, editors. Diabetes mellitus: a fundamental and clinical text. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 20–26. [Google Scholar]
- 25.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y, Shioi T, Kasahara H, Jobe S M, Wiese R J, Markham B E, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leger H, Sock E, Renner K, Grummt F, Wegner M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol. 1995;15:3738–3747. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy M R. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 29.MacFarlane W, Read M, Gilligan M, Bujalska I, Docherty K. Glucose modulates the binding activity of the beta-cell transcription factor IUF1 in a phosphorylation-dependent manner. Biochem J. 1994;303:625–631. doi: 10.1042/bj3030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshak S, Totary H, Cerasi E, Melloul D. Purification of the beta-cell glucose-sensitive factor that transactivates the insulin gene differentially in normal and transformed islet cells. Proc Natl Acad Sci USA. 1996;93:15057–15062. doi: 10.1073/pnas.93.26.15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massari M E, Jennings P A, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C P, McGehee R E, Jr, Habener J F. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murre C, Baltimore D. The helix-loop-helix motif: structure and function. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 861–879. [Google Scholar]
- 34.Mutoh H, Fung B P, Naya F J, Tsai M J, Nishitani J, Leiter A B. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naya F J, Stellrecht C M, Tsai M J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 36.Offield M F, Jetton T L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L, Wright C V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 37.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peers B, Leonard J, Sharma S, Teitelman G, Montminy M R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 39.Peers B, Sharma S, Johnson T, Kamps M, Monteminy M. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: importance of the FPWMK motif and of the homeodomain. Mol Cell Biol. 1995;15:7091–7097. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peshavaria M, Henderson E, Sharma A, Wright C V, Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudnick A, Ling T Y, Odagiri H, Rutter W J, German M S. Pancreatic beta cells express a diverse set of homeobox genes. Proc Natl Acad Sci USA. 1994;91:12203–12207. doi: 10.1073/pnas.91.25.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sepulveda J L, Belaguli N, Nigam V, Chen C Y, Nemer M, Schwartz R J. GATA-4 and Nkx-2.5 coactive Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serup P, Petersen H V, Pedersen E E, Edlund H, Leonard J, Petersen J S, Larsson L I, Madsen O D. The homeodomain protein IPF-1/STF-1 is expressed in a subset of islet cells and promotes rat insulin 1 gene expression dependent on an intact E1 helix-loop-helix factor binding site. Biochem J. 1995;310:997–1003. doi: 10.1042/bj3100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirokawa J M, Courey A J. A direct contact between the dorsal rel homology domain and Twist may mediate transcriptional synergy. Mol Cell Biol. 1997;17:3345–3355. doi: 10.1128/mcb.17.6.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoffers D A, Zinkin N T, Stanojevic V, Clarke W L, Habener J F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 47.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 48.van Dijk M A, Murre C. extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 49.Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol. 1996;10:1327–1334. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- 50.Walker M D, Edlund T, Boulet A M, Rutter W J. Cell-specific expression controlled by the 5′ flanking regions of the insulin and chymotrypsin genes. Nature. 1983;306:557–581. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- 51.Watada H, Kajimoto Y, Kaneto H, Matsuoka T, Fujitani Y, Miyazaki J, Yamasaki Y. Involvement of the homeodomain-containing transcription factor PDX-1 in islet amyloid polypeptide gene transcription. Biochem Biophys Res Commun. 1996;229:746–751. doi: 10.1006/bbrc.1996.1875. [DOI] [PubMed] [Google Scholar]
- 52.Watada H, Kajimoto Y, Umayahara Y, Matsuoka T, Kaneto H, Fujitani Y, Kamada T, Kawamori R, Yamasaki Y. The human glucokinase gene beta-cell-type promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HIT-T15 cells. Diabetes. 1996;45:1478–1488. doi: 10.2337/diab.45.11.1478. [DOI] [PubMed] [Google Scholar]
- 53.Zappavigna V, Falciola L, Citterich M H, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]