Abstract
The objectives of the study were to use tumor size data from 10 phase II/III atezolizumab studies across five solid tumor types to estimate tumor growth inhibition (TGI) metrics and assess the impact of TGI metrics and baseline prognostic factors on overall survival (OS) for each tumor type. TGI metrics were estimated from biexponential models and posttreatment longitudinal data of 6699 patients. TGI‐OS full models were built using parametric survival regression by including all significant baseline covariates from the Cox univariate analysis followed by a backward elimination step. The model performance was evaluated for each trial by 1000 simulations of the OS distributions and hazard ratios (HR) of the atezolizumab‐containing arms versus the respective controls. The tumor growth rate estimate was the most significant predictor of OS across all tumor types. Several baseline prognostic factors, such as inflammatory status (C‐reactive protein, albumin, and/or neutrophil‐to‐lymphocyte ratio), tumor burden (sum of longest diameters, number of metastatic sites, and/or presence of liver metastases), Eastern Cooperative Oncology Group performance status, and lactate dehydrogenase were also highly significant across multiple studies in the final multivariate models. TGI‐OS models adequately described the OS distribution. The model‐predicted HRs indicated good model performance across the 10 studies, with observed HRs within the 95% prediction intervals for all study arms versus controls. Multivariate TGI‐OS models developed for different solid tumor types were able to predict treatment effect with various atezolizumab monotherapy or combination regimens and could be used to support design and analysis of future studies.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
The association between tumor growth inhibition (TGI) metrics and overall survival (OS) for atezolizumab was previously investigated in patients with non‐small cell lung cancer from a phase II trial for model development and a phase III trial as external evaluation.
WHAT QUESTION DID THIS STUDY ADDRESS?
Whether the TGI‐OS platform could be generalized for atezolizumab by the inclusion of 10 clinical studies across five solid tumor types.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
The TGI‐OS models predicted the treatment effects of atezolizumab‐containing and control arms based on the comparison of hazard ratios. The tumor growth rate was the most significant predictor of OS across tumor types, and inflammatory status and tumor burden were also strong predictors.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS?
Identification of patient‐level baseline prognostic factors and early on‐treatment information can be leveraged to predict longer term survival benefit in cancer immunotherapy studies in multiple cancer types and support early development decisions with combination treatments.
INTRODUCTION
The use of tumor dynamics model‐based approaches has become increasingly attractive to evaluate treatment response for decision‐making through the course of clinical development in oncology. 1 , 2 , 3 Model‐based tumor dynamics metrics (including early shrinkage, time to regrowth, on‐treatment growth rate, or the full dynamic profile) have been demonstrated to predict overall survival (OS) in different types of solid tumors, including colorectal cancer, 4 , 5 , 6 breast cancer, 7 , 8 non‐small cell lung cancer (NSCLC), 9 , 10 , 11 locally advanced and metastatic urothelial carcinoma (mUC), 12 , 13 renal cell carcinoma (RCC), 14 , 15 and several other tumor types 16 , 17 , 18 , 19 for a variety of treatments. Leveraging tumor dynamics as a biomarker to predict OS in phase II trials with cancer immunotherapy (CIT) is not a novel concept, but longitudinal tumor response to CIT treatment may elicit different patterns compared with treatments with other mechanisms of action, such as delayed responses or increased tumor burden before regression. 10 , 12 , 13 , 16
Atezolizumab is a humanized immunoglobulin G1 monoclonal antibody that targets human programmed death‐ligand 1 (PD‐L1) on tumor‐infiltrating immune cells (ICs) and tumor cells (TCs) and inhibits PD‐L1 interaction with programmed death 1 (PD‐1) and B7.1 receptors, thereby sending inhibitory signals to T cells. 20 , 21 , 22 Atezolizumab is approved to treat locally advanced or metastatic NSCLC, mUC, extensive‐stage small‐cell lung cancer (SCLC), locally advanced or metastatic triple‐negative breast cancer (TNBC), and unresectable hepatocellular carcinoma (HCC) by the US Food and Drug Administration (US FDA) and/or the European Medicines Agency. 23 , 24
The association between tumor growth inhibition (TGI) metrics and OS for atezolizumab was previously investigated in patients with NSCLC who progressed during or following prior platinum chemotherapy, using atezolizumab and control (docetaxel) data from a phase II trial (POPLAR) for model development and a phase III trial (OAK) as external evaluation. 10 A TGI‐OS model, with on‐treatment tumor growth rate constant (KG) as estimated using time profiles of the sum of longest diameters (target lesions per response evaluation criteria in solid tumours [RECIST] 1.1), albumin (ALB), and number of metastatic sites as independent prognostic factors, was able to predict the OS hazard ratio (HR) in subpopulations of patients with varying baseline PD‐L1 expression in both trials. This model will be referred to herein as the “historical” OS model. In POPLAR and OAK, slower KG in the atezolizumab arm when compared with the docetaxel (control) arm predicted the OS benefit, whereas the other TGI metrics (i.e., time to growth, early change in tumor size, and tumor shrinkage rate constant), as well as classical clinical end points of overall response rate (ORR) and progression‐free survival (PFS), did not predict the observed difference in OS in the two studies. 10 Although the link between tumor dynamics and OS was shown for other CITs, 12 , 13 , 16 , 25 this is the only analysis where the difference in TGI metrics (namely KG) across treatment arms in randomized studies was shown to predict treatment effect (HR of investigational treatment vs. control) on OS. Among the various tumor dynamic‐based approaches to predict OS, the use of KG estimates seems to be quite promising as demonstrated in many studies. 6 , 10 , 15 , 18 , 25
Limitations of the previous atezolizumab study 10 are the use of only one tumor type and the model development was based on data from a single clinical trial. To determine whether the TGI‐OS platform can be generalized, the inclusion of other studies and tumor types should be explored. The objectives of our analysis were to develop multivariate TGI‐OS models to predict the OS distribution and the benefit of atezolizumab‐containing treatments compared with controls in 10 clinical studies stratified by five solid tumor types. The goal is to identify appropriate patient‐level baseline prognostic factors as well as individualized, early on‐treatment information in the form of KG to predict longer term survival benefit in CIT studies in multiple cancer types and to identify patients who are most likely to benefit from these therapies. Ultimately, a successful confirmation of the TGI‐OS platform can facilitate application across other CITs to support phase II study design, end‐of‐phase II decisions, and phase III planning and analysis particularly with combination treatments. 1 , 2 , 3
METHODS
The clinical trial protocols of the 10 trials have been previously described 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 and are summarized in Table 1. Across all trials, patients in the atezolizumab‐containing arm received either atezolizumab intravenously at 840 mg every 2 weeks (IMpassion130, triple negative breast cancer [TNBC]) or 1200 mg every 3 weeks (the other nine trials). The 10 trials were conducted in five solid tumor types—NSCLC, SCLC, TNBC, RCC, and mUC—in accordance with the Declaration of Helsinki after approval by institutional review boards or independent ethics committees. All patients provided written informed consent.
TABLE 1.
Clinical trial descriptions, number of patients, and tumor assessments used in the analysis
| Trial name | ClinicalTrials.gov identifier | Tumor type | Atezolizumab‐containing arm (A) | Control arm (C) | Number of patients enrolled (A/C) | Number of TGI‐evaluable patients (A/C) | Tumor assessment schedule | Number of tumor size assessments among TGI‐evaluable patients | Number of postbaseline tumor size assessments (A/C) |
|---|---|---|---|---|---|---|---|---|---|
| OAK | NCT02008227 | NSCLC | Atezolizumab | Docetaxel | 421/401 | 388/363 | q6w for 36 weeks, then q9w | 5277 | 2546/1653 |
| POPLAR | NCT01903993 | NSCLC | Atezolizumab | Docetaxel | 142/135 | 129/123 | q6w for 36 weeks, then q9w | 1309 | 669/388 |
| IMpower130 | NCT02367781 | NSCLC | Atezolizumab + carboplatin + nab‐paclitaxel | Carboplatin + nab‐paclitaxel | 472/232 | 441/214 | q6w for 48 weeks, then q9w | 4451 | 2868/929 |
| IMpower150 | NCT02366143 | NSCLC |
Arm A: Atezolizumab + carboplatin + paclitaxel Arm B: Atezolizumab + carboplatin + paclitaxel + bevacizumab |
Carboplatin + paclitaxel + bevacizumab | 402 (Arm A)/400 (Arm B)/400 (Control) | 377 (Arm A)/360 (Arm B)/369 (Control) | q6w for 48 weeks, then q9w | 8007 | 2312 (Arm A)/2668 (Arm B)/1921 (Control) |
| IMpower131 | NCT02367794 | NSCLC | Arm B: Atezolizumab + carboplatin + nab‐paclitaxel | Carboplatin + nab‐paclitaxel | 343/340 | 311/317 | q6w for 48 weeks, then q9w | 6197 | 2057/1312 |
| IMpower132 | NCT02657434 | NSCLC | Atezolizumab + cisplatin/carboplatin + pemetrexed | Cisplatin/carboplatin + pemetrexed | 292/286 | 241/239 | q6w for 48 weeks, then q9w | 2168 | 968/721 |
| IMpower133 | NCT02763579 | SCLC | Atezolizumab + carboplatin + etoposide | Placebo + carboplatin + etoposide | 198/196 | 189/187 | q6w for 48 weeks, then q9w | 1969 | 862/731 |
| IMvigor211 | NCT02302807 | mUC | Atezolizumab | Chemotherapy (vinflunine vs. taxane:paclitaxel and docetaxel) | 459/443 | 382/362 | q9w for 54 weeks, then q12w | 2822 | 1204/874 |
| IMpassion130 | NCT02425891 | TNBC | Atezolizumab + nab‐paclitaxel | Placebo + nab‐paclitaxel | 452/438 | 439/422 | q8w for 12 months, then q12w | 4171 | 1843/1467 |
| IMmotion151 | NCT02420821 | RCC | Atezolizumab + bevacizumab | Sunitinib | 454/461 | 425/421 | At 12 weeks after randomization, followed by q6w until Week 78, then q12w after 78 weeks | 7076 | 3355/2873 |
| Overall | 7367 (4266 in NSCLC) | 6699 (3872 in NSCLC) | 43,447 (27,409 in NSCLC) | 34,221 (21,012 in NSCLC) |
Atezolizumab is given at 1200 mg q3w except in IMpassion130, where atezolizumab is given 840 mg q2w.
Abbreviations: mUC, metastatic urothelial carcinoma; NSCLC, non‐small cell lung cancer; q#w, every # weeks; RCC, renal cell carcinoma; SCLC, extensive‐stage small cell lung cancer; TNBC, triple‐negative breast cancer.
Tumor lesions were measured using computed tomography or magnetic resonance imaging at baseline and at regular intervals afterward (approximately every 6–9 weeks for 1 year and then every 9–12 weeks thereafter until disease progression, death, or loss of follow‐up). Longitudinal tumor size data, defined as the sum of the longest diameters of target lesions at each visit according to RECIST 1.1, were used for the estimation of TGI metrics. Patients with at least baseline and one postbaseline tumor size measurements were defined as evaluable, and data from patients who only had baseline tumor assessments were excluded from the analysis.
TGI modeling methods have been previously published. 10 Briefly, the biexponential TGI model proposed by Stein et al. 18 was fit to the longitudinal tumor size data set by tumor type. In the model, TS0 is the model‐estimated tumor size at the start of treatment (time = 0), KG is the tumor growth rate constant (1/week), and KS is the tumor shrinkage rate constant (1/week). The model was implemented as a nonlinear mixed effect model using NONMEM version 7.4. In each of the models by tumor type, a log‐normal distribution was used to characterize the interindividual variability of KG and KS by treatment, with a common log‐normal distribution for TS0 within the same tumor type, and an additive residual error was described by a normal distribution. TGI model evaluation was conducted using standard goodness‐of‐fit plots. Individual post hoc parameter estimates from the model were used as TGI metrics in the subsequent TGI‐OS modeling.
The TGI‐OS model was developed and evaluated as previously described. 10 The impact on OS from a predefined list of potential covariates, which consisted of individual baseline prognostic factors for each tumor type (Table S1) and TGI metrics, was first explored using the Kaplan–Meier method. Baseline prognostic factors investigated across the five tumor types included variables associated with tumor burden (baseline tumor size [SLD], number of metastatic sites [MET], liver metastasis), inflammatory status (ALB, neutrophil‐to‐lymphocyte ratio [NLR], C‐reactive protein [CRP]), and general prognostic factors (Eastern Cooperative Oncology Group performance status [ECOG], lactate dehydrogenase [LDH], alkaline phosphatase). In addition, race (White vs. non‐White or Asian vs. non‐Asian) and tumor‐specific variables, such as hemoglobin, time since initial diagnosis, calcium, and sarcomatoid histology for RCC, were also tested in tumor‐specific TGI‐OS models. PD‐L1, which is expressed on TCs and tumor‐infiltrating ICs on a wide variety of cancer expressions and is targeted by atezolizumab, was scored by immuno‐histochemistry as percentage of PD‐L1–expressing TC (TC3 ≥ 50%, TC2 ≥ 5%, and <50%, TC1 ≥ 1% and <5%, and TC0 < 1%) and as percentage of PD‐L1–expressing tumor area for IC (IC3 ≥ 10%, IC2 ≥ 5%, and <10%, IC1 ≥ 1% and <5%, and IC0 < 1%). 36
Univariate screening of the covariates was evaluated using Cox regression analyses, and all significant covariates with a significance level of p < 0.05 per the log‐likelihood ratio test were included in the full model. If several covariates relating to the same variable were significant in the Cox analysis, such as dichotomizing race to White versus non‐White or Asian versus non‐Asian, only the one with the best likelihood improvement was retained in the full model. The continuous covariates were not normalized by median values.
The full model parameters were estimated based on a parametric survival regression. The probability density function that best described the observed survival times was selected among normal, log‐normal, Weibull, logistic, log‐logistic, and exponential by using the difference in Akaike information criterion of the alternative models. Backward stepwise elimination of the full model was performed using a significance level of p < 0.01, and this resulted in the final model, in which all covariates were significant. The TGI‐OS models were developed independently by tumor type, and model development and evaluation were implemented in R version 3.6.3 (R Foundation for Statistical Computing).
The model performances were evaluated using a simulation‐based approach. Baseline prognostic factors as well as KG were resampled from observed values in the analysis data set for each tumor type. Model parameters were sampled from the estimated mean values and uncertainty in parameter estimates from the TGI‐OS model for each of the simulated study replicate. Censoring was simulated by sampling patient study duration from a uniform distribution based on observed censoring data as shown in Figure S1. Simulation results of the TGI‐OS model were further summarized as OS HR by comparing the atezolizumab‐containing arm(s) to the respective control arm of each clinical trial. OS distributions and HR of the atezolizumab‐containing arm versus control of each trial were simulated 1000 times.
The performance of the TGI‐OS model for NSCLC was further evaluated using external validation by randomly splitting the data from 70% of the patients into a training set and 30% into a testing set and using the model developed from the training set to predict the OS outcome of the patients in the testing set. The model performance was evaluated based on the concordance index (c‐index). 37
RESULTS
Data from one phase II and nine phase III atezolizumab trials were included in the analysis. Among these 10 trials, treatment consisted of atezolizumab monotherapy, atezolizumab in combination with chemotherapy (e.g., carboplatin), and/or targeted therapy (e.g., bevacizumab), and the active control arm generally included the corresponding combination treatment, chemotherapy (e.g., docetaxel), or targeted therapy (e.g., sunitinib), as shown in Table 1. A total of 6699 of the 7367 patients randomized (90.0%) to the 10 trials were considered TGI evaluable. The numbers of randomized and TGI‐evaluable patients by trial are listed in Table 1. Of the five tumor types included in the analysis, the majority of the data (57%) were in NSCLC. A total of 43,447 baseline and posttreatment tumor assessments from the TGI‐evaluable patients were used for TGI model development, with an average of 6.49 tumor assessments per patient. The median duration of tumor assessments ranged between 123 days (mUC) and 380 days (RCC), and the majority of OS data with censoring was up to 405 days of follow‐up, except in RCC, where the median OS had not been reached (Figure S1). The difference between the median duration of tumor assessment and OS duration ranged from 129 days (mUC) to 407 days (TNBC), with a median of 275 days across tumor types. Within each trial, the atezolizumab‐containing treatment arm is generally associated with longer duration of tumor assessment, as shown in Figure S1, as the patients who had slower disease progression were more likely to continue clinical visits.
The biexponential TGI models were fitted to tumor type‐specific data. An example model was included in the Supplementary Material. The TGI model was sufficiently flexible to capture different patterns of tumor dynamics observed for NSCLC as well as for the other four solid tumor types. Example model fits of individual tumor size data are illustrated in Figure S2, and the goodness‐of‐fit plots for the longitudinal tumor data are shown in Figure S3, indicating no bias across time or tumor size as well as demonstrating good correlations between observed and predicted values among the five tumor‐specific models.
The final TGI model parameter estimates by tumor type are shown in Table S2. All parameters are estimated precisely regardless of treatment type or tumor type, with a relatively low standard error <10% for KG and <13% for KS. Overall, individual parameter estimates were not reduced to the population mean, and particularly for KG, the estimated shrinkage values 38 were generally below 20%, indicating the data were informative for individual parameter estimations.
A comparison of KG distributions between treatments for NSCLC is shown in Figure 1. Across tumor types, mUC has faster tumor shrinkage with larger KS, and SCLC has the slowest tumor regrowth with smaller KG, irrespective of treatment type. The typical TGI profiles stratified by treatment and trial are shown in Figure 2.
FIGURE 1.
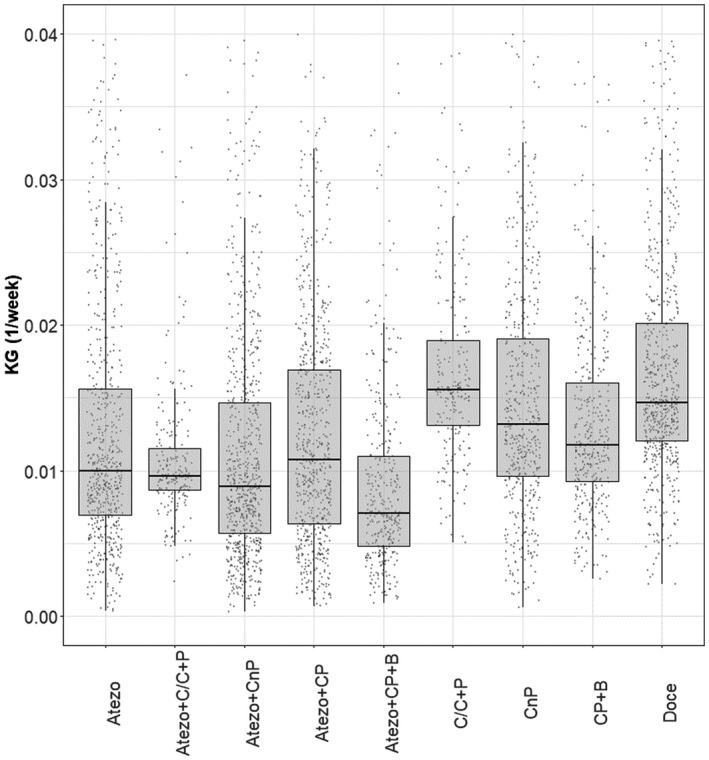
Model‐estimated KG comparison between treatments in patients with non‐small cell lung cancer. Dots indicate individual model‐estimated KG, ends of boxes indicate 25th and 75th percentiles (i.e., interquartile range), horizontal lines indicate medians, and ends of whiskers indicate 1.5 times the interquartile range. Atezo, atezolizumab; B, bevacizumab; C/C, cisplatin/carboplatin; CnP, carboplatin + nab‐paclitaxel; CP, carboplatin + paclitaxel; Doce, docetaxel; KG, tumor growth rate constant; P, pemetrexed
FIGURE 2.
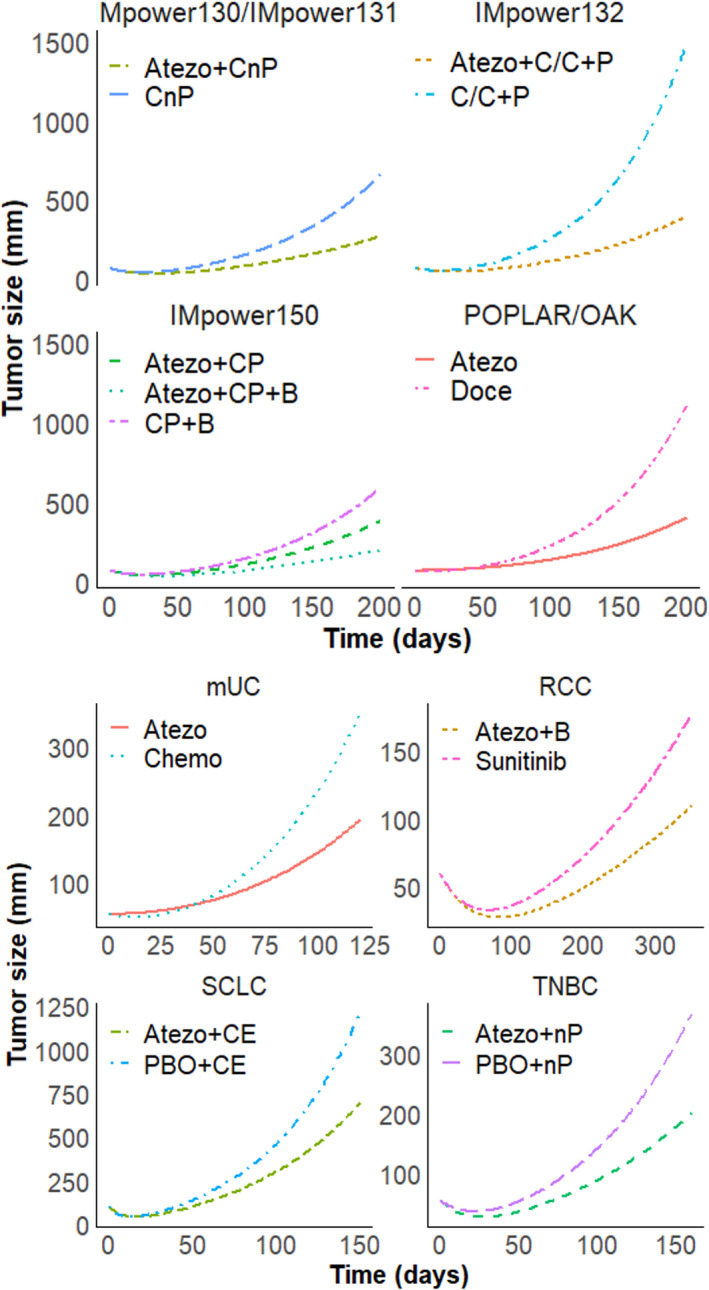
Typical tumor growth inhibition profiles by trial/tumor type, stratified by arm, with non‐small cell lung cancer on top panels, and extensive‐stage small cell lung cancer, triple‐negative breast cancer, renal cell carcinoma, and metastatic urothelial carcinoma on bottom panels. Atezo, atezolizumab; B, bevacizumab; C/C, cisplatin/carboplatin; CE, carboplatin+etoposide; CnP, carboplatin + nab‐paclitaxel; CP, carboplatin + paclitaxel; Doce, docetaxel; nP, nab‐paclitaxel; P, pemetrexed; PBO, placebo
Along with the predefined lists of baseline prognostic factors, individual KG estimates were considered as covariates for the TGI‐OS model development. Previously, KG has been shown to be a strong predictor of OS in atezolizumab trials for NSCLC. 10 Exploration of the association between KG and OS data was conducted using Kaplan–Meier analysis and is shown in Figure S4. In all tumor types, a trend of inverse correlation is observed between tumor growth and survival, with the slowest KG (lowest quartile of log(KG)) having the longest OS.
TGI‐OS models for each tumor type were developed using univariate screening, followed by backward elimination. The parameter estimates of the final TGI‐OS models based on a log‐normal probability density function are shown in Table 2 for NSCLC and in Table S3 for other tumor types. KG was the most significant predictor of OS across all five tumor types.
TABLE 2.
Parameter estimates from tumor growth inhibition–overall survival models in non‐small cell lung cancer
| Parameter | Estimate | SE | z | p |
|---|---|---|---|---|
| Intercept | 3.47 | 0.173 | 20.1 | 8.41E−90 |
| log(KG) | −0.616 | 0.0224 | −27.4 | 1.35E−165 |
| C‐reactive protein (mg/L) | −0.00385 | 0.000348 | −11.1 | 1.75E−28 |
| ECOG (1 vs. 0) | −0.233 | 0.0298 | −7.81 | 5.61E−15 |
| Number of metastatic sites | −0.0764 | 0.0139 | −5.51 | 3.53E−08 |
| Race (Asian vs. non‐Asian) | 0.244 | 0.0443 | 5.52 | 3.42E−08 |
| Albumin (g/L) | 0.0135 | 0.00304 | 4.43 | 9.48E−06 |
| IC or TC > 0 vs. IC and TC = 0 | 0.119 | 0.0286 | 4.16 | 3.20E−05 |
| Lactate dehydrogenase (U/L) | −0.00014 | 4.00E−05 | −3.53 | 0.00041 |
| Neutrophil‐to‐lymphocyte ratio | −0.009 | 0.00262 | −3.44 | 0.000582 |
| Lines of therapy (2+ vs. 1) | −0.109 | 0.0341 | −3.2 | 0.00138 |
| Liver metastasis (yes vs. no) | −0.118 | 0.0401 | −2.94 | 0.00332 |
| Log(scale) | −0.264 | 0.0161 | −16.3 | 4.95E−60 |
Survival time was analyzed in days.
Abbreviations: ECOG, Eastern Cooperative Oncology Group performance status (reference group is 0); IC, tumor‐infiltrating immune cells; log(KG), log of tumor growth rate constant (1/week) from the tumor growth inhibition model; p, Wald test p value; scale, standard deviation of log(OS); SE, standard error of parameter estimate; TC, tumor‐infiltrating tumor cells (reference group is IC and TC = 0); z, Wald statistic.
Several baseline prognostic factors were highly significant across multiple tumor types (Table 3), such as inflammatory status (CRP, ALB, and/or NLR in all tumor types), tumor burden (SLD, MET, and/or liver metastasis in all tumor types), ECOG (four tumor types, excluding RCC), LDH (two tumor types), and race in terms of Asian versus non‐Asian (two tumor types).
TABLE 3.
Significant baseline prognostic factors in the final tumor growth inhibition–overall survival models by tumor type
| Tumor burden | Inflammatory status | General factors | Others | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor type | SLD | MET | Liver | ALB | NLR | CRP | ECOG | LDH | ALP | Race | ICTC | Line | HGB | RCC1YR | CA | SRCMT |
| NSCLC | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| SCLC | ✓ | ✓ | ✓ | ✓ | ||||||||||||
| TNBC | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||||
| RCC | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||
| mUC | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||||
Abbreviations: ALB, albumin; ALP, alkaline phosphatase; CA, calcium; CRP, C‐reactive protein; ECOG, Eastern Cooperative Oncology Group performance status score; HGB, hemoglobin; ICTC, tumor‐infiltrating immune cells (IC) or tumor cells (TC) > 0; LDH, lactate dehydrogenase; Line, lines of therapy; Liver, liver metastasis; MET, number of metastatic sites; mUC, metastatic urothelial carcinoma; NLR, neutrophil‐to‐lymphocyte ratio; NSCLC, non‐small cell lung cancer; Race: Asian vs. non‐Asian; RCC, renal cell carcinoma; RCC1YR, time since initial diagnosis; SCLC, extensive‐stage small cell lung cancer; SLD, baseline tumor burden; SRCMT, sarcomatoid histology; TNBC, triple‐negative breast cancer.
According to TGI‐OS models, survival probability increases when CRP, NLR, SLD, or the number of metastatic sites decreases, and ALB increases. In addition, IC/TC PD‐L1 expression groups and line of treatment were significant prognostic factors for NSCLC, in which the data combined both first‐line and later lines of treatment. Four tumor‐specific variables were also significant prognostic factors for RCC. Statistical summaries of the significant prognostic factors by trial and by treatment for NSCLC and by tumor type for the other models are shown in Table S4. Notably, 40.8% (N = 196) of patients in the IMpower132 trial for NSCLC had missing IC or TC PD‐L1 expression information, but when combined with the rest of the NSCLC data, only 4.9% of patients (203 of 4179) had missing IC or TC information.
The predictive ability of the TGI‐OS models was evaluated by comparing the 95% prediction intervals (PIs) of simulated survival distributions with observed distributions, stratified by treatment type (Figure S5). The overlapping of the curves suggests that the models can reasonably reproduce the observed survival distributions in all treatment types across the five tumor types.
The TGI‐OS models were also evaluated by simulating OS HR by atezolizumab treatment arm versus appropriate control in each trial, conditional on the individual baseline prognostic factors and estimated KG, as shown in Figure 3. The model‐predicted HRs indicate good model performance across all 11 comparisons from 10 trials, with observed HRs within the 95% PIs for all trial arms.
FIGURE 3.
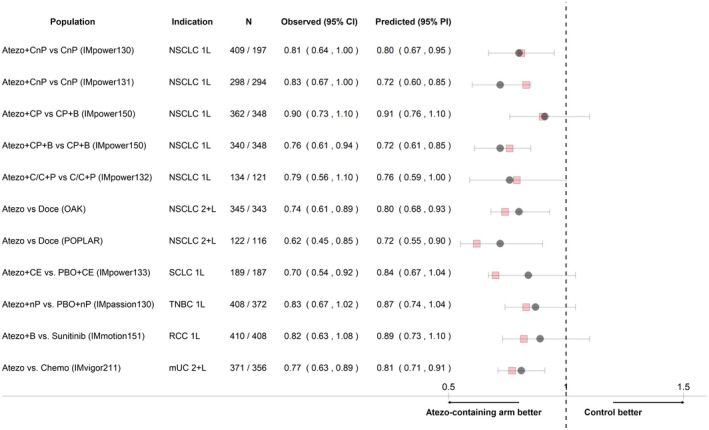
Hazard ratios of atezolizumab‐containing arm versus control by study based on simulations of the tumor growth inhibition–overall survival models. N indicates number of tumor growth inhibition–evaluable patients in each group and arm (atezolizumab/control) with nonmissing covariates. Squares indicate observed hazard ratios, circles indicate median model‐predicted hazard ratios, and bars indicate 95% prediction intervals (PIs) based on 1000 replicates. 1L, first line; 2+L, second or later line; Atezo, atezolizumab; B, bevacizumab; C/C, cisplatin/carboplatin; CE, carboplatin+etoposide; Chemo, chemotherapy; CI, confidence interval; CnP, carboplatin+nab‐paclitaxel; CP, carboplatin+paclitaxel; Doce, docetaxel; mUC, metastatic urothelial carcinoma; nP, nab‐paclitaxel; NSCLC, non‐small cell lung cancer; P, pemetrexed; PBO, placebo; RCC, renal cell carcinoma; SCLC, extensive‐stage small cell lung cancer; TNBC, triple‐negative breast cancer
The results of the external validation by randomly splitting the data show that the c‐index of the training (0.753) and of the testing sets (0.754) are the same or similar to the c‐index of the model developed using the full set of data (0.754), indicating a good and consistent predictive accuracy of the model. The model evaluation using OS HR based on the testing set is shown in Figure S6. External validation for the other tumor types was not conducted due to the relatively small sample sizes.
DISCUSSION
Model‐based estimates of on‐treatment growth rate (KG) have been found to predict for OS in a variety of tumor types and treatments. 6 , 10 , 15 , 18 , 25 More recently, KG was found to be the only tumor dynamic metric able to predict survival benefit in one phase II study and one phase III study of atezolizumab versus chemotherapy in second and later lines of therapy for NSCLC, whereas other model‐based metrics or classical end points (ORR, PFS) did not. 10 US FDA scientists found that KG was inversely associated with OS in 24 randomized NSCLC clinical trials with CPIs or targeted therapies, 25 although they did not develop a predictive model, suggesting that KG estimates from early tumor dynamic data might serve as an earlier end point in clinical studies. 1 , 2 , 3
The present analysis aimed to confirm the use of KG in predicting long‐term survival benefit as well as to identify baseline prognostic factors that are common or specific across five solid tumor types using data from 10 atezolizumab randomized clinical trials.
Because the majority of data in the analysis was collected from patients with NSCLC (N = 3872 TGI‐evaluable patients), a tumor‐specific model was constructed from a large NSCLC data set of 27,409 tumor assessments, enabling a robust evaluation of the impact of a diverse set of patient characteristics. Comparing to the historical atezolizumab TGI‐OS model that was developed from a phase II trial, 10 in addition to ALB and the number of metastatic sites, the updated model incorporated the additional independent baseline prognostic factor effects of CRP, LDH, NLR, ECOG, race, presence of liver metastases, and PD‐L1 expression (IC or TC > 0), which were investigated in the previous analysis but deemed not to be significant. This phenomenon was probably attributed to the narrow range of the covariate values available or to the small sample size in each subgroup for the previous analysis, thereby causing a decrease in power in detecting the effect on OS from these covariates. The updated NSCLC model also differentiated the effects between the first‐line and later lines of treatment, mainly by using a more comprehensive data set for model development, as opposed to the previous analysis, where only data for second and later lines of therapy were available. The directions of effect from the significant covariates on OS are consistent with the general knowledge of the trends between these prognostic factors and patient survival (e.g., higher ECOG score is associated with worse prognosis). In contrast, tumor burden, age, sex, body weight, histology (nonsquamous vs. squamous), and smoking history were not independent significant baseline prognostic factors in the historical or in the updated multivariate TGI‐OS model.
PD‐L1 expression was determined to be a significant covariate in the TGI‐OS model for NSCLC. PD‐LI expression is the most widely accepted predictive biomarker for treatments with immune checkpoint inhibitors; however, its role is not clear, particularly in the combination treatment setting. 39 Because PD‐L1 expression information in terms of IC/TC groups was missing in a large number of patients in one of the NSCLC trials (IMpower132), a sensitivity analysis was conducted by excluding IC/TC from the TGI‐OS model development. The results of the sensitivity analysis showed that the same set of covariates besides IC/TC were selected for the final model after backward elimination, and parameter estimates were similar to the model incorporating IC/TC groups (Table S5), with similar model evaluations using OS HR (Figure S7). On the other hand, because IC/TC information is specific to immune checkpoint inhibitors targeting PD‐L1 receptors, the TGI‐OS model without the IC/TC group in the sensitivity analysis could be applied to NSCLC trial data based on treatments with other mechanisms of action. The c‐index of the model without IC/TC is 0.752, which is similar to that of the model with IC/TC (0.754), further confirming the good predictive accuracy of both models.
The most impactful covariate (second to KG) in the updated NSCLC model was CRP, which was not available previously for the development of the historical model that only used data from one clinical trial. This is consistent with results from a separate analysis based on data from two of the trials included in the current analysis. 40 CRP was also a significant baseline prognostic factor in the TNBC model. CRP is an inflammation biomarker, routinely used in clinical practice for monitoring patients with cancer, and studies show an association between elevated CRP levels with a poor response and worse survival in epithelial cancers, such as liver, lung, colorectal, and breast cancers. 41 , 42 , 43 , 44
Another biomarker that was shown to be a significant covariate in the TGI‐OS models from four of five tumor types is ALB, where hypoalbuminemia is a known risk factor for poor prognosis in patients with cancer 45 and may be associated with cancer cachexia and impaired response to checkpoint inhibitors. 46 A similar association between ALB and OS have been identified in patients with locally advanced urothelial carcinoma or mUC treated with durvalumab 13 and in Asian patients with NSCLC treated with motesanib. 9
In the five tumor‐type specific TGI‐OS models developed in the current analysis (i.e., NSCLC, SCLC, TNBC, mUC, and RCC), KG was the most significant covariate in predicting OS, showing that increasing KG is associated with decreasing survival time. This is similar to an analysis conducted by the US FDA using data from 24 NSCLC trials. 25 In the current analysis, atezolizumab‐containing treatments had lower median KG, that is, slower growth, than treatments from the respective control arms, and this is consistent with the observation that the atezolizumab‐containing arms have shown significant or numerical improvements in OS over control in all of the trials. 26 , 27 , 29 , 33 , 34 Therefore, the exposure‐driven treatment effects were assumed to have been captured by the estimated KG, and the subsequent survival model can be considered treatment independent. 1 , 2 The biexponential TGI model was able to provide an adequate fit to the observed tumor size data, indicating that the individual post hoc estimates of KG described the individual longitudinal profiles well. No covariates were investigated for the TGI model (and KG), as the objective of the analysis was to simulate OS, conditional on TGI metrics and baseline prognostic factors.
Tumor dynamics has been studied as an early efficacy marker for other CITs as well as for chemotherapy and targeted therapy. Among CIT treatments, tumor growth patterns were investigated with pembrolizumab, ipilimumab, nivolumab, durvalumab, and bevacizumab across melanoma, NSCLC, mUC, and kidney cancer types, and all reported a strong association between survival and tumor response metrics, such as early tumor size change, KG, or KS, 13 , 15 , 16 , 47 , 48 supporting the validity of TGI‐OS correlation in CIT. Although tumor growth rate in terms of KG was the most significant TGI metrics in predicting OS for atezolizumab‐containing treatments, other TGI metrics should be explored when applying the TGI‐OS framework to molecules and study designs (e.g., patient population or trial follow‐up duration) not investigated previously.
A similar model‐based framework has been applied to other NSCLC 9 , 11 , 25 , 40 and breast cancer 8 analyses as well as in other tumor types such as prostate 15 , 18 and ovarian cancer. 19 In most cases, the analysis focuses on a single tumor type. The strength of the current analysis lies in its conglomeration of five different tumor types and thereby providing a wealth of support for the TGI‐OS platform in various solid tumor types. In addition, it is the first study where TGI‐OS models have been shown to predict OS treatment benefit across a wide variety of trials and tumor types. Although the analysis data originated from atezolizumab trials, the analysis is not restricted to atezolizumab monotherapy because various treatment types were included in the analysis due to the different treatments used in the control arms and in combination with atezolizumab. However, whether the TGI‐OS platform will apply to other anticancer treatments with different mechanisms of action remains to be determined, and external validation studies with other data sets are warranted. Furthermore, the association between tumor growth rates and OS could be explored using novel modeling approaches, such as machine learning. 40 , 49
One limitation of the current work is that the two‐stage approach may suffer from selection bias due to patients with progressing‐only disease dropping out early in the trial. Joint modeling has been shown to address this issue based on a simulation study. 50 However, in an analysis conducted using observed clinical data, the difference in model predictions between the joint model and two‐stage approaches might not be clinically meaningful. 51 Recent research indicates that competing risks joint models may be needed to correct for dropouts (by inducing informative censoring) when simultaneously modeling longitudinal biomarker and terminal event data. 52 These questions require further evaluation with regard to the objectives of the modeling effort. The impact of follow‐up on TGI metrics estimates and OS HR predictions at the study level to support decisions based on early data cuts is being assessed using both two‐stage and joint models. We contend that TGI metrics derived based on two‐stage models are more actionable to support trial decisions, 1 whereas current SLD (or SLD slope)‐based joint models are more suitable to perform patient‐level dynamic predictions 12 and enable personalized health care.
In conclusion, our study results provide further support that tumor dynamic model‐based metrics (such as KG) could help support early decisions between alternative treatments, select the most promising combinations in future CIT clinical trials, and predict the probability of success of a phase III clinical trial. 1 , 6 , 25
CONFLICT OF INTEREST
P.C., K.Y., S.V., N.W., A.L., B.W., M.B., N.S., J.Y.J., and R.B. are employees and stockholders of Genentech, Inc. M.M. is employed by Certara Strategic Consulting.
AUTHOR CONTRIBUTIONS
P.C., M.M., K.Y., B.W., M.B., N.S., J.Y.J., and R.B. wrote the manuscript and designed the research. P.C., M.M., and K.Y. performed the research. S.V., N.W., and A.L. analyzed the data.
Supporting information
Supplementary Material
Supinfo
ACKNOWLEDGMENTS
We thank the patients, family members, and staff who participated in the clinical trials. The authors also thank Francois Mercier (Department of Biostatistics, Roche, Basel, Switzerland) for helpful discussions on joint models.
Chan P, Marchand M, Yoshida K, et al. Prediction of overall survival in patients across solid tumors following atezolizumab treatments: A tumor growth inhibition–overall survival modeling framework. CPT Pharmacometrics Syst Pharmacol. 2021;10:1171–1182. 10.1002/psp4.12686
The results of the analysis were previously presented in part at the American Conference on Pharmacometrics meeting in November 2020.
Funding information
This study was sponsored by F. Hoffmann‐La Roche, Ltd.
REFERENCES
- 1. Bruno R, Bottino D, de Alwis DP, et al. Progress and opportunities to advance clinical cancer therapeutics using tumor dynamic models. Clin Cancer Res. 2020;26:1787‐1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruno R, Mercier F, Claret L. Evaluation of tumor size response metrics to predict survival in oncology clinical trials. Clin Pharmacol Ther. 2014;95:386‐393. [DOI] [PubMed] [Google Scholar]
- 3. Venkatakrishnan K, Friberg LE, Ouellet D, et al. Optimizing oncology therapeutics through quantitative translational and clinical pharmacology: challenges and opportunities. Clin Pharmacol Ther. 2015;97:37‐54. [DOI] [PubMed] [Google Scholar]
- 4. Claret L, Girard P, Hoff PM, et al. Model‐based prediction of phase III overall survival in colorectal cancer on the basis of phase II tumor dynamics. J Clin Oncol. 2009;27:4103‐4108. [DOI] [PubMed] [Google Scholar]
- 5. Claret L, Pentafragka C, Karovic S, et al. Comparison of tumor size assessments in tumor growth inhibition‐overall survival models with second‐line colorectal cancer data from the VELOUR study. Cancer Chemother Pharmacol. 2018;82:49‐54. [DOI] [PubMed] [Google Scholar]
- 6. Maitland ML, Wilkerson J, Karovic S, et al. Enhanced detection of treatment effects on metastatic colorectal cancer with volumetric CT measurements for tumor burden growth rate evaluation. Clin Cancer Res. 2020;26:6464‐6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruno B, Lindbom L, Schaedeli Stark F, et al. Simulations to assess phase II noninferiority trials of different doses of capecitabine in combination with docetaxel for metastatic breast cancer. CPT Pharmacometrics Syst Pharmacol. 2012;1:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lim HS, Sun W, Parivar K, Wang D. Predicting overall survival and progression‐free survival using tumor dynamics in advanced breast cancer patients. AAPS J. 2019;21:22. [DOI] [PubMed] [Google Scholar]
- 9. Claret L, Bruno R, Lu JF, Sun YN, Hsu CP. Exploratory modeling and simulation to support development of motesanib in Asian patients with non‐small cell lung cancer based on MONET1 study results. Clin Pharmacol Ther. 2014;95:446‐451. [DOI] [PubMed] [Google Scholar]
- 10. Claret L, Jin JY, Ferté C, et al. A model of overall survival predicts treatment outcomes with atezolizumab versus chemotherapy in non–small cell lung cancer based on early tumor kinetics. Clin Cancer Res. 2018;24:3292‐3298. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y, Sung C, Dartois C, et al. Elucidation of relationship between tumor size and survival in non‐small‐cell lung cancer patients can aid early decision making in clinical drug development. Clin Pharmacol Ther. 2009;86:167‐174. [DOI] [PubMed] [Google Scholar]
- 12. Tardivon C, Desmee S, Kerioui M, et al. Association between tumor size kinetics and survival in patients with urothelial carcinoma treated with atezolizumab: Implication for patient follow‐up. Clin Pharmacol Ther. 2019;106:810‐820. [DOI] [PubMed] [Google Scholar]
- 13. Zheng Y, Narwal R, Jin C, et al. Population modeling of tumor kinetics and overall survival to identify prognostic and predictive biomarkers of efficacy for durvalumab in patients with urothelial carcinoma. Clin Pharmacol Ther. 2018;103:643‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Claret L, Mercier F, Houk BE, Milligan PA, Bruno R. Modeling and simulations relating overall survival to tumor growth inhibition in renal cell carcinoma patients. Cancer Chemother Pharmacol. 2015;76:567‐573. [DOI] [PubMed] [Google Scholar]
- 15. Stein WD, Figg WD, Dahut W, et al. Tumor growth rates derived from data for patients in a clinical trial correlate strongly with patient survival: a novel strategy for evaluation of clinical trial data. Oncologist. 2008;13:1046‐1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Y, Wang X, Suryawanshi S, Bello A, Roy A. Linking tumor growth dynamics to survival in ipilimumab‐treated patients with advanced melanoma using mixture tumor growth dynamic modeling. CPT Pharmacometrics Syst Pharmacol. 2019;8:825‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hansson EK, Amantea MA, Westwood P, et al. PKPD modeling of VEGF, sVEGFR‐2, sVEGFR‐3, and sKIT as predictors of tumor dynamics and overall survival following sunitinib treatment in GIST. CPT Pharmacometrics Syst Pharmacol. 2013;2:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stein WD, Gulley JL, Schlom J, et al. Tumor regression and growth rates determined in five intramural NCI prostate cancer trials: the growth rate constant as an indicator of therapeutic efficacy. Clin Cancer Res. 2011;17:907‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zecchin C, Gueorguieva I, Enas NH, Friberg LE. Models for change in tumour size, appearance of new lesions and survival probability in patients with advanced epithelial ovarian cancer. Br J Clin Pharmacol. 2016;82:717‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen DS, Irving BA, Hodi FS. Molecular pathways: next‐generation immunotherapy–inhibiting programmed death‐ligand 1 and programmed death‐1. Clin Cancer Res. 2012;18:6580‐6587. [DOI] [PubMed] [Google Scholar]
- 21. Chen DS, Mellman I. Oncology meets immunology: the cancer‐immunity cycle. Immunity. 2013;39:1‐10. [DOI] [PubMed] [Google Scholar]
- 22. Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti‐PD‐L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. European Medicines Agency Tecentriq: EPAR‐product information. https://www.ema.europa.eu/en/documents/product‐information/tecentriq‐epar‐product‐information_en.pdf. Accessed January 18, 2021.
- 24. Food and Drug Administration . Tecentriq: FDA full prescribing information. https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/761034s033s034s035s036s037s038lbl.pdf. Accessed January 18, 2021.
- 25. Gong Y, Mason J, Shen Y‐L, et al. An FDA analysis of the association of tumor growth rate and overall and progression‐free survival in metastatic non‐small cell lung cancer (NSCLC) patients. J Clin Oncol. 2020;38:9541. [Google Scholar]
- 26. Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non‐small‐cell lung cancer (POPLAR): a multicentre, open‐label, phase 2 randomised controlled trial. Lancet. 2016;387:1837‐1846. [DOI] [PubMed] [Google Scholar]
- 27. Horn L, Mansfield AS, Szczesna A, et al. First‐line atezolizumab plus chemotherapy in extensive‐stage small‐cell lung cancer. N Engl J Med. 2018;379:2220‐2229. [DOI] [PubMed] [Google Scholar]
- 28. Jotte R, Cappuzzo F, Vynnychenko I, et al. Atezolizumab in combination with carboplatin and Nab‐paclitaxel in advanced squamous NSCLC (IMpower131): results from a randomized phase III trial. J Thorac Oncol. 2020;15:1351‐1360. [DOI] [PubMed] [Google Scholar]
- 29. Papadimitrakopoulou V, Cobo M, Bordoni R, et al. OA05.07 IMpower132: PFS and safety results with 1L atezolizumab + carboplatin/cisplatin + pemetrexed in stage IV non‐squamous NSCLC. J Thorac Oncol. 2018;13:S332‐S333. [Google Scholar]
- 30. Powles T, Duran I, van der Heijden MS, et al. Atezolizumab versus chemotherapy in patients with platinum‐treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open‐label, phase 3 randomised controlled trial. Lancet. 2018;391:748‐757. [DOI] [PubMed] [Google Scholar]
- 31. Rini BI, Powles T, Atkins MB, et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open‐label, phase 3, randomised controlled trial. Lancet. 2019;393:2404‐2415. [DOI] [PubMed] [Google Scholar]
- 32. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non‐small‐cell lung cancer (OAK): a phase 3, open‐label, multicentre randomised controlled trial. Lancet. 2017;389:255‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. West H, McCleod M, Hussein M, et al. Atezolizumab in combination with carboplatin plus nab‐paclitaxel chemotherapy compared with chemotherapy alone as first‐line treatment for metastatic non‐squamous non‐small‐cell lung cancer (IMpower130): a multicentre, randomised, open‐label, phase 3 trial. Lancet Oncol. 2019;20:924‐937. [DOI] [PubMed] [Google Scholar]
- 34. Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first‐line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378:2288‐2301. [DOI] [PubMed] [Google Scholar]
- 35. Schmid P, Adams S, Rugo HS, et al. IMpassion130 Trial Investigators. Atezolizumab and nab‐paclitaxel in advanced triple‐negative breast cancer. N Engl J Med. 2018;379:2108‐2121. [DOI] [PubMed] [Google Scholar]
- 36. Fehrenbacher L, von Pawel J, Park K, et al. Updated efficacy analysis including secondary population results for OAK: a randomized phase III study of atezolizumab versus docetaxel in patients with previously treated advanced non‐small cell lung cancer. J Thorac Oncol. 2018;13:1156‐1170. [DOI] [PubMed] [Google Scholar]
- 37. Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361‐387. [DOI] [PubMed] [Google Scholar]
- 38. Savic RM, Karlsson MO. Importance of shrinkage in empirical bayes estimates for diagnostics: problems and solutions. AAPS J. 2009;11:558‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doroshow DB, Bhalla S, Beasley MB, et al. PD‐L1 as a biomarker of response to immune‐checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18(6):345‐362. [DOI] [PubMed] [Google Scholar]
- 40. Hopkins AM, Kichenadasse G, Garrett‐Mayer E, et al. Development and validation of a prognostic model for patients with advanced lung cancer treated with the immune checkpoint inhibitor atezolizumab. Clin Cancer Res. 2020;26:3280‐3286. [DOI] [PubMed] [Google Scholar]
- 41. Allin KH, Nordestgaard BG, Flyger H, Bojesen SE. Elevated pre‐treatment levels of plasma C‐reactive protein are associated with poor prognosis after breast cancer: a cohort study. Breast Cancer Res. 2011;13:R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carr BI, Akkiz H, Guerra V, et al. C‐reactive protein and hepatocellular carcinoma: analysis of its relationships to tumor factors. Clin Pract (Lond). 2018;15:625‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kersten C, Louhimo J, Algars A, et al. Increased C‐reactive protein implies a poorer stage‐specific prognosis in colon cancer. Acta Oncol. 2013;52:1691‐1698. [DOI] [PubMed] [Google Scholar]
- 44. Xiao X, Wang S, Long G. C‐reactive protein is a significant predictor of improved survival in patients with advanced non‐small cell lung cancer. Medicine (Baltimore). 2019;98:e16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gupta D, Lis CG. Pretreatment serum albumin as a predictor of cancer survival: a systematic review of the epidemiological literature. Nutr J. 2010;9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Coss CC, Clinton SK, Phelps MA. Cachectic cancer patients: immune to checkpoint inhibitor therapy? Clin Cancer Res. 2018;24:5787‐5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feng Y, Agrawal S, Gupta M, et al. Association between immune‐checkpoint inhibitor induced tumor shrinkage and overall survival in advanced melanoma and NSCLC. J Clin Oncol. 2014;32:3053. [Google Scholar]
- 48. Wang M, Chen C, Jemielita T, et al. Are tumor size changes predictive of survival for checkpoint blockade based immunotherapy in metastatic melanoma? J Immunother Cancer. 2019;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chan P, Zhou X, Wang N, et al. Application of machine learning for tumor growth inhibition–overall survival modeling platform. CPT Pharmacometrics Syst Pharmacol. 2020;10:59‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Desmée S, Mentré F, Veyrat‐Follet C, Guedj J. Nonlinear mixed‐effect models for prostate‐specific antigen kinetics and link with survival in the context of metastatic prostate cancer: a comparison by simulation of two‐stage and joint approaches. AAPS J. 2015;17:691‐699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Claret L, Bruno R, Wang B, et al. A comparison of two stage and joint tumor growth inhibition‐progression free survival modeling approach to simulate clinical outcome in oncology [abstract]. 24th Annual Meeting of the Population Approach Group Europe; Jun 2‐5, 2015; Crete, Greece. Abstract 3391.
- 52. Vonesh EF, Greene T. Biased estimation with shared parameter models in the presence of competing dropout mechanisms [published online ahead of print 2021]. Biometrics. 10.1111/biom.13438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supinfo


