Abstract
Background
detection of anaplastic lymphoma receptor tyrosine kinase gene (ALK) rearrangements in patients with non‐small‐cell lung cancer (NSCLC) has become a routine pathological diagnosis worldwide.
Methods
there are three major conventional diagnostic methods for ALK fusions: fluorescent in situ hybridization (FISH); immunohistochemistry (Ventana IHC (D5F3)); and polymerase chain reaction (PCR). Next‐generation sequencing (NGS) technology as is a new tool for ALK status detection with great potential. These four methods are highly consistent in detecting ALK status (coincidence rate >96%). However, discrepancies in ALK status have been found in some patients among these methods, which causes confusion for clinicians.
Results and conclusion
in this study, we analyzed two patients whose ALK statuses were not consistent using these four methods. We explored the potential reasons for deviation of the test results and found a novel EML4‐ALK break site, which had been not described previously.
This novel EML4‐ALK break site case is a typical example of a patient's clinical characteristics and pathologic findings indicating that the patient was ALK‐positive. This result was confirmed by IHC and NGS. The patient responded well to the ALK inhibitor crizotinib after 1 month of treatment. In fact, the novel break site in this case is in exon 20 ofALK gene, which makes the kinase domain is not integral. This response indicated that even the ALK gene fusion domain is not structural integrity, the patient still could benefit from crizotinib. The disease progressed after 4 months of crizotinib treatment, which indicate that the missing of part exon 20 of ALK gene affect curative effect of crizotinib.
INTRODUCTION
Lung cancer has been the leading cause of cancer‐related mortality for both men and women worldwide for the last decade, and it is associated with a 5‐year survival rate of <15%. 1 , 2 NSCLC accounts for ~80%–85% of lung cancer. 1 , 3 , 4 NSCLC is a heterogeneous disease characterized by oncogenic driver alterations that can be targeted with precision tyrosine kinase inhibitors (TKIs). Anaplastic lymphoma kinase (ALK) is a transmembrane tyrosine kinase receptor that is physiologically expressed only in the nervous system during embryogenesis; its expression decreases postnatally. The incidence of ALK rearrangement is reported to range from ~3%–13% in unselected or selected patients with NSCLC. 5 , 6 , 7 , 8
ALK mutations were first described in NSCLC in 2007 when a subset (7%) of Japanese patients were found to have echinoderm microtubule associated protein like‐4 (EML4) rearrangement with ALK leading to a fusion oncogene EML4‐ALK. 9 The EML4‐ALK fusion gene represents an outstanding molecular target. EML4‐ALK translocation can result in constitutive ALK kinase activity and represents an oncogenic addiction pathway in lung cancer. 9 , 10 , 11 Rearrangements of the ALK gene with partner genes other than EML4 have been described, including, KIF5B, KLC1, TFG, TPR, HIP1, STRN, DCTN1, SQSTM1, NPM1, BCL11A, and BIRC6. 12 , 13 , 14 , 15 , 16
EML4‐ALK fusion protein serves as a therapeutic target for an ALK tyrosine kinase inhibitor, which has shown promising results when used to treat NSCLC patients carrying ALK rearrangement. 17 , 18 , 19 , 20 Furthermore, a previous study demonstrated that crizotinib is effective at treating tumors harboring ALK fused with other partner genes, including NPM1 and BCL11A. 19
ALK rearrangements may involve distinct breakpoints and multiple fusion partners, and therefore, routine ALK testing presents a significant technical challenge. The fluorescent in situ hybridization (FISH) break‐apart assay is considered to be a “gold standard” for the evaluation of ALK status, although immunohistochemistry (IHC) (Ventana IHC [D5F3]) and reverse transcription‐polymerase chain reaction (RT‐PCR) have also been evaluated for this purpose, with the former being Food and Drug Administration (FDA)‐approved in June 2015. 21 , 22 FISH relies on a spatial separation of 5′‐ and 3′‐portions of the ALK gene on rearrangement and produces characteristic amend ALK‐specific signals in case of translocation. Despite the FISH assay being the most reliable approach to ALK testing, it has a number of critical disadvantages. FISH requires a significant amount of time from extensively trained personnel and cannot be subjected to a reasonable automation; furthermore, it demonstrates relatively high failure rates in some sample series and may provide poorly interpretable results in a noticeable fraction of NSCLC cases. 23 , 24 , 25
The development of highly sensitive ALK diagnostic antibodies has offered an opportunity to detect ALK‐driven tumors by a standard IHC method. The principle of IHC is based on the fact that activating ALK rearrangements are accompanied by significant overexpression of the catalytic portion of this tyrosine kinase. Ventana IHC (D5F3) can produce highly reliable results when performed in reference laboratories; however, its interlaboratory reproducibility and performance in heterogeneous lung cancer tissue collections remains to be evaluated. 25 , 26 , 27 , 28 , 29
RT‐PCR is also a common approach for ALK status testing. This platform can detect certain variants of the rearrangement. Therefore, this platform could provide definite evidence for the presence of ALK fusion. Another advantage of RT‐PCR is that it is highly sensitive. 30 It can detect as little as 1% of ALK‐driven NSCLC cells in the presence of normal tissues. This platform can sensitively detect certain variants of the rearrangement and can provide definite evidence for the presence of ALK fusion. Furthermore, PCR is highly sensitive. 30 It can detect as little as 1% of ALK‐driven NSCLC cells in the presence of normal tissues. The disadvantage of this method is PCR only could analyze certain types of fusion defined by the commercial PCR kits. 31 Analyze multiple types of fusion and unknown fusion type the method of next‐generation sequencing (NGS) has great potential as a powerful tool for cancer diagnostics, including ALK status detection.
Clinicians are often confused by the methods used for ALK detection when the results are not consistent among different platforms. Here, we analyze the ALK status of two patients in which these four methods give inconsistent results. By analyzing the ALK status detected by these four platforms, we found a novel EML4‐ALK break site.
CASE PRESENTATION 1
A 53‐year‐old Chinese woman was referred to the hospital because of headache, emesis, and a decrease of muscle force in May 2015. She was an ex‐smoker with 3‐pack‐year history of regular smoking during 1933–1995. Computed tomography (CT) scans of her chest found a space‐occupying lesion of the lung. Magnetic resonance imaging (MRI) found a space‐occupying lesion of the brain, showing cerebral and vertebral metastasis. Despite stability after chemotherapy and radiotherapy from July to September of 2015, the disease progressed after January 2016. Epidermal growth factor receptor (EGFR) and EML4‐ALK were both wild‐types tested by quantitative RT‐PCR (qRT‐PCR) in August 2017 (Tables 1 and 2). Based on these results, the patient was treated with chemotherapy from October 2015 to January 2016. The disease progressed again with larger intrapulmonary and intracranial lesions based on a CT conducted in March 2016 (Figure 1(a)). Needle biopsy of the lung revealed that the tumor was a mucinous adenocarcinoma in the right side of lung (Figure 2(a)). This sample was identified as thyroid transcription factor 1 (TTF1) (Figure 2(b)) and napsin A expression‐positive by IHC staining. Ventana IHC (D5F3) analyses of ALK protein expression were ALK‐positive (Figure 2(c)), whereas ALK FISH analyses were negative (Figure 2(d)). CT‐guided needle biopsy of a liver metastasis confirmed carcinoma, and the tissue was subjected to genomic profiling through a clinical grade NGS assay (Illumina Hiseq 4000 NGS platforms). NGS results revealed that this tumor specimen harbored a new EML4‐ALK fusion variant involving rearrangement and fusion of exons 1–19 of EML4 to partial exons 20–29 of ALK (E19; A20ins). This fusion type was previously recorded, but the break site of this fusion had not been reported before (Figure 3). Furthermore, this fusion comprises ~20% of the sequencing runs where tumor purity in the biopsy sample was estimated to be 80% (Table 2).
TABLE 1.
Patients' characteristics
| Patient no./gender | Age (y) | Smoking history | PFS (months) | Assessment | Follow‐up |
|---|---|---|---|---|---|
| 1. Female | 53 | Ever smoker | 3 | Partial response | Alive |
| 2. Female | 58 | Ever smoker | 3 | Stable disease | Alive |
Abbreviation: PFS, progression‐free survival.
TABLE 2.
Pathologic characteristics and molecular test results in both patients
| No. | Sample types | TTF1 (IHC) | Naspin A (IHC) | EGFR/KRAS mutations | ALK (IHC) | ALK (RT‐PCR) | ALK (FISH) | ALK (NGS) | Tumor percentage |
|---|---|---|---|---|---|---|---|---|---|
| P1 | LP | + | + | WT/WT | + | − | − (12%) | EML4‐ALK | 80 |
| P2 | LP | + | + | WT/WT | + | − | + (20%) | PRKAR1A‐ALK | 40 |
Abbreviations: LB, lymph nodes biospy; LP, lung puncture; +, positive; −, negative.
FIGURE 1.
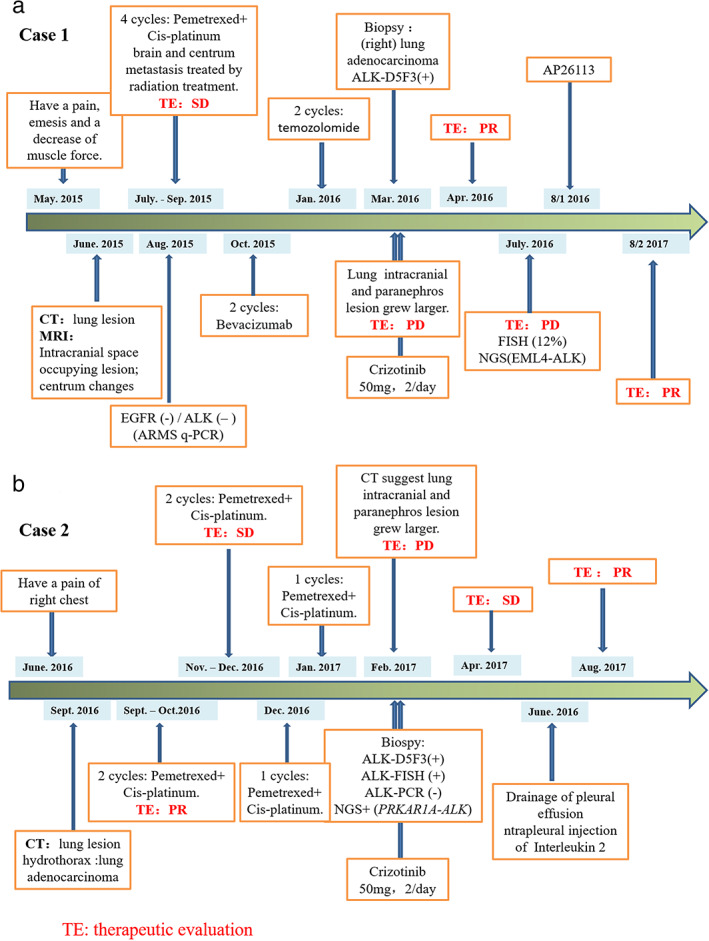
Procedures for medical treatment of these two patients. a is for case 1, b is for case 2
FIGURE 2.
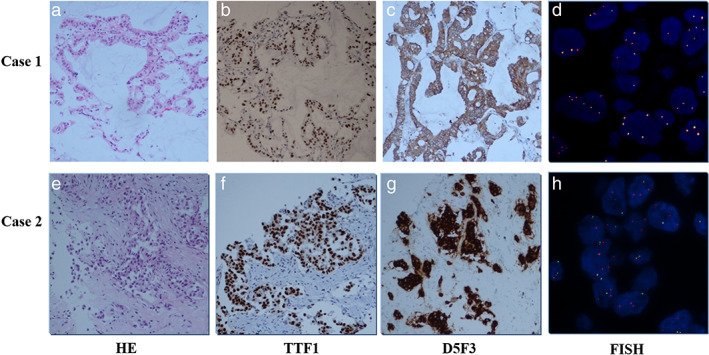
Hematoxylin & eosin (HE) staining, TTF1 staining, Ventana IHC (D5F3), and FISH staining slides from patients
FIGURE 3.
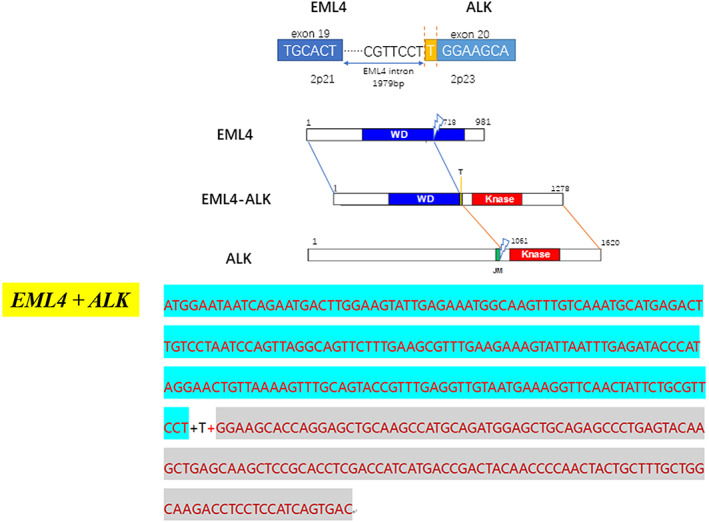
Schematic structure of the genomic DNA and RNA sequence (up); mRNA sequence of novel EML4‐ALK fusion gene(low)
Based on these results, treatment was initiated with 100 mg crizotinib per day starting in March 2016. She tolerated the crizotinib dose well with slight nausea, vomiting, diarrhea, and visual impairment during the first 2 weeks. By her 1‐month follow‐up visit, CT scan showed significant reduction of the tumor burden in the chest (Figure 4(a)). She achieved a confirmed partial response (PR) after 1 month on crizotinib that lasted for 4 months when several new liver lesions were noted. She then received AP26113 starting on August 1, 2017. The disease has remained controlled for ~11 months on this treatment.
FIGURE 4.
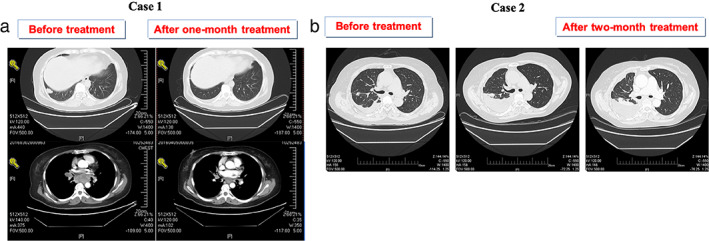
CT images before and after treatment of crizotinib
CASE PRESENTATION 2
A 58‐year‐old Chinese woman was referred to hospital because of pain in the right side of her chest in June 2016. She was an ex‐smoker with a 20‐pack‐year history of regular smoking from 1986 to 2016. A CT scan of the chest in September 2016 found a space‐occupying lesion in the lung. Lymph nodes in her right clavicle turned out lung adenocarcinoma. From September 30, 2016 to December 4, 2016, she was treated with four cycles of chemotherapy (pemetrexed + cisplatinum). After two cycles of therapy, the assessment was partial response (PR) and when the fourth cycle finished, the patient's condition was stable. The patient accepted one cycle of chemotherapy on December 30, 2016 and January 20, 2017 because the disease progressed. On February 14, 2017, a CT scan of the chest suggested disease progress. Genetic testing of a formalin‐fixed and paraffin‐embedded (FFPE) tissue specimen was ALK‐positive at the DNA (FISH and NGS) and protein levels (D5F3) (Figure 2(e)–(h)). NGS testing indicated that this case had a rare fusion of PRKAR1A‐ALK, which explains why the (amplification‐refractory mutation system) ARMS‐PCR result was negative, because this fusion type is not included in the EML4‐ALK Fusion Gene Detection Kit (AmoyDx). This fusion comprised only 1.27% of the sequencing runs where tumor purity in the biopsy sample was estimated to be 40% (Table 2). Based on these genetic testing results, the patient took 100 mg crizotinib per day starting in February 2017. She achieved a confirmed stable disease (SD) after 2 months (Figure 4(b)), but developed intrathoracic progression after 3 months of crizotinib treatment. The disease has remained under control up to July 2018 by drainage of pleural effusion and intrapleural injection of interleukin‐2 treatment.
DISCUSSION
Lung cancer is a leading cause of cancer mortality worldwide. ALK rearrangements have been identified in 5%–6% of lung adenocarcinomas. 21 The most reliable detection method of ALK status is the FISH break‐apart assay, but IHC and RT‐PCR have also been used successfully. Each of these methods have merits, however, NGS is more promising for accurately detecting ALK status.
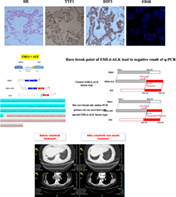
In case 1, we detected the ALK rearrangement by ARMS‐PCR in August 2015. Owing to the negative PCR result, the patient was treated with chemotherapy, but received few benefits, which reminded us to reevaluate the ALK fusion status. Pathologists decided to use Ventana IHC (D5F3) analyses of ALK protein expression. The result of this test was ALK‐positive. The histologic sample from this patient was then successfully tested by ALK‐FISH. The percentage of ALK‐positive nuclei was 12% (FISH‐negative). Finally, we choose the NGS method to confirm the IHC‐positive result. The NGS result showed that this is a new fusion breakpoint of EML4 at intron 19 and ALK in exon 20. This breakpoint means that RT‐PCR primers do not match with this novel EML4‐ALK fusion type, which explains why the ARMS‐PCR result was negative (Figure 5).
FIGURE 5.
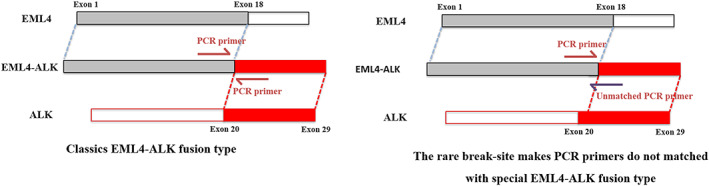
Rare breakpoint of EML4‐ALK lead to negative result of q‐PCR
Despite the high concordance of IHC, FISH, and PCR methods for detecting ALK status, many studies have found discrepancies among the different methods. 28 , 31 This case is a typical example of a patient's clinical characteristics and pathologic findings indicating that the patient was ALK‐positive. This result was confirmed by IHC and NGS.
In case 1, the patient responded well to the ALK inhibitor crizotinib after 1 month of treatment. In fact, this novel break site in this case is in exon 20 of ALK gene, which makes the kinase domain is not integral. This response indicated that even the ALK gene fusion domain is not structural integrity, the patient still could benefit from crizotinib. The disease progressed after 4 months of crizotinib treatment, which indicates that the missing of part exon 20 of ALK gene affects curative effect of crizotinib.
ALK translocations result in increased tyrosine kinase activity, leading to increased cell proliferation and survival and ultimately, tumorigenesis. In NSCLC, ALK as an oncogenic driver, gene fusions that can induce both ALK expression and activation are required. Previous studies have found that regardless of the involved partners, all chimeras retain the ALK gene kinase domain responsible for the constitutive activation of ALK signaling pathways. 32 Exons 20–29 are the functional kinase domain of the ALK gene. EML4 is the most common ALK rearrangement fusion partner; the fusion gene contains the complete ALK gene kinase domain. However, the break site in this case is in exon 20, which makes the kinase domain not integral. We doubt that the incomplete structure of the ALK function domain made the patient drug‐resistant to crizotinib after 4 months. The mechanisms of resistance to crizotinib require further research.
The tumor of the patient in case 2 harbored a PRKAR1A‐ALK interchromosomal rearrangement and both ALK‐FISH and ALK‐IHC were positive. The ARMS‐PCR result was negative because the PRKAR1A‐ALK fusion type is not included in the EML4‐ALK Fusion Gene Detection Kit (AmoyDx) (Table S1). Research by Ali et al. 33 demonstrated that the PRKAR1A‐ALK fusion was both oncogenic and imparted sensitivity to crizotinib. Almost all patients treated with crizotinib eventually develop tumor progression; case 2 achieved a confirmed SD after 1 month, but developed intrathoracic progression after 3 months of crizotinib treatment. The short progression‐free survival (PFS) may be because of the very low percentage (1.27%) of the sequencing runs identified where tumor purity in the biopsy sample was estimated to be 40%.
This study discusses four methods for detecting ALK status. The gold standard for detection of predictive ALK rearrangements is currently FISH. However, the ALK‐FISH assay is fraught with technical challenges, including FISH signal instability and scoring difficulties. Additionally, ALK‐FISH may be missing ALK translocation because of variations of partners for ALK gene rearrangement and fusion patterns or a low proportion of ALK‐rearranged tumor cells in the tested tumor samples as in case 1. An alternative method for determining ALK in NSCLC is to identify ALK protein overexpression using IHC; this method is mentioned in version 3 of the National Comprehensive Cancer Network (NCCN) guideline. 34 The RT‐PCR assay, also mentioned in the NCCN guideline, can sensitively detect certain ALK fusion gene variants, but it cannot detect unknown ALK fusion types. NGS has displayed an impressive capability to detect PCR‐negative ALK‐rearranged NSCLC and is, therefore, a clinically practical alternative for diagnosis of ALK rearrangements in addition to FISH, IHC, and RT‐PCR. It is worth noting that clinical symptoms and basic pathologic findings are extremely important for the detection of ALK status.
Crizotinib has been shown to provide a valuable first‐ and second‐line treatment option and is now the first‐line standard of care for patients with advanced ALK‐positive NSCLC. Unfortunately, drug resistance develops after an initial benefit, through a variety of mechanisms. Broadly, there are three major mechanisms of resistance to targeted drugs. First, genetic alteration in the target; second, activation of bypass tracks or phenotypic change in the tumor such as development of epithelial mesenchymal transition; and last, limited penetration to “sanctuary” sites such as the central nervous system. In a significant proportion of patients, the exact mechanism of resistance is unknown. In our paper, two cases developed drug resistance after 3 months. As ALK fusion gene is the oncogenic driver gene of NSCLC and most of the patients can get benefits from the TKI, when few patients have drug‐resistant then analysis of the function of certain fusion type is necessary.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Supporting information
Table S1 EML4‐ALK fusion types detected with the qRT‐PCR kit
ACKNOWLEDGMENT
This work was supported by the National Natural Science Foundation of China (Grant No. 81972485).
Du X, Zhang J, Gao H, Tai Y. A novel break site of EML4‐ALK report and a rare PRKAR1A‐ALK report analyzed by different ALK detection platforms in non‐small cell lung cancer patients. Thorac Cancer. 2021;12:2773–2779. 10.1111/1759-7714.14123
Funding information National Natural Science Foundation of China, Grant/Award Number: 81972485
Contributor Information
Hongjun Gao, Email: gaohj_307@163.com.
Yanhong Tai, Email: taiyanhong29@163.com.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. [DOI] [PubMed] [Google Scholar]
- 3. Wen M, Wang X, Sun Y, Xia J, Fan L, Xing H, et al. Detection of EML4‐ALK fusion gene and features associated with EGFR mutations in Chinese patients with non‐small‐cell lung cancer. Onco Targets Ther. 2016;9:1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. [DOI] [PubMed] [Google Scholar]
- 5. Sun YH, Ren Y, Fang Z, Li C, Fang R, Gao B, et al. Lung adenocarcinoma from East Asian never‐smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol. 2010;28:4616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaw AT, Yeap BY, Mino‐Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non‐small‐cell lung cancer who harbor EML4‐ALK. J Clin Oncol. 2009;27:4247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inamura K, Takeuchi K, Togashi Y, Nomura K, Ninomiya H, Okui M, et al. EML4‐ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3:13–7. [DOI] [PubMed] [Google Scholar]
- 8. Horn L, Pao W. EML4‐ALK: honing in on a new target in non‐small‐cell lung cancer. J Clin Oncol. 2009;27:4232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4‐ALK fusion gene in non‐small‐cell lung cancer. Nature. 2007;448:561–U3. [DOI] [PubMed] [Google Scholar]
- 10. Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology (vol 13, pg 685, 2012). Nat Rev Cancer. 2013;13:821–1. [DOI] [PubMed] [Google Scholar]
- 11. Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, et al. A mouse model for EML4‐ALK‐positive lung cancer. Proc Natl Acad Sci U S A. 2008;105:19893–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iyevleva AG, Raskin GA, Tiurin VI, Sokolenko AP, Mitiushkina NV, Aleksakhina SN, et al. Novel ALK fusion partners in lung cancer. Cancer Lett. 2015;362:116–21. [DOI] [PubMed] [Google Scholar]
- 13. Mano H. ALKoma: a cancer subtype with a shared target. Cancer Discov. 2012;2:495–502. [DOI] [PubMed] [Google Scholar]
- 14. Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. [DOI] [PubMed] [Google Scholar]
- 15. Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B‐ALK, a novel fusion Oncokinase identified by an immunohistochemistry‐based diagnostic system for ALK‐positive lung cancer. Clin Cancer Res. 2009;15:3143–9. [DOI] [PubMed] [Google Scholar]
- 16. Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, et al. KLC1‐ALK: a novel fusion in lung cancer identified using a formalin‐fixed paraffin‐embedded tissue only. PLos One. 2012;7(2):e31323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4‐ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–95. [DOI] [PubMed] [Google Scholar]
- 19. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non‐small‐cell lung cancer. N Engl J Med. 2010;363:1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, et al. Effect of crizotinib on overall survival in patients with advanced non‐small‐cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12:1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16:E342–51. [DOI] [PubMed] [Google Scholar]
- 22. Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–26. [DOI] [PubMed] [Google Scholar]
- 23. Iwama E, Okamoto I, Harada T, Takayama K, Nakanishi Y. Development of anaplastic lymphoma kinase (ALK) inhibitors and molecular diagnosis in ALK rearrangement‐positive lung cancer. Onco Targets Ther. 2014;7:375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thunnissen E, Bubendorf L, Dietel M, Elmberger G, Kerr K, Lopez‐Rios F, et al. EML4‐ALK testing in non‐small cell carcinomas of the lung: a review with recommendations. Virchows Arch. 2012;461:245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tembuyser L, Tack V, Zwaenepoel K, Pauwels P, Miller K, Bubendorf L, et al. The relevance of external quality assessment for molecular testing for ALK positive non‐small cell lung cancer: results from two pilot rounds show room for optimization. PLoS One. 2014;9:e112159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, et al. Multiplex reverse transcription‐PCR screening for EML4‐ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–24. [DOI] [PubMed] [Google Scholar]
- 27. Camidge DR, Doebele RC. Treating ALK‐positive lung cancer‐early successes and future challenges. Nat Rev Clin Oncol. 2012;9:268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shan L, Lian F, Guo L, Yang X, Ying J, Lin D. Combination of conventional immunohistochemistry and qRT‐PCR to detect ALK rearrangement. Diagn Pathol. 2014;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conde E, Hernandez S, Prieto M, Martinez R, Lopez‐Rios F. Profile of Ventana ALK (D5F3) companion diagnostic assay for non‐small‐cell lung carcinomas. Expert Rev Mol Diagn. 2016;16:707–13. [DOI] [PubMed] [Google Scholar]
- 30. Tuononen K, Sarhadi VK, Wirtanen A, Rönty M, Salmenkivi K, Knuuttila A, et al. Targeted resequencing RevealsALKFusions in non‐small cell lung carcinomas detected by FISH, immunohistochemistry, and real‐time RT‐PCR: a comparison of four methods. Biomed Res Int. 2013;2013:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma D, Wang Z, Yang L, Mu X, Wang Y, Zhao X, et al. Responses to crizotinib in patients with ALK‐positive lung adenocarcinoma who tested immunohistochemistry (IHC)‐positive and fluorescence in situ hybridization (FISH)‐negative. Oncotarget. 2016;7:64410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. [DOI] [PubMed] [Google Scholar]
- 33. Ali SM, Hensing T, Schrock AB, Allen J, Sanford E, Gowen K, et al. Comprehensive genomic profiling identifies a subset of Crizotinib‐responsive ALK‐rearranged non‐small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist. 2016;21:762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wood DE, Kazerooni EA, Baum SL, Eapen GA, Ettinger DS, Hou L, et al. NCCN lung cancer screening, version 3.2018 clinical practice guidelines in oncology. Natl Compr Cancer Network. 2018;16:412–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 EML4‐ALK fusion types detected with the qRT‐PCR kit


