Abstract
Objective
This study was performed to investigate the relationship among UHRF1 expression, its biological function and immune infiltration in human hepatocellular carcinoma (HCC).
Methods
Gene Expression Profiling Interactive Analysis (GEPIA), Oncomine, and The Cancer Genome Atlas (TCGA) databases were used to analyze UHRF1 expression between HCC and normal tissues. Subsequently, GEPIA, TCGA-Portal, Kaplan–Meier Plotter, Protein Atlas and SurvExpress databases were utilized for survival analysis. UHRF1 co-expression genes were identified via the cBioPortal and LinkedOmics databases. Further, gene ontology (GO) analysis as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed. Protein–protein interaction (PPI) networks was constructed by STRING database and Cytoscape 3.7.1. Single-sample gene set enrichment analysis (ssGSEA) and CIBERSORT algorithm were employed to assess the correlation between UHRF1 and tumor immune infiltrates on TCGA database. TIMER 2.0 database was used to explore the correlation of UHRF1 expression and immune infiltration level in HCC. Additionally, RT-qPCR was used to analyze the expression of UHRF1 and the relative genes in HCC cell lines.
Results
Expression level of UHRF1 was upregulated in HCC tissues compared with paired normal tissues (P < 0.05 in GEPIA; P = 1.78E−6 in Oncomine; and P < 0.0001 in TCGA). Its high expression was significantly related with a shorter overall survival in five databases (P < 0.05). Function enrichment analysis demonstrated that functions of UHRF1 concentrated in cell division process and cell cycle (P < 0.05). High UHRF1 expression exhibited dysregulated immune infiltration (ie, neutrophils, eosinophils, dendritic cells resting, macrophages M2, macrophages M0) and poor survival of high UHRF1 expression was tight correlated with immune infiltration status. Moreover, TP53 mutation can lead to high expression of UHRF1 (P = 4.2E−10).
Conclusion
UHRF1 might function as an oncogene via inducing dysregulated immune infiltration in HCC and was identified as a novel prognostic biomarker and potential therapeutic target for HCC.
Keywords: UHRF1, biomarker, prognosis, immune infiltrates, HCC
Introduction
Liver cancer is the third leading cause of cancer death worldwide in 2020. Although treatments of liver cancer get improved recently, majority of patients hold a grim prognosis, with approximately 906,000 new cases and 830,000 deaths.1 The most common histological type of primary liver cancer is hepatocellular cancer (HCC).2 However, the early accurate diagnosis biomarkers of HCC are scarce, which is the key to the management of HCC. Thus, it is urgent to identify new accurate prognostic biomarkers and therapeutic targets to improve survival of HCC patients.
Ubiquitin-like with plant homeodomain (PHD) and RING finger domains 1 (UHRF1) is a protein containing five domains: N-terminal ubiquitin-like, TTD, PHD, SRA and RING domains.3 These domains correlate with the function of UHRF1 in DNA methylation and histone H3 ubiquitination.4,5 UHRF1 plays essential roles in epigenetic information integration; its aberrations can induce the hypermethylation of tumor suppressor genes (TSGs) such as p16INK4A, P73 and BRCA1, leading to genomic instability and carcinogenesis.6–10 L Sabatino et al demonstrated that UHRF1 negatively regulated PPARγ thus promoted the invasion and migration of colorectal cancer cells.11 Moreover, recent studies have revealed that the aberrant regulation of UHRF1 involved in tumorigenesis and progression in various cancers; its overexpression correlated with poor prognosis. Therefore, UHRF1 could be a promising therapeutic target for cancers. However, the underlying signaling pathways of UHRF1 in cancers still remain elusive that require further investigation.
In this study, we aim to get a better understanding of the biological roles of the UHRF1 in HCC. First, we obtained UHRF1 gene expression data from GEPIA, Oncomine and TCGA databases, then analyze the prognostic significance of UHRF1 expression in HCC. Subsequently, the main biological functions and the protein–protein interaction (PPI) network were constructed through functional enrichment analysis and Cytoscape software. Finally, the Tumor Immune Estimation Resource (TIMER 2.0) database analyses was performed to explore the correlation between tumor-infiltrating immune cells and UHRF1 in HCC. In addition, we used these methods to evaluate the relationship between UHRF1 and tumor infiltrating immune cells. Successively, these results enable us to contribute to the current literature on the potential positive impact of UHRF1 in HCC. The results suggest that there may be correlation and mechanism between UHRF1 and tumor immune interaction.
Materials and Methods
Data Acquisition and Survival Analysis
The datasets of UHRF1 expression in HCC and paired normal tissue were downloaded from GEPIA, Oncomine, and TCGA databases. Subsequently, the effect of UHRF1 on survival rate was assessed using GEPIA, TCGA-Portal, Kaplan–Meier Plotter, Protein Atlas and SurvExpress databases.
Gene Enrichment Analysis and PPI Network Analysis
LinkedOmics and cBioPortal were used to identify UHRF1 co-expression genes. P-value < 0.05 and Pearson’s correlation coefficient (r) >|0.5| were chosen as the screening criteria. A Venn diagram screening 152 genes was outlined online. GO and KEGG pathway enrichment analyses were utilized to explore the main functions of UHRF1 co-expression genes. PPI network with a protein–protein confidence score ≥0.4 was established using the STRING database. Cytoscape software 3.7.1 was utilized to visualize the network. Additionally, the correlations between UHRF1 and the top 9 hub genes were predicted by TIMER 2.0 and GEPIA database.
Cell Culture, RNA Extraction, cDNA Synthesis and Real-Time PCR Analysis
Normal cell line (LO2) and two HCC cell lines SMMC7721 and Huh7 were acquired from ATCC (Manassas, USA). The cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, GIBCO, Carlsbad, CA) and 2 mm L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin in Dulbecco modified Eagle medium (Invitrogen, CA), 37 °C, 5% CO2.
Total RNA was extracted from three cell lines using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Then, the RNA was transcribed into cDNA with primeScript RT Master Mix (Takara). Subsequently, the expression of UHRF1 and other 9 related genes in SMMC7721 and Huh7 cell lines were analyzed using SYBR premix ex Taq II Kit (Takara) according to the manufacturer’s agreement. Fold changes were assessed using the 2−ΔΔCt method, with β-actin as an endogenous control. And the RT-qPCR experiments were carried out three repeats in SMMC7721 and Huh7 cell lines, each result was used to verify the relationship between UHRF1 and other genes. The PCR primers used in this study are shown in Table 1.
Table 1.
Sequences for RT-qPCR
| Group Name | Sequences |
|---|---|
| UHRF1: F’ | 5′- CCACATCGTCCTCACAGC-3′ |
| UHRF1: R’: | 5′- GGTCCACATCATCCTCATAGC-3′ |
| β-actin: F’ | 5′-CGGC ATTGTCACCAACTG-3′ |
| β-actin: R’ | 5′-CGCTCGGTCAGGATCTTC-3′ |
| CDC20: F’ | 5′-ATGCGCCAGAGGGTTATCAG-3′ |
| CDC20: R’ | 5′-AGGATGTCACCAGAGCTTGC-3′ |
| CDK1: F’ | 5′-CCCTTTAGCGCGGATCTACC-3′ |
| CDK1: R’ | 5′-CATGGCTACCACTTGACCTGT-3′’ |
| TOP2A:F’ | 5′-CATTGAAGACGCTTCGTTATGG-3′ |
| TOP2A:R’ | 5′-CAGAAGAGAGGGCCAGTTGTG-3′ |
| CCNA2:F’ | 5′-TGGAAAGCAAACAGTAAACAGCC-3′ |
| CCNA2:R’ | 5′-GGGCATCTTCACGCTCTATTT-3′ |
| PLK1:F’ | 5′-GGCAACCTTTTCCTGAATGA-3′ |
| PLK1:R’ | 5′-AATGGACCACACATCCACCT-3′ |
| BUB1:F’ | 5′-AGCCCAGACAGTAACAGACTC-3′ |
| BUB1:R’ | 5′-GTTGGCAACCTTATGTGTTTCAC-3′ |
| AURKB:F’ | 5′-CGCAGAGAGATCGAAATCCAG-3′ |
| AURKB:R’ | 5′-AGATCCTCCTCCGGTCATAAAA-3′ |
| BUB1B: F’ | 5′-TCTCAGAAACAGAATCCACGATCC-3′ |
| BUB1B: R’ | 5′-TGCTAAATCTGCTATACCAAACAGG-3′ |
| CCNB1:F’ | 5′-AATGGGAAGGGAGTGAGTGC-3′ |
| CCNB1:R’ | 5′-GCATTAATTTTCGAGTTCCTGGTG-3′ |
Immune Infiltration Analysis and Statistical Analyses
Statistical analysis was conducted by the R software (version 4.03). A total of 622 HCC samples from TCGA were divided into high-expression group and low-expression group according to median expression level of UHRF1. Then, CIBERSORT as well as ssGSEA analysis were used to assess the correlation between the proportions of immune cells and UHRF1 expression in HCC. Furthermore, TIMER 2.0 database was employed to analyze the associations between UHRF1 expression and tumor-infiltrating immune cells infiltration levels. Kaplan–Meier survival analysis was utilized to determine whether immune infiltration status make an impact on prognosis of HCC patients. The Pearson Correlation was performed using GraphPad Prism software (GraphPad Software, San Diego, California, USA).
Results
UHRF1 Expression in HCC Tissues and Survival Analysis via Online Databases
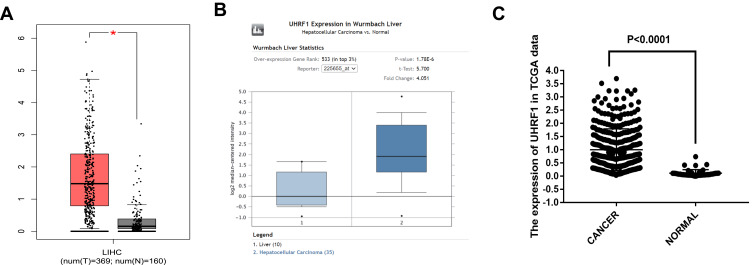
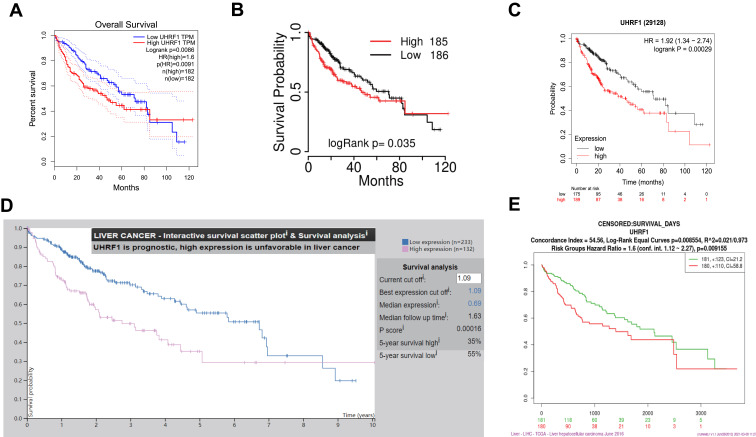
Assessment of GEPIA database containing 369 specimens of HCC tissue and 160 tumor-adjacent normal tissue specimens demonstrated that UHRF1 mRNA expressions were significantly higher in HCC than in normal tissue (Figure 1A). Moreover, data in Oncomine and TCGA also confirmed that UHRF1 was overexpressed in HCC (Figure 1B and C). Furthermore, Kaplan–Meier survival analysis revealed that patients with higher expression of UHRF1 was significantly associated with worse prognosis (Figure 2A–E, p < 0.05).
Figure 1.
UHRF1 expression in HCC. (A) Expression of UHRF1 between HCC and paired normal liver tissues based on the GEPIA database. *P < 0.05. (B) The UHRF1 expression level of tumor tissues and normal tissues in HCC from Oncomine database. *P < 0.05. (C) UHRF1 expression was upregulated in HCC using TCGA dataset analysis.
Figure 2.
Survival analysis of UHRF1 in HCC. (A) OS curve of UHRF1 in HCC patients in GEPIA dataset. (B) OS curve of UHRF1 in HCC patients in TCGA-portal. (C) OS curve of UHRF1 in HCC patients in Kaplan–Meier Plotter. (D) OS curve of UHRF1 in HCC patients in Protein Atlas database. (E) OS curve of UHRF1 in HCC patients in SurExpress dataset.
Functional Enrichment Analysis of UHRF1
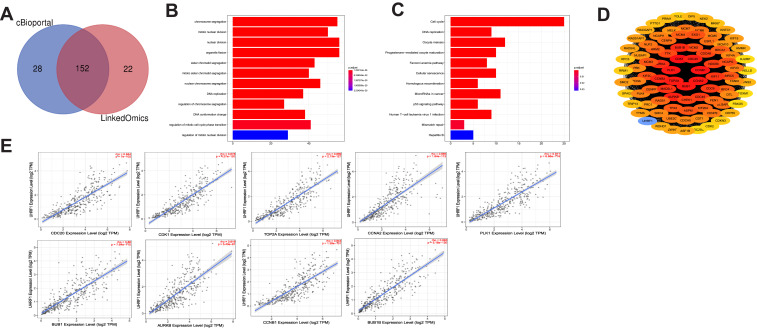
To investigate the potential biological functions associated with UHRF1 in HCC, we identified 152 UHRF1 co-expression genes using the LinkedOmics and cBioportal online bioinformatics databases, which were shown by Venn diagram (Figure 3A). Then we explored the enrichment of the functions and pathways of UHRF1 by performing GO annotations and KEGG pathway enrichment analysis. In Figure 3B, UHRF1 interactive genes were mainly enriched in chromosome segregation, nuclear division and organelle fission. KEGG pathway analysis showed that these genes were enriched in signal pathways such as cell cycle, oocyte meiosis, microRNAs signal pathways and cellular senescence (Figure 3C).
Figure 3.
Enrichment analysis of GO and KEGG. (A) Analysis from LinkedOmics and cBioportal databases identified 152 UHRF1 co-expression genes. (B) GO enrichment analysis. (C) KEGG enrichment analysis. (D) The PPI network of UHRF1 co-expression genes. (E) Analysis from TIMER 2.0 database revealed that the expression of UHRF1 was significantly correlated with the top 9 hub genes.
Identification and Validation of PPI Network and the Hub Genes
As illustrated in Figure 3D, PPI network analysis using STRING database and Cytoscape 3.7.1 showed the PPI network contains 89 nodes. The relationships between UHRF1 expression and top 9 highest connected genes in PPI network were analyzed using the TIMER 2.0 database, and the results demonstrated that UHRF1 expression was positively correlated with CDC20 (rho = 0.844, p = 7e-102), CDK1 (rho = 0.878, p = 4.27e-120), TOP2A (rho = 0.889, p = 2.13e-127), CCNA2 (rho = 0.865, p = 1.84e-112), PLK1 (rho = 0.871, p = 6.66e-116), BUB1 (rho = 0.86, p = 7.64e-110), AURKB (rho = 0.819, p = 5.43e-91), CCNB1 (rho = 0.843, p = 1.38e-101), BUB1B (rho = 0.893, p = 5.18e-130) (Figure 3E).
Next, GEPIA database was utilized to further verify the relationship between UHRF1expression and its relative genes, as shown in Figure S1, the expression of UHRF1 and the 9 highest related genes were positively correlated, moreover, in order to determine the relationship between UHRF1 and 9 highest related genes, we performed RT-qPCR in SMMC7721 and Huh7 cell lines (experiments were performed with three independent times in each cell line), and the results of each experiment were analyzed by Pearson correlation. Finally, it was found that UHRF1 showed a positive correlation with the nine highest related genes (Figure S2). Taken together, the UHRF1 positively correlated with CDC20, CDK1, TOP2A, CCNA2, PLK1, BUB1, AURKB, CCNB1 and BUB1B in HCC.
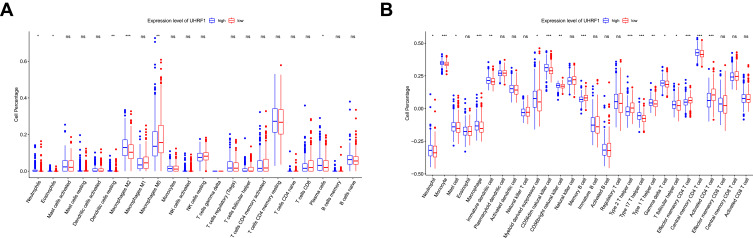
Correlation Between UHRF1 and Immune Cells Infiltration
We further evaluated the status of immune cell infiltration, in order to dissect the relationship between UHRF1 expression and the status of immune cells infiltration in HCC. As shown in Figure 4, the immune infiltration between the high and low UHRF1 expression groups was compared in 622 HCC samples from TCGA database. We conducted CIBERSORT algorithm and found that UHRF1 expression was significantly related to the infiltration level of neutrophils, eosinophils, macrophages M2, macrophages M0 and dendritic cells resting (Figure 4A, p < 0.05). Additionally, ssGSEA analysis showed that UHRF1 expression had significant correlations with immune cells including neutrophils, monocyte, macrophage, CD56dim natural killer cell and type 2T/17T helper cell, especially correlated with three differentiated types of CD4+ T cell (Figure 4B, p < 0.05).
Figure 4.
The proportion of immune cells was associated with the expression of UHRF1 in TCGA dataset. ns: no significance, *P < 0.05, **P < 0.01, ***P < 0.001. (A) Analysis of the proportion of immune cells from CIBERSORT algorithm. (B) Analysis of the proportion of immune cells from ssGSEA.
Prognosis Analysis of UHRF1 Expression with Immune Infiltration Status
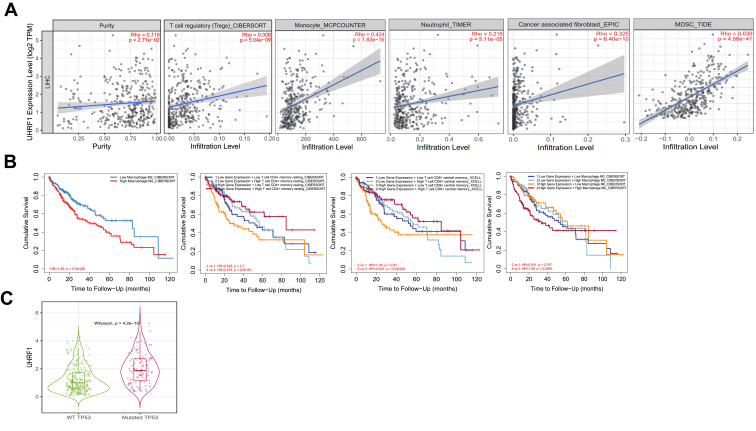
In TIMER 2.0 database, we next uncovered that UHRF1 expression was positively correlated with tumor purity (Rho = 0.119, p = 2.71e-02). Moreover, we confirmed that UHRF1 expression was significantly correlated to the infiltration of immune cells, including regulatory T cell (r = 0.308, p = 5.04e-09), monocyte (r = 0.424, p = 1.83e-16), neutrophil (r = 0.216, p = 5.11e-05), cancer associated fibroblast (r = 0.325, p = 6.40e-10) and myeloid-derived suppressor cell (r = 0.639, p = 4.58e-41) (Figure 5A).
Figure 5.
Correlation analysis and prognosis analysis between UHRF1 and immune infiltration. (A) The correlation between tumor immune infiltrated cell levels and UHRF1 expression in HCC. (B) Cumulative survival of UHRF1 expression with immune infiltration status in HCC. (C) The expression of UHRF1 was positively correlated with the mutation of TP53.
Additionally, we found that the high expression of UHRF1 in HCC resulted in the increase of M2 cell infiltration, which significantly shortened the overall survival of patients with HCC. Then, we performed the Kaplan–Meier analysis to compare the cumulative survival between high and low UHRF1 expression level with different immune cell infiltration. The results indicated that high UHRF1 expression with high immune infiltration status of memory resting CD4+ T cell and central memory CD8+ T cell achieved a better prognosis than with low immune infiltration status. However, high UHRF1 expression with high immune infiltration status of macrophage M0 was associated with worse prognosis (Figure 5B). We then evaluated that the mutation of TP53 was positively correlated with the expression of UHRF1 (Figure 5C).
Discussion
Despite the advances in HCC diagnosis and treatment in the past decades, the prognosis of advanced patients still remains poor. Accumulating researches have focused on HCC immunotherapy and several immune-checkpoint inhibitors have improved the survival rate of advanced HCC.12,13 However, due to therapy resistance, a large number of patients gradually developed no response to immunotherapy. Therefore, it is crucial to explore sensitive diagnosis and prognosis biomarkers for HCC.
Epigenetic modification changes such as aberrant DNA methylation and histone modifications were always correlated tightly with tumorigenesis.14 Accordingly, UHRF1 (also known as ICBP90 and Np95) is characterized as a multidomain protein that served as a hub regulator in DNA methylation and histone modifications.3,15 With the SRA domain and TTD domain, UHRF1 recognized hemi-methylated DNA and hemi-methylated histone H3, and then recruited DNMT1 to ensure maintenance of DNA methylation.5 Thus, the dysregulated of UHRF1 was involved in abnormal chromatin structure and gene expression. Previous research indicated that UHRF1 expression was significantly upregulated in many kinds of cancer.16,17 Wang et al found that UHRF1 was overexpressed in CRC and contributed to tumorigenesis of CRC.10 Similarly, QIN and his colleagues demonstrated that UHRF1 depletion inhibited cell proliferation and migration in gallbladder cancer.18 Moreover, UHRF1-depletion activated caspase 3 and PARP1, thus induced apoptosis.19 Several researches have also cleared that high UHRF1 expression was associated with clinical pathological parameters and poor overall survival.11,20 In this study, we conducted bioinformatic analysis on GEPIA, GEO and TCGA databases. UHRF1 was remarkably overexpressed in HCC tissues compared to normal liver tissues and high expression was significantly associated to worsened survival. In addition, functional enrichment analysis was performed to investigate the functions and regulation networks of UHRF1 co-expression genes in HCC. We uncovered that the biological processes of target genes mainly concentrated in Cell division processes including chromosome segregation, nuclear division and organelle fission. Through KEGG analyses, we knew that the target genes might occur in cell cycle and microRNAs signal pathways. Therefore, we assumed that the biological function of UHRF1 might be mediated through these underlying molecular mechanisms. The PPI network showed that UHRF1 was positively correlated with the 9 highest related genes, which was further confirmed by GEPIA database and RT-qPCR results, these genes may collaborate to promote the progression of HCC, further investigations are required to confirm the effects of UHRF1 in future.
Immunotherapy has gradually become a promising choice of treatments in advanced HCC. Recently, it was reported that immune infiltration levels affected the efficacy of immunotherapy.21,22 However, to our knowledge, there is little known about the correlation of UHRF1 and immune infiltrates. In order to analyze whether the expression of UHRF1 is related to the immune microenvironment of HCC, we downloaded the data of 622 patients with live cancer from TCGA database. Our analysis showed that the expression of UHRF1 is related to different immune marker sets and immune infiltration levels in HCC. CIBERSORT analysis showed that the high expression of UHRF1 was positively correlated with the infiltration levels of neutrophils, eosinophils, macrophages M2, macrophages M0 and dendritic cells resting. Similarly, further ssGESA analysis showed that UHRF1 expression was significantly correlated with immune cells such as neutrophils, monocytes, macrophages, CD56dim natural killer cell and 2T/17T helper cells, especially with three differentiated CD4 + T cells. Moreover, we performed analyses on the TIMER 2.0 database to uncover the potential correlation between the high expression of UHRF1 and the level of immune cell infiltration. As our results showed, UHRF1 was positively correlated with the infiltration level of T cell, monocyte, neutrophil, cancer associated fibroblast and myeloid-derived suppressor cell, indicating the potential contribution of UHRF1 to immune microenvironments of HCC and resistance to immunotherapy. Likewise, in TIMER 2.0 database, we also found that the high expression of UHRF1 and immune infiltration level jointly affected the prognosis of HCC patients. Consistent with our results, a study from Zhang et al demonstrated a critical role for circUHRF1 in resistance to anti-PD1 therapy via inducing NK cell dysfunction in nude mice.23 Besides, UHRF1 might regulate tumor immune evasion through IFN-γ mediated PD-L1 pathway.24
It is well known that TP53 mutation is one of the most common mutations in HCC, which affects the progression and prognosis of HCC,25 in TIMER 2.0 database, we found UHRF1 expression was significantly higher in TP53 mutations than that of TP53 wild types. This may partly explain that the mutation of TP53 can promote the progression of HCC.
There are some limitations to our study. First, this study was based on public databases. Validation from external samples is necessary for verifying the expression of UHRF1. Second, taking normal tissues as control, analyzing and predicting the receiver operating characteristic curve of HCC can further more convincingly prove that UHRF1 can be used as a marker for the diagnosis and treatment of HCC. Third, further studies of the biological function of UHRF1 expression in cell lines and animal models need to be performed. Taken together, UHRF1 could be a potential target of immunotherapy in HCC and more efforts are needed to clarify this biological impact.
Conclusion
In summary, our results demonstrated that UHRF1 was upregulated in HCC and its overexpression might cause worse overall survival in HCC patients. UHRF1 might take part in key pathways associated with cell division process and cell cycle. Further, the adverse effect of high UHRF1 expression on the survival outcome of HCC patients might be due to dysregulated immune infiltrates. Our results demonstrated that UHRF1 might shed new light on tumor pathogenesis and molecular treatment in HCC.
Acknowledgments
This work was financially supported by the Shantou Science and Technology Bureau (NO. 2006241552607), “Dengfeng Project” for the construction of high-level hospitals in Guangdong Province – the First Affiliated Hospital of Shantou University Medical College Supporting Funding (NO.202003-17).
Abbreviation
TTD, Tandem Tudor Domain; HDAC1, histone deacetylase 1; SRA, Set and Ring Associated; RING, Really Interesting New Gene; UBL, ubiquitin-like; PPARγ, peroxisome proliferator activated receptor gamma.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 3.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- 4.Bronner C, Alhosin M, Hamiche A, Mousli M. Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes. 2019;10(1):65. doi: 10.3390/genes10010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishiyama A, Yamaguchi L, Sharif J, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502(7470):249–253. doi: 10.1038/nature12488 [DOI] [PubMed] [Google Scholar]
- 6.Alhosin M, Abusnina A, Achour M, et al. Induction of apoptosis by thymoquinone in lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent pathway which targets the epigenetic integrator UHRF1. Biochem Pharmacol. 2010;79(9):1251–1260. doi: 10.1016/j.bcp.2009.12.015 [DOI] [PubMed] [Google Scholar]
- 7.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455(7214):818–821. doi: 10.1038/nature07249 [DOI] [PubMed] [Google Scholar]
- 8.Jin W, Chen L, Chen Y, et al. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat. 2010;123(2):359–373. doi: 10.1007/s10549-009-0652-2 [DOI] [PubMed] [Google Scholar]
- 9.Venza M, Visalli M, Biondo C, et al. Epigenetic marks responsible for cadmium-induced melanoma cell overgrowth. Toxicol in Vitro. 2015;29(1):242–250. doi: 10.1016/j.tiv.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Yang YZ, Shi CZ, et al. UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann Surg Oncol. 2012;19(8):2753–2762. doi: 10.1245/s10434-011-2194-1 [DOI] [PubMed] [Google Scholar]
- 11.Sabatino L, Fucci A, Pancione M, et al. UHRF1 coordinates peroxisome proliferator activated receptor gamma (PPARG) epigenetic silencing and mediates colorectal cancer progression. Oncogene. 2012;31(49):5061–5072. doi: 10.1038/onc.2012.3 [DOI] [PubMed] [Google Scholar]
- 12.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, Phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–2502. doi: 10.1016/s0140-6736(17)31046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M, He M, Guo Y, et al. The Influence of immune heterogeneity on the effectiveness of immune checkpoint inhibitors in multifocal hepatocellular carcinomas. Clin Cancer Res. 2020;26(18):4947–4957. doi: 10.1158/1078-0432.CCR-19-3840 [DOI] [PubMed] [Google Scholar]
- 14.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10(3):192–206. doi: 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- 15.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397 [DOI] [PubMed] [Google Scholar]
- 16.Beck A, Trippel F, Wagner A, et al. Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma. Clin Epigenetics. 2018;10:27. doi: 10.1186/s13148-018-0462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang ZY, Zhu B, Zhao XW, et al. Regulation of UHRF1 by microRNA-378 modulates medulloblastoma cell proliferation and apoptosis. Oncol Rep. 2017;38(5):3078–3084. doi: 10.3892/or.2017.5939 [DOI] [PubMed] [Google Scholar]
- 18.Qin Y, Wang J, Gong W, et al. UHRF1 depletion suppresses growth of gallbladder cancer cells through induction of apoptosis and cell cycle arrest. Oncol Rep. 2014;31(6):2635–2643. doi: 10.3892/or.2014.3145 [DOI] [PubMed] [Google Scholar]
- 19.Tien AL, Senbanerjee S, Kulkarni A, et al. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J. 2011;435(1):175–185. doi: 10.1042/BJ20100840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei C, Lu N, Wang L, et al. Upregulation of UHRF1 promotes the progression of melanoma by inducing cell proliferation. Oncol Rep. 2018;39(6):2553–2562. doi: 10.3892/or.2018.6356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Liu H, Shen Z, et al. Tumor-infiltrating Neutrophils is Prognostic and Predictive for Postoperative Adjuvant Chemotherapy Benefit in Patients With Gastric Cancer. Ann Surg. 2018;267(2):311–318. doi: 10.1097/SLA.0000000000002058 [DOI] [PubMed] [Google Scholar]
- 22.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 23.Zhang PF, Gao C, Huang XY, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110. doi: 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Zhang Y, Song H. A disparate role of RP11-424C20.2/UHRF1 axis through control of tumor immune escape in liver hepatocellular carcinoma and thymoma. Aging. 2019;11(16):6422–6439. doi: 10.18632/aging.102197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain SP, Schwank J, Staib F, et al. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26(15):2166–2176. doi: 10.1038/sj.onc.1210279 [DOI] [PubMed] [Google Scholar]