Abstract
Host cell factor 1 (HCF-1) is a nuclear protein required for progression through G1 phase of the cell cycle and, via its association with VP16, transcriptional activation of the herpes simplex virus immediate-early genes. Both functions require a six-bladed β-propeller domain encoded by residues 1 to 380 of HCF-1 as well as an additional amino-terminal region. The β-propeller domain is well conserved in HCF homologues, consistent with a critical cellular function. To date, the only known cellular target of the β-propeller is a bZIP transcription factor known as LZIP or Luman. Whether the interaction between HCF-1 and LZIP is required for cell proliferation remains to be determined. In this study, we used directed mutations to show that all six blades of the HCF-1 β-propeller contribute to VP16-induced complex assembly, association with LZIP, and cell cycle progression. Although LZIP and VP16 share a common tetrapeptide HCF-binding motif, our results reveal profound differences in their interaction with HCF-1. Importantly, with several of the mutants we observe a poor correlation between the ability to associate with LZIP and promote cell proliferation in the context of the full HCF-1 amino terminus, arguing that the HCF-1 β-propeller domain must target other cellular transcription factors in order to contribute to G1 progression.
The transcription of eukaryotic genes is predominantly controlled through the assembly of transcription factors into macromolecular complexes on cis-acting DNA target sequences. The frequency with which multicomponent regulatory complexes (sometimes referred to as enhancesomes) are used predicts the existence of specialized proteins that regulate gene expression by coordinating the recruitment and ordered assembly of the various components into an active complex. One example of this new class of regulatory protein is host cell factor 1 (HCF-1; also known as C1 factor). First identified through its role in assembly of the VP16-induced complex (VIC), a multiprotein-DNA complex that coordinates the activation of the herpes simplex virus immediate-early genes (31, 41), HCF-1 is likely to perform a similar function in the assembly of cellular transcription complexes.
VP16 (also known as Vmw65 or αTIF) is a potent transcriptional activator encoded by herpes simplex virus and packaged into the infective virion particle. Once released into the newly infected cell, VP16 binds directly to HCF-1, allowing translocation to the nucleus and interaction with the POU domain transcription factor Oct-1 and a DNA sequence element known as the TAATGARAT motif that is found upstream of each viral immediate-early gene. VIC assembly is highly selective and is achieved through a combination of specific protein-protein and protein-DNA contacts (12, 14, 23, 32, 39, 44). Both HCF-1 and Oct-1 belong to multiprotein families, and VP16 has evolved mechanisms to recruit single members of each family. VP16 can distinguish Oct-1 from other POU proteins, including the very similar Oct-2, through recognition of differences on the solvent-exposed surface of the POU homeodomain (23, 32). Similarly, VP16 preferentially targets HCF-1 rather the closely related protein HCF-2 through recognition of a limited number of amino acid differences in blades 5 and 6 of the β-propeller domain (14). Once the VIC is formed, the carboxy-terminal activation domain of VP16 activates transcription by recruiting coactivators (42) and by making direct contacts with components of the general transcription machinery (16, 43).
The function of HCF-1 is poorly understood but is likely to yield information of general significance. HCF-1 comprises a series of polypeptides derived from a >2,000-amino-acid precursor through proteolytic processing (19, 48). Cleavage occurs at a series of six centrally located 26-amino-acid repeats (called HCFPRO repeats) producing multiple amino- and carboxy-terminal fragments that remain tightly, but noncovalently, associated following cleavage (49). VP16 interacts with a discrete amino-terminal domain (HCFVIC) composed of six kelch-like repeats that fold into a six-bladed β-propeller. The HCF-1 β-propeller is in itself sufficient for VIC assembly in vitro and in vivo (13, 34, 47), although the carboxy terminus of HCF-1 contributes to the efficiency of complex formation and to translocation of VP16 to the nucleus (21, 22).
HCF-1 is expressed in all mammalian cell types and is essential for cell proliferation (reviewed in reference 14). The role in cell cycle progression was demonstrated through studies of tsBN67 cells, a temperature-sensitive hamster cell line that undergoes a G0/G1 arrest at the nonpermissive temperature (39.5°C) (8). The cell cycle arrest is reversible, and cells will reenter the proliferative cycle if returned to the permissive temperature (33.5°C). The tsBN67 phenotype is due to a single proline-to-serine change in the β-propeller domain of HCF-1 (8, 47). The mutation apparently does not alter the stability or processing of HCF-1 but prevents association with VP16. The fact that a single mutation in HCF-1 could disrupt both known functions (transactivation by VP16 and cell proliferation) led to the idea that VP16 may have copied a preexisting interaction between HCF-1 and an unknown cellular protein involved in G1 progression (5, 8, 47). This hypothesis is supported by the strong conservation of the amino acid sequence of the β-propeller during evolution (14, 27) and the fact that HCF from invertebrates such as insects and nematodes can readily support VIC formation (18, 27, 46).
To date, the only known cellular target of the HCF-1 β-propeller is a ubiquitous basic leucine zipper transcription factor known as LZIP or Luman (5, 28). Mammalian LZIP and a related protein from Drosophila, called dCREB-A or BBF-2, interacts with HCF-1 through a short tetrapeptide motif known as the HCF-binding motif (HBM) that is also found in VP16 (5, 29). The interaction can be disrupted by point mutations in the HBM (5, 9, 29) or by competition with short peptides derived from VP16 that span the motif (10, 34, 50). Both LZIP and dCREB-A function as potent transcriptional activators (1, 29, 35), indicating that HCF-1 is likely to be involved in the regulation of cellular as well as viral transcription.
In this study, we used mutagenesis to define the surfaces on the HCF-1 β-propeller involved in the association with VP16 and LZIP. Our results show that all six blades of the β-propeller contribute to recognition of the relatively simple HBM. Surprisingly, we find major differences between LZIP and VP16 in terms of their sensitivities to individual HCF-1 mutants, implying a significant contribution by the nonconserved sequences flanking the HBM. In general, mutations that disrupt VIC assembly affect the HCF-1–VP16 interaction; however, we identify a mutation in the sixth blade of the β-propeller that prevents complex formation without disrupting association with VP16. Last, our mutational analysis reveals a poor correlation between association with LZIP and ability to complement the tsBN67 proliferation defect, arguing that the HCF-1–LZIP interaction may not be required for G1 progression.
MATERIALS AND METHODS
Plasmid construction and site-directed mutagenesis.
Mammalian expression vectors encoding the wild-type and P134S mutant β-propeller domains of HCF-1 (residues 1 to 380; pCGNHCF-1N380 and pCGNHCF-1N380P134S, respectively), the amino terminus of HCF-1 (residues 1 to 902; pCGNHCF-1N902), VP16 (residues 5 to 411; pCGTVP16ΔC), and (residues 1 to 154; pCGTLZIP) have been described previously (14, 47).
Mutations were generated by oligonucleotide-directed mutagenesis following the Altered-Sites (Promega Inc.) or Quick-change (Stratagene Inc.) protocol. Where possible, a diagnostic restriction site was included to serve as a marker for screening the mutants. The sequence changes of the mutants described in this study are as follows: P30S, TGagcCGGC (NgoMI/NaeI+); P79S, TTagCCCgGG (SmaI/XmaI+); P197S, TCtTAagcC (AflII+); P252S, CaagctTaC (HindIII+); P319S, TCtCtCGaGC (XhoI+); C82D, TCGTcgacGA (SalI+); R137D, CagatCTCG (BglII+); R200D, ACCtCcGGAc (BspEI+); R255A, TCCggaCA (BspEI+); R322D, CCCGgGCTgacGC (SmaI/XmaI+); K105D, GGgAcTAtAGC (SfcI+); R228D, GcgacCTaGG (PstI−/AvrII+); RK344A2, ACgctgcaG (PstI+); EWK289A3, GctgcagcaTG (PstI+); and S338A, GgccGGcCG (NgoMI/NaeI+). Uppercase letters represent wild-type sequences, and lowercase letters represent mutations. Missense codons are indicated with bold typeface. Diagnostic restriction sites are underlined and identified in parentheses; superscript plus and minus signs indicate sites generated and sites destroyed, respectively. Each mutation was verified by DNA sequence analysis.
Transfections, coimmunoprecipitations, immunoblotting, and electrophoretic mobility shift assays.
Human 293T cells were transfected with Lipofectamine (Life Technologies), using 20 μl lipid reagent per 6-cm-diameter dish. Whole cell and nuclear extracts were prepared after 24 h by lysing cells in high-salt lysis buffer (420 mM KCl, 10 mM Tris-HCl [pH 7.9], 5% glycerol, 0.25% NP-40, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.2 mM sodium vanadate, 50 μM sodium fluoride, 1 mM dithiothreitol). Nuclei were extracted at 4°C for 30 min and removed by centrifugation. For immunoprecipitations, 100 μl of extract was incubated with 2.5 μl of antihemagglutinin (αHA) antibody (12CA5)-coupled protein G-agarose beads at 4°C for 1 h. The beads were washed four times in 1 ml of wash buffer (200 mM KCl, 10 mM Tris-HCl [pH 7.9], 5% glycerol, 0.5 mM EDTA) before separation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Immunoblotting was performed by semidry transfer and detected by enhanced chemiluminescence (SuperSignal; Pierce). The αHA antibody and αT7 antibody (Novagen) were diluted 1:5,000 and 1:10,000, respectively.
For cell-free expression, HCF-1-encoding fragments were shuffled into pNCITE, a derivative of pCITE2a+ that includes the HA epitope at the amino terminus of the expressed protein. Full-length Oct-1 was expressed by using a non-epitope-tagged version of pCITE (gift of Ethan Ford and Nouria Hernandez, Cold Spring Harbor Laboratory). In vitro transcription and translation reactions were performed in the presence of [35S]methionine, using the TNT Quick Coupled transcription-translation system (Promega, Inc.). HA-tagged HCF-1 polypeptides were radiolabeled to a lower specific activity by including a 20-fold excess of unlabeled methionine in the translation reaction. Coimmunoprecipitation assays with in vitro-translated proteins were performed as described above except that the antibody beads were pretreated with unprogrammed lysate to reduce nonspecific binding by Oct-1. The binding reactions and washes were adjusted to 100 mM KCl and 0.05% NP-40. Electrophoretic mobility shift assays were performed as described previously (40, 41); complex formation was performed at 30°C, and electrophoresis was carried out at room temperature. Luciferase reporter assays were performed under standard conditions. Extracts were prepared by using a commercial lysis buffer (Promega, Inc.) and measured with an LB9507 luminometer (EG&G Berthold, Inc.).
Complementation of tsBN67 cells.
Subconfluent tsBN67 cells were incubated at 33.5°C for 20 h and transfected with 1 μg of each HCF-1 expression vector together with 0.5 μg of pSV2neo, using Lipofectamine (Life Technologies). The DNA mixes were sterilized by ethanol precipitation prior to transfection. After 2 days at 33.5°C, transfected cells were split into two 15-cm-diameter dishes, and Geneticin (0.8 mg/ml) was included in the medium to select for stable transfectants. Following 1.5 to 2 weeks of incubation at 39.5°C, the plates were stained with crystal violet and colonies of proliferating cells were counted. In some cases, complementation was confirmed by subcloning individual colonies.
RESULTS
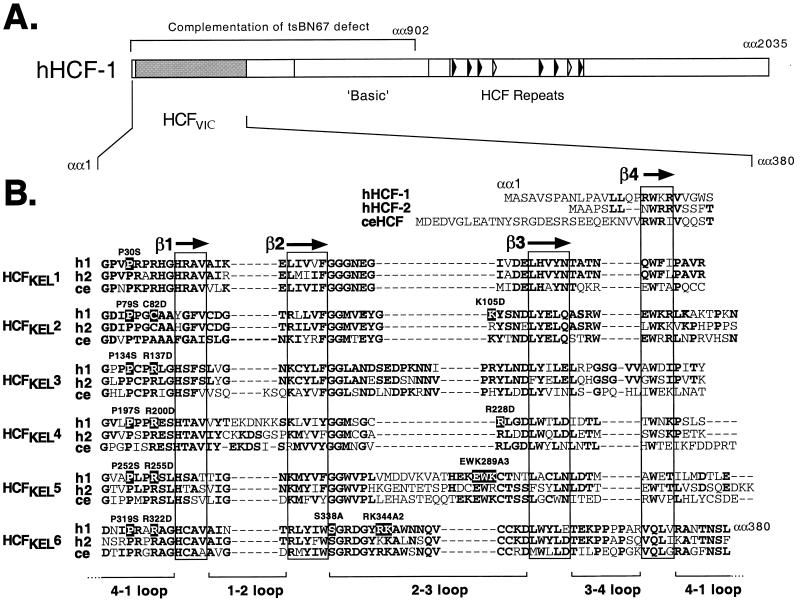
VP16 associates with an amino-terminal domain of HCF-1, called the HCFVIC domain (13, 22, 34, 47) (Fig. 1A). Spanning approximately 380 residues, the HCFVIC domain is comprised of six degenerate sequence repeats that were first identified in the Drosophila actin-associated protein Kelch and are referred to as HCFKEL1 to HCFKEL6 (47). A sequence alignment of the HCFVIC domains from HCF-1, HCF-2, and Caenorhabditis elegans HCF is shown in Fig. 1B, illustrating the extensive sequence conservation across the domain (14, 27). By analogy to kelch repeat proteins of known structure, the HCFKEL repeats are predicted to fold into four-stranded β-sheets that stack pseudosymmetrically around a central axis, forming a structure known as a β-propeller (2). This compact arrangement is decorated by loops of variable length clustered on one surface (36, 37, 45). We follow a standard nomenclature for mutants; for example, P134S indicates proline 134 changed to serine.
FIG. 1.
(A) Primary structure of HCF-1. The amino-terminal β-propeller (HCFVIC) domain is shown as a shaded box. The eight HCFPRO repeats are located near the center of the polypeptide and represented by filled (functional) and open (nonfunctional) arrowheads. The first 902 residues of HCF-1 comprising the β-propeller, amino-terminal self-association domain, and poorly defined basic region are required for complementation of the tsBN67 cell proliferation defect. (B) Alignment of the six kelch repeats that make up the β-propeller domain of HCF-1 (h1), HCF-2 (h2) and C. elegans HCF (ce) (14, 27). The predicted β-strands in each repeat unit are boxed. Residues that are conserved in two or more HCF proteins are shown in bold. The residues that have been mutated in this study are numbered and highlighted. αα, amino acids.
All six kelch repeats contribute to VIC formation.
The tsBN67 cell cycle arrest phenotype results from a single proline-to-serine substitution at position 134 in the fourth residue in HCFKEL3 (Fig. 1B). In addition to blocking G1 progression, this single mutation abolishes the interaction of HCF-1 with VP16 and LZIP but does not alter the overall stability or folding of the β-propeller domain itself (5, 8, 47). Remarkably, there is a proline residue at the equivalent position in each of the six kelch repeats of HCF-1, HCF-2 and C. elegans HCF (14, 27, 47), suggesting a critical role in domain function. By analogy to other β-propeller domains, the universally conserved proline lies within a loop connecting the fourth strand (β4) of one β-sheet (or kelch repeat unit) to the first strand (β1) on the next sheet. These 4-1 loops are positioned on the “top” surface of the structure, lining the central cavity, and are thus ideally positioned to contribute to protein-protein interactions.
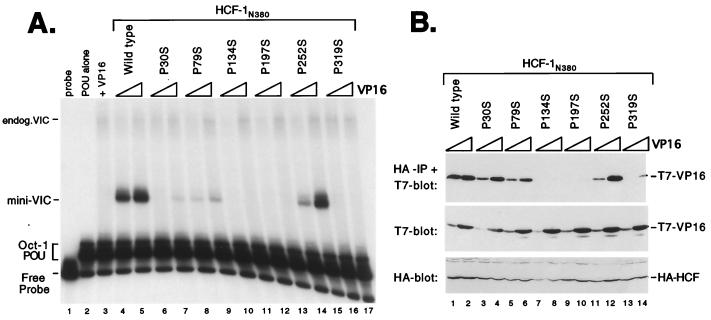
To address the significance of this conserved proline, we generated serine substitutions in each of the remaining kelch repeats of HCF-1 and assayed the resulting mutant HCFVIC domains for association with VP16. Figure 2 shows the results of this analysis. Human 293T cells were cotransfected with expression plasmids encoding wild-type or mutant HCF-1 β-propeller domains, together with increasing amounts of VP16 expression plasmid. Whole cell extracts were prepared from the transfected cells and mixed with a labeled DNA probe containing a VP16-responsive TAATGARAT element derived from the ICP0 promoter together with Escherichia coli-expressed Oct-1 POU domain protein. Wild-type HCF-1 (HCF-1N380 [Fig. 2A, lanes 4 and 5]) gave rise to a strong VIC (labeled mini-VIC), while the P134S mutant failed to support complex formation (lanes 10 and 11) as we have reported previously (14, 47). The VIC incorporating the recombinant HCF-1 β-propeller (mini-VIC) has a significantly faster gel mobility than the complex produced by full-length HCF-1 present in the extract (endogenous VIC). In addition to P134S, substitutions P30S (lanes 6 and 7), P79S (lanes 8 and 9), P197S (12 and 13), and P319S (lanes 16 and 17) significantly reduced, or in some cases eliminated, VIC formation. In striking contrast, P252S (lanes 14 and 15) retained substantial complex-forming activity. Immunoblotting with αHA and αT7 monoclonal antibodies showed that the mutant versions of HCF-1N380 were expressed at levels similar to those for the wild type and that the levels of T7-epitope tagged VP16 increased in proportion to the amount of expression plasmid (Fig. 2B). Thus, five of the six HCFKEL repeats contribute to VIC assembly.
FIG. 2.
Proline-to-serine substitutions at position 4 in each HCFKEL repeat. (A) HCF-1 polypeptides were coexpressed with VP16ΔC by transfection of 293T cells. Extracts were prepared after 40 h and assayed for VIC formation in an electrophoretic mobility shift assay. The first three lanes are controls showing probe alone (lane 1), Oct-1 POU domain protein alone (lane 2), and cell extract transfected with 1 μg of VP16ΔC (lane 3). Each HCF-1N380 expression plasmid (1 μg) was cotransfected with 0.1 and 1.0 μg of VP16ΔC expression plasmid. The HCF-1N380 proteins used are indicated above the lanes. Positions of the free probe, Oct-1 POU domain complex, and VIC containing native (endogenous) human HCF-1 (endog.VIC) or truncated HCF-1 (mini-VIC) are indicated. (B) Coimmunoprecipitation of VP16. To measure the direct association between HCF-1N380 and VP16ΔC, the extracts shown in panel A were subject to coimmunoprecipitation assay. Extracts were immunoprecipitated (IP) with an αHA antibody (12CA5) coupled to agarose beads and resolved on an SDS–12% polyacrylamide gel, and coimmunoprecipitated VP16 was detected by immunoblotting with an αT7 epitope tag antibody. Direct immunoblotting of the extracts (lower two panels) showed that each HA-tagged HCF-1 polypeptide was expressed at equivalent levels and that the expression of T7-tagged VP16 was proportional to amount of input plasmid. Irrelevant cross-reacting polypeptides are indicated with an asterisk.
Using a coimmunoprecipitation assay (Fig. 2B), we compared the ability of each mutant β-propeller domain to associate with VP16. Using the same transfected cell extracts, HA-tagged HCF-1 polypeptides were recovered by immunoprecipitation with an αHA antibody, and the coimmunoprecipitated VP16 was detected by immunoblotting with an αT7 antibody. Consistent with the inability to support VIC formation, P134S, P197S, and P319S were severely compromised for association with VP16. In contrast, P30S, P79S, and P252S retained significant activity, suggesting that the P30S and P79S mutants affect a different aspect of complex assembly. P252S was unique in being essentially wild type for association and VIC formation.
The conserved arginine at position 7 in each HCFKEL repeat is critical for activity.
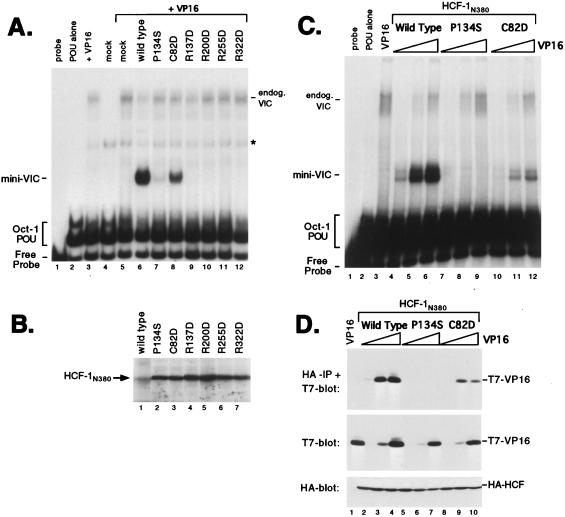
With the exception of HCFKEL2, the amino acid residue at position 7 of each HCFKEL repeat corresponds to an arginine residue, again implying a critical role in domain function. In HCFKEL2 (or, interestingly, in the analogous position in HCF-2), this position is occupied by a cysteine residue. To address the importance of this semiconserved position with respect to association with VP16, we made a set of radical substitution mutants, replacing the residue at position 7 with an aspartic acid. We chose this substitution because C. elegans HCF uses an alanine at position 7 in HCFKEL2 (27), suggesting that a noncharged residue may be more readily tolerated. Unfortunately, we were unable to generate a mutation at R33 in HCFKEL1. Our analysis of these point mutants is shown in Fig. 3A. Exchanging the cysteine at position 82 for aspartic acid led to only a moderate reduction in complex formation (Fig. 3A, compare lane 6 with lane 8). In contrast, R137D (lane 9), R200D (lane 10), R255D (lane 11), and R322D (lane 12) disrupted VIC formation. The experiment shown in Fig. 3A and B used HCF-1N380 polypeptides expressed by in vitro translation. Equivalent results were obtained for HCF-1 expressed by transient transfection, and we have chosen to show the in vitro translation experiment simply because P134S (lane 7) retains some activity when expressed in a cell-free system and is actually more active than R137D, R200D, R255D, and R322D. This implies that the arginine-to-aspartic acid substitution is extremely severe, perhaps resulting in a gross disruption of the β-propeller structure.
FIG. 3.
Radical substitution of the conserved arginine residue at position 7 of each HCFKEL repeat has a severe effect on VIC formation. (A) Electrophoretic mobility shift assay of HCF-1 proteins expressed by in vitro translation. The first three lanes are controls showing probe alone (lane 1), Oct-1 POU domain protein alone (lane 2), bacterially produced glutathione S-transferase–VP16ΔC (lane 3), or unprogrammed reticulocyte lysate (lane 4). In the remaining lanes, unprogrammed lysate (lane 5) or lysates expressing recombinant HCF-1N380 polypeptides were mixed with glutathione S-transferase–VP16ΔC (lanes 6 to 12). The HCF-1N380 proteins used are indicated above the lanes. Positions of the free probe, Oct-1 POU domain complex, and VIC containing rabbit HCF-1 from the lysate (endog.VIC) or truncated HCF-1N380 (mini-VIC) are indicated. A nonspecific complex is indicated with an asterisk. (B) In vitro translation products were resolved on an SDS–12% polyacrylamide gel and detected by fluorography. (C) Electrophoretic mobility shift assay. Extracts were prepared from transfected 293T cells expressing wild-type or mutant versions of HCF-1N380 together with 0.01 μg (lanes 4, 7, and 10), 0.1 μg (lanes 5, 8, and 11), and 1.0 μg (lanes 6, 9, and 12) of VP16ΔC expression plasmid. The HCF-1N380 proteins used are indicated above the lanes. (D) The extracts shown in panel C were used in a coimmunoprecipitation assay as described for Fig. 2B. The upper panel shows the αHA-immunoprecipitated (IP) proteins probed with αT7 antibody; the lower two panels show direct immunoblots of the extracts. Each HCF-1N380 expression plasmid (1 μg) was cotransfected with 0.01 μg (lanes 2, 5, and 8), 0.1 μg (lanes 3, 6, and 9), and 1.0 μg (lanes 4, 7, and 10) of VP16ΔC expression plasmid. The transfections were as follows: VP16 alone (1.0 μg, lane 1), wild-type HCF-1N380 (lanes 2 to 4), P134S (lanes 5 to 7), and C82D (lanes 8 to 10).
To better evaluate the relative activity of the C82D mutant, we titrated the amount of VP16 (Fig. 3C and D). 293T cells were transfected with a constant amount of each HCF-1N380 expression plasmid and increasing amounts of VP16 expression plasmid. VIC formation was measured by electrophoretic mobility shift assay (Fig. 3C). The amount of complex formed by wild-type HCF-1N380 (Fig. 3C, lanes 4 to 6) and C82D (lanes 10 to 12) increased in proportion to the amount of VP16 expressed. Quantitation revealed a five- to sixfold reduction in complex formation by C82D. We also assayed direct association by coimmunoprecipitation (Fig. 3D). C82D showed a reduction in binding comparable to that for VP16 (compare lanes 2 and 3 with lanes 8 to 10). Overall, these results indicate that the arginine residue at position 7 in five of the six HCFKEL repeat is essential for VIC assembly and that the variant cysteine at position 82 in HCFKEL2 is less critical.
Residues within the 2-3 loops also contribute to VIC assembly.
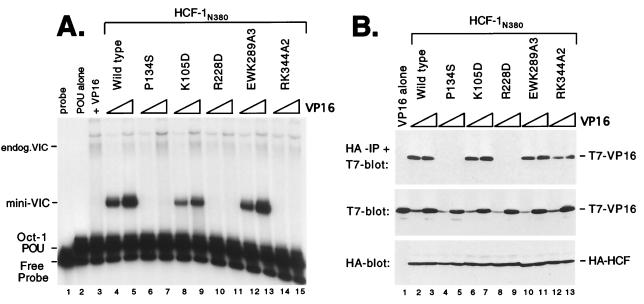
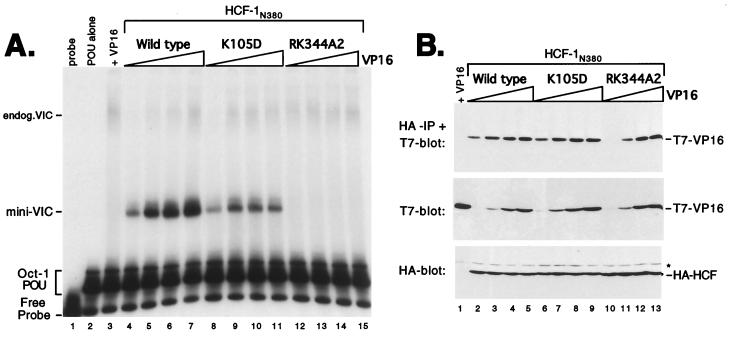
Next we examined the role of the 2-3 loops (connecting β2 to β3), which also contribute to the predicted top surface of the β-propeller. In HCF-1, the 2-3 loops show relatively little sequence homology to each other and also differ significantly in length (Fig. 1B). To determine whether the 2-3 loops contribute to interaction with VP16, we generated substitutions in the 2-3 loops of HCFKEL2 (K105D), HCFKEL4 (R228D), HCFKEL5 (EWK289A3), and HCFKEL6 (RK344A2). Each mutant was expressed in transfected 293T cells and assayed for association with VP16 by the electrophoretic mobility shift and coimmunoprecipitation assays (Fig. 4). Substitutions in HCFKEL2 (K105D) and HCFKEL5 (EWK289A3) had a minor affect on both VIC assembly (Fig. 4A, lanes 8, 9, 12, and 13) and association with VP16 (Fig. 4B, lanes 6, 7, 10, and 11), suggesting that these residues do not make a significant contribution to complex assembly. In contrast, mutation in HCFKEL4 (R228D) and HCFKEL6 (RK344A2) disrupted VIC assembly (Fig. 4A, lanes 10, 11, 14, and 15). This result indicates that residues in the 2-3 loops of HCFKEL4 and HCFKEL6 are critical for VIC assembly. In contrast to a previous report (34), the K105D mutation had only a minor effect on VIC assembly.
FIG. 4.
Residues within the 2-3 loops also participate in association with VP16. (A) HCF-1 polypeptides were expressed with VP16ΔC by cotransfection of 293T cells and assayed for VIC formation. The first three lanes are controls showing probe alone (lane 1), Oct-1 POU domain protein alone (lane 2), and cell extract transfected with 1.0 μg of VP16ΔC (lane 3). Each HCF-1N380 expression plasmid (1.0 μg) was cotransfected with 0.1 and 1.0 μg of VP16ΔC expression plasmid. The HCF-1N380 proteins used are indicated above the lanes. (B) Coimmunoprecipitation assay using extracts from panel A. The upper panel shows the αHA-immunoprecipitated proteins (IP) probed with αT7 antibody; the lower two panels show direct immunoblots of the extracts. The samples were as indicated above the lanes.
To determine whether the R228D and RK344A2 prevent VIC assembly through a failure to recruit VP16, we measured direct association by coimmunoprecipitation. R228D was severely compromised for association with VP16 (Fig. 4B, lanes 8 and 9), thus accounting for the lack of VIC assembly. In this respect, R228D resembles the inactivating R-to-D mutations at position 7 in the 4-1 loops (Fig. 3). In contrast, the behavior of RK344A2 was more similar to that of P30S and P79S (Fig. 2). Although unable to mediate VIC assembly, RK344A2 remained competent for association with VP16 (Fig. 4B, lanes 12 and 13). This interesting result demonstrates that association of HCF-1 with VP16 is not in itself sufficient for VIC assembly.
To further compare K105D and RK344A2, we performed a broader titration of VP16 levels (Fig. 5). Even at the lowest concentration of VP16 (50 ng), both wild-type HCFN380 and K105D supported complex assembly (Fig. 5A, lanes 4 and 8). For wild-type HCF-1, complex formation increased in proportion to input VP16 (lanes 4 to 7). In contrast, the amount of complex formed by K105D reached a maximum at 150 ng of VP16 plasmid (lane 9) and then remained constant (lanes 10 and 11). At present, we do not understand this plateau effect. Finally, at all levels of VP16 assayed, RK344A2 failed to assemble a VIC (lanes 12 to 15). Even with 5 μg of VP16 expression plasmid, we were unable to detect complex formation by this mutant (data not shown).
FIG. 5.
RK344A2 associates with VP16 and yet fails to support VIC formation. (A) Transfected 293T cells extracts were prepared and assayed by electrophoretic mobility shift assay as described for Fig. 2. The first three lanes are controls showing probe alone (lane 1), Oct-1 POU domain protein alone (lane 2), and cell extract transfected with 1.35 μg of VP16ΔC (lane 3). Each HCF-1N380 expression plasmid (1.0 μg) was cotransfected with 0.05, 0.15, 0.45, or 1.35 μg of VP16ΔC expression plasmid. The HCF-1N380 proteins used are indicated above the lanes. (B) Coimmunoprecipitation assay using extracts from panel A. The upper panel shows the αHA-immunoprecipitated (IP) proteins probed with αT7 antibody; the lower two panels show direct immunoblots of the extracts. The samples are as follows: VP16 alone (lane 1) or increasing amounts of VP16 expression plasmid cotransfected with wild-type HCF-1N380 (lanes 2 to 5), K105D (lanes 6 to 9), and RK344A2 (lanes 10 to 13).
When assayed for direct association with VP16 (Fig. 5B), we found no significant difference between K105D and wild-type HCF-1N380 (compare lanes 2 to 5 with lanes 6 to 9). RK344A2, on the other hand, showed a small reduction in its ability the recovery VP16 (best illustrated by comparing lanes 2 and 10); however, this difference is not large enough to account for the loss of VIC formation. In summary, the RK344A2 mutant is unable to form a VIC but retains the ability to associate with VP16.
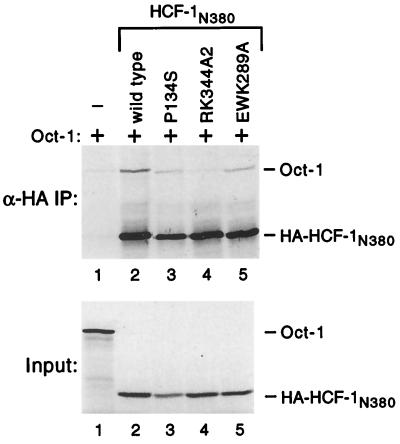
Mutagenesis of VP16 has shown that residues involved in association with Oct-1 lie very close to those required for association with HCF-1 (24), and it is possible that the RK344A2 mutation interferes with the Oct-1 POU domain, thus explaining the loss of VIC formation. To address this directly, we performed coimmunoprecipitation experiments using in vitro-translated HA-tagged HCF-1N380 and untagged in vitro-translated full-length Oct-1 (Fig. 6). We typically recovered five- to sixfold more Oct-1 with wild-type HA-tagged HCF-1N380 than with antibody beads alone (Fig. 6A, upper panel, compare lanes 1 and 2), suggesting that HCF-1 and Oct-1 can indeed interact weakly in the absence of VP16. Interestingly, the RK344A2 mutant was unable to coprecipitate Oct-1 above the level of beads alone (compare lanes 1 and 4). Oct-1 could be recovered with HCF-1N380 P134S (lane 3) and EWK389A3 (lane 5), although the efficiency was slightly less than the wild-type level.
FIG. 6.
The RK344A2 mutation interferes with coimmunoprecipitation of Oct-1. Full-length Oct-1 and HA-tagged HCF-1N380 were expressed in vitro and mixed, and coassociation was assayed by coimmunoprecipitation using αHA antibody beads. Immunoprecipitates (IP) were fractionated by SDS-PAGE (10% gel), and radiolabeled proteins detected by fluorography (upper panel). The input translations (prior to mixing) are shown in the lower panel.
VP16 and LZIP show different sensitivities to individual point mutants in the HCF-1 β-propeller.
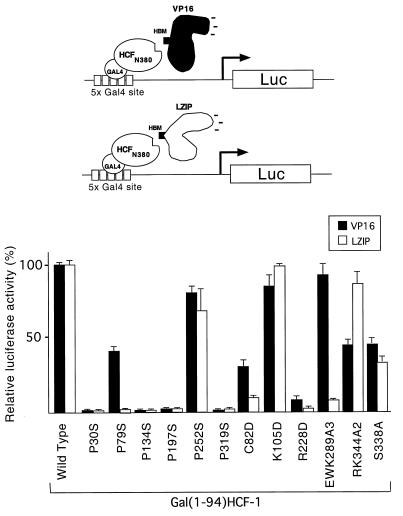
VP16 and LZIP contain a tetrapeptide sequence (EHAY in VP16 [residues 361 to 364] and DHTY in LZIP [residues 78 to 81]), HBM, that is essential for association with HCF-1 (5, 29). In both VP16 and LZIP, individual mutation of the acidic, histidine, or tyrosine residue was sufficient to prevent association with HCF-1 (5, 29). This similarity suggested that the two proteins interact with HCF-1 equivalently. To examine this further, we used a cell-based recruitment assay to compare the sensitivities of VP16 and LZIP binding to each of the HCFVIC domain mutants (Fig. 7). We used this recruitment assay because LZIP is expressed at very low levels in transfected cells, making it difficult to quantitate association with cotransfected HCF-1 mutants by coimmunoprecipitation. The HCF-1 β-propeller domain was expressed in 293T cells as a Gal4 fusion together with either VP16 (residues 5 to 490) or the amino terminus of LZIP (residues 1 to 154). Recruitment was measured in terms of activation of a Gal4-responsive luciferase reporter gene and the amounts of each expression plasmid chosen to give a linear response (data not shown). The recruitment of VP16 was generally consistent with the results of the immunoprecipitation assay described above. The one exception was P30S, which immunoprecipitated VP16 relatively efficiently (Fig. 2B, lanes 3 and 4) but failed to interact in the recruitment assay. This result was highly reproducible, and the reason for this single inconsistency is unclear.
FIG. 7.
Individual mutations in the HCFVIC domain have different effects on association with VP16 and LZIP. A cell-based recruitment assay was performed with transiently transfected 293T cells. A Gal4-responsive luciferase reporter gene (p5xGal-E1B-luc) was transfected into 293T cells by electroporation together with 1 μg of wild-type or mutant version of pCGNGal(1-94)HCF-1N380 and 500 ng of pCGTVP16ΔC or pCGTLZIPN154, as indicated. The values are the average of three independent transfections, and the standard deviation from the mean is indicated by error bars. Fold activation was calculated relative to the activity of wild-type pCGNGal(1-94)HCF-1N380 cotransfected with pUC119.
Although VP16 and LZIP use a related sequence motif (the HBM) to associate with HCF-1, we observed striking differences in their sensitivities to individual point mutations in the HCF-1 β-propeller. P79S, C82D, and EWK2893A disrupted the recruitment of LZIP more than VP16, in each case reducing the association to less than 10% of the wild-type level. RK344A2 showed the reverse phenotype, having a minimal effect on recruitment of LZIP while reducing association with VP16 to approximately 50% of that of wild-type HCF-1. Of the mutations assayed, K105D was unique in having a minimal effect on both interactions. These results indicate that the HCF-1 β-propeller domain recognizes LZIP and VP16 differently. Because point mutations within the HBM tetrapeptide (5, 29) behave similarly, it is likely that differential recognition is mediated by the nonconserved sequences flanking the HBM.
Limited correlation between complementation of the tsBN67 proliferation defect and association with VP16 or LZIP.
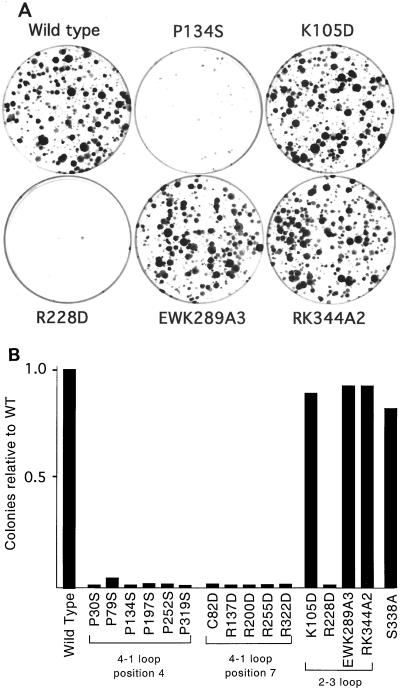
Inactivation of the HCFVIC domain in tsBN67 cells leads to a G1/G0 cell cycle arrest. This defect can be complemented by stable expression of an amino-terminal fragment (residues 1 to 902) comprising the HCFVIC domain, amino-terminal self-association domain (HCFSASN), and basic region (Fig. 1A) (8, 14, 47). The association of LZIP with HCF-1 is prevented by the tsBN67 mutation, suggesting that LZIP is a candidate target of HCF-1 in controlling cell proliferation (5). To address this, we asked whether there is a close correlation between the ability of HCFVIC to interact with LZIP and to support the growth of tsBN67 cells at the nonpermissive temperature. Each mutant was constructed in an expression plasmid encoding the amino-terminal 902 residues of HCF-1 (pCGNHCF-1N902) and transfected into tsBN67 cells together with a selectable marker. After nearly 2 weeks at the nonpermissive temperature, the number of proliferating (or complemented) colonies was determined. Under the conditions used, wild-type HCF-1N902 gave a large number of colonies (∼120 to 200/dish), whereas an empty vector produced no more than one or two revertant colonies. Representative plates are shown in Fig. 8A, and complementation by each of the mutants is summarized in Fig. 8B. These fell into two categories: those capable of supporting cell proliferation and those that were incapable. Mutations at positions 4 (P30S, P79S, P134S, P197S, P252S, and P319S) and 7 (C82D, R137D, R200D, R255D, and R322D) of each HCFKEL repeat abolished complementation. Although complementation appears to be an all-or-none event, subcloning reveals small differences in the relative growth rates of rescued colonies, presumably reflecting differences in the capability of individual mutations to promote G1 progression (data not shown).
FIG. 8.
K105D, EWK289A3, and RK344A2 complement the tsBN67 cell proliferation defect. (A) Hamster tsBN67 cells were stably transfected with 1 μg of pCGNHCF-1N902 wild type, P134S, K105D, EWK298A2, and RK344A2. Following transfection, cells were incubated at 39.5°C with G418 for 2 weeks, and proliferating (rescued) colonies were stained with 0.5% crystal violet. (B) Quantitation of the number of rescued colonies after 2 weeks of selection at the nonpermissive temperature. Numbers are expressed relative to the value for wild-type HCF-1 (pCGNHCF-1N902).
The failure of P252S to support tsBN67 cell proliferation is especially interesting because this mutation has a relatively minor effect on interaction with VP16 and LZIP (∼80 and 60%, respectively, of the wild-type level [Fig. 7]). It is useful to contrast P252S with S338A, which showed a greater reduction in association with VP16 and LZIP (<45% activity [Fig. 7]) and yet was sufficient to complement the tsBN67 growth defect. These results show that the ability to interact with VP16 serves as a poor indicator of the ability to support cell proliferation. Perhaps most striking outcome of this analysis was the behavior of EWK289A3. Although this mutant was severely compromised for interaction with LZIP (Fig. 7), it was fully sufficient to rescue tsBN67 cell growth. This results argues that interaction with LZIP is not in itself required for HCF-dependent cell proliferation and points to the existence of other cellular targets for the HCF-1 β-propeller.
DISCUSSION
In this study, we have used mutagenesis to probe the structure of the HCF-1 β-propeller domain and have identified a number of mutants that disrupt the three known functions of the domain: VIC assembly, recruitment of HCF-1 to the cellular transcription factor LZIP, and complementation of the tsBN67 proliferation defect. The properties of each mutant (summarized in Table 1) can be grouped into three functional categories: inactive, partially active, and fully active. Eight of the mutants (P134S, P197S, P319S, R137D, R200D, R255D, R228D, and R322D), including the previously characterized tsBN67 mutant (P134S), were inactive for all three functions. This may indicate a shared role for each of these evolutionarily conserved residues or that these substitutions simply bring about a global change in domain structure. Separating these two options will require further analysis; however, the fact that all of these mutants were expressed at near-wild-type levels and were equally soluble argues against a global defect in folding of the β-propeller.
TABLE 1.
Phenotypes associated with amino acid substitutions in the β-propeller domain of HCF-1
| Construct | VIC formation | VP16a association | VP16b association | LZIPb association | tsBN67 rescue |
|---|---|---|---|---|---|
| Wild type | ++++ | ++++ | ++++ | ++++ | ++++ |
| P30S | −/+ | ++ | − | − | − |
| P79S | + | ++ | ++ | − | − |
| P134S | − | − | − | − | − |
| P197S | − | − | − | − | − |
| P252S | +++ | +++ | +++ | +++ | − |
| P319S | − | −/+ | − | − | − |
| C82D | + | + | + | −/+ | − |
| R137D | − | − | NDc | ND | − |
| R200D | − | − | ND | ND | − |
| R255D | − | − | ND | ND | − |
| R322D | − | − | ND | ND | − |
| K105D | +++ | ++++ | +++ | ++++ | ++++ |
| R228D | − | − | −/+ | − | − |
| EWK289A3 | ++++ | ++++ | ++++ | −/+ | ++++ |
| RK344A2 | − | ++ | ++ | +++ | ++++ |
| S338A | +++ | +++ | ++ | ++ | +++ |
Determined by coimmunoprecipitation.
Determined by cell-based recruitment assay.
ND, not determined.
Of the remaining eight mutants, two (K105D and S338A) showed little or no phenotype, while six (P30S, P79S, C82D, P252S, EWK289A2, and RK344A2) showed differential effects on each of the functions tested. This latter class of mutants is likely to be the most informative. Four mutants (P79S, C82D, EWK289A3, and RK344A2) showed significant differences in the ability to interact with LZIP compared to VP16. This was most striking in EWK289A3, which was similar to wild type for association with VP16 but inactive for association with LZIP. These results point to an important role for residues flanking the core HBM tetrapeptide. It should be noted that while mutagenesis of VP16 implicates three aspartic acids (residues 385 to 387) on the carboxy-terminal side of the HBM (361 to 364) in HCF-1 binding (24), both LZIP and its Drosophila homologue dCREB-A/BBF-2 lack an equivalent acidic stretch.
Association of HCF-1 with VP16 is not sufficient for VIC formation.
HCF-1's role in VIC formation is poorly understood. HCF-1 is not essential for specific recognition of Oct-1 or the TAATGARAT element by high levels of VP16 in vitro (38, 44) but is required for VIC formation at lower protein concentrations and in vivo (8, 24). Deletion and mutagenesis studies of VP16 have mapped the primary determinants for interaction with Oct-1, HCF-1, and the TAATGARAT element to a small region between residues 331 and 391 of VP16 (9, 11, 24, 38). Although this region is unstructured in the VP16 crystal structure, it is thought that part of this loop may fold as an amphipathic α-helix, capable of packing against helices 1 and 2 on the exposed surface of the Oct-1 homeodomain (25, 26). HCF-1 may facilitate the Oct-1-VP16 interaction by stabilizing this α-helix (24). The predicted α-helix (VP16 residues 376 to 387) lies between the HBM and three aspartic acid residues implicated in HCF-1 association (24). By clasping each end of the recognition α-helix or by constraining the loop within a narrow groove, it seems reasonable to imagine that HCF-1 stabilizes the more ordered conformation and thus facilitates VIC formation.
Studies of other β-propeller proteins provide precedent for the idea that β-propeller domains serve as docking sites for flexible arms that extend from unrelated proteins. In the targeting of membrane proteins to coated pits, adapter molecules such as β-arrestin and arrestin 3 use a short unstructured peptide to interact with the seven-bladed β-propeller of the clathrin terminal domain (20, 40). The flexible arm of arrestin uses three hydrophobic residues and several acidic residues to interact with clathrin, and this is reminiscent of the conserved acidic and hydrophobic residues of the HCF-binding tetrapeptide. The arrestin peptide itself interacts with hydrophobic and positively charged residues lining a shallow groove formed by blades 1 and 2 of the clathrin β-propeller (7, 40). This contrasts with our analysis of HCF-1, which implicated all six blades of the β-propeller. Thus, the disordered loop presented by VP16 and LZIP may fit into a centrally located groove involving residues from each of the HCFKEL repeats. Alternatively, it may be that some of the mutations tested in this study affect the overall structure of the domain and thus indirectly influence a more limited functional surface. Indeed, exchanges between the β-propeller domains of HCF-1 and HCF-2 highlight the most divergent repeat, HCFKEL5, as the primary determinant for specific recognition of LZIP and VP16 (14).
Using in vitro-translated proteins, we have shown that an HA-tagged version of the HCF-1 β-propeller can immunoprecipitate full-length Oct-1. This association is sensitive to the RK344A2 mutation in HCF-1, suggesting that perhaps the mutant fails to support VP16-induced complex formation by preventing efficient recruitment of Oct-1. Based on the current data, we cannot say whether the association is direct or is mediated by other components present in the reticulocyte lysate. However, we did find that Oct-1 was not coprecipitated in the presence of the DNA-intercalating agent ethidium bromide (data not shown), suggesting that DNA fragments in the lysate might contribute in some way to the association.
The β-propeller as a molecular work surface.
β-Propeller proteins perform a remarkable variety of functions (reviewed in reference 37). Some have enzymatic activity, while others serve as scaffolds upon which protein-protein interactions are built. Perhaps as a result of this inherent flexibility, β-propeller-containing proteins are implicated in such diverse functions as protein trafficking (40), signal transduction (6), regulation of chromatin structure (30), transcriptional activation, and transcriptional repression (4, 15, 17). One emerging theme that unites these different examples is the ability of a single β-propeller domain to associate with a variety of target molecules (17, 33, 40). Because the β-propeller fold does not require a strict sequence pattern, it may be particularly amenable to the evolution of shallow grooves capable of trapping solvent-exposed flexible arms that are presented by partner proteins. At the same time, the compact nature of the domain provides an opportunity for regulation, either by steric hindrance or through the ability to bring prospective partners into close proximity to each other. For instance, a number of different regulatory cofactors interact with the β-propeller subunit of heterotrimeric G proteins and use steric hindrance to prevent the GDP-binding Gα subunit from accessing its binding site on the β-propeller (3, 6). The HCFVIC domain provides a good example of how the compact architecture of the β-propeller fold can be used to promote protein-protein interactions. Thus, HCF-1 is able to regulate viral immediate-early gene transcription, and ultimately the viral life cycle, by simply enabling VP16 and Oct-1 to come together on the TAATGARAT element.
The HCF-1 β-propeller serves as an interaction site for multiple cellular components.
The results presented in this study argue that HCF-1 does not need to recruit LZIP in order to promote G1 progression. This is best illustrated by the behavior of mutants P252S and EWK289A3. Although the P252S mutation can still interact with LZIP (∼80% of the wild-type level [Fig. 7]), it is unable to overcome the tsBN67 block to cell proliferation. One interpretation of this result is that the P252S mutation effects an undescribed interaction with another cellular component necessary for cell cycle progression. The behavior of EWK289A3 also brings into question the relevance of LZIP to understanding HCF-dependent proliferation: this mutant is able to complement the tsBN67 proliferation defect and yet appears to be severely impaired for association with LZIP. This observation argues that the HCF-1–LZIP interaction must not be essential for tsBN67 cell growth. While it is conceivable that on natural target promoters the weakened association between EWK289A3 and LZIP is stabilized through additional protein-protein or protein-DNA interactions, we favor the hypothesis that the cell cycle arrest reflects the disruption of an interaction between the HCF-1 β-propeller and an additional cellular protein. To this end, we have recently identified an unrelated cellular polypeptide that interacts with the HCF-1 β-propeller mutants in a manner more closely paralleling the results of the tsBN67 complementation assay. Specifically, the new candidate interacts in a yeast-based assay with EWK289A3 and RK344A2 but not with P252S or P134S (S. S. Mahajan, M. D. Little, and A. C. Wilson, unpublished data). Its role in promoting cell proliferation is currently under investigation.
ACKNOWLEDGMENTS
We thank Michael Garabedian, Richard Freiman, and Naoko Tanese for thoughtful comments on the manuscript; we also thank Muktar Mahajan and Kristy Johnson for help with the mutagenesis.
This work was supported by a development award from the Kaplan Comprehensive Cancer Center and an institutional award from the American Cancer Society (IRG-14-39).
REFERENCES
- 1.Abel T, Bhatt R, Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Doolittle R F. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 3.Clapham D E. The G-protein nanomachine. Nature. 1996;379:297–299. doi: 10.1038/379297a0. [DOI] [PubMed] [Google Scholar]
- 4.Dynlacht B D, Weinzierl R O, Admon A, Tjian R. The dTAFII80 subunit of Drosophila TFIID contains beta-transducin repeats. Nature. 1993;363:176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- 5.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudet R, Bohm A, Sigler P B. Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 7.Goodman O B, Jr, Krupnick J G, Gurevich V V, Benovic J L, Keen J H. Arrestin/clathrin interaction. Localization of the arrestin binding locus to the clathrin terminal domain. J Biol Chem. 1997;272:15017–15022. doi: 10.1074/jbc.272.23.15017. [DOI] [PubMed] [Google Scholar]
- 8.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 9.Greaves R F, O'Hare P. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J Virol. 1990;64:2716–2724. doi: 10.1128/jvi.64.6.2716-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigh A, Greaves R, O'Hare P. Interference with the assembly of a virus-host transcription complex by peptide competition. Nature. 1990;344:257–259. doi: 10.1038/344257a0. [DOI] [PubMed] [Google Scholar]
- 11.Hayes S, O'Hare P. Mapping of a major surface-exposed site in herpes simplex virus protein Vmw65 to a region of direct interaction in a transcription complex assembly. J Virol. 1993;67:852–862. doi: 10.1128/jvi.67.2.852-862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C C, Herr W. Differential control of transcription by homologous homeodomain coregulator. Mol Cell Biol. 1996;16:2967–2976. doi: 10.1128/mcb.16.6.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes T A, La Boissiere S, O'Hare P. Analysis of functional domains of the host cell factor involved in VP16 complex formation. J Biol Chem. 1999;274:16437–16443. doi: 10.1074/jbc.274.23.16437. [DOI] [PubMed] [Google Scholar]
- 14.Johnson K M, Mahajan S S, Wilson A C. Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J Virol. 1999;73:3930–3940. doi: 10.1128/jvi.73.5.3930-3940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnstone R W, Wang J, Tommerup N, Vissing H, Roberts T, Shi Y. Ciao 1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J Biol Chem. 1998;273:10880–10887. doi: 10.1074/jbc.273.18.10880. [DOI] [PubMed] [Google Scholar]
- 16.Klemm R D, Goodrich J A, Zhou S, Tjian R. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc Natl Acad Sci USA. 1995;92:5788–5792. doi: 10.1073/pnas.92.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komachi K, Johnson A D. Residues in the WD repeats of Tup1 required for interaction with α2. Mol Cell Biol. 1997;17:6023–6028. doi: 10.1128/mcb.17.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein-DNA complexes with the HSV αTIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 20.Krupnick J G, Goodman O B, Jr, Keen J H, Benovic J L. Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J Biol Chem. 1997;272:15011–15016. doi: 10.1074/jbc.272.23.15011. [DOI] [PubMed] [Google Scholar]
- 21.La Boissiere S, Hughes T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaBoissiere S, Walker S, O'Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai J-S, Cleary M A, Herr W. A single amino acid exchange transfers VP16-induced positive control from Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 24.Lai J-S, Herr W. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol Cell Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Stark M R, Johnson A D, Wolberger C. Crystal structure of the MATa1/MATα2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Gong W, Huang C C, Herr W, Cheng X. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 1999;13:1692–1703. doi: 10.1101/gad.13.13.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Hengartner M O, Herr W. Selected elements of herpes simplex virus accessory factor HCF are highly conserved in Caenorhabditis elegans. Mol Cell Biol. 1999;19:909–915. doi: 10.1128/mcb.19.1.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cell factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, Luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Balbas M A, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 32.Pomerantz J L, Kristie T M, Sharp P A. Recognition of the surface of a homeo domain protein. Genes Dev. 1992;6:2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- 33.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 34.Simmen K A, Newell A, Robinson M, Mills J S, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. Protein interactions in the herpes simplex type 1 VP16 induced complex: VP16 peptide inhibition and mutational analysis of the host cell factor requirements. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolik S M, Rose R E, Goodman R H. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 37.Springer T A. Folding of the N-terminal ligand-binding region of integrin α subunits into a β-propeller domain. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stern S, Herr W. The herpes simplex virus trans-activator VP16 recognizes the Oct-1 homeo domain: evidence for a homeo domain recognition subdomain. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 39.Stern S, Tanaka M, Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 40.ter Haar E, Musacchio A, Harrison S C, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson C C, McKnight S L. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 42.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 43.Walker S, Greaves R, O'Hare P. Transcriptional activation by the acidic domain of Vmw65 requires the intergrity of the domain and additional determinants distinct from those necessary for TFIIB binding. Mol Cell Biol. 1993;13:5233–5244. doi: 10.1128/mcb.13.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker S, Hayes S, O'Hare P. Site-specific conformational alteration of the Oct-1 POU domain-DNA complex as the basis for differential recognition by Vmw65 (VP16) Cell. 1994;79:841–852. doi: 10.1016/0092-8674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 45.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein heterotrimer Giα1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 46.Wilson A C, Cleary M A, Lai J-S, LaMarco K, Peterson M G, Herr W. Combinatorial control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- 47.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell-cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 49.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 50.Wu T J, Monokian G, Mark D F, Wobbe C R. Transcriptional activation by herpes simplex virus type 1 VP16 in vitro and its inhibition by oligopeptides. Mol Cell Biol. 1994;14:3484–3493. doi: 10.1128/mcb.14.5.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]