Abstract
Background: Idiopathic pulmonary fibrosis (IPF) is a chronic progressive disease with unknown etiology and unfavorable prognosis. Ferroptosis is a form of regulated cell death with an iron-dependent way that is involved in the development of various diseases. Whereas the prognostic value of ferroptosis-related genes (FRGs) in IPF remains uncertain and needs to be further elucidated.
Methods: The FerrDb database and the previous studies were screened to explore the FRGs. The data of patients with IPF were obtained from the GSE70866 dataset. Wilcoxon's test and univariate Cox regression analysis were applied to identify the FRGs that are differentially expressed between normal and patients with IPF and associated with prognosis. Next, a multigene signature was constructed by the least absolute shrinkage and selection operator (LASSO)-penalized Cox model in the training cohort and evaluated by using calibration and receiver operating characteristic (ROC) curves. Then, 30% of the dataset samples were randomly selected for internal validation. Finally, the potential function and pathways that might be affected by the risk score-related differently expressed genes (DEGs) were further explored.
Results: A total of 183 FRGs were identified by the FerrDb database and the previous studies, and 19 of them were differentially expressed in bronchoalveolar lavage fluid (BALF) between IPF and healthy controls and associated with prognosis (p < 0.05). There were five FRGs (aconitase 1 [ACO1], neuroblastoma RAS viral (v-ras) oncogene homolog [NRAS], Ectonucleotide pyrophosphatase/phosphodiesterase 2 [ENPP2], Mucin 1 [MUC1], and ZFP36 ring finger protein [ZFP36]) identified as risk signatures and stratified patients with IPF into the two risk groups. The overall survival rate in patients with high risk was significantly lower than that in patients with low risk (p < 0.001). The calibration and ROC curve analysis confirmed the predictive capacity of this signature, and the results were further verified in the validation group. Risk score-related DEGs were found enriched in ECM-receptor interaction and focal adhesion pathways.
Conclusion: The five FRGs in BALF can be used for prognostic prediction in IPF, which may contribute to improving the management strategies of IPF.
Keywords: idiopathic pulmonary fibrosis, ferroptosis, prognostic, signature, model
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, age-related interstitial pulmonary disease (ILD) with unknown etiology, significant morbidity, and unfavorable survival (1, 2). The prevalence of IPF is on the rise worldwide and about 40,000 new cases of IPF are diagnosed per year in Europe alone (3, 4). The life expectancy of patients with IPF is similar to that of patients with lung cancer, with median survival rates of 50 and 20% at 3- and 5-years after diagnosis, respectively (5, 6), which imposed a substantial socio-economic burden on the world (7). There was no medicine authorized for this devastating disease before 2010. Two medications, nintedanib and pirfenidone, have been proven to be safe and effective in slowing disease progression in recent years (8). Despite the advances in treatment technology, there is still no cure for IPF, and neither nintedanib nor pirfenidone improves the pulmonary function and living quality, and both the therapies have tolerability issues (9, 10). In addition, IPF was found at increased risk of severe coronavirus disease 2019 (COVID-19) due to sharing the same risk factors for severe COVID-19 (11), and as an independent risk for the development of lung cancer (12). Hence, as a devastating disease with unknown etiology and difficult treatment, and the risk factor of other fatal diseases, there is an additional need to investigate the novel biomarkers for its diagnosis, treatment, and prognosis.
Ferroptosis, a newly identified form of iron-dependent cell death unlike other cell death forms, is primarily driven by the iron excessive accumulation, lipid peroxidation, and subsequent plasma membrane rupture, which ultimately caused cell death and is often accompanied by the disturbance of lipid and iron metabolism (13). Ferroptosis has been verified to play a crucial part in the development and disease of organisms (14). Meanwhile, many genes have also been defined as markers or regulatory factors of ferroptosis [also known as ferroptosis-related genes (FRGs)] and are involved in the occurrence and development of various benign and malignant lung diseases, such as chronic obstructive pulmonary disease (COPD) (15), acute lung injury (16), asthma (17), and lung cancer (18). It is worth noting that the disturbance of certain molecular mechanisms related to metabolism and fibrosis, such as oxidative stress, lipid, and iron metabolism, in the bronchoalveolar lavage fluid (BALF) of IPF patients have been investigated in several studies recently (19–21). Elevated iron levels have been detected in the lung tissue of patients with IPF, and iron accumulation is associated with significant increases in airway fibrosis and altered pulmonary function (22). In addition, increased numbers of iron-laden macrophage clusters in BALF enable cause macrophages to produce reactive oxygen species (ROS) and result in lipid peroxidation in IPF (23). These findings indicated the potential role of ferroptosis in the pathogenesis of IPF due to the disturbance of lipid and iron metabolism contributes to the occurrence of ferroptosis. Li X et al. further elucidated that the radiation-induced lung fibrosis was significantly alleviated by ferroptosis inhibitor (24). Erastin, an inducer of ferroptosis, has been verified to promote the main pathogenesis of IPF called fibroblast-to-myofibroblast differentiation, and this effect can be suppressed by the Fer-1 (an inhibitor of ferroptosis) (25, 26). In addition, Tsubouchi K demonstrated that lipid peroxidation regulated by GPX4 is involved in the pathogenesis of IPF (27). However, no studies have been conducted on the relationship between ferroptosis and the prognosis of IPF, and the prognostic value of FRGs in IPF remains largely unknown.
Herein, we aimed to investigate whether FRGs in BALF are associated with the prognosis of IPF. First, we evaluated the expression levels of FRGs in BALF of IPF patients by downloading the mRNA expression profiles and corresponding clinical data from the Gene Expression Omnibus (GEO). Subsequently, a multigene prognostic model with FRGs was constructed and validated in the training and testing cohort, respectively. Finally, a functional enrichment analysis was performed to investigate the potential biological functions and signaling pathways affected by these risk-related differently expressed genes (DEGs). We hope to find some biomarkers that can contribute to the diagnosis, treatment, or prognosis of IPF in these ways.
Materials and Methods
Exploration of FRGs
The FerrDb database (http://www.zhounan.org/ferrdb/) is the first ferroptosis-related database that integrates the latest makers and regulators of ferroptosis and ferroptosis-disease associations (28). A total of 183 FRGs were identified by the Driver, Suppressors, and Marker model of the FerrDb database and the previous studies (14, 29–31). The details of FRGs are shown in Supplementary Table 1.
Data Collection
The GSE70866 dataset is the only one that simultaneously includes the prognostic data and mRNA expression profiles in BALF of IPF (20 healthy controls and 176 IPF patients), which were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) and normalized using the R “limma” package. This study does not need to be granted by the local ethics committee as the data of GEO are publicly available, and we strictly adhere to the data access policies and publication guidelines of the GEO. The clinical information of the GSE 70866 cohort is shown in Supplementary Table 2.
Identification of FRGs
The mRNA expression profiles of FRGs were extracted and the DEGs of FRGs in BALF between normal and IPF were identified by the “limma” package of R with the criteria of a false discovery rate (FDR) <0.05 and P < 0.05. The results were visualized by the “heatmap” package.
Identification of the Prognostic-Related FRGs
A univariate Cox regression of the “survival” package was conducted to investigate the FRGs associated with the prognosis of IPF in the GEO database. The value of p < 0.05 was confirmed as the filtering criteria. The results were visualized by the forest map. Next, a Venn diagram was performed to identify the intersection of DEGs of FRGs and prognostic FRGs.
Protein–Protein Interaction (PPI) Network Construction
The protein–protein interaction (PPI) network for the overlapping genes was constructed using the STRING database (https://www.string-db.org) (32) to explore the interactions between these genes.
Construction and Internal Validation of the Prognostic FRGs Signature
The patients with IPF from the GSE70866 dataset were randomly separated into two groups at a ratio of 7:3, the former for the training group and the latter for internal validation. The least absolute shrinkage and selection operator (LASSO)-penalized Cox regression analysis was utilized in the present study to establish a prognostic model in the training group to minimize the risk of overfitting by using the “glmnet” package of R (33, 34). First, the range of 19 FRGs associated with prognosis was narrowed through 10-fold cross-validation by using the LASSO algorithm with penalty parameter adjustment, and the datasets were sub-sampled 1,000 times and the FRGs repeated >900 times were chosen for the next analysis. Then, eight FRGs with non-zero regression coefficients were retained for the multivariate Cox regression analysis. Finally, the risk scores of the patients were calculated according to the normalized expression levels and the corresponding regression coefficients of the five FRGs, and patients with IPF in the training group were classified into high- and low-risk groups depending on the median risk score.
The “rms” package of R was utilized to visualize the risk score in the nomogram and generate the calibration curve to assess the prediction accuracy of the nomogram. Principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) were carried out with the “Rtsne” and “ggplot2” R package to explore the distribution of the two risk groups. Then, the “survival,” “suvminer,” and “timeROC” R package were utilized to generate Kaplan–Meier (K–M) survival and time-dependent receiver operational characteristic (ROC) curves to further evaluate the predictive capacity of the prognostic model, respectively. Additionally, the “heatmap” package was performed to visualize the gene expression levels in each sample in the two risk groups. Subsequently, the “survival” package was performed to evaluate whether clinical parameters and risk scores are independent prognostic factors for overall survival (OS). Finally, the results were further identified in the internal validation group.
Functional Enrichment Analysis
The patients with IPF were classified as the high- and low-risk group base on the median risk score in the training and test cohort, and the risk-related DEGs between the two risk groups were screened out as much as possible with FDR <0.05 and |log2FC| > 0.3 in the two cohorts by using the “limma” package. Then, Gene Ontology (GO) as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed by the “clusterProfiler” package to explore the biological functions and signaling pathways affected by the risk-related DEGs with the criteria of FDR <0.05 and p < 0.05 (35).
Expression of the Prognostic Model FRGs in the Lung Tissue of IPF
The datasets containing the mRNA expression profiles of IPF lung tissue were retrieved from the GEO database. Four datasets (GSE110147, GSE72073, GSE53845, and GSE24206) with a total of 28 healthy control and 84 patients with IPF were selected for analyzing and visualizing the expression of the prognostic model FRGs between the tissue of normal and IPF by using the “limma” and “beeswarm” package of R. The value of p < 0.05 was considered statistically significant.
Statistical Analysis
The Perl, R (4.0.1), and SPSS 18.0 were used for data processing and statistical analysis. The Wilcoxon and univariate Cox regression tests were utilized to evaluate the expression levels of FRGs in BALF between IPF and healthy controls and identified the genes associated with prognosis, respectively. A prognostic model was generated by the LASSO-penalized Cox regression analysis and further evaluated the predictive capacity of which by K–M survival and ROC curve analysis. The univariate and multivariate Cox regression analysis were utilized to identify independent predictors of OS. The probability p < 0.05 was considered significant.
Results
Identification of FRGs in IPF
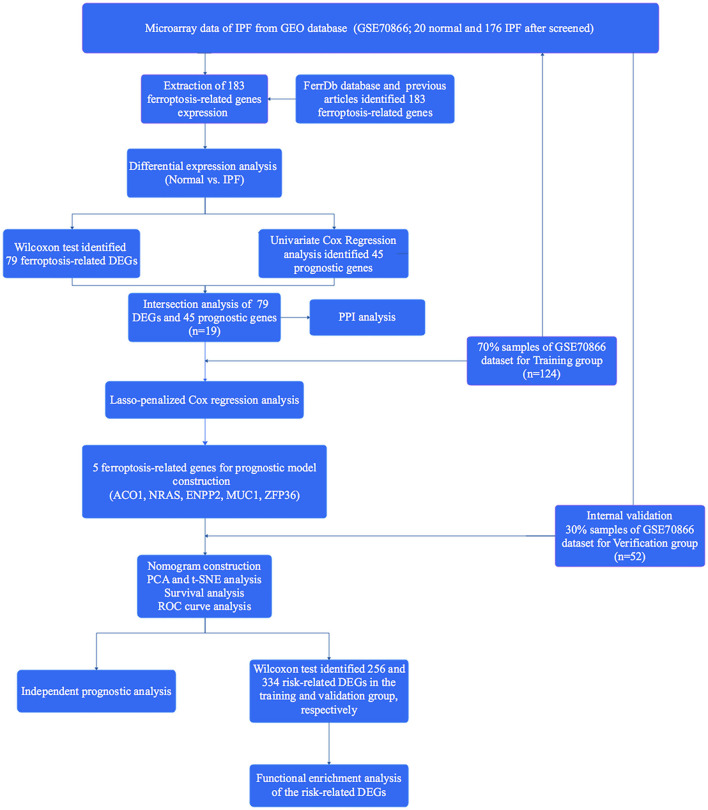
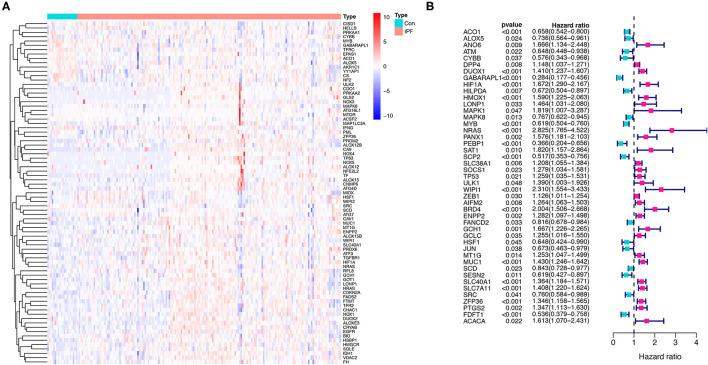
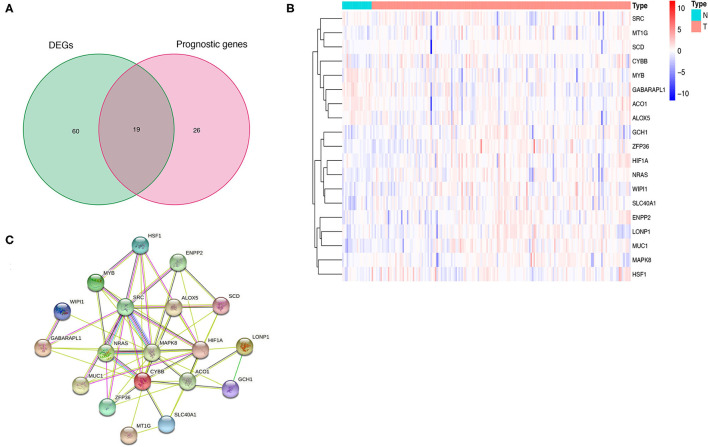
A total of 176 patients with IPF and 20 healthy controls from the cohort of GSE70866 dataset were enrolled in this study, and 183 FRGs were explored from the FerrDb database and the literature report (Supplementary Table 1). The detailed information of the candidate patients is presented in Supplementary Table 2. Figure 1 illustrates the flow of our research. In the present study, we found that 79 FRGs (43.17%) were significantly differentially expressed in BALF between IPF and normal population (Figure 2A), and 45 (24.59%) FRGs were associated with OS of IPF (Figure 2B). In addition, 19 ferroptosis-related prognostic DEGs were identified using the Venn diagram (all P < 0.05, Figures 3A,B).
Figure 1.
Flow diagram of the present study.
Figure 2.
Identification of the FRGs in the patients with IPF from the Gene Expression Omnibus (GEO) database. (A) The expression of the 79 ferroptosis-related DEGs in IPF. (B) The 45 ferroptosis-related prognostic genes in IPF. p < 0.05 was considered significant. FRGs, ferroptosis-related genes; IPF, idiopathic pulmonary fibrosis; and DEGs, differentially expressed genes.
Figure 3.
Identification and expression of the overlapping genes between DEGs and prognostic genes in IPF. (A) Venn diagram of 19 overlapping genes. (B) Expression of the 19 overlapping genes in IPF. (C) The correlation network of the overlapping genes. IPF, idiopathic pulmonary fibrosis; DEGs, differentially expressed genes; PPI, protein–protein interaction.
PPI Network Construction Based on Prognostic FRGs
The PPI network based on the 19 prognostic ferroptosis-related DEGs was generated by the STRING online platform (Figure 3C), which demonstrated that there is a significant correlation between these FRGs in BALF of IPF.
Construction of a Prognostic Model in the Training Group
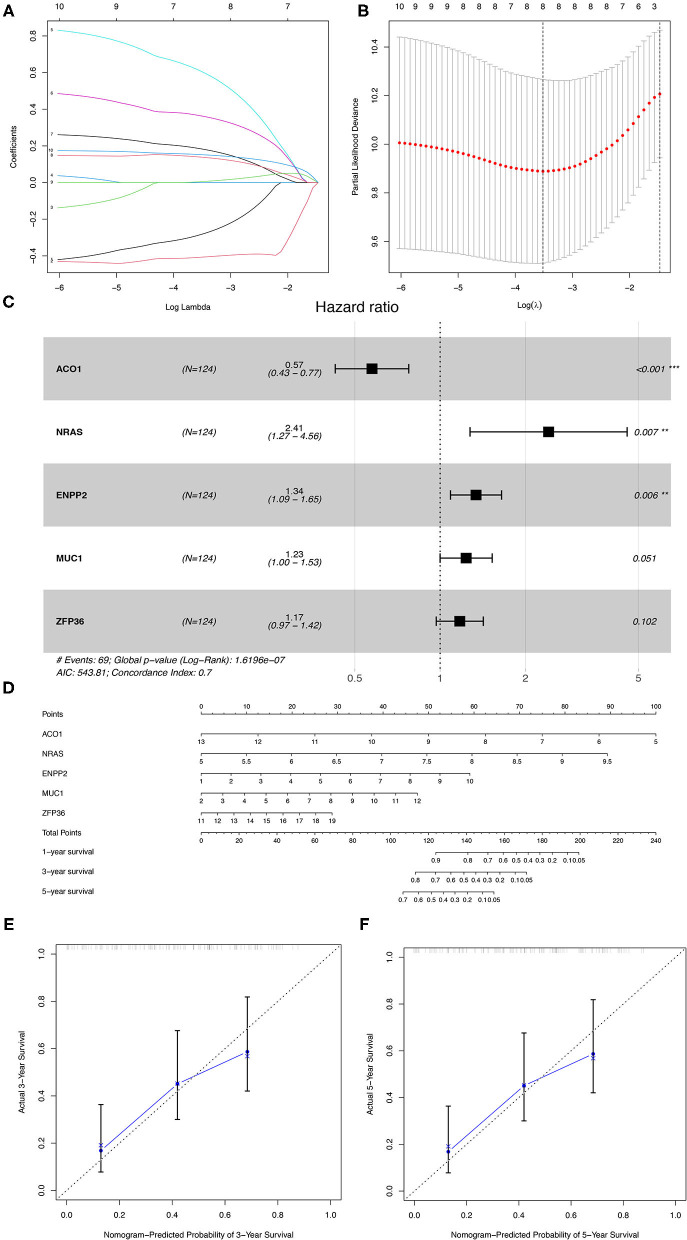
The 176 patients with IPF in the GSE70866 dataset were randomly categorized into training and testing cohorts at a ratio of 7:3, and the LASSO-penalized Cox regression analysis was used to set up a prognostic signature in the training group. The result showed that only five genes (aconitase 1 [ACO1], neuroblastoma RAS viral (v-ras) oncogene homolog [NRAS], ectonucleotide pyrophosphatase/phosphodiesterase 2 [ENPP2], mucin 1 [MUC1], and ZFP36 ring finger protein [ZFP36]) remained in LASSO regression from the 19 prognostic FRGs (Figures 4A–C), among which, NRAS, ENPP2, MUC1, and ZFP36 are the risk factors, and ACO1 is a protective factor for IPF prognosis (Figure 4C, Table 1). Then, the nomogram of five FRGs for predicting 1-, 3-, and 5-year OS of IPF was visualized in Figure 4D. At the same time, the calibration curves showed satisfactory agreement between the predicted and observed values at the probabilities of 3- and 5-year survival (Figures 4E,F). These results revealed a good accuracy of the nomogram in predicting the 3- or 5-year survival in patients with IPF.
Figure 4.
The construction of the prognostic model of IPF in the training group. (A) LASSO coefficients profiles of the 19 overlapping genes. (B) LASSO regression with 10-fold cross-validation obtained eight prognostic FRGs. (C) The multivariate Cox regression analysis identified five prognostic FRGs for prognostic model construction. (D) The nomogram of the prognostic model based on the five FRGs. The calibration curves of the nomogram for predicting 3- (E) and 5-years survival (F) of the patients with IPF. IPF, idiopathic pulmonary fibrosis; FRGs, ferroptosis-related genes. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 1.
Multivariate Cox regression analysis of the five FRGs.
| Gene | Description | coef | HR(95%CI) | P-value |
|---|---|---|---|---|
| ACO1 | Aconitase 1, soluble | −0.553 | 0.575 (0.427,0.775) | <0.001 |
| NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | 0.880 | 2.412(1.275,4.564) | 0.007 |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | 0.291 | 1.338(1.087,1.646) | 0.006 |
| MUC1 | Mucin 1, cell surface associated | 0.211 | 1.235(0.999,1.526) | 0.051 |
| ZFP36 | ZFP36 ring finger protein | 0.159 | 1.173 (0.969,1.420) | 0.102 |
FRGs, ferroptosis-related genes; coef, regression coefficients; HR, hazard ratio; 95% CI, 95% confidence interval.
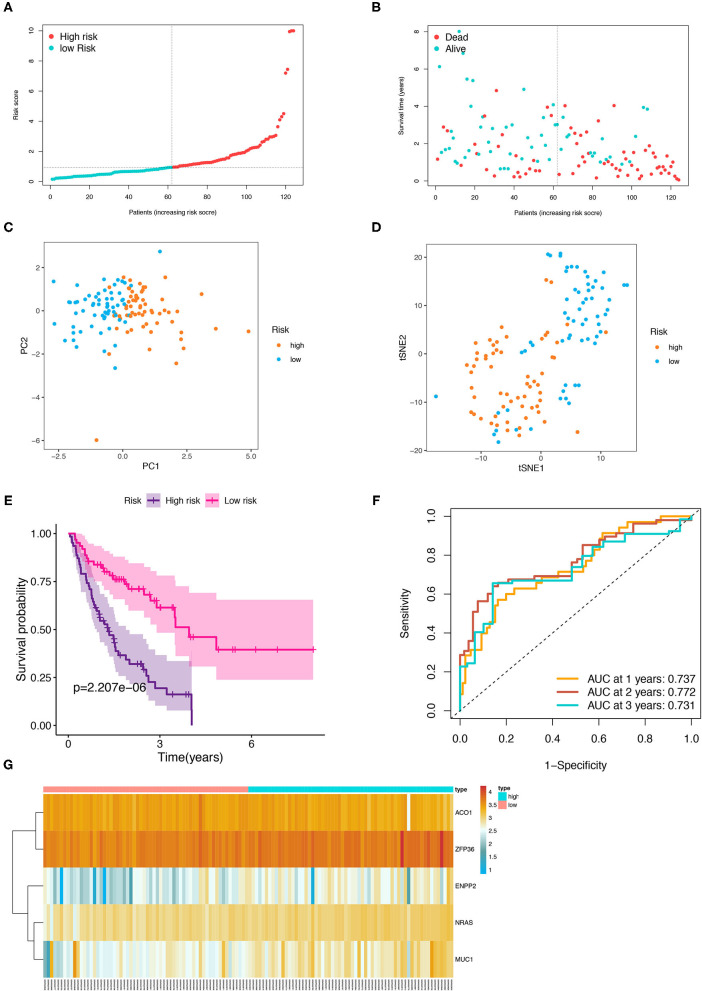
Then, the patients with IPF in the training cohort were categorized into a high- and low-risk group according to the median risk score (Figure 5A), and the high-risk group was demonstrated to be more likely to encounter death earlier (Figure 5B). At the same time, the patients in both subgroups were demonstrated to be distributed in a discrete direction by PCA and t-SNE analysis (Figures 5C,D). Subsequently, the predictive power of the prognostic model was further evaluated by the survival and a ROC curve analysis. The K–M plot revealed that the patients with IPF in the high-risk group had a worse prognosis than those in the low-risk group (p < 0.001, Figure 5E). In addition, the time-dependent ROC curve analysis elucidated that the 1-, 2-, and 3-year area under the curve (AUC) were 0.737, 0.772, and 0.731, respectively (Figure 5F). The expression levels of the five FRGs in each sample between the two risk groups are presented in Figure 5G. In addition, three of the five FRGs (NRAS, MUC1, and ZFP36) were also found differentially expressed between the normal and IPF tissue in the combination of the four GEO datasets (Supplementary Figure 1, p < 0.05).
Figure 5.
Prognostic analysis of the five-FRGs signature in the training group. Distribution of IPF samples (A) and OS status (B) based on the median risk score. PCA (C) and t-SNE (D) analysis of the training group. Kaplan–Meier survival (E) and time-dependent ROC curve (F) analysis of the five-FRGs signature in training group. (G) Heatmap of the expression profiles of the five-FRGs in low- and high-risk groups. FRGs, ferroptosis-related genes; IPF, idiopathic pulmonary fibrosis; OS, overall survival; PCA, principal component analysis; t-SNE, t-distributed stochastic neighbor embedding; ROC, receiver operating characteristic.
Validation of the Prognostic Signature in the Testing Group
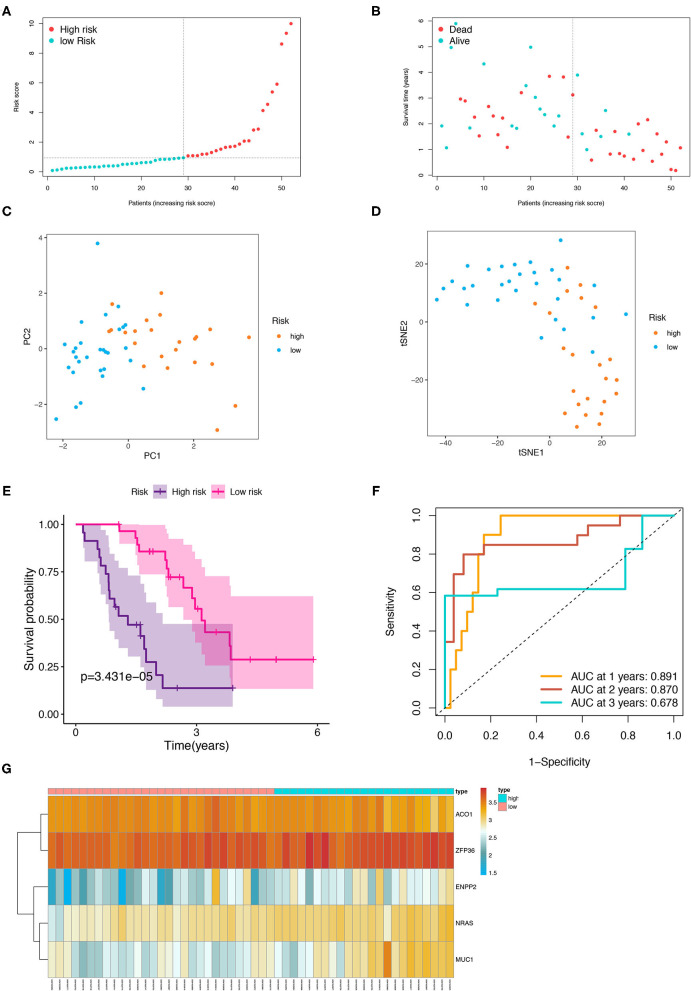
The patients from the testing cohort were also separated in the two risk groups based on the median risk score to evaluate the robustness of the prognostic model constructed in the training cohort (Figure 6A). Similar to the results of the training cohort, the patients with the high-risk tended to reach the end of life earlier in the validation group (Figure 6B). The PCA and t-SNE analysis showed that the distribution in the discrete direction of patients in the two subgroups was similar to that in the training group (Figures 6C,D). Meanwhile, a worse prognosis was also found in the patients with IPF with high-risk in the testing group (p < 0.001, Figure 6E), and an ROC curve also elucidated a satisfactory accuracy of the prognostic model to identify the 1-, 2-, and 3-year OS of IPF in the testing group (Figure 6F, AUC at 1 year: 0.891; 2 years: 0.870; and 3 years: 0.678). The heatmap showed that the expression trend of the five FRGs between the high- and low-risk samples of the testing group was similar to that of the training group (Figure 6G).
Figure 6.
Internal validation of the five-FRGs signature. Distribution of IPF samples (A) and OS status (B) based on the median value of the risk scores. PCA (C) and t-SNE (D) analysis of the testing group. Kaplan–Meier survival (E) and time-dependent ROC curve (F) analysis of the five-FRGs signature in the testing group. (G) Heatmap of the expression profiles of the five-FRGs in low- and high-risk groups of the testing cohort. FRGs, ferroptosis-related genes; IPF, idiopathic pulmonary fibrosis; OS, overall survival; PCA, principal component analysis; t-SNE, t-distributed stochastic neighbor embedding; ROC, receiver operating characteristic.
Independent Prognostic Analysis
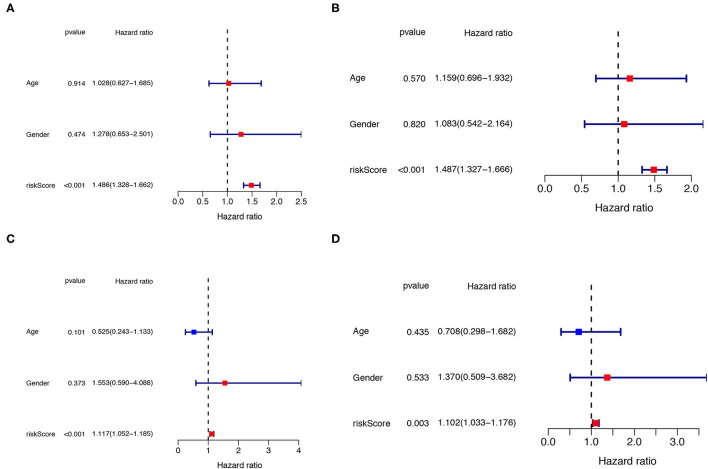
The univariate and multivariate Cox regression analyses were performed to determine whether the clinical parameters (such as, gender and age; the cut-off value of age is 65 years) and risk score are independent prognostic factors for OS in IPF. The results demonstrated that the risk score remained as an independent risk factor for the outcome of IPF in both the training (Figures 7A,B) and testing groups (Figures 7C,D) after controlling for gender and age.
Figure 7.
Independent prognostic analysis of clinical parameters and risk score. The univariate (A) and multivariate (B) Cox regression analysis of the associations between the risk score, the clinical parameters and OS of IPF patients in the training cohort. The univariate (C) and multivariate (D) Cox regression analysis of the associations between the risk score, clinical parameters and OS of IPF patients in the testing cohort. IPF, idiopathic pulmonary fibrosis; OS, overall survival.
Functional Enrichment Analysis
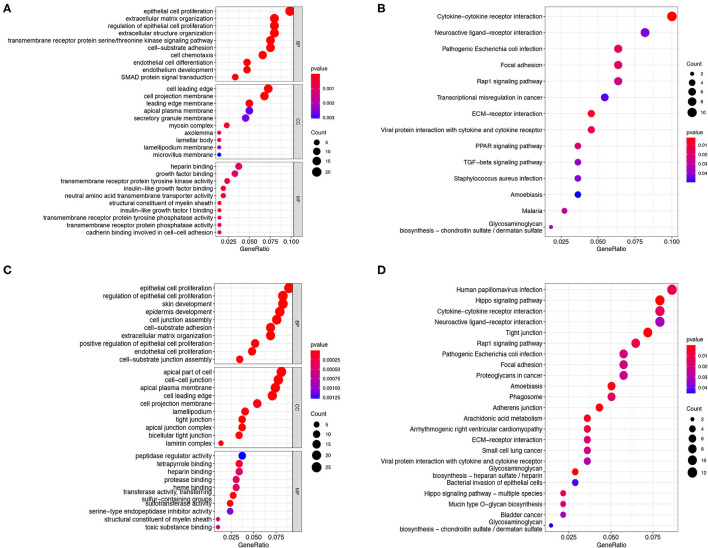
There were 256 and 334 risk-related DEGs identified in the training and validation group by using Wilcoxon's test, respectively (Supplementary Table 3). The GO and KEGG analysis on the risk-related DEGs were performed in the present study to further investigate the potential functions and pathways that are associated with the risk score. The results indicated that the risk-related DEGs mainly focused on epithelial cell proliferation, SMAD protein signal transduction, extracellular matrix organization, cell projection membrane, growth factor binding, cadherin binding involved in cell–cell adhesion, etc., in the training group (Figure 8A). In addition, except for some functions mentioned above, the risk-related DEGs were also found to be enriched in the regulation of cell-junction, tight junction, and protease binding in the validation group (Figure 8C). KEGG pathway analysis also indicated that the ECM-receptor interaction, Rap1 signaling pathway, focal adhesion, and cytokine-cytokine receptor interaction pathway were enriched in both the cohorts (Figures 8B,D). In addition, the transforming growth factor-beta (TGF-β) signaling pathway was also identified enriched in the training cohorts (Figure 8B).
Figure 8.
GO and KEGG analysis. The GO enrichment and KEGG pathways of the risk score-related DEGs in the training cohort (A,B) and testing cohort (C,D). GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genome; DEGs, differentially expressed genes.
Discussion
Idiopathic pulmonary fibrosis is a progressive and devastating disease with limited treatment options, unsatisfactory therapeutic effects, and poor prognosis (1, 2). Despite significant innovations in the treatment and diagnosis of IPF compared with the past, medication has not enhanced the survival rate. Early assessment of disease progression and prognosis, and timely appropriate treatment have important clinical significance due to the variable and unpredictable of the clinical course of IPF (36). Hence, there is a current and urgent need for reliable, safe, and feasible markers that can accurately predict prognosis, to make the management of IPF patients more accurate, personalized, and timely.
Ferroptosis is considered as an effective cancer treatment because it overcomes the resistance of malignant cells to chemotherapy and promotes the clearance of defective cells. Recently, various studies have illustrated that ferroptosis regulate the development of pulmonary fibrosis to some extent (24–27, 37), however, as the most common pathological type of pulmonary fibrosis, the correlation between ferroptosis and the OS of patients with IPF has not been investigated. In this study, we found that approximately half of FRGs (79/183, 43.17%) in BALF were differentially expressed between IPF and healthy controls, and 19 of them were correlated with the OS of the IPF patients. In addition, a novel prognostic model integrating the five FRGs was first established and verified in an internal cohort. Then, we further verified that the risk score is an independent prognostic factor for OS of IPF and revealed the potential function and pathways that are associated with the risk score-related DEGs by the functional enrichment analysis. The findings showed above strongly indicate a potentially important role of ferroptosis in IPF.
In our study, the predictive model was established based on the five FRGs (ACO1, NRAS, ENPP2, MUC1, and ZFP36). Cytosolic aconitase 1/iron regulatory protein 1 (ACO1/IRP1) is a bifunctional protein expressed in cytoplasm that performs its function as aconitase or regulating intracellular iron homeostasis according to iron availability (38, 39). Wang J et al. found that IRP1/ACO1 activated by nitric oxide enables impair cellular iron homeostasis and triggers iron accumulation during neuroinflammation, leading to neuronal death (40). In the present study, we elucidated that ACO1 is downregulated in patients with IPF and as a protective factor. NRAS (N-ras), a member of the RAS family, and commonly mutated in almost all human cancers (41). NRAS has been verified associated with the erastin resistance in hepatocellular carcinoma (42), however, whether ferroptosis pathway is involved in this effect needs to be further clarified. N-Ras was also found to promote renal fibrosis by participating in TGF-β1 induced proliferation and the synthesis of collagen and fibronectin (43), whereas the function of NRAS in IPF has not been studied. We found that the overexpressed NRAS is associated with the poor prognosis in IPF. ENPP2 encodes the protein autotaxin (ATX), an enzyme primarily responsible for the production of extracellular lysophosphatidic acid, has been illustrated to protect cardiomyocytes from ferroptosis induced by erastin (44). In contrast, ENPP2/ATX was found to be upregulated and promote the development of fibrosis in the BALF of pulmonary fibrosis patients (45). Similar to the results of the previous study (45), we found that ENPP2 is upregulated in the BALF of IPF and associated with poor prognosis. Hence, it may be doubtful whether ENPP2 is involved in the pathogenesis of IPF through regulating ferroptosis. Mucin 1 (MUC1) is a glycoprotein and abnormally overexpressed in most respiratory diseases, such as IPF (46), which was also found involved in the stabilization of the system X cystine/glutamate antiporter (xCT) complex, an important intracellular antioxidant element involved in the ferroptosis (47, 48) and in breast cancer cells (49). Herein we elucidated the potential risk role of FRGs MUC1 in the IPF prognosis. ZFP36 ring finger protein has two tandem repeat CCCH zinc fingers domains, which mainly regulate the fate of cytoplasmic mRNA and modulate the response of the cell to oxidative stress, apoptosis, and lipid peroxidation by affecting the post-transcriptional effects on specific target mRNAs (50, 51). Overexpression of ZFP36 was found to result in resistance to ferroptosis by triggering autophagy inactivation and blocking autophagy ferritin degradation (52). Interestingly, ZFP36 was identified as a specific and shared biological biomarker in COPD, asthma, and IPF (53), and was verified as a risk-predicting factor in our study. In summary, all the five FRGs were found to be upregulated in BALF of IPF and were correlated with unfavorable prognosis in our study except for ACO1. However, whether these genes play a role in the prognosis of patients with IPF by affecting the ferroptosis process need to be further clarified.
It is worth mentioning that we further evaluated the expression of the five FRGs in the tissue of IPF. The results demonstrated that three of the five FRGs (NRAS, MUC1, and ZFP36) are dysregulated in the lung of IPF patients. Contrary to our findings, MUC1 and ZFP36 were found to be downregulated in the lung tissues of patients with IPF. The possible reasons are as follows: First, the authors of the IPF-BALF dataset (GSE70866) pointed that none of the patients received pirfenidone or nintedanib before BAL examination in their study (54). However, all the lung tissue of the GSE110147, GSE72073, GSE53845, and GSE24206 datasets were obtained from the patients with IPF at the time of biopsy or lung transplantation, and whether patients received treatment were not mentioned in the literature (55–58). It could be that the drug treatment changes the expression of these genes. The anti-fibrotic effects of pirfenidone are proved that partially mediated by the inhibition of MUC1 bioactivation (59). Second, the sample size of the tissue datasets (28 normal and 84 IPF) is less than the BALF dataset (28 normal and 176 IPF), which could also contribute to the difference in the results. Third, the gene expression levels were also related to race, tissue sampling site, disease severity, and complications. Therefore, more rigorous studies need to be designed to determine the expression of these genes in lung tissue and BALF, as well as the mechanism of their involvement in the occurrence and development of pulmonary fibrosis.
Function analysis found that the risk score-related DEGs are mainly enriched in epithelial cell proliferation and extracellular matrix organization, etc. In addition, these DEGs were identified enriched in the cytokine–cytokine receptor interaction, TGF-β signaling pathways, focal adhesion, and ECM–receptor interaction signaling pathways, all of them are the most crucial pathways involved in the development of IPF (60–62). These results indicated that the FRGs may regulate the development and progression of IPF by modulating these major pathways.
The advantage of this study is that the FRGs were explored more comprehensively by using the FerrDb database and the previous studies. In addition, the data of IPF from an online open database were systematically analyzed and the predicting value about the FRGs in IPF prognosis was evaluated and summarized for the first time. Meanwhile, there are some limitations present in this study. First, the sample size of normal control from the GEO database was not large enough. Second, only internal validation of the model was performed in the study due to the lack of similar datasets available for external validation. Third, molecular biology experiments were not carried out to further investigate the above results. Therefore, the prospective cohort study and molecular biology experiments need to be designed and performed to further verify the accuracy of the prognostic model and the mechanism of the five FRGs in the pathogenesis of IPF.
Conclusion
To sum up, 19 FRGs associated with the OS were identified in BALF of IPF and a novel prognostic model was constructed based on the five FRGs in the present study. Then, the model was verified as an independent risk factor of OS of IPF patients in both the derivation and validation cohorts. These findings have potential reference value for guiding the treatment and prognosis evaluation of the patients with IPF.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/.
Author Contributions
HR, MC, and ML: conception and design. HR and MC: administrative support. ML, KW, YZ, MF, AL, JZ, TY, PS, DL, and GZ: provision of study materials or patients. ML, KW, YZ, MF, AL, JZ, and DL: experiments operations, collection, and assembly of data. ML, KW, YZ, AL, JZ, TY, PS, DL, GZ, MC, and HR: data analysis and interpretation. All authors: manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Key Research and Development Projects of Shaanxi Province (Nos. 2018SF-071, 2019KW-034, 2018KW-039, and 2021SF-041), Fundamental Research Funds for the Central Universities in Xi'an Jiaotong University (Nos. xzy012021064 and 1191320149); the National Natural Science Foundation of China (No. 81802291), China Postdoctoral Science Foundation Grant (No. 2019M653666).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Abbreviations
- IPF
Idiopathic pulmonary fibrosis
- ILD
Interstitial pulmonary disease
- COPD
Chronic obstructive pulmonary disease
- BALF
Bronchoalveolar lavage fluid
- FRGs
Ferroptosis-related genes
- GEO
Gene Expression Omnibus
- DEGs
Differentially expressed gene
- LASSO
Least absolute shrinkage and selection operator
- PCA
Principal component analysis
- t-SNE
t-distributed stochastic neighbor embedding
- K-M
Kaplan–Meier
- ROC
receiver operational characteristic
- AUC
Area under curve
- OS
Overall survival
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein-protein interactions
- ACO1, Aconitase 1
soluble
- NRAS
Neuroblastoma RAS viral (v-ras) oncogene homolog
- ENPP2
Ectonucleotide pyrophosphatase/phosphodiesterase 2
- MUC1, Mucin 1
cell surface associated
- ZFP36
ZFP36 ring finger protein
- ATX
autotaxin.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.693959/full#supplementary-material
References
- 1.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. (2011) 183:788–824. 10.1164/rccm.2009-040GL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. (2018) 378:1811–23. 10.1056/NEJMra1705751 [DOI] [PubMed] [Google Scholar]
- 3.Navaratnam V, Fleming KM, West J, Smith CJ, Jenkins RG, Fogarty A, et al. The rising incidence of idiopathic pulmonary fibrosis in the UK. Thorax. (2011) 66:462–7. 10.1136/thx.2010.148031 [DOI] [PubMed] [Google Scholar]
- 4.Hutchinson J, Fogarty A, Hubbard R, McKeever T. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. Eur Respir J. (2015) 46:795–806. 10.1183/09031936.00185114 [DOI] [PubMed] [Google Scholar]
- 5.Olson AL, Swigris JJ, Lezotte DC, Norris JM, Wilson CG, Brown KK. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am J Respir Crit Care Med. (2007) 176:277–84. 10.1164/rccm.200701-044OC [DOI] [PubMed] [Google Scholar]
- 6.Vancheri C, Failla M, Crimi N, Raghu G. Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur Respir J. (2010) 35:496–504. 10.1183/09031936.00077309 [DOI] [PubMed] [Google Scholar]
- 7.Diamantopoulos A, Wright E, Vlahopoulou K, Cornic L, Schoof N, Maher TM. The burden of illness of idiopathic pulmonary fibrosis: a comprehensive evidence review. Pharmacoeconomics. (2018) 36:779–807. 10.1007/s40273-018-0631-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spagnolo P, Kropski JA, Jones MG, Lee JS, Rossi G, Karampitsakos T, et al. Idiopathic pulmonary fibrosis: disease mechanisms and drug development. Pharmacol Ther. (2020) 222:107798. 10.1016/j.pharmthera.2020.107798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreuter M, Bonella F, Wijsenbeek M, Maher TM, Spagnolo P. Pharmacological treatment of idiopathic pulmonary fibrosis: current approaches, unsolved issues, and future perspectives. Biomed Res Int. (2015) 2015:329481. 10.1155/2015/329481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galli JA, Pandya A, Vega-Olivo M, Dass C, Zhao H, Criner GJ. Pirfenidone and nintedanib for pulmonary fibrosis in clinical practice: tolerability and adverse drug reactions. Respirology. (2017) 22:1171–8. 10.1111/resp.13024 [DOI] [PubMed] [Google Scholar]
- 11.George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med. (2020) 8:807–15. 10.1016/S2213-2600(20)30225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozawa Y, Suda T, Naito T, Enomoto N, Hashimoto D, Fujisawa T, et al. Cumulative incidence of and predictive factors for lung cancer in IPF. Respirology. (2009) 14:723–8. 10.1111/j.1440-1843.2009.01547.x [DOI] [PubMed] [Google Scholar]
- 13.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. (2021) 31:107–25. 10.1038/s41422-020-00441-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. (2017) 171:273–85. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida M, Minagawa S, Araya J, Sakamoto T, Hara H, Tsubouchi K, et al. Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nat Commun. (2019) 10:3145. 10.1038/s41467-019-10991-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Li X, Cheng Y, Yang M, Wang R. Inhibition of ACSL4 attenuates ferroptotic damage after pulmonary ischemia-reperfusion. FASEB J. (2020) 34:16262–75. 10.1096/fj.202001758R [DOI] [PubMed] [Google Scholar]
- 17.Tang W, Dong M, Teng F, Cui J, Zhu X, Wang W, et al. TMT-based quantitative proteomics reveals suppression of SLC3A2 and ATP1A3 expression contributes to the inhibitory role of acupuncture on airway inflammation in an OVA-induced mouse asthma model. Biomed Pharmacother. (2021) 134:111001. 10.1016/j.biopha.2020.111001 [DOI] [PubMed] [Google Scholar]
- 18.Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, et al. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. (2017) 551:639–43. 10.1038/nature24637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landi C, Bargagli E, Carleo A, Bianchi L, Gagliardi A, Prasse A, et al. A system biology study of BALF from patients affected by idiopathic pulmonary fibrosis (IPF) and healthy controls. Proteomics Clin Appl. (2014) 8:932–50. 10.1002/prca.201400001 [DOI] [PubMed] [Google Scholar]
- 20.Carleo A, Bargagli E, Landi C, Bennett D, Bianchi L, Gagliardi A, et al. Comparative proteomic analysis of bronchoalveolar lavage of familial and sporadic cases of idiopathic pulmonary fibrosis. J Breath Res. (2016) 10:026007. 10.1088/1752-7155/10/2/026007 [DOI] [PubMed] [Google Scholar]
- 21.Carleo A, Landi C, Prasse A, Bergantini L, d'Alessandro M, Cameli P, et al. Proteomic characterization of idiopathic pulmonary fibrosis patients: stable versus acute exacerbation monaldi. Arch Chest Dis. (2020) 90:180–190. 10.4081/monaldi.2020.1231 [DOI] [PubMed] [Google Scholar]
- 22.Ali MK, Kim RY, Brown AC, Donovan C, Vanka KS, Mayall JR, et al. Critical role for iron accumulation in the pathogenesis of fibrotic lung disease. J Pathol. (2020) 251:49–62. 10.1002/path.5401 [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Arisi I, Puxeddu E, Mramba LK, Amicosante M, Swaisgood CM, et al. Bronchoalveolar lavage (BAL) cells in idiopathic pulmonary fibrosis express a complex pro-inflammatory, pro-repair, angiogenic activation pattern, likely associated with macrophage iron accumulation. PLoS ONE. (2018) 13:e0194803. 10.1371/journal.pone.0194803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Zhuang X, Qiao T. Role of ferroptosis in the process of acute radiation-induced lung injury in mice. Biochem Biophys Res Commun. (2019) 519:240–5. 10.1016/j.bbrc.2019.08.165 [DOI] [PubMed] [Google Scholar]
- 25.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. (2012) 18:1028–40. 10.1038/nm.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Y, Wang N, Liu N, Dong H. Lipid peroxidation and GPX4 inhibition are common causes for myofibroblast differentiation and ferroptosis. DNA Cell Biol. (2019) 38:725–33. 10.1089/dna.2018.4541 [DOI] [PubMed] [Google Scholar]
- 27.Tsubouchi K, Araya J, Yoshida M, Sakamoto T, Koumura T, Minagawa S, et al. Involvement of GPx4-regulated lipid peroxidation in idiopathic pulmonary fibrosis pathogenesis. J Immunol. (2019) 203:2076–87. 10.4049/jimmunol.1801232 [DOI] [PubMed] [Google Scholar]
- 28.Zhou N, Bao J. FerrDb: A Manually Curated Resource for Regulators and Markers of Ferroptosis and Ferroptosis-Disease Associations. Database (Oxford: ). (2020). 10.1093/database/baaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. (2019) 575:688–92. 10.1038/s41586-019-1705-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. (2019) 35:830–49. 10.1016/j.ccell.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 31.Liang JY, Wang DS, Lin HC, Chen XX, Yang H, Zheng Y, et al. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int J Biol Sci. (2020) 16:2430–41. 10.7150/ijbs.45050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. (2011) 39:D561–568. 10.1093/nar/gkq973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. (1997) 16:385–95. [DOI] [PubMed] [Google Scholar]
- 34.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for cox's proportional hazards model via coordinate descent. J Stat Softw. (2011) 39:1–13. 10.18637/jss.v039.i05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. (2019) 10:1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez FJ, Safrin S, Weycker D, Starko KM, Bradford WZ, King Jr TE, et al. The clinical course of patients with idiopathic pulmonary fibrosis. Ann Intern Med. (2005) 142:963–7. 10.7326/0003-4819-142-12_Part_1-200506210-00005 [DOI] [PubMed] [Google Scholar]
- 37.Rashidipour N, Karami-Mohajeri S, Mandegary A, Mohammadinejad R, Wong A, Mohit M, et al. Where ferroptosis inhibitors and paraquat detoxification mechanisms intersect, exploring possible treatment strategies. Toxicology. (2020) 433:152407. 10.1016/j.tox.2020.152407 [DOI] [PubMed] [Google Scholar]
- 38.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. (2006) 2:406–14. 10.1038/nchembio807 [DOI] [PubMed] [Google Scholar]
- 39.Volz K. The functional duality of iron regulatory protein 1. Curr Opin Struct Biol. (2008) 18:106–11. 10.1016/j.sbi.2007.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Song N, Jiang H, Wang J, Xie J. Pro-inflammatory cytokines modulate iron regulatory protein 1 expression and iron transportation through reactive oxygen/nitrogen species production in ventral mesencephalic neurons. Biochim Biophys Acta. (2013) 1832:618–25. 10.1016/j.bbadis.2013.01.021 [DOI] [PubMed] [Google Scholar]
- 41.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol. (2018) 15:709–20. 10.1038/s41571-018-0105-0 [DOI] [PubMed] [Google Scholar]
- 42.Dietrich P, Gaza A, Wormser L, Fritz V, Hellerbrand C, Bosserhoff AK. Neuroblastoma RAS viral oncogene homolog (NRAS) is a novel prognostic marker and contributes to sorafenib resistance in hepatocellular carcinoma. Neoplasia. (2019) 21:257–68. 10.1016/j.neo.2018.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martínez-Salgado C, Fuentes-Calvo I, García-Cenador B, Santos E, López-Novoa JM. Involvement of H- and N-Ras isoforms in transforming growth factor-beta1-induced proliferation and in collagen and fibronectin synthesis. Exp Cell Res. (2006) 312:2093–106. 10.1016/j.yexcr.2006.03.008 [DOI] [PubMed] [Google Scholar]
- 44.Bai YT, Chang R, Wang H, Xiao FJ, Ge RL, Wang LS. ENPP2 protects cardiomyocytes from erastin-induced ferroptosis. Biochem Biophys Res Commun. (2018) 499:44–51. 10.1016/j.bbrc.2018.03.113 [DOI] [PubMed] [Google Scholar]
- 45.Oikonomou N, Mouratis MA, Tzouvelekis A, Kaffe E, Valavanis C, Vilaras G, et al. Pulmonary autotaxin expression contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol. (2012) 47:566–74. 10.1165/rcmb.2012-0004OC [DOI] [PubMed] [Google Scholar]
- 46.Ballester B, Milara J, Cortijo J. The role of mucin 1 in respiratory diseases. Eur Respir Rev. (2021) 30:200149. 10.1183/16000617.0149-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miess H, Dankworth B, Gouw AM, Rosenfeldt M, Schmitz W, Jiang M, et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene. (2018) 37:5435–50. 10.1038/s41388-018-0315-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu H, Tang LJ, Luo XJ, Ai KL, Peng J. Insights into the novel function of system Xc- in regulated cell death. Eur Rev Med Pharmacol Sci. (2021) 25:1650–62. 10.26355/eurrev_202102_24876 [DOI] [PubMed] [Google Scholar]
- 49.Hasegawa M, Takahashi H, Rajabi H, Alam M, Suzuki Y, Yin L, et al. Functional interactions of the cystine/glutamate antiporter, CD44v and MUC1-C oncoprotein in triple-negative breast cancer cells. Oncotarget. (2016) 7:11756–69. 10.18632/oncotarget.7598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pillai VG, Bao J, Zander CB, McDaniel JK, Chetty PS, Seeholzer SH, et al. Human neutrophil peptides inhibit cleavage of von Willebrand factor by ADAMTS13: a potential link of inflammation to TTP. Blood. (2016) 128:110–9. 10.1182/blood-2015-12-688747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiedje C, Diaz-Muñoz MD, Trulley P, Ahlfors H, Laaß K, Blackshear PJ, et al. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. (2016) 44:7418–40. 10.1093/nar/gkw474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Guo M, Li Y, Shen M, Kong D, Shao J, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. (2020) 16:1482–505. 10.1080/15548627.2019.1687985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maghsoudloo M, Azimzadeh Jamalkandi S, Najafi A, Masoudi-Nejad A. Identification of biomarkers in common chronic lung diseases by co-expression networks and drug-target interactions analysis. Mol Med. (2020) 26:9. 10.1186/s10020-019-0135-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasse A, Binder H, Schupp JC, Kayser G, Bargagli E, Jaeger B, et al. BAL cell gene expression is indicative of outcome and airway basal cell involvement in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2019) 199:622–30. 10.1164/rccm.201712-2551OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meltzer EB, Barry WT, D'Amico TA, Davis RD, Lin SS, Onaitis MW, et al. Bayesian probit regression model for the diagnosis of pulmonary fibrosis: proof-of-principle. BMC Med Genomics. (2011) 4:70. 10.1186/1755-8794-4-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DePianto DJ, Chandriani S, Abbas AR, Jia G, N'Diaye EN, Caplazi P, et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax. (2015) 70:48–56. 10.1136/thoraxjnl-2013-204596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geng J, Huang X, Li Y, Xu X, Li S, Jiang D, et al. Down-regulation of USP13 mediates phenotype transformation of fibroblasts in idiopathic pulmonary fibrosis. Respir Res. (2015) 16:124. 10.1186/s12931-015-0286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cecchini MJ, Hosein K, Howlett CJ, Joseph M, Mura M. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir Res. (2018) 19:153. 10.1186/s12931-018-0857-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballester B, Milara J, Cortijo J. Pirfenidone anti-fibrotic effects are partially mediated by the inhibition of MUC1 bioactivation. Oncotarget. (2020) 11:1306–20. 10.18632/oncotarget.27526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. (1999) 96:319–28. 10.1016/S0092-8674(00)80545-0 [DOI] [PubMed] [Google Scholar]
- 61.Selman M, Pardo A. Alveolar epithelial cell disintegrity and subsequent activation: a key process in pulmonary fibrosis. Am J Respir Crit Care Med. (2012) 186:119–21. 10.1164/rccm.201206-0997ED [DOI] [PubMed] [Google Scholar]
- 62.Liu G, Philp AM, Corte T, Travis MA, Schilter H, Hansbro NG, et al. Therapeutic targets in lung tissue remodelling and fibrosis. Pharmacol Ther. (2021) 225:107839. 10.1016/j.pharmthera.2021.107839 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/.