Abstract
The structure of Janus kinases (JAKs) is unique among protein tyrosine kinases in having tandem, nonidentical kinase and pseudokinase domains. Despite its conservation in evolution, however, the function of the pseudokinase domain remains poorly understood. Lack of JAK3 expression results in severe combined immunodeficiency (SCID). In this study, we analyze two SCID patients with mutations in the JAK3 pseudokinase domain, which allows for protein expression but disrupts the regulation of the kinase activity. Specifically, these mutant forms of JAK3 had undetectable kinase activity in vitro but were hyperphosphorylated both in patients' Epstein-Barr virus-transformed B cells and when overexpressed in COS7 cells. Moreover, reconstitution of cells with these mutants demonstrated that, although they were constitutively phosphorylated basally, they were unable to transmit cytokine-dependent signals. Further analysis showed that the isolated catalytic domain of JAK3 was functional whereas either the addition of the pseudokinase domain or its deletion from the full-length molecule reduced catalytic activity. Through coimmunoprecipitation of the isolated pseudokinase domain with the isolated catalytic domain, we provide the first evidence that these two domains interact. Furthermore, whereas the wild-type pseudokinase domain modestly inhibited kinase domain-mediated STAT5 phosphorylation, the patient-derived mutants markedly inhibited this phosphorylation. We thus conclude that the JAK3 pseudokinase domain is essential for JAK3 function by regulating its catalytic activity and autophosphorylation. We propose a model in which this occurs via intramolecular interaction with the kinase domain and that increased inhibition of kinase activity by the pseudokinase domain likely contributes to the disease pathogenesis in these two patients.
Cytokines are critical regulators of cellular growth and differentiation, a subset of which bind to members of the type I cytokine receptor superfamily and initiate their actions by ligand-induced receptor oligomerization (23). Although cytokine receptors lack intrinsic kinase activity, they associate with and activate cytoplasmic protein tyrosine kinases (PTKs), which then phosphorylate downstream signaling molecules such as the signal transducers and activators of transcription (STATs). Activated STATs, in turn, translocate to the nucleus and regulate gene expression (7, 17, 35).
The Janus kinase (JAK) family of nonreceptor PTKs are critical elements in cytokine signaling (7, 17). Of four mammalian members (JAK1, JAK2, JAK3, and TYK2) identified so far (35), JAK3 is unique in its predominant expression in hematopoietic cells (13, 20, 45) and its ability to specifically associate with the common gamma chain (γc) of cytokine receptors through its N terminus (1, 5, 31, 37). JAK3 is activated by interleukin 2 (IL-2), IL-4, IL-7, IL-9, and IL-15 (35), which all utilize γc as a component of their receptors (23). Mutation of JAK3 results in autosomal recessive severe combined immunodeficiency (JAK3-SCID) in humans and mice, illustrating the importance of this JAK in the proper development and function of the immune system (4, 27, 34, 36, 38, 41). Interestingly, patients with JAK3-SCID present with a clinical phenotype virtually identical to X-linked SCID, which is caused by mutations of γc (3, 33). Both forms of SCID are characterized by the absence of circulating mature T lymphocytes and natural killer (NK) cells, normal to elevated numbers of nonfunctional B cells (T−B+SCID), and marked hypoplasia of lymphoid tissues. Thus, JAK3-SCID and X-SCID can both be thought of as cytokine signaling disorders (3, 22).
JAKs are structurally unique among metazoan PTKs in having a C-terminal catalytic domain immediately preceded by another putative domain with many features of a tyrosine kinase catalytic domain, referred to as the Janus homology 2 (JH2) domain (9, 14, 44). This feature is conserved in mammalian, teleost, and Drosophila JAKs, suggesting that it serves an important function for this kinase family (35). However, many of the canonical residues that are essential for phosphotransferase activity are altered in the JH2 domain. Thus, based on primary structure, it would be predicted that this domain lacks tyrosine kinase activity, and indeed, this has been borne out experimentally (44). The JH2 domain has therefore also been termed the pseudokinase or kinase-like domain. Several lines of evidence suggest an alternative function, namely, a regulatory function, for the JH2 domain, but no actual mechanism by which this putative regulation occurs has been proposed. For TYK2, deletion of the JH2 domain abrogated the in vitro catalytic activity and the ability to transmit alpha/beta interferon-dependent signals, suggesting that the JH2 domain was required for catalytic function of TYK2 (42). In contrast, however, a similar mutant of JAK2 was able to transmit growth hormone-dependent signals (10). Indeed, in the context of CD16-JAK2 chimeras, a construct lacking JH2 had increased catalytic activity compared to the one containing the JH2 domain (39). This finding was interpreted as showing that the JH2 domain might serve to tonically inhibit kinase activity. In addition, it was reported that a mutation within the JH2 domain of the Drosophila JAK, Hopscotch, produced a presumably hyperactive JAK that caused leukemia in flies (26). The equivalent mutation in JAK2 also resulted in a kinase with an increased ability to phosphorylate STAT5 in an overexpression system (26). However, the catalytic activity of these mutants was not specifically assessed. Thus, the data to this point did not indicate a simple negative or positive regulatory function of the JH2 domain. Given these discrepancies and the potential differences among the JAKs, it was not clear what would be the consequence of mutations of the JAK3 JH2 domain. Furthermore, none of these studies provided an in-depth biochemical and functional analysis of JAKs with mutations in the JH2 domain, nor did they offer a mechanism by which the JH2 domain might regulate catalytic function.
The study of naturally occurring JAK3 variants with mutations in the JH2 domain offers an advantage in elucidating the functional characteristics of this region. Since the initial description of JAK3-SCID patients, who were devoid of JAK3 expression (27, 38), we recently identified two new patients with mutations involving the JH2 domain that allowed for expression of JAK3 protein but who nonetheless had presented with a SCID phenotype (4). In an effort to understand the basis of their immunodeficiency and the function of the JH2 domain, we analyzed in detail the biochemical and signaling properties of JAK3 alleles with mutations in the JH2 domain. Our results demonstrate that the JH2 domain is essential for the proper function of JAK3. Not only is it critical for full catalytic activity of the full-length molecule and the ability to transmit cytokine-dependent signals but it also plays an autoinhibitory role in regulating the kinase activity and basal phosphorylation of JAK3. It achieves this regulation most likely through interactions with the catalytic domain. Moreover, mutations in the JH2 domain result in the increased inhibition of JH1 function by the JH2 domain, which likely contributes to the disease pathogenesis in these two patients.
MATERIALS AND METHODS
Cytokines and antibodies.
Human IL-2 was obtained from C. Reynolds (National Cancer Institute, Frederick, Md.). Rabbit polyclonal antisera against JAK3 and STAT5a and 7G7 monoclonal antibody (MAb) (anti-Tac) were described previously (5). The anti-JAK3 N and C terminus antibodies work comparably well in both immunoprecipitation and Western blotting (data not shown). Rabbit antiserum against STAT3 was obtained from Andrew Larner (Cleveland Clinic Foundation Research Institute, Cleveland, Ohio). The 4G10 antiphosphotyrosine MAb and anti-JAK2 polyclonal antisera were from Upstate Biotechnology (Lake Placid, N.Y.). Rabbit polyclonal anti-glutathione S-transferase (anti-GST), anti-IL2Rγc, anti-extracellular signal-regulated kinase (anti-ERK), and anti-Flag MAb (M2) were purchased from Pharmacia (Piscataway, N.J.), Santa Cruz Biotechnology (Santa Crutz, Calif.), Transduction Laboratories (Lexington, Ky.) and Kodak Scientific Imaging System (Rochester, N.Y.), respectively.
Plasmid and mutagenesis.
The wild-type JAK3 cDNA was cloned into pME18s as previously described (5). The mutant JAK3 cDNA containing the C759R mutation from patient 5 (previously designated LP) (4) was generated with the Transformer site-directed mutagenesis kit (Clontech) with oligonucleotides designed to change the codon for cysteine at amino acid (aa) position 759 to one for arginine. The JAK3 mutant found in patient 6 (previously designated NK) with a seven-amino-acid deletion (aa 586 to 592) in the JH2 domain (4) and the E639K mutant with glutamic acid-to-lysine mutation at codon 639 (26) were also constructed with the same mutagenesis kit. The catalytically inactive mutants, K855A and C759R/K855A, were generated as previously described (5). The ΔJH2 mutant carrying the deletion of the entire JH2 domain (aa 519 to 792) was generated by replacing the HindIII-BglII region (nucleotides [nt] 1653 to 2881) of plasmid pME18sJ3(H3) (5) with a PCR-generated HindIII-BglII fragment spanning nt 2471 to 2881. For plasmids Flag-JH2, Flag-JH1, and Flag-JH2-1 (see Fig. 6A), PCR-generated fragments containing JAK3 sequences encoding aa 520 to 813, 801 to 1124, and 520 to 1124, respectively, were cloned into pFlag-CMV2 (Kodak). The plasmids Flag-JH2(C759R) and Flag-JH2(Δ586-592) were generated in a similar way as Flag-JH2 except that the mutant sequences were used. All the constructs were verified by automatic DNA sequencing with an ABI PRISM Dye Terminator Cycle Sequencing kit (Perkin-Elmer). GST-γc expression plasmid pGEX-γc was constructed as previously described (47). Chimeric IL-2R αγγ (Tacγc) cDNA was kindly provided by Warren Leonard (National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Md.). STAT5A cDNA was a gift from Lothar Hennighausen (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health). HA-tagged ERK2 cDNA was provided by Andrew Larner (Cleveland Clinic Foundation Research Institute).
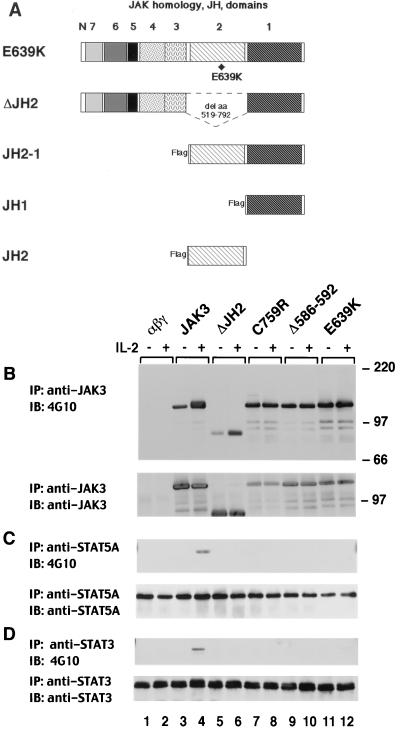
FIG. 6.
The JH2 pseudokinase domain regulates JAK3 kinase activity and substrate phosphorylation. (A) Schematic representation of the JH2 mutants. E639K is a JAK3 mutant corresponding to the hyperactivating mutation reported in Drosophila Hopscotch (26). ΔJH2 contains a complete deletion of the JH2 domain, from aa 519 to aa 792. (B to D) αβγ cells expressing various forms of JAK3 were left unstimulated (−) or stimulated with IL-2 for 15 min (+); cell lysates were immunoprecipitated (IP) with anti-JAK3 (B), anti-STAT5A (C), or anti-STAT3 (D) and then subjected to immunoblotting (IB) analysis with anti-phosphotyrosine (B to D, top panels). Membranes were reprobed with anti-JAK3 (B), anti-STAT5A (C), or anti-STAT3 (D) to confirm equal loading (bottom panels).
Cells and transfection.
Epstein-Barr virus (EBV)-transformed B cells from a healthy donor (HBC) and patients with JAK3-SCID (designated patient 5 and patient 6) have been previously described (4) (http://www.uta.fi/laitokset/imt/bioinfo/dbases/JAK3base.html). These cells were cultured in RPMI medium (Biofluids, Inc., Rockville, Md.) supplemented with 10% fetal calf serum (Biofluids), 2 mM l-glutamine (Biofluids), and antibiotics. COS7 cells and 3T3αβγ cells (30) were maintained as previously described (5). COS7 cells were transiently transfected by a DEAE-dextran method (Promega, Madison, Wis.) following the manufacturer's protocol. 3T3αβγ cells reconstituted with various versions of JAK3 were established as previously described (5). Clones expressing similar levels of JAK3 were chosen for subsequent analysis, and flow cytometry was performed to ensure that the cells had comparable levels of receptor expression.
Cell stimulation, immunoprecipitation, immunoblotting, and immune-complex kinase assay.
3T3αβγ cells (10 × 106 to 15 × 106) or EBV-transformed B cells from patients or healthy donors were acid washed and serum starved for 4 h and then stimulated with 1,000 U of IL-2 per ml for 15 min at 37°C. Subsequently, cells were washed once with ice-cold phosphate-buffered saline (PBS) containing 0.1 mM Na3VO4 and 2 mM EDTA and then lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 2 mM EDTA, 0.5% Triton X-100 (Fisher), 200 μM Na3VO4, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, and 2.5 μM p-nitrophenyl p-guanidinobenzoate (NPGB). Clarified lysates were immunoprecipitated with antibodies against STAT5A, STAT3, or JAK3, followed by immunoblotting with 4G10 and the indicated immunoprecipitation antibody as previously described (19). For in vitro kinase assays, JAK3 immunoprecipitates were washed three times with lysis buffer and once with buffer containing 100 mM NaCl and 10 mM HEPES (pH 7.5) and resuspended in kinase reaction buffer (20 mM Tris [pH 7.5], 5 mM MgCl2, 5 mM MnCl2, 1 μM ATP) containing 10 μCi of [γ-32P]ATP (Amersham) with or without 1 μg of GST-γc fusion protein as an exogenous substrate. The reactions were performed at 0°C (without GST-γc) or room temperature (with GST-γc), and the radioactivity incorporated by JAK3 and GST-γc was assessed as described previously (47).
RESULTS
Defective IL-2-dependent signaling in B cells derived from SCID patients.
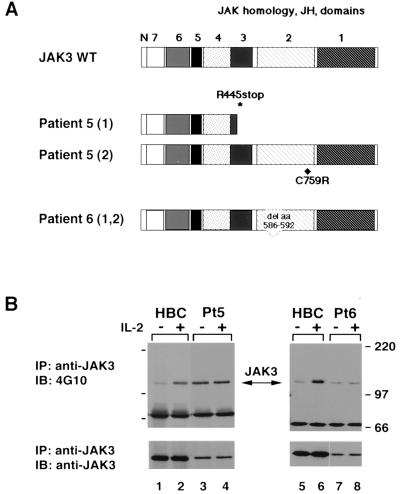
Two JAK3-SCID patients with mutations in the JH2 domain were recently identified (4) (Fig. 1A). To analyze the function of the mutant JAK3 proteins, we first studied their abilities to transmit IL-2-dependent signals. IL-2 stimulation of lymphocytes results in the phosphorylation and activation of JAK3, which then leads to the phosphorylation and activation of STATs (STAT3 and STAT5) (15, 18, 19, 24). The phosphorylation of wild-type JAK3 in EBV-transformed B lymphocytes from a healthy individual is shown in Fig. 1B (lanes 2 and 6). In contrast, when B cells from JAK3-SCID patients 5 and 6 were stimulated with IL-2, no inducible phosphorylation of JAK3 was observed (Fig. 1B, lanes 4 and 8). However, higher constitutive phosphorylation was noted (lanes 3 and 7) when compared to normal, unactivated JAK3 (lanes 1 and 5), particularly when the level of expression of the mutant proteins was taken into account (Fig. 1B, lower panel). Additionally, in these cells, IL-2 stimulation failed to induce STAT5 phosphorylation (data not shown). Thus, the cells from these two SCID patients were defective in their ability to transmit IL-2 signals.
FIG. 1.
JAK3 from SCID patients' cells is constitutively phosphorylated and unresponsive to IL-2 stimulation. (A) Illustration of patient alleles. Patient 5 is a compound heterozygote bearing two different alleles, one encoding a missense mutation substituting a cysteine for an arginine at codon 759 and the other encoding a premature stop codon at residue 445. Both patient 6-derived alleles encode a deletion of aa 586 to 592 in the JH2 domain of JAK3. (B) EBV-transformed B cells (1.5 × 107 cells) from a healthy individual (HBC) and the JAK3-SCID patients 5 (Pt5) and 6 (Pt6) were serum starved for 4 h and then were left unstimulated (−) or were stimulated with IL-2 (1,000 U/ml) for 15 min (+); cell lysates were immunoprecipitated (IP) with anti-JAK3. Tyrosine phosphorylation was assayed by blotting (IB) with antiphosphotyrosine (4G10) (top panel). The membrane was reprobed with anti-JAK3 to confirm equal loading (bottom panel).
Mutations in the JH2 domain are responsible for the defects in IL-2-dependent responses.
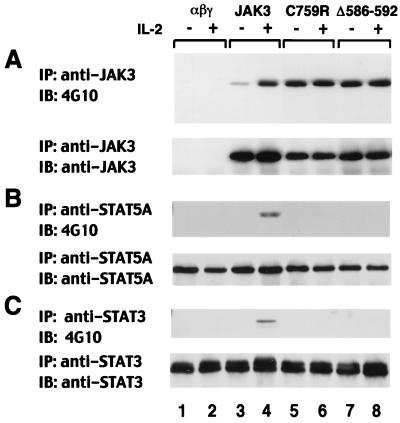
Since the patients' B cells failed to transmit IL-2-dependent signals and contained mutations in the JH2 domain of JAK3, it was likely that these JH2 mutations were responsible for the observed defects. Other possibilities remained, however. One explanation might have been that there is a relatively low level of expression of the mutant JAK3 proteins in the patients' B cells (Fig. 1B). Alternatively, other unidentified defects might exist in these cell lines. Finally, patient 5 cells have two mutant alleles that encode parts of JAK3, one causing a truncation of the JAK3 protein. Although expression of the truncated protein is barely detectable by our JAK3 antibody, it could conceivably interfere with the function of the missense mutant at a low expression level. To exclude these possibilities and to ascertain that the mutations in the JH2 domain of JAK3 resulted in the disruption of IL-2 signaling observed, IL-2R expressing fibroblasts (3T3αβγ) that lack endogenous JAK3 (5, 30) were reconstituted with wild-type JAK3 or a patient-derived mutant (C759R or Δ586-592). Their response to IL-2 stimulation was then analyzed by measuring the presence and absence of inducible phosphorylation of JAK3, STAT5A, and STAT3. As shown in Fig. 2A, in spite of its basal phosphorylation, wild-type JAK3 was inducibly phosphorylated in response to IL-2 (lanes 3 and 4), while patient 5- and 6-derived mutants C759R and Δ586-592 were constitutively hyperphosphorylated and not further phosphorylated upon IL-2 treatment (lanes 5 to 8). Moreover, IL-2 failed to induce STAT5A and STAT3 phosphorylation in the presence of these patient-derived mutants (Fig. 2B and C, lanes 5 versus 6 and 7 versus 8), whereas this was readily detectable upon ectopic expression of wild-type JAK3 (lanes 3 versus 4). These results thus provide a formal demonstration that the mutations in the JH2 domain are directly responsible for the defects in IL-2 signaling previously seen in patient-derived B cell lines.
FIG. 2.
Defective IL-2 responsiveness in IL-2R-expressing fibroblasts reconstituted with patient-derived mutants. NIH 3T3 fibroblasts reconstituted with the IL-2R complex (αβγ cells) were stably transfected with cDNAs encoding wild-type JAK3 and patient 5- and 6-derived mutants C759R and Δ586-592, respectively. Serum-starved cells were left unstimulated (−) or were stimulated with IL-2 for 15 min (+); cell lysates were immunoprecipitated (IP) with anti-JAK3 (A), anti-STAT5A (B), or anti-STAT3 (C) and then subjected to immunoblotting (IB) with antiphosphotyrosine (4G10) (top panels). Membranes were reprobed with anti-JAK3 (A), anti-STAT5A (B), or anti-STAT3 (C) to confirm equal loading (bottom panels).
Mutations in the JH2 domain do not disrupt the ability of JAK3 to bind the common gamma chain, γc.
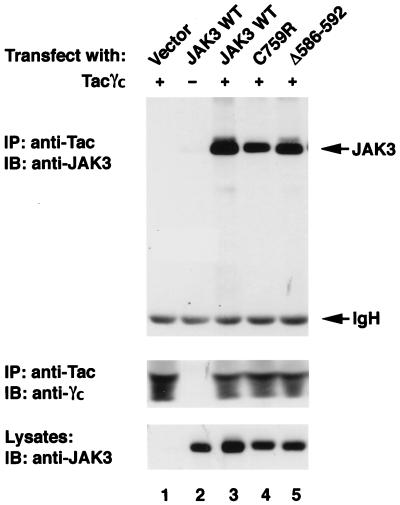
It is well established that JAK3-γc interaction is vital for IL-2 signaling (5, 31, 37). Several naturally occurring mutations of γc which affect its association with JAK3 cause X-SCID or X-linked combined immunodeficiency (32, 37, 40). Although our previous studies indicate that the JH2 domain of JAK3 is dispensable for receptor association (5), it was still possible that global deformity of the protein structure by the patients' specific mutations disrupted the binding of the mutant JAK3 to γc and thus abolished IL-2 signaling. To address this point, the mutant alleles were coexpressed with a chimeric γc molecule containing the cytoplasmic domain of γc fused to the extracellular domain of the IL-2Rα chain (Tacγc) (5, 37). The ability of the mutant JAK3 to associate with this chimeric receptor was then analyzed. As shown in Fig. 3, the expression levels of the various forms of JAK3 (bottom) and of Tacγc (middle) did not vary significantly among the transfected cells. Consistent with previous reports (5, 37), anti-Tac MAb coprecipitated JAK3 from cells expressing both Tacγc and JAK3 (lane 3) but not Tacγc (lane 1) or JAK3 (lane 2) alone. Importantly, both of the mutant proteins bearing the mutations found in the JAK3-SCID patients bound γc comparably to binding by wild-type JAK3 (lanes 4 and 5 versus lane 3). Thus, the data indicate that the conformation of the mutant JAK3 proteins is not completely disrupted by the JH2 mutations; one key function of JAK3, the ability to bind to receptor, is retained.
FIG. 3.
Patients' JH2 mutations do not disrupt the ability of JAK3 to bind γc. COS7 cells were transfected with 3 μg of the indicated cDNAs. Lysates were immunoprecipitated (IP) with anti-Tac and blotted with anti-JAK3 (top panel) or anti-γc (middle panel). Expression levels of various JAK3 proteins were analyzed by immunoblotting (IB) with anti-JAK3 (bottom panel). WT, wild type.
Effect of the JH2 mutations on in vitro kinase activity of JAK3.
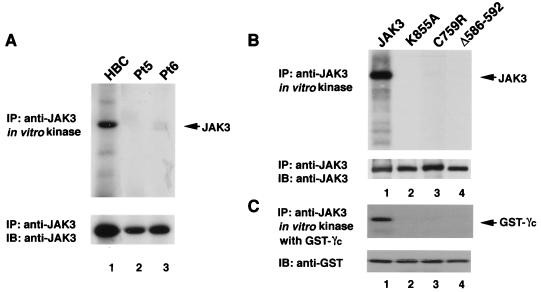
Given that the mutations in the JH2 domain did not affect the ability of patient-derived mutants to bind γc (Fig. 3) but abrogated their ability to transmit IL-2-dependent signals, we next considered the possibility that these patients' mutations interfered with the catalytic activity of JAK3. To address this issue, EBV-transformed B cells from a healthy donor and both patients were treated with IL-2 and lysed. JAK3 was then immunoprecipitated from the lysates and subjected to in vitro kinase assays as previously described (47). As shown in Fig. 4A, wild-type JAK3 had vigorous catalytic activity as measured by autophosphorylation (top panel, lane 1). In contrast, both patient-derived JH2 mutants completely lacked catalytic activity (top panel, lanes 2 and 3). Again, to exclude the possibility of interference by other confounding factors, each mutant JAK3 construct was also expressed in COS7 cells; the lack of in vitro kinase activity was also evident under these circumstances (Fig. 4B and C, top panels, lanes 3 and 4). JH2 mutants C759R and Δ586-592 not only failed to autophosphorylate but also failed to phosphorylate an exogenous substrate, the cytoplasmic region of a human IL-2R γc subunit (a physiologic substrate of JAK3) fused to GST (47). Shown as a control is a JAK3 mutant with the mutation of the ATP binding site (K855A) that also abrogated catalytic activity (lane 2). Equal amounts of JAK3 and GST-γc were shown to be loaded on each lane when the filter was immunoblotted with appropriate antibodies (Fig. 4B and C, lower panels).
FIG. 4.
Patient-derived JH2 mutants lack in vitro kinase activity. (A) EBV-transformed B cells (1.5 × 107 cells) from HBC (lanes 1) and patients 5 (Pt5) and 6 (Pt6) were treated with IL-2 (15 min), lysed, immunoprecipitated (IP) with anti-JAK3, and then subjected to an in vitro kinase assay. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. Incorporated radioactive phosphate was visualized by autoradiography (top panel). Expression levels of JAK3 were determined by immunoblotting (IB) with anti-JAK3 (bottom panel). (B and C) Wild-type JAK3 (lanes 1), catalytically inactive JAK3 (K855A) (lanes 2), and patient 5- and 6-derived mutants (lanes 3, C759R; lanes 4, Δ586-592) were expressed in COS7 cells. Cell lysates were immunoprecipitated with anti-JAK3 and subjected to an in vitro kinase assay. Autophosphorylation (B) and phosphorylation of an exogenous substrate, GST-γc (C), are shown in the top panels. Membranes were probed with relevant antibody to verify equal expression and loading (bottom panels).
Patient-derived JH2 mutants are phosphorylated when expressed in COS7 cells.
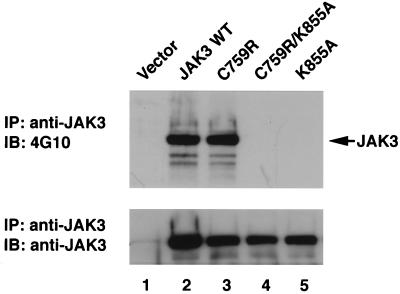
The preceding data indicated that the patient-derived mutants lack in vitro kinase activity regardless of whether they were obtained from the patient-derived B-cell lines or from transfected COS7 cells. Although this helped explain why these mutant JAKs failed to transmit cytokine-dependent signals, the results did not help explain the finding of their constitutive phosphorylation (Fig. 1B and 2A). To further investigate this issue, we asked whether the mutant JAKs were also phosphorylated in COS7 cells as they were in the patients' EBV-transformed B cells. We therefore immunoblotted JAK3 proteins immunoprecipitated from COS7 cells with antiphosphotyrosine antibody. As shown in Fig. 5, wild-type JAK3 was phosphorylated when overexpressed in COS7 cells (top panel, lane 2) whereas catalytically inactive JAK3 (K855A) (47) was not (top panel, lane 5). Similar to the results obtained from the patient-derived B-cell lines, the patient 5-derived C759R mutant was phosphorylated at least as well as wild-type JAK3 (lane 3) despite its lack of activity measured in vitro (Fig. 4). The same result was also obtained with the patient 6-derived mutant Δ586-592 (data not shown). The levels of the various JAK3 mutant proteins were assayed by reblotting (Fig. 5, lower panel).
FIG. 5.
Patient 5-derived JH2 mutant C759R is hyperphosphorylated when expressed in COS7 cells. COS7 cells were transfected with 3 μg of the indicated cDNAs. The cells were lysed, immunoprecipitated (IP) with anti-JAK3, and blotted (IB) with antiphosphotyrosine antibody 4G10 (top panel). The membrane was then stripped and reprobed with anti-JAK3 to show the levels of various JAK3 proteins (bottom panel).
To exclude the possibility that the phosphorylation of the JH2 mutants observed in cells was not the result of autophosphorylation but rather was because these mutant kinases were more efficient substrates for another kinase, we prepared a double mutant with the C759R mutation in the JH2 domain and the K855A mutation of the ATP binding site. Mutation of the latter site clearly abrogated kinase activity whether measured by in vitro kinase assay or by phosphorylation in COS7 cells (Fig. 4 and 5). If the hyperphosphorylation observed with C759R (patient 5-derived mutant) was due to cross-phosphorylation by a different kinase, it would be predicted that the double mutant C759R/K855A would be phosphorylated to a similar extent as the C759R allele itself. However, as shown in lane 4 of Fig. 5 (top panel), this was not the case; the double JH2-ATP binding site mutant (C759R/K855A) was not phosphorylated when overexpressed in COS7 cells. This indicated that the phosphorylation of the mutant C759R required endogenous phosphotransferase activity and was not likely due to its simply being a more accessible substrate for another kinase. Taken together, then, these data indicated that the JH2 mutants were catalytically inactive kinases when measured by in vitro kinase assays, regardless of the cellular systems used (patients' EBV-transformed B cells or COS7 cells). However, they were basally phosphorylated in cells, a feature that is paradoxically dependent on their own catalytic potential. The precise reason why the JH2 mutants have no kinase activity in vitro but can still incorporate phosphate in cells remains unclear (see Discussion).
Multiple distinct JH2 mutants fail to transmit IL-2-dependent signals.
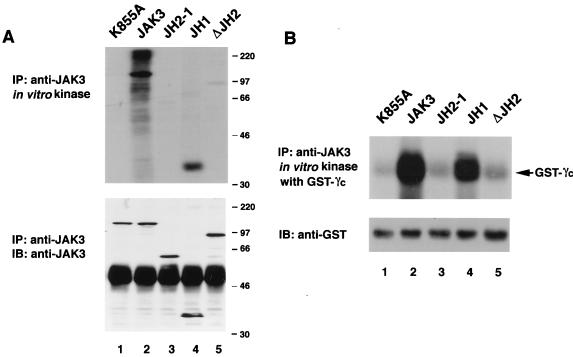
The preceding experiments therefore indicated that patient 5-derived JH2 mutant C759R is a dysregulated kinase. To ascertain whether the aberrant kinase activity of the C759R and Δ586-592 mutants was the result of their unique mutations or disruption of the general regulatory function of the JH2 domain, we generated two additional JH2 mutants (Fig. 6A) and evaluated their in vitro kinase activity as well as their ability to transmit IL-2-dependent signals. As illustrated in Fig. 6A, the mutant designated E639K contains a JAK3 mutation that corresponds to the gain-of-function Drosophila Hopscotch mutation (26), whereas the mutant ΔJH2 contains a complete deletion of the JH2 domain, from aa 519 to aa 792. To examine the function of these mutants, wild-type and mutant versions of JAK3 were stably expressed in 3T3αβγ fibroblasts (5, 30). The cells were then stimulated with IL-2, and as shown in Fig. 6B, this induced tyrosine phosphorylation of transfected wild-type JAK3 (top panel, lane 4). In contrast, a high level of basal phosphorylation was noted for mutant E639K and there was no IL-2 inducible phosphorylation (top panel, lanes 11 and 12). This result is similar to that seen for mutants derived from patients 5 and 6 (Fig. 2 and 6B, top panel, lanes 7 to 10). Moreover, E639K did not allow for inducible phosphorylation of STAT5A and STAT3 (Fig. 6C and D, top panels, lanes 11 and 12), whereas this was readily detected in cells expressing wild-type JAK3 (Fig. 6C and D, top panels, lanes 3 and 4). Interestingly, IL-2 stimulation induced phosphorylation of the ΔJH2 mutant (Fig. 6B, top panel, lanes 5 and 6) but failed to induce phosphorylation of STAT5 and STAT3 in cells expressing this mutant (Fig. 6C and D, top panels, lanes 5 and 6), suggesting a potential role of the JH2 domain in regulating the ability to phosphorylate given substrates. To confirm these results, we also performed an in vitro kinase assay on these mutants, and as expected, their kinase activity correlated with their responsiveness to IL-2 (data not shown; for ΔJH2, also see Fig. 7, top panels, lanes 5). Thus, multiple distinct mutations in the JH2 domain of JAK3 all interfere with the catalytic activity and the ability to mediate cytokine-dependent signals.
FIG. 7.
The JH2 pseudokinase domain inhibits kinase activity of the isolated catalytic domain of JAK3. (A) JAK3 autophosphorylation. (B) In vitro kinase assay using GST-γc as the exogenous substrate. COS7 cells were transfected with 3 μg of the indicated cDNAs. Lysates were immunoprecipitated (IP) with anti-JAK3 and then subjected to an in vitro kinase assay in the absence (A) or presence (B) of GST-γc. Incorporated radioactive phosphate was visualized by autoradiography (top panels). Immunoblotting (IB) with anti-JAK3 (A) or anti-GST (B) was performed to confirm equal loading and expression.
Regulation of catalytic activity by addition of the pseudokinase domain to the kinase domain.
The data presented thus far argued strongly for an essential role of the pseudokinase domain in the proper function of JAK3. To examine this question, we expressed cDNAs encoding the isolated JAK3 kinase domain (JH1), the JAK3 kinase domain with the pseudokinase domain (JH2-1), and the full-length JAK3 molecule. Next, we compared the catalytic activity of these polypeptides. As shown in Fig. 7A, the isolated kinase domain (JH1) had substantial in vitro kinase activity in the absence of the pseudokinase domain (lane 4), albeit reduced compared to full-length JAK3 as assessed by autophosphorylation (25% of that of full-length JAK3) (lane 2). Interestingly, addition of the pseudokinase domain (JH2-1) abrogated kinase activity (lane 3). As a control, the activity of the ΔJH2 mutant is also shown, and it, too, demonstrated minimal catalytic activity (1% of that of full-length JAK3) (lane 5). These results were further confirmed when GST-γc was used as a substrate (Fig. 7B). It was impossible, however, to test the abilities of JH1 and JH2-1 constructs to transmit cytokine receptor-mediated signals, since these kinase mutants were unable to bind γc (5). The level of expression of the different mutants is shown in Fig. 7A (lower panel). The finding that both the deletion of the JH2 domain from the full-length molecule (designated ΔJH2) and its addition to the JH1 domain (JH2-1) resulted in defective kinase activity initially seemed contradictory. However, this could be explained if both the N terminus and the JH2 domain interact with the JH1 domain and the removal of either deforms the structure and interferes with the kinase activity (see below and Discussion). These data therefore strongly support the previous contention that the pseudokinase domain is required for the proper activity of JAK3 in the context of the full-length molecule but also inhibits catalytic activity in the absence of the N terminus.
Interaction between the JH2 and JH1 domains of JAK3.
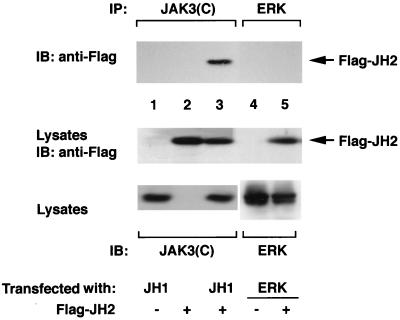
The structure of the JAKs is presently unknown, but there is no reason to suspect, a priori, that the kinase and pseudokinase domains necessarily interact. However, the preceding data clearly suggest an autoinhibitory role for the pseudokinase domain on the JAK3 catalytic activity. The simplest model for this regulation would be direct interaction between the kinase and pseudokinase domains. We therefore sought to determine whether more direct evidence for such a physical interaction could be obtained. To address this issue, COS7 cells were transfected with cDNAs encoding the isolated kinase domain (Flag-JH1) along with a cDNA encoding the JH2 domain (Flag-JH2). As a control, a different kinase, ERK2, was also expressed. Lysates were immunoprecipitated with an antiserum against the JAK3 C terminus (Fig. 8, lanes 1 to 3) or ERK (lanes 4 and 5) and then were subjected to immunoblotting analysis with anti-Flag to reveal any coprecipitated Flag-tagged JH2. As shown in Fig. 8, the anti-JAK3 C terminus coprecipitated Flag-JH2 from cells expressing both the JH1 and JH2 domains (lane 3) but not the JH1 (lane 1) or JH2 (lane 2) domain alone, suggesting a specific interaction between the JH2 and JH1 domains. This specific interaction was further supported by the finding that a control kinase, ERK2, failed to coprecipitate Flag-JH2 when coexpressed (lanes 4 and 5). The lysates were blotted with anti-Flag and demonstrated that comparable levels of Flag-JH2 were present in all of the lanes (middle panel). Control immunoblotting with anti-JAK3 and anti-ERK also confirmed the expression of these proteins in transfected cells (lower panel).
FIG. 8.
Interaction between the JH2 and JH1 domains of JAK3. COS7 cells were transfected with 3 μg of the indicated cDNAs. Lysates were immunoprecipitated (IP) with an antiserum against the C terminus of JAK3 (lanes 1 to 3) or anti-ERK (lanes 4 and 5) and subjected to immunoblotting (IB) with anti-Flag. The expression levels of Flag-tagged JH2 (middle panel) and JH1 and ERK2 (bottom panel) were analyzed by immunoblotting with the indicated antibodies.
Mutant JH2 domains markedly inhibit STAT5A phosphorylation mediated by the JH1 domain of JAK3.
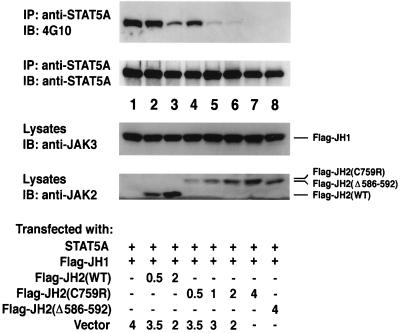
To confirm the association between the pseudokinase domain and the catalytic domain, we next asked whether expression of the isolated JH2 domain would inhibit the function of the isolated kinase domain as measured by STAT5A phosphorylation. To this end, COS7 cells were cotransfected with STAT5A, epitope-tagged JH1, and increasing amounts of epitope-tagged JH2. Lysates were then immunoprecipitated with anti-STAT5A and blotted with antiphosphotyrosine and anti-STAT5A. As shown in Fig. 9, expression of Flag-JH2 (top panel, lanes 2 and 3) reduced the level of STAT5A phosphorylation mediated by the JH1 domain without reducing the levels of expression of Flag-JH1 (lower panel) and STAT5A (middle panel). In contrast, overexpression of the parental vector failed to generate such effects (top panel, lane 1). Strikingly, whereas the wild-type JH2 only modestly inhibited the JH1-dependent phosphorylation of STAT5A (lanes 2 and 3 versus lane 1), both the patient-derived mutants, JH2(C759R) and JH2(Δ586-592), markedly inhibited STAT5A phosphorylation (lanes 4 to 8 versus lane 1), even though they were not expressed quite as well as the wild type (lanes 4 to 8 versus lanes 2 and 3). Of note, the mutant JH2 domains migrate more slowly on the gel than the wild type, suggesting their structures are different from the wild-type JH2 domain. These data further support the model in which the JH2 domain is able to regulate the catalytic activity by physical interaction with the JH1 domain and strongly argue that the mechanism of the disease pathogenesis in these patients is likely enhanced inhibition of JH1 activity by the mutated JH2 domain.
FIG. 9.
Overexpression of the patient-derived JH2 domain inhibits JH1-mediated STAT5A phosphorylation to a greater extent than that of the wild type. COS7 cells were cotransfected with STAT5A (0.5 μg) and Flag-JH1 (0.5 μg) together with the indicated amounts of various JH2 constructs and the parental vector (total amount of transfected DNA was 5 μg in all cases). Lysates were immunoprecipitated (IP) with anti-STAT5A and blotted (IB) with anti-phosphotyrosine (4G10) (first panel) and anti-STAT5A (second panel). The expression levels of Flag-JH1 and various JH2 proteins were analyzed by immunoblotting with anti-JAK3 (third panel) and an antibody that recognizes the JH2 domains of JAK2 and JAK3 (bottom panel).
DISCUSSION
The pseudokinase, or JH2, domain is a unique and highly conserved feature that defines membership in the family of JAKs, PTKs that appear to have a specialized and essential function in transmitting cytokine receptor-mediated signals. Despite its conservation, the function of this kinase-like domain has been poorly understood. Having identified JAK3-SCID patients with mutations in this domain, we were offered an opportunity to investigate the function of the JH2 domain and the mechanism of disease pathogenesis in these patients.
In this report, we have shown that two patients' alleles encoded kinases with mutations in the JH2 domain; these kinases lacked in vitro catalytic activity and were unable to transmit cytokine-dependent signals. This was true both in immortalized lymphocytes from SCID patients and in a reconstitution system in which fibroblasts expressing the IL-2R constituents were transfected with these JAK3 mutants. Additionally, we show that other distinct mutations in this region also generated defective kinases. We also demonstrate that the JH2 domain interacted with the JH1 domain and regulated kinase activity both positively and negatively. Finally, we show that the mutant JH2 domain inhibits JH1 function to a greater extent than does the wild type. These findings, therefore, establish the importance of the JH2 domain in regulating JAK3 kinase activity and demonstrate an additional mechanism for autosomal SCID pathogenesis.
Previous studies have found that SCID results from mutations in a host of different genes, one form of which, characterized by the absence of T and NK cells, involves defects in JAK3 or γc (3). The majority of previously characterized mutations in JAK3 or γc result in SCID either by abolishing the expression of JAK3 or γc or by disrupting the JAK3-γc interaction (2, 27, 32, 38, 40). Our study provides an alternative mechanism and suggests that disruption of the regulatory function of the JH2 domain of JAK3 can also lead to a SCID phenotype. Our findings that the patients' alleles encoded products that lacked kinase activity are consistent with the clinical presentation of these patients, the degree of their immunological deficits being identical to that of those whose mutations resulted in the complete absence of JAK3 (4). Based on their immunodeficiency, it would be expected that these patients would have dysfunctional kinases, and that was determined to be the case. Intriguingly though, these JAK3 mutants were constitutively phosphorylated in the patient cells and in COS7 cells despite the lack of in vitro kinase activity. Our data argue against the possibility of these mutants being phosphorylated by other kinases as we constructed a double mutant with one patient's mutation (C759R) in conjunction with a mutation of the ATP binding site (C759R/K855A), which was not phosphorylated when overexpressed in COS7 cells. We interpret these data to show that the phosphorylation of JAK3 mutants seen in the cells likely represents autophosphorylation. That these mutants have no kinase activity in vitro but can autophosphorylate in vivo is perplexing. One explanation that might reconcile these seemingly contradictory results would be that wild-type and mutant JAK3s were phosphorylated on different sites. Phosphorylation on an inhibitory tyrosine residue has been shown to be an important regulatory mechanism for several tyrosine kinases (16), a prominent example being the c-Src kinase, in which phosphorylation at Y527 abrogates the catalytic activity (46). The relevant phosphorylation sites in JAKs have begun to be identified but have not been fully characterized. The best-characterized sites are those within the JAK activation loop (8, 12, 25, 47). We have recently identified tyrosine residues 980 and 981 in the activation loop that, when phosphorylated, can activate and inhibit kinase activity, respectively (47). However, these sites were phosphorylated to the same degree in the mutant JAK3 as in the wild type (data not shown). Thus, at present, we cannot explain the function of the JH2 mutants based on the differences in phosphorylation sites. Nonetheless, this hypothesis would provide a good explanation for the findings observed through studying these JH2 mutants. The question remains how and why the mutant kinases are phosphorylated. It is also possible that the patients' mutations result in the changes in JAK3 oligomerization or interaction with a phosphatase, which then lead to phosphorylation on an inhibitory tyrosine. Regardless, resolution of this issue requires complete characterization of JAK3 phosphorylation sites, which is under way.
Our results regarding the complex effects of the JH2 mutations may also help to reconcile some of the seemingly discrepant reports on the regulatory function of the JH2 domain. Previous studies have oppositely concluded that the JH2 domain has a positive (42) or negative (10) regulatory role. In light of our data, these discrepancies can be explained as follows. First, apparent discrepancies may be the result of drawing conclusions based on different assays. For instance, a JAK2 allele with the mutation corresponding to the gain-of-function mutation in the JH2 domain of Hopscotch was reported to be activating because this JAK2 mutant was hyperphosphorylated when overexpressed (26). Kinase activity was not measured, nor was its ability to transmit cytokine-dependent signals. However, we found that the equivalent mutation in JAK3 (E639K) was not activating, as demonstrated by the lack of in vitro autophosphorylation and phosphorylation of exogenous substrates and the failure to transmit IL-2-dependent signals. Thus, our data provide a cautionary note and suggest that the utilization of multiple assays is required to obtain insights into the consequences of a particular mutation. Second, it is entirely possible and indeed likely that the JH2 domain has the capacity to provide both positive and negative regulatory functions, the function being revealed by the precise constructs generated. On one hand, it appears that the JH2 domain is critical for catalytic activity of the full-length molecule and the ability to transmit cytokine-dependent signals, since removal of the pseudokinase domain of JAK3 abrogated kinase activity and IL-2-mediated signaling (Fig. 6 and 7). This is consistent with the findings with TYK2, in which the deletion of the JH2 domain also generated a defective kinase (42). On the other hand, the JH2 domain also inhibits the catalytic activity and the basal phosphorylation of the kinase, as demonstrated by the lack of catalytic activity of the JH2-1 construct and the inhibition of the JH1-mediated phosphorylation of STAT5A by the JH2 domain (Fig. 7 and 9). These results are consistent with the findings obtained with a growth hormone receptor-JAK2 chimera, in which deletion of the JH2 domain led to more robust signaling (10). In light of our data obtained with JAK3, we suggest a model in which the N terminus and the JH2 domain both interact with the JH1 domain. Removal of either likely deforms the structure and interferes with the kinase activity. This would explain how the JH2 domain could regulate the kinase activity both positively and negatively, depending upon the construct made. We are presently investigating the ability of the N terminus to interact with the JH1 domain. Nonetheless, one must consider that inherent differences among different JAKs that limit extrapolation of the results obtained with one JAK to another may exist. For instance, while conserved Y1007 in the activation loop of JAK2 is required for ligand-induced JAK2 kinase activity, the corresponding Y981 in JAK3 inhibits JAK3 kinase activity (8, 47). The JAK3 E639K mutant is another example of this point. Thus, various JAKs may be regulated in subtly different ways; what appears to be true for JAK3 may not be necessarily the case for other JAKs.
A previous study by Fujitani et al. (11) showed that the JH2 domain of JAK2 was able to mediate the direct interaction with STAT5, suggesting a potential function of the JH2 domain as a docking site for STATs and possibly other signaling molecules. Consistent with this notion, we found that mutant JAK3 ΔJH2 was inducibly phosphorylated upon IL-2 stimulation but was unable to mediate IL-2 induced phosphorylation of STAT5 and STAT3. Though the ΔJH2 mutant contains minimal in vitro kinase activity, it is possible that deletion of the JH2 domain in this mutant also disrupted JAK-STAT interaction and further contributed to the failure of STAT activation seen in the cells expressing this mutant.
Our analysis of the present mutants also provides a useful prelude to structural information on JAKs. The findings that the kinase-like domain interacted with the kinase domain and inhibited its catalytic activity suggest a model in which these two domains may be closely positioned to each other in the three-dimensional structure. The crystal structure of the nonreceptor tyrosine kinase c-Src has recently been determined (46). In this kinase, the SH2 domain interacts with the phosphorylated C-terminal tail, whereas the SH3 domain interacts with the SH2-kinase domain linker. Both interactions appear to inhibit kinase activity by restricting ATP access to the active site and by destabilizing the active structure of the kinase domain. This is consistent with earlier biochemical studies indicating that both the SH2 domain and the SH3 domain are involved in negative regulation of the c-Src catalytic activity. Though JAK3 does not contain obvious SH2 or SH3 domains, modeling of the JAK3 kinase domain suggests that many similarities between the structures of c-Src and JAK3 may exist (data not shown). By analogy with c-Src (46), one might speculate that, in the absence of ligand, the interaction between the JH2 domain and the kinase domain interferes with substrate access to the kinase domain. Ligand-induced conformational change may disrupt this inhibitory intramolecular interaction, resulting in an active conformation, permitting access to the binding sites for ATP and protein substrates. The JH2 domain could also be required for stabilizing the active conformation. Perhaps, the mutations in the JH2 domain that we investigated in this study generate a kinase with a partially open conformation that accommodates initial substrate binding; however, the subsequent autophosphorylation locks them in an even more stable inactive or closed conformation, preventing further activation. This model, if proved to be true, might help explain their hyperphosphorylation in vivo in the absence of ligand but lack of kinase activity and failure to transmit cytokine-dependent signals. Regardless of the model, our data clearly argue for direct interactions between the pseudokinase and catalytic domains. The crystal structure of the JAKs, when available, will prove whether the JH1 and JH2 domains truly interact and if the interaction is an important aspect of kinase regulation.
In summary, our data demonstrate that naturally occurring mutations in the JH2 domain disrupt the regulatory function of the JH2 domain and result in nonfunctional kinases. We also show that the JH2 domain of JAK3 can regulate the kinase activity both positively and negatively, likely through interaction with the JH1 kinase domain. These data allow an understanding of disease pathogenesis in some cases of autosomal SCID, in which patients express the JAK3 protein and offer insights into the molecular mechanism involved in regulating the catalytic activity of JAK3. Given the importance of JAK3 in the proper development and function of the immune system and its restricted expression in hematopoietic cells (13, 20, 27, 38), targeting JAK3 kinase activity is an attractive possibility for generating novel immunosuppressants (43). In addition, JAKs also appear to be important in the pathogenesis of some malignancies (6, 21, 28, 29); thus, specific inhibition of JAK function might be useful in treating these diseases. The complex regulatory function of the JH2 domain of the JAKs, therefore, may also have pragmatic importance. Clearly, the JH2 domain is a unique aspect of the structure of the JAKs, and it will be of great interest to determine if this singular feature can be exploited pharmacologically.
ACKNOWLEDGMENTS
We thank Stuart Frank, David Frucht, Massimo Gadina, Henry Metzger, Juan Rivera, Pam Schwarzberg, Harold Varmus, Roberta Visconti, and Owen Witte for helpful discussions. We also thank Lothar Hennighausen and Warren J. Leonard for reagents.
This work was supported in part by NATO grant CRG.CRG 973041 (L.D.N.) and Telethon grant E. 668 (L.D.N.).
REFERENCES
- 1.Boussiotis V A, Barber D L, Nakarai T, Freeman G J, Gribben J G, Bernstein G M, D'Andrea A D, Ritz J, Nadler L M. Prevention of T cell anergy by signaling through the gamma c chain of the IL-2 receptor. Science. 1994;266:1039–1042. doi: 10.1126/science.7973657. [DOI] [PubMed] [Google Scholar]
- 2.Cacalano N A, Migone T S, Bazan F, Hanson E P, Chen M, Candotti F, O'Shea J J, Johnston J A. Autosomal SCID caused by a point mutation in the amino-terminus of JAK3: mapping of the JAK3-receptor interaction domain. EMBO J. 1999;18:1549–1558. doi: 10.1093/emboj/18.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Candotti F, O'Shea J J, Villa A. Severe combined immune deficiencies due to defects of the common gamma chain-JAK3 signaling pathway. Springer Semin Immunopathol. 1998;19:401–415. doi: 10.1007/BF00792599. [DOI] [PubMed] [Google Scholar]
- 4.Candotti F, Oakes S A, Johnston J A, Giliani S, Schumacher R F, Mella P, Fiorini M, Ugazio A G, Badolato R, Notarangelo L D, Bozzi F, Macchi P, Strina D, Vezzoni P, Blaese R M, O'Shea J J, Villa A. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood. 1997;90:3996–4003. [PubMed] [Google Scholar]
- 5.Chen M, Cheng A, Chen Y Q, Hymel A, Hanson E P, Kimmel L, Minami Y, Taniguchi T, Changelian P S, O'Shea J J. The amino terminus of JAK3 is necessary and sufficient for binding to the common gamma chain and confers the ability to transmit interleukin 2-mediated signals. Proc Natl Acad Sci USA. 1997;94:6910–6915. doi: 10.1073/pnas.94.13.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danial N N, Pernis A, Rothman P B. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 7.Darnell J E J, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Witthuhn B A, Matsuda T, Kohlhuber F, Kerr I M, Ihle J N. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol. 1997;17:2497–2501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski J J. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- 10.Frank S J, Yi W, Zhao Y, Goldsmith J F, Gilliland G, Jiang J, Sakai I, Kraft A S. Regions of the JAK2 tyrosine kinase required for coupling to the growth hormone receptor. J Biol Chem. 1995;270:14776–14785. doi: 10.1074/jbc.270.24.14776. [DOI] [PubMed] [Google Scholar]
- 11.Fujitani Y, Hibi M, Fukada T, Takahashi-Tezuka M, Yoshida H, Yamaguchi T, Sugiyama K, Yamanaka Y, Nakajima K, Hirano T. An alternative pathway for STAT activation that is mediated by the direct interaction between JAK and STAT. Oncogene. 1997;14:751–761. doi: 10.1038/sj.onc.1200907. [DOI] [PubMed] [Google Scholar]
- 12.Gauzzi M C, Velazquez L, McKendry R, Mogensen K E, Fellous M, Pellegrini S. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J Biol Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 13.Gurniak C B, Berg L J. Murine JAK3 is preferentially expressed in hematopoietic tissues and lymphocyte precursor cells. Blood. 1996;87:3151–3160. [PubMed] [Google Scholar]
- 14.Harpur A G, Andres A C, Ziemiecki A, Aston R R, Wilks A F. JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene. 1992;7:1347–1353. [PubMed] [Google Scholar]
- 15.Hou J, Schindler U, Henzel W J, Wong S C, McKnight S L. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity. 1995;2:321–329. doi: 10.1016/1074-7613(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard S R, Mohammadi M, Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. J Biol Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- 17.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 18.Johnston J A, Bacon C M, Finbloom D S, Rees R C, Kaplan D, Shibuya K, Ortaldo J R, Gupta S, Chen Y Q, Giri J D, O'Shea J J. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc Natl Acad Sci USA. 1995;92:8705–8709. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston J A, Kawamura M, Kirken R A, Chen Y Q, Blake T B, Shibuya K, Ortaldo J R, McVicar D W, O'Shea J J. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature. 1994;370:151–153. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura M, McVicar D W, Johnston J A, Blake T B, Chen Y Q, Lal B K, Lloyd A R, Kelvin D J, Staples J E, Ortaldo J R, O'Shea J J. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacronique V, Boureux A, Valle V D, Poirel H, Quang C T, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 22.Leonard W J. Dysfunctional cytokine receptor signaling in severe combined immunodeficiency. J Investig Med. 1996;44:304–311. [PubMed] [Google Scholar]
- 23.Leonard W J. Type I cytokines and interferons. In: Paul W E, editor. Fundamental immunology. 4th ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1999. pp. 741–774. [Google Scholar]
- 24.Lin J X, Migone T S, Tsang M, Friedmann M, Weatherbee J A, Zhou L, Yamauchi A, Bloom E T, Mietz J, John S, Leonard W J. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 25.Liu K D, Gaffen S L, Goldsmith M A, Greene W C. Janus kinases in interleukin-2-mediated signaling: JAK1 and JAK3 are differentially regulated by tyrosine phosphorylation. Curr Biol. 1997;7:817–826. doi: 10.1016/s0960-9822(06)00369-1. [DOI] [PubMed] [Google Scholar]
- 26.Luo H, Rose P, Barber D, Hanratty W P, Lee S, Roberts T M, D'Andrea A D, Dearolf C R. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17:1562–1571. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macchi P, Villa A, Gillani S, Sacco M G, Frattini A, Porta F, Ugazio A G, Johnston J A, Candotti F, O'Shea J J, Vezzoni P, Notarangelo L D. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 28.Meydan N, Grunberger T, Dadi H, Shahar M, Arpaia E, Lapidot Z, Leeder J S, Freedman M, Cohen A, Gazit A, Levitzki A, Roifman C M. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature. 1996;379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]
- 29.Migone T S, Lin J X, Cereseto A, Mulloy J C, O'Shea J J, Franchini G, Leonard W J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 30.Minami Y, Oishi I, Liu Z J, Nakagawa S, Miyazaki T, Taniguchi T. Signal transduction mediated by the reconstituted IL-2 receptor. Evidence for a cell type-specific function of IL-2 receptor beta-chain. J Immunol. 1994;152:5680–5690. [PubMed] [Google Scholar]
- 31.Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu Z J, Oishi I, Silvennoinen O, Witthuhn B A, Ihle J N, Tanaguchi T. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 32.Morelon E, Dautry-Varsat A, Le Deist F, Hacein-Bay S, Fischer A, de Saint Basile G. T-lymphocyte differentiation and proliferation in the absence of the cytoplasmic tail of the common cytokine receptor gamma c chain in a severe combined immune deficiency X1 patient. Blood. 1996;88:1708–1717. [PubMed] [Google Scholar]
- 33.Noguchi M, Yi H, Rosenblatt H M, Filipovich A H, Adelstein S, Modi W S, McBride O W, Leonard W J. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 34.Nosaka T, van Deursen J M, Tripp R A, Thierfelder W E, Witthuhn B A, McMickle A P, Doherty P C, Grosveld G C, Ihle J N. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 35.O'Shea J J, Leonard W J. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 36.Park S Y, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 37.Russell S M, Johnston J A, Noguchi M, Kawamura M, Bacon C M, Friedmann M, Berg M, McVicar D W, Witthuhn B A, Silvennoinen O, Goldman A S, Schmalsteig F C, Ihle J N, O'Shea J J, Leonard W J. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 38.Russell S M, Tayebi N, Nakajima H, Riedy M C, Roberts J L, Aman M J, Migone T S, Noguchi M, Markert M L, Buckley R H, O'Shea J J, Leonard W J. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 39.Sakai I, Kraft A S. The kinase domain of Jak2 mediates induction of bcl-2 and delays cell death in hematopoietic cells. J Biol Chem. 1997;272:12350–12358. doi: 10.1074/jbc.272.19.12350. [DOI] [PubMed] [Google Scholar]
- 40.Schmalstieg F C, Leonard W J, Noguchi M, Berg M, Rudloff H E, Denney R M, Dave S K, Brooks E G, Goldman A S. Missense mutation in exon 7 of the common gamma chain gene causes a moderate form of X-linked combined immunodeficiency. J Clin Investig. 1995;95:1169–1173. doi: 10.1172/JCI117765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomis D C, Gurniak C B, Tivol E, Sharpe A H, Berg L J. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 42.Velazquez L, Mogensen K E, Barbieri G, Fellous M, Uze G, Pellegrini S. Distinct domains of the protein tyrosine kinase tyk2 required for binding of interferon-alpha/beta and for signal transduction. J Biol Chem. 1995;270:3327–3334. doi: 10.1074/jbc.270.7.3327. [DOI] [PubMed] [Google Scholar]
- 43.Waldmann T A, O'Shea J J. The use of antibodies against the IL-2 receptor in transplantation. Curr Opin Immunol. 1998;10:507–512. doi: 10.1016/s0952-7915(98)80215-x. [DOI] [PubMed] [Google Scholar]
- 44.Wilks A F, Harpur A G, Kurban R R, Ralph S J, Zurcher G, Ziemiecki A. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witthuhn B A, Silvennoinen O, Miura O, Lai K S, Cwik C, Liu E T, Ihle J N. Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature. 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 46.Xu W, Harrison S C, Eck M J. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y J, Hanson E P, Chen Y Q, Magnuson K, Chen M, Swann P G, Wange R L, Changelian P S, O'Shea J J. Distinct tyrosine phosphorylation sites in JAK3 kinase domain positively and negatively regulate its enzymatic activity. Proc Natl Acad Sci USA. 1997;94:13850–13855. doi: 10.1073/pnas.94.25.13850. [DOI] [PMC free article] [PubMed] [Google Scholar]