Abstract
Background
Antisense peptide nucleic acids (PNAs) constitute an alternative to traditional antibiotics, by their ability to silence essential genes.
Objectives
To evaluate the antibacterial effects of antisense PNA-peptide conjugates that target the gene encoding the alpha subunit (NrdA) of the Escherichia coli ribonucleotide reductase (RNR).
Methods
Bacterial susceptibility of a series of NrdA-targeting PNAs was studied by MIC determination and time–kill analysis. Western-blot analysis, gene complementation and synergy with hydroxyurea were employed to determine the efficiency of NrdA-PNA antisense treatment. The effect on chromosome replication was addressed by determining the DNA synthesis rate, by flow cytometry analysis, by quantitative PCR and by fluorescence microscopy. The use of DNA repair mutants provided insight into the bactericidal action of NrdA-PNA.
Results
Treatment with NrdA-PNA specifically inhibited growth of E. coli, as well as NrdA protein translation at 4 μM. Also, the DNA synthesis rate was reduced, preventing completion of chromosome replication and resulting in formation of double-stranded DNA breaks and cell death.
Conclusions
These data present subunits of the NrdAB RNR as a target for future antisense microbial agents and provide insight into the bacterial physiological response to RNR-targeting antimicrobials.
Introduction
Since the ‘golden era’ of antibiotic drug discovery (1930–60s), decades of antibiotic administration in hospital settings, as well as in agriculture, has led to the emergence of MDR bacteria. Both the CDC and the WHO have declared such bacteria an emerging global health threat and emphasize the urgent need for advancing new classes of antibiotics with novel modes of action. Peptide nucleic acids (PNAs) can potentially fulfil this requirement.
PNA molecules are chemically related to DNA, but with the entire deoxyribose backbone of DNA replaced by a structurally similar achiral, uncharged and flexible pseudo-peptide backbone, consisting of 2-amino-ethyl-glycin units. PNA is a potent DNA structural mimic that can form strong sequence specific complexes with RNA and DNA via Watson-Crick base-pairing.1 The pseudo-peptide backbone of PNA is stable and resistant to biological degradation in human serum, bacterial cell extracts and nuclear extracts from mouse tumour cells and by proteinase/peptidases.2 PNAs designed as antisense oligonucleotides against transcripts encoding essential bacterial proteins hold promise as antimicrobials. Such PNAs act by sterically hindering translation and the optimal effect is seen when targeting the sequence between the start codon and the ribosome binding site of the mRNA.3 Antibacterial PNAs have previously targeted essential cellular processes, including protein synthesis,4 fatty acid biosynthesis,5 cell division,6 resistance genes,7 efflux pumps,8 biofilm formation and toxin-antitoxin systems.9 The outer membrane poses a barrier for successful PNA uptake in Gram-negative bacteria.10,11 Therefore, PNAs are conjugated to bacteria-penetrating peptides (BPPs) such as (KFF)3K5 (PNA-peptide).
Ribonucleotide reductases (RNRs) catalyse the rate-limiting step of the de novo pathway for DNA precursor synthesis, by reducing ribonucleotide di/triphosphates to deoxyribonucleotides di/triphosphates.12 RNRs are grouped into three classes (reviewed elsewhere12). Escherichia coli carries genes encoding an iron-dependent class Ia RNR that is essential for viability during aerobic growth (nrdAB), a manganese-dependent class Ib RNR (nrdEFHI) and a strictly anaerobic class III RNR (nrdDG). nrdA and nrdB are transcribed as a polycistronic mRNA with the nrdA gene located upstream of the nrdB gene, which is followed by a third non-essential gene, yfaE.12
Here we designed and tested antisense PNA-peptide conjugates against E. coli nrdA. The anti-NrdA-PNA inhibited bacterial growth, while displaying typical hallmarks of deoxyribonucleotidetriphoshate (dNTP) starvation; this includes decreased DNA synthesis rate, increased hydroxyurea (HU) sensitivity and inability to complete chromosome replication. We propose that, during NrdA-PNA treatment, DNA replication is reduced due to dNTP starvation, which eventually causes an accumulation of double-stranded DNA breaks, beyond the repair capability of cells, leading to cell death.
Materials and methods
Bacterial growth conditions
Cells were grown at 37°C in non-cation-adjusted Mueller–Hinton broth (MHB-I; Sigma–Aldrich), LB or AB minimal medium13 supplemented with 10 mg/L thiamine, 0.2% glucose and 0.5% casamino acids (AB medium). Antibiotics were used at the following concentrations: ampicillin, 150 mg/L; chloramphenicol; 20 mg/L; tetracycline, 10 mg/L; kanamycin, 50 mg/L; ciprofloxacin, 0.0125 mg/L; HU, 50 mM; rifampicin, 300 mg/L; and cefalexin, 36 mg/L.
Bacterial strains and plasmid construction
All strains used in this study are listed in Table S1 (available as Supplementary data at JAC Online). The relevant mutations were transferred to MG1655 by P1 transduction. The nrdAB-yfaE region was PCR amplified from MG1655 using primer pair 5’-aagcttcgttgtatctggca and 5’-acctcgagctctagattaaggaggccatatgaatcagaatctgctggt. This PCR product does not contain any sequences upstream of the nrdA start codon. The PCR product was digested with XhoI and HindIII and ligated into plasmid pFH2102, digested with the same enzymes, resulting in plasmid pCC:nrdAByfaE. In this plasmid, also carrying the lacI and bla genes, nrdAB-yfaE is under control of the IPTG-inducible promoter lacPA1-04/03.14,15 Cultures of pCC:nrdAByfaE were grown in the presence of ampicillin.
PNA design, synthesis and handling
PNAs were designed complementary to the nrdA mRNA region around and between the Shine-Dalgarno sequence and the start codon of nrdA (Table 1). Optimal pairing parameters for a 10 bp target region were estimated using Oligo Walk 5.0 (https://rna.urmc.rochester.edu/servers/oligowalk2/help.html), followed by a nrdA specificity check using NCBI Blastn software (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch; Table S2). PNAs were synthesized by continuous standard solid-phase peptide synthesis, with tBoc-protected monomers, on an MBHA-resin and purified and characterized as described previously.16 For a detailed description of PNA handling, materials and concentration determination, see Goltermann and Nielsen.17
Table 1.
PNA-peptide conjugates used and MIC values
| PNA | Sequence incl. BPP (5’-3’) | Target in mRNA sequence | MIC (μM) |
|---|---|---|---|
| PNA5561 (NrdA-PNA) | H-(KFF)3K-eg1-ATG TAT GTC G-NH2 | CAGGUACGACAUACAUGAAUCAG | 4 |
| PNA5645 (2-base mismatch-PNA) | H-(KFF)3K-eg1-ATT TAT GGC G-NH2 | NA | 8 |
| PNA5866 (4-base mismatch-PNA) | H-(KFF)3K-eg1-ATTGAT TGC G -NH2 | NA | 16 |
| PNA 5562 | H-(KFF)3K-eg1-ATG TCG TAC C-NH2 | CAGGUACGACAUACAUGAAUCAG | 8 |
| PNA5563 | H-(KFF)3K-eg1-TGT ATG TCG T-NH2 | CAGGUACGACAUACAUGAAUCAG | 8 |
NA, not applicable.
Sequences of NrdA-targeting PNA-peptides, their binding positions on mRNA target and their corresponding MIC values. The nrdA start codon is highlighted in bold, the PNA binding position is double underlined and mismatch bases are underlined. PNA-peptides were synthesized as described previously.48
MIC determination
Activities of antisense PNA-peptides were determined by broth microdilution on MG1655, using a CLSI protocol modified for PNA use. Because PNA-peptides can adhere to polystyrene surfaces, the modified protocol includes the use of low-binding plastic materials, non-cation-adjusted Mueller–Hinton broth and PNA-peptide stocks prepared in 0.4% BSA/0.02% acetic acid. MICs were determined using a BIOTEK™ Synergy H1 microplate reader.17 MICs of chloramphenicol and nalidixic acid in AB medium were determined as described by Oddo et al.18
Time–kill assays
Bacterial cells were grown exponentially in MHB-I and diluted to 1–5 × 105 cfu/mL. PNA-peptide was added and incubation continued at 37°C, while continuously shaking at 225 rpm. Samples were collected in 1 h intervals for 5 h, diluted in 0.9% NaCl, plated on LB agar plates and cfu counted after incubation at 37°C overnight. Survival fractions were calculated as the number of cfu relative to the initial inoculum.
Western-blot analysis
WT cells were grown exponentially in AB medium. At OD450 = 0.05, cells were treated with PNA-peptide (8 μM) and harvested after 3 h. Cells were washed in 10 mM Tris-HCl pH 8/10 mM MgCl2. Samples were heated at 95°C for 5 min and sonicated using a Branson 450 Sonifier. Protein content was determined using Bradford Reagent (Sigma–Aldrich) and normalized. Total protein (1.25 μg) was separated using SDS-PAGE (Precast Gel: 10%–20% Tris-HCL; Bio-Rad) in an XCell4 SureLock™ Midi-Cell (ThermoFisher Scientific) and transferred to a PVDF membrane (GE Healthcare, Whatman™), using a semi-dry blotting apparatus (JKA Biotech, Denmark). NrdA protein was detected with a rabbit polyclonal anti-NrdA antibody (MyBiosource) and goat anti-rabbit immunoglobulins/HRP (Agilent) as the secondary antibody. The PVDF membrane was incubated with ECL chemiluminescence substrate (Bio-Rad) and the signal detected using an ImageQuant LAS400 (GE Healthcare, Life Sciences).
Flow cytometry and quantitative PCR
Cell cycle parameters and the ori/ter ratio were determined by flow cytometry using an Apogee A10 Bryte instrument and by quantitative PCR as described previously.19–22 Samples used for quantitative PCR and flow cytometry were taken from cultures prior to and following rifampicin/cefalexin treatment. Briefly, 1 mL of cells was centrifuged and the cell pellet fixed in 70% ethanol and 100 μL of 10 mM Tris buffer pH 7.5 and stored at 4°C. For determination of number of origins per cell, 1 mL of culture was incubated at 37°C for 4 h with rifampicin (300 mg/L) and cefalexin (36 mg/L) prior to fixation. Rifampicin inhibits RNA synthesis and indirectly initiation of DNA replication, but ongoing replication is allowed to finish. Cefalexin inhibits cell division. Thus, the number of chromosomes per cell represents the number of origins of replication prior to rifampicin/cefalexin treatment.
Macromolecular synthesis
Cells were grown exponentially at 37°C in AB medium without casamino acids and diluted to OD450 = 0.05. After 40 min of further growth, each culture was treated with PNA-peptide (12 μM), nalidixic acid (10×MIC, 40 mg/L) or chloramphenicol (10×MIC, 80 mg/L) or left untreated. In order to ensure complete shutdown of DNA and protein synthesis, 10×MIC was used. Samples were collected both prior to and following drug treatment. DNA and protein synthesis rates were determined by incorporation of methyl-3H-labelled thymidine and arginine, respectively. For each pulse-labelling a 500 μL aliquot of culture was added to 10 μL (0.375 μCi) of precursor and incubated for 4 min at 37°C, before adding 5 mL of ice-cold TCA (5%)/1 mM NaCl. All samples were filtered through a glass filter (ADVANTEC®, 25 mm, lot no. 80302708), washed with 5 mL of TCA solution and dried overnight in a scintillation vial (PV1AS, MERIDIAN, BIOTECHNOLOGIES Ltd). Samples were counted on a HINDEX 300SL scintillation counter (scintillation fluid ULTIMA GOLD™; PerkinElmer; lot no. 77–18151).
In vivo origin and terminus visualization by fluorescence microscopy
Strain ALO4223 was grown exponentially in AB medium, diluted to OD450 = 0.05 and treated with PNA-peptide (8 μM) for 2 h. Fluorescence microscopy analysis was performed using an AxioImager Z1 microscope (Carl Zeiss MicroImaging, Inc), with a × 100 objective and a Hamamatsu ORCA-ER C4742-80-12AG camera, as described previously.20 The images were analysed, processed and intensity adjusted using Image J Software.23 Note that foci co-localization and resolution limitations may lead to underestimation of especially the number of oriC copies.
In vivo double-stranded DNA break visualization by fluorescence microscopy
Strain SNR14350 was grown exponentially in AB medium, diluted to OD450 = 0.05 and incubated with PNA-peptide (8 μM) or ciprofloxacin (0.0125 mg/L) for 4 h. Synthesis of Gam-GFP was induced by addition of anhydrotetracycline (10 mg/L) at the same time as drug addition. Fluorescence microscopy was performed using a Nikon Eclipse Ti-E inverted microscope as described by Haugan et al.24 The images were analysed, processed and intensity adjusted using Image J Software.23 Note that Gam-GFP foci co-localization and resolution limitations may lead to underestimation of double-stranded DNA breaks.
Statistical analysis
GraphPad Prism (v.5, GraphPad Software) was used for graph illustrations and statistical analysis. One-way analysis of variance (ANOVA) was performed to evaluate the level of significance observed using Dunnett’s multiple comparison test comparing treated with non-treated samples. Statistical significance was set to a P value of ≤0.01.
Results
NrdA-PNA inhibits nrdA translation and bacterial growth
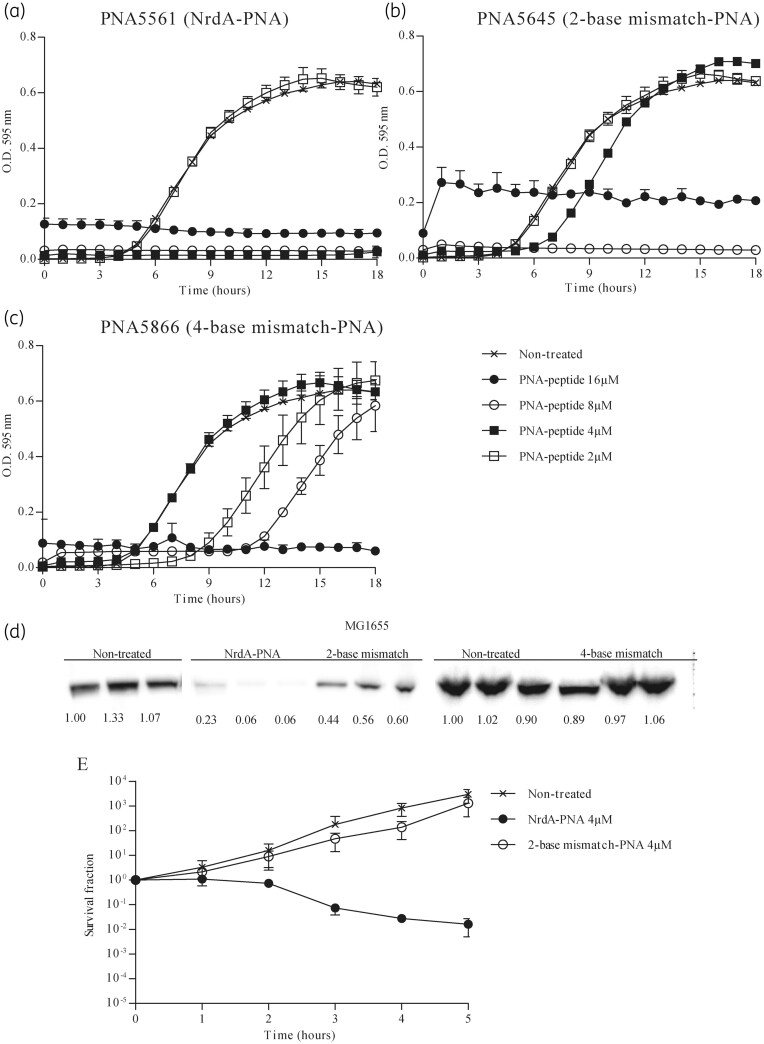
In order to target nrdA mRNA translation, PNA5561, PNA5562 and PNA5563 were designed to anneal to a 10 nt region near the start codon and the ribosomal binding site region of the E. coli nrdA mRNA (Table 1). All PNAs were conjugated to the (KFF)3K BPP. When bacterial susceptibility was tested for WT E. coli cells, all three NrdA-PNA constructs inhibited cell growth, with PNA5561 showing the lowest MIC at 4 μM (Figure 1a, Figure S1 and Table 1). PNA5561 is referred to as NrdA-PNA for the remainder of this paper. A 2-base mismatch-PNA (PNA5645), with 2 bases swapped, exhibited an increased MIC of 8 μM, whereas a 4-base mismatch-PNA (PNA5866), with 4 bases swapped, exhibited a higher MIC of 16 μM (Figure 1b and c and Table 1), suggesting that a PNA with 2 mismatches out of 10 bases may retain some affinity for the target. Western-blot analysis, using an NrdA-specific antibody, revealed an ∼90% decreased cellular NrdA protein level in cells treated with 8 μM NrdA-PNA (Figure 1d). The 2-base mismatch-PNA caused an ∼50% decrease in the NrdA level, confirming that the 2-base mismatch-PNA does indeed have a target effect, although with a lower efficiency. The 4-base mismatch-PNA did not inhibit NrdA protein levels at 8 μM, suggesting loss of target affinity (Figure 1d). We chose to include the 2-base mismatch-PNA (PNA5645) in further studies to evaluate the effect of reduced target affinity, while always comparing with untreated cells as a control.
Figure 1.
NrdA-PNA inhibits nrdA translation and bacterial growth. Bacterial growth inhibition of PNA-peptide conjugates: (a) NrdA-PNA (PNA5561), (b) 2-base mismatch-PNA (PNA5645) and (c) 4-base mismatch-PNA (PNA5866). Non-treated (crosses), 16 μM (filled circles), 8 μM (open circles), 4 μM (filled squares) and 2 μM (open squares). Overnight cultures of WT E. coli were diluted to 105 cfu/mL in MHB-I and incubated with appropriate concentrations of PNA-peptide conjugates and incubated at 37°C for 18 h, while turbidity was measured at OD595. (d) Effect of PNA-peptide on NrdA translation. Protein extracts of cells growing exponentially in AB medium were exposed to 8 μM of the indicated PNA-peptide at OD450 = 0.05. Samples were collected after 3 h of PNA-peptide exposure, proteins separated in SDS-PAGE gels and the NrdA protein detected by immunoblotting. Shown below each band is the band intensity relative to WT. (e) Bactericidal effect of NrdA-PNA. WT cells were grown exponentially in MHB-I medium, diluted to 1–5 × 105 cfu/mL and treated with 4 μM PNA-peptide for 5 h. Non-treated (crosses), NrdA-PNA (4 μM, filled circles) and 2-base mismatch-PNA (4 μM, open circles). Cell viability is displayed as survival fractions in cfu/mL. For details, see the Materials and methods section. Shown is the mean±SD based on three independent determinations.
Treatment of WT cells with NrdA-PNA caused a gradual loss of viability over 5 h, with a decrease in cfu of about 2 log, demonstrating bactericidal activity (Figure 1e). The 2-base mismatch-PNA had no effect on viability when used at the same concentration.
NrdA-PNA activity depends on the nrdAB transcription level and is enhanced by HU
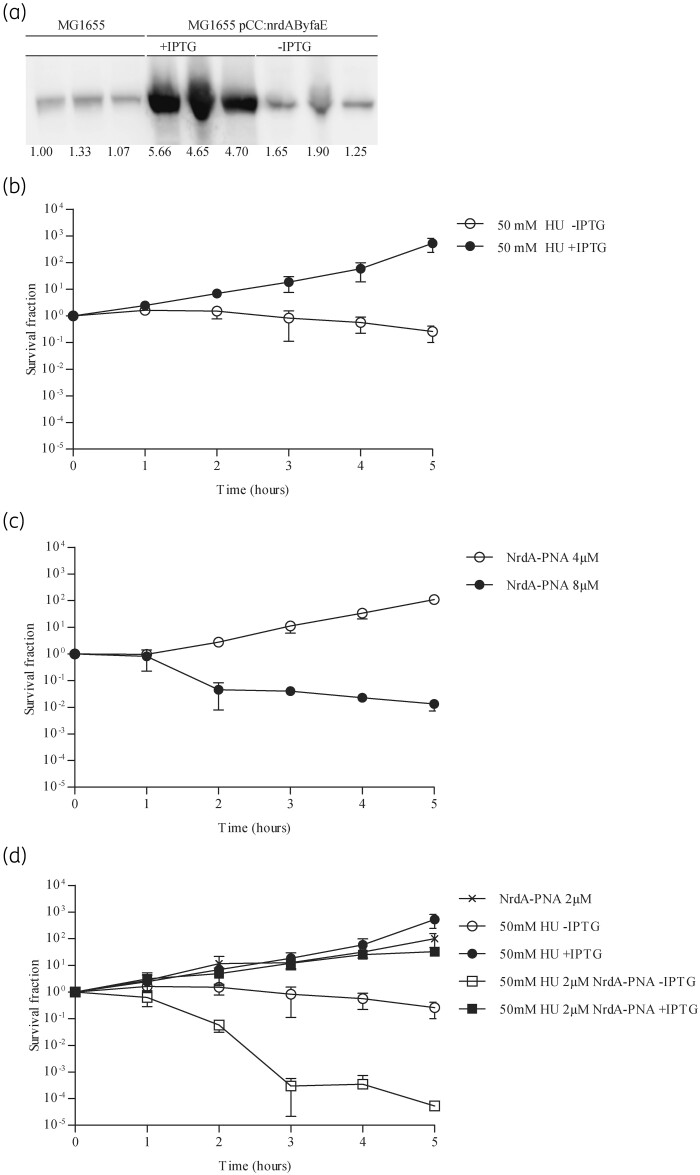
In order to verify that the bactericidal effect of NrdA-PNA was mediated through inhibition of nrdA translation we cloned the complete nrdAB-yfaE operon into a pBR322-based vector under the control of an IPTG-inducible promoter (pCC:nrdAByfaE). In this construct, none of the sequences upstream of the nrdA start codon, which the NrdA-PNA targets, is present. Expression of NrdA was confirmed by Western blot (Figure 2a). Plasmid expressed NrdAB conferred resistance to 50 mM HU confirming that the enzyme was functional (Figure 2b). Cells overexpressing NrdAB were less susceptible to NrdA-PNA (compare Figure 2c with Figure 1e).
Figure 2.
NrdA-PNA activity is specifically dependent on nrdAB transcription level and RNR activity. (a) Plasmid pCC:nrdAByfaE directs overproduction of the NrdA protein. Protein extracts were prepared from WT cells carrying pCC:nrdAByfaE growing exponentially in AB medium containing 150 mg/L ampicillin and induced by 1 mM IPTG. Samples were collected at OD450 = 0.4–5, proteins separated in SDS-PAGE gels and NrdA protein detected by immunoblotting. Shown below each band is the intensity relative to WT. (b) Excess NrdAB confers HU resistance. Exponentially growing cultures of WT carrying pCC:nrdAByfaE in MHB-I containing 150 mg/L ampicillin were treated with 50 mM HU plus (filled circles) or minus (open circles) 1 mM IPTG. Cell viability is displayed as survival fractions in cfu/mL. Shown is the mean±SD based on three independent determinations. (c) WT cells expressing NrdAB are less susceptible to NrdA-PNA. Exponentially growing cultures of WT carrying pCC:nrdAByfaE in MHB-I containing 150 mg/L ampicillin were diluted to 1–5 × 105 cfu/mL in MHB-I supplemented with 1 mM IPTG and challenged with NrdA-PNA at 4 μM (open circles) and 8 μM (filled circles). Cell viability is displayed as survival fractions in cfu/mL. Shown is the mean±SD based on three independent determinations. (d) Sub-MIC levels of NrdA-PNA sensitize E. coli to HU. Exponentially growing cultures of WT carrying pCC:nrdAByfaE in MHB-I, in the presence of ampicillin, were diluted to 1–5 × 105 cfu/mL in MHB-I and treated with sub-MIC levels of NrdA-PNA (2 μM, crosses), HU minus IPTG (50 mM, open circles), HU plus 1 mM IPTG (50 mM, filled circles) or combined (50 mM HU and 2 μM PNA) with (filled squares) or without (open squares) 1 mM IPTG. Cell viability is displayed as survival fractions in cfu/mL. For details, see the Materials and methods section. Shown is the mean±SD based on three independent determinations.
We further tested whether NrdA-PNA acts in synergy with HU, both being specific inhibitors of RNR, albeit with different modes of action. At a sub-MIC concentration of 2 μM NrdA-PNA, only a moderate effect on bacterial growth was observed, whereas treatment with 50 μM HU resulted in a slight <1 log reduction in cfu/mL over a period of 5 h (Figure 2d). When cells were treated with the combination of 50 μM HU and 2 μM NrdA-PNA, cell viability rapidly decreased more than 3 log after 5 h (Figure 2d). In addition, overproduction of NrdAB restored the viability of cells treated with both drugs, confirming that NrdA-PNA and HU inhibit NrdA by different mechanisms (Figure 2d).
Collectively, the lowered NrdA protein level, the NrdA complementation analysis and the HU sensitivity strongly suggest that NrdA-PNA acts specifically via reduction of nrdA translation.
NrdA-PNA specifically inhibits DNA synthesis
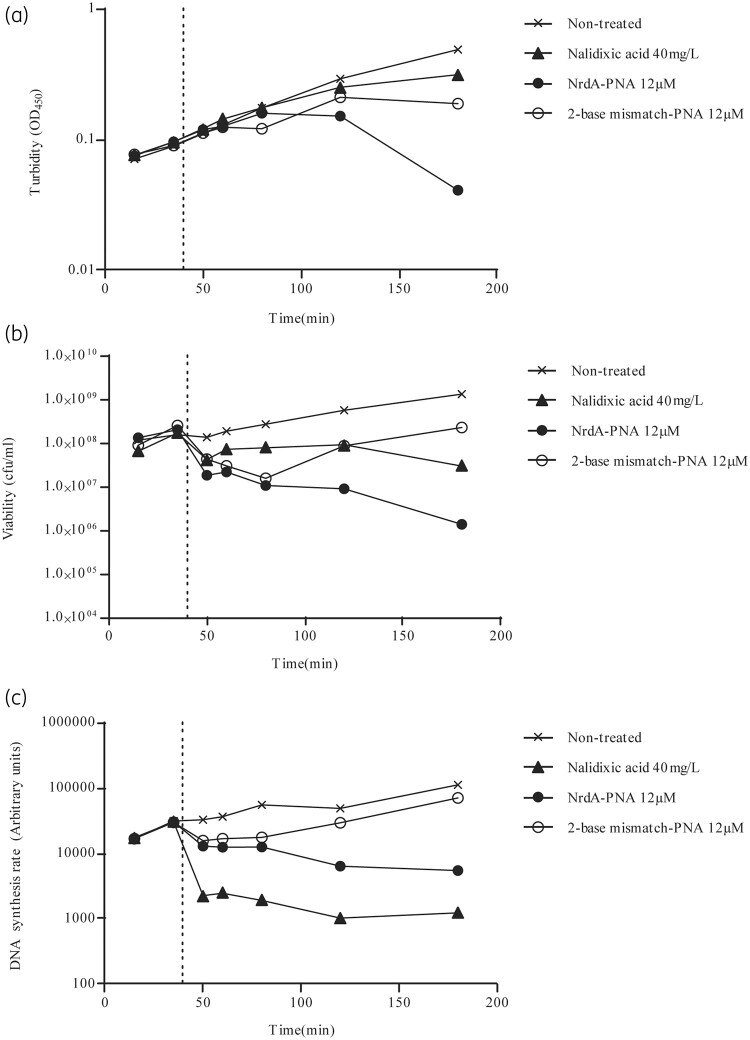
Because RNR catalyses the rate-limiting step of de novo dNTP synthesis,12 we proceeded to determine the effect of NrdA-PNA on cellular DNA synthesis. When exponentially growing WT cells were treated with 12 μM NrdA-PNA, growth ceased as determined by no further increase in optical density (Figure 3a and Figure S2A) and cell viability was reduced (Figure 3b and Figure S2B) as was the overall DNA synthesis rate (Figure 3c). As expected, the 2-base mismatch-PNA caused a smaller decrease in cell growth, viability (Figure 3a and b) and rate of DNA synthesis (Figure 3c), consistent with a reduced, but not complete, loss of activity for this compound. Treatment with the known DNA synthesis inhibitor nalidixic acid resulted in a faster cessation of DNA replication (Figure 3c), whereas the effects on optical density and cell counts were more moderate relative to NrdA-PNA (Figure 3a and b). NrdA-PNA treatment did not affect protein synthesis (Figure S2C), showing that inhibition of DNA replication is not a consequence of inhibition of protein synthesis.25
Figure 3.
DNA synthesis rate of PNA-treated cells. WT E. coli cells were grown exponentially in AB medium without casamino acids. At the time indicated by a dotted line, cultures were treated with NrdA-PNA (12 μM, filled circles), 2-base mismatch-PNA (12 μM, open circles) or nalidixic acid (40 mg/L, filled triangles) or left non-treated (crosses). Samples were collected for determination of turbidity (a), viability (b) and DNA synthesis rate (c). Figures show a single experiment.
NrdA-PNA-treated cells cannot complete chromosome replication
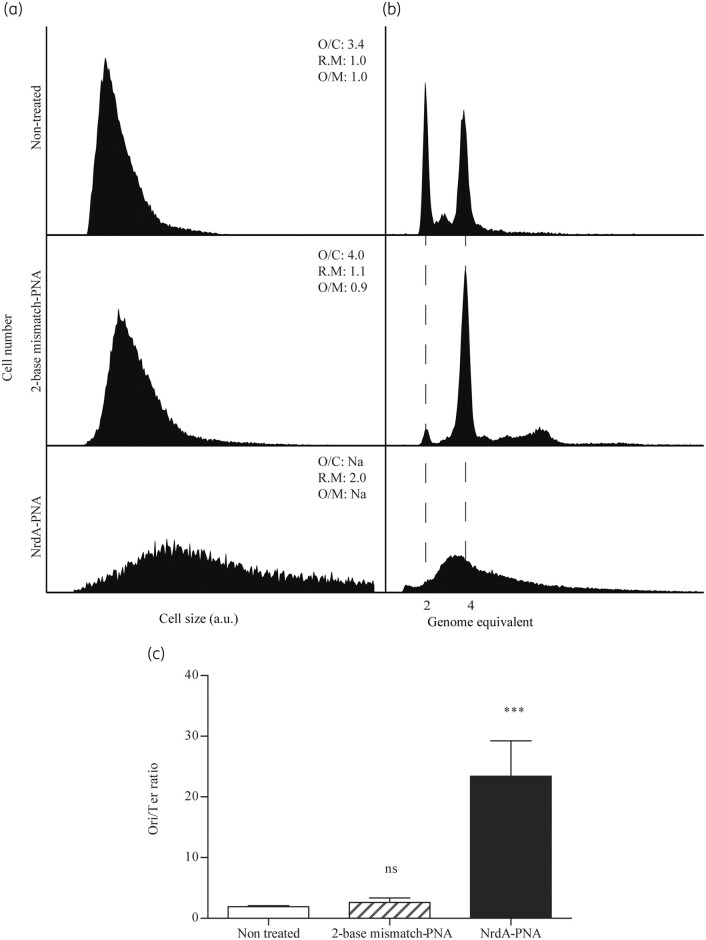
We proceeded to analyse PNA-treated cells using flow cytometry,19 in order to assess the effect of NrdA-PNA treatment on chromosome replication and cell cycle progression. WT cells were uniform in cell size (Figure 4a) and contained mainly two or four fully replicated chromosomes (Figure 4b), following rifampicin and cefalexin treatment. These fully replicated chromosomes correspond to the number of chromosomal origins (oriC) carried in each cell prior to drug treatment.19 The overall number of oriC was 3.4 and the ori/ter ratio was 2.6 in non-treated cells (Figure 4c and Table S3). Cells treated for 120 min with 8 μM NrdA-PNA were different. The average cell size was doubled relative to untreated cells and the ori/ter ratio showed a dramatic increase to ∼23 (Figure 4c and Table S3). Furthermore, the average number of chromosomal origins per cell could not be determined, because replication runout was incomplete (Figure 4b), indicating that replication fork progression was affected. Cells treated with the 2-base mismatch-PNA for 120 min were uniform in cell size and similar to non-treated cells. The ori/ter ratio of the 2-base mismatch-PNA was similar to that of untreated cells and they were able to complete replication and ended up with a higher number of origins per cell relative to non-treated cells (Figure 4a and b and Table S3). Overall, the reduction in DNA synthesis rate, the inability to complete chromosome replication in the presence of rifampicin and cefalexin and the greatly elevated ori/ter ratio strongly suggest that NrdA-PNA treatment reduces the elongation rate and/or stalls individual replication forks.
Figure 4.
Cell cycle analysis of NrdA-PNA treated cells. WT E. coli cells were grown exponentially in AB medium and diluted to OD450 = 0.05 and treated with NrdA-PNA (8 μM, bottom panels) or 2-base mismatch-PNA (8 μM, middle panels) or left non-treated (top panels) for 2 h prior to sample collection and flow cytometry and quantitative PCR analysis. (a) Cell size distribution of exponentially growing cells. (b) DNA content of cells treated for 4 h with rifampicin and cefalexin for chromosome replication to complete. The number of fully replicated chromosomes reflects the cellular number of origins at time of drug treatment, indicated by hatched lines.19,21,22 Panels represent 30–50 000 cell events for each condition. The average oriC/cell (O/C), relative mass (R.M) and oriC/mass (O/M) relative to WT are inserted in the panels. (c) ori/ter ratios were determined by quantitative PCR of oriC and terminus regions on exponentially growing cells. Shown is the mean±SD based on three determinations. All ratio values were normalized to the ori/ter ratio of a stationary-phase MG1655 culture, when cells contain fully replicated chromosomes and hence an ori/ter ratio of 1. ns, not significant; ***, P<0.01.
Morphology of NrdA-PNA-treated cells
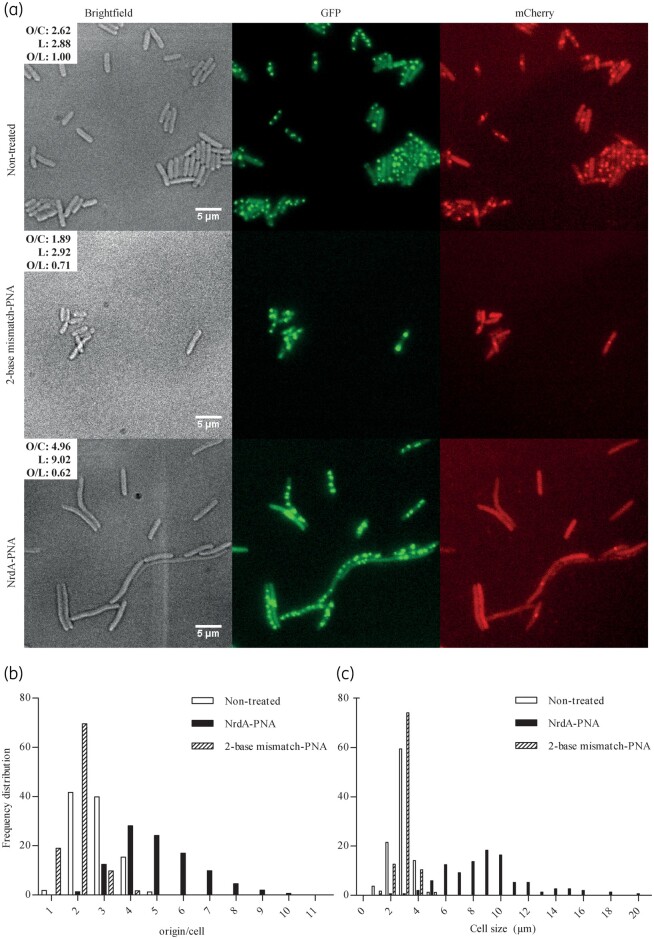
The increase in ori/ter ratio upon NrdA-PNA treatment may result from either an increase in initiation of replication from oriC or an inability of ongoing replication forks to reach the terminus and complete chromosome replication. We proceeded to visualize origins and termini in vivo, using a strain (ALO4223) with the replication origins labelled with GFP and termini with mCherry as described previously.20 Non-treated cells were homogeneous (Figure 5a), with the population containing two (42%), three (40%) or four (15%) replication origin foci and an average number of oriC foci/cell of 2.6 ± 0.8 (Figure 5b). The average cell length was 2.9 ± 0.7 μm (Figure 5c). Cells treated with 8 μM of the 2-base mismatch-PNA resembled untreated cells, with a slight decrease in average oriC foci/cell to 1.9 ± 0.5 and similar cell length of 3.0 ± 0.5 μm (Figure 5). Treatment with 8 μM NrdA-PNA resulted in a heterogeneous population of cells, with the appearance of filaments (Figure 5a). The average number of oriC foci/cell increased to 5.0 ± 1.5, accompanied by an increase in cell length to an average of 9.0 ± 3.0 μm. The number of termini foci was unaffected, with cells containing either one or two termini foci (similar to non-treated cells), despite the increase in oriC foci and cell length (Figure 5b and c). Because the ratio of oriC foci/cell length did not increase relative to untreated cells (Table S3), the dramatic ∼23-fold increase in ori/ter ratio observed upon NrdA-PNA treatment (Figure 4c) did not result from increased initiations from oriC, but from an inability of replication forks started at oriC to reach the terminus. This may cause a delay in cell division, leading to filamentous cells, with a high number of replication origins.
Figure 5.
Localization of oriC and terminus regions and cell morphology of PNA-treated cells. Strain ALO4223 was grown exponentially in AB medium, diluted to OD450 = 0.05 and treated with NrdA-PNA (8 μM) or 2-base mismatch-PNA (8 μM) or left non-treated for 2 h. (a) In vivo localization of oriC and ter as GFP-oriC foci and mCherry-ter foci. Scale bar is 5 μm. (b) oriC foci/cell and (c) cell length (μm) were determined for each cell in three biological triplicate experiments (non-treated, n = 316; NrdA-PNA, n = 428; 2-base mismatch-PNA, n = 496). Data are shown as frequency distributions of origins/cell (b) and cell size (μm) (c). The average oriC foci/cell (O/C), average cell length in μm (L) and average oriC foci/average cell length in μm (O/L) values are shown in the micrographs.
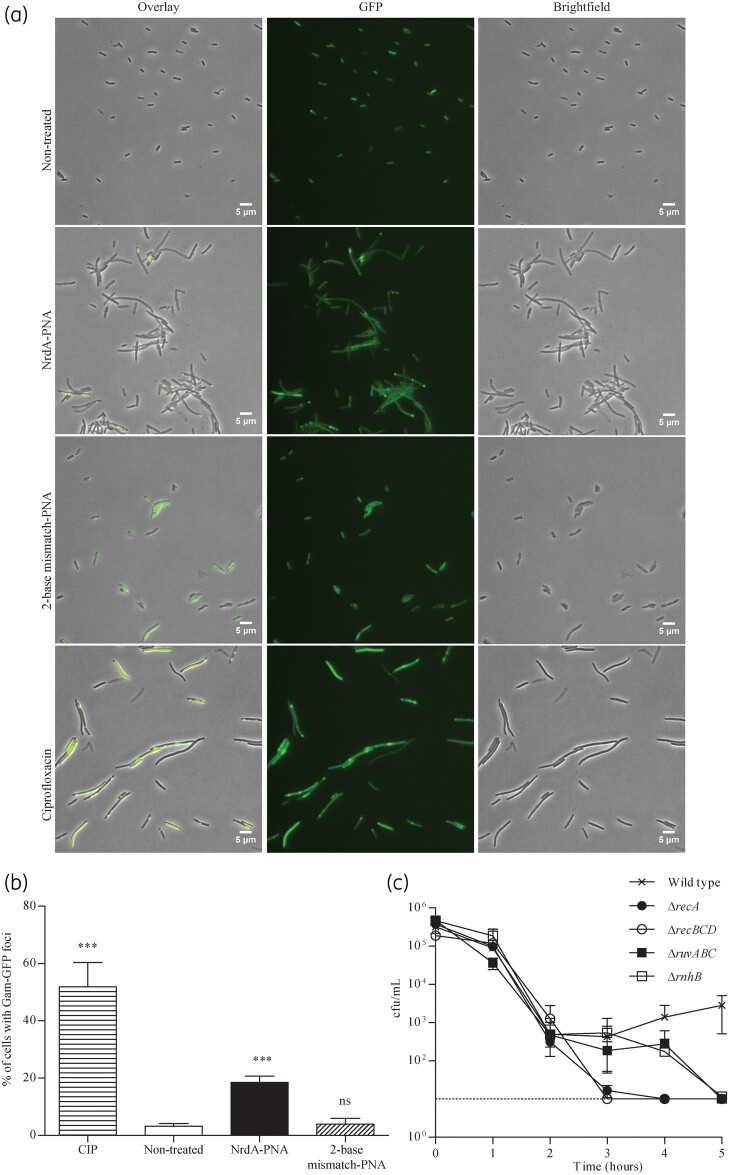
NrdA-PNA causes double-stranded DNA breaks
Stalled replication forks are known to be susceptible to breakage and collapse,26,27 resulting in double-stranded DNA breaks.28 In order to visualize NrdA-PNA-induced DNA strand breaks in vivo, we used cells expressing a Mu Gam-GFP fusion protein (SMR14350).29 The bacteriophage Mu Gam protein binds double-stranded DNA ends with high specificity, forming complexes that can be visualized by fluorescence microscopy. Gam-GFP-expressing cells were treated with NrdA-PNA (8 μM), 2-base mismatch-PNA (8 μM) or ciprofloxacin, an antibiotic well known to cause double-stranded DNA breaks (Figure 6a and b). Gam-GFP foci were observed in 3.2 ± 0.8% of the WT population (Figure 6a and b), in agreement with previous reports that double-stranded DNA breaks occur spontaneously at a low frequency in WT cells.29 Treatment with NrdA-PNA increased the frequency of foci-containing cells to 18.5 ± 1.8% and cells were filamentous (Figure 6a and b). The 2-base mismatch-PNA-treated cells were similar to non-treated cells, with 3.9 ± 1.6% of the population cells containing Gam-GFP foci (Figure 6a and b). Treatment with ciprofloxacin resulted in 51.9 ± 6.9% of the cells in the population containing Gam-GFP foci (Figure 6a and b).
Figure 6.
Double-stranded DNA breaks and DNA repair stress in PNA-treated cells. Strain SMR14350 was grown exponentially in AB medium, diluted to OD450 = 0.05 and treated with NrdA-PNA (8 μM), 2-base mismatch-PNA (8 μM) or ciprofloxacin (0.0125 mg/L) or left non-treated for 4 h. (a) In vivo visualization of double-stranded DNA breaks by Gam-GFP foci. Scale bar is 5 μm. (b) Gam-GFP foci/cell was determined for three independent experiments [non-treated, n = 3687; NrdA-PNA, n = 3860; 2-base mismatch-PNA, n = 2811; ciprofloxacin (CIP), n = 2553]. Data are given as percentage of cells with Gam-GFP foci. Shown is the mean±SD based on three determinations. ns, not significant; ***, P<0.01. (c) Susceptibility of cells deficient in major DNA damage repair pathways towards NrdA-PNA. All the cells were derivatives of MG1655 and were grown exponentially in MHB-I medium, diluted to about 1–5 × 105 cfu/mL and treated with 4 μM NrdA-PNA for 5 h. WT (crosses), ΔrecA (filled circles), ΔrecBCD (open circles), ΔruvABC (filled squares) and ΔrnhB (open squares). Cell viability is displayed as cfu/mL. For details, see the Materials and methods section. Shown is the mean±SD based on three determinations.
Repair of double-stranded DNA breaks is highly dependent on DNA repair enzymes RecABC-RuvABC.30 Cells carrying mutations in recA, recBCD or ruvABC were all sensitized to NrdA-PNA relative to WT cells (Figure 6c) and, for recA and recBCD strains, the viable counts decreased below the detection limit (10 cfu/mL) after 3 h of exposure. This is consistent with recombination-deficient cells being defective in the repair of double-stranded DNA breaks. Cells lacking RnhB that catalyses the first step of ribonucleotide excision repair were also sensitized to NrdA-PNA (Figure 6c), suggesting that increased incorporation of ribonucleotides in the bacterial DNA is the source of double-stranded DNA breaks and that these subsequently cause cell death.
Discussion
In E. coli the class Ia NrdAB RNR is essential during aerobic growth. Here we designed an antisense PNA to inhibit translation of nrdA, the first gene of the nrdAB operon. This greatly reduced the NrdA protein level and resulted in cessation of DNA replication, inhibition of bacterial proliferation and cell death.
NrdA-PNA specifically targets RNR synthesis
Several observations support the conclusion that NrdA-PNA targets the mRNA and inhibits nrdA translation, which in turn reduces the NrdA protein level. First, Western blots directly showed that NrdA-PNA treatment reduced the NrdA protein levels to about 10% of WT levels and cells were sensitized to HU, a known RNR inhibitor. Furthermore, NrdA-PNA negatively affected DNA synthesis and, finally, NrdA-PNA-treated cells had a dramatic increase in the ori/ter ratio. This increase is most likely the result of a decreased replication elongation rate, which in turn is the consequence of reduced RNR activity and hence reduced dNTP levels. This may also explain why ongoing rounds of replications cannot be completed in the presence of rifampicin. These data are in agreement with previous observations on the consequences of dNTP limitation.31–35
Note that NrdA-PNA is predicted to bind the nanE gene transcript. NanE is a 2-epimerase, involved in the acetylmannosamine degradation pathway, utilizing amino sugar as a carbon source (Table S2).36 Because nanE is not essential, and poorly expressed under the conditions tested,37 we assumed that the effect of NrdA-PNA is through nrdA.
When two bases were swapped in the PNA sequence, NrdA translation was lowered to about 50% of the WT level and a 2-fold increase in MIC was observed. The 2-base mismatch-PNA did not match other sequences in the coding region or in the vicinity of the ribosomal binding site of essential genes (Table S2).3 Because the mismatch-PNA reduces NrdA levels, we attribute the residual antimicrobial activity to the reduction in RNR activity. When two additional bases were swapped resulting in a 4-base mismatch-PNA the MIC value increased further, suggesting that affinity to the nrdA mRNA target was lost.
NrdA-PNA as a bactericidal agent
In E. coli, ribonucleotides are readily mis-incorporated into DNA and are a common source of DNA lesions.38–41 Removal of genomic ribonucleotides in bacteria is performed by RNase HI (rnhA), RNase HII (rnhB) and the nucleotide excision repair pathway.39,42 Because NrdA-PNA treatment results in cessation of synthesis of the NrdA subunit of RNR and therefore presumably dNTP shortage, we propose that cells incorporate an increased number of ribonucleotides into the DNA. These genomic ribonucleotides are, if left unrepaired, a source of double-stranded DNA breaks, which in turn result in cell filamentation, nucleoid defects and non-viability.43 This explains why both rnhB and recABCD-ruvABC mutants are hyper-susceptible to NrdA-PNA; these mutants are compromised with regard to removing ribonucleotides from DNA and with regard to homologous recombination, respectively. Another explanation for the formation of double-stranded DNA breaks is reversal of replication forks in response to dNTP shortage.28 Replication fork reversal results in a four-arm structure, with a double-stranded DNA end, similar to a Holliday junction.30
Double-stranded DNA ends caused by either ribonucleotide mis-incorporation or replication fork reversal require RecABC-RuvABC enzymes for repair/resolution, explaining why cells deficient in these functions are hyper-susceptible to NrdA-PNA.
RNRs as targets for antibacterial agents
RNR enzymes are essential and distributed across all branches of life, making RNRs attractive drug targets. Today, RNRs are targeted by anti-cancer drugs, which prevent uncontrolled proliferation by limiting dNTP levels.44 Clinically available drugs include radical scavengers (HU)45 and nucleoside/nucleotide and base analogues.46 Despite the potential of RNR as an antimicrobial drug target, no known RNR-targeting antibiotics are—to our knowledge—approved for use in the clinic, even though small-molecule RNR inhibitors have been described.47 The ability of a PNA to potentially discriminate between RNR species provides a major advantage for targeting RNR inhibitors to specific cell types/species. We therefore suggest that PNA bacterial RNR inhibitors may have the potential for future bacterial drug discovery approaches.
PNAs are promising antibiotics and can potentially be the source of a continuous and unlimited arsenal of easily modified and optimized broad- or narrow-spectrum drugs. However, PNAs are not taken up by cells10 and conjugation to a BPP is required for uptake.5 The downsides of using such BPP-PNA conjugates are that resistance can be achieved through mutations in the transporter molecules,6 that the BPP is susceptible to enzymatic degradation and that the BPP exhibits cytotoxic properties.48,49 Furthermore, the PNA backbone exhibits solubility issues and a tendency to form aggregates and non-specifically adhere to surfaces and macromolecular complexes.50 Fortunately, second-generation peptide carriers exhibit improved properties6 and the PNA scaffold is highly tunable, as shown by incorporation of lysine, guanidinium or small hydrophilic (R)-diethylene glycol groups into the PNA backbone, which improves binding efficiency and solubility, prevents aggregation and improves cellular uptake.9 These modifications exemplify the durability and flexibility of PNA-peptide technology.
Supplementary Material
Acknowledgements
Thanks to Anurag K. Sinha for valuable discussions and suggestions. Thanks to Jolanta Barbara Ludvigsen for PNA synthesis. Strains GM7390 and GM7346 were generously provided by Martin Marinus.
Funding
This research was funded through a Challenge programme (NNF16OC0021700), Tandem (NNF16OC0023482) grant, both from the Novo Nordisk Foundation and the Danish National Research Foundation (DNRF120). Furthermore, we would like to thank the Kirsten and Freddy Johansens Foundation (ALO).
Transparency declarations
None to declare.
Supplementary data
Tables S1 to S3 and Figures S1 an S2 are available as Supplementary data at JAC Online.
References
- 1. Egholm M, Buchardt O, Christensen L. et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 1993; 365: 566–8. [DOI] [PubMed] [Google Scholar]
- 2. Demidov VV, Potaman VN, Frank-Kamenetskii MD. et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol 1994; 48: 1310–3. [DOI] [PubMed] [Google Scholar]
- 3. Dryselius R, Aswasti SK, Rajarao GK. et al. The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides 2003; 13: 427–33. [DOI] [PubMed] [Google Scholar]
- 4. Good L, Nielsen PE.. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc Natl Acad Sci USA 1998; 95: 2073–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Good L, Awasthi SK, Dryselius R. et al. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol 2001; 19: 360–4. [DOI] [PubMed] [Google Scholar]
- 6. Ghosal A, Vitali A, Stach JE. et al. Role of SbmA in the uptake of peptide nucleic acid (PNA)-peptide conjugates in E. coli. ACS Chem Biol 2013; 8: 360–7. [DOI] [PubMed] [Google Scholar]
- 7. Wang H, He Y, Xia Y. et al. Inhibition of gene expression and growth of multidrug-resistant Acinetobacter baumannii by antisense peptide nucleic acids. Mol Biol Rep 2014; 41: 7535–41. [DOI] [PubMed] [Google Scholar]
- 8. Oh E, Zhang Q, Jeon B.. Target optimization for peptide nucleic acid (PNA)-mediated antisense inhibition of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Antimicrob Chemother 2014; 69: 375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wojciechowska M, Rownicki M, Mieczkowski A. et al. Antibacterial peptide nucleic acids-facts and perspectives. Molecules 2020; 25: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Good L, Sandberg R, Larsson O. et al. Antisense PNA effects in Escherichia coli are limited by the outer-membrane LPS layer. Microbiology (Reading) 2000; 146: 2665–70. [DOI] [PubMed] [Google Scholar]
- 11. Eriksson M, Nielsen PE, Good L.. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J Biol Chem 2002; 277: 7144–7. [DOI] [PubMed] [Google Scholar]
- 12. Torrents E. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol 2014; 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clark DJ, Maaløe O.. DNA replication and the division cycle in Escherichia coli. J Mol Biol 1967; 23: 99–112. [Google Scholar]
- 14. Lanzer M, Bujard H.. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA 1988; 85: 8973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Freiesleben U, Krekling MA, Hansen FG. et al. The eclipse period of Escherichia coli. EMBO J 2000; 19: 6240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christensen L, Fitzpatrick R, Gildea B. et al. Solid-phase synthesis of peptide nucleic acids. J Pept Sci 1995; 1: 175–83. [DOI] [PubMed] [Google Scholar]
- 17. Goltermann L, Nielsen PE.. PNA antisense targeting in bacteria: determination of antibacterial activity (MIC) of PNA-peptide conjugates. Methods Mol Biol 2020; 2105: 231–9. [DOI] [PubMed] [Google Scholar]
- 18. Oddo A, Thomsen TT, Kjelstrup S. et al. An amphipathic undecapeptide with all d-amino acids shows promising activity against colistin-resistant strains of Acinetobacter baumannii and a dual mode of action. Antimicrob Agents Chemother 2016; 60: 592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boye E, Lobner-Olesen A.. Bacterial growth control studied by flow cytometry. Res Microbiol 1991; 142: 131–5. [DOI] [PubMed] [Google Scholar]
- 20. Charbon G, Bjorn L, Mendoza-Chamizo B. et al. Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res 2014; 42: 13228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lobner-Olesen A, Skarstad K, Hansen FG. et al. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 1989; 57: 881–9. [DOI] [PubMed] [Google Scholar]
- 22. Skarstad K, Boye E, Steen HB.. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J 1986; 5: 1711–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schindelin J, Rueden CT, Hiner MC. et al. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 2015; 82: 518–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haugan MS, Charbon G, Frimodt-Moller N. et al. Chromosome replication as a measure of bacterial growth rate during Escherichia coli infection in the mouse peritonitis model. Sci Rep 2018; 8: 14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riber L, Lobner-Olesen A.. Inhibition of Escherichia coli chromosome replication by rifampicin treatment or during the stringent response is overcome by de novo DnaA protein synthesis. Mol Microbiol 2020; 114: 906–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bierne H, Michel B.. When replication forks stop. Mol Microbiol 1994; 13: 17–23. [DOI] [PubMed] [Google Scholar]
- 27. Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays 1995; 17: 733–41. [DOI] [PubMed] [Google Scholar]
- 28. Guarino E, Salguero I, Jimenez-Sanchez A. et al. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J Bacteriol 2007; 189: 5782–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shee C, Cox BD, Gu F. et al. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife 2013; 2: e01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michel B, Sinha AK, Leach DRF.. Replication fork breakage and restart in Escherichia coli. Microbiol Mol Biol Rev 2018; 82: e00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Odsbu I, Morigen, Skarstad K.. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PLoS One 2009; 4: e7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guarino E, Jimenez-Sanchez A, Guzman EC.. Defective ribonucleoside diphosphate reductase impairs replication fork progression in Escherichia coli. J Bacteriol 2007; 189: 3496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Itsko M, Schaaper RM.. dGTP starvation in Escherichia coli provides new insights into the thymineless-death phenomenon. PLoS Genet 2014; 10: e1004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Si F, Li D, Cox SE. et al. Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr Biol 2017; 27: 1278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin CM, Viguera E, Guzman EC.. Rifampicin suppresses thymineless death by blocking the transcription-dependent step of chromosome initiation. DNA Repair (Amst) 2014; 18: 10–7. [DOI] [PubMed] [Google Scholar]
- 36. Vimr ER, Kalivoda KA, Deszo EL. et al. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 2004; 68: 132–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalivoda KA, Steenbergen SM, Vimr ER. et al. Regulation of sialic acid catabolism by the DNA binding protein NanR in Escherichia coli. J Bacteriol 2003; 185: 4806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cronan GE, Kouzminova EA, Kuzminov A.. Near-continuously synthesized leading strands in Escherichia coli are broken by ribonucleotide excision. Proc Natl Acad Sci USA 2019; 116: 1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schroeder JW, Randall JR, Matthews LA. et al. Ribonucleotides in bacterial DNA. Crit Rev Biochem Mol Biol 2015; 50: 181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yao NY, Schroeder JW, Yurieva O. et al. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Natl Acad Sci USA 2013; 110: 12942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet 2012; 28: 592–7. [DOI] [PubMed] [Google Scholar]
- 42. Vaisman A, McDonald JP, Huston D. et al. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet 2013; 9: e1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kouzminova EA, Kadyrov FF, Kuzminov A.. RNase HII saves rnhA mutant Escherichia coli from R-loop-associated chromosomal fragmentation. J Mol Biol 2017; 429: 2873–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aye Y, Li M, Long MJ. et al. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 2015; 34: 2011–21. [DOI] [PubMed] [Google Scholar]
- 45. Sterkers Y, Preudhomme C, Lai JL. et al. Acute myeloid leukemia and myelodysplastic syndromes following essential thrombocythemia treated with hydroxyurea: high proportion of cases with 17p deletion. Blood 1998; 91: 616–22. [PubMed] [Google Scholar]
- 46. Shelton J, Lu X, Hollenbaugh JA. et al. Metabolism, biochemical actions, and chemical synthesis of anticancer nucleosides, nucleotides, and base analogs. Chem Rev 2016; 116: 14379–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tholander F, Sjoberg BM.. Discovery of antimicrobial ribonucleotide reductase inhibitors by screening in microwell format. Proc Natl Acad Sci USA 2012; 109: 9798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yavari N, Goltermann L, Nielsen PE.. Uptake, stability, and activity of antisense anti-acpP PNA-peptide conjugates in Escherichia coli and the role of SbmA. ACS Chem Biol 2021; 16: 471–9. [DOI] [PubMed] [Google Scholar]
- 49. Vaara M, Porro M.. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob Agents Chemother 1996; 40: 1801–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tackett AJ, Corey DR, Raney KD.. Non-Watson-Crick interactions between PNA and DNA inhibit the ATPase activity of bacteriophage T4 Dda helicase. Nucleic Acids Res 2002; 30: 950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.