Carbapenems are critically important antimicrobials for the treatment of clinical infections caused by MDR Enterobacteriaceae, with colistin reserved for those cases in which even carbapenems are ineffective. Therefore, co-localization of carbapenem and colistin resistance genes on the same plasmid is worrisome and, to date, has been described only in China in an Escherichia coli from a diseased chicken (blaNDM-4 and mcr-1 on an IncHI2/ST3 plasmid)1 and in an E. coli from a diseased pet cat (blaNDM-5 and mcr-1 on an IncX3-X4 hybrid plasmid).2 Here, we report a novel case of co-localization of blaOXA-162 carbapenemase and mcr-1 on a transferable IncHI2 plasmid in E. coli from a healthy broiler chicken in Europe.
Within the framework of the specific monitoring of ESBL-, AmpC- or carbapenemase-producing indicator commensal E. coli according to Commission Implementing Decision 2013/652/EU ‘on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria’, four presumptive carbapenemase-producing E. coli from chicken caeca and meat were collected in Romania in 2016 (Table S1, available as Supplementary data at JAC Online). The four isolates were tested for antimicrobial susceptibility by broth microdilution using EUVSEC and EUVSEC2 Sensititre panels (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions and obtained MIC values were interpreted according to EUCAST epidemiological cut-off values (http://eucast.org/). Confirmed carbapenemase-producing isolates were sequenced (WGS) using Illumina MiSeq (CA, USA) as previously described3 and PacBio (Menlo Park, CA, USA) technologies. PacBio sequences were de novo assembled using the Hierarchical Genome Assembly Process version 4.0 (HGAPv4) with the ‘aggressive option’ and the final polished sequence was obtained using the Resequencing analysis (default settings). De novo assembly of Illumina reads and analyses of assembled sequences were performed using the CGE tools with default settings (http://www.genomicepidemiology.org/). Accession numbers are in Table S2. BLASTn analyses, alignments and mapping were carried out using CLC Genomics workbench v20.0.4 (Qiagen, Aarhus, Denmark) and plasmids were annotated using RAST.4 Previously described IncHI2 plasmids, including the IncHI2 prototype plasmid R478 and representative IncHI2 plasmids with and without mcr-1 (Table S3), were used for comparison, which was visualized using BRIG.5 Plasmid transferability was assessed by filter-mating experiments. Briefly, strain CFSAN083827 was used as the donor and E. coli MT102 and Salmonella Typhimurium SL5324 were used as recipients in independent experiments. Transconjugants were selected on selective medium containing colistin (2 mg/L), cefotaxime (0.5 mg/L) and rifampicin (50 mg/L) and verified using PCR for mcr6 and blaOXA-48,7 followed by WGS (Illumina MiSeq) and bioinformatics analyses as described above.
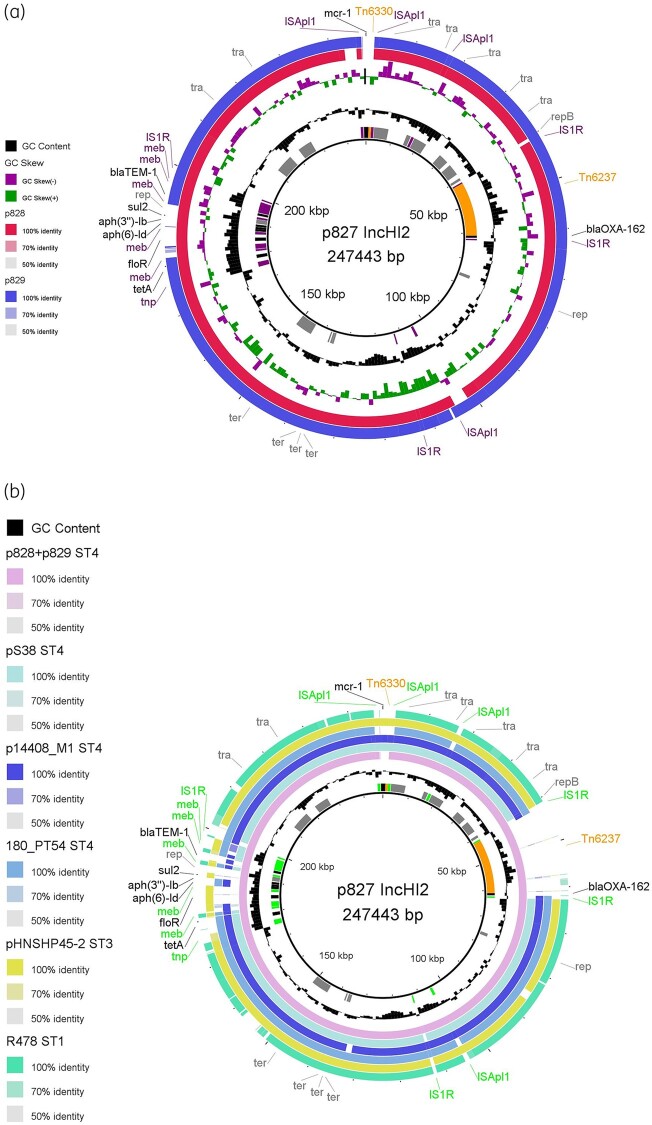
The carbapenemase phenotype (meropenem MIC >0.12 mg/L) was confirmed for three isolates that also exhibited resistance to antimicrobials other than β-lactams and one of them also showed colistin resistance. See Table S1. Genomic characterization showed that the three isolates had different serotypes, STs, plasmid contents, antimicrobial resistance genes and parC mutations mediating fluoroquinolone resistance. The carbapenemase phenotype was mediated by blaOXA-162, which was carried on IncHI2/pST4 plasmids. These IncHI2 plasmids, here designated pCFSAN083827_1, pCFSAN083828_1 and pCFSAN083829_1, had a GC content of 46.8%, 47.1% and 46.3%, respectively, and differed by size (Table S1). blaOXA-162 was carried on the 21.9 kb transposon Tn6237 (Figure 1a). In all three plasmids, Tn6237 was almost entirely conserved (>99.5% nucleotide identity and 100% sequence length) compared with pRA35.8 Sequence alignment showed that Tn6237 was identical across the three plasmids, with the exception of one SNP in parB. In pCFSAN083828_1 and pCFSAN083829_1 a C was found at position 341, which is similar to pRA35, whereas pCFSAN083827_1 had a T. This leads to a L (leucine) in pCFSAN083827_1 and P (proline) in pCFSAN083828_1 and pCFSAN083829_1 at amino acid position 114. The parB sequence of pCFSAN083827_1 has no 100% identical sequence in GenBank, whereas that of pCFSAN083828_1 and pCFSAN083829_1 has.
Figure 1.
BLAST atlases of IncHI2 plasmids. (a) IncHI2 plasmids from this study. pCFSAN083828_1 (p828, due to layout limitations) and pCFSAN083829_1 (p829) were compared with pCFSAN083827_1 (p827; reference sequence). Mobile element (meb), insertion sequence (IS) and transposase (tnp) genes are shown in purple, whereas IncHI2 backbone genes are shown in grey. Antimicrobial resistance genes are shown in black and it can be seen that only pCFSAN083829_1 is missing aph(3'')-Ib, aph(6)-Id, floR and sul2. Notably, only pCFSAN083827_1 carries mcr-1 located on Tn6330, whereas all three plasmids carry blaOXA-162 located on Tn6237. (b) Comparative plasmid analysis. The three IncHI2/ST4 plasmids from this study were compared with other IncHI2 plasmids of ST1, 3 and 4 (Table S1) using pCFSAN083827_1 as the reference sequence. Tn6237 is only found in the plasmids from this study, whereas replication (rep), transfer (tra) and tellurium resistance (ter) genes were nearly entirely conserved among all analysed plasmids. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Notably, plasmid pCFSAN083827_1 carried the colistin resistance gene mcr-1 on a 4.7 kb transposon, i.e. Tn6330,9 flanked by ISApl1 (Figure 1a). In pCFSAN083827_1, Tn6330 was entirely conserved (100% nucleotide identity and 100% sequence length) and was located next to a region of conjugative transfer genes (Figure 1a). Comparison of the three plasmids from this study with selected IncHI2 plasmids showed that Tn6237 (carrying blaOXA-162) was only present in the plasmids from this study, whereas Tn6330 was also found on other IncHI2 plasmids inserted at the same position (Figure 1b). The replication, transfer and tellurium resistance genes were nearly entirely conserved among all analysed IncHI2 plasmids (Figure 1b). The filter-mating experiment confirmed that pCFSAN083827_1 was transferable from isolate CFSAN083827 to the E. coli and Salmonella sp. recipients and transferred all of its antimicrobial resistance genes (Table S1).
Previously, mcr-1-carrying IncHI2 plasmids have been widely observed among isolates from chickens, other production animals and food products from Europe10 and, indeed, pCFSAN083827_1 shared similarities with those plasmids, including, among others, the Tn6330 insertion site (Figure 1b). However, the presence of Tn6237 with blaOXA-162 on an mcr-1-carrying IncHI2 plasmid is unique. This study highlights the importance of monitoring antimicrobial resistance in a One Health perspective by leveraging on WGS technologies that allow fine resolution of mobilizable elements mediating resistance.
Funding
We acknowledge funding from the European Commission to the European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR).
Transparency declarations
None to declare.
Disclaimer
B. Guerra and P.-A. Beloeil are currently employed by the European Food Safety Authority (EFSA) in its BIOCONTAM Unit that provides scientific and administrative support to EFSA’s scientific activities in the area of Microbial Risk Assessment. The positions and opinions presented in this article are those of the authors alone and are not intended to represent the views or scientific works of EFSA.
Supplementary data
Tables S1, S2 and S3 are available as Supplementary data at JAC Online.
Supplementary Material
References
- 1. Liu BT, Song FJ, Zou M. et al. High incidence of Escherichia coli strains coharboring mcr-1 and blaNDM from chickens. Antimicrob Agents Chemother 2017; 61: e02347–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun J, Yang RS, Zhang Q. et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol 2016; 1: 16176. [DOI] [PubMed] [Google Scholar]
- 3. Hendriksen RS, Leekitcharoenphon P, Lukjancenko O. et al. Genomic signature of multidrug-resistant Salmonella enterica serovar Typhi isolates related to a massive outbreak in Zambia between 2010 and 2012. J Clin Microbiol 2015; 53: 262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brettin T, Davis JJ, Disz T. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 2015; 5: 8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alikhan NF, Petty NK, Ben Zakour NL. et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rebelo AR, Bortolaia V, Kjeldgaard JS. et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 2018; 23: pii=17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poirel L, Walsh TR, Cuvillier V. et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 2011; 70: 119–23. [DOI] [PubMed] [Google Scholar]
- 8. Beyrouthy R, Robin F, Delmas J. et al. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of blaOXA-48 into the Escherichia coli chromosome. Antimicrob Agents Chemother 2014; 58: 3785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li R, Xie M, Zhang J. et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 2017; 72: 393–401. [DOI] [PubMed] [Google Scholar]
- 10. Matamoros S, Van Hattem JM, Arcilla MS. et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep 2017; 7: 15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.