Abstract
Objectives
As part of an active MRSA surveillance programme in our neonatal ICU, we identified nares surveillance cultures from two infants that displayed heterogeneity in methicillin resistance between isolated subclones that lacked mecA and mecC.
Methods
The underlying mechanism for the modified Staphylococcus aureus (MODSA) methicillin-resistance phenotype was investigated by WGS.
Results
Comparison of finished-quality genomes of four MODSA and four MSSA subclones demonstrated that the resistance changes were associated with unique truncating mutations in the gene encoding the cyclic diadenosine monophosphate phosphodiesterase enzyme GdpP or a non-synonymous substitution in the gene encoding PBP2.
Conclusions
These two cases highlight the difficulty in identifying non-mecA, non-mecC-mediated MRSA isolates in the clinical microbiology laboratory, which leads to difficulties in implementing appropriate therapy and infection control measures.
Introduction
Expression of mecA or mecC is the most commonly recognized mechanism for MRSA.1 Non-mecA and non-mecC-mediated resistance through other mechanisms has also been described.1,2 When these involve isolates with mutations to native genes, such as those encoding penicillin-binding proteins (PBPs),3–6 GdpP1,2,4,7,8 or the cation multidrug efflux transporter AcrB,4 the isolates are generally referred to as modified Staphylococcus aureus (MODSA). Alternatively, resistance caused by overproduction of β-lactamases is typically classified as borderline resistant S. aureus (BORSA).1 Methods of detection for MODSA or BORSA are limited. Here we describe the use of WGS to determine the mechanisms of methicillin resistance for two patients with heterogeneous MSSA/MODSA surveillance cultures, highlighting the challenges of identifying these organisms in a routine clinical microbiology laboratory (CML).
Materials and methods
Ethics statement
The study protocol was reviewed and approved by the Mount Sinai School of Medicine Institutional Review Board for the collection and bacterial genome sequencing of discarded clinical specimens by the Pathogen Surveillance Program (protocol HS# 13–00981), as defined by DHHS regulations. A waiver of authorization for use and disclosure of protected health information (PHI) and a waiver of informed consent was approved based on the criteria that the use or disclosure of PHI involved no more than minimal risk to the privacy of individuals and because the research could not practically be conducted without the waiver and without access to and use of the PHI. The research conformed to the principles of the Helsinki Declaration.
Phenotypic testing
Isolates were processed per the CML Standard Operating Procedure for screening for MRSA. Swabs were plated on chromID® MRSA agar (CA) (bioMérieux, Marcy-l’Étoile, France) and on trypticase soy agar supplemented with 5% sheep blood (SBA) (BD Diagnostics, Sparks, MD, USA) and incubated at 37°C for 24 h. β-Haemolytic colonies on SBA were identified via conventional biochemical testing and subcultured for storage on BBL Nutrient Agar slants (BD Diagnostics). Isolates that were catalase- and coagulase-positive were confirmed and tested for antimicrobial susceptibility by automated broth microdilution assays on the MicroScan WalkAway Plus system, PM34 (Siemens, Sacramento, CA, USA).
Following collection of residual CML isolates from the original SBA plates, subcultures were prepared for whole-genome analysis for research purposes and plated on CA for MRSA confirmation testing prior to DNA isolation and sequencing.
Additional testing of MSSA/MODSA subcultures derived from the original agar slants for isolates with discrepant results between clinical and research assays was done using latex agglutination (Staphaurex™, Thermo Fisher Scientific, Switzerland), mannitol salt agar (BD Diagnostics) and Cepheid Xpert® SA Nasal Complete (Cepheid, Sunnyvale, CA, USA). Phenotypic testing of subcultures was performed using the Vitek® GP 2 ID panel or the Gram-Positive Susceptibility PM34 panel on the MicroScan platform (Beckman Coulter, CA, USA), as well as cefoxitin disc (cefoxitin 30 μg; BD Sensi-Disc, Becton Dickinson, Germany) and oxacillin Etest (bioMérieux) using Mueller–Hinton agar supplemented with 2% NaCl (BD Diagnostics) as per CLSI performance standards.9
DNA preparation and sequencing
For genome sequencing, selected subclones were cultured on SBA (Thermo Fisher Scientific) under non-selective conditions. The Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) was used for DNA extraction, as previously described.10 Following quality control, DNA extraction and library preparation, long-read sequencing was performed on the Pacific Biosciences RS-II platform to a depth of >200×. Additional Illumina short-read sequencing was performed to aid final assembly polishing. Briefly, libraries were prepared for DNA isolated from the same subclones using the Nextera DNA Flex Library Prep Kit (Illumina) and sequenced in a paired-end format (2 × 150 nt) on the NextSeq platform to a minimum depth of 86-fold.
Genome assembly, annotation and comparison
PacBio long-read sequencing data were processed using a custom genome assembly pipeline, as previously described.10 Complete and circularized genome assemblies were then polished using the arrow algorithm based on minimap2-aligned PacBio raw subreads.11,12 Next, Illumina paired-end reads were aligned to PacBio assemblies using BWA-MEM followed by a second round of assembly polishing with Pilon.13,14 Finally, high-quality curated genomes were annotated for genes using PROKKA, MLST ST using PubMLST and staphylococcal protein A (spa) and staphylococcal chromosomal cassette mec (SCCmec) types using custom scripts.15,16 Pairwise comparisons between the genomes of susceptible and resistant samples were performed using NucDiff to identify genomic variants that correlated with sample phenotypes and the functional effect of these genomic variants was annotated using ANNOVAR.17,18
Sequence data
Complete genome sequences are available in GenBank, with accession numbers CP075570 to CP075582 (see also the Supplementary data available at JAC Online).
Results
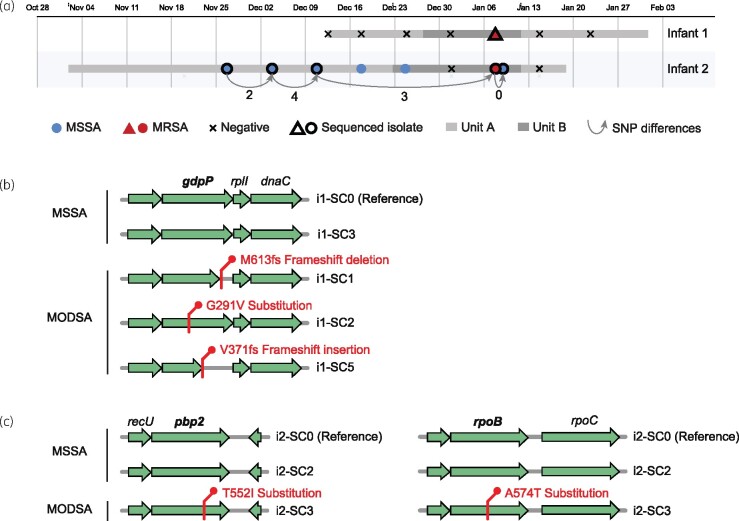
During neonatal ICU surveillance for MRSA colonization of the nares in late 2018, isolates from two infants (i1 and i2) yielded green colonies on CA plates. Automated broth microdilution testing confirmed both isolates to be MRSA. However, when the same isolates were independently subcultured and sequenced for research purposes, both subcultures (named i1-SC0 and i2-SC0) were identified as MSSA on CA plates and the resulting finished-quality genome sequences did not contain SCCmec elements. The genome obtained from i2 also differed by no more than four single nucleotide variants (SNVs) from genomes obtained from three earlier isolates and one later isolate from the same infant that were typed as MSSA (Figure 1a), indicating that the sequential isolates were clonally related. Each infant carried a different strain, based on molecular typing of the genomes from i1 (MLST ST8, spa type t024) and i2 (MLST ST72, spa type t4359).
Figure 1.
Genetic variants uniquely identified in MODSA strains. (a) Epidemiological timeline for MSSA/MRSA surveillance of infants 1 and 2. Stays in different units are indicated by horizontal bars shaded in grey. Results from periodic surveillance are indicated by symbols, with the two distinct S. aureus strain types identified in each infant shown as triangles and circles, respectively. The two isolates with conflicting clinical (MRSA) and research (MSSA) assay results are highlighted in red. Arrows connecting sequenced isolates from infant 2 denote the number of core-genome SNV differences. (b) Variants seen in MODSA strains in infant 1 that were not present in MSSA strains from the same patient. (c) Same as (b) but for infant 2. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
To investigate the cause of the discrepancy between the clinical and research assays, we performed additional evaluation of the original surveillance cultures. Agar slants made from the original clinical isolates were plated again on SBA and CA, and incubated at 37°C. After 24 h, plates were reviewed and two colonies from the CA and three colonies from the SBA were selected from different morphotypes and subcultured on CA and SBA, for a total of five subcultures per infant (SC1–5). All subcultures, including the original subcultures (SC0) obtained for research purposes, were then tested using Staphaurex™, mannitol salt agar and the Cepheid Xpert MRSA assay. All subcultures from both infants were confirmed to be S. aureus and tested negative by the Cepheid Xpert MRSA assay for mecA, mecC and SCCmec. Repeat phenotypic testing was performed for all 12 isolates using the Vitek® GP 2 panel or the PM34 MicroScan panel, as well as cefoxitin disc and oxacillin Etest using Mueller–Hinton agar supplemented with 2% NaCl as per CLSI methods. For i1, three of six subcultures tested consistently as MRSA and three as MSSA (Table 1). For i2, only one of six subcultures tested as MRSA based on cefoxitin disc and oxacillin Etest, although not by automated oxacillin testing (Table 1).
Table 1.
Phenotypic antimicrobial susceptibility testing results
| Test | SC0 | SC1 | SC2 | SC3 | SC4 | SC5 |
|---|---|---|---|---|---|---|
| Infant 1 (i1) | ||||||
| cefoxitin (ABM) | NEG | POS | POS | NEG | NEG | POS |
| cefoxitin disc | S | S | S | S | S | S |
| oxacillin (ABM) | S (1) | R (>4) | R (>4) | S (0.5) | S (0.5) | R (>4) |
| oxacillin Etest | S (1) | R (8) | R (8) | S (0.5) | S (0.5) | R (8) |
| Infant 2 (i2) | ||||||
| cefoxitin (ABM) | NEG | NEG | NEG | POS | NEG | NEG |
| cefoxitin disc | S | S | S | S | S | S |
| oxacillin (ABM) | S (0.5) | S (≤0.25) | S (0.5) | R* (0.5) | S (≤0.25) | S (0.5) |
| oxacillin Etest | S (1.5) | S (0.25) | S (0.19) | R (8) | S (0.25) | S (0.25) |
ABM, automated broth microdilution assays on the Vitek® or MicroScan platform; SC, subclone; S, susceptible; R, resistant; R*, converted via expert rule to resistant; POS, positive; NEG, negative.
MIC is indicated between brackets where available.
The finding of susceptible and resistant subclones within isolates from the same individuals suggested the presence of genetic heterogeneity. As such, we performed additional WGS and assembly of finished-quality genomes for three resistant and one susceptible subclone for i1, as well as one resistant and one susceptible subclone of i2. All genomes matched the MLST and spa types of the original SC0 i1 and i2 susceptible subclones. Comparison of complete genomes with the SC0 reference identified only 8 mutations for the i1 subclones and 10 mutations for the i2 subclones (see the Supplementary data available at JAC Online), indicating that they were clonally related. In i1, the only gene that was mutated in a pattern consistent with the observed phenotypic differences was gdpP. Each of the three resistant isolates had a different gdpP mutation; namely a truncating frameshift deletion (M613fs), a substitution (G291V) or a truncating frameshift insertion (V371fs) (Figure 1b). For i2, the resistant isolate was found to have a T552I substitution in the transpeptidase domain of PBP2, as well as an A574T substitution in RpoB (Figure 1c).
Discussion
In this study we identified two S. aureus isolates of a mixed MODSA MSSA phenotype that did not contain mecA or mecC. The resistant isolates from i1 contained variants in the gene encoding GdpP. It has been previously described that GdpP hydrolyses the secondary messenger cyclic diadenosine monophosphate (c-di-AMP), such that inactivation or deletion of gdpP leads to an increased level of c-di-AMP, which produces a reduced susceptibility to β-lactams.2,7,8 In a study by Ba et al.,1 an array of truncating mutations of the gdpP gene was identified in 17 out of 20 mec-negative MRSA isolates. Although diverse mutations in the gdpP gene have been reported, the isolates from our patient i1 are unusual in that concurrent isolation and identification from an individual patient of three different subclones with different gdpP mutations in the same strain has not been described previously.
In i2, the subclone that expressed phenotypic methicillin resistance had unique mutations in pbp2 and rpoB. S. aureus possesses four PBPs that contribute to the assembly of cell wall peptidoglycan. However, functional PBP2 has an important role in the expression of resistance to methicillin and the T552I substitution observed in the transpeptidase domain of PBP2 has previously been identified in mec-negative MRSA isolates.3,5 There is also evidence to suggest that rpoB mutations may contribute to MRSA.19,20 Aiba et al.19 demonstrated that introduction of an rpoB mutation was accompanied by tolerance to bactericidal concentrations of methicillin. Moreover, Panchal et al.20 demonstrated that insertion of a mutant rpoB gene into an MSSA strain led to conversion to MRSA. Thus, mutations in both genes may contribute to the MODSA phenotype observed in this strain.
In conclusion, we have described two different strains of MODSA, for which resistance is mediated by two distinct mechanisms. Furthermore, subpopulations of the same strain of S. aureus in the same host but with different phenotypic and genotypic expressions, as opposed to subpopulations of differing strains, are not recognized commonly. Lastly, it is important to recognize that there are multiple mechanisms leading to MRSA that might be missed when testing only for the presence of mec genes.
Funding
This work was supported by a grant from the National Institute for Allergy and Infectious Diseases (R01AI119145).
Transparency declarations
None to declare.
Supplementary data
Supplementary data are available at JAC Online.
Supplementary Material
References
- 1. Ba X, Kalmar L, Hadjirin NF. et al. Truncation of GdpP mediates β-lactam resistance in clinical isolates of Staphylococcus aureus. J Antimicrob Chemother 2019; 74: 1182–91. [DOI] [PubMed] [Google Scholar]
- 2. Argudin MA, Roisin S, Nienhaus L. et al. Genetic diversity among Staphylococcus aureus isolates showing oxacillin and/or cefoxitin resistance not linked to the presence of mec genes. Antimicrob Agents Chemother 2018; 62: e00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ba X, Harrison EM, Edwards GF. et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J Antimicrob Chemother 2014; 69: 594–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banerjee R, Gretes M, Harlem C. et al. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother 2010; 54: 4900–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leski TA, Tomasz A.. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 2005; 187: 1815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomasz A, Drugeon HB, de Lencastre HM. et al. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother 1989; 33: 1869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corrigan RM, Abbott JC, Burhenne H. et al. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 2011; 7: e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Griffiths JM, O'Neill AJ.. Loss of function of the GdpP protein leads to joint β-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 2012; 56: 579–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Eighth Edition: M100. 2018.
- 10. Sullivan MJ, Altman DR, Chacko KI. et al. A complete genome screening program of clinical methicillin-resistant Staphylococcus aureus isolates identifies the origin and progression of a neonatal intensive care unit outbreak. J Clin Microbiol 2019; 57: e01261-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.PacificBiosciences. PacificBiosciences/GenomicConsensus. https://github.com/PacificBiosciences/GenomicConsensus.
- 12. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018; 34: 3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv2013; 1303.3997.
- 14. Walker BJ, Abeel T, Shea T. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jolley KA, Bray JE, Maiden MCJ.. Open-access bacterial population genomics: IGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [DOI] [PubMed] [Google Scholar]
- 17. Khelik K, Lagesen K, Sandve GK. et al. NucDiff: in-depth characterization and annotation of differences between two sets of DNA sequences. BMC Bioinformatics 2017; 18: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aiba Y, Katayama Y, Hishinuma T. et al. Mutation of RNA polymerase β-subunit gene promotes heterogeneous-to-homogeneous conversion of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2013; 57: 4861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Panchal VV, Griffiths C, Mosaei H. et al. Evolving MRSA: high-level β-lactam resistance in Staphylococcus aureus is associated with RNA polymerase alterations and fine tuning of gene expression. PLoS Pathog 2020; 16: e1008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.