Abstract
The docking protein FRS2 was implicated in the transmission of extracellular signals from the fibroblast growth factor (FGF) or nerve growth factor (NGF) receptors to the Ras/mitogen-activated protein kinase signaling cascade. The two members of the FRS2 family, FRS2α and FRS2β, are structurally very similar. Each is composed of an N-terminal myristylation signal, a phosphotyrosine-binding (PTB) domain, and a C-terminal tail containing multiple binding sites for the SH2 domains of the adapter protein Grb2 and the protein tyrosine phosphatase Shp2. Here we show that the PTB domains of both the α and β isoforms of FRS2 bind directly to the FGF or NGF receptors. The PTB domains of the FRS2 proteins bind to a highly conserved sequence in the juxtamembrane region of FGFR1. While FGFR1 interacts with FRS2 constitutively, independent of ligand stimulation and tyrosine phosphorylation, NGF receptor (TrkA) binding to FRS2 is strongly dependent on receptor activation. Complex formation with TrkA is dependent on phosphorylation of Y490, a canonical PTB domain binding site that also functions as a binding site for Shc (NPXpY). Using deletion and alanine scanning mutagenesis as well as peptide competition assays, we demonstrate that the PTB domains of the FRS2 proteins specifically recognize two different primary structures in two different receptors in a phosphorylation-dependent or -independent manner. In addition, NGF-induced tyrosine phosphorylation of FRS2α is diminished in cells that overexpress a kinase-inactive mutant of FGFR1. This experiment suggests that FGFR1 may regulate signaling via NGF receptors by sequestering a common key element which both receptors utilize for transmitting their signals. The multiple interactions mediated by FRS2 appear to play an important role in target selection and in defining the specificity of several families of receptor tyrosine kinases.
The fibroblast growth factor (FGF) and nerve growth factor (NGF) receptor families belong to a large group of protein tyrosine kinases that play a crucial role in controlling cell growth, differentiation, and survival among other activities (reviewed in references 2 and 12). Like all receptor tyrosine kinases (RTKs), the FGF and NGF receptors comprise an extracellular ligand binding domain, a single transmembrane region, a cytoplasmic domain composed of a protein tyrosine kinase core, and sequences containing phosphotyrosine residues which function as recognition domains for a variety of signaling proteins (30, 33). The FGF and NGF receptors are expressed in different tissues of the central nervous system and play a crucial role in the control of signaling processes essential for neuronal development and survival (2, 12). Activation of the FGF and NGF receptors results in a strong activation of the Ras/mitogen-activated protein kinase (MAPK) cascade by means of recruitment of the Grb2-Sos complex to the plasma membrane (19, 22, 34). However, both the FGF and NGF receptors lack the consensus pYXN binding site for the SH2 domain of Grb2 and therefore utilize docking proteins to indirectly recruit the Grb2-Sos complex, a step essential for activation of the Ras/MAPK kinase signaling cascade. Activation of the FGF and NGF receptors results in tyrosine phosphorylation of the docking proteins Shc and FRS2 (19). Of particular interest is FRS2, which unlike Shc is tyrosine phosphorylated by a limited repertoire of RTKs shown to be involved in neuronal signaling, namely, the NGF, glial cell-derived neurotrophic factor, brain-derived neurotrophic factor, and FGF receptors (10, 14, 19, 31, 32).
Many docking proteins have the common structure of an N-terminal membrane targeting signal, either a pleckstrin homology (PH) domain or a myristylation signal and a phosphotyrosine-binding (PTB) domain that mediates direct association with activated RTKs (30). The remaining sequence of a docking protein contains multiple tyrosine residues which are phosphorylated by the ligand-activated RTK, leading to the recruitment of a variety of signaling molecules to the cell membrane. Thus docking proteins play a role in the control of membrane targeting of signaling proteins and expansion of the repertoire of signaling pathways that are activated by RTKs by providing additional recruitment sites for signaling proteins. For example, members of the insulin receptor substrate (IRS) family of docking proteins have an N-terminal PH domain followed by a PTB domain and a large C-terminal sequence containing numerous tyrosine phosphorylation sites (40). Dok has a modular structure similar to that of the IRS proteins (5, 39) and is utilized by Eph receptors in the signaling process that controls axon guidance (17). The docking protein Gab1 contains a PH domain and a specific receptor recognition domain. It was shown that Gab1 functions downstream of the insulin, epidermal growth factor (EGF), and HGF receptors (16, 37) as well as in signaling via certain cytokine receptors (35).
Two members of the FRS2 family have been identified (19, 38; this study). Both FRS2α and FRS2β contain a membrane-anchoring N-myristylation signal, a PTB domain, and a C-terminal region which contains tyrosine residues which, when phosphorylated, form the binding sites for the SH2 domain of Grb2 and the N-SH2 domain of Shp2. We have previously demonstrated that FRS2α is tyrosine phosphorylated and forms a complex with Grb2 and Shp2 in response to FGF or NGF stimulation (19, 29). The interaction of Shp2 results in its own tyrosine phosphorylation and complex formation between Shp2 and Grb2. Thus, by recruitment of Grb2-Sos complexes directly and indirectly via Shp2, FRS2 plays a major role in mediating signals from the activated receptors to the Ras/MAPK signaling cascade (14, 19).
In this report we characterize the mechanism by which FRS2 is specifically targeted to and phosphorylated by the FGF and NGF receptors. We found that FRS2α interacts directly and constitutively with FGFR1. The interaction is mediated by the PTB domain of FRS2α and is not dependent on receptor activation. The region of binding is mapped to the juxtamembrane domain of FGFR1, a highly conserved sequence throughout the mammalian FGF receptor family. This sequence is distinct from all the currently established sequences recognized by PTB domains of docking proteins. Unlike its interaction with FGFR1, the binding of the PTB domain of FRS2α is preferentially to the activated tyrosine-phosphorylated NGF receptor, TrkA. The PTB domain binds specifically to the juxtamembrane region of a sequence containing phosphotyrosine at amino acid 490 bearing the well-established recognition motif (NPQpY) for the PTB domain of adapter protein Shc (9, 28). The PTB domain of FRS2α is capable of recognizing both ligands bearing the classical NPXpY motif of TrkA and a totally distinct sequence in the FGF receptor. The PTB domain of FRS2β binds constitutively to the same sequences on the FGF receptor as FRS2α and to TrkA in a phosphorylation-dependent manner. We have also shown that mutations in the binding site on FGFR1 for the PTB domain of FRS2α leading to diminished binding of FRS2α in cells resulted in abrogated MAPK phosphorylation induced by FGF receptor activation. Finally, coexpression of increasing levels of the kinase-inactive FGFR1 with wild-type TrkA resulted in diminished TrkA-induced phosphorylation of FRS2α, suggesting that signaling via NGF receptors may be regulated by a docking protein that serves as a common target for TrkA and FGF receptors.
MATERIALS AND METHODS
Antibodies.
Anti-FGFR1, anti-FRS2α, anti-Grb2, anti-Shc, anti-Sos1, and anti-P-Tyr were previously described (19, 27). Anti-TrkA, anti-Erk, and anti-His tag antibodies were from Santa Cruz Biotechnology. Anti-phospho-Erk antibodies were from New England Biolabs.
Cell lines.
L6 myoblasts stably transfected with FGFR1 were previously described (27). The cells were cultured in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum, 10 mM l-glutamine, and 100 μg each of penicillin and streptomycin/ml, all from Gibco BRL. Human 293 cells were cultured in the same medium. PC12 cells stably expressing TrkA and FRS2α (PC12-TrkA-FRS2α) or FGFR1 and FRS2α (PC12-Flg-FRS2α) were previously described (19). The cells were grown in DMEM supplemented with 10% fetal calf serum and 10% horse serum from Gibco BRL.
Expression constructs.
The FGFR1 coding sequences (wild type or kinase-inactive mutant) were cloned into the mammalian expression vector pRK5 as previously described (27). The FRS2α constructs used in this study, including the full-length FRS2α and the PTB domain cloned in pRK5 were previously described (19). For expression in mammalian cells, Lipofectamine (Gibco BRL) was used for transfection, following the protocol recommended by the manufacturer.
For expression of glutathione S-transferase (GST) fusion proteins, PCR-amplified sequences corresponding to the regions of interest of the proteins were cloned into the pGEX2T vector (Pharmacia). The GST fusion proteins of the PTB domains of FRS2α (amino acids 1 to 158), FRS2β (amino acids 9 to 130), and Shc (amino acids 4 to 109) and the SH2 domains of phospholipase C-γ (PLC-γ; amino acids 538 to 760) were expressed and purified with glutathione-conjugated agarose beads (Sigma). The juxtamembrane domain of FGFR1 (amino acids 399 to 470) was expressed as a C-terminal hexahistidine-tagged fusion protein with the pET22b vector (Novagen) and purified through a nickel Hi-Trap chelating column (Pharmacia). The integrity of the His-tagged fusion protein was ascertained by N-terminal microsequencing.
Mutagenesis of FGFR1. (i) Deletion mutagenesis.
C-terminal deletion mutants of FGFR1 were generated by PCR. The hemagglutinin (HA) epitope tag was added at the C terminus of each mutant to allow immunoprecipitation and immunoblotting of the mutant receptors from cell lysates with standard reagents. The DNA sequence encoding the HA tag followed by a stop codon was introduced in frame by the 3′ oligonucleotide primer used for PCR amplification on the FGFR1 cDNA template. The transmembrane and extracellular domains were retained to allow proper targeting of the mutant receptors to the plasma membrane. Deletion mutants truncated at amino acids 410, 420, 421, 423, 427, 432, 445, 450, 469, 578, 594, and 765 of FGFR1 were generated. The HA-tagged full-length wild-type and kinase-inactive FGFR1 (Y653/654F-KM) were also constructed. All FGFR1 mutants were cloned in the vector pRK5.
(ii) Alanine scan mutagenesis.
Amino acids 419 to 430 in the juxtamembrane region of FGFR1 were mutated to alanine with the Quickchange site-directed mutagenesis kit (Stratagene). The mutations were confirmed by DNA sequencing.
Binding assays.
To assay the binding of FRS2α to FGFR1 in cells, cDNA of full-length FRS2α protein or its PTB domain and FGFR1 were cotransfected into 293 cells. The levels of expression of FRS2α or the FGFRs were optimized and verified to be equivalent by immunoblotting on total cell lysates before the assay was carried out. For binding to GST fusion proteins, the GST fusion proteins were purified and immobilized on glutathione-agarose beads. The binding assays are carried out in the cell lysis buffer (20 mM HEPES [pH 7.5], 137 mM NaCl, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2, 1 mM EGTA, 1 μg of leupeptin and 1 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride, 0.2 mM Na3VO4) and washed in a similar buffer, except that the concentration of Triton X-100 was reduced to 0.2%. Specifically bound proteins were eluted and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by detection by immunoblotting with specific antibodies.
To assay for the direct interaction between the PTB domain of FRS2 and FGFR1, the PTB domain of FRS2α expressed as a GST fusion protein (GST-PTB-FRS2α) and the juxtamembrane region of FGFR1 (amino acids 399 to 470) expressed as a histidine-tagged fusion protein were used. Equivalent amounts of GST-PTB-FRS2α immobilized on glutathione beads were incubated with increasing concentrations of the juxtamembrane region of FGFR1. Specifically bound proteins were eluted and resolved by SDS-PAGE, followed by immunoblotting with antibodies against the hexahistidine epitope.
To assay for the binding of FRS2α to the TrkA receptor in vivo, we used stably transfected PC12 cells that coexpress both FRS2α and TrkA. For binding to GST fusion proteins we used the same protocol described above for FGFR1. For peptide competition assays, the peptides corresponding to amino acids 412 to 433 (MAVHKLAKSIPLRRQVTVSADS) of human FGFR1 (Biosynthesis Inc.), amino acids 808 to 822 (PRHPAQLANGGKLRR) of human FGFR1 (hFGFR1), and amino acids 489 to 497 (LQGHIIENPQpYFSDACVH) of human TrkA prepared according to standard procedures were used.
RESULTS
FRS2α binds directly and constitutively to the FGF receptor.
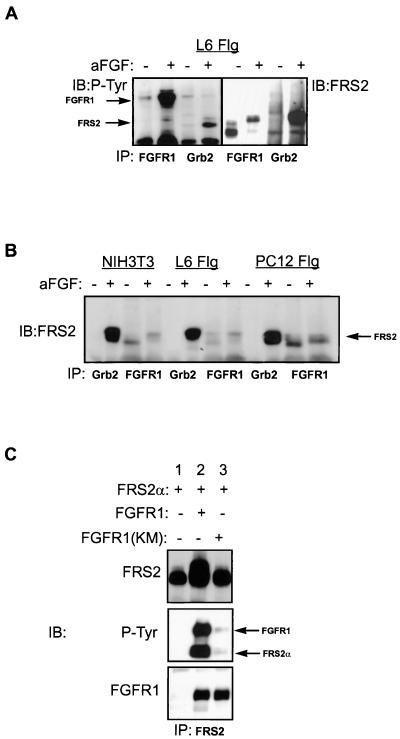
We have previously shown that following FGF or NGF stimulation, FRS2α is phosphorylated on multiple tyrosine residues that function as docking sites for Grb2-Sos and Shp2–Grb2-Sos complexes via the SH2 domains of Grb2 and Shp2, respectively (14, 19). However, the mechanism underlying FGF- or NGF-induced tyrosine phosphorylation of FRS2α is not yet understood. To ascertain the interaction between FRS2α and the FGF receptor, we used three cell lines (Fig. 1): NIH 3T3 cells which express endogenous FRS2α, FRS2β, and FGF receptors, L6 myoblasts stably transfected with FGFR1, and PC12 cells stably transfected with both FGFR1 and FRS2α (PC12-Flg-FRS2α). Quiescent cells grown to 80% confluency were treated with aFGF and heparin (100 ng/ml and 5 μg/ml, respectively) for 5 min at 37°C. The cells were solubilized, and the cell lysates were subjected to immunoprecipitation with either anti-FGFR1 or anti-Grb2 antibodies. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with anti-FRS2α or anti-pTyr antibodies (Fig. 1A and B). FRS2α from the lysates of quiescent L6 myoblasts (Fig. 1A) migrated on SDS-polyacrylamide gels with an apparent molecular mass of 70 kDa and upon phosphorylation in response to anti-FGF treatment migrated with an apparent molecular mass 90 kDa. FRS2α is coimmunoprecipitated with both nonactivated and ligand-activated FGFR1 but is coimmunoprecipitated with Grb2 only upon FGF stimulation. Thus, unlike Grb2, FGFR1 interacts with FRS2 constitutively, independent of ligand activation and the type of cells in which both are expressed (Fig. 1B). To further address the question of whether the kinase activity of FGFR1 is dispensable for FRS2α binding, FRS2α was coexpressed with wild-type FGFR1 or the kinase-inactive (KM) FGFR1 mutant in 293 cells by transient transfection. The lysates were immunoprecipitated with anti-FRS2α antibodies and immunoblotted with anti-FRS2α or anti-FGFR1 antibodies. This experiment showed that FRS2α coimmunoprecipitated equally with the wild-type and kinase-inactive FGFR1 (Fig. 1C). The wild-type FGFR1 is activated and autophosphorylated when transiently overexpressed in 293 cells and in turn phosphorylates FRS2α (Fig. 1C). Taken together, these experiments demonstrate that FRS2α associates as a complex with FGFR1 constitutively, independent of receptor activation and tyrosine phosphorylation.
FIG. 1.
Constitutive complex formation between FGFR1 and FRS2α. (A) L6 myoblasts (L6 Flg) stably transfected with FGFR1 were stimulated with aFGF and heparin (+) (100 ng/ml and 5 μg/ml, respectively) or with medium alone (−) for 5 min at 37°C. The lysates were immunoprecipitated (IP) with anti-FGFR1 or anti-Grb2 antibodies. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting (IB) with anti-P-Tyr (left) or anti-FRS2α (right) antibodies. (B) NIH 3T3 (mouse fibroblast), L6 (rat myoblast), and PC12 (rat pheochromocytoma) cells stably transfected with FGFR1 were stimulated (+) with aFGF and heparin (100 ng/ml and 5 μg/ml, respectively) or with medium alone (−) for 5 min at 37°C. The lysates were immunoprecipitated with anti-FGFR1 or anti-Grb2 antibodies. The immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with anti-FRS2α antibodies. (C) 293 cells were transiently transfected with FRS2α alone (lane 1), together with the wild-type FGFR1 (lane 2) or with the kinase-inactive FGFR1 (KM; lane 3). The lysates were immunoprecipitated with anti-FGFR1 antibodies, followed by SDS-PAGE and immunoblotting with anti-FRS2α (top), anti-P-Tyr (middle), or anti-FGFR1 antibodies (bottom).
The PTB domain of FRS2 mediates complex formation with FGF receptor.
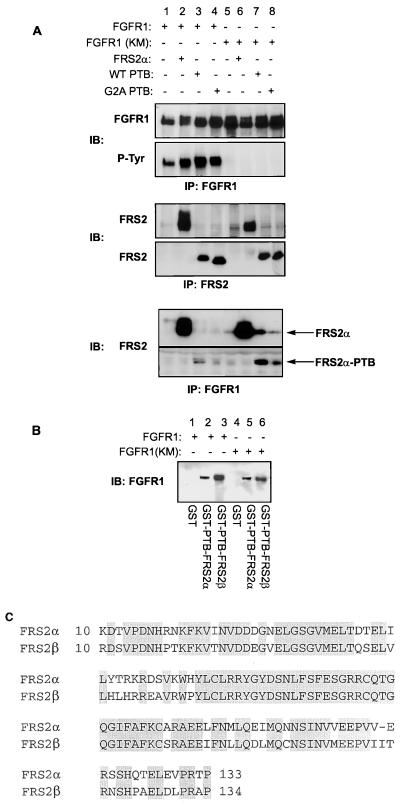
Several docking proteins associate with activated RTKs via their PTB domains; for example, the IRS proteins bind to the insulin receptor via their PTB domains (40) and Shc binds to EGF receptor (3) and TrkA via its PTB domain (9). To investigate the interaction between FRS2α and the FGF receptor, intact FRS2α or the amino-terminal portion of FRS2α (amino acids 1 to 158) including the PTB domain and either the natural myristylation sequence or the nonmyristylated G2A mutant sequence was coexpressed with FGFR1 or its kinase-inactive mutant in 293 cells. Full-length FRS2α or its PTB domain with the natural myristylation signal coimmunoprecipitated with wild-type FGFR1 as well as with the kinase-inactive FGFR1 mutant (Fig. 2A, lanes 2, 3, 6, and 7). In contrast the coimmunoprecipitation of the G2A mutant was significantly reduced compared to that of the wild-type protein (Fig. 2A, lanes 4 and 8). This result provided the probable basis for our previous observation that the nonmyristylated G2A mutant of FRS2α is not tyrosine phosphorylated upon aFGF stimulation and, unlike wild-type FRS2α, is deficient in promoting FGF-induced neurite outgrowth in PC12 cells (19). To further confirm that the PTB domains of FRS2α and FRS2β bind directly to the FGF receptor, we transiently expressed wild-type FGFR1 or its kinase-inactive mutant in 293 cells and subjected the lysates to a pulldown assay with immobilized PTB domains of FRS2α and FRS2β expressed as GST fusion proteins. This experiment showed that the levels of binding of the PTB domains of both FRS2 proteins to the wild-type FGFR1 and its kinase-inactive mutant are comparable (Fig. 2B). The PTB domains of FRS2α and FRS2β are highly conserved, with 75% identity in the primary sequence (Fig. 2C).
FIG. 2.
Association between FGFR1 and the PTB domain of FRS2α in transfected cells and in vitro. (A) N-myristylation of FRS2α enhances complex formation with FGFR1 in transfected cells. 293 cells were transiently transfected with wild-type FGFR1 (lanes 1 to 4) or the kinase-inactive (KM; lanes 5 to 8) FGFR1 mutant together with the full-length FRS2α (lanes 2 and 6), the N-terminal region of FRS2α consisting of the N-myristylation signal and the PTB domain (lanes 3 and 7), or the G2A mutant with a mutation in the N-myristylation sequence and the PTB domain of FRS2α (lanes 4 and 8). The expression level of FGFR1 and its kinase activity were analyzed by immunoprecipitation (IP) with anti-FGFR1 followed by immunoblotting (IB) with anti-FGFR1 and anti-P-Tyr antibodies. The expression levels of the various FRS2α constructs were analyzed by immunoprecipitation and immunoblotting with anti-FRS2 antibodies which were directed against the N-terminal region of FRS2α. The interactions between wild-type FGFR1 or its kinase-inactive mutant and the various FRS2 mutants were analyzed by immunoprecipitating the lysates with anti-FGFR1 antibodies followed by immunoblotting with anti-FRS2 antibodies. (B) The PTB domains of the FRS2 proteins interact with FGFR1. Lysates from 293 cells transiently transfected with the wild-type (lanes 1 to 3) or kinase-inactive (lanes 4 to 6) FGFR1 were subjected to a pulldown assay with the PTB domains of FRS2α (lanes 2 and 5) or FRS2β (lanes 3 and 6) expressed as GST fusion proteins and immobilized on glutathione-agarose beads. GST alone was used as a control (lanes 1 and 4). The specifically bound proteins were eluted and resolved by SDS-PAGE, and the presence of FGFR1 was detected by immunoblotting with anti-FGFR1 antibodies. (C) Alignment of the PTB domains of FRS2α and FRS2β by the Clustal method with DNASTAR. The two PTB domains exhibit 75% sequence identity. Identical residues are shaded.
The PTB domains of FRS2 proteins associate with FGFR1 at the juxtamembrane region.
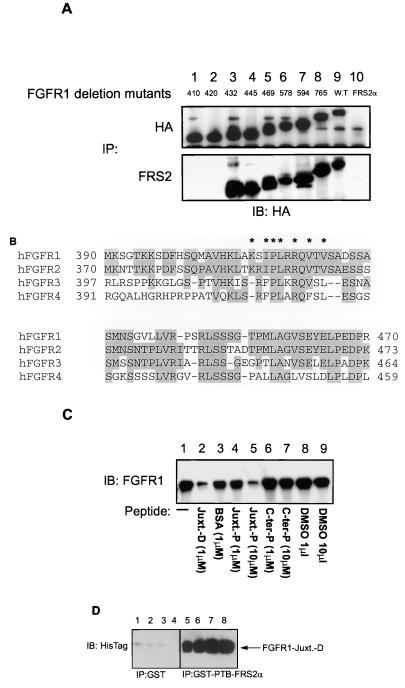
Our results clearly showed that the interaction between the PTB domains of the FRS2 proteins and FGFR1 does not require receptor activation or its tyrosine phosphorylation. We mapped the region on the FGF receptor involved in binding with FRS2α by comparing the binding properties of C-terminal deletion mutants of FGFR1. The HA epitope tag was added to each of the FGFR1 deletion mutants at the C-terminal end to facilitate immunoprecipitation and immunoblotting of the different mutant receptors with standard reagents. To assay for the binding of these deletion mutants of FGFR1 to FRS2α, FRS2α and the individual deletion mutants of FGFR1 were coexpressed in 293 cells by transient transfection, followed by immunoprecipitation of the various deletion mutants or FRS2α (Fig. 3A) from the lysates. The immunoprecipitates were resolved by SDS-PAGE, and the coimmunoprecipitated deletion mutants of FGFR1 were detected by anti-HA immunoblotting (Fig. 3A). The mutants with amino acids at 410 or 420 deleted did not coimmunoprecipitate with FRS2α (Fig. 3A, lanes 1 and 2), whereas those with amino acids beyond 432 deleted did (Fig. 3A, lanes 3 to 9). This result shows that the region of interaction is located at the juxtamembrane domain of the receptor, in the vicinity of amino acids 420 to 432 bearing the sequence SIPLRRQVTVSADS. This region is highly homologous among the four members of the mammalian FGFR family (Fig. 3B) and to a lesser extent among the FGF receptors of other organisms. To verify whether this region alone is sufficient to mediate the interaction with the PTB domain of the FRS2 proteins, a synthetic peptide encompassing residues 412 to 433 (MAVHKLAKSIPLRRQVTVSADS) was assayed for its potential to inhibit the binding between FGFR1 and the PTB domain of the FRS2 proteins. As shown in Fig. 3C, 10 μl of the peptide containing the FRS2α binding site of FGFR1 inhibited the binding of the PTB domains of both FRS2 proteins to FGFR1. A shorter peptide corresponding to residues 416 to 432 of hFGFR1 also inhibits the interaction between FGFR1 and the PTB domain of FRS2α (data not shown). To further demonstrate that the interaction between the PTB domain of FRS2 and the juxtamembrane region of FGFR1 is indeed direct, the PTB domain of FRS2α expressed as a GST fusion protein and the juxtamembrane region of FGFR1 (amino acids 399 to 470) expressed as a histidine-tagged fusion protein were used. Equivalent amounts of GST-PTB-FRS2α immobilized on glutathione beads were incubated with increasing concentrations of the juxtamembrane region of FGFR1 protein (2.5, 5, 10, and 20 μM; Fig. 3D, lanes 5 to 8). As a control, similar concentrations of the juxtamembrane region of FGFR1 were incubated with GST alone (lanes 1 to 4). Specifically bound protein was eluted and resolved by SDS-PAGE, followed by immunoblotting with antibodies against the hexahistidine epitope. As shown in Fig. 3D the N-terminal region of FGFR1 binds directly to the PTB domain of FRS2α.
FIG. 3.
The PTB domains of the FRS2 proteins bind to the juxtamembrane region of FGFR1. (A) Mapping of the binding site of FRS2α on FGFR1 by deletion mutagenesis. Deletion mutagenesis of FGFR1 was carried out by insertion of the DNA sequence encoding the HA epitope, followed by the stop codon after codons for amino acids 410, 420, 432, 445, 469, 578, 594, and 765 (lanes 1 to 8). Each of these deletion mutants and the full-length FGFR1 were tagged with the HA epitope at the C-terminal end. 293 cells were transiently transfected with FRS2α either alone (lane 10) or together with each of the HA-tagged FGFR1s (wild type [lane 9] or the deletion mutants [lanes 1 to 8]). The lysates were subjected to immunoprecipitations (IP) with anti-HA (top) or anti-FRS2α (bottom) antibodies followed by SDS-PAGE and immunoblotting (IB) with anti-HA antibodies. (B) Alignment of amino acid sequences of the juxtamembrane domains of FGF receptors. The primary sequences of human FGFR1, FGFR2, FGFR3, and FGFR4 were aligned by the Clustal method with DNASTAR. Identical residues are shaded. ∗, amino acids critical for the interaction between the PTB domains of the FRS2 proteins and FGFR1. (C) FRS2α binds to the juxtamembrane region of FGFR1. 293 cells were transiently transfected with FGFR1. The lysates were subjected to a pulldown assay with the GST-PTB-FRS2α beads either alone (lane 1) or in the presence of the recombinant protein of the juxtamembrane of FGFR1 (amino acids 399 to 470) expressed as a histidine-tagged fusion protein (1 μM; lane 2), a bovine serum albumin control (1 μM; lane 3), FGFR1 juxtamembrane peptide (MAVHKLAKSIPLRRQVTVSADS; 1 and 10 μM; lanes 4 and 5, respectively), FGFR1 C-terminal peptide (PRHPAQLANGGKLRR; 1 and 10 μM; lanes 6 and 7, respectively), and the peptide solvent dimethyl sulfoxide (1 and 10 μl; lanes 8 and 9, respectively). The specifically bound proteins were resolved by SDS-PAGE, and the presence of FGFR1 was detected by immunoblotting with anti-FGFR1 antibodies. Similar results were observed when the PTB domain of FRS2α was used for the pulldown experiments (data not shown). (D) The interaction between the PTB domain of FRS2 and the juxtamembrane region of FGFR1 is direct. Equivalent amounts of the PTB domain of FRS2α expressed as a GST fusion protein (GST-PTB-FRS2α) and immobilized on glutathione-agarose beads were incubated with increasing concentrations (2.5, 5, 10 and 20 μM; lanes 5 to 8, respectively) of the juxtamembrane region of FGFR1 expressed as a hexahistidine-tagged fusion protein (Juxt. D; amino acids 399 to 470). As a control, similar concentrations of the Juxt. D protein were incubated with GST beads (lanes 1 to 4). Specifically bound proteins were eluted and resolved by SDS-PAGE, followed by detection and Western blotting with antibodies against the hexahistidine epitope.
Mapping of the FGFR1 binding domain by alanine scanning mutagenesis.
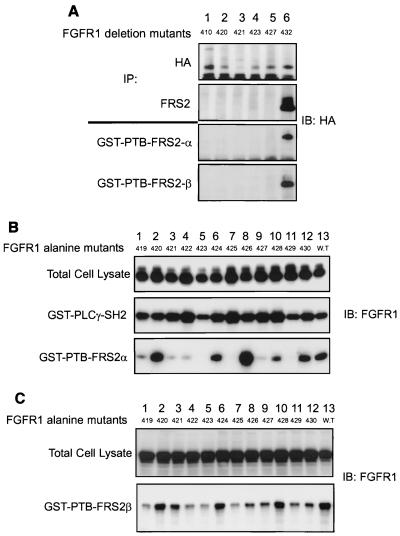
In our initial screening the binding domain for FRS2α was mapped to the vicinity of amino acids 420 to 432 of FGFR1 (Fig. 3A). We generated additional deletion mutants of FGFR1 with the HA tag and a stop codon introduced after codons for amino acids 421, 423, and 427 to further define the binding domain on the FGF receptor. None of these deletion mutants coprecipitated with FRS2α when the two were expressed together in 293 cells (Fig. 4A). Moreover, when lysates of 293 cells expressing these deletion mutants were subjected to a pulldown experiment using the PTB domain of the FRS2 proteins, only the mutant with amino acids beyond 432 deleted was detected (Fig. 4A).
FIG. 4.
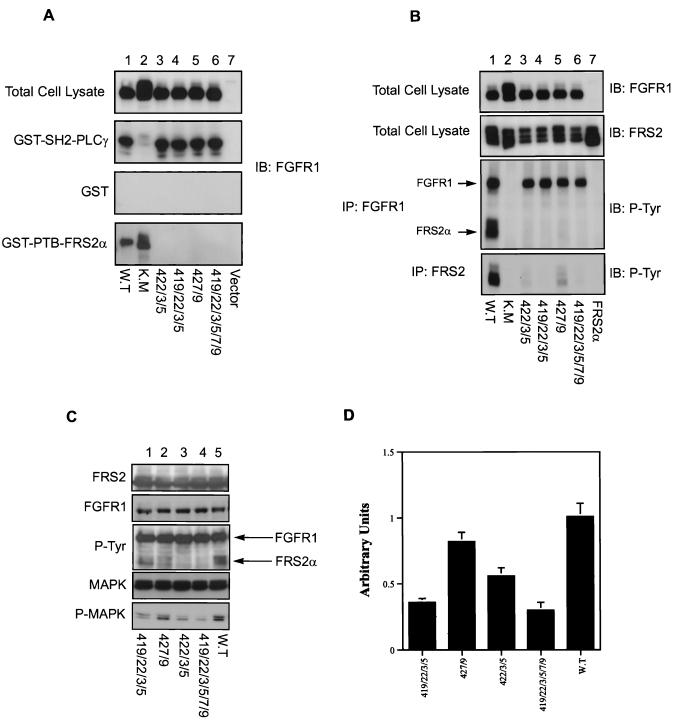
FRS2 proteins bind to similar regions on FGFR1 via their PTB domains. (A) Binding of FRS2 proteins to deletion mutants of FGFR1. HA-tagged deletion mutants of FGFR1 truncated at amino acids 410, 420, 421, 423, 427, or 432 (lanes 1 to 6, respectively) were cotransfected with FRS2α expression plasmid in 293 cells. The lysates were subjected to immunoprecipitations (IP) with anti-HA or anti-FRS2α antibodies, followed by SDS-PAGE and immunoblotting (IB) with anti-HA antibodies. The same series of deletion mutants were expressed in 293 cells, and the lysates were subjected to a pulldown assay with GST-PTB-FRS2α or GST-PTB-FRS2β beads. The specifically bound proteins were eluted and resolved by SDS-PAGE, and the presence of various deletion mutants of FGFR1 was detected by immunoblotting with anti-HA antibodies. (B) Alanine scanning of the PTB domain binding site on human FGFR1. 293 cells were transfected with FGFR1 mutants, each with a single alanine substitution at amino acids 419 to 430 (lanes 1 to 12, respectively), or with wild-type FGFR1 (lane 13). The expression levels of the alanine mutants were shown to be equivalent by immunoblotting total cell lysate with anti-FGFR1 antibodies. The lysates were subjected to a pulldown assay with GST-SH2-PLC-γ or GST-PTB-FRS2α beads. The specifically bound proteins were eluted and resolved by SDS-PAGE, and the presence of alanine mutants of FGFR1 was detected by immunoblotting with anti-FGFR1 antibodies. (C) The PTB domains of the FRS2 proteins bind to similar binding sites on FGFR1. 293 cells were transiently transfected with FGFR1 mutants, each with a single alanine substitution at amino acids 419 to 430 (lanes 1 to 12, respectively), or with wild-type FGFR1 (lane 13). The expression levels of the alanine mutants were shown to be equivalent by immunoblotting the total cell lysate with anti-FGFR1 antibodies. The lysates were subjected to a pulldown assay with GST-PTB-FRS2β beads. The specifically bound proteins were resolved by SDS-PAGE, and the presence of the alanine mutants of FGFR1 was detected by immunoblotting with anti-FGFR1 antibodies.
To further understand the interaction between FGFR1 and the PTB domain of the FRS2 proteins, alanine scanning mutagenesis of FGFR1 was carried out. Residues 419 to 430 of FGFR1 were individually mutated to alanine, and the ability of each point mutant to bind the FRS2 proteins was assayed. 293 cells were transiently transfected with each of the alanine point mutants of FGFR1. The lysates prepared were subjected to binding assays with the PTB domain of the FRS2 proteins expressed as GST fusion proteins and immobilized on glutathione-agarose beads. The bound FGFR1 was detected by immunoblotting with anti-FGFR1 antibodies. The mutation of residues 419, 421, 422, 423, 425, 427, and 429 (Fig. 4B, lanes 1, 3, 4, 5, 7, 9, and 11, respectively) to alanine significantly diminished the binding of FGFR1 to the PTB domain of FRS2α, compared to that of the wild-type receptor (lane 13). Similar results were obtained for the PTB domain of FRS2β (Fig. 4C). These amino acids appear to be critical for the interaction between the PTB domains of the FRS2 proteins and FGFR1 (Fig. 3B). The integrity of the various FGFR1 mutants and their overall structures were not affected since each of the mutants was tyrosine phosphorylated and pulled down by the SH2 domains of PLC-γ expressed as a GST fusion protein and bound on glutathione-agarose beads (Fig. 4B). The results obtained in the alanine scan experiment are consistent with those from deletion mutagenesis, confirming that the binding site for the PTB domains of FRS2 proteins on FGFR1 resides in the juxtamembrane region within amino acids 419 to 430. Therefore, we generated FGF receptor mutants with several alanine substitutions in the binding domain and studied their interaction with FRS2 as well as their effect on MAPK activation, as assayed by immunoblotting with phospho-MAPK antibodies. The results showed that the combined alanine mutants do not bind to the PTB domains of the FRS2 proteins (Fig. 5A). The alanine mutants of FGFR1 also do not interact with or stimulate tyrosine phosphorylation of FRS2α in vivo (Fig. 5B). The mutations at the N-terminal region of the FGF receptor binding domain (amino acids 419, 422, 423, and 425A; Fig. 5C, lanes 1 and 3) have a stronger inhibitory effect on MAPK activation than mutations in the C-terminal region (amino acids 427 and 429A; Fig. 5C, lane 2), in contrast to what is found for the wild-type receptor (Fig. 5C, lane 5). The mutations at the N-terminal region caused about 60% inhibition of MAPK activation compared to that for the wild-type receptor, while the mutations at the C-terminal region caused 20% inhibition of MAPK activation (Fig. 5D; the standard deviations from two separate experiments are shown). Taken together, these results indicated that the major interaction of the PTB domain of FRS2 occurs at the N-terminal region of the binding domain involving residues 419, 422, 423, and 425 of human FGFR1.
FIG. 5.
Alanine substitutions of amino acids in the FRS2 binding site on FGFR1 abrogates FRS2 phosphorylation and MAPK activation. (A) 293 cells were transfected with FGFR1 with combined alanine substitutions at amino acids 422, 423, and 425 (lane 3); 419, 422, 423, and 425 (lane 4); 427 and 429 (lane 5); 419, 422, 423, 425, 427, and 429 (lane 6); and wild-type FGFR1 (lane 1) or its kinase-inactive mutant (lane 2) as a control. The expression of each receptor was shown to be equivalent by immunoblotting (IP) the total cell lysate with anti-FGFR1 antibodies. The lysates were subjected to a pulldown assay with GST-SH2-PLC-γ, GST alone, or GST-PTB-FRS2α beads. The specifically bound proteins were resolved by SDS-PAGE, and the presence of the alanine mutants of FGFR1 was detected by immunoblotting with anti-FGFR1 antibodies. (B) 293 cells were transfected with FRS2α (lanes 1 to 7) and FGFR1 with combined alanine substitutions as described for panel A. The expression levels of the FGF receptors were shown to be equivalent by immunoblotting total cell lysate with anti-FGFR1 and anti-FRS2α antibodies. The lysates were subjected to immunoprecipitation (IP) with anti-FGFR1 antibodies or anti-FRS2α antibodies followed by immunoblotting with anti-P-Tyr antibodies. (C) 293 cells were transfected with FRS2α and ERK1 (lanes 1 to 5) and FGFR1 with the indicated combined alanine substitutions (lanes 1 to 4) or the wild-type FGFR1 (lane 5). The cells were lysed, and equivalent amounts of the total cell lysates were resolved by SDS-PAGE and immunoblotted with anti-FRS2, anti-FGFR1, anti-P-Tyr, anti-MAPK, or anti-phospho-MAPK (P-MAPK) antibodies. (D) Densitometric quantitation of MAPK phosphorylation. Standard deviations from two separate experiments are shown. WT, wild type.
The PTB domain of FRS2 interacts only with activated TrkA.
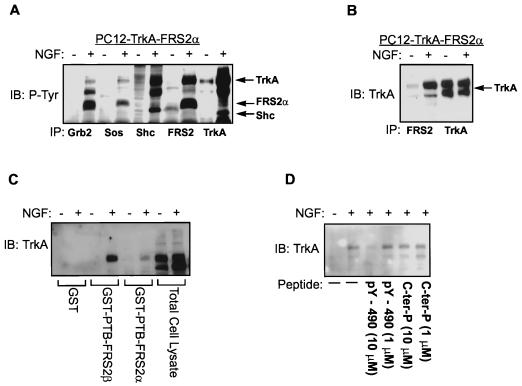
To study the interaction between FRS2 and the NGF receptor, we used parental PC12 cells and PC12 cells overexpressing both TrkA and FRS2α (PC12-Trk-FRS2α). Quiescent PC12 cells were stimulated with NGF, and the lysates were immunoprecipitated with anti-Grb2, anti-Sos1, anti-Shc, anti-FRS2, or anti-TrkA antibodies. The immunoprecipitates were resolved and immunoblotted with anti-P-Tyr antibodies. As shown in Fig. 6A, FRS2α was coimmunoprecipitated with Grb2, Sos1, and TrkA in NGF-stimulated PC12-TrkA-FRS2α cells. TrkA was coimmunoprecipitated with either Shc or FRS2. To verify that the interaction between FRS2α and TrkA is dependent on receptor activation and autophosphorylation, lysates from NGF-stimulated PC12-TrkA-FRS2α cells were immunoprecipitated with anti-FRS2α antibodies followed by immunoblotting with anti-TrkA antibodies. This experiment demonstrated that TrkA coprecipitated with FRS2α only from lysates of NGF-stimulated cells (Fig. 6B).
FIG. 6.
FRS2 binds via its PTB domain to Y490 site on NGF receptor in a phosphorylation-dependent manner. (A) Ternary complex of FRS2α–Grb2-Sos1 and tyrosine-phosphorylated TrkA. PC12 cells stably transfected with TrkA and FRS2α were stimulated (+) with 100 ng of NGF/ml or with medium alone (−) for 5 min at 37°C. The lysates were subjected to immunoprecipitations (IP) with anti-Grb2, anti-Sos1, anti-Shc, anti-FRS2, or anti-TrkA antibodies. The immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting (IB) with anti-P-Tyr antibodies. (B) FRS2α binds to tyrosine-phosphorylated TrkA. PC12 cells stably transfected with TrkA and FRS2α were stimulated with 100 ng of NGF/ml or with medium alone for 5 min at 37°C. The lysates were subjected to immunoprecipitations with anti-FRS2α or anti-TrkA antibodies. The immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-TrkA antibodies. (C) FRS2α binds to tyrosine-phosphorylated TrkA via the PTB domain. PC12 cells stably transfected with TrkA and FRS2α were stimulated with 100 ng of NGF/ml or medium alone for 5 min at 37°C. The lysates were subjected to a pulldown assay with immobilized GST (control), GST-PTB-FRS2α, or GST-PTB-FRS2β. The specifically bound proteins were resolved by SDS-PAGE, and the associated TrkA was detected by immunoblotting with anti-TrkA antibodies. (D) The PTB domains of FRS2 proteins bind to the NPQpY sequence of TrkA. PC12 cells stably transfected with TrkA and FRS2α were stimulated with 100 ng of NGF/ml or medium alone for 5 min at 37°C. The lysates were subjected to a pulldown assay with immobilized GST-PTB-FRS2β. The phosphopeptide encompassing the NPQpY motif of TrkA was added at 10 or 1 μM. A control peptide corresponding to the C-terminal region of human FGFR1 (C-ter-P) was similarly added at 10 or 1 μM. The specifically bound proteins were resolved by SDS-PAGE, and the associated TrkA was detected by immunoblotting with anti-TrkA antibodies.
It has been established that Shc interacts with TrkA via its PTB domain (9, 28). We further investigated whether the interaction between FRS2α and TrkA is also mediated through the PTB domain. Lysates of NGF-stimulated or unstimulated PC12-TrkA-FRS2α cells were subjected to a pulldown assay with the immobilized PTB domains of the FRS2 proteins. The PTB domains of both FRS2α and FRS2β pulled down TrkA only from lysates of NGF-stimulated cells (Fig. 6C), indicating that only activated and tyrosine-phosphorylated TrkA binds to the PTB domains of the FRS2 proteins. The juxtamembrane region of TrkA contains an NPXpY motif, which has been defined as a consensus binding motif for several PTB domains and which serves as a docking site for the PTB domain of Shc (3, 18, 21, 44). Since the PTB domains of the FRS2 proteins bind only to the activated TrkA receptor, we tested whether a peptide corresponding to the Shc binding site on TrkA, which includes pY-490 and surrounding amino acids (LQGHIIENPQpYFSDACVH) could inhibit the interaction between the activated TrkA and the PTB domains of FRS2 proteins. As shown in Fig. 6D, the pY-490 peptide diminished the association between TrkA and the PTB domain of the FRS2 proteins in a pulldown experiment using lysates from NGF-stimulated and unstimulated PC12-TrkA-FRS2α cells and immobilized PTB domains of the FRS2 proteins. The inhibition occurred with 10 μM or higher concentrations of the peptide, suggesting that the PTB domains of FRS2 and Shc target similar binding sites on TrkA, involving pY-490.
FGFR1 and TrkA interact with similar sites on the PTB domain of FRS2.
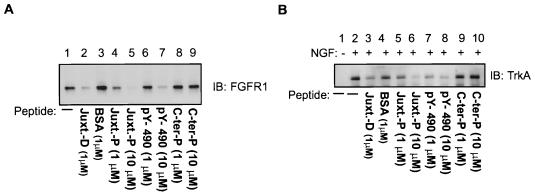
To gain an insight into the mechanism by which the PTB domains of the FRS2 proteins recognize the sequences of FGFR1 and TrkA, which show no apparent homology, we performed peptide competition assays with synthetic peptides based upon the binding sites on the receptors, as described above. The FGFR1 peptide consists of amino acids 412 to 433 (MAVHKLAKSIPLRRQVTVSADS) from human FGFR1, and the TrkA phosphopeptide consists of amino acids 489 to 497 (LQGHIIENPQpYFSDACVH) from human TrkA. FGFR1 was transfected in 293 cells, and lysates were subjected to a pulldown assay with the PTB domains of the FRS2 proteins expressed as GST fusion proteins and immobilized on glutathione-agarose beads. The bound FGFR1 was detected by elution off the beads, SDS-PAGE, and immunoblotting with anti-FGFR1 antibodies. As shown in Fig. 7A, in the presence of 10 μM FGFR1 peptide, the binding of FGFR1 to the immobilized PTB domains of the FRS2 proteins was virtually abolished (lane 5). Similar inhibition of the binding was observed with 10 μM TrkA phosphopeptide (lane 7). Similar results were obtained when this experiment was performed with the kinase-inactive FGFR1 (data not shown).
FIG. 7.
FGFR1 and TrkA interact with similar sites on the PTB domain of FRS2. (A) 293 cells were transiently transfected with FGFR1. The lysates were subjected to a pulldown assay with the GST-PTB-FRS2β beads either alone (lane 1) or in the presence of the recombinant protein of the juxtamembrane of FGFR1 (amino acids 399 to 470) expressed as a histidine-tagged fusion protein (1 μM; lane 2), bovine serum albumin (control; 1 μM; lane 3), FGFR1 juxtamembrane peptide (1 and 10 μM; lanes 4 and 5, respectively), TrkA phosphopeptide (1 and 10 μM; lanes 6 and 7, respectively), and a control peptide (1 and 10 μM; lanes 8 and 9, respectively). The specifically bound proteins were resolved by SDS-PAGE, and the presence of FGFR1 was detected by immunoblotting (IB) anti-FGFR1 antibodies. (B) A similar experiment was carried out with lysates of PC12 cells stably transfected with TrkA and FRS2α. Cells were stimulated (+) with 100 ng of NGF/ml or medium alone (−) for 5 min at 37°C. The specifically bound proteins were resolved by SDS-PAGE, and the associated TrkA was detected by immunoblotting with anti-TrkA antibodies.
We further investigated the ability of the peptides to compete for the binding of TrkA to the PTB domains of the FRS2 proteins. Quiescent PC12-TrkA-FRS2α cells were stimulated with NGF (100 ng/ml, 5 min) and lysed, and the lysates were subjected to a pulldown assay with immobilized PTB domains of the FRS2 proteins, as described above. Bound TrkA was detected by elution off the beads, SDS-PAGE, and immunoblotting with anti-TrkA antibodies. In the presence of 10 μM FGFR1 or TrkA peptide, the binding of TrkA to the PTB domain of FRS2 was abolished (Fig. 7B, lanes 6 and 8). From the collective data above we conclude that FGFR1 and TrkA bind to the PTB domains of the FRS2 proteins at similar or overlapping binding sites. However, FRS2 binds preferentially to tyrosine-phosphorylated, activated TrkA, while FRS2 interacts with FGFR1 in a phosphorylation-independent manner.
Expression level of FGFR1 modulates NGF-induced tyrosine phosphorylation of FRS2α.
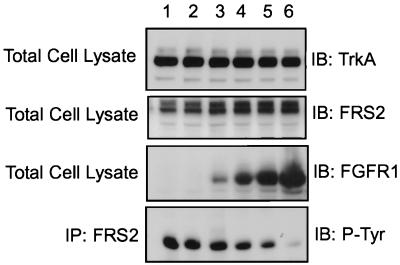
Since FRS2 proteins interact with both FGFR1 and the NGF receptor, we have examined the possibility that overexpression of FGFR1 will influence NGF-induced tyrosine phosphorylation of FRS2α. To test this hypothesis, 293 cells were transfected with constant levels of the TrkA and FRS2α cDNAs and increasing levels of DNA of the kinase-inactive mutant of FGFR1. The cells were then lysed, and FRS2α was immunoprecipitated. The immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting with anti-pTyr antibodies. The results showed that increased expression of the kinase-inactive mutant of FGFR1 diminished the tyrosine phosphorylation of FRS2α in response to NGF stimulation (Fig. 8). We also found that in PC12 cells overexpressing FGFR1, the level of NGF-induced tyrosine phosphorylation of endogenous FRS2 is somewhat reduced (data not shown). These observations suggest that FGF receptors may regulate signaling through NGF receptors by sequestering a common docking protein, such as FRS2, that both receptors utilize for transmitting their signals.
FIG. 8.
Inhibition of TrkA-mediated FRS2α phosphorylation by overexpression of kinase-inactive FGFR1 mutant. 293 cells were transfected with constant levels of TrkA and FRS2α expression plasmids (lanes 1 to 6; 1 and 0.5 μg of the respective plasmids) and increasing levels of expression plasmid encoding the kinase-inactive FGFR1 mutant (0, 0.5, 1, 3, 6, and 18 μg of plasmids in lanes 1 to 6, respectively). The cells were lysed, and equivalent amounts of the total cell lysates were resolved by SDS-PAGE and immunoblotted (IB) with anti-TrkA, anti-FRS2α, or anti-FGFR1 antibodies, respectively. Immunoprecipitation (IP) of FRS2α was followed by immunoblotting with anti-P-Tyr antibodies.
DISCUSSION
FRS2α is a major intracellular substrate of the ligand-activated FGF and NGF receptors and is rapidly and highly tyrosine phosphorylated in cells upon FGF or NGF stimulation. Structurally, FRS2α bears an N-terminal myristylation site, a PTB domain, four potential Grb2(SH2) recognition sites, and two Shp2(SH2) recognition sites (19). Tyrosine-phosphorylated FRS2α forms a complex with Grb2-Sos and Shp2, which is itself tyrosine phosphorylated and bound to the Grb2-Sos complex (14). Thus FRS2α mediates the recruitment of the Grb2-Sos complexes directly and indirectly via Shp2 in signaling via the FGF and NGF receptors. The overexpression of wild-type FRS2α enhances FGF-induced MAPK activation and neurite outgrowth in PC12 cells. Conversely, overexpression of FRS2α mutated to abolish the recruitment of Shp2 and Grb2-Sos complexes abrogates FGF-induced MAP kinase activation and neurite outgrowth (14, 19). In addition, microinjection of anti-FRS2α antibodies inhibits FGF-induced DNA synthesis (19). These observations establish that FRS2α serves as a physiological link between ligand-activated FGF and NGF receptors and the Ras/MAPK signaling pathway (14, 19).
To achieve regulation and specificity in signaling, many RTKs utilize a class of proteins called docking proteins. These proteins contain multiple phosphotyrosine residues that serve as recruitment sites for a variety of signaling molecules. Docking proteins commonly possess an N-terminal PTB domain involved in direct interaction with RTKs (30). It was previously demonstrated that certain docking proteins bind to a consensus NPXpY sequence motif in several RTKs. For example, the activated insulin receptor possesses an NPXpY sequence in the juxtamembrane region which is the recognition site for the PTB domains of IRS1 and Shc (13). A similar NPEpY motif is also present in the EGF receptor, to which the PTB domain of Shc binds (3, 20). Upon ligand stimulation the NGF receptor becomes autophosphorylated on an NPQY motif in the juxtamembrane region which serves as a high-affinity binding site for the PTB domain of Shc. This motif is conserved in TrkB, TrkC (36), and glial cell-derived neurotrophic factor receptor, Ret (1).
Since the FRS2 proteins are major intracellular substrates and downstream effectors of the FGF and NGF receptors, we set out to investigate the mechanism of interaction between the FRS2 proteins and these two receptors. The interaction between FRS2 and FGF receptor represents a novel paradigm. The association of FRS2 with the FGF receptor is constitutive and does not require tyrosine phosphorylation of the receptor. We have shown in this report that the interaction occurred specifically through the PTB domain of FRS2 binding to the juxtamembrane region of FGF receptor despite the absence of an NPXY motif. We further mapped the binding site and showed that it resides at amino acids 419 to 430 bearing the sequence KSIPLRRQVTVS. This sequence is conserved throughout the mammalian FGF receptor family (Fig. 3B), and amino acids K419, I421, P422, L423, R425, V427, and V429, when mutated to alanine, strongly diminished the interaction between FGFR1 and the PTB domains of the FRS2 proteins. The inefficient recruitment of FRS2α by FGFR1 bearing combined substitutions of alanine for these amino acids resulted in decreased tyrosine phosphorylation of FRS2α. We have earlier shown that tyrosine-phosphorylated FRS2α bound directly to the SH2 domains of Grb2 and Shp2, leading to potentiation of FGF-induced MAPK activation (14, 19). Elimination of Grb2 and Shp2 binding by mutation of the tyrosine residues responsible for Grb2 and Shp2 binding to phenylalanine led to reduction in FRS2α-mediated MAPK activation by 30 to 40% (14, 19). Indeed, the reduced tyrosine phosphorylation of FRS2α by FGFR1 due to inefficient binding to FGFR1 with alanine substitutions for amino acids in the binding domain resulted in reduced MAPK activation to a similar extent. The incomplete inhibition of MAPK activation by the FRS2 and FGFR1 mutants could be attributed to recruitment of Grb2-Sos complexes by other docking proteins, such as Shc and Gab1, that are tyrosine phosphorylated in response to FGF stimulation.
Interestingly, the sequence of the binding domain does not seem to acquire the β-turn conformation which is found in PTB domain-interacting ligands (reviewed in references 7 and 15). The PTB recognition site on FGFR1 contains a putative protein kinase C phosphorylation site bearing the sequence RQVT (amino acids 425 to 428) (11). The phosphorylation of this threonine is not required for interaction with the PTB of FRS2α since the T428A mutant FGFR1 was still able to interact with FRS2α. In addition, a bacterially produced protein of the juxtamembrane domain of FGFR1 (amino acids 399 to 470) which was not phosphorylated and a synthetic peptide corresponding to the binding site on FGFR1 bind directly to the PTB domains of FRS2 proteins and compete effectively with the binding of the full-length FGFR1.
Unlike what is found for FGFR1, the binding of FRS2 to the NGF receptor (TrkA) is mediated through the classical NPQpY motif of TrkA. FRS2 binds in vivo a phosphopeptide corresponding to the binding site on TrkA, resulting in reduced interaction between the activated receptor and the PTB domains of the FRS2 proteins. Indeed, mutation of the tyrosine residue in this motif (Y490) to phenylalanine on the TrkA receptor completely abrogated its signaling capacity. PC12 cells stably transfected with the Y490F mutant TrkA do not exhibit NGF-induced MAPK activation and neurite outgrowth (28, 41). Recently, it was demonstrated that the PTB domain of FRS2α bound directly to the Shc binding site on TrkA and TrkB (10, 26).
Our results suggest that the PTB domains of the FRS2 proteins are capable of recognizing diverse sequences specifically. As the synthetic peptides corresponding to the sequences of the binding regions of FGFR1 and TrkA prevent the interaction between these receptors and the PTB domains of the FRS2 proteins, it is likely that the recognition of the diverse ligand sequences occurs through similar or overlapping sites on the PTB domains. Thus, contrary to the previously established paradigms of interaction between RTKs and PTB domains of docking proteins, the phosphorylation of the FGF receptor is not obligatory for recognition by the PTB domains of FRS2 proteins. However, the binding of the PTB domains of the FRS2 proteins to TrkA is dependent upon tyrosine phosphorylation of the NPXY motif.
An emerging notion on the recognition of ligands by PTB domains is that PTB domains exist as a family of structurally conserved protein modules with diverse ligand-binding specificities that is not restricted to recognition of the β-turn-forming sequence NPXpY. The PTB domains of Shc and IRS1 preferentially bind to targets containing the NPXpY motif. Those of X11 and FE65 recognize as their target the β-amyloid precursor protein (βAPP) on an NPTY motif, but the phosphorylation of the tyrosine residue is not obligatory (4, 43). Indeed the replacement of the tyrosine residue with alanine results in no significant loss of binding affinity (42). The binding of the PTB domain of FE65 to its ligand βAPP requires the presence of an extra 28 residues flanking the NPXY site, but unlike that of X11 this PTB domain does not require the asparagine residue of the NPXY motif (42). An example of the diverse binding specificity to the same PTB domain is that of the Numb protein and its ligands. Numb is a protein involved in asymmetric cell division in Drosophila melanogaster. It binds to the adapter protein Lnx through an NPXY sequence, where tyrosine phosphorylation decreases the binding affinity (8). It also binds to the Numb-associating kinase via the sequence GFSNMSFEDFP, which does not contain a tyrosine residue (6). In a degenerate phosphopeptide library screen, it was found that the PTB domain of Numb preferentially bound GPY motifs and that the affinity of the nonphosphorylated version of this peptide is significantly lower than that of the phosphorylated peptide (24). Recently, competitive binding studies showed that the PTB domain of Numb binds to its three ligands with similar affinities (23). Moreover, since the peptides compete with each other, it is likely that they bind overlapping sites in the PTB domain. The solution structure of the PTB domain of Numb in a complex with the GPpY peptide shows that rather than the typical type I β-turn conformation observed in other PTB domain-peptide structures, the GPpY peptide assumes a helical-turn conformation that determines the contact between this peptide and the PTB domain (23). Based on the above examples, it seems that PTB domains can recognize a wide range of ligands and that their specificities will be defined by their spatial contacts. Our results show that the PTB domains of the FRS2 proteins can be classified in the group of PTB domains which can interact with multiple ligands that have no sequence homology.
FRS2 proteins recognize multiple receptors in phosphorylation-dependent and -independent manners. This property endows FRS2 proteins with a capacity to modulate a signal common to the receptors for FGF, NGF, and probably other neurotropic factors. The experiments presented in this report demonstrate that the relative levels of expression of these receptors and their activation states may influence the degree of recruitment of a limiting element (i.e., FRS2α or -β) that is crucial for activation of a common signal transduction pathway (i.e., Grb2/Sos/Ras). This may provide a plausible mechanism for transmodulation of signals between several different RTKs. The increased expression of FGF receptors in the absence of ligand activation could lead to changes in the distribution of FRS2 binding to ligand-activated TrkA and serve to downregulate the signals originating from TrkA via FRS2α.
Other studies have shown that activation of other members of the neurotrophin receptor family of tyrosine kinases, such as Ret (32) and TrkA and TrkB (10, 14) induce tyrosine phosphorylation of FRS2 in PC12 cells and cortical neurons, respectively. These observations are of particular interest since they suggest that transmodulation between RTKs could be a general mechanism that controls the strength of signals initiated by various tyrosine kinases and the fate of the cell in which these tyrosine kinases are expressed. It is therefore important to establish more physiologically relevant systems for exploring the possibility that tyrosine phosphorylation of a limiting docking protein could be an important step that defines specificity and provides a control element crucial for regulation of several families of RTKs that control neuronal cell differentiation and survival.
ACKNOWLEDGMENT
This work was supported by a fellowship grant from HFSPO.
REFERENCES
- 1.Arighi E, Alberti L, Torriti F, Ghizzoni S, Rizzeti M G, Pelicci G, Pasini B, Bongarzone I, Piutti C, Pierotti M A, Borrello M G. Identification of Shc docking site on Ret tyrosine kinase. Oncogene. 1997;14:773–782. doi: 10.1038/sj.onc.1200896. [DOI] [PubMed] [Google Scholar]
- 2.Barde Y. Trophic factors and neuronal survival. Neuron. 1989;2:1525–1534. doi: 10.1016/0896-6273(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 3.Batzer A, Blaikie P, Nelson K, Schlessinger J, Margolis B. The phosphotyrosine interaction domain of Shc binds an LXNPXY motif on the epidermal growth factor receptor. Mol Cell Biol. 1995;15:4403–4409. doi: 10.1128/mcb.15.8.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borg J P, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B. p62(dok): a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 6.Chien C T, Wang S, Rothenberg M, Jan L Y, Jan T N. Numb-associated kinase interacts with the phosphotyrosine binding domain of Numb and antagonizes the function of Numb in vivo. Mol Cell Biol. 1998;18:598–607. doi: 10.1128/mcb.18.1.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowburn D. Peptide recognition by PTB and PDZ domains. Curr Opin Struct Biol. 1997;7:835–838. doi: 10.1016/s0959-440x(97)80155-8. [DOI] [PubMed] [Google Scholar]
- 8.Dho S E, Jacob S, Wolting C D, French M B, Rohrschneider L R, McGlade C J. The mammalian Numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing Numb binding protein, Lnx. J Biol Chem. 1998;273:9179–9187. doi: 10.1074/jbc.273.15.9179. [DOI] [PubMed] [Google Scholar]
- 9.Dikic I, Batzer A G, Blaikie P, Obermeier A, Ullrich A, Schlessinger J, Margolis B. Shc binding to nerve growth factor receptor is mediated by the phosphotyrosine interaction domains. J Biol Chem. 1995;270:15125–15129. doi: 10.1074/jbc.270.25.15125. [DOI] [PubMed] [Google Scholar]
- 10.Easton J B, Moody N M, Zhu X, Middlemas D S. Brain-derived neurotrophic factor induces phosphorylation of fibroblast growth factor receptor substrate 2. J Biol Chem. 1999;274:11321–11327. doi: 10.1074/jbc.274.16.11321. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie L L, Chen G, Paterno G. Cloning of a fibroblast growth factor receptor 1 splice variant from Xenopus embryos that lacks a protein kinase C site important for the regulation of receptor activity. J Biol Chem. 1995;270:22758–22763. doi: 10.1074/jbc.270.39.22758. [DOI] [PubMed] [Google Scholar]
- 12.Goldfarb M. Function of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 13.Gustafson T A, He W, Craparo A, Schaub C D, O'Neill T J. Phosphotyrosine-dependent interaction of Shc and insulin receptor substrate-1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol Cell Biol. 1995;15:2500–2508. doi: 10.1128/mcb.15.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadari Y R, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison S. Peptide-surface association: the case of PDZ and PTB domains. Cell. 1996;86:341–343. doi: 10.1016/s0092-8674(00)80105-1. [DOI] [PubMed] [Google Scholar]
- 16.Holgado-Madruga M, Emlet D R, Moscatello D K, Godwin A K, Wong A J. A Grb2-associated docking protein in EGF- and insulin-receptor signaling. Nature. 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 17.Holland S J, Gale N W, Gish G D, Roth R A, Songyang Z, Cantley L C, Henkemeyer M, Yancopoulos G D, Pawson T. Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J. 1997;16:3877–3888. doi: 10.1093/emboj/16.13.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kavanaugh W M, Turck C W, Williams L T. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- 19.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 20.Lai K M V, Olivier J P, Gish G D, Henkemeyer M, McGlade J, Pawson T. A Drosophila Shc gene product is implicated in signaling by the DER receptor tyrosine kinase. Mol Cell Biol. 1995;15:4810–4818. doi: 10.1128/mcb.15.9.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laminet A A, Apel G, Conroy L, Kavanaugh W M. Affinity, specificity and kinetics of the Shc phosphotyrosine binding domain with asparagine-X-X-phosphotyrosine motifs of growth factor receptors. J Biol Chem. 1996;271:264–269. doi: 10.1074/jbc.271.1.264. [DOI] [PubMed] [Google Scholar]
- 22.Lange-Carter C A, Johnson G L. Ras-dependent growth factor regulation of MEK kinase in PC12 cells. Science. 1994;265:1458–1461. doi: 10.1126/science.8073291. [DOI] [PubMed] [Google Scholar]
- 23.Li S C, Zwahlen C, Vincent S J F, McGlade C J, Kay L E, Pawson T. Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat Struct Biol. 1998;5:1075–1083. doi: 10.1038/4185. [DOI] [PubMed] [Google Scholar]
- 24.Li S C, Songyang Z, Vincent S J F, Zwahlen C, Wiley S, Cantley L, Kay L E, Forman-Kay J, Pawson T. High-affinity binding of the Drosophila Numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc Natl Acad Sci USA. 1997;94:7204–7209. doi: 10.1073/pnas.94.14.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandiyan V, O'Brien R, Zhou M, Margolis B, Lemmon M A, Sturtevant J M, Schlessinger J. Thermodynamic studies of Shc phosphotyrosine-interaction-domain recognition of the NPXpY-motif. J Biol Chem. 1996;271:4770–4775. doi: 10.1074/jbc.271.9.4770. [DOI] [PubMed] [Google Scholar]
- 26.Meakin S O, MacDonald J I S, Gryz E A, Kubu C J, Verdi J M. The signaling adaptor FRS2 competes with Shc for binding to the nerve growth factor receptor TrkA. J Biol Chem. 1999;274:9861–9870. doi: 10.1074/jbc.274.14.9861. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadi M, Dionne C A, Li W, Spivak-Kroizman T, Li N, Honegger M A, Schlessinger J, Jaye M. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature. 1992;358:681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- 28.Obermeier A, Lammers R, Weismuller K H, Jung G, Schlessinger J, Ullrich A. Identification of Trk binding sites for Shc and phosphatidylinositol 3′-kinase and formation of a multimeric signaling complex. J Biol Chem. 1993;268:22963–22966. [PubMed] [Google Scholar]
- 29.Ong S H, Lim Y P, Low B C, Guy G R. Shp2 associates directly with tyrosine phosphorylated p90 (SNT) protein in FGF-stimulated cells. Biochem Biophys Res Commun. 1997;238:261–266. doi: 10.1006/bbrc.1997.7272. [DOI] [PubMed] [Google Scholar]
- 30.Pawson T, Scott J D. Signaling through scaffolding, anchoring and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 31.Rabin S, Cleghon V, Kaplan D R. SNT, a differentiation-specific target of neurotrophic factor-induced tyrosine kinase activity in neurons and PC12 cells. Mol Cell Biol. 1993;13:2203–2213. doi: 10.1128/mcb.13.4.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizzo C, Califano D, Colucci-D'Amato G L, De Vita G, D'Alessio A, Dathan N A, Fusco A, Monaco C, Santelli G, Vecchio G, Santoro N, de Franciscis V. Ligand stimulation of a Ret chimeric receptor carrying the activating mutation responsible for the multiple endocrine neoplasia type 2B. J Biol Chem. 1996;271:29497–29501. doi: 10.1074/jbc.271.46.29497. [DOI] [PubMed] [Google Scholar]
- 33.Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Cell. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 34.Schlessinger J. How receptor tyrosine kinases activate Ras. Trends Biochem Sci. 1993;18:273–275. doi: 10.1016/0968-0004(93)90031-h. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi-Tezuka M, Yoshida Y, Fukada T, Ohtani T, Yamanaka Y, Nishida K, Nakajima K, Hibi M, Hirano T. Gab1 acts as an adapter molecule linking the cytokine receptor gp130 to ERK mitogen-activated protein kinase. Mol Cell Biol. 1998;18:4109–4117. doi: 10.1128/mcb.18.7.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Geer P, Pawson T. The PTB domain: a new protein module implicated in signal transduction. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- 37.Weidner K M, Di Cesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature. 1996;384:173–176. doi: 10.1038/384173a0. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Lee K, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adaptor proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]
- 39.Yamanashi Y, Baltimore D. Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 40.Yenush L, White M F. The IRS-signaling system during insulin and cytokine action. Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- 41.Yoon S O, Soltoff S P, Chao M V. A dominant role of the juxtamembrane region of the TrkA nerve growth factor receptor during neuronal cell differentiation. J Biol Chem. 1997;272:23231–23238. doi: 10.1074/jbc.272.37.23231. [DOI] [PubMed] [Google Scholar]
- 42.Zambrano N, Buxbaum J D, Minopoli G, Fiore F, De Candia P, De Renzis S, Faranio R, Sabo S, Cheetham J, Sudol M, Russo T. Interaction of the phosphotyrosine interaction/phosphotyrosine binding-related domains of Fe65 with wild type and mutant Alzheimer's β-amyloid precursor protein. J Biol Chem. 1997;272:6399–6405. doi: 10.1074/jbc.272.10.6399. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Lee C H, Mandiyan V, Borg J P, Margolis B, Schlessinger J, Kuriyan J. Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou S, Margolis B, Chaudhuri M, Shoelson S E, Cantley L C. The phosphotyrosine interaction domain of Shc recognizes tyrosine-phosphorylated NPXY motif. J Biol Chem. 1995;270:14863–14866. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]