Abstract
The ability to stimulate recombination in a site-specific manner in mammalian cells may provide a useful tool for gene knockout and a valuable strategy for gene therapy. We previously demonstrated that psoralen adducts targeted by triple-helix-forming oligonucleotides (TFOs) could induce recombination between tandem repeats of a supF reporter gene in a simian virus 40 vector in monkey COS cells. Based on work showing that triple helices, even in the absence of associated psoralen adducts, are able to provoke DNA repair and cause mutations, we asked whether intermolecular triplexes could stimulate recombination. Here, we report that triple-helix formation itself is capable of promoting recombination and that this effect is dependent on a functional nucleotide excision repair (NER) pathway. Transfection of COS cells carrying the dual supF vector with a purine-rich TFO, AG30, designed to bind as a third strand to a region between the two mutant supF genes yielded recombinants at a frequency of 0.37%, fivefold above background, whereas a scrambled sequence control oligomer was ineffective. In human cells deficient in the NER factor XPA, the ability of AG30 to induce recombination was eliminated, but it was restored in a corrected subline expressing the XPA cDNA. In comparison, the ability of triplex-directed psoralen cross-links to induce recombination was only partially reduced in XPA-deficient cells, suggesting that NER is not the only pathway that can metabolize targeted psoralen photoadducts into recombinagenic intermediates. Interestingly, the triplex-induced recombination was unaffected in cells deficient in DNA mismatch repair, challenging our previous model of a heteroduplex intermediate and supporting a model based on end joining. This work demonstrates that oligonucleotide-mediated triplex formation can be recombinagenic, providing the basis for a potential strategy to direct genome modification by using high-affinity DNA binding ligands.
One of the major goals of gene therapy for human disease is the targeted modification of the genome. Although methods for delivery of genes into mammalian cells are well developed, the frequency of homologous recombination is limited (22), in contrast to the more favorable situation in yeast. Most of the time, transfected DNA integrates in a nonhomologous manner, and, as a result, the simple introduction of new genes is of only modest value for gene therapy because it is difficult to achieve appropriate regulation of the gene when it is not in its cognate site. Techniques to select for homologous recombination of a sequence into the matching chromosomal locus have been developed (39), and these techniques have been invaluable in enabling the generation of mammalian cells and mice in which specific genes have been knocked out and replaced with desired sequences. However, such selection techniques require growth of cells in culture and are, in general, not applicable to gene therapy approaches. Because of this, investigators have recognized that methods to enhance the frequency of homologous recombination in mammalian cells will be critical in developing gene replacement strategies for research and gene therapy applications.
Recently, efforts have been directed toward modification of the recipient site to create a substrate prone to homologous recombination. It has been shown that a site-specific endonuclease, I-SceI, can induce double-strand breaks within extrachromosomal and genomic DNA designed to carry the rare 18-bp recognition site. This strategy has been used to induce intermolecular and intramolecular recombination in both Xenopus oocytes (51) and mammalian cells (3, 10, 14, 27, 44, 47). However, this has limited general application, as it involves the prior introduction of the recognition site within the genome.
Besides double-strand breaks, other types of DNA damage have been shown to be recombinagenic, but in a non-site-specific way. These include DNA damage from UV radiation (54), chemical carcinogens (64), and photoreactive molecules such as psoralen (48). Psoralens are DNA-damaging agents that intercalate into DNA and form covalent monoadducts and interstrand cross-links upon exposure to near-UV light (UVA). Photo-induced psoralen interstrand cross-links, and to a lesser extent monoadducts, can induce recombination in bacteria (11, 37), yeast (48), and mammalian cells and viruses (21, 60).
In previous work, members of our group demonstrated that psoralen damage can be introduced into DNA within mammalian cells in a site-specific manner by linking psoralen to an oligonucleotide that is designed to form a triple helix (62). Triplex DNA can be formed when oligonucleotides bind in the major groove of the double helix in a sequence-dependent manner at polypurine-polypyrimidine stretches in duplex DNA (1, 12, 33, 41, 43). The specificity of oligonucleotide-mediated triplex formation has also been used to inhibit transcription and to cleave DNA at unique sites (24). Using this strategy, we found that triple-helix-targeted psoralen damage can generate site-specific mutations and mediate gene knockout within viral genomes replicating in monkey or human cells (17, 31, 62, 63) and within chromosomal genes in mouse and hamster cells (38, 58).
Prompted by the ability of triple-helix-forming oligonucleotides (TFOs) to mediate genome modification in the form of mutation, we went on to test the ability of third-strand-targeted psoralen adducts to provoke homologous recombination at selected sites within cells. Using a simian virus 40 (SV40)-based shuttle vector carrying two mutant copies of the supF reporter gene, we demonstrated the ability of psoralen-conjugated TFOs to enter cells, to find and bind to their target site, and to stimulate intramolecular recombination within an episomal substrate (18). Other groups have also investigated recombination stimulated by triplex-directed psoralen adducts (50, 59). These studies have established directed psoralen adducts as potential tools to sensitize selected genomic sites to modification (7, 59), although the mechanism of this sensitization has not been worked out.
Based on these results, we sought in the present work to test whether triple-helix formation itself could stimulate homologous recombination. Previous work examining targeted mutagenesis had revealed that high-affinity third-strand binding, alone, could induce mutations in a target gene via a repair-dependent pathway (63). In that work, a substantial portion of the targeted mutations were deletions. An analysis of the deletion end points, presented here, reveals a pattern consistent with a pathway of strand break induction and end joining at regions of microhomology, suggesting that triplex formation might be able to generate intermediate structures containing strand breaks prone to homologous recombination. Using an SV40-vector-based assay, we report here the induction of homologous recombination in mammalian cells mediated by triple-helix formation even in the absence of an associated psoralen adduct or any other covalent damage. We find that the third-strand-stimulated recombination is dependent on nucleotide excision repair (NER), as triplex-induced recombination was not detected in cells deficient in the NER damage recognition factor, XPA, but was observed in a derivative line expressing the XPA cDNA. In comparison, we found that the recombination induced by the psoralen-triplex lesions was reduced only about 50% in the absence of NER, suggesting a partial but not exclusive role for NER in processing the targeted psoralen adducts into recombinagenic intermediates. The induced recombination, however, was unaffected in cells deficient in mismatch repair, prompting reevaluation of our previous model of an extensive heteroduplex intermediate and leading to a new model based on end joining.
MATERIALS AND METHODS
Vectors.
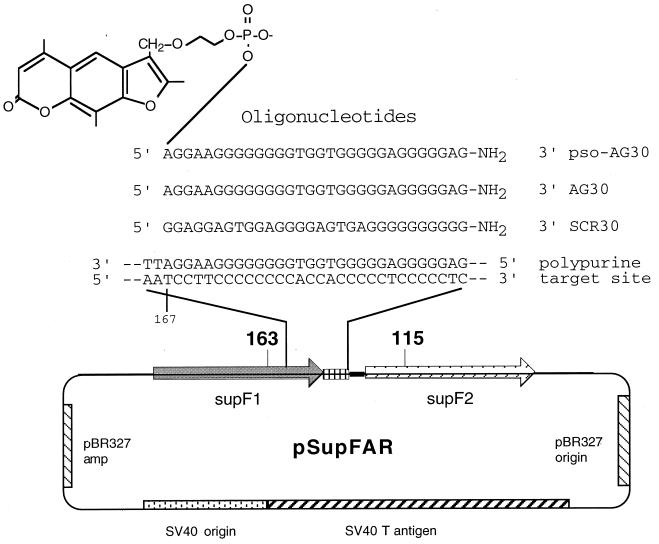
Plasmid pSupFAR was constructed to contain two mutant supF tRNA genes in tandem (Fig. 1). The two genes differ only at positions 115 and 163. This vector is similar to pSupF2 used in previous work of members of our group (18), except that the polymorphisms at positions 101 and 177 have been eliminated to simplify product analysis. The supF genes were derived from the genes in the vectors pSupFG1 (62) and pSP189 (42). pSupFG1 carries a novel supF gene modified to create a polypurine binding site for high-affinity triple-helix formation. The pSupFG1 derivative, pGW47, containing a C-to-G mutation at bp 163 within the supFG1 gene (yielding the gene designated here as supF1), was digested with EagI. A mutant variant of the supF gene, carrying a G-to-A mutation at nucleotide position 115 (designated supF2), was generated as a synthetic 104-bp oligonucleotide fragment and inserted into the EagI-digested pGW47 DNA to yield the pSupFAR shuttle vector containing the two mutant supF genes as direct repeats separated by 9 bp. The orientation of the cloned fragment was confirmed by DNA sequencing. The vector also contains the SV40 origin of replication and large T-antigen coding sequence along with the β-lactamase gene and replication origin from pBR327, as described previously (18).
FIG. 1.
Schematic representation of the pSupFAR vector. The SV40-based shuttle vector contains two mutant supF genes in the form of a tandem dimer. The upstream mutant supF gene, supF1, contains a C-to-G point mutation at nucleotide position 163; the downstream mutant supF gene, supF2, contains a G-to-A point mutation at nucleotide position 115. The two supF genes are separated by a 9-bp sequence that contains an EagI restriction site used for cloning. At the 3′ end of supF1 is an engineered polypurine sequence (bp 167 to 196), creating a high-affinity third-strand binding site. A purine-rich oligonucleotide with a length of 30 nucleotides (AG30) was designed to form a triple helix in the anti-parallel triplex motif at this site, as shown. As a control, SCR30, containing the same base composition but a scrambled sequence, was used. In some experiments, the AG30 and the SCR30 oligonucleotides were conjugated at their 5′ end to 4′-hydroxymethyl-4,5′,8-trimethylpsoralen via the 4′ hydroxymethyl position, as shown in the case of pso-AG30. In this case, by formation of the triple helix, psoralen intercalation is targeted to the duplex-triplex junction at bp 166 to 167 of supF1. Upon photoactivation with UVA, both monoadducts and interstrand cross-links are generated at the thymidines in these base pairs.
Oligonucleotides.
Unconjugated and psoralen-linked oligonucleotides were synthesized by the Keck Facility at Yale University, by standard phosphoramidite chemistry with materials from Glen Research (Sterling, Va.). All oligomers contained phosphodiester backbones and were synthesized to contain a 3′ propylamine group to minimize susceptibility to degradation by 3′ exonucleases (26). The oligomers were purified by either gel electrophoresis or high-pressure liquid chromatography, followed by Centricon-3 filtration in distilled water (Amicon, Beverly, Mass.). The psoralen was incorporated into the oligonucleotide synthesis as a psoralen phosphoramidite, resulting in an oligonucleotide linked at its 5′ end via a two-carbon linker arm to 4′-hydroxymethyl-4,5′,8-trimethylpsoralen. The oligonucleotides used in this study were pso-AG30 (5′ psoralen-AGGAAGGGGGGGGTGGTGGGGGAGGGGGAG-3′), AG30 (same as pso-AG30 but without 5′ psoralen), pso-SCR30 (5′ psoralen-GGAGGAGTGGAGGGGAGTGAGGGGGGGGGG-3′), and SCR30 (same as pso-SCR30 but without 5′ psoralen).
Cells.
Monkey COS-7 cells were obtained from the American Type Culture Collection (1651-CRL). Transformed XPA fibroblasts from patient XP2OS and the XP2OS cells transfected with a vector expressing XPA cDNA [XP2OS(pCAH19WS)] were obtained from K. Kraemer (34). XPF and XPG fibroblasts (from patients with xeroderma pigmentosum, groups F and G) were obtained from the National Institute of General Medical Science Human Genetic Mutant Cell Repository (Camden, N.J.), repository no. GM08437 and GM03021B, respectively. The XPF cells were obtained as an SV40-transformed fibroblast line; the XPG cells are a primary fibroblast culture obtained at passage 10. The human colon cancer cell line, HCT116, deficient in the DNA mismatch repair (MMR) factor MLH1, along with a subline corrected by whole-chromosome transfer to restore MMR function (HCT116.3-6, corrected with chromosome 3) and a control subline (HCT116.2-3, with chromosome 2), were obtained from T. Kunkel (28). A similar set of MSH2-deficient and corrected cell lines, HC (MSH2 deficient), HC.2-4 (corrected with chromosome 2), and HC.7-2 (control with chromosome 7), were also obtained from T. Kunkel (55). The cells were grown in growth media (Dulbecco's modified Eagle's medium [Life Technologies, Gaithersburg, Md.], 10% fetal calf serum [Life Technologies], and 1% penicillin–1% streptomycin (Sigma, St. Louis, Mo.)] at 37°C in a humidified incubator in the presence of 5% CO2.
Intracellular targeting and recombination assay.
In the case of the COS cells, subconfluent cells were detached by trypsinization and washed three times in growth medium. The cells were resuspended at a density of 107 cells/ml, and pSupFAR plasmid DNA was added at a concentration of 3 μg of DNA/106 cells. The cell-DNA mixture was then incubated on ice for 10 min. Transfection of the cells was carried out by electroporation with a gene pulser (Bio-Rad, Hercules, Calif.) at a setting of 25 μF/250 W/250 V in 0.4-cm gap transfection cuvettes (Bio-Rad). After 10 min of incubation at room temperature, cells were washed twice in growth medium to remove excess extracellular plasmid DNA and replated in culture in growth media into 100-mm-diameter dishes. After 24 h, cells were transfected with selected oligonucleotides by electroporation at a concentration of 2 μM in the cell suspension, as described above, and UVA irradiation, if indicated, was administered 2 h later. In the case of human cell lines (XPA, XPA corrected, XPF, and XPG), the shuttle vector DNA was transfected by using Lipofectamine (Life Technologies), with a ratio of 3 μg of plasmid DNA to 30 μl of Lipofectamine, premixed with 600 μl of Dulbecco's modified Eagle's medium, and was added to cells with 2.4 ml of medium in 60-mm-diameter dishes, as directed by the manufacturer. After 24 h, the oligonucleotides were transfected into the cells with Lipofectamine, with an oligonucleotide-to-lipid ratio of 6 μg to 60 μl in a 600-μl volume.
For all cell types, 48 h after transfection the cells were harvested for plasmid DNA isolation by a modified alkaline lysis procedure (62), and the resulting vector DNA was subjected to digestion with DpnI (to eliminate any unreplicated molecules that had not acquired the mammalian methylation pattern) and used to transform bacteria for genetic analysis of supF gene function as previously described (62). Selected colonies were purified, and the plasmids were isolated for PCR or DNA sequence analysis.
Product analysis.
Screening of the recombinants by PCR amplification of the sequences containing the supF gene(s) in the vectors was performed with the following primers: 5′ GGCGACACGGAAATGTTGAA 3′ (forward) and 5′ TTAGCTTTCGCTAAGGATCCGG 3′ (reverse). DNA sequencing was carried out as described previously (62). The sequencing primer was the same as the forward primer used for screening of the recombinants, listed above.
In vitro triplex binding, photoadduct formation, and cotransfection shuttle vector assay.
In a reaction volume of 10 μl, 3 μg (50 nM) of the pSupFAR DNA was incubated with a 100:1 molar excess of oligonucleotide (5 μM) for 2 h at 37°C in 10% sucrose, 20 mM MgCl2, 10 mM Tris (pH 8.0), and 1 mM spermidine (triplex binding buffer). If indicated, irradiation of samples with 1.8 J/cm2 of UVA was performed using 365-nm lamps supplied by Southern New England Ultraviolet Co. (Branford, Conn.). A radiometer (International Light, Newburyport, Mass.) was used to measure the lamp output (typical UVA irradiance of 5 to 7 mW/cm2 at 320 to 400 nm). A window glass filter was used to eliminate any UVB contamination during the UVA irradiation. Cotransfection of the preformed oligonucleotide-plasmid DNA complexes into the human repair-deficient and -proficient cell lines was carried out using Lipofectamine (Life Technologies), as described above. After 48 h, shuttle vector isolation and analysis was performed as described above.
RESULTS
Design of vector to detect induced recombination.
The vector pSupFAR was constructed to investigate the ability of triplex-directed DNA damage to trigger recombination between tandem supF genes flanking the third-strand binding site (Fig. 1). supF encodes an amber suppressor tyrosine tRNA whose function can be screened by a colorimetric assay in host bacteria carrying a lacZ nonsense mutation. To facilitate the detection of recombinants via a phenotypic screen, the upstream supF1 gene in the vector carried a C-to-G mutation at bp 163, whereas the downstream supF2 gene was engineered to contain a G-to-A mutation at position 115. Both of these changes disrupt supF activity. Hence, recombination between supF1 and supF2 has the potential to reconstruct a fully functional supF gene, which can be identified by the resulting blue colonies on indicator plates. (This vector differs slightly from the pSupF2 vector used in our previous study of psoralen-triplex-induced recombination (18) in that the sequence differences between the two supF genes at positions 101 and 177 have been eliminated, a situation which in the previous work reduced the yield of detectable wild-type recombinants).
The 30-bp polypurine-polypyrimidine site in the vector at the 3′ end of supF1 is a good target for triplex formation by the G-rich oligonucleotide, AG30 (Fig. 1). This oligomer was shown to bind to the target sequence with high affinity (equilibrium dissociation constant [Kd], 10−8 M) (63). In some of the experiments reported here, a derivative of this TFO was used; the derivative was synthesized to contain a psoralen conjugate at its 5′ end, yielding pso-AG30, such that the third-strand binding positions the tethered psoralen for intercalation and adduct formation at bp 166 to 167 of the supF1 gene (Fig. 1). The binding of the modified pso-AG30 to this site was determined to have a Kd of 3 × 10−9 M (62). The level of binding by AG30 and pso-AG30 was previously found to be sufficient for intracellular triplex formation and targeted mutagenesis in a shuttle vector carrying the supFG1 gene (62, 63). As a control, a scrambled sequence oligonucleotide, SCR30, was used. It is a G-rich 30-mer and has the same base composition as AG30, but it has 14 mismatches in the third-strand binding code relative to the polypurine site in the pSupFAR vector, whereas AG30 has only two mismatches. No binding of SCR30 to the supF target site is detectable in gel mobility shift assays up to a concentration of 10−6 M.
Triplex-induced recombination in COS cells.
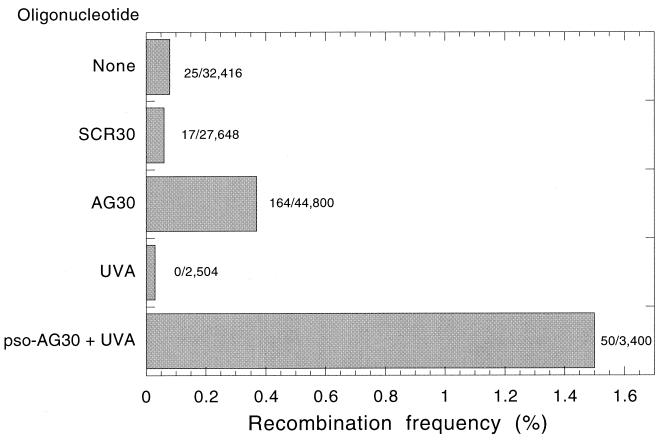
To test the possibility that intermolecular triple-helix formation, by itself, might promote recombination, we examined the activity of AG30 in the pSupFAR recombination assay. COS cells were transfected with pSupFAR and subsequently transfected with either AG30, pso-AG30, or, as a sequence control, SCR30. In the case of pso-AG30, 2 h after oligonucleotide transfection, the cells were irradiated with long wavelength UVA light (365 nm) to activate the tethered psoralen for photoadduct formation. The treated cells were maintained in culture for 48 h before rescue of the vector DNA for analysis.
Recombinant colonies were identified by their nonparental blue phenotype among the parental white ones, the colonies were counted, and the recombination frequency was determined (Fig. 2). Cells not exposed to any oligonucleotide yielded a background frequency of 0.08% blues. When cells were treated with pso-AG30 followed by UVA, blue colonies were generated at a frequency of 1.5%, consistent with the previous work of members of our group (18). When AG30, lacking any psoralen conjugate, was used, recombinants were generated at a frequency of 0.37%, fivefold above background. SCR30 yielded only background levels of recombinants (0.06%). Hence, triplex formation, itself, is able to provoke recombination, at a level in this assay about 25% of that induced by targeted psoralen-triplex lesions.
FIG. 2.
Triplex-induced recombination in COS cells. Cells pretransfected with the pSupFAR vector were subsequently transfected with the indicated oligonucleotides. After 48 h, shuttle vector DNA was isolated from the cells for analysis of supF gene function and quantification of recombination events. The number of blue colonies (representing recombinants) out of the total number of colonies is presented to the right of each bar, with the bars indicating recombination frequency. One sample received only irradiation with UVA light (1.8 J/cm2 of broad band long wavelength UV light centered at 365 nm) and no oligonucleotide, as indicated. In the case of pso-AG30 plus UVA, the irradiation was administered 2 h after TFO transfection.
Analysis of nonparental products.
To ascertain the nature of the oligomer-induced events, we used PCR analysis to determine the supF gene copy number in the nonparental plasmids. Plasmid DNA from selected blue colonies, along with DNA from parental white ones for comparison, was amplified with primers flanking the tandem supF genes in pSupFAR. Analysis of the blue colonies produced by both AG30 and pso-AG30 revealed that all (n = 22 and 12, respectively) were recombinants in which only a single, wild-type supF gene was present (data not shown; this interpretation was confirmed by DNA sequencing of 10 randomly selected recombinant vectors), whereas the white colonies contained the parental plasmid with two supF genes. Hence, all the blue recombinants tested appear to have arisen from nonconservative events involving the loss of sequences in which a functional supF gene was generated.
Repair dependence of triplex-induced recombination.
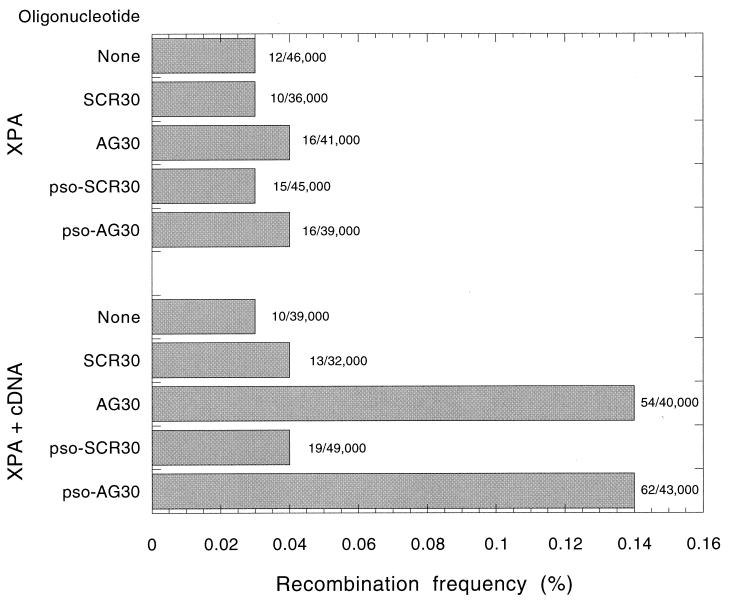
To address the mechanism by which an intermolecular triple helix could promote recombination, we carried out the shuttle vector recombination assay in a repair-deficient human cell line (XP2OS) derived from a patient with xeroderma pigmentosum, group A (34). The XPA protein is a critical damage recognition factor in the NER pathway (49), and in previous work of members of our group, cells without XPA function were found to lack TFO-induced mutagenesis (63). In comparison, an otherwise isogenic subclone corrected to normal NER function by transfection with a vector expressing the wild-type XPA cDNA was used (34). In these experiments, the cells were transfected with pSupFAR on day 1 and with the oligomers on day 2, with cationic lipids being used both days. (The transfection protocol differed from that used for COS cells because of the poor survival of the XPA cells after electroporation). The oligonucleotides tested were AG30, pso-AG30, and SCR30. However, UVA irradiation was not performed, so in this experiment the comparison between AG30 and pso-AG30 reflects the possible effect of the psoralen conjugate only as an intercalator, not as a covalent adduct or interstrand cross-link.
The data (Fig. 3) reveal that AG30 and pso-AG30 (without UVA and hence without cross-linking) were effective in stimulating recombination in pSupFAR, both at frequencies of 0.14%, but only in the XPA cells that had been corrected to normal or near normal NER activity by expression of the XPA cDNA. In the mutant XPA cell line, no TFO-induced recombination was detected above the background (0.03%). Hence, the results show that the ability of an intermolecular triple helix to stimulate recombination requires an intact NER pathway, presumably to generate a recombinagenic intermediate. Interestingly, pso-AG30 without UVA (no covalent adducts) was not more effective than AG30, suggesting that the potential intercalation at the duplex-triplex junction by psoralen does not have a major influence on this process, and that the primary lesion that induces recombination is the triple helix itself. As in the COS cell experiments, all the analyzed blue colonies produced by AG30 were single gene recombinants (n = 14).
FIG. 3.
Triplex-induced recombination in human repair-proficient and repair-deficient cells. The cells, either XPA (XP20S, deficient in the damage recognition factor XPA) or XPA corrected [XP20S(pCAH19WS) cells, a subline of XP2OS expressing wild-type XPA cDNA], were pretransfected with the pSupFAR vector and subsequently transfected with the indicated oligonucleotides. After 48 h, shuttle vector DNA was recovered and analyzed as described for Fig. 2.
Note that the TFO-induced recombination frequencies in the XPA-corrected cells are slightly lower than that seen with AG30 in the COS cells (Fig. 2). This difference could have several explanations, as follows: (i) the NER correction may be incomplete; (ii) the COS cells have a more robust recombination activity; or (iii) the transfection of the TFOs into the COS cells was more effective. None of these would invalidate the basic observations and conclusions.
Partial NER dependence of psoralen-triplex-induced recombination.
For comparison, we examined the role of the NER pathway in the recombination induced by psoralen-triplex lesions. In these experiments, the triple helices were formed in vitro on the vector DNA and the complexes were UVA irradiated to generate covalent psoralen adducts prior to transfection into the cells. This protocol was carried out to avoid any potential confounding variability in cellular oligonucleotide uptake and intracellular triplex formation that might arise because two unrelated, nonisogenic cell lines were included in the comparison. In this way, all the cell lines were challenged with a uniform DNA substrate. Also, this protocol avoids any possible toxicity to the repair-deficient cells from the UVA.
In prior studies (20, 61, 62), we demonstrated that under the conditions used here, the TFO-tethered psoralen covalently reacts with both strands of the duplex (via photoaddition at both the pyrone and furan rings to T's in opposite strands) to generate interstrand cross-links. In this structure, all three strands of the triplex are covalently linked via the psoralen. The TFO is connected through the linker arm attached to the 5′ end of the TFO and to the 4′-hydroxymethyl group of the psoralen. The duplex strands are linked via cyclobutane bonds between thymidines and the pyrone and furan rings of the psoralen.
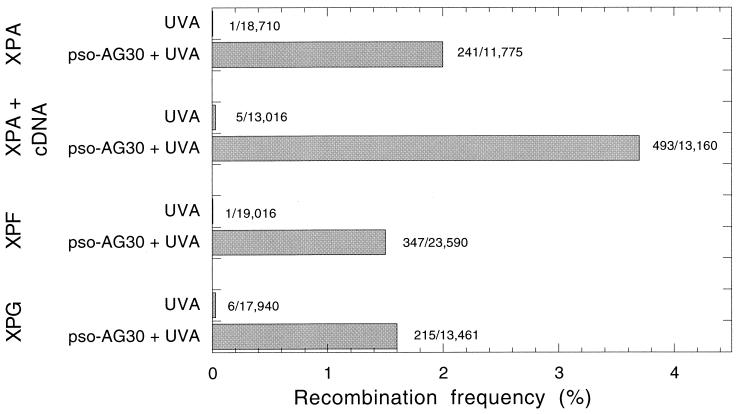
The preformed pSupFAR-triplex-psoralen adduct complexes were transfected into a series of repair-deficient and -proficient cells, including not only the XPA cells and those of their corrected subclone, but also those of cell lines derived from xeroderma pigmentosum patients in complementation groups F and G. (The XPA and XPF cell lines were SV40 transformed; the XPG cells were a primary fibroblast culture at passage 10.) XPF and XPG proteins play central roles in the endonuclease incision steps in NER, at positions 5′ and 3′ to the lesion, respectively (49). The data (Fig. 4) show that the covalent psoralen-triplex lesions stimulate recombination to a substantial degree even in the three NER-deficient cell types, XPA (2.0%), XPF (1.5%), and XPG (1.6%). However, a higher overall frequency of induced recombination was observed in the corrected XPA cells (3.7%). These results suggest that some but not all of the recombination induced by the psoralen-triplex compound lesion depends on NER. This conclusion is consistent with the emerging concept that non-NER pathways exist in cells that can metabolize psoralen cross-links. Furthermore, the observation that, as in the COS cells (Fig. 2), the covalent psoralen-triplex lesions (Fig. 4) are more effective than the triplex alone (Fig. 3) in stimulating recombination is also in keeping with the possibility that the psoralen cross-links are subject to metabolism by repair pathways in addition to NER.
FIG. 4.
Recombination induced by triplex-directed psoralen adducts in human repair-proficient and repair-deficient cells. Cells included either XPA (XP2OS, deficient in the NER damage recognition factor XPA) or XPA corrected [XP2OS(pCAH19WS) cells, a subline of XP2OS expressing wild-type XPA cDNA], along with cells from patients with xeroderma pigmentosum groups F and G, both of which are deficient in specific endonuclease activities required for NER. As indicated, the pSupFAR vector DNA was incubated in vitro with pso-AG30, followed by UVA irradiation to generate triplex-directed, psoralen photoadducts. The vector-triplex-psoralen adduct complexes were transfected into the indicated cells. Control samples included vector DNA irradiated with UVA in the absence of any oligonucleotide and then transfected directly into the various cell lines, as indicated. After 48 h, shuttle vector DNA was recovered and analyzed for recombination as described for Fig. 2.
Triplex-induced recombination is mismatch repair independent.
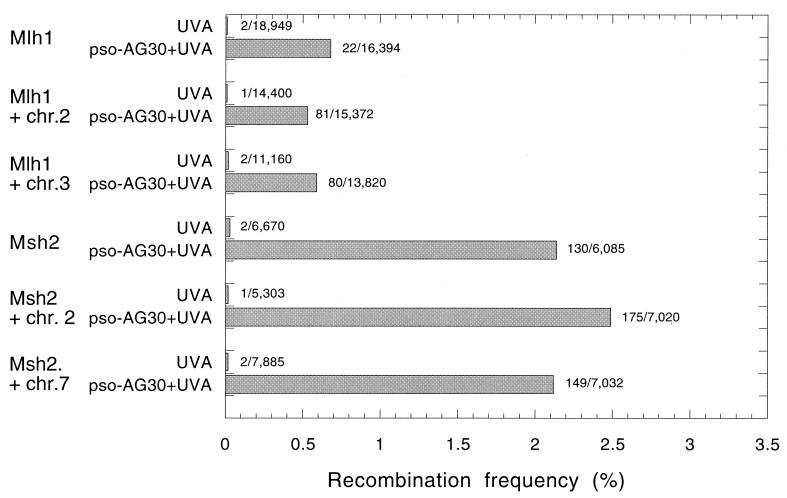
To further extend our mechanistic understanding of the TFO-induced recombination, we tested the ability of pso-AG30 plus UVA to promote recombination in cells deficient in DNA MMR. Two human cancer-derived cell lines, HCT116 and HC, defective in MLH1 and MSH2, respectively were tested in comparison to subclones corrected to MMR proficiency by chromosome transfer, along with control sublines carrying noncorrecting chromosomes (28, 55). No differences in the yields of recombinants were detected whether or not the cells were MMR proficient or deficient (Fig. 5). (A difference was seen between the level of recombination in the HCT116-derived cells versus the HC cells. However, these differences do not correlate with MMR capacity. They may reflect differences in other, as yet unidentified repair or recombination factors, but this remains to be determined.) In these experiments, analysis of the induced blue colonies revealed single gene recombinants regardless of whether they were produced in MMR mutant or wild-type cells (data not shown).
FIG. 5.
Recombination induced by triplex-directed psoralen adducts in human DNA MMR-deficient and -proficient cells. Cells included the human colon cancer cell line, HCT116, deficient in the MMR factor MLH1, along with a subline corrected by whole-chromosome [chr.] transfer to restore MMR function (HCT116.3-6, corrected with chromosome 3) and a control sub-line (HCT116.2-3, with chromosome 2). A similar set of MSH2-deficient and corrected cell lines, HC (MSH2 deficient), HC.2-4 (corrected with chromosome 2), and HC.7-2 (control with chromosome 7) were also compared. The pSupFAR vector DNA was incubated in vitro with pso-AG30, followed by UVA irradiation to generate triplex-directed, psoralen photoadducts. The vector-triplex-psoralen adduct complexes were transfected into the indicated cells. Control samples included vector DNA irradiated with UVA in the absence of any oligonucleotide and then transfected directly into the various cell lines, as indicated. After 48 h, shuttle vector DNA was recovered and analyzed for recombination as described for Fig. 2.
These results call into question a previous model invoking a heteroduplex intermediate that is resolved by mismatch repair (18). The possibility remains that such an intermediate is, in fact, formed but is resolved by a mismatch repair pathway that is MSH2 and MLH1 independent. However, another possibility is that the triplexes promote recombination via a pathway in which strand breaks are produced (either by NER in the case of the triplexes alone or NER plus some other repair activity in the case of the psoralen-triplex lesion), and that these strand breaks lead to end-joining events at regions of microhomology.
Triplex-induced deletion mutations.
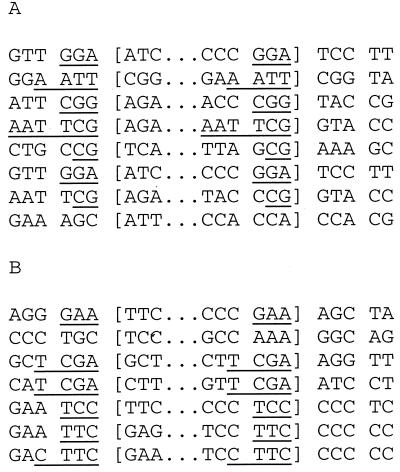
If such a pathway of strand break production and end joining is provoked by triplex formation, we reasoned that the triplex-induced deletions found in our previous work (62, 63) might also provide evidence consistent with an end-joining mechanism. In our previous COS cell experiments, such deletions typically constituted about 15 to 25% of the observed mutations, yielding deletion frequencies in the range of 0.1 to 0.5% (for triplex alone and for covalent psoralen-triplexes, respectively), values which are comparable to the frequencies of induced recombinants seen in the work reported here (compare the COS cell data in Fig. 2). In Fig. 6, an analysis of randomly selected deletion mutations generated by psoralen-triplexes (Fig. 6A) and by intermolecular triple helices alone (Fig. 6B) is shown. In both data sets, most of the deletion end points appear to have been joined at sites of 3 to 4 bp of homology, in keeping with an end-joining model. However, unlike the supF recombinants in the experiments described above, the joints in the deletions otherwise occur within surrounding regions of nonhomology. Also, there is one example in each set that has no homology around the deletion. These products are consistent with previous analyses of joints made in mammalian cells with ends produced intentionally by restriction enzymes (46).
FIG. 6.
Sequence analysis of deletion mutations generated in the supF gene in COS cells by triplex-associated psoralen adducts (A) and by triple helix formation alone (B). The deleted sequences are indicated within the brackets. Underlined nucleotides indicate stretches of microhomology at the deletion end points.
DISCUSSION
The work presented here demonstrates that intermolecular triple-helix formation can provoke a pathway of DNA metabolism that can lead to recombination. Cells pretransfected with an SV40-based shuttle vector carrying two mutant copies of the supF gene were subsequently transfected with specific TFOs, and it was found that such oligomers could provoke recombination within the vector in a sequence-dependent manner. The reaction was sequence specific, because only the AG30 or pso-AG30 oligonucleotides, which bind with high affinity to the target site, were found to generate recombinant products in the cells. The scrambled sequence oligomer, SCR30, was not active in the assay.
Experiments with NER-deficient cells lacking XPA function and with a corrected, otherwise isogenic subline reveal that NER is required for production of the recombinants by the triple helices. This finding is consistent with our previous observation that high-affinity third-strand binding can stimulate DNA repair activity in human cell extracts and can induce mutations in a target gene in human cells in an NER-dependent manner (63). The combined results provide strong support for the hypothesis that the formation of an intermolecular triple helix by an oligonucleotide is recognized by the NER pathway as a form of DNA damage, putting triplexes in a broad category of NER-processed lesions with pyrimidine dimers and bulky adducts such as those from acetyl-aminofluorene and benzo[a]pyrene. It is likely that further studies will reveal common structural features shared among triplexes and the other types of DNA damage that promote NER recognition. Furthermore, by extension from these results, it is expected that other classes of noncovalent DNA binding ligands, such as polyamides (65) and peptide nucleic acids (15, 16), may create alterations of DNA structure that will be recognized and metabolized by NER. As a result, TFOs, polyamides, peptide nucleic acids, and similar ligands may prove to be useful reagents in strategies aimed at harnessing the cell's repair mechanisms to promote genome modification. However, the particular properties of these various ligands in this regard remain to be determined.
The necessity for NER in the triplex-induced recombination also supports our previous study of gene correction with TFOs covalently linked to short “donor” fragments of DNA (8). In that work, the site-specific binding of the TFO domain of the hybrid molecule was designed to enhance the homology search and thereby accelerate recombination or gene conversion. The results reported here further support the notion that the TFO domain can also promote recombination of the donor fragment with the target site by recruiting the NER apparatus to the target to create strand breaks and recombinagenic intermediates.
The role of the NER pathway in recombination stimulated by triplex-directed psoralen cross-links was also examined. In the case of the triplex-psoralen lesion, however, there was only a partial requirement for NER. About 40 to 50% of the induced recombination was found to be independent of NER function. This observation is in keeping with a study by Hall and Scherer (21) in which reactivation of psoralen and UVA-treated herpes simplex virus was seen in XPA-deficient cells when the cells were infected at high multiplicity, suggesting that a recombinational repair pathway was still active in these cells even in the absence of XPA function. This observation is also consistent with recent studies providing further evidence for non-NER repair of psoralen cross-links in mammalian cells. Li et al. found that psoralen cross-links but not monoadducts in one plasmid can stimulate repair synthesis in a second, undamaged plasmid in a human whole-cell extract (35). This activity was found to be separate from NER, as it was observed in extracts of several NER-deficient cell lines (XPA-, XPC-, and XPG-deficient cells).
Interestingly, the activity observed by Li et al. (35), while present in extracts from XPA cells, was absent from XPF cell extracts. This finding fits with earlier observations of an extreme hypersensitivity of XPF (ERCC4) mutant CHO cells to cross-linking agents (4, 25). Evidence suggests that, of the known NER factors, the XPF-ERCC1 heterodimer complex appears to have a special role in cross-link repair (4). Yet, our results suggest that even in XPF-deficient cells, pso-TFO-mediated cross-links can promote recombination, raising the additional possibility that either (i) another factor or set of factors may act on pso-TFO lesions in a recombinational repair pathway or (ii) the ability of pso-TFO-directed cross-links to block replication (rather than provoke repair) may play a role in stimulating recombination.
In other work looking more directly at cross-link repair in vitro, Reardon et al. showed that psoralen monoadducts and cross-links can be removed from a DNA substrate in human cell extracts (45). However, Bessho et al. (2) reported that purified NER factors reconstituted in vitro that can otherwise carry out excision repair of pyrimidine dimers cannot excise psoralen cross-links. In fact, they observed the production of dual incisions on one strand on the same side of the lesion rather than the expected flanking incisions (2), representing essentially a situation of incomplete repair. While such a strand break product may constitute an intermediate potentially prone to recombinational repair, this observation suggests that the known NER factors are insufficient for full psoralen cross-link repair in mammalian cells. Interestingly, the situation in Escherichia coli appears to be different, as NER factors do make dual incisions flanking the psoralen cross-link and are required for mediating cross-link repair, in a model elaborated by the work of Cole (11), Sladek et al. (53), and others (9, 56, 57).
Such subtle but important differences in the way psoralen lesions are repaired may help to explain why triplex-directed psoralen adducts were relatively ineffective at stimulating recombination in Xenopus oocytes compared with human or monkey cells (52). In the Xenopus system, the dual supF-carrying plasmid containing triplex-psoralen adducts showed little recombination (52), while similar substrates containing restriction enzyme-induced double-strand breaks recombined at high efficiency (51).
Experiments with MMR-deficient cells provided the unexpected result that MMR activity, at least the activity dependent on MSH2 and MLH1, is not required for the induced recombination in the pSupFAR shuttle vector. This result calls into question some aspects of our previous working model (18), in which we proposed that the psoralen-triplex-induced recombination takes place via a single-strand annealing pathway (18, 36). In such a pathway, damage-induced strand breaks in the substrate DNA would be followed by exonuclease activity to expose mostly homologous single-stranded regions that can anneal to form heteroduplexes. We had proposed the creation of an extensive heteroduplex intermediate, requiring resolution via MMR to generate the observed recombination products.
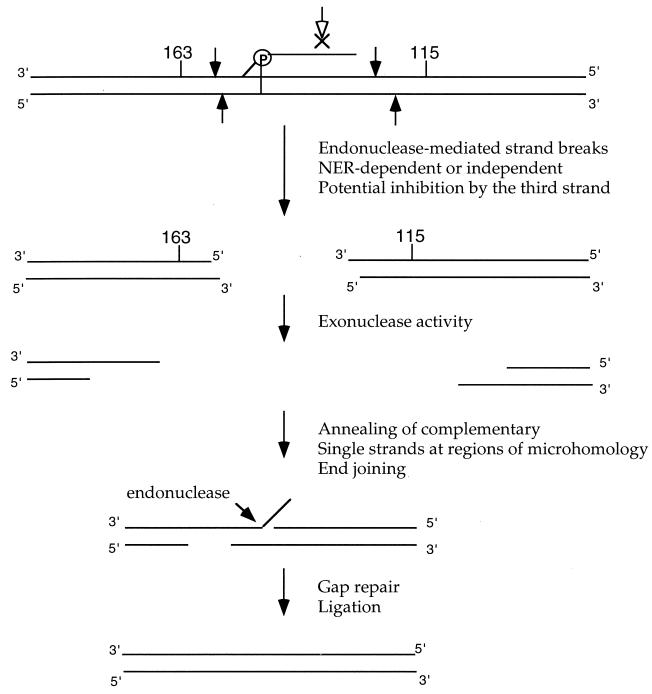
The lack of an effect of MSH2 and MLH1 in our assay is subject to several interpretations, as follows. (i) A heteroduplex intermediate is formed but is metabolized by an MSH2- and MLH1-independent pathway. The formation of such heteroduplexes in extrachromosomal recombination substrates carrying duplicated genes has been demonstrated (5, 13, 32, 40), so we cannot rule out this possible explanation. (ii) The triplex-targeted damage is metabolized through an end-joining pathway in which double-strand breaks are produced, but followed first by a more limited exonuclease activity and then by annealing of the ends at relatively short segments of homology. Such a pathway, diagrammed in Fig. 7, could eliminate the mutations at positions 163 and 115 and construct a functional supF gene without necessitating the production of a heteroduplex. A similar mechanism could account for the joints formed in the deletion mutations shown in Fig. 5, as these appear to have occurred by joining at short (3 to 4 bp) stretches of homology. In this model, however, a distinction should be made between the deletion and the recombination events. The deletions are proposed to arise from triplex-induced strand breaks that undergo illegitimate recombination and end joining at regions of microhomology within otherwise heterologous sequences. The recombination products, in contrast, because they are identified by reconstruction of a functional supF gene from two tandem copies of the gene, occur by end joining within large regions of homology. Nonetheless, it is possible that, in the pSupFAR recombination assay, some of the induced breaks are joined, like the deletions, at microhomologies within heterologous regions, but these products would not be detected in the screen for wild-type supF function. (iii) The strand breaks are metabolized to expose single-stranded regions which are capable of strand invasion into homologous sites within another plasmid vector in the cells. The resulting structures could serve as primers for DNA synthesis, promoting further strand displacement and leading to strand exchange, crossing over and recombination. Such a mechanism of break-induced replication was proposed by Li et al. (35) to account for the observation that psoralen cross-links in one plasmid could stimulate repair synthesis in a coincubated undamaged plasmid in human cell extracts. Distinguishing between these pathways will require further studies with defined substrates, with the caveat that the episomal target vector described here may be metabolized by a different set of pathways than would a chromosomal substrate.
FIG. 7.
Model for intramolecular recombination induced by triplex-directed DNA damage. The model illustrates a proposed end-joining pathway in which the third strand-directed psoralen interstrand cross-link is processed into strand breaks, either by NER or by an NER-independent mechanism. (The potential ability of a triple helix to block repair endonuclease activity is also indicated as a complicating factor [61].) The resulting ends are subject to exonuclease digestion, eliminating the mutant sequences and exposing regions of homology capable of joint formation to reconstruct a functional supF gene.
Another caveat is that the current work was performed in the presence of SV40 T antigen. T antigen has helicase activity that can unwind triplexes, but such activity was found to require a 3′ single-stranded tail (29), which is not present in our triplex substrates. T antigen can also bind and inactivate p53 and so may influence DNA repair (19). However, we have carried out preliminary experiments using a modified shuttle vector in which the T-antigen gene was substituted with a version mutated within the p53 binding domain (30). Essentially similar results were obtained in the recombination assay in both p53 wild-type and mutant human cells (23) (data not shown), suggesting that the p53 status of the cells does not have a major influence on the pso-TFO-induced recombination in this assay. In addition, it should be noted that the SV40 genome is rapidly chromatinized upon transfection into primate cells (6), so the shuttle vector target approximates the nucleosomal structure of a chromosomal site.
The ability of triple-helix formation, without any other associated DNA damage, to promote recombination reported here provides a novel tool with potential utility for genome modification. Although these experiments were carried out in a model system, with a nonnaturally occurring target sequence, the results should be applicable to any third strand that can bind with high affinity to a genomic site. Recent works demonstrating triplex-mediated targeted mutagenesis of chromosomal loci in mammalian cells support the feasibility of this approach (38, 58). Further, since a number of nucleotide analogs that can significantly enhance triplex formation under physiologic conditions have recently been described (31), this may turn out to be a realistic strategy for genome modification at a variety of sites. More generally, this work raises the possibility that other sequence-specific DNA binding ligands capable of triggering repair may have utility in the genetic manipulation of mammalian cells.
ACKNOWLEDGMENTS
We thank L. Narayanan, P. Chan, L. Lacroix, K. Vasquez, M. Lin, D. Segal, F. P. Gasparro, L. Cabral, R. Franklin, and S. J. Baserga for their assistance. We also thank K. Kraemer and T. Kunkel for providing cell lines.
This work was supported by the Leukemia Society of America and the NIH (GM54731). A.F.F. was supported in part by the Anna Fuller Foundation.
REFERENCES
- 1.Beal P A, Dervan P B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 2.Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol Cell Biol. 1997;17:6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenneman M, Gimble F S, Wilson J H. Stimulation of intrachromosomal homologous recombination in human cells by electroporation with site-specific endonucleases. Proc Natl Acad Sci USA. 1996;93:3608–3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brookman K W, Lamerdin J E, Thelen M P, Hwang M, Reardon J T, Sancar A, Zhou Z Q, Walter C A, Parris C N, Thompson L H. ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol Cell Biol. 1996;16:6553–6562. doi: 10.1128/mcb.16.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll D, Lehman C W, Jeong-Yu S, Dohrmann P, Dawson R J, Trautman J K. Distribution of exchanges upon homologous recombination of exogenous DNA in Xenopus laevis oocytes. Genetics. 1994;138:445–457. doi: 10.1093/genetics/138.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cereghini S, Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984;3:1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan P P, Glazer P M. Triplex DNA: fundamentals, advances, and potential applications for gene therapy. J Mol Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- 8.Chan P P, Lin M, Faruqi A F, Powell J, Seidman M M, Glazer P M. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J Biol Chem. 1999;274:11541–11548. doi: 10.1074/jbc.274.17.11541. [DOI] [PubMed] [Google Scholar]
- 9.Cheng S, Van Houten B, Gamper H B, Sancar A, Hearst J E. Use of psoralen-modified oligonucleotides to trap three-stranded RecA-DNA complexes and repair of these cross-linked complexes by ABC excinuclease. J Biol Chem. 1988;263:15110–15117. [PubMed] [Google Scholar]
- 10.Choulika A, Perrin A, Dujon B, Nicolas J-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole R S. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc Natl Acad Sci USA. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooney M, Czernuszewicz G, Postel E H, Flint S J, Hogan M E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 13.Deng W P, Nickoloff J A. Mismatch repair of heteroduplex DNA intermediates of extrachromosomal recombination in mammalian cells. Mol Cell Biol. 1994;14:400–406. doi: 10.1128/mcb.14.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Norden B, Nielsen P E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 16.Faruqi A F, Egholm M, Glazer P M. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc Natl Acad Sci USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruqi A F, Krawczyk S H, Matteucci M D, Glazer P M. Potassium-resistant triple helix formation and improved intracellular gene targeting by oligodeoxyribonucleotides containing 7-deazaxanthine. Nucleic Acids Res. 1997;25:633–640. doi: 10.1093/nar/25.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruqi A F, Seidman M M, Segal D J, Carroll D, Glazer P M. Recombination induced by triple helix-targeted DNA damage in mammalian cells. Mol Cell Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford J M, Hanawalt P C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 20.Gasparro F P, Havre P A, Olack G A, Gunther E J, Glazer P M. Site-specific targeting of psoralen photoadducts with a triple helix-forming oligonucleotide: characterization of psoralen monoadduct and crosslink formation. Nucleic Acids Res. 1994;22:2845–2852. doi: 10.1093/nar/22.14.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall J D, Scherer K. Repair of psoralen-treated DNA by genetic recombination in human cells infected with herpes simplex virus. Cancer Res. 1981;41:5033–5038. [PubMed] [Google Scholar]
- 22.Hanson K D, Sedivy J M. Analysis of biological selections for high-efficiency gene targeting. Mol Cell Biol. 1995;15:45–51. doi: 10.1128/mcb.15.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havre P A, Yuan J, Hedrick L, Cho K R, Glazer P M. p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer Res. 1995;55:4420–4424. [PubMed] [Google Scholar]
- 24.Helene C. The anti-gene strategy: control of gene expression by triplex-forming-oligonucleotides. Anti-Cancer Drug Design. 1991;6:569–584. [PubMed] [Google Scholar]
- 25.Hoy C A, Salazar E P, Thompson L H. Rapid detection of DNA-damaging agents using repair-deficient CHO cells. Mutat Res. 1984;130:321–332. doi: 10.1016/0165-1161(84)90018-9. [DOI] [PubMed] [Google Scholar]
- 26.Ing N H, Beekman J M, Kessler D J, Murphy M, Jayaraman K, Zendegui J G, Hogan M E, O'Malley B W, Tsai M J. In vivo transcription of a progesterone-responsive gene is specifically inhibited by a triplex-forming oligonucleotide. Nucleic Acids Res. 1993;21:2789–2796. doi: 10.1093/nar/21.12.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 28.Koi M, Umar A, Chauhan D P, Cherian S P, Carethers J M, Kunkel T A, Boland C R. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 29.Kopel V, Pozner A, Baran N, Manor H. Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res. 1996;24:330–335. doi: 10.1093/nar/24.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus E, Strong L C, Tainsky M A. pZ402, an improved SV40-based shuttle vector containing a T-antigen mutant unable to interact with wild-type p53. Gene. 1998;211:229–234. doi: 10.1016/s0378-1119(98)00121-8. [DOI] [PubMed] [Google Scholar]
- 31.Lacroix L, Lacoste J, Reddoch J F, Mergny J L, Levy D D, Seidman M M, Matteucci M D, Glazer P M. Triplex formation by oligonucleotides containing 5-(1-propynyl)-2′-deoxyuridine: decreased magnesium dependence and improved intracellular gene targeting. Biochemistry. 1999;38:1893–1901. doi: 10.1021/bi982290q. [DOI] [PubMed] [Google Scholar]
- 32.Lehman C W, Jeong-Yu S, Trautman J K, Carroll D. Repair of heteroduplex DNA in Xenopus laevis oocytes. Genetics. 1994;138:459–470. doi: 10.1093/genetics/138.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letai A G, Palladino M A, Fromm E, Rizzo V, Fresco J R. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988;27:9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- 34.Levy D D, Saijo M, Tanaka K, Kraemer K H. Expression of a transfected DNA repair gene (XPA) in xeroderma pigmentosum group A cells restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis. 1995;16:1557–1563. doi: 10.1093/carcin/16.7.1557. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Peterson C A, Lu X, Wei P, Legerski R J. Interstrand cross-links induce DNA synthesis in damaged and undamaged plasmids in mammalian cell extracts. Mol Cell Biol. 1999;19:5619–5630. doi: 10.1128/mcb.19.8.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin F L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin P F, Bardwell E, Howard-Flanders P. Initiation of genetic exchanges in lambda phage-prophage crosses. Proc Natl Acad Sci USA. 1977;74:291–295. doi: 10.1073/pnas.74.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majumdar A, Khorlin A, Dyatkina N, Lin F-L M, Powell J, Liu J, Fei Z, Khripine Y, Watanabe K A, George J, Glazer P M, Seidman M M. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 39.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 40.Maryon E, Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991;11:3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moser H E, Dervan P B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 42.Parris C N, Seidman M M. A signature element distinguishes sibling and independent mutations in a shuttle vector plasmid. Gene. 1992;117:1–5. doi: 10.1016/0378-1119(92)90482-5. [DOI] [PubMed] [Google Scholar]
- 43.Praseuth D, Perrouault L, Le Doan T, Chassignol M, Thuong N, Helene C. Sequence-specific binding and photocrosslinking of alpha and beta oligodeoxynucleotides to the major groove of DNA via triple-helix formation. Proc Natl Acad Sci USA. 1988;85:1349–1353. doi: 10.1073/pnas.85.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reardon J T, Spielmann P, Huang J C, Sastry S, Sancar A, Hearst J E. Removal of psoralen monoadducts and crosslinks by human cell free extracts. Nucleic Acids Res. 1991;19:4623–4629. doi: 10.1093/nar/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth D B, Wilson J H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saffran W A, Cantor C R, Smith E D, Magdi M. Psoralen damage-induced plasmid recombination in Saccharomyces cerevisiae: dependence on RAD1 and RAD52. Mutat Res. 1992;274:1–9. doi: 10.1016/0921-8777(92)90038-5. [DOI] [PubMed] [Google Scholar]
- 49.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 50.Sandor Z, Bredberg A. Triple helix directed psoralen adducts induce a low frequency of recombination in an SV-40 shuttle vector. Biochim Biophys Acta. 1995;1263:235–240. doi: 10.1016/0167-4781(95)00109-t. [DOI] [PubMed] [Google Scholar]
- 51.Segal D J, Carroll D. Endonuclease-induced, targeted homologous extrachromosomal recombination in Xenopus oocytes. Proc Natl Acad Sci USA. 1995;92:806–810. doi: 10.1073/pnas.92.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal D J, Faruqi A F, Glazer P M, Carroll D. Processing of targeted psoralen cross-links in Xenopus oocytes. Mol Cell Biol. 1997;17:6645–6652. doi: 10.1128/mcb.17.11.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sladek F M, Munn M M, Rupp W D, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J Biol Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- 54.Tsujimura T, Maher V M, Godwin A R, Liskay R M, McCormick J J. Frequency of intrachromosomal homologous recombination induced by UV radiation in normally repairing and excision repair-deficient human cells. Proc Natl Acad Sci USA. 1990;87:1566–1570. doi: 10.1073/pnas.87.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 56.Van Houten B, Gamper H, Hearst J E, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J Biol Chem. 1988;263:16553–16560. [PubMed] [Google Scholar]
- 57.Van Houten B, Gamper H, Holbrook S R, Hearst J E, Sancar A. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc Natl Acad Sci USA. 1986;83:8077–8081. doi: 10.1073/pnas.83.21.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vasquez K M, Wang G, Havre P A, Glazer P M. Chromosomal mutations induced by triplex-forming oligonucleotides in mammalian cells. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasquez K M, Wilson J H. Triplex-directed modification of genes and gene activity. Trends Biochem Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 60.Vos J M, Hanawalt P C. DNA interstrand cross-links promote chromosomal integration of a selected gene in human cells. Mol Cell Biol. 1989;9:2897–2905. doi: 10.1128/mcb.9.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G, Glazer P M. Altered repair of targeted psoralen photoadducts in the context of an oligonucleotide-mediated triple helix. J Biol Chem. 1995;270:22595–22601. doi: 10.1074/jbc.270.38.22595. [DOI] [PubMed] [Google Scholar]
- 62.Wang G, Levy D D, Seidman M M, Glazer P M. Targeted mutagenesis in mammalian cells mediated by intracellular triple-helix formation. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang G, Seidman M M, Glazer P M. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y Y, Maher V M, Liskay R M, McCormick J J. Carcinogens can induce homologous recombination between duplicated chromosomal sequences in mouse L cells. Mol Cell Biol. 1988;8:196–202. doi: 10.1128/mcb.8.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White S, Szewczyk J W, Turner J M, Baird E E, Dervan P B. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]