Abstract
Understanding the tissue-specific genetic controls of protein levels is essential to uncover mechanisms of post-transcriptional gene regulation. We generated a genomic atlas of protein levels in three tissues relevant to neurological disorders (brain, cerebrospinal fluid (CSF), and plasma), by profiling thousands of proteins from participants with and without Alzheimer disease (AD). We identified 274, 127, and 32 protein quantitative trait loci (pQTLs) for CSF, plasma, and brain, respectively. Cis-pQTLs were more likely to be tissue-shared, but trans-pQTLs tended to be tissue-specific. Between 48.0 to 76.6% of pQTLs did not colocalize with expression, splicing, DNA-methylation, or histone-acetylation QTLs. Using Mendelian randomization (MR), we nominated proteins implicated in neurological diseases, including AD, Parkinson’s disease or stroke. This first multi-tissue study will be instrumental to map signals from genome-wide association studies (GWAS) onto functional genes, to discover pathways, and to identify drug targets for neurological diseases.
Introduction
Genetic studies have been successful in identifying genetic regions associated with complex traits, including diabetes, cardiovascular disease, and neurodegenerative diseases among others1–4. However, the studies have fallen short in promoting understanding of the biological mechanisms underlying those traits. Most GWAS identify multiple disease loci rather than functional variants or genes, which makes it difficult to biologically interpret association results and to identify novel biomarkers and drug targets. By leveraging gene-expression and genetic data generated by multiple studies5,6, including the Genotype-Tissue Expression (GTEx) project, it has been possible to identify the functional variants or genes driving some GWAS signals. GTEx and others5,7–9 have shown that there are more tissue-specific expression QTLs (eQTLs) in trans than in cis, and that to identify disease relevant functional genes, it is important to interrogate the tissue of interest for the specific trait in question.
However, eQTL mapping has not been able to fully identify the functional variants and genes driving GWAS signals. One explanation is that many genetic variants alter protein levels without affecting transcript levels10. Several published studies analyzed the genetic architecture of protein levels, but most are focused on a single tissue, mainly plasma10–13. A few studies with smaller sample sizes investigate CSF14,15 or brain tissue16. These studies suggest that a sizeable proportion of pQTLs are not eQTLs and that additional GWAS signals can be mapped to protein levels. Integration of pQTLs with MR has identified pathways and biomarkers for complex traits as well as potential therapeutics that could be repurposed10.
In this study, we combined high-throughput proteomics from multiple tissues with genetic data to determine the genetic architecture of protein levels in neurologically relevant tissues (brain, CSF, and plasma). This integration led to the identification of tissue-shared and tissue-specific pQTLs that are critical for the understanding of the biology of complex traits, particularly neurological diseases.
Results
Discovery of multi-tissue pQTLs
We measured the abundance of 1,305 proteins using an aptamer-based platform17 in CSF (n=971), plasma (n=636), and brain (n=458) samples (Extended Data Fig.1, Table S1). We included multiple technical and biological replicates to confirm the replicability and reproducibility of our proteomic measurements (Extended Data Fig.2). We performed stringent quality control (QC) steps for the proteomic data (See Methods). After QC, 835 CSF samples and 713 proteins, 529 plasma samples and 931 proteins, and 380 brain samples and 1079 proteins were included in the analyses (Tables 1, S2). The cohort included individuals with AD and cognitively normal individuals of European ancestry (Table 1). To identify pQTLs within each tissue (Fig. 1a), we performed genome-wide association analyses of 14.06 million imputed autosomal common variants (minor allele frequency (MAF) ≥ 0.02) against protein levels in each tissue. We defined cis-signals as those where the single nucleotide polymorphism (SNP) fell within 1Mb upstream or downstream of the gene start site, which may not include all enhancers for the corresponding gene. Trans-signals were defined as those where the SNP fell outside of the 2Mb window. To correct for multiple tests, we used a stringent genome-wide threshold of p<5×10−8 for cis-pQTLs and 5×10−8/(number-of-independent-proteins) for trans-pQTLs. There were 169, 230, and 75 independent proteins in CSF, plasma, and brain, respectively (see Methods). Therefore, p-value thresholds were set at 2.96×10−10 for CSF, 2.17×10−10 for plasma, and 6.67×10−10 for brain.
Table 1.
Demographics of the baseline cohort.
| CSF | Plasma | Brain | |
|---|---|---|---|
| N | 835 | 529 | 380 |
| Age [mean+/−sd] | 69.4+/−9.3 | 69.8+/−9.4 | 83.3+/−10 |
| Female (%) | 53% | 54% | 57% |
| % CDR=0 | 74.37% | 68.24% | 11.57% |
| APOE e4 (%) | 38% | 41% | 48% |
Characteristics of the baseline cohort after QC, including age, gender, Alzheimer disease status (as Clinical Dementia Rating (CDR)) and APOE e4 allele percentage. For CSF, age denotes age at lumbar puncture; For plasma, age denotes age at plasma draw; For brain, age denotes age at death. Values are reported in years (mean ± standard deviation [sd]). For basic demographics of the entire cohort before QC, please see Table S1.
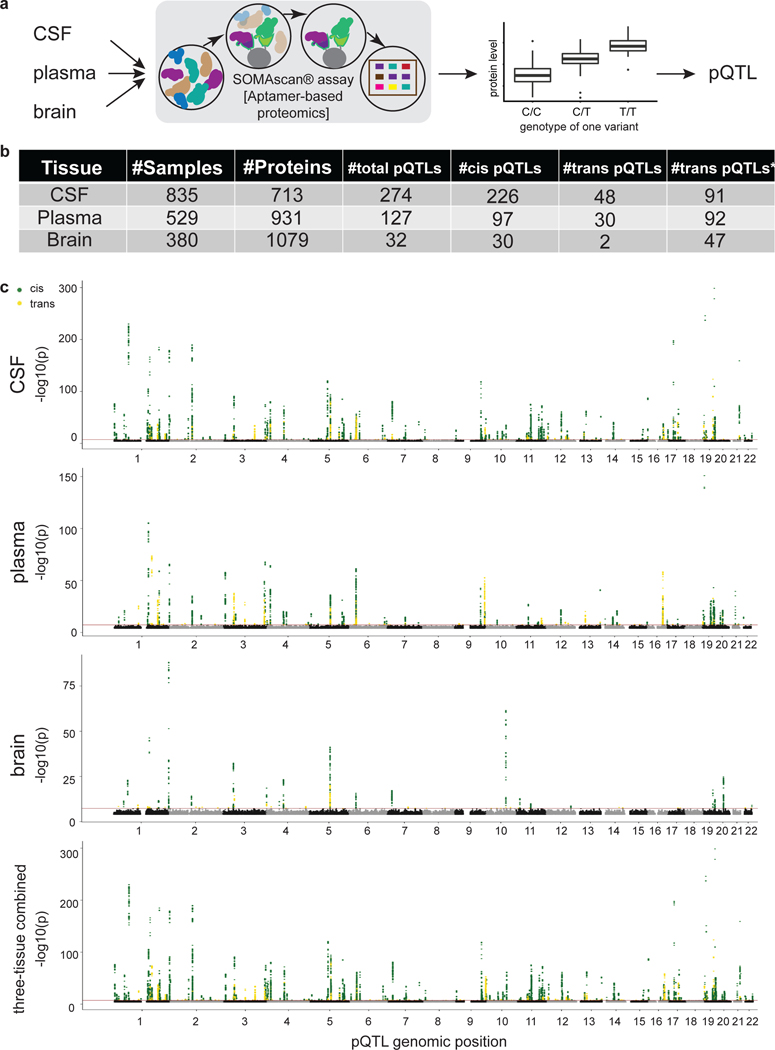
Fig. 1. Study design and overview of the significant pQTLs within each tissue.
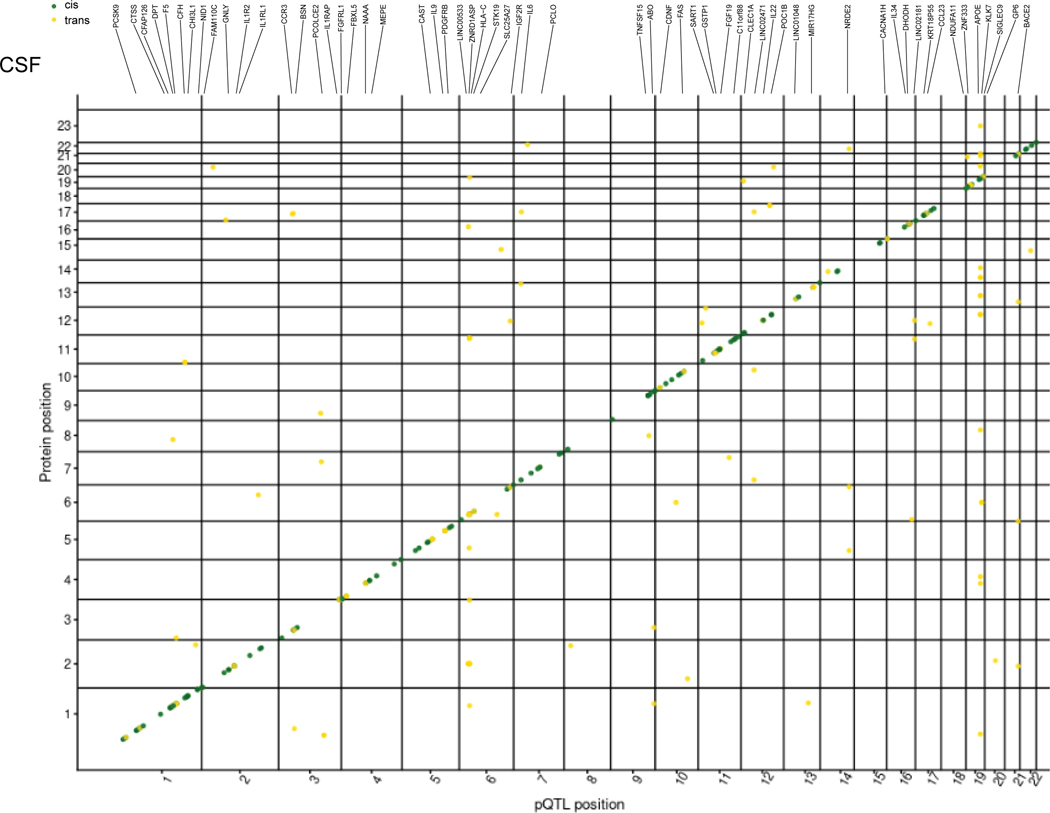
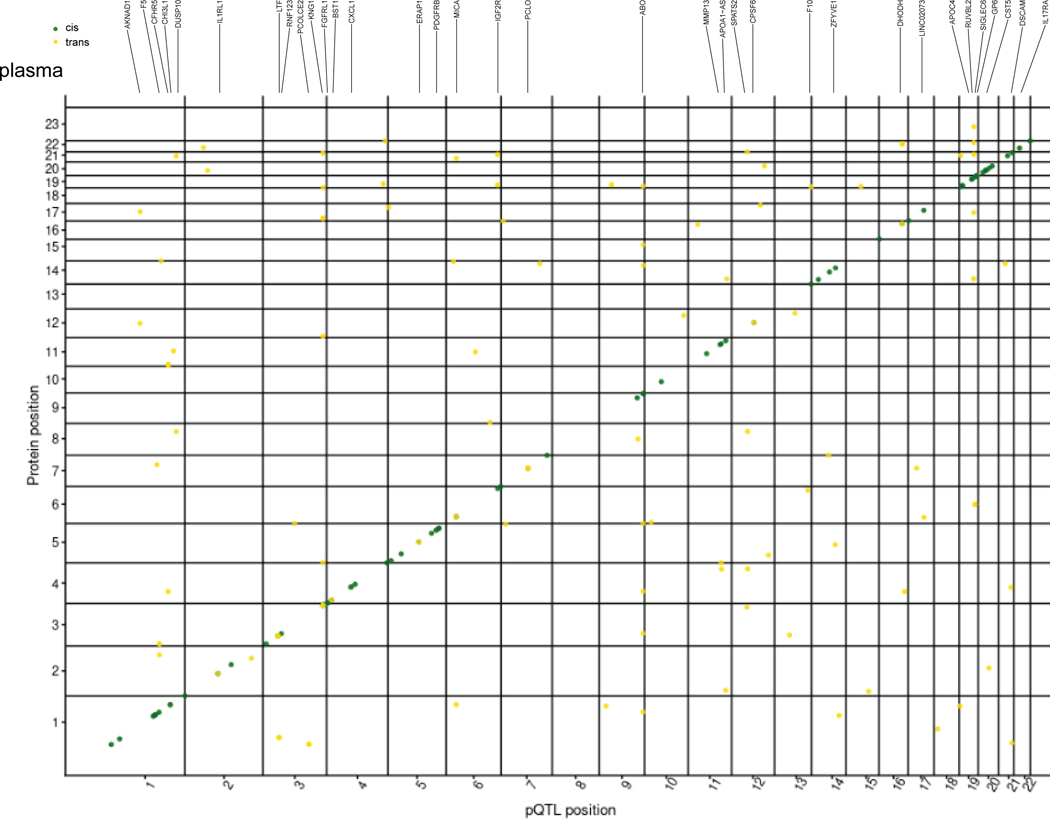
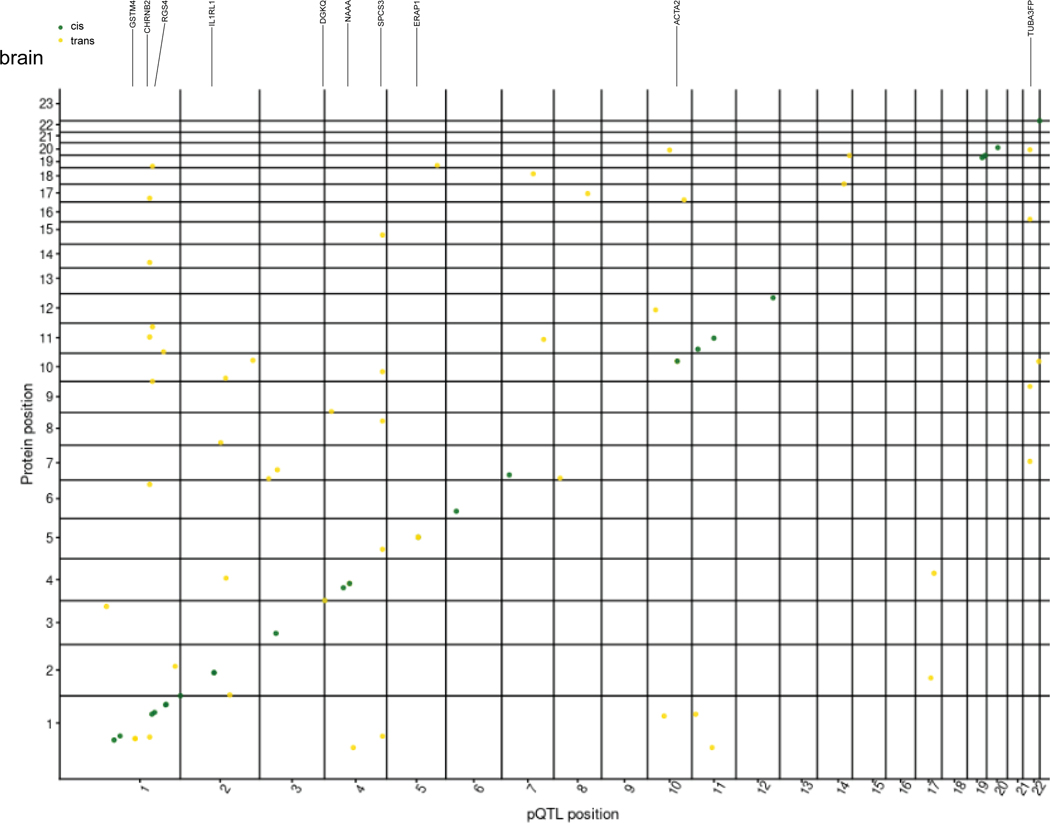
(a) Schematic of study design. CSF, plasma, and brain tissues were profiled using a high-throughput aptamer-based proteomics platform. We identified common genetic variants associated with each protein within each tissue after integrating both the genotype for each variant and protein level. The box-plot of pQTL is just for illustration purpose, showing the median (line), quartiles (box) and whiskers extending to ±1.5 times the interquartile range. (b) Table of sample size after QC and total number of pQTLs (split by cis, P < 5×10−8, and trans P < 5×10−8/number_PCs) for each tissue. For trans-pQTLs, the p-value cutoff for CSF is 3×10−10 (5×10−8/169), for plasma it is 2×10−10 (5×10−8/230), and for brain it is 7×10−10 (5×10−8/75). Trans* represents replication of trans-pQTLs given genome-wide significance (p-value < 5×10−8). (c) Stacked Manhattan plots for all three tissues mapping genomic locations of these pQTL within each tissue (cis: dark-green; trans: gold). The X-axis denotes the positions of the common variants. The darkred line represents P = 5×10−8.
In total, we identified 274 significant independent pQTLs for 184 CSF proteins, 127 independent pQTLs for 100 plasma proteins, and 32 independent pQTLs for 27 brain proteins (Fig. 1b,c, Tables S3, S4, S5). The number of significant pQTLs was proportional to the sample size, rather than the number of proteins. Of the 274 significant associations in CSF, 82% were cis associations and 18% were trans associations. In plasma, 76% were cis associations and 24% were trans associations. Lastly, in brain, 94% were cis associations and 6% were trans associations (Fig. 1b,c). We next investigated the distance between trans-pQTLs from the transcription start site of their respective genes to discover how many trans-pQTLs are inter-chromosomal or intra-chromosomal. We found that 91.5 to 98.9% of the trans-pQTLs were inter-chromosomal (Table S6).
Disentangling independent local pQTLs
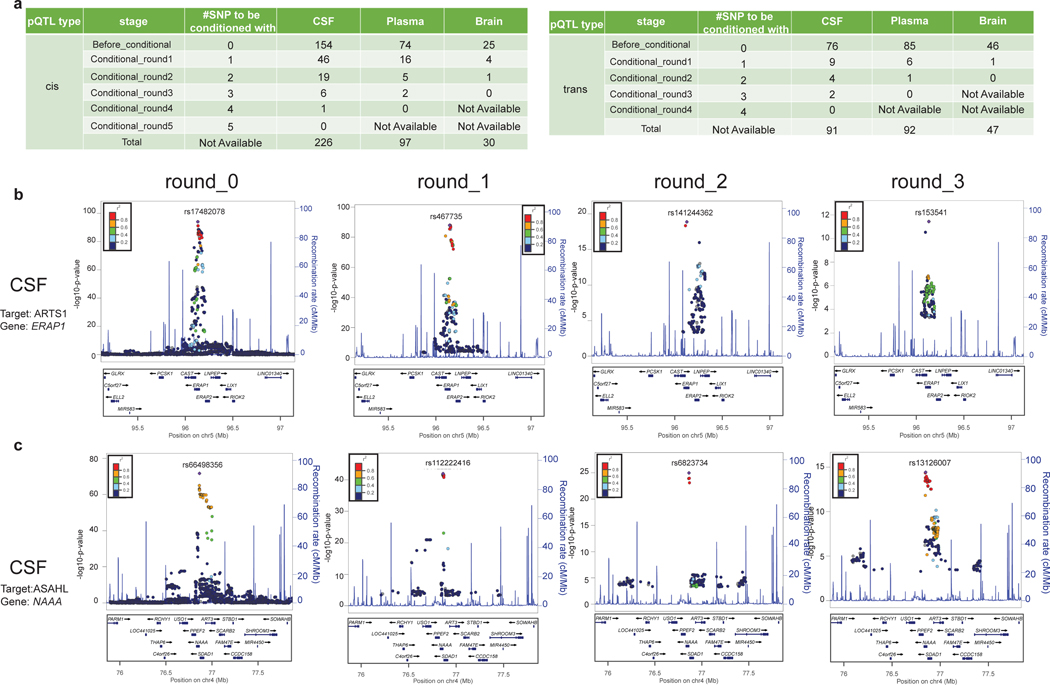
To identify independent local pQTLs where more than one association exists within 1Mb up- or downstream of the region’s top SNP, we performed conditional analyses. After the first round of conditional analyses, 55 CSF, 22 plasma, and 5 brain loci still had SNPs with independent and genome-wide significance (Fig. 2a). We performed a second round of conditional analyses including the top SNPs from the initial and first conditional analyses. In the second round, 23 CSF, 6 plasma, and 1 brain loci still showed independent and genome-wide significant associations (Fig. 2a). We continued performing conditional analyses until no SNPs passed the genome-wide threshold. We found 1 protein with up to 5 independent signals, 10 proteins with 4 independent signals, and 30 proteins with 3 independent signals. This highlights the complexity of regions with multiple independent local pQTLs that regulate protein levels.
Fig. 2. Identification of conditionally independent local pQTLs.
(a) Tables of conditionally independent pQTLs (cis and trans) locally (2 Mb window) after each round for each tissue. Before conditional, no SNPs were used as a covariate given one region. For round_1 conditioning, the top SNP from before-conditioning stage given the same region was used as an additional covariate in the default model. For round_2 conditioning, the top SNP from before-conditioning stage and top SNP from round_1 stage was used as an additional covariate in the default model. Both SNPs were within the same region. For each round we added the previous independent top hits from the prior rounds until no variants passed genome-wide significance threshold given the same region. (b) Regional association plots of the ERAP1 region associated with CSF ARTS1 protein: (round_0) before conditional analyses, centered on rs17482078; (round_1) after conditioning on the prior top SNP (rs17482078, centered on rs467735; (round_2) after conditioning on the prior top SNPs (rs17482078 and rs467735, centered on rs141244362; (round_3) after conditioning on the prior top SNPs (rs17482078 and rs467735 and rs141244362, centered on rs153541. No genome-wide significant SNPs was observed in round_4 after conditioning on all prior top SNPs. (c) Regional association plots of the NAAA region associated with CSF ASAHL protein: (round_0) before conditional analyses, centered on rs66498356; (round_1) after conditioning on the prior top SNP (rs66498356, centered on rs112222416; (round_2) after conditioning on the prior top SNPs (rs66498356 and rs112222416, centered on rs6823734; (round_3) after conditioning on the prior top SNPs (rs66498356 and rs112222416and rs6823734, centered on rs13126007. No genome-wide significant SNP was observed in round_4 after conditioning on all prior top SNPs. The SNPs for each regional plot are denoted as a purple diamond. Each dot represents individual SNPs, and dot colors in the regional plots represent linkage disequilibrium with the named SNP at the center. Blue vertical lines in the regional plots show recombination rate as marked on the right-hand Y-axis.
As an example of these complex regions, the main signal for CSF ARTS1 (Fig. 2b) was primarily associated with the nonsynonymous variant p.R725Q (rs17482078, p-value = 8.64×10−95). After conditioning on this signal, there was still a genome-wide signal tagged by rs467735 (conditional p-value=9.82×10−89) that was associated with gene expression (GTEx; multi-tissue p-value=0). We found two additional independent signals for this region. Similarly, there are four independent variants for CSF ASAHL (Fig. 2c). Together, these results indicate that proteins are highly regulated and include several independent mechanisms, even in the same region. These mechanisms may affect not only gene expression (eQTLs), but also protein levels by affecting cleavage, cell secretion, receptor binding, or clearance in the case of nonsynonymous variants. One example of such regulation by nonsynonymous variants is IL6R, which had a strong cis-pQTL that was primarily associated with the coding-variant rs2228145 p.D358A (and rs12730935 which was in perfect linkage disequilibrium; plasma=1.6×10−105 CSF= 1.3×10−104), as this variant leads to increased cleavage by ADAM10 and ADAM17 of membrane bound IL6R18.
Replication and meta-analyses using independent datasets
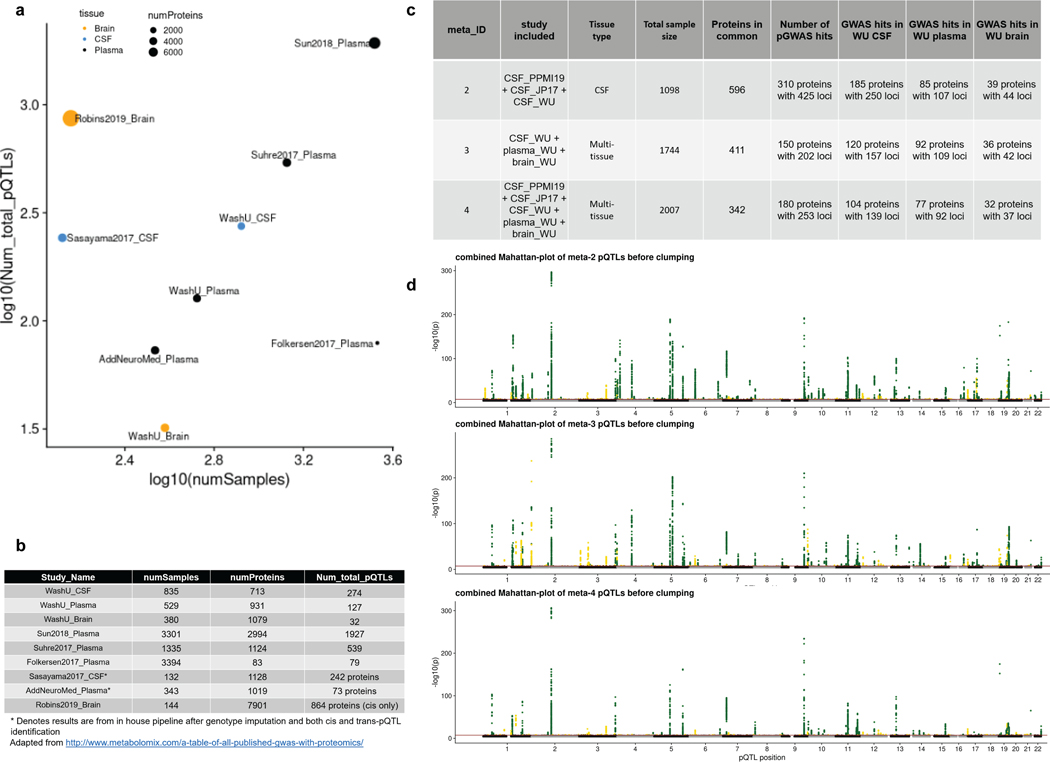
To replicate our pQTL findings, we analyzed several publicly available datasets (Fig. 3a,b,c, Extended Data Fig.3a,b). We also performed meta-analyses and cross-tissue replication. For CSF (Fig. 3a), we found 274 independent pQTLs, of which 223 (81.3%) were novel. We leveraged two CSF studies in which similar proteomic and genetic data were available. Sasayama et al. generated aptamer-based proteomic and array-based genotype data from 132 samples of Japanese origin14 and the Parkinson’s Progression Markers Initiative19 (n=131; released December 2019, PPMI19 hereafter) also had proteomics and genotype data available. We found that 51 pQTLs (49 cis and 2 trans, 18.6%) identified in our study have genome-wide significance with effects in the same direction as one or both studies. We also performed a meta-analysis of the Sasayama and PPMI19 studies (Table S7) to identify additional genome-wide and nominal associations. We found that 153 (55.8%) of our significant signals had genome-wide significance in our meta-analysis of the PPMI19 and Sasayama studies and 27 (9.9%) additional pQTLs showed at least a nominal association (p-value < 0.05) in the same direction. We identified an additional 16 (5.8%) pQTLs that have been reported as pQTLs in other tissues (plasma studies from AddNeuroMed 20, INTERVAL10, KORA11, SCALLOP12, the Phenoscanner database21, a brain mass spectrometry-based pQTL study16, and in our plasma or brain pQTL data). We were unable to test for replication of 5 (1.8%) pQTLs as protein levels were not available in these other studies. In summary, we were able to replicate more than 90.1% of the CSF pQTLs, which is higher than in previous studies10. Twenty-two (8.1%) pQTLs are still pending replication, as current CSF studies with smaller sample sizes do not provide enough statistical power. However, based on our validation with plasma and brain pQTLs, we estimate that more than 90% of those pQTLs are real. This is supported by the fact that we have been able to replicate 96.8% and 96.9% of plasma and brain pQTLs (see below).
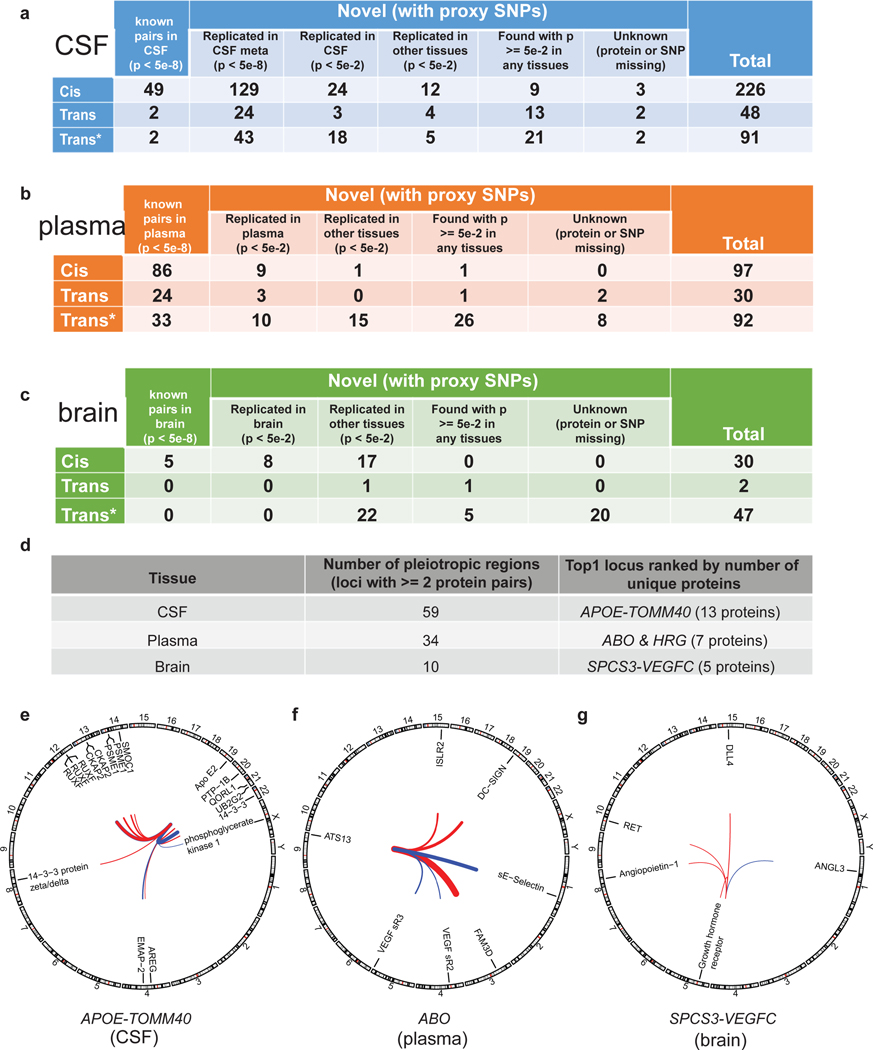
Fig. 3. Overview of the replication of the pQTLs and identification of pleiotropic regions within each tissue.
(a-c) Tables of replication of these pQTLs within CSF, plasma, and brain, given different p-value thresholds for different datasets. Overall, we classified pQTLs into five mutually exclusive groups: 1) known pQTLs in the matched-tissue (single-study) with a p-value less than 5×10−8; 2) replicated pQTLs in the matched-tissue with a p-value less than 5×10−2 but greater than or equal to 5×10−8 [*NOTE: for CSF, we split this group into two sub-groups: 2a) replicated only in the meta-analysis of two external CSF studies with a p-value less than 5×10−8; 2b) replicated pQTLs in the matched-tissue with a p-value less than 5×10−2 but greater than or equal to 5×10−8]; 3) replicated pQTLs in the other tissues with a p-value less than 5×10−2; 4) pQTLs found in any tissues (matched or not) with a p-value greater than or equal to 5×10−2; 5) unknown (either protein or SNP missing). For CSF, we further split the 2nd group into 2a) replicated pQTLs in the matched-tissue (meta-analysis, Table S6) with a p-value less than 5×10−8 and 2b) replicated pQTLs in the matched-tissue (meta-analysis and/or single-study) with a p-value less than 5×10−2 but greater than or equal to 5×10−8. Trans* represents replication of trans-pQTLs given genome-wide significance (p-value < 5×10−8) but not necessarily passing study-wide significance. Actual p-values (two-sided) without multiple comparison adjustments for each variant–protein pair were estimated using an additive linear regression model. (d) Table of all pleiotropic regions within each tissue given genome-wide significance threshold for both cis and trans-pQTLs and the name of top-1 locus ranked by number of unique proteins. (e) Circos plot of top-1 locus (mapped to APOE-TOMM40) associated with 13 unique CSF proteins. (f) Circos plot of top-1 locus (mapped to ABO or HRG) associated with 7 unique plasma proteins. (g) Circos plot of top-1 locus (mapped to SPCS3-VEGFC) associated with 5 unique brain proteins. Outermost numbers denote chromosomes. Lines link the genomic location of this locus with genes encoding significantly associated proteins. Associations denote genome-wide significance. Line thickness is proportional to effect size of linear regression (red, positive; blue, negative).
For plasma (Fig. 3b), we found 127 independent pQTLs, of which 17 were novel. For replication of plasma pQTLs, we used the five studies mentioned above. We were able to replicate 96.8% of our 127 pQTLs. We were unable to test 2 (1.6%), as they were not measured in those studies. For brain (Fig. 3c), we found 32 independent pQTLs, of which 27 were novel. As there were no published studies using the same aptamer-based proteomic method, we matched our proteins with a mass spectrometry-based pQTL dataset16. We were able to replicate 5 (15.6%) signals at genome-wide significance, 8 (25%) signals at a nominal (p-value < 0.05) association, and 18 (56.3%) pQTLs that showed at least a nominal association in brain or CSF. Only 1 (3.1%) pQTL was not replicated.
To increase statistical power and to identify additional genome-wide significant pQTLs, we performed meta-analyses that included all CSF cohorts as well as multi-tissue analyses. We first performed a CSF meta-analysis including the 596 common proteins shared among our study, PPMI19, and the Sasayama study (Extended Data Fig.3c,d). Due to the increased sample-size, we identified 425 pQTLs for 310 proteins, compared to 250 pQTLs for 185 proteins identified in our CSF cohort alone. This represents a nearly twofold increase in pQTL signals by increasing the sample size by 25%, suggesting that more pQTLs will be identified with larger sample sizes. We observed a similar increase in the number of pQTLs when performing a multi-tissue meta-analysis. For these analyses, we included 342 proteins that passed QC in all three tissue types as well as the PPMI19 and Sasayama study. We found 253 pQTLs compared to the 139 that were found in our CSF cohort alone (Extended Data Fig.3c,d).
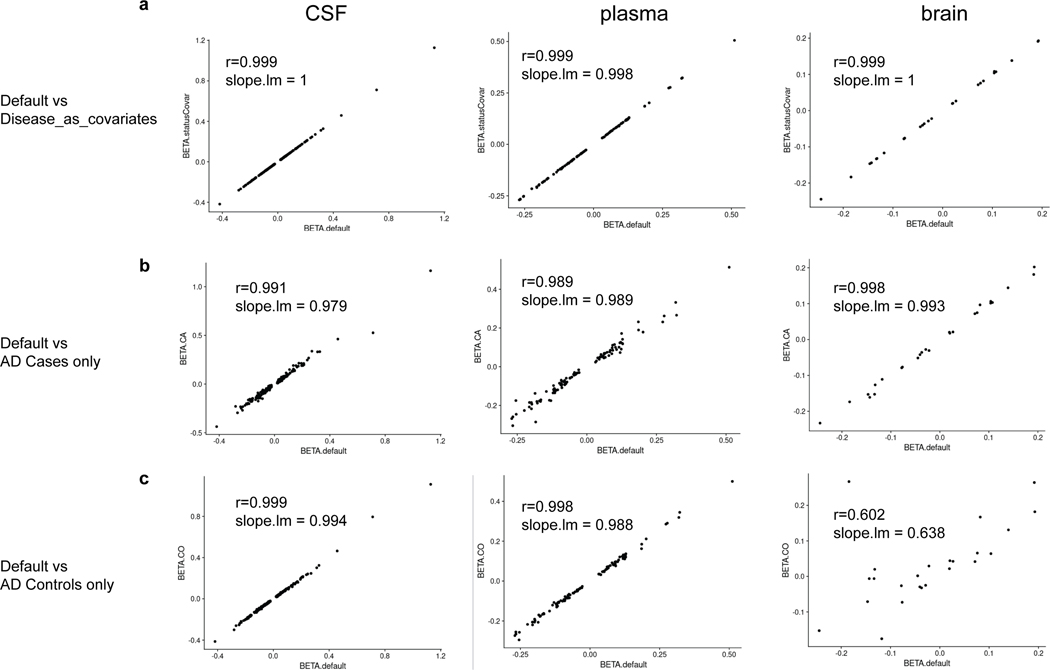
Because our study includes cognitively normal older individuals and AD cases, we performed additional analyses to determine if any of the pQTLs are disease- or age-specific. To investigate a disease-specific effect, we first included disease status as a covariate. Next, we performed case-only and control-only analyses. Finally, we compared the effect sizes of the genome-wide significant pQTLs from the initial analyses with the beta of these analyses. We found an extremely high correlation (Pearson’s r>0.98, Extended Data Fig.4, Tables S8, S9, S10) indicating that the associations of the genetic variants with protein levels are not disease-specific. To investigate an age-specific effect, we performed separate analyses in participants younger than or older than the average age of our cohort and compared the effect sizes of the genome-wide significant pQTLs from the initial analyses with the beta of these analyses. We found an extremely high correlation (Pearson’s r>0.98, Table S11), indicating that few pQTLs are age-specific.
Pleiotropic loci
We found that some loci were associated with the levels of more than one protein, and up to 13 different proteins in the case of genetic variants in the APOE region. Prior findings reported that the APOE locus is a pleiotropic region using plasma, and we found this in CSF as well. Genetic variants in the APOE gene region were associated with 13 different CSF proteins, including apo E2 and 14-3-3 (Fig. 3d,e, Extended Data Fig.5, Table S12). The genes encoding these proteins were located on different chromosomes indicating that this is not just cis regulation. Variants in the APOE locus are the strongest genetics risk factors for AD. Several studies have found that the 14-3-3 protein is a marker of non-specific neuronal death22,23, and our results indicated that 14-3-3 protein was also regulated by the APOE locus. For CSF, we found 59 pleiotropic regions where a single locus was associated with 2 or more proteins. In plasma, we replicated the known pleiotropy of the ABO locus for 7 different proteins, including E-Selectin (Fig. 3f, Extended Data Fig.6, Table S13), which was implicated in stroke risk by a recent study24. Further studies are needed to establish how these genetic variants are associated with multiple proteins. For example, genetic variants in the SPCS3-VEGFC region regulated brain levels of 5 different proteins including Angiopoietin-1 and Growth hormone receptor (Fig. 3g, Extended Data Fig.7, Table S14). We found an additional 32 pleiotropic regions in plasma and 9 in brain.
Several published studies on eQTLs and pQTLs have not identified pleiotropic loci as they only analyzed cis-associations16,25–27. Our results indicate that one protein is regulated in coordination with other independent proteins, which are likely part of the same signaling pathway. To understand the biological processes of health and disease, it is important to identify which proteins are regulated by the same genetic factors. This study identifies tissue-specific pleiotropic effects highlighting the complex mechanisms that regulate protein levels. Identification of additional tissue-specific cis-, trans-, and pleiotropic regions will lead to discovery of novel pathways relevant to pathogenesis.
Tissue-specificity investigation
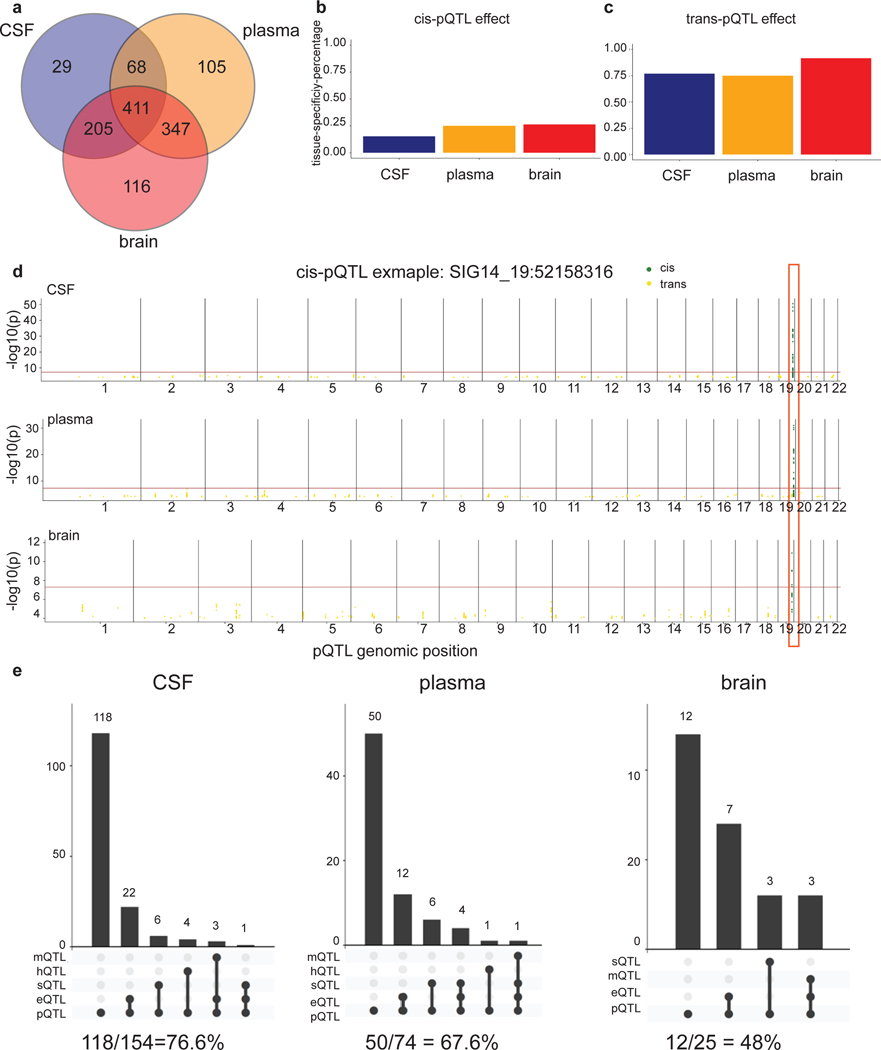
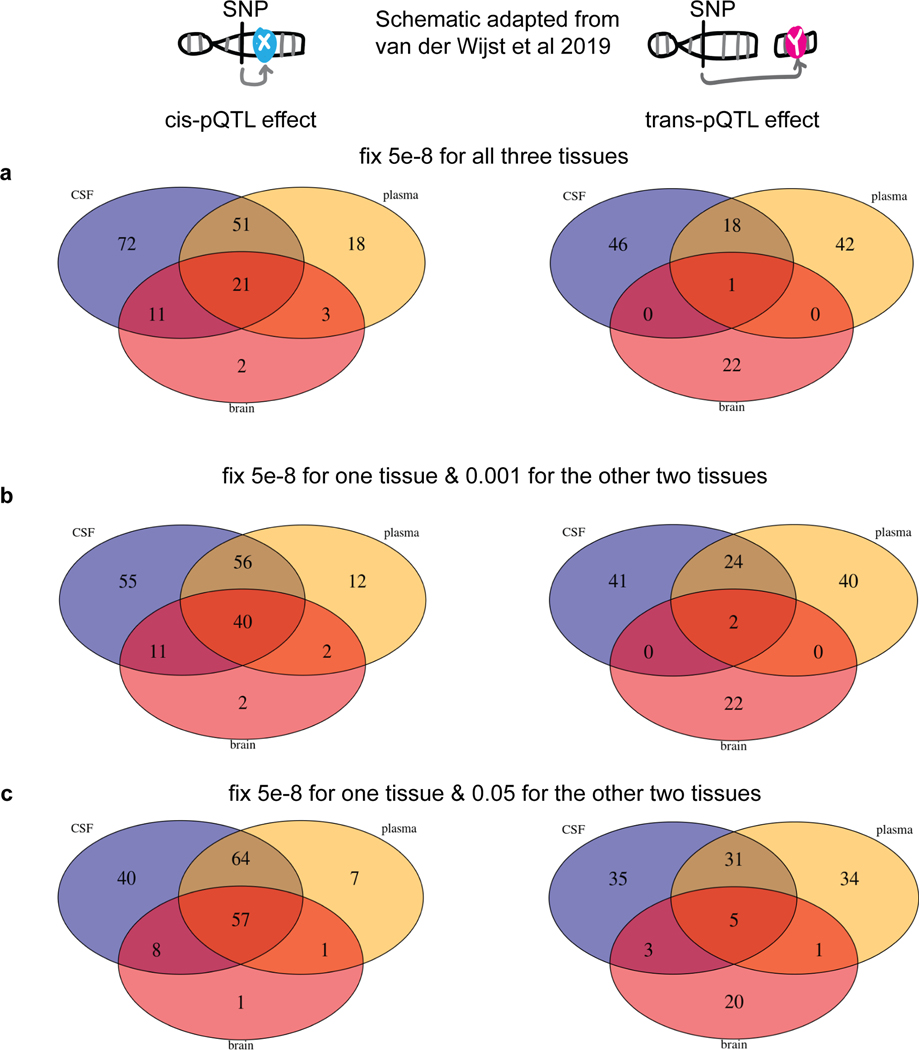
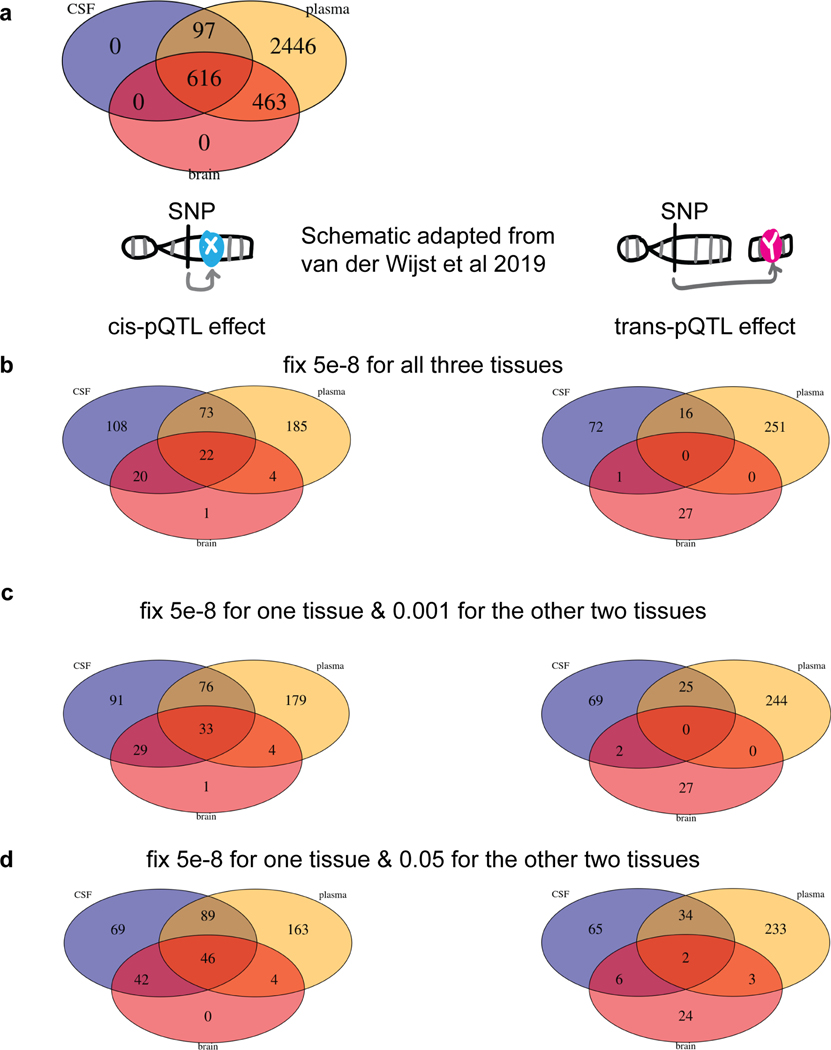
Our unique study design, which included protein measurements in multiple tissues linked to genetic data, enables us to investigate the overlap of the genetic architecture of protein levels across tissues. To identify tissue specificity, we performed mashr28analyses on our multi-tissue pQTL results using a p-value threshold of < 0.05 with proteins (n=411) that passed QC in all three tissues (Fig. 4a). Given a local false sign rate (LFSR) < 0.05 and Z-score of at least a 2-fold difference, we found that 15 to 26% of cis-pQTLs were tissue specific (Fig. 4b) while 77 to 91% of trans-pQTLs were tissue specific (Fig. 4c). This analysis indicated that cis-pQTLs were more likely to be shared across tissues than trans-pQTLs. For example, SIG14 only had a cis-pQTL shared across all three tissues (Fig. 4d). We performed additional analyses by comparing the proportion of pQTLs shared across tissues with different p-value thresholds, reaching the same conclusion (Supplementary materials, Extended Data Fig.8,9; Tables S15, S16, S17). CSF and plasma each shared more than 70% of brain pQTLs, suggesting that CSF and plasma were informative tissues for studying brain-related disorders such as AD.
Fig. 4. Summary of the tissue-specificity analyses and colocalization of pQTLs with other molecular QTLs.
(a) Venn diagrams of proteins passing QC across all three tissues. (b) Bar plot of tissue specificity percentage inferred from mashr on all cis-pQTLs across all three tissues given p-value < 0.05 threshold. (c) Bar plot of tissue specificity percentage inferred from mashr on all trans-pQTLs across all three tissues given p-value < 0.05 threshold. (d) Manhattan plots of the SIG14-chr19:52158316 within each tissue as an example of three-tissue-shared cis-pQTL. The darkred line represents P = 5×10−8. Actual p-values (two-sided) without multiple comparison adjustments for each variant–protein pair were estimated using an additive linear regression model. (e) Upset plots for colocalization investigation on pQTLs vs expression-QTLs vs splicing-QTLs vs DNA-methylation-QTLs vs histone-acetylation-QTLs for each tissue in cis and the bottom panel with the percentage of remaining pQTLs not colocalized.
Functional annotation and biological mechanisms of pQTLs
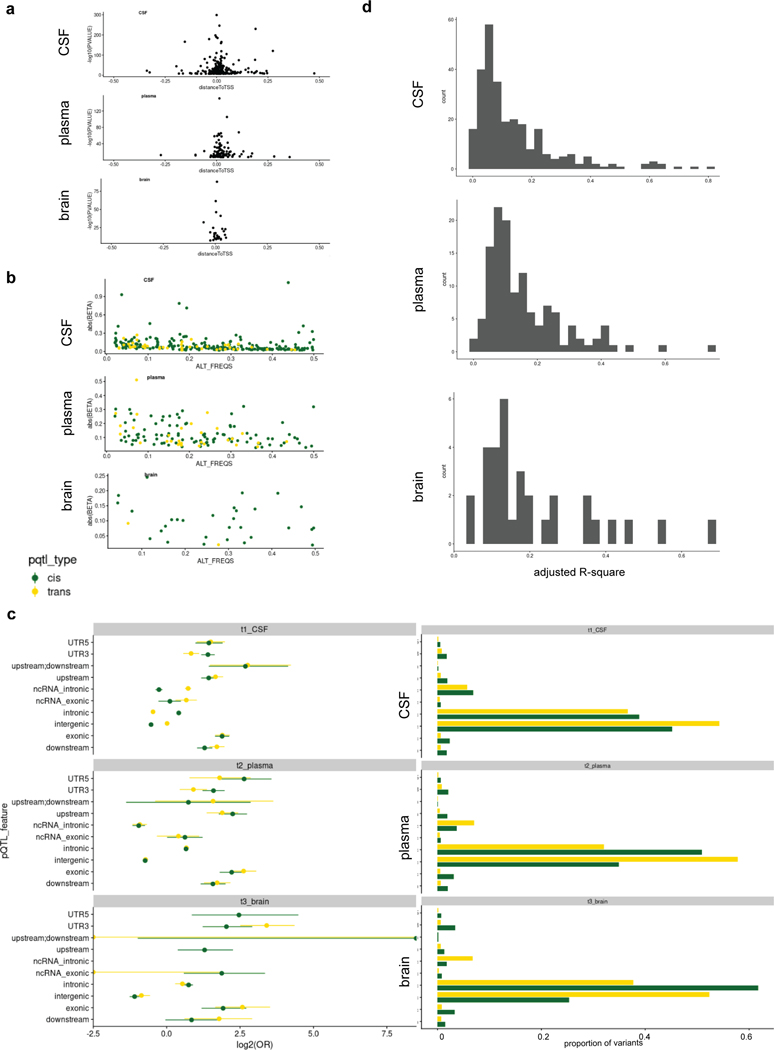
Previous studies discovered that most eQTLs are non-coding variants leading to the hypothesis that most eQTLs modulate transcription factor binding or chromatin structure7. However, it remains elusive if this is the case for pQTLs. For this reason, we performed bioinformatic functional annotation and statistical analyses to determine if pQTLs are enriched in specific regions, such as untranslated regions (UTRs), downstream or upstream of genes, introns, exons, splice sites, non-coding RNA (ncRNA) splice sites, ncRNA_introns, ncRNA_exons, or intergenic regions. We found that the strength of the association (negative log10 p-value) for cis-signals decreased with distance from the transcription start site (Extended Data Fig.10a), similar to what has been previously reported for cis-eQTLs7. This effect was found in all three tissues, suggesting that this is a common biological event. There was an inverse relationship between absolute value of the effect size (beta) and MAF (Extended Data Fig.10b), which is consistent with previous protein-level genome-wide association studies10,29.However, both cis- and trans-pQTLs were strongly enriched for synonymous and non-synonymous exonic variants (Odds Ratio = 3.71, 5.25, 4.19 for CSF, plasma, and brain, respectively; Extended Data Fig.10c; Supplementary Fig. 1). For 42 to 53% of cis-pQTLs (95 out of 226 in CSF, 44 out of 97 in plasma, and 16 out of 30 in brain), the association can be explained by a coding-variant, whereas for cis-eQTLs5 only 2 to 5% of signals are located in coding regions. This indicates that pQTLs are significantly enriched for coding-variants. These results suggest a role for additional regulatory mechanisms (including post-transcriptional changes), as protein levels may not correlate with mRNA levels.
The enrichment of coding-variants for pQTLs in cis and trans (Extended Data Fig.10c, Tables S3, S4, S5) suggests that protein levels are likely to be regulated post-transcriptionally rather than by regulating mRNA level30. Cis-pQTLs could lead to changes in protein levels by affecting the signal or the cleavage peptide. For trans-pQTLs, these variants could alter function of a receptor of the protein, affect the machinery that cleaves proteins from the membrane, or regulate the function of a gene encoding a transcription factor. This coding-variant enrichment observation may be confounded by aptamer-binding effects inherent to the aptamer-based platform. In multiple cases, the most significant signal was a coding-variant in a gene that affects protein cleavage or secretion (cis-signal; as in the case of IL6R or YKL-40; Extended Data Fig.10c, Table S3), or a coding-variant in the receptor of the protein that is likely to modify protein-receptor binding (trans-signal; as in the case of variants in the APOC4 gene region associated with the BAFF Receptor; Extended Data Fig.10c, Table S3). In line with the hypothesis that coding-variants have a greater effect size and that pQTLs are enriched for coding-variants, we found that pQTLs explained a large proportion of the variation in protein levels. The median variation in protein levels explained by pQTLs (adjusted R-squared calculated using the top variant as the only predictor and again using a nested model accounting for the covariates) was 9 to 14.9% (interquartile range: 13.2 to 15%; Extended Data Fig.10d). However, there were some extreme cases in which the top pQTL explained more than half of the variability in protein levels. For example, rs2075803 (p.K100E) explained 81% of CSF Siglec-9, rs5498 (p.K469E) explained 74.4% of plasma sICAM-1, and rs5498 (p.K469E) explained 67.4% of brain PPAC (Extended Data Fig.10d, Tables S3, S4, S5). CSF Siglec-9, plasma sICAM-1, and brain PPAC have been replicated in other studies16,29,31,32 using a different proteomic approach, which indicates that these findings may not be driven by platform.
Colocalization of pQTLs with other molecular QTLs
eQTL mapping and colocalization analyses have been instrumental in identifying the functional genes in genetic studies of complex traits5,33. However, it is known that changes in transcript level do not necessarily translate to changes in protein level. To identify the most likely gene underlying the GWAS signals, it is vital to go beyond eQTL and use other types of molecular QTLs. To determine if pQTLs provide additional information to that of eQTLs, we performed colocalization analyses on pQTLs with eQTLs.
Colocalization analyses indicated that only 13.3 to 33.3% of pQTLs had a posterior probability of hypothesis-4 (PP.H4)>0.8 for the GTEx cortex eQTLs or 14.2 to 17.5% for GTEx whole blood eQTLs (Tables S18, S19). These results were further replicated on the much larger study of blood eQTLs from the eQTLgen consortium9 (N=31,684 samples; Table S20). In brain, 28% of our pQTLs were colocalized in GTEx, which is within a similar scale to the Robins et al. (2019) study16 in which the authors used mass spectrometry-based proteomics to measure 7,901 proteins in 144 samples. This may also be explained by the fact that there are only 323 brain samples in GTEx compared to 380 in this study, which represents the largest brain pQTL analysis performed so far. In plasma, 16% of our pQTLs colocalized with eQTLs, similar to previous studies10. No previous studies have analyzed the colocalization between pQTLs and eQTLs in CSF.
Although eQTL mapping has been the most common approach to identify the genes driving the GWAS signals, there are other types of molecular QTLs (splicing, DNA-methylation, or histone-acetylation) that could be useful. We also wanted to determine the overlap of pQTLs found in this study with splicing, DNA-methylation, and histone-acetylation QTLs (Fig. 4e). Our colocalization analyses indicated that just 3.1 to 16.7% of pQTLs were splicing-QTLs (Tables S21, S22). Between 1.03 to 10% of pQTLs colocalized with DNA-methylation QTLs, and fewer than 2% colocalized with histone-acetylation QTLs (Tables S23, S24). Overall, between 48 (brain) to 76.6% (CSF) of pQTLs identified in this study did not colocalize with QTLs for expression, splicing, DNA-methylation, or histone-acetylation, suggesting that protein level may help explain some missing heritability of disease when integrated with other molecular traits.
Mendelian randomization and drug repositioning
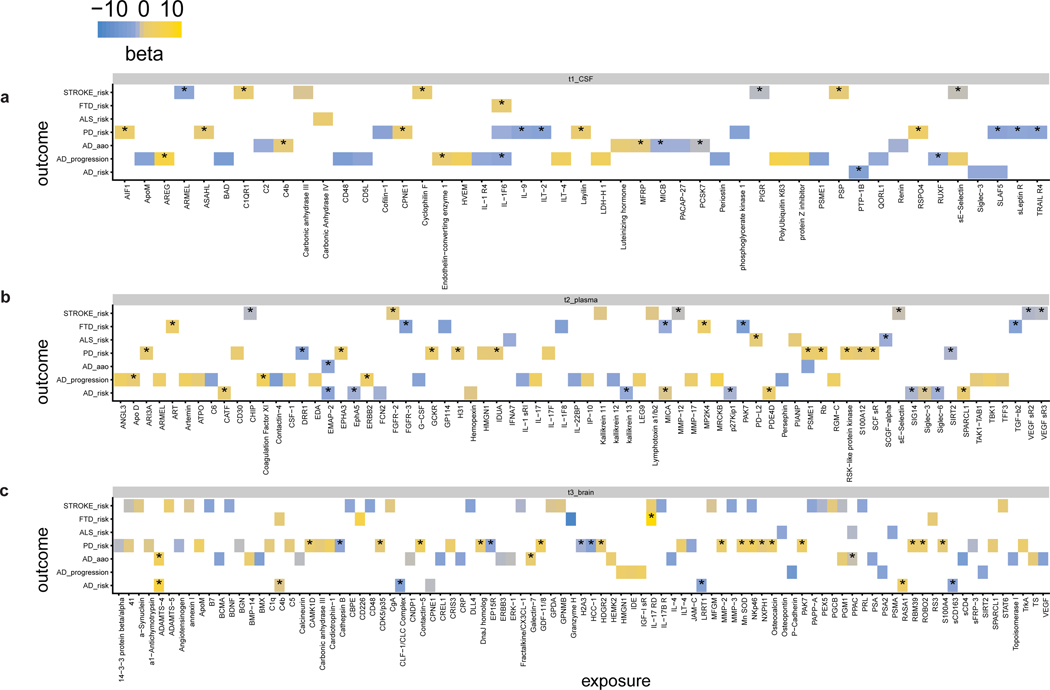
As a large proportion of pQTLs are not eQTLs or other types of QTLs, it is possible that the pQTLs identified in this study can help to identify the gene explaining GWAS signals. Using the MR framework, it is possible to identify functional genes and to prioritize proteins involved with complex traits for further analyses13,34. To identify proteins implicated in AD and other neurological traits, we performed MR analysis by using pQTLs as instrumental variables and AD risk, progression, and onset; Parkinson’s disease (PD); Frontotemporal dementia (FTD); Amyotrophic lateral sclerosis (ALS); and stroke risk (Table S25, S26) as outcomes. Only pQTLs with an F-statistic≥10 (Table S25) were included, and all pleotropic regions identified in this study were excluded. After multiple testing correction, we identified three proteins involved in AD risk in CSF, 13 in plasma, and 7 in brain. Other proteins found to be implicated in other traits by MR can be found in Table 2, Fig. 5, and Table S27. The Steiger test was used to confirm the direction of inference from protein to trait (Table S28). Colocalization provides additional supporting evidence of inference by reducing the possibility that linkage disequilibrium (LD) has influenced MR findings35. We found that 42.5% of protein-disease associations colocalized with GWAS loci, including plasma Siglec-3 (also known as CD33) and AD risk (Tables S29, S30). Integrating MR results (using cis- and trans-pQTLs as instrumental variables) with data from drug databases can be used to repurpose known drugs for diseases without treatment. In this study, we identified 25 drugs (Table S31) that are known to interact with proteins that were identified by MR as being associated with AD, PD, ALS, or stroke.
Table 2.
Number of significant protein-trait associations from Mendelian randomization analyses.
| Outcome | CSF | Plasma | Brain |
|---|---|---|---|
| AD risk | 3 | 13 | 7 |
| AD progression | 18 | 25 | 6 |
| AD Age at Onset | 8 | 2 | 20 |
| PD risk | 13 | 15 | 35 |
| ALS risk | 1 | 4 | 3 |
| FTD risk | 1 | 8 | 5 |
| Stroke risk | 7 | 8 | 24 |
| Asthma-risk (non-neuro) | 14 | 4 | 2 |
Within each tissue, the table contains the number of significant proteins (FDR < 0.05/24) and associations with the seven neurological traits: 1) AD risk3; 2) AD progression45; 3) AD Age At Onset46; 4) PD risk38; 5) ALS risk47; 6) FTD risk48; 7) Stroke risk49; and a non-neurological trait: asthma risk50.
Fig. 5. Mendelian randomization prioritized proteins in the associated relationship with seven neurological traits.
MR results were calculated using the TwoSampleMR R package44, and the effects for each protein-disease pair are visualized using Heatmap of MR inference of (a) CSF, (b) plasma, and (c) brain protein effect on seven neurological-related traits. The p-value threshold for significance is 0.05 after multiple testing correction accounting for both tissues and diseases. The color represents whether the effect size is positive (yellow) or negative (blue). Alzheimer disease (AD); Parkinson’s disease (PD); Amyotrophic lateral sclerosis (ALS); Frontotemporal dementia (FTD). Stroke is the general risk, not a specific subset of the stroke. The asterisk sign* represents colocalization with a PP.H4 > 0.8 for the protein-disease pair. The summary statistics are curated from published datasets (see Table S27 & S28 for details).
Some specific examples of how this data can be leveraged to map additional GWAS loci and identify drug targets include plasma CD33 for AD risk, plasma IDUA for PD risk, CSF Carbonic Anhydrase IV for ALS risk, and CSF/plasma E-Selectin for stroke risk. We found that variants in the CD33 locus, associated with plasma CD33 protein levels, are also known to be associated with AD risk3. MR analyses indicated that CD33 is implicated in AD (Table S29). CD33 is a microglia-specific gene36 and likely affects microglia activity and innate immune response. Our analyses also indicated that CD33 is a target of AVE9633, an anti-CD33 antibody used to treat acute myeloid leukemia, and therefore this antibody could be used in the context of AD. An ongoing clinical trial has been using antibodies targeting CD33 as a potential therapy for AD37. In summary, this analysis validated CD33 as a key protein to AD; it is also part of the TREM2-MS4A4A pathway.
Another example is plasma IDUA for PD risk. The IDUA locus is the third most significant locus in the largest GWAS38 studying PD risk. This locus contains more than one independent signal associated with PD38 and it was unclear which gene was functional. We found a SNP at this locus that is associated with plasma IDUA protein level and PD risk (rs35220088; p= 2.47×10−6 and 3.52×10−9, respectively). The MR and colocalization analyses indicated that IDUA, encoded by IDUA, is functional (FDR=9.37×10−6; Table S27, PP.H4 = 0.998; Table S30). IDUA degrades glycosaminoglycans in the lysosome. Our investigation identified chondroitin sulfate as a drug that could play a role in the IDUA pathway.
The third example is CSF Carbonic Anhydrase IV for ALS risk, which is supported by MR analysis (FDR = 7.61×10−6; Table S27). We further inferred Acetazolamide as a potential drug to treat ALS risk. Acetazolamide, used to treat glaucoma, epilepsy, and altitude sickness, is known to inhibit carbonic anhydrases39. Moreover, a recent in vitro study40 supported this drug as a potential treatment for ALS.
The fourth example is CSF or plasma E-Selectin protein. E-Selectin is a known stroke biomarker24, and our MR plus colocalization analyses indicated that this protein is genetically associated with the disease (Tables S27, S30). Carvedilol was identified from our drug repositioning analysis and has been reported to provide neuroprotection in animal models41.
In summary, our study is useful for mapping additional GWAS loci and identifying drug targets. We integrated our MR results with drug databases to identify potential drugs that could be repurposed for neurological diseases. It is known that compounds targeting proteins that are supported by genetic data are more likely to work than those without such support42. This study goes beyond AD or PD, and the data generated here can be applied to other traits.
Limitations
Our study has several limitations. An important note is that we used an aptamer-binding platform to profile proteomics, which might be confounded by recognition difference instead of real protein abundance change, as already addressed by Sun et al10. Mass spectrometry can be a complementary platform30 that will not be affected by this problem. Our observation of an enrichment of coding-variants in pQTLs could be largely biased by the platform. However, we found 83 to 90% of our non-coding pQTL and 10 to 17% coding pQTL, respectively and depending on tissues, were also seen in a mass spectrometry-based pQTL study16 (Table S32). The percentages for coding variants were similar (Proportional-test: p-value > 0.05 in all three tissues), indicating that this enrichment was not exclusively due to the bias of a single technology.
Our data suggest that pQTLs are located in coding regions more often than are eQTLs, and our results could be biased, because coding pQTLs have stronger effects than non-coding pQTLs. Therefore, non-coding pQTLs may remain undetected due to a lack of statistical power. However, the sample size of our study is similar to or larger than GTEx for brain-relevant tissues, and eQTL studies from GTEx do not see the same amount of coding-variant enrichment5. Our data suggest that around half of pQTLs do not colocalize with other molecular QTLs, but this data cannot be extrapolated to all coding genes. GTEx and eQTLgen include eQTLs for more than 34,000 RNA molecules, but not all of those RNA molecules code for proteins or are expressed in all tissues. It is clear that we cannot anticipate an eQTL-pQTL colocalization for a large proportion of coding genes. Another reason for a potential lack of eQTL-pQTL colocalization in brain is because our brain pQTLs are derived from parietal lobe cortex, and the brain eQTLs from GTEx are from whole cortex. The gene regulation may be region-specific, and thus complicates the overlapping between two QTL types. The small proportion of overlap between eQTL and pQTL data can also be caused by measurements of mRNA expression and protein level in different cohorts, sample sizes, or other technical details.
A standard limitation of MR analyses is the inability to fully account for horizontal pleiotropy; though, confidence in our results may be bolstered by the fact that we performed MR after removing all pleotropic regions reported in this study within each tissue. Another limitation of MR interpretation is that our observation on associations between proteins and neurological diseases may not be disease-specific, as we observed several associations (Table 2, S33) between proteins and asthma risk, an example of non-neurological diseases.
Additional studies performed in more tissues and using approaches that include an even larger number of proteins are necessary to better understand the genetic architecture of protein levels and to replicate these findings. This is the first study to address the single-tissue bias when comparing pQTLs to results of multi-tissue eQTLs.
Discussion
In this study we generated a detailed map of multi-tissue pQTLs that will be crucial for understanding the tissue-specific genetic architecture of protein levels. By leveraging this data, we have been able to map some additional GWAS loci, identify additional causal proteins implicated in disease pathogenesis, identify novel biomarkers, and reposition drugs. If we want to fully understand the genetic architecture of complex traits, we need to understand the genetic architecture of protein levels. Until now, multi-tissue pQTL mapping has been constrained by the limited availability of large-scale proteomic analyses in multiple tissues. We present the largest brain and CSF pQTL analyses to date, the first neurologically-relevant multi-tissue pQTL study, and a unique resource for leveraging multi-tissue pQTL to understand neurological traits. These data can be further used to perform MR analysis on other complex traits with available GWAS. All results can be downloaded from https://www.niagads.org/datasets/ng00102 and interactively accessed through a PheWeb43-based website, the Online Neurodegenerative Trait Integrative Multi-Omics Explorer (ONTIME; https://ontime.wustl.edu/).
To achieve personalized medicine, these results highlight the need to implement additional functional genomic approaches beyond gene expression towards understanding the biology of complex traits and to identify potential drug targets for those traits. We predict that this and other studies including additional omic layers (e.g. epigenomics, metabolomics) will be instrumental in advancing the field.
Methods
Ethics declarations
This project was approved the ethical committee of the Washington University School of Medicine in St. Louis
Aptamer-based proteomics data sample collection
This study included 1,537 participants from Washington University School of Medicine in St. Louis. All participants provided informed consent to allow their data and biospecimens to be included. The study was approved by an Institutional Review Board at Washington University School of Medicine in St. Louis.
Samples from participants include three tissue types: CSF, plasma, and brain (parietal lobe cortex). CSF samples were collected the morning after an overnight fast, processed, and stored at −80°C. Plasma samples were collected the morning after an overnight fast, immediately centrifuged, and stored at −80°C. Brain tissues (~500mg) were collected from fresh frozen human parietal lobes.
For CSF tissue, there were 971 unique participants (including 249 AD cases, 717 cognitively normal controls, and 5 with unknown status) and 1,300 samples (329 participants provided one baseline and one longitudinal sample) in total. Age is distributed with a mean of 69.0 years and standard deviation of 9.3 years. 53% of the samples are from women.
For plasma tissue, there were 636 unique participants (including 230 AD individuals, 401 cognitively normal controls and 5 with unknown status) and 648 samples in total. Age is distributed with a mean of 70.4 years and standard deviation of 9.8 years. 56% of the samples are from women.
For brain tissue, there were 458 unique participants (including 297 AD cases, 27 cognitively normal controls and 134 with unknown or other status (e.g. FTD, other neurological diseases)) and 459 samples in total. Age is distributed with a mean of 82.2 years and standard deviation of 12 years. 60% of the samples are from women.
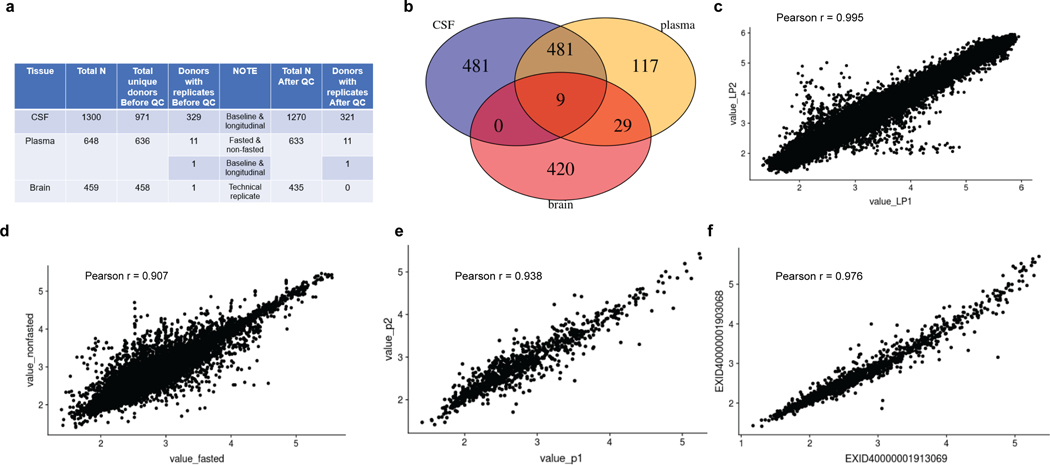
The donor overlap across three tissues are shown as a Venn diagram in Extended Data Fig.2b: 9 donors were shared across all three tissues; 481 donors were shared by both CSF and plasma; 29 donors were shared by plasma and brain; 481, 117 and 420 were exclusively for CSF, plasma, and brain, respectively. The Venn Diagram was drawn using VennDiagram51 R package (v1.6.20).
These recruited participants were evaluated by clinical personnel from Washington University. AD severity was determined by the Clinical Dementia Rating (CDR)52 scale at the time of lumbar puncture (LP, CSF samples) or blood draw (plasma).
For brain samples, case–control status was determined by postmortem neuropathological analysis of study participant brains based on CERAD53 and/or Khachaturian54 criteria. All AD cases have a Braak stage of IV or higher and controls are III or lower. The neuropathological diagnosis was performed studying several brain regions, not only parietal.
Proteomic data QC process
We used a multiplexed, aptamer-based platform17, to measure the relative concentrations (relative fluorescent units, RFU) of proteins from CSF, plasma, and brain tissues, using 1,305 modified aptamers in total. The assay covers a dynamic range of 108, and measures all three major categories: secreted, membrane, and intracellular proteins (Table S34, Supplementary Fig 2, 3). The proteins cover a wide range of molecular functions, such as protein binding and the MAPK cascade. The coverage of proteins on the platform has taken into account proteins known to be relevant to human disease, including neurodegenerative diseases55 and cardiovascular diseases56; thus it has been widely used for biomarker discovery.
Aliquots of 150 μl of tissue were sent to the Genome Technology Access Center at Washington University in St. Louis for protein measurement. Assay details have been previously described by Gold et al.17 In brief, modified single-stranded DNA aptamers are used to bind specific protein targets that are then quantified by a DNA microarray. Protein concentrations are quantified as RFU.
Quality control (QC) was performed at the sample and aptamer levels using control aptamers (positive and negative controls) and calibrator samples. At the sample level, hybridization controls on each plate were used to correct for systematic variability in hybridization. The median signal over all aptamers was used to correct for within-run technical variability. This median signal was assigned to different dilution sets within each tissue. For CSF samples, a 20% dilution rate was used. For plasma samples, three different dilution sets (40%, 1%, and 0.005%) were used. For brain samples, a 20% dilution rate was used.
To QC the proteomics datasets (Extended Data Fig.1a-d), the protein/analyte outliers were first removed by applying four criteria:
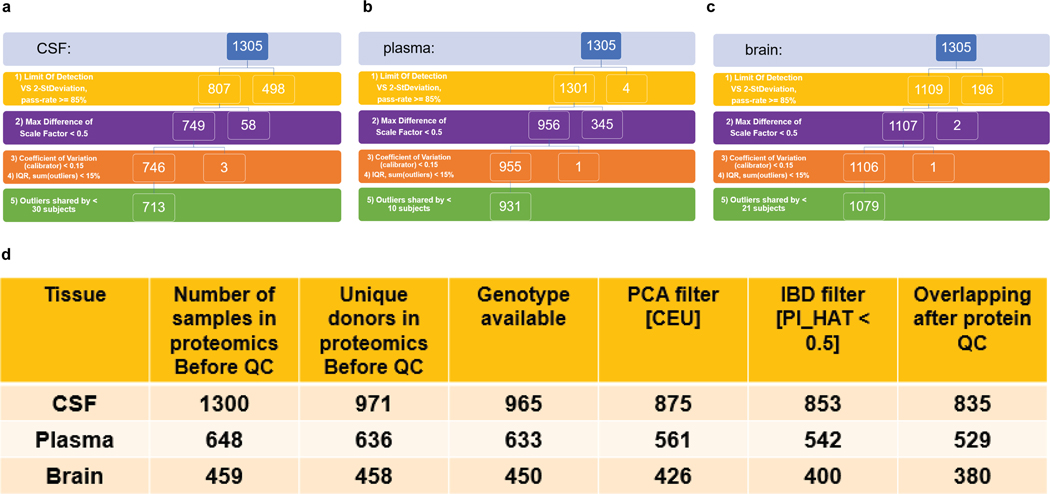
Minimum detection filtering. Limit of detection (LOD) was defined as the summation of average expression level of the new NP-buffer (used as dilution buffer of CSF samples since plate-42) and K fold of standard deviation (K = 2). If the analyte for a given sample was less than the LOD, this sample was an outlier. Collectively, if the number of outliers given an analyte was less than 15% of total sample size, the analyte was kept.
Flagging analytes based on the scale factor difference. The scale factor difference was calculated as the absolute value of the maximum difference between the calibration scale factor per aptamer and the median for each of the plates run. If the value for this analyte was less than 0.5, the analyte passed this criterion (NOTE: SOMAlogic SQS report used 0.4).
CV of calibrators lower than 0.15. The coefficient of variation (CV) for each aptamer was calculated by dividing the standard deviation by the mean of each calibrator at the raw protein level. If the analyte had a CV of less than 0.15, this analyte passed the CV QC.
Interquartile range (IQR) strategy. Outliers were identified if the subject was located outside of either end of distribution using a 1.5-fold of IQR given the log10 transformation of the protein level. Collectively, if the number of outliers given an analyte was greater than 15% of total sample size (aka none-outliers given an analyte was fewer than 85% of total sample size) this analyte was filtered. Analytes were kept after passing all the criteria above for the downstream statistical analysis.
An orthogonal approach was used to call subject outliers based on IQR. Collectively, if the number of outliers given an analyte was greater than 15% of total number of analytes passed QC (aka none-outliers given an analyte was fewer than 85% of total number of analytes passed QC), this subject was labeled as an outlier. Furthermore, the analytes were removed if shared by most (~80%) of the subject outliers. After this second removal of analytes, subject outliers were called again. Outlier subjects were again removed.
We processed the proteomics data using SomaDataIO (v1.8.0)17 and Biobase (v2.42.0)57. Proteins were mapped to UniProt58 identifiers and Entrez gene symbols. Mapping to Ensembl gene IDs and genomic positions (start and end coordinates) was performed using gencode v30 liftover to hg19/GRCh37.
Reproducibility investigation via comparisons between biological or technical replicates
To measure the reproducibility of the aptamer-based platform, we compared the replicates for the same subject given each tissue.
For plasma samples, we included 11 subjects with two measures, one as fasted and the other as non-fasted. After QC, we kept 931 proteins in 633 samples. The overall Pearson’s correlation coefficient between fasted and non-fasted samples is 0.907, with a 95% confidence interval from 0.904 to 0.911 (Extended Data Fig.2a,d).
For plasma samples, we included one subject with two biological replicates: one collected in 1997, the other in 2007. Both samples passed QC. The overall Pearson’s correlation coefficient between these two biological replicates is 0.938, with a 95% confidence interval from 0.9299 to 0.9453 (Extended Data Fig.2a,e).
For brain samples, we included one subject with two technical replicates. After QC, we kept 1,079 proteins and 435 samples. Out of these 435 samples, only one replicate of the subject remained. Thus, we compared two technical replicates using the values before QC across all 1,305 proteins. The overall Pearson’s correlation coefficient between these two replicates is 0.976, with a 95% confidence interval from 0.9762 to 0.9812 (Extended Data Fig.2a,f).
For CSF samples, we designed 329 subjects with two measures, one as baseline (LP date1) and the other as longitudinal (LP date 2). After QC, we kept 713 proteins and 1,270 samples. Out of these 1,270 samples, 321 subjects with two measures remained in the analysis (Extended Data Fig.2a,c). The average time difference between the two LP dates was 6.14 years, and the standard deviation was 2.98 years. The overall Pearson’s correlation coefficient between two LP dates was 0.995, with a 95% confidence interval from 0.99518 to 0.99526 (Extended Data Fig.2c).
The overall high correlations within each tissue indicated that the aptamer-based technology was highly reproducible.
Genomic data QC process
Most of the samples with proteomic profiling were collected with genotyping data (Extended Data Fig.1d). For CSF, 965 out of 971 unique subjects have both genotype and proteomic data. For plasma, 633 out of 636 unique subjects have both genotype and proteomic data. For brain tissue, 450 out of 458 unique subjects have both genotype and proteomic data.
Samples were genotyped on multiple genotyping platforms from Illumina. Stringent quality thresholds were applied to the genotype data for each platform separately. SNPs were kept if they passed all of the following criteria: i) genotyping successful rate ≥ 98% per SNP or per individual; ii) MAF ≥0.01; iii) Hardy-Weinberg equilibrium (HWE) (p ≥1 × 10−6).
After removing low quality SNPs and individuals, genotype imputation was performed using the Impute2 program59 with haplotypes derived from the 1,000 Genomes Project (released June 2012). Genotype imputation was performed separately based on the genotype platform used. SNPs with an info-score quality of less than 0.3 reported by Impute2, with a MAF < 0.02, or out of HWE were removed. After Imputation and QC, the different imputed PLINK files were merged. A total of 14,059,245 imputed and directly-genotyped SNPs and 1,530 individuals were used for final analyses. To adjust for population substructure (Extended Data Fig.1d), principal component analysis (PCA) was conducted using the PLINK1.9 (v1.90b6.4)60 subcommand pca. HapMap samples (CEU: Caucasian Europeans from Utah; JPT: Japanese in Tokyo; YRI: Yoruba in Ibadan, Nigeria) were included in the analyses to remove outliers and confirm self-reported ethnicity. Samples within the CEU cluster were kept. To identify unanticipated duplicates and cryptic relatedness using pair-wise genome-wide estimates of proportion identity by descent (IBD) (Extended Data Fig.1d), we used the subcommand IBD from PLINK1.9 (v1.90b6.4)60. When duplicate samples or a pair of samples with cryptic relatedness (PI_HAT ≥ 0.5) were identified, we removed one sample from the pair. A total of 835 CSF, 529 plasma, and 380 brain samples passed filters on both genomic and proteomic QC.
pQTL identification
To test for the association between genetic variants and protein levels we performed a linear regression (additive model), including age, sex, principal component factors from population stratification, and genotype platform as covariates:
cis-pQTL mapping.
We conducted cis-pQTL mapping within each of the three tissues. Only proteins passing QC were included in the analyses. Protein level was 10-based logarithm transformed to approximate the normal distribution. For these tests, data distribution was assumed to be normal, but this was not formally tested. Within each tissue, cis-pQTL were identified by linear regression, as implemented in PLINK2 (v2.00a2LM)60, adjusting for sex, age, the first two genotype-based principal components (PCs) [genetically very homogeneous], and genotyping platforms (e.g. Omni1, Omini2.5, NeuroX, etc.). We restricted our search to variants within 1 Mb upstream and downstream of the gene start site by which each protein is coded. Actual p-values for each variant-protein pair were estimated using an additive linear regression model. To identify the list of all significant variant–protein pairs, variants with an actual p-value below the genome-wide significance (5×10−8) level were considered significant and included in the final list.
trans-pQTL mapping.
PLINK2 (v2.00a2LM)60 was used to test all autosomal variants (MAF ≥0.02) using the same QC pipelines as cis-pQTL mapping, but was restricted to variants and proteins encoded by the genes locating outside the 2Mb window in each tissue independently using an additive linear model. For trans-pQTL mapping, we tested variants for association with the same protein list as for cis-pQTL mapping. We included the covariates of the first two genotype PCs [genetically very homogeneous], age, sex, and genotyping platforms when performing association tests. The correlation between variant and protein levels was evaluated using an additive linear regression model. For these tests, data distribution was assumed to be normal, but this was not formally tested. To identify the list of all significant variant-protein pairs, variants with an actual p-value below the study-wide significance (5×10−8/number_PCs) were considered significant and included in the final list. The number of PCs was derived as the minimum principal component number that cumulatively explain 95% of the variance for each tissue after QC. For CSF, plasma, and brain, the number of PCs is 169, 230, and 75, respectively. Thus, the p-value thresholds are 2.96×10−10, 2.17×10−10, and 6.67×10−10, respectively.
Disease specific analyses:
To investigate a disease-specific effect on pQTLs, we performed linear regression on the same protein-loci pairs (before conditioning on top variants) identified from the above default model using three additional models: 1) joint analysis including disease status as another covariate (CO vs non-CO); 2) AD case (CA) only using the same covariates as the default model; 3) cognitive unimpaired (CO) only using the same covariates as the default model. Using scatterplots, we visualized the correlation between each of the additional models and our default model. Using model 1 for comparison, we observed a Pearson correlation coefficient of 0.999, 0.999, 0.999 for CSF, plasma, and brain, respectively. Using model 2 for comparison, we observed a Pearson correlation coefficient of 0.991, 0.989, 0.998 for CSF, plasma, and brain, respectively. Using model 3 for comparison, we observed a Pearson correlation coefficient of 0.999, 0.998, 0.602 (p-value = 0.002) for CSF, plasma, and brain, respectively. The relatively low correlation seen when using model 3 for comparison with controls only in brain samples was due to a much smaller sample size.
Age-specific analyses:
To confirm that none the of findings were age specific, we performed separate analyses in participants younger and older than the average age of our cohort and compared the β1 of SNP dosage for all significant pQTLs to identify any age-specific effect.
Variance explained by the top variant on certain protein:
We calculated the adjusted R-squared value using the linear regression model with the top variant as the only predictor and the log10-based certain protein abundance as the outcome. To account for variance explained by covariates, we also calculated the adjusted R-squared value using a nested model and the estimates were similar to using the top variant as the only predictor (Supplementary Fig. 4).
Conditional analysis on independent local pQTLs
To identify independent significant local pQTLs, we performed a conditional analysis on all pQTLs from round_0 using PLINK2 (v2.00a2LM)60 with the --condition or --condition-list option. We used the same significance threshold of p-value = 5×10−8 used for the univariable analysis on identifying independent local pQTLs within a window size of 2Mb.
Conditional analyses were performed as follows: Before conditional (row-1), no SNPs were used as a covariate given one region. For round_1 (row-2) conditioning, the top SNP from the before-conditioning stage given the same region was used as an additional covariate in the default model. For round_2 (row-3) conditioning, the top SNP from the before-conditioning stage and the top SNP from round_1 stage were used as an additional covariate in the default model. Both SNPs were within the same region. This iteration was continued for each round by adding one more top SNP from the prior round until no variants passed the genome-wide significance threshold given the same region. For CSF and plasma samples, in total 4 rounds of conditional analyses were performed. For brain samples, 3 rounds of conditional analyses were performed. The results were visualized using locusZoom v1.361.
Replication strategy for CSF pQTL
To identify all previously reported pQTLs from large-scale protein-level GWAS (pGWAS), we performed CSF replication analyses. We first searched the reprocessed pQTL results using Sasayama et al., 2017 (SOMAscan-based, CSF); PPMI19 (SOMAscan-based, CSF); and meta-analysis of these two prior studies. Next, we checked summary statistics from INTERVAL (SOMAscan-based, plasma). Finally, we queried other plasma pGWAS findings from EBI-NHGRI using phenoscannerV221 with proxy SNPs (r2 > 0.5).
Reprocessing pQTL using Sasayama 2017 CSF SOMAscan individual data:
To replicate CSF pQTLs, we performed linear regression on all proteins using the individual level proteomic and genotype data from Sasayama and colleagues published in 201714. We decided to reprocess the pQTL analyses because the original studies used unimputed genotype data. We performed imputation in the Sasayama data to have a similar number of SNPs across studies. For proteomics QC, only the IQR strategy was used, as neither calibrator nor positive/negative control values were provided. (QC Positive is a technical sample provided with the SOMAscan platform for use as a positive control. Similarly, QC Negative is a technical sample used as a negative control.) Protein outliers were identified if the subject was located outside of either end of distribution using 1.5-fold of IQR given the log10 transformation of the protein level. Collectively, if the number of outliers given an analyte was greater than 85% of total sample size, this analyte was filtered. Next, an orthogonal approach was used to call subject outliers based on IQR. Collectively, if the number of outliers given an analyte was greater than 85% of total number of analytes passed QC, this subject was labeled as an outlier. Overall, 1,128 proteins and 133 subjects passed protein data QC. Genotype data QC and imputation was performed as described above. A total of over 5 million (5,187,563) imputed and directly-genotyped SNPs and 154 individuals were used for final analyses. Population substructure analyses was performed as described above, except samples kept were within the JPT cluster. A total of 132 CSF samples from study by Sasayama and colleagues passed filters on both genomics and proteomics QC. We performed linear regression (additive model), including first two principal component factors from population stratification as covariates.
Reprocessing pQTL using PPMI19 CSF SOMAscan data:
To replicate CSF pQTL, we performed linear regression on all 709 shared proteins using the proteomic and genotype data from PPMI cohort19. We performed protein QC, genotype imputation and QC, and analyses using the same protocols as those used for the data generated in this study and described above. A total of over 7 million (7,392,620) whole-genome sequenced SNPs and 132 CSF samples from study by PPMI19 passed filters on both genomics and proteomics QC. We performed a linear regression (additive model), including age, sex, and first two principal component factors from population stratification as covariates.
Meta-analyses on pQTL using summary statistics from single studies:
To replicate more pQTLs, we performed fixed effect meta-analyses using METAL62 based on inverse-variance weighting. Overall, we performed four different combinations of meta-analyses: 1) meta1_PPMI19_JP17: meta-analysis on both the CSF studies by Sasayama et al. 2017 and by PPMI19; 2) meta2_WUcsf_PPMI19_JP17: meta-analysis on all three CSF studies by Sasayama et al. 2017, by PPMI19, and by Washington University cohort (this study); 3) meta3_WUcsf_WUplasma_WUbrain: meta-analysis on all three-tissue findings from CSF, plasma, and brain by Washington University cohort (this study); 4) meta4_ WUcsf_WUplasma_WUbrain_ PPMI19_JP17: meta-analysis on both the CSF studies by Sasayama et al. 2017 and by PPMI19 plus all three-tissue findings from CSF, plasma, and brain by Washington University cohort (this study).
Replication strategy for plasma pQTL
To identify all previously reported pQTL from large-scale protein-level GWAS, we performed plasma replication analyses using the following strategies. We first searched the reprocessed pQTL results using AddNeuroMed (SOMAscan-based, plasma). Next, we checked summary statistics from INTERVAL (SOMAscan-based, plasma). Finally, we queried other plasma pGWAS findings from EBI-NHGRI using phenoscannerV2 with proxy SNPs (r2 > 0.5).
Reprocessing pQTL using AddNeuroMed plasma SOMAscan data:
To replicate plasma pQTL, we performed linear regression on all proteins using the proteomic and genotype data from the AddNeuroMed consortium 25. A total of over 7 million (7,313,640) imputed and directly-genotyped SNPs and 343 plasma samples from the study by AddNeuroMed passed filters on both genomics and proteomics QC. We performed a linear regression (additive model), including age, gender, first two principal component factors from population stratification, centers, status, visit cohorts, and batch effects as covariates.
Replication strategy for brain pQTL
To identify all previously reported pQTL from large-scale protein-level GWAS, we performed the brain replication analyses using the following strategies. We first searched the processed pQTL results using results from the published brain findings16. Next, we queried all plasma pGWAS findings from EBI-NHGRI using phenoscannerV2 and from our CSF findings.
For each locus, we investigated whether the sentinel SNP or a proxy (r2 > 0.5) was associated with the same target protein (or aptamer) in our study at different defined significance thresholds. For the known category in our primary assessment, we used a p-value threshold of 5×10−8. For the replicated category in our primary assessment, we used a p-value threshold of 5×10−2.
Identification of tissue-specific/shared pQTLs
We first performed mashr28 analyses on our multi-tissue pQTL results given 411 proteins shared by all tissues from the Washington University dataset. We defined the tissue-specificity as LFSR < 0.05 [LFSR is analogous to a false discovery rate] and Z-score to be at least 2-fold difference.
Annotation of pQTL
All significant pQTL (hg19) were annotated using ANNOVAR63 version (2018–04-16) with the option -geneanno in gene-based annotation mode. Genomic features and variants affecting the nearest genes were used for downstream analyses. The bar plots were drawn using the ggplot264 R package.
Testing for genomic feature enrichment
We used Fisher’s exact test (two-sided) for testing the enrichment. The null set was set using p-value > 5e-8 and permutated with the same amount of variants as the positive-set. The various groupings were determined by ANNOVAR version (2018-04-16). We also tested whether our pQTL were enriched for functional and regulatory characteristics using GARFIELD v265.GARFIELD is short for GWAS Analysis of Regulatory or Functional Information Enrichment with LD correction, and it is a method to test feature enrichment by integrating GWAS findings and a large set of regulatory or functional annotations (1005 features in total) mainly from ENCODE and Roadmap epigenomics consortia. It takes into account the LD while annotating and calculating enrichments. The enrichment is quantified as odds ratio (OR).
Identification of pleiotropic regions
To identify unique non-overlapping regions associated with a given an aptamer, we first defined a 2Mb region 1Mb upstream and 1Mb downstream of each significant variant for that aptamer. Within the 2Mb region containing the variant with the smallest p-value any overlapping regions were then merged into the same locus. Owing to the complexity of the major histocompatibility (MHC) region, we assigned genetic region spanning from 25.5 to 34.0Mb on chromosome 6 as one region. To identify whether a region was associated with multiple aptamers, we next used an LD-based clumping approach (LD block from the 1000 Genome Project implemented into the RHOGE66 R package (v0.1), as we also used 1000 Genome Project as our imputation reference panel). Variants were combined into a single region per LD (EUR) defined loci. Any loci associated with more than one protein were identified as pleiotropic regions. The cytoband ID was also annotated. Circos plots were generated using functions from R package circlize67. To measure variance explained by each pleiotropic region, we also calculated the adjusted R-squared value (Supplementary Fig 5-8).
Performing MR using TwoSampleMR R package
Mendelian randomization is a method of using measured variation in genes of known function to examine the causal effect of a modifiable exposure on disease. This method obtains unbiased estimates of the effects of a putative causal variable without conducting a traditional randomized trial. We used the R package TwoSampleMR v0.4.2244. For single SNP remained after clumping, the most basic method, the Wald ratio, was used. This package also implements the harmonization steps before performing MR, and these steps are: 1) Correcting the wrong effect/non-effect alleles; 2) Correcting the strand issues; 3) Fixing the palindromic SNPs; 4) Removing the SNPs with incompatible alleles. The SNPs selected for the analysis were the based on a suggestive threshold of 1×10−5. The beta-coefficients and standard errors (SEs) for the selected variants (pQTL) from this study were used as input of instrumental variables. These instrumental variables were also extracted from the summary statistics from the latest GWAS for the outcome on neurological disease related traits. (Details see Table S26; Briefly, AD-risk GWAS was published in 20193; AD-progression GWAS in 201845; AD-age at onset GWAS in 201746; PD-risk GWAS in 201938; ALS-risk GWAS in 201647; FTD-risk GWAS in 201448; Stroke-risk GWAS in 201849). To check the specificity of protein-neurological disease associations, we also chose asthma-risk GWAS50 as an outcome of non-neurological disease. To test the directionality of exposure causing outcome is valid, we used the directionality_test function from the same R package. The method confirms whether the exposure (protein) and outcome (trait) directions are correct or not.
Colocalization analyses
We performed Bayesian colocalization analysis using the coloc.abf function from the coloc R package68,69 v3.1. We used the default priors with p1= 1×10−4, p2 = 1×10−4, and p12 = 1×10−5. Evidence for colocalization was assessed using the posterior probability (PP) for hypothesis 4 (indicating that there is an association for both protein and disease and that they are driven by the same causal variant(s)). We used PP.H4 > 0.8 as a threshold to suggest that associations were highly likely to colocalize.
For colocalization of pQTLs with disease status:
We downloaded and used the full GWAS summary statistics for each disease/trait from their original publications as the same for MR analysis.
For colocalization of cis-pQTLs with cis-eQTLs, cis-sQTLs from GTEx v8 release:
We downloaded and used the significant cis-eQTLs and cis-sQTLs summary statistics for two single tissues, cortex and whole blood, from GTEx5 (https://gtexportal.org/home/datasets). For cis-sQTLs we used gene-level sQTL results, rather than exon-level sQTLs.
For colocalization analysis of plasma pQTLs with eQTLs from eQTLgen:
We downloaded and used the significant cis- and trans-eQTL summary statistics for blood, from eQTLgen9 (https://www.eqtlgen.org/index.html). In both cases we analyzed cis- and trans-QTLs.
For colocalization of cis-pQTLs with cis-DNA-methylation-QTLs, cis-histone-acetylation-QTLs from ROSMAP:
We downloaded and used the significant cis-DNA-methylation-QTL summary statistics for brain tissue, from ROSMAP70 (http://mostafavilab.stat.ubc.ca/xQTLServe/). We downloaded the significant cis-histone-acetylation-QTL summary statistics (assigning to up to 10Mb upstream of the transcription start site given the same gene) for brain tissue, from ROSMAP70 as well. To ensure that DNA-methylation-QTLs affecting pQTLs are mediated by eQTLs, we further subset the DNA-methylation-QTLs-pQTLs colocalization result with eQTLs-pQTLs colocalization result.
For colocalization of cis-pQTLs with cell-type-specific cis-eQTLs from ROSMAP:
We identified the neuron-, oligodendrocyte-, microglia-, and astrocyte-eQTL data using a pseudo-bulk strategy on snRNA-seq (N=48) from ROSMAP data71. In total, we recreated the expression matrices on five cell-types (microglia, excitatory neurons, inhibitory neurons, oligodendrocytes, and astrocytes). We next identified cis-eQTLs for each cell type using fastqtlv2.072 after integrating with the whole-genome sequencing data from ROSMAP (N=39). Using both the nominal and permutation modes, we followed the significant eGene calling approach from the GTEx pipeline. We used different priors because the pseudo-bulk derived cell-type specific eQTLs were underpowered compared with bulk-level pQTLs with p1 as 1×10−4, p2 as 1×10−2, and p12 as 1×10−3. The results can be found in Supplementary Fig 9 & Table S35).
Overlap of proteins with pQTLs and drug targets
To obtain information on drugs that target proteins with pQTLs from this study, we used the DrugBank database (as of 1/3/2020)73. This is a manually curated database that maintains profiles for >15,000 drugs (including FDA-approved and experimental drugs). For our analysis, we focused on the protein target for each drug. For each protein assayed, we identified all drugs in the DrugBank with a matching protein target based on UniProt ID, annotated via https://www.uniprot.org/database/DB-0019. We further integrated the MR results on proteins as drug targets.
Randomization
Data collection for the proteomics was randomized. All samples were randomly assigned to a profiling pool prior to proteomics measurement, considering age, sex, and disease status. Data collection for the genotype was not randomized or blocked.
Sample sizes
No statistical methods were used to predetermine sample sizes within each tissue type, but our sample sizes are much larger or comparable to those in previous publications13,14,16.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability Statement
Three tissues from Knight ADRC dataset for discovery: Both summary statistics and individual-level data have been uploaded to NIAGADS (The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site) repository at https://www.niagads.org/datasets/ng00102. Summary statistics (pQTL) data is freely available, as the data exceeds 500Gb, so please email niagads@pennmedicine.upenn.edu to set up an FTP transfer of the data. Summary association results can also be explored through Online Neurodegenerative Trait Integrative Multi-Omics Explorer, ONTIME (https://ontime.wustl.edu/), a PheWeb (v1.1.14)-based browser.
CSF-Sasayama2017 dataset for replication:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83711
Plasma-AddNeuroMed dataset for replication: https://www.synapse.org/#!Synapse:syn4988768
Drug targets were queried using DrugBank database collected via UniProtKB (as of 1/3/2020) at https://www.uniprot.org/database/DB-0019.
Extended Data
Extended Data Fig. 1. QC pipeline.
QC on both proteins (a to c) and samples (d) were described as follows: (a) Flowchart of CSF protein level QC, starting from 1305; after step-1, Limit Of Detection VS 2-StDeviation, 807 proteins were kept with a pass-rate >= 85%; after step-2, given Max Difference of Scale Factor < 0.5, 749 proteins were kept; after step-3, given Coefficient of Variation (of calibrator) < 0.15 & step-4, given IQR, sum(outliers) < 15%, 746 proteins were kept. After step-5, 713 proteins that shared by < 30 samples (shared by ~80% of the subject outliers) were kept. (b) Flowchart of plasma protein level QC, starting from 1305; after step-1, 1301 proteins were kept with a pass-rate >= 85%; after step-2, 956 proteins were kept; after step-3 & step-4, 955 proteins were kept. After step-5, 931 proteins that shared by < 10 samples were kept. (c) Flowchart of brain protein level QC, starting from 1305; after step-1, 1109 proteins were kept with a pass-rate >= 85%; after step-2, 1107 proteins were kept; after step-3 & step-4, given IQR, sum(outliers) < 15%, 1106 proteins were kept. After step-5, 1079 proteins that shared by < 21 samples were kept. (d) Table of sample size after each step of QC in genotype and proteomics. Within each tissue (1st column), we profiled proteomics from 1300 CSF, 648 plasma and 459 samples (2nd column). From unique donors in proteomics data (3rd column), we first kept donors with genotyping array data (4th column). We next kept only the donors with a European ancestry after checking principal components (5th column). Moreover, we kept donors that were not close with each other (PI_HAT < 0.05) after checking identity by descent (6th column). Finally, the samples remained only passing both the genotype and protein data QC (7th column).
Extended Data Fig. 2. Reproducibility of proteomic data.
(a) Table of total sample size for each tissue before and after QC, including the biological and technical replicates. (b) Venn diagram on the designed donor overlap across tissues. (c) Scatterplot of 321 subjects with both longitudinal and baseline samples from CSF indicates a Pearson correlation coefficient of 0.995 (95% confidence interval from 0.995 to 0.995). (d) Scatterplot of 11 subjects with both fasted and nonfasted samples from plasma indicates a Pearson correlation coefficient of 0.907 (95% confidence interval from 0.904 to 0.911). (e) Scatterplot of one subject with both longitudinal and baseline samples from plasma indicates a Pearson correlation coefficient of 0.938 (95% confidence interval from 0.930 to 0.945). (f) Scatterplot of one subject with two technical replicates from brain indicates a Pearson correlation coefficient of 0.976 (95% confidence interval from 0.976 to 0.981). All statistical tests used were two-sided from (c) to (f).
Extended Data Fig. 3. Overview of the sample size and number of pQTLs from pQTL studies mentioned in this paper and the summary statistics from the meta-analyses.
(a) Scatter plot of sample size (log10-scaled) and number of total pQTLs after clumping or unique proteins when no clumping was performed (log10-scaled). Dot color represents the tissue type; dot size represents total number of proteins profiled. (b) Table of these nine datasets listed the exact numbers for drawing the scatter plot. (c) Table of three different combinations of meta-analyses: 2) meta2_WUcsf_PPMI19_JP17: meta-analysis on all three CSF studies by Sasayama and colleagues published in 2017, by PPMI released in 2019, and by Washington University cohort (this study); 3) meta3_WUcsf_WUplasma_WUbrain: meta-analysis on all three-tissue findings from CSF, plasma and brain respectively by Washington University cohort (this study); 4) meta4_ WUcsf_WUplasma_WUbrain_ PPMI19_JP17: meta-analysis on both the CSF studies by Sasayama and colleagues published in 2017 and by PPMI released in 2019 plus all three-tissue findings from CSF, plasma and brain respectively by Washington University cohort (this study). The columns include number of proteins in common, number of protein-level GWAS hits after meta-analysis, number of protein-level GWAS hits before meta-analysis using only the common proteins within each tissue for each combination. (d) Stacked Manhattan plots for all three different combinations of meta-analyses. The darkred line represents P = 5×10−8.
Extended Data Fig. 4. Disease stratified analysis on comparing pQTLs effect size.
To investigate of disease status effect on pQTLs, we performed linear regression on the same protein-loci pairs (before conditioning on top variants) identified from above default model using three additional models: (a) joint analysis but with disease status as another covariate (CO vs non-CO). Pearson correlation coefficient was 0.999 (p-value < 2.2×10−16, 95%CI = 0.999 to 0.999), 0.999 (p-value = 4.3×10−202, 95%CI =0.999 to 0.999), 0.999 (p-value = 9.5×10−52, 95%CI = 0.999 to 0.999) for CSF, plasma, and brain respectively. Sample size for this joint analysis was 835, 529, and 380 for CSF, plasma, and brain respectively. (b) AD case (CA) only using the same covariates as default model. Pearson correlation coefficient of 0.991 (p-value = 3.9×10−160, 95%CI =0.988 to 0.993), 0.989 (p-value = 1.8×10−83, 95%CI =0.983 to 0.992), 0.998 (p-value = 2.4×10−29, 95%CI =0.995 to 0.999) for CSF, plasma, and brain respectively. Sample size for this AD case (CA) only analysis was 217, 168, and 248 for CSF, plasma, and brain respectively. (c) Cognitive unimpaired (CO) only using the same covariates as default model. Pearson correlation coefficient of 0.999 (p-value = 5.2×10−234, 95%CI =0.998 to 0.999), 0.998 (p-value = 1.17×10−122, 95%CI =0.997 to 0.999), 0.602 (p-value = 0.002, 95%CI =0.262 to 0.809) for CSF, plasma, and brain respectively. Sample size for this cognitive unimpaired (CO) only analysis was 614, 357, and 24 for CSF, plasma, and brain respectively. The relatively low correlation in default model comparison with control only in brain samples was due to much smaller sample size as a control for brain samples. All statistical tests used were two-sided from (a) to (c).
Extended Data Fig. 5. Global view of pleiotropic regions in CSF.
In total, 59 Pleiotropic regions passing genome-wide significance threshold (5×10−8) in CSF (sample size = 835). Unique non-overlapping regions associated with a given SOMAmer were first defined as 1-Mb region upstream and downstream of each significant variant for that SOMAmer. Within the region (2Mb) containing the variant with the smallest P value, any overlapping regions were then merged into the same locus. Next, an LD-based clumping approach was adapted to identify whether a region was associated with multiple SOMAmers. Variants were combined into a single region per LD (EUR) defined loci. Any loci associated with more than one protein were identified as pleiotropic regions. Genomic locations of pQTLs were visualized by a squared-Manhattan plot. Dark-green represents cis-pQTLs; gold represents trans-pQTLs. X-axis indicates the positions of the top variant; and Y-axes indicates the gene encoding the protein. All pleiotropic genomic regions are annotated at the top of each plot along the X-axis.
Extended Data Fig. 6. Global view of pleiotropic regions in plasma.
In total, 34 pleiotropic regions passing genome-wide significance threshold (5×10−8) in plasma (sample size = 529). Genomic locations of pQTLs were visualized by a squared-Manhattan plot, same as Extended Data Fig.5.
Extended Data Fig. 7. Global view of pleiotropic regions in brain.
In total, 10 pleiotropic regions passing genome-wide significance threshold (5×10−8) in brain (sample size = 380). Genomic locations of pQTLs were visualized by a squared-Manhattan plot, same as Extended Data Fig.5.
Extended Data Fig. 8. Tissue specificity exploration with permissive thresholds.
To determine whether our tissue-specificity results were biased by statistical power, we performed similar analyses with two more permissive p-values on the 411 proteins. (a) Venn diagrams of all pQTLs across all three tissues by fixing genome-wide significance threshold (5×10−8) for all three tissues. (b) Venn diagrams of all pQTLs across all three tissues by fixing genome-wide significance threshold for one tissue and 0.001 for the other two tissues. For example, when checking CSF pQTLs shared in plasma or brain, we chose 5×10−8 as threshold for CSF and 0.001 for plasma or brain. (c) Venn diagrams of all pQTLs across all three tissues by fixing genome-wide significance threshold for one tissue and 0.05 for the other two tissues. For example, when checking CSF pQTLs shared in plasma or brain, we chose 5×10−8 as threshold for CSF and 0.05 for plasma or brain.
Extended Data Fig. 9. Tissue specificity exploration with plasma result from INTERVAL study.
To further demonstrate that tissue-specificity findings are not a product of different sample size, we performed similar comparisons by analyzing the plasma pQTLs from the INTERVAL study on 616 proteins that passed QC in our CSF, brain and plasma INTERVAL. (a) Venn diagrams of proteins passing QC across all three tissues: CSF and brain results are from WashU cohort, plasma result is from INTERVAL study. (b) Venn diagrams of all pQTLs across all three tissues by fixing genome-wide significance threshold (5×10−8) for all three tissues. (c) Venn diagrams of all pQTLs across all three tissues by fixing genome-wide significance threshold for one tissue and 0.001 for the other two tissues. For example, when checking CSF pQTLs shared in plasma or brain, we chose 5×10−8 as threshold for CSF and 0.001 for plasma or brain. (d) Venn diagrams of all pQTLs across all three tissues by fixing genome-wide significance threshold for one tissue and 0.05 for the other two tissues. For example, when checking CSF pQTLs shared in plasma or brain, we chose 5×10−8 as threshold for CSF and 0.05 for plasma or brain.
Extended Data Fig. 10. Properties of pQTLs.
(a) Dot plots of -log10(P) from all significant associations (via linear regression) against the distance of sentinel SNPs from TSS within each tissue. (b) Dot plots of absolute effect size associated with MAF within each tissue. (c) Forest plot of enrichment on the predicted functional annotation classes of pQTLs versus null sets of variants from permutation within each tissue (Data are presented as mean values of Odds Ratio +/− 95% confidence interval from Fisher’s Exact Test) and Bar plots of the proportion of variants annotate in each class. (Note: Features on exonic_splicing/ncRNA_splicing/splicing/UTR5_UTR3 are not shown due to not all tissues have these features). (d) Histograms of variance explained by conditionally independent variants within each tissue. For CSF, the mean = 0.141, standard deviation = 0.144, mode = 0.061; For plasma, the mean = 0.157, standard deviation = 0.125, mode = 0.188; For brain, the mean = 0.208, standard deviation = 0.151, mode = 0.092.
Supplementary Material
Acknowledgments
We thank all the participants and their families, as well as the many involved institutions and their staff.
Funding: This work was supported by grants from the National Institutes of Health (R01AG044546 (CC), P01AG003991(CC, JCM), RF1AG053303 (CC), RF1AG058501 (CC), U01AG058922 (CC), R01NS118146 (BAB), and R01AG057777 (OH)), the Alzheimer Association (NIRG-11-200110 (CC), BAND-14-338165 (CC), AARG-16-441560 (CC) and BFG-15-362540 (CC)). This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, and the Departments of Neurology and Psychiatry at Washington University School of Medicine. The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50AG05681(JCM), P01AG03991(JCM), and P01AG026276(JCM).
Footnotes
Competing Interests Statement
CC receives research support from: Biogen, EISAI, Alector and Parabon. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. CC is a member of the advisory board of Vivid genetics, Halia Therapeutics and ADx Healthcare. The remaining authors declare no competing interests.
References
- 1.Altshuler D, Daly MJ & Lander ES Genetic Mapping in Human Disease. Science 322, 881–888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris AP et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics 44, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunkle BW et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nature Genetics 51, 414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claussnitzer M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguet F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv 787903 (2019) doi: 10.1101/787903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Wijst MGP et al. Single-cell eQTLGen Consortium: a personalized understanding of disease. arXiv:1909.12550 [q-bio] (2019). [Google Scholar]
- 7.Aguet F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamazon ER, Zwinderman AH, Cox NJ, Denys D. & Derks EM Multi-tissue transcriptome analyses identify genetic mechanisms underlying neuropsychiatric traits. Nature Genetics 1 (2019) doi: 10.1038/s41588-019-0409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Võsa U. et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 447367 (2018) doi: 10.1101/447367. [DOI] [Google Scholar]
- 10.Sun BB et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suhre K. et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nature Communications 8, 14357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folkersen L. et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLOS Genetics 13, e1006706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deming Y. et al. Genetic studies of plasma analytes identify novel potential biomarkers for several complex traits. Scientific Reports 6, 18092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasayama D. et al. Genome-wide quantitative trait loci mapping of the human cerebrospinal fluid proteome. Hum Mol Genet 26, 44–51 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Kauwe JSK et al. Genome-Wide Association Study of CSF Levels of 59 Alzheimer’s Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation. PLOS Genetics 10, e1004758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robins C. et al. Genetic control of the human brain proteome. bioRxiv 816652 (2019) doi: 10.1101/816652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gold L. et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLOS ONE 5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haddick PCG et al. A Common Variant of IL-6R is Associated with Elevated IL-6 Pathway Activity in Alzheimer’s Disease Brains. J. Alzheimers Dis. 56, 1037–1054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marek K. et al. The Parkinson Progression Marker Initiative (PPMI). Progress in Neurobiology 95, 629–635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovestone S. et al. AddNeuroMed—The European Collaboration for the Discovery of Novel Biomarkers for Alzheimer’s Disease. Annals of the New York Academy of Sciences 1180, 36–46 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Kamat MA et al. PhenoScanner V2: an expanded tool for searching human genotype–phenotype associations. Bioinformatics (2019) doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayaratnam S, Khoo AKL & Basic D. Rapidly progressive Alzheimer’s disease and elevated 14–3-3 proteins in cerebrospinal fluid. Age Ageing 37, 467–469 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Foote M. & Zhou Y. 14–3-3 proteins in neurological disorders. Int J Biochem Mol Biol 3, 152–164 (2012). [PMC free article] [PubMed] [Google Scholar]
- 24.Ibanez L. et al. Overlap in the Genetic Architecture of Stroke Risk, Early Neurological Changes, and Cardiovascular Risk Factors. Stroke 50, 1339–1345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lourdusamy A. et al. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet 21, 3719–3726 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker RL et al. Genetic Control of Expression and Splicing in Developing Human Brain Informs Disease Mechanisms. Cell 179, 750–771.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orozco LD et al. Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration. Cell Reports 30, 1246–1259.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Urbut SM, Wang G, Carbonetto P. & Stephens M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nature Genetics 51, 187–195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillary RF et al. Genome and epigenome wide studies of neurological protein biomarkers in the Lothian Birth Cohort 1936. Nat Commun 10, 3160–3160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suhre K, McCarthy MI & Schwenk JM Genetics meets proteomics: perspectives for large population-based studies. Nature Reviews Genetics 1–19 (2020) doi: 10.1038/s41576-020-0268-2. [DOI] [PubMed] [Google Scholar]
- 31.Yao C. et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nature Communications 9, 3268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paré G. et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 4, e1000118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndungu A, Payne A, Torres JM, Bunt M. van de & McCarthy MIA Multi-tissue Transcriptome Analysis of Human Metabolites Guides Interpretability of Associations Based on Multi-SNP Models for Gene Expression. The American Journal of Human Genetics 0, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cruchaga C. et al. Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet 21, 4558–4571 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kibinge NK, Relton CL, Gaunt TR & Richardson TG Characterizing the Causal Pathway for Genetic Variants Associated with Neurological Phenotypes Using Human Brain-Derived Proteome Data. Am J Hum Genet 106, 885–892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del-Aguila JL et al. A single-nuclei RNA sequencing study of Mendelian and sporadic AD in the human brain. Alzheimer’s Research & Therapy 11, 71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alector Inc. First in Human Study for Safety and Tolerability of AL003. - Full Text View - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03822208.
- 38.Nalls MA et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. The Lancet Neurology 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bethea JW Clinical Anesthesia, 6th Edition. Anesthesiology 112, 767–768 (2010). [Google Scholar]
- 40.Camerino GM et al. Elucidating the Contribution of Skeletal Muscle Ion Channels to Amyotrophic Lateral Sclerosis in search of new therapeutic options. Scientific Reports 9, 3185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savitz SI et al. The novel beta-blocker, carvedilol, provides neuroprotection in transient focal stroke. J Cereb Blood Flow Metab 20, 1197–1204 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nature Genetics 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Gagliano Taliun SA et al. Exploring and visualizing large-scale genetic associations by using PheWeb. Nature Genetics 52, 550–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemani G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del-Aguila JL et al. Assessment of the Genetic Architecture of Alzheimer’s Disease Risk in Rate of Memory Decline. J. Alzheimers Dis. 62, 745–756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang K. et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nature Neuroscience 20, 1052–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Rheenen W. et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nature Genetics 48, 1043–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrari R. et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol 13, 686–699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malik R. et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics 50, 524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demenais F. et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet 50, 42–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H. VennDiagram: Generate High-Resolution Venn and Euler Plots. (2018). [Google Scholar]
- 52.Morris JC The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43, 2412–2414 (1993). [DOI] [PubMed] [Google Scholar]
- 53.Mirra SS et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–486 (1991). [DOI] [PubMed] [Google Scholar]
- 54.Khachaturian ZS Diagnosis of Alzheimer’s disease. Arch. Neurol. 42, 1097–1105 (1985). [DOI] [PubMed] [Google Scholar]
- 55.Sattlecker M. et al. Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement 10, 724–734 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Williams SA et al. Plasma protein patterns as comprehensive indicators of health. Nature Medicine 1–7 (2019) doi: 10.1038/s41591-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huber W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Meth 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consortium UniProt. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howie BN, Donnelly P. & Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruim RJ et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willer CJ, Li Y. & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang K, Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164–e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickham H. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, 2009). doi: 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
- 65.Iotchkova V. et al. GARFIELD classifies disease-relevant genomic features through integration of functional annotations with association signals. Nature Genetics 51, 343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mancuso N. et al. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. The American Journal of Human Genetics 100, 473–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu Z, Gu L, Eils R, Schlesner M. & Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Wallace C. Statistical Testing of Shared Genetic Control for Potentially Related Traits. Genetic Epidemiology 37, 802–813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giambartolomei C. et al. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLOS Genetics 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathys H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ongen H, Buil A, Brown AA, Dermitzakis ET & Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wishart DS et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34, D668–D672 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Three tissues from Knight ADRC dataset for discovery: Both summary statistics and individual-level data have been uploaded to NIAGADS (The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site) repository at https://www.niagads.org/datasets/ng00102. Summary statistics (pQTL) data is freely available, as the data exceeds 500Gb, so please email niagads@pennmedicine.upenn.edu to set up an FTP transfer of the data. Summary association results can also be explored through Online Neurodegenerative Trait Integrative Multi-Omics Explorer, ONTIME (https://ontime.wustl.edu/), a PheWeb (v1.1.14)-based browser.
CSF-Sasayama2017 dataset for replication:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83711
Plasma-AddNeuroMed dataset for replication: https://www.synapse.org/#!Synapse:syn4988768
Drug targets were queried using DrugBank database collected via UniProtKB (as of 1/3/2020) at https://www.uniprot.org/database/DB-0019.