Significance
The cellular functions of Hsp70 molecular chaperones rely on their ability to bind many clients and discriminate folded from unfolded substrates. We find that the Escherichia coli Hsp70 substrate binding domain (SBD) “reads” sequence motifs without regard for backbone orientation and selects binding modes that optimize residue compatibility with pockets in the SBD binding cleft. Multiple potential binding sites on the unfolded substrate interacted with the SBD independently in roughly the same order of apparent affinities observed for the peptide models of the sites. Our results show in detail how an Hsp70 chaperone binds to an unfolded protein substrate, which will inform efforts to predict Hsp70 binding sites and shed light on chaperone functions.
Keywords: Hsp70 molecular chaperone, DnaK, NMR, crystallography, substrate binding
Abstract
Heat shock protein 70 (Hsp70) chaperones bind many different sequences and discriminate between incompletely folded and folded clients. Most research into the origins of this “selective promiscuity” has relied on short peptides as substrates to dissect the binding, but much less is known about how Hsp70s bind full-length client proteins. Here, we connect detailed structural analyses of complexes between the Escherichia coli Hsp70 (DnaK) substrate-binding domain (SBD) and peptides encompassing five potential binding sites in the precursor to E. coli alkaline phosphatase (proPhoA) with SBD binding to full-length unfolded proPhoA. Analysis of SBD complexes with proPhoA peptides by a combination of X-ray crystallography, methyl-transverse relaxation optimized spectroscopy (methyl-TROSY), and paramagnetic relaxation enhancement (PRE) NMR and chemical cross-linking experiments provided detailed descriptions of their binding modes. Importantly, many sequences populate multiple SBD binding modes, including both the canonical N to C orientation and a C to N orientation. The favored peptide binding mode optimizes substrate residue side-chain compatibility with the SBD binding pockets independent of backbone orientation. Relating these results to the binding of the SBD to full-length proPhoA, we observe that multiple chaperones may bind to the protein substrate, and the binding sites, well separated in the proPhoA sequence, behave independently. The hierarchy of chaperone binding to sites on the protein was generally consistent with the apparent binding affinities observed for the peptides corresponding to these sites. Functionally, these results reveal that Hsp70s “read” sequences without regard to the backbone direction and that both binding orientations must be considered in current predictive algorithms.
Heat shock protein 70 (Hsp70) molecular chaperones are central players in protein homeostasis (1, 2). In performing their many cellular functions, they bind to a wide array of protein clients without relying on a defined consensus binding motif; nonetheless, they select for substrates that are incompletely folded, are misfolded, or present unstructured regions (3). We refer to the ability of Hsp70s chaperones to bind a broad spectrum of sites but to distinguish sequence properties diagnostic for incompletely folded substrates as “selective promiscuity.” An ongoing challenge is to develop a comprehensive, predictive understanding of binding site selection by Hsp70s despite their highly permissive binding. Such predictive understanding will shed light on the functional diversity of Hsp70 functions, including facilitating protein folding, inhibiting aggregation, delaying folding to enable membrane translocation, and handing off to downstream chaperone partners.
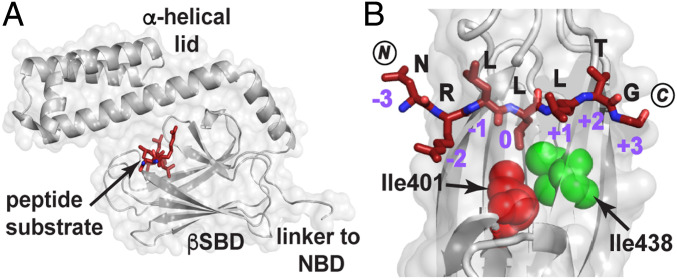
Substrate binding to Hsp70s is achieved via their C-terminal substrate binding domain (SBD), the affinity of which is governed by the nucleotide binding state of the adjacent N-terminal nucleotide-binding domain (4, 5). The SBD is composed of a β-sandwich subdomain (βSBD) and an α-helical lid (Fig. 1A) followed by a flexible C-terminal tail. Considerable biochemical data have shown that there is a pattern of substrate residues in Hsp70 binding sites (6–10). They comprise short linear motifs of five to seven residues enriched in hydrophobic residues and often flanked by positively charged residues. Structural analyses, primarily carried out on the Escherichia coli Hsp70 DnaK, have shed light on how the canonical binding site accommodates such sequences (Fig. 1B). Crystal structures of the DnaK SBD bound to short peptides showed that only five “core” residues of the sequence directly interact with five pockets in the canonical substrate binding cleft of the βSBD (dubbed –2, –1, 0, +1, and +2) (11, 12). All peptides bind in an extended conformation and form an extensive hydrogen bond network with the residues in the cleft. The central pocket, termed the 0th site, is the most restrictive in terms of side chains accommodated and is most frequently occupied by leucine based on structural data, although isoleucine and less commonly, valine or proline can be bound here as well (11–13). Comparisons of X-ray structures and analyses based on peptide array data (9, 10) have led to the conclusion that the –1 and –2 positions show the next highest binding bias, while positions +1 and +2 are quite nonselective, although cysteine, aspartic acid, and glutamic acid are disfavored in all the βSBD pockets. Strikingly, the ability of βSBD subsites to accommodate different residues does not arise from induced fit, as the overall conformation of the βSBD backbone is maintained in all solved structures of SBD–peptide complexes.
Fig. 1.
How the DnaK SBD binds a model peptide. (A) The crystal structure of the SBD of DnaK (gray) bound to the model peptide NR (NRLLLTG; maroon; PDB ID code 1DKZ) showing the peptide bound to a cleft on the β-subdomain covered by the α-helical lid (11). (B) Top view of the SBD substrate binding cleft showing the mode of binding of NR to the five pockets created by the topography of this domain. Note in particular the deep 0th pocket here occupied by L4. The SBD Ile401 (red) and Ile438 (green) are shown as spheres. Residues 507 to 603 and residues 404 to 429 are not shown to better visualize the bound peptide. (Structures here and elsewhere are depicted using PyMol [Schrodinger LCC].) NBD, nucleotide binding domain.
There are several remarkable features of substrate binding by Hsp70s and several major gaps in current knowledge that present barriers to understanding how these chaperones interact with their substrates. 1) Different modes of binding within the canonical substrate binding site are distinguished by small energy differences. The model peptide NRLLLTG (NR) was shown in two different crystals to bind with a “register shift,” where the sequence shifts laterally to allow a different leucine residue to occupy the central 0th position (11, 12). 2) While not widely considered in efforts to predict binding site preferences of Hsp70s, evidence shows that binding can occur with the substrate positioned either N to C, as originally reported by Zhu et al. (11), or C to N (sometimes referred to as “reverse binding”), as observed first in the structure of a complex of the Hsp70 family member HscA with a fragment of its intrinsic substrate IscU (14). In subsequent work, Tapley et al. (15) used fluorescence energy transfer to show that the IscU peptide bound in the same reverse or C to N orientation to DnaK, and Zahn et al. (12) observed this orientation for several proline-rich analogs of the antimicrobial pyrrocidin and an NR variant, NRLILTG, bound to the DnaK SBD. The initial hypothesis was that the presence of proline residues in the model substrates led to favoring of the C to N orientation (14, 15), but the result from Zahn et al. (12) indicates that this is not the only explanation for the orientational variation in chaperone–substrate binding. 3) Efforts to understand the details of the Hsp70–substrate interaction have relied nearly exclusively on short peptides as model substrates. The details of the chaperone–substrate interaction have been assumed to be mechanistically similar whether a sequence binds to Hsp70 as a short peptide or as part of a whole protein. However, this assumption does not consider a few key factors. First, site accessibility directly impacts the ability of Hsp70s to bind hydrophobic stretches within protein substrates. A sequence that is easily accessible as a peptide might not be readily available for the chaperone binding in the context of a large protein since even unfolded protein states can have significant elements of ordered structure (16). Second, it is not clear whether regions flanking the five core substrate residues influence the binding preferences of Hsp70s. Lastly, multiple chaperone binding sites can be available in the same polypeptide chain, and when this situation obtains, there may be cooperativity or nonadditivity in the binding outcome.
In this study, we sought a deeper understanding of the origins of selective promiscuity in Hsp70 binding, and we addressed the gaps in understanding listed above. Our strategy was to explore in depth the binding of DnaK to a 471-residue-long unfolded protein substrate, the cytoplasmic precursor to the periplasmic protein alkaline phosphatase (proPhoA), which has proven to be biophysically tractable (17–20) and is also physiologically relevant in this system (21, 22). We dissected the interactions of the DnaK SBD with each of the potential binding sites on proPhoA identified in peptide array data (9). We used an integrated approach by determining the mode of binding of peptides corresponding to putative chaperone binding sites with the DnaK SBD crystallographically as well as deducing the nature of their interaction in solution by methyl-TROSY NMR spectroscopy and their preferred bound orientation by cross-linking and paramagnetic relaxation enhancement (PRE) NMR experiments. We then correlated our findings with NMR data for SBD binding to the full-length protein substrate. Our results provide insights into the ability of the Hsp70 SBD to accommodate different substrate sequences and demonstrate that optimizing the characteristics and order of residues occupying SBD binding pockets determines binding site selection regardless of the backbone orientation of a given sequence. Importantly, our results reveal that while binding of the Hsp70 SBD to the several potential binding sites on full-length proPhoA is to a great extent hierarchical in terms of affinity of the sites and largely displays additive behavior for these sites as independent entities, this picture is incomplete. We uncovered a high-affinity binding site that was unexpected based on peptide array data (9). These results afford a fuller picture of the nature of Hsp70–protein substrate complexes than previously available, and this picture will markedly influence efforts to predict Hsp70 binding sites and will inform our understanding of Hsp70 functional actions on its substrates.
Results
The DnaK SBD Binds Multiple Sites in the Unfolded Protein Substrate proPhoAS4.
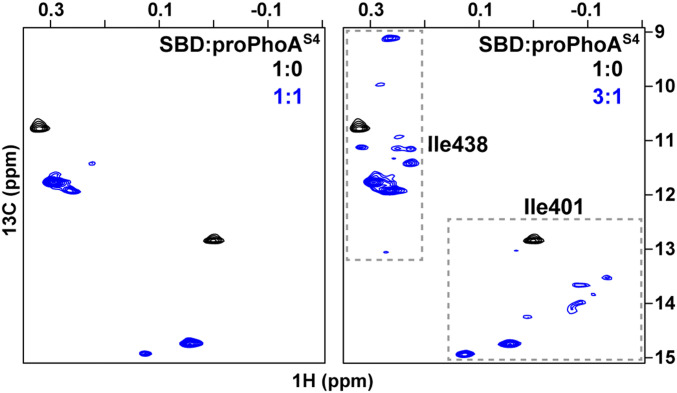
Binding of DnaK to proPhoA was anticipated based on peptide array results (9) and genetic studies (21, 22). The peptide array results suggested that proPhoA contains multiple potential binding sites. To assess directly how DnaK binds to this large protein, we examined by NMR the complex between unfolded proPhoA and a DnaK SBD construct that is selectively labeled (1H,13C) at Ile δ1-methyl positions in a 2H, 12C background (23). Past work shows that the δ1-methyl resonance positions of Ile401 and Ile438 of the SBD in 1H-13C-HMQC (heteronuclear multiple quantum coherence) spectra report on occupancy of the substrate binding cleft (SI Appendix, Fig. S1A). The side chains of these two isoleucines are in direct contact with the substrate residue occupying the 0th site in the SBD binding cleft (Fig. 1B), and their δ1-methyl chemical shifts are exquisitely sensitive to the identity of the bound sequence (13, 24, 25). At a low ratio of DnaK SBD to proPhoAS4 (a variant with the four native cysteine residues mutated to serine to improve the properties of the unfolded protein) (SI Appendix, SI Text and Fig. S1B), we observed two sets of Ile401 and Ile438 resonances. Upon increasing the SBD to proPhoAS4 ratio, multiple resonances were seen for these two isoleucines, which suggested, consistent with the peptide array data (9), that multiple sequences within proPhoA can be bound by the chaperone (Fig. 2). We utilize the DnaK SBD to obtain better-quality NMR spectra than those for the complex between full-length DnaK and proPhoA. The SBD recapitulates binding of full-length DnaK, as shown by 1) the ability of a strong SBD binding peptide p5 (26) to displace bound protein from either full-length DnaK or the SBD (SI Appendix, Fig. S2A), which argues that binding is mediated by the canonical substrate binding cleft in the SBD, and 2) overlap of the Ile401 and Ile438 δ1-methyl resonances observed for full-length DnaK or SBD in complexes with peptides containing the potential binding sites of proPhoA (Fig. 3 and SI Appendix, Fig. S2B).
Fig. 2.
Multiple sequences in proPhoAS4 are bound by the DnaK SBD. The Ile401 and Ile438 region of 1H-13C-HMQC spectra of ILV-13CH3–labeled DnaK SBD in the presence of unlabeled, unfolded proPhoAS4 at low (Left) and high (Right) chaperone to substrate ratios showing several resonances arising from proPhoAS4 binding.
Fig. 3.
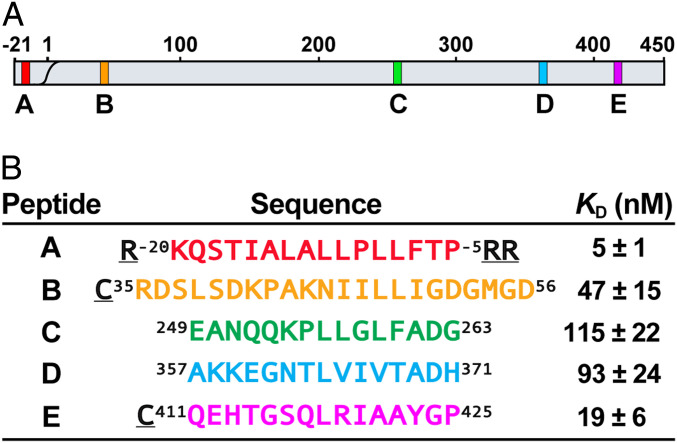
Potential DnaK binding sites on proPhoA and designed peptide models. (A) Schematic of the proPhoA sequence with strong DnaK binding sites identified from a proPhoA peptide array (9) labeled as A, B, C, D, and E and colored in red, orange, green, blue, and magenta, respectively (this color code is used in all figures). (B) Model peptides and their SBD apparent binding affinities (KD) (SI Appendix, Fig. S4). (Underlined residues are not the part of proPhoA sequence.)
Experimental Approach to Probe the Details of proPhoA Binding to the DnaK SBD.
We designated the strongest binders among the 13-residue-long peptides in the peptide array as sites A to E and postulated that they reflect five potential DnaK binding sites in the unfolded protein (Fig. 3A) (9). In keeping with many previous observations, the DnaK binding sequences identified in the peptide array are highly hydrophobic (6–10) and have high predicted aggregation propensity (27) (SI Appendix, Fig. S3A). Except for site A, which corresponds to the signal sequence of the secretory precursor of PhoA, all other sites adopt β-strand conformations in the native structure of mature PhoA, and site E is part of the dimer interface (SI Appendix, Fig. S3B). Thus, these sites would only be exposed in nonnative states of protein substrate and would benefit from chaperone binding to minimize aggregation or misfolding. To elucidate whether and how DnaK binds to the potential binding sites in the context of full-length unfolded proPhoA, we designed peptides containing the seven-residue core DnaK binding sequences A to E as predicted from the peptide array data plus an additional four to eight flanking residues on their N and C termini (Fig. 3B). As expected, all these peptides bound to the DnaK SBD in solution; their apparent dissociation constants (KD) ranged from 5 to 115 nM, with the order A > E > B > D > C (Fig. 3B and SI Appendix, Fig. S4). In addition, for some experiments we used shorter versions of peptides containing only the core binding sequences. We observed no differences in the mode of binding (orientation and binding register) between the longer and shorter versions of the peptides that encompass the core binding sequences, arguing that the flanking sequences in large measure did not participate in binding.
We deployed an integrated structural dissection of the binding of the DnaK SBD to all peptides representing potential proPhoA chaperone binding sites. Crystal structures of the SBD complexed with the peptides were solved (SI Appendix, Table S1), and the positions of the δ1-methyl resonances of SBD residues Ile401 and Ile438 were followed in 1H-13C-HMQC NMR spectra for all complexes. Because previous results (12, 15) and our crystallographic data opened the possibility that binding might occur in either an N to C or a C to N orientation, two assays that reported on the orientation of the SBD-bound peptide in solution were applied to the SBD–peptide complexes (in the next paragraph), and the results were then used to deduce the best-fit binding mode of each peptide. The peptide data were then used to identify the binding sites and mode of binding of the SBD to the proPhoAS4 protein substrate.
We designed two assays that report on the binding orientation of proPhoA peptides in complexes with SBD: 1) PREs (28) and 2) disulfide-mediated cross-linking (SI Appendix, Fig. S5A). For both experiments, the peptides representing binding sites on proPhoA were modified with an N-terminal cysteine residue; the modified peptides showed no difference from their wild-type counterparts in binding to the DnaK SBD (SI Appendix, Fig. S5B). For PREs, the thiol moieties of the cysteines were labeled with a spin label MTSL [(1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate], and resonance broadening was monitored in 1H-15N-TROSY-HSQC (heteronuclear single quantum coherence) spectra of the SBD bound to the peptides. The flattened trapezoidal shape of the SBD lends itself to discrimination of the peptide orientation based on which side was affected when the paramagnetically labeled peptide was bound. For the second approach, we used two SBD variants with cysteine residues introduced on either side of the binding cleft at position 425 or 458 (referred to as SBDCys425 and SBDCys458) (15); the Cys-containing SBD mutants showed no difference in their substrate peptide binding (SI Appendix, Fig. S5 C and D). We monitored preferential disulfide bond formation between the N-terminal cysteine residues on the bound peptides and either of the Cys-SBD variants; formation of a disulfide bond to SBDCys425 indicates an N to C–bound orientation and to SBDCys458 reports on C to N binding mode. These strategies for orientation determination were validated by checking two well-known Hsp70 binding peptides (SI Appendix, SI Text and Fig. S6).
Peptides Encompassing proPhoA Sites C and D Bind to the SBD in Opposite Orientations, both with Leucine Occupying the SBD 0th Pocket.
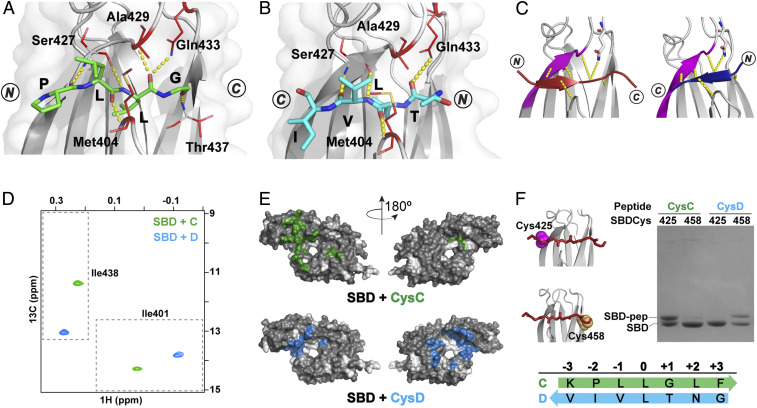
We solved crystal structures of SBD complexes with peptides encompassing proPhoA binding sites C (peptide C, 249EANQQKPLLGLFADG263) and D (peptide Dsh, R361GNTLVIV367SR; Note: here and elsewhere, we refer to residues in the peptide and protein substrates by single letter codes and to those in the DnaK SBD by three-letter codes, and the underlined residues were added to the peptides and are not present in the protein sequence). Leucine residues, L257 in peptide C and L364 in Dsh, occupy the SBD 0th pocket in both cases, but strikingly, the peptides bind in opposing backbone orientations; C adopts an N to C orientation analogous to that seen in the structure of NR-bound SBD (11), while Dsh adopts the opposite C to N orientation (Fig. 4 A and B).
Fig. 4.
Peptides containing proPhoA sites C and D bind to the DnaK SBD in opposite orientations. (A and B) Crystal structures (top view) of the SBD (gray) in complex with proPhoA peptide C (A), which binds in an N to C orientation (green), and Dsh (B), which binds in a C to N orientation (blue). Residues 507 to 603 are not shown. The side chains of SBD residues contacting the peptide are in red sticks, and hydrogen bonds between the peptide backbone and the SBD are in yellow. (C) Schematic showing that the bound peptide interacts with the β3-strand of the SBD (magenta) in a parallel or antiparallel strand–strand arrangement for the N to C [Left; peptide in maroon, PDB ID code 1DKZ (11)] or the C to N binding mode [Right; peptide in blue, PDB ID code 4EZY (12)]. (D) The Ile401 and Ile438 region of 1H-13C-HMQC spectra of ILV-13CH3–labeled DnaK SBD in the presence of unlabeled peptides C (green) and D (blue). (Peptides and SBD are at 40 μM.) (E) PRE NMR of SBD complexes with CysC and CysD peptides. SBD residues significantly broadened in the presence of spin-labeled peptides are mapped on the SBD structure (in green, CysC; in blue, CysD). Residues for which no data are available are shown in white. (F) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of SBDCys425 and SBDCys458 (cysteine residues in magenta and yellow spheres in the structures in Left) cross-linked to CysC and CysD peptides.
The hydrogen bonds formed between the SBD and peptide C were the same as those observed for the NR peptide, which bound in the same N to C orientation (11). 1) Three hydrogen bonds form between the peptide residue occupying the 0th position and SBD residues—from Ala429 NH and Gln433 side chain NH2 to the L257 C = O and from the Ser427 C = O to L257 NH); 2) additional hydrogen bonds form between the peptide backbone C = O in SBD positions –2, –1, and +1 and the NH of SBD residues Ser427, Met404, and Ser437, respectively (Fig. 4A and SI Appendix, Table S2). The hydrogen bond observed in the SBD–NR structure in position +3 cannot be seen in the structures of SBD bound to C due to a lack of peptide electron density at this position.
Remarkably, while Dsh adopts a C to N orientation, the peptide backbone forms a comparable hydrogen bond network via interactions with the backbone groups of SBD residues Met404, Ser427, and the side chain of Gln433 as it does in complexes with the peptides bound in the N to C orientation (Fig. 4B and SI Appendix, Table S2). These backbone interactions—stabilized primarily by the β3-strand of the SBD—provide an explanation for the observation that peptides may bind the SBD in either the N to C binding mode, in which there is a parallel β-strand interaction with SBD strand-β3, or the C to N binding mode, in which there is an antiparallel β-strand interaction with SBD strand-β3 (Fig. 4C and SI Appendix, Table S2).
While we did not obtain crystals for the complex between the longer version of peptide D (357AKKEGNTLVIVTADH371) and the DnaK SBD, the coincidence of the Ile401 and Ile438 resonances for the SBD complexes with the shorter and longer versions of peptide D in solution NMR experiments indicates that these peptides bind the SBD in the same manner (SI Appendix, Fig. S7). For both C and D peptides, we observed a single set of resonances for peptide-bound isoleucine-leucine-valine (ILV)-13CH3-SBD Ile401 and Ile438 residues in 1H-13C-HMQC spectra, indicative of only one mode of binding populated in solution (Fig. 4D). Intriguingly, the chemical shifts of Ile401 and Ile438 were far apart for ILV-13CH3-SBD bound to the two peptides despite the fact that leucine occupies the central pocket in both cases; we independently made a similar observation for two control peptides, CysNR and CysNRIle, both of which bind with leucine in the central pocket but one of which (CysNR) binds in an N to C orientation and the other (CysNRIle) from C to N (SI Appendix, SI Text and Fig. S6F). We thus propose that the binding mode populated in solution for C and D is the same as that seen in the respective crystal structures and that the distinct positions of Ile401 and Ile438 chemical shifts report on the opposite binding orientations of these two peptides. Consistent with this interpretation, PRE experiments with the MTSL-labeled CysC and CysD peptides resulted in resonance broadening on the opposite sides of the SBD domain as expected for N to C and C to N binding orientations, respectively (Fig. 4E). Moreover, CysC formed a disulfide bond with SBDCys425, consistent with an N to C orientation, while CysD cross-linked exclusively to SBDCys458 (Fig. 4F). We conclude that peptide C binds in the N to C orientation with the “PLLGL” motif occupying the five SBD pockets and peptide D binds via the “NTLVI” sequence in the opposite orientation.
Prior to this study, most sequences reported to bind DnaK in a C to N orientation contained at least one proline residue (12, 15), which led the field to correlate the presence of prolines in the bound sequences to the preference for C to N binding. The fact that peptide D has no prolines and that peptide C places proline in one of the SBD pockets yet binds in the N to C mode suggests that the role of proline in biasing the binding backbone orientation might have been overestimated. We hypothesize that the fit of residues to preferred SBD pockets is the dominant factor in determining preferred orientation. Here, if L364 occupied the 0th position in the N to C orientation, T363 would occupy the −1 position. Past analyses (9, 10) predict binding of threonine at the −1 position to be disfavored relative to the hydrophobic valine residue that occupies this pocket when the peptide is in the C to N orientation. Moreover, our results are not consistent with current prediction algorithms (9, 10), which would lead one to expect binding of the sequence “LVIVT” with isoleucine in the central 0th binding pocket.
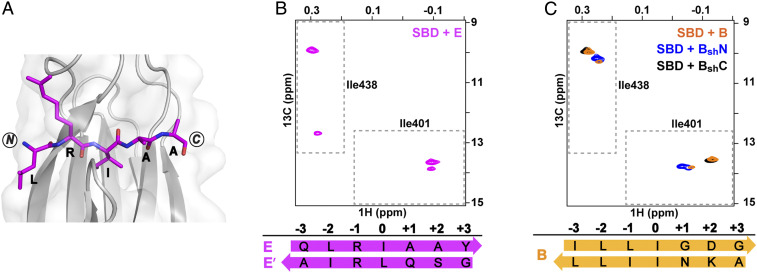
The Peptide Corresponding to proPhoA Site E Populates More than One SBD Binding Mode Including both Backbone Orientations.
The crystal structure of the DnaK SBD bound to the peptide containing the proPhoA site E sequence (C411QEHTGSQLRIAAYGP425) revealed N to C binding mode, where the SBD 0th pocket is occupied by I420 and pockets –2 and –1 are occupied by L418 and R419, respectively (Fig. 5A). While this X-ray structure yielded no surprises, NMR results for SBD bound to this peptide brought to light a behavior not previously observed and pointed out that there can be energetically competitive, alternative substrate binding modes populated in solution. In the 1H-13C-HMQC spectra of ILV-13CH3-SBD in complex with peptide E, the diagnostic Ile401 and Ile438 residues show a major set of resonances plus an additional set of less intense resonances, consistent with the presence of two modes of binding (Fig. 5B). We have concluded that the major Ile401 and Ile438 resonances seen for peptide E–bound SBD in solution correspond to the binding mode observed in the crystal structure (i.e., N to C). We propose that the peptide isoleucine occupies the SBD 0th binding pocket, as the chemical shift of Ile438 at ∼10 ppm in the 13C dimension is distinct from the resonance positions observed for bound sequences with leucine (Fig. 4D) or valine in the 0th pocket (13).
Fig. 5.
Peptides containing proPhoA sites E and B both populate more than one binding mode, one N to C, and another minor state, C to N. (A) Crystal structure of the DnaK SBD (gray) complexed with proPhoA peptide E (magenta). (B) The Ile401 and Ile438 region of 1H-13C-HMQC spectra of ILV-13CH3 ILV-13CH3-DnaK SBD in the presence of unlabeled peptide E (magenta). Two binding modes are populated; the minor C to N binding mode is denoted E’ (SI Appendix, Fig. S8). (C) The Ile401 and Ile438 region of 1H-13C-HMQC spectra of ILV-13CH3 DnaK SBD in the presence of unlabeled peptides B (orange), BshN (blue), and BshC (black). The major resonances for peptide B represent an N to C binding mode with I50 in the central pocket; the minor resonances can be assigned to C to N binding mode with I46 in the 0th pocket (SI Appendix, Fig. S9).
The positions of the minor Ile401 and Ile438 resonances for the SBD in complex with peptide E were similar to those seen for the SBD bound to peptides D and CysNRIle (Fig. 4D and SI Appendix, Fig. S6F), which led us to hypothesize that the second binding mode might correspond to peptide bound in the C to N orientation with leucine in the central pocket. We proposed that the minor species represents the peptide bound with L418 (the only leucine in this sequence) in the 0th SBD pocket in a C to N backbone orientation, which places favorable residues in the SBD pockets (i.e., I420 in position –2 and R419 in position –1). To test this hypothesis, we designed a mutant peptide EshV (R415GSQVRIA421SR) corresponding to site E with an L418V substitution. Our interpretation for the minor species predicts that V418 occupies the central 0th pocket and that the Ile401 and Ile438 resonances of the SBD bound to EshV should shift significantly when compared with the original peptide E. Consistent with this prediction, the Ile438 resonance moved from 12.7 to 8.7 ppm in the 13C dimension (SI Appendix, Fig. S8A), as has been previously observed for a substrate valine in the SBD central pocket (13), indicating that the new sequence “GSQVRIA” binds the SBD in the C to N orientation and places valine in the central pocket. In contrast, the Ile401 and Ile438 resonances of the major species for the mutant peptide moved only slightly, as expected since peptide residue I420 remains in the 0th site of the N to C orientation. We attribute the small differences in the positions of Ile401 and Ile438 resonance positions to the presence of a different substrate residue in the SBD –2 position (“V” in EshV vs. “L” in E) and +2 and +3 positions (“SR” vs. “AY,” respectively).
PRE experiments confirmed that peptide E binds to the SBD predominantly in the N to C orientation (SI Appendix, Fig. S8B). Importantly, we were able to assign a few key resonances from the minor species in 1H-15N-TROSY-HSQC spectra, and the resonance intensity loss seen in the presence of the paramagnetic label could be explained only if the minor species corresponded to the peptide bound in a C to N binding mode (SI Appendix, Fig. S8C). Consistent with this interpretation was the observation that an N-terminal cysteine on peptide E formed a disulfide bond with both SBDCys425 and SBDCys458, as expected if the peptide adopts both backbone orientations when bound to the SBD (SI Appendix, Fig. S8D). Thus, the site E peptide populates two distinct SBD binding modes in solution: one in the N to C orientation with I420 occupying the SBD central pocket and another in the C to N orientation with L418 in the 0th position.
proPhoA Peptide B Can Bind the DnaK SBD in Either Backbone Orientation and in Each Mode Places Isoleucine in the Central Pocket.
In solution, a peptide encompassing proPhoA site B (C35RDSLSDKPAKNIILLIGDGMGD56) populates two SBD binding modes, as evident from two distinct sets of resonances for Ile401 and Ile438 in 1H-13C-HMQC spectra of the ILV-13CH3-SBD–peptide B complex (Fig. 5C). The Ile438 resonances were in the same spectral region as those observed for the major species of the SBD complexed with peptide E (Fig. 5B), which led us to suggest that peptide B binds to the SBD with isoleucine in the 0th pocket. Confounding the analysis of the binding modes for this peptide is the presence of three isoleucines in the sequence; in order to determine which occupies the central pocket, we divided the sequence into two shorter peptides BshN (R43AKNIILL49SR) and BshC (CR47ILLIGDG53SR) and recorded the 1H-13C-HMQC spectra of ILV-13CH3-SBD bound to each peptide. The major Ile401 and Ile438 resonances of the SBD bound to peptide B overlapped with the only resonances observed for the complex with BshC (Fig. 5C), arguing that I50 occupies the 0th position. If isoleucine I47 was bound in the central pocket, the Ile401 and Ile438 chemical shifts for BshC-bound SBD would differ from the ones observed for peptide B because of the different peptide residues placed in –1 and –2 positions (or +1 and +2 positions for the C to N binding mode) for these two peptides (i.e., “NI” for B vs. “CR” for BshC). Given that the orientation of the B peptide is predominantly N to C based on PREs and cross-linking results (SI Appendix, Fig. S9 A and B), we concluded that the major species for Ile438 and Ile401 in the 1H-13C-HMQC spectra correspond to a bound state with I50 in the central pocket and L49 and L48 in –1 and –2 positions, respectively.
The minor set of Ile401 and Ile438 resonances in the spectra of the SBD–peptide B complex overlap with the minor resonances for the SBD bound to BshN, which in addition had a distinct major population not seen for the peptide B–SBD complex (Fig. 5C). This minor binding mode populated by peptides B and BshN was determined to be C to N by both PRE and disulfide cross-linking experiments (SI Appendix, Fig. S9 A–C). Coincidentally, BshN crystallized in complex with the SBD in a C to N backbone orientation with I46 in the 0th pocket and I47 and L48 in –1 and –2 positions (SI Appendix, Fig. S9D). Hence, we could conclude that this is also the minor binding mode for peptide B in solution. Significantly, this conclusion is consistent with an emerging correlation; Ile438 resonances resulting from substrate isoleucine bound in the central pocket cluster together at around 10 ppm in the 13C dimension in 1H-13C-HMQC spectra (SI Appendix, Fig. S11).
Provocatively, the major binding mode for peptide B contains an aspartic acid residue in the core binding sequence “LLIGD,” previously considered to be unfavorable for Hsp70 binding (9, 10). We speculate that in this case, the favorable energetic contributions from fitting a hydrophobic isoleucine into the 0th pocket and two leucine residues in the restrictive –1 and –2 positions outweigh the negative contribution from the aspartate in the more permissive +2 position. Interestingly, in both major and minor binding modes, leucines/isoleucines occupy the most restrictive 0, –1, and –2 SBD pockets (N to C LLIGD and C to N “LIINK”).
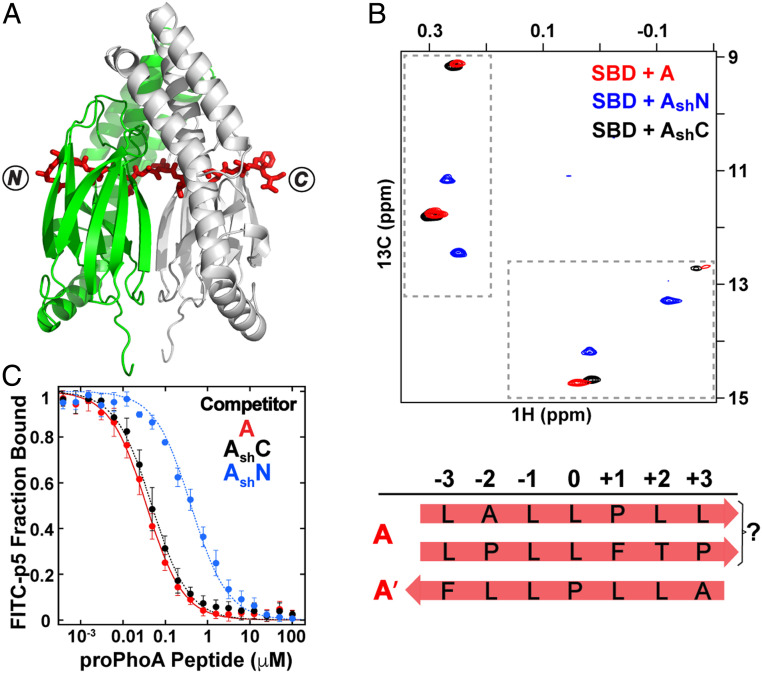
The Intriguing Story of proPhoA Peptide A.
Peptide A (R-20KQSTIALALLPLLFTP−5RR) encompasses a strongly chaperone binding sequence based on the original peptide array (9) and displays the highest apparent binding affinity to the SBD of all the peptides tested in this study (Fig. 3B and SI Appendix, Fig. S4). It is intriguing that it contains within it the signal sequence of proPhoA, which directs this precursor of PhoA to the periplasm (29–31). Consistent with its biological role, this region also represents the most hydrophobic sequence in proPhoA (SI Appendix, Fig. S3A), which makes sense in terms of its affinity.
Crystals of a complex of the DnaK SBD with peptide A yielded an electron density map that proved challenging to interpret. The best fit resulted in an unexpected and surprising final structure. Like other SBD–proPhoA peptide structures, these crystals resulted in one SBD per asymmetric unit and a twofold axis of symmetry within the unit cell. However, surprisingly, the packing of the SBD–peptide A crystal was distinct from the other SBD–proPhoA peptide crystals, with different unit cell dimensions. Moreover, it was observed that the electron density for peptide A appeared continuous across the twofold axis of symmetry, indicating that it breaks the symmetry axis to bind both SBD molecules within the unit cell (Fig. 6A). To accommodate this dual binding, the two SBD molecules are arranged in a way that aligns the substrate binding pockets, which explains the unique unit cell dimensions. Moreover, the electron densities observed for the main-chain carbonyl groups were unusually large—too large to be consistent with the peptide being positioned in one backbone direction. In addition, the electron densities for the peptide side chains were also difficult to fit to a specific residue and looked more like an average of two (such as T/L, I/L, and A/P). Furthermore, the position and orientation of the carbonyl groups as well as the residue side-chain electron densities and the interactions with the SBD including the hydrogen bonds could only be satisfied when peptide A was built extending through the two SBDs within the unit cell in N to C and C to N backbone directions simultaneously. In the final structure, we observe the SBD bound to peptide A via two sequences with opposing backbone orientations: C to N with I(−16) in the central pocket of one SBD and N to C with L(−9) in the 0th pocket of the other SBD (SI Appendix, Fig. S10A). Interestingly, the constraints imposed on the complex within the crystal environment led to occupancy of the –1 position of the SBD binding site by an alanine in one case and a proline in the other, neither of which had been considered to bind to DnaK favorably previously (10).
Fig. 6.
The peptide containing proPhoA site A spans an SBD dimer in crystals and in solution binds 1:1 to the SBD, populating more than one binding mode. (A) Crystal structure of the DnaK SBD in complex with peptide A. Two SBDs (green and gray) are aligned at the twofold axis, and peptide A (red sticks) extends through both SBDs with two sequences in the binding clefts, one in an N to C and one in a C to N backbone direction. (B) The Ile401 and Ile438 region of 1H-13C-HMQC spectra of ILV-13CH3–labeled DnaK SBD in the presence of unlabeled peptides A (red), AshN (blue), and AshC (black). The binding modes were assigned as 1) N to C with a leucine in the central pocket and 2) C to N with P(−10) in the central pocket. (C) Competition binding assays for peptides A, AshN, and AshC to the SBD bound to a fluorescein isothiocyanate (FITC)-labeled peptide (SI Appendix, SI Materials and Methods and Fig. S10).
In solution, two sets of resonances are observed in the 1H-13C-HMQC spectrum of SBD bound to peptide A, indicating its occupancy of two binding modes. However, neither of the two are consistent with presence of an isoleucine in the central pocket as seen in one of the binding modes in the crystal structure (Fig. 6B). The major Ile401 and Ile438 chemical shifts are in the same region of the 1H-13C-HMQC spectra as seen for the SBD in complexes with peptides C, D, and CysNR, where leucine is bound in the central pocket. The minor Ile401 and Ile438 resonances have very distinct positions in the 13C dimension (9 and 12.7 ppm, respectively), previously seen only when a substrate valine occupies the 0th pocket (13).
To identify which of the five leucines in the sequence of peptide A occupies the 0th site in solution, we divided peptide A into two shorter peptides, AshN (CR−20KQSTIALA−13SR) and AshC (CR−12LLPLLFTP−5SR) (SI Appendix, Fig. S10C). Strikingly, the two sets of Ile401 and Ile438 resonances of peptide A–bound SBD overlapped with the two sets of resonances seen for the SBD–AshC complex, indicating that both SBD complexes with peptide A arise exclusively from binding of the C-terminal portion of the sequence and that the first seven residues do not participate in the binding in the context of the long peptide A (Fig. 6B). Consistent with this interpretation, peptide AshC displayed the same apparent high SBD binding affinity as peptide A (higher than that of the other proPhoA peptides) (Fig. 6C and SI Appendix, Fig. S10 B–D). PRE experiments argue that peptide A predominantly adopts the N to C backbone orientation when bound to the SBD, while in cross-linking experiments, both binding orientations were observed (SI Appendix, Fig. S10 E and F). Nonetheless, a definitive determination of the SBD binding mode of peptide A was made difficult because the C-terminal fragment of peptide A contains four leucine residues. Considering that alanine and proline are expected to be disfavored for binding in the –1 SBD position (10), we propose that the mode of binding observed in the SBD–peptide A crystals, which has L(−9) in the central pocket, is unlikely in solution. This still leaves two potential N to C binding modes, “ALLPL” and “PLLFT.” At present, we cannot unequivocally assign the favored mode of binding of peptide A to the SBD other than to conclude that the C-terminal half of the peptide is the site of interaction, and leucine occupies the central pocket.
As mentioned earlier, the minor A–bound SBD species had unusual Ile401 and Ile438 chemical shifts in 1H-13C-HMQC spectra, previously observed only when a valine in the substrate occupies the SBD central pocket (13). In this case, there has to be another explanation for this chemical shift pattern, as the A/AshC sequences lack valine. We hypothesized that proline might occupy the 0th pocket, and to test this hypothesis, we collected the 1H-13C-HMQC spectrum of SBD in complex with a small antimicrobial peptide drosocin carrying an N-terminal cysteine (CSHPRPIRV), previously shown in crystal structures to bind the SBD with proline in the central pocket (12). The Ile401 and Ile438 chemical shifts of SBD-bound drosocin were very close to the ones observed for the minor A–bound SBD species, supporting our hypothesis (SI Appendix, Fig. S10G). As the minor binding mode was determined to be C to N by PREs and disulfide cross-linking experiments (SI Appendix, Fig. S10 F and H), we could confidently assign it to “LLPLL” with L(−9) and L(−8) in the SBD –1 and –2 positions, respectively.
Notably, the results for peptide A corroborate our general conclusion that the positions of the Ile401 and Ile438 resonances report on the type of substrate residue occupying the SBD central pocket, as we observe distinct chemical shift patterns for these two residues when leucine, isoleucine, or proline occupies the 0th pocket. Furthermore, the fact that the major SBD Ile401 and Ile438 resonances for peptide A were similar to the resonances seen for the SBD bound to peptides C and CysNR supports the notion that an Ile438 chemical shift at ∼11 to 12 ppm in the 13C dimension can be correlated with a substrate bound with leucine in the 0th position in an N to C binding orientation. Likewise, the Ile438 chemical shift at ∼13 ppm corresponds to a substrate bound in a C to N binding orientation with leucine in the 0th position as observed for peptides D and CysNRIle and the minor species for peptide E (SI Appendix, Fig. S11). Thus, Ile401 and Ile438 signatures in 1H-13C-HMQC spectra report on the binding mode of any potential DnaK binding sequence, including the orientation and the residue bound in the central pocket.
From Peptides to Protein: The DnaK SBD Binds an Unfolded Protein Substrate via Largely Independent Peptide-Like Sites, with Binding Preference Largely Determined by Site Affinities and with a Variety of Alternative Binding Modes Including Both N to C and C to N Orientations.
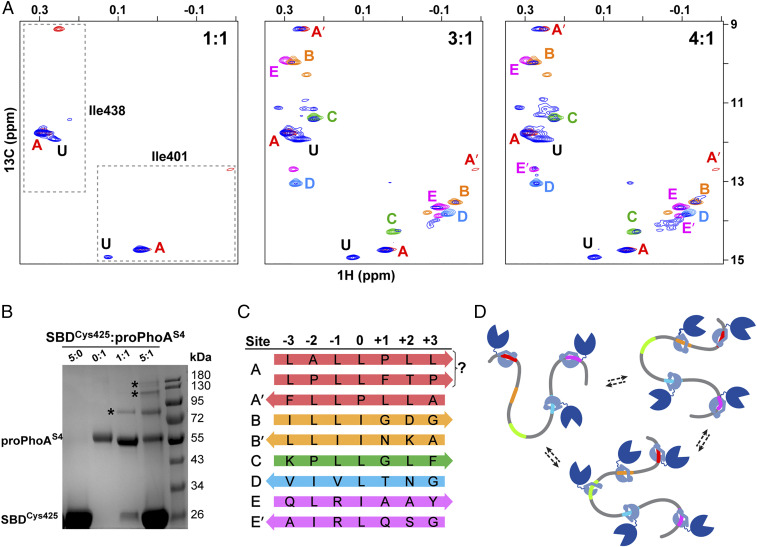
The detailed analysis of how peptides corresponding to potential binding sites in proPhoA bind to the DnaK SBD armed us with spectral signatures, structural models, and anticipated site affinities, all of which can now be used to tackle the binding behavior of the chaperone when presented with the full-length unfolded protein substrate. To explore the nature of DnaK SBD binding to full-length unfolded proPhoAS4, we titrated the substrate with increasing concentrations of ILV-13CH3-DnaK SBD and compared diagnostic Ile401 and 438 resonances in the 1H-13C-HMQC spectrum with results obtained with the peptides harboring the binding sites (Fig. 7A). At a high chaperone to substrate ratio, the resonances of ILV-13CH3-SBD bound to proPhoAS4 overlap closely with their positions in spectra of the SBD bound to the peptides that correspond to potential binding sites A to E. Notably, the close correspondence of the chemical shifts of Ile401 and Ile438 of the SBD complexes with the binding site peptides to their chemical shifts when bound to proPhoAS4 argues that the atomic details observed in the peptide complexes characterize the mode of SBD binding to these sites in the full-length substrate. Most importantly, our analysis of the isoleucine signatures for ILV-13CH3-SBD bound to peptides corresponding to the potential binding sites on proPhoAS4 allowed us to deduce the actual sequences bound by the SBD, including the pocket occupancy (“the register”) and the backbone orientation of the bound sequences (Fig. 7C). These details have not previously been obtained for the interaction of DnaK with any protein substrate.
Fig. 7.
From peptide to protein, the DnaK SBD binds multiple sites on proPhoA with differing affinities, largely consistent with studies of peptides, and one previously unidentified high-affinity site. (A) Titration of unlabeled proPhoAS4 with increasing amounts of ILV-13CH3 DnaK SBD. The Ile401 and Ile438 resonances of the SBD in complex with proPhoAS4 are shown in blue, and resonances for complexes with the proPhoA peptides A to E are color coded as in Fig. 3. At a 1:1 ratio of SBD to proPhoAS4, the resonances for the protein complex coincide with those for peptide A (red), and an additional resonance is seen (labeled U) that does not correspond to any of the peptides tested. At a 3:1 ratio of SBD to proPhoAS4, additional resonances appear that overlay on those for the SBD complexes with peptides C and D and major species for complexes with peptides B and E. Finally, at a 4:1 ratio of SBD to proPhoAS4, all the resonances seen for the peptides appear. (B) SDS-PAGE of SBDCys425 cross-linked to proPhoAS4 using Sulfo-GMBS. Addition of the cross-linker to SBD or proPhoAS4 alone does not result in higher–molecular mass species (lanes 1 and 2). At a 1:1 ratio of SBD to proPhoAS4, a 1:1 complex is observed (lane 3), while at a 5:1 ratio of SBD to proPhoAS4, complexes with multiple SBD bound to one proPhoA molecule are seen. (Controls are shown in SI Appendix, Fig. S13.) (C) Summary of DnaK SBD binding sites on proPhoAS4, their occupancy of SBD binding pockets, and their orientation. The prime symbol indicates an alternative minor binding mode. (D) Cartoon depicting DnaK binding to proPhoA. Site A, which has the highest binding affinity to DnaK, is occupied by the chaperone at the lowest chaperone to substrate ratios and remains bound as the chaperone to substrate ratio increases. Other binding sites on proPhoA are occupied based largely on the affinities they manifest as peptides but in a dynamic manner, with several or all of them occupied by chaperone simultaneously.
In addition, the titration results reveal the progressive occupancy of binding sites on proPhoAS4 as the concentration of chaperone increases (Fig. 7A). At substoichiometric concentration, the SBD binds predominantly to site A, for which the corresponding peptide showed the highest apparent binding affinity (Fig. 3B and SI Appendix, Fig. S4). As the chaperone concentration is increased, Ile401 and Ile438 resonances corresponding to the chaperone bound to sites B to E appear, and roughly, the binding follows the order expected based on the binding experiments for the peptides corresponding to each site (a control experiment in which all individual peptides were presented to the SBD at the same concentration is in SI Appendix, Fig. S12).
The data in Fig. 7A make it clear that the picture described above is not complete, as the 1H-13C-HMQC spectrum of ILV-13CH3-SBD bound to proPhoAS4 reveals a set of strong Ile401 and Ile438 resonances (labeled “U”) and additional minor resonances that do not coincide with their positions in complexes with any of the candidate binding site peptides A to E. Moreover, the Ile401 and Ile438 resonances reporting on ILV-13CH3-SBD binding to the “unknown” site appear simultaneously with the peaks corresponding to the SBD bound to site A, indicating that the chaperone binds to this site with high affinity. The peptide array data (9) give some indication that there could be additional binding sites but none that would be expected to be comparable in affinity with site A. The Limbo prediction algorithm (10) points to a few potential additional strong binding sites, although these are not evident in the array data, and we are currently carrying out a variety of experiments to identify this intriguing site.
An additional conclusion clearly supported by the data in Fig. 7A is that multiple chaperones can bind simultaneously to the substrate. With the addition of excess SBD, new sites get populated, while sites A and U stay occupied. To further expand on this observation and determine the stoichiometry of the SBD–proPhoAS4 complex in solution, we designed a cross-linking experiment in which we utilized SBDCys425 and a bifunctional reagent that covalently conjugates free thiol groups with lysine amino groups, Sulfo-GMBS (N-γ-maleimidobutyryl-oxysulfosuccinimide ester) and performed a cross-linking reaction between the SBD Cys425 and proPhoAS4. Four of the five SBD binding sites on proPhoAS4 (A, B, C, and D) contain at least one lysine residue either within the binding site or within three residues, which should in principle enable the formation of cross-links between proPhoAS4 and SBDCys425 in the preformed complex. Therefore, if multiple SBDs occupy the sites on proPhoAS4 simultaneously, higher–molecular mass cross-linked species are anticipated in addition to a 1:1 SBD–proPhoA complex, which has a molecular mass of ∼74 kDa. Indeed, when the SBD was present in fivefold excess over the protein substrate, we observed distinct species corresponding to complexes up to molecular masses that would fit at least three SBDs bound to a proPhoAS4 molecule (Fig. 7B).
Discussion
A major puzzle in understanding how Hsp70 chaperones bind their clients is the molecular origin of their selectivity for certain sites on clients that are unfolded or partially folded or present unstructured regions in concert with their promiscuity in binding many, many different sequences. A simple explanation has been offered, namely that these chaperones look for hydrophobic sequences that are normally sequestered in folded proteins. This is certainly part of the story; however, there are many details about their binding mechanism that will bear on the functional roles of Hsp70s. The results described here underline the plasticity of the binding site of the E. coli Hsp70 DnaK and help explain its promiscuity. Bridging our in-depth study of the DnaK SBD complexes with peptides corresponding to potential binding sites on the unfolded substrate proPhoAS4 and analysis of the SBD complex with the full-length proPhoAS4 has provided a picture showing how the chaperone interacts with its binding sequences within an unfolded protein (Fig. 7C). It is clear that there are different binding preferences for the pockets within the substrate binding cleft of the SBD. These biases are most evident for the central 0th SBD binding pocket, with relatively strong residue preferences also characterizing pockets –2 and –1, and these biases help explain the chaperone’s selectivity. These conclusions from direct analysis of substrate-bound SBD complexes are broadly consistent with previous work, which was based on statistical calculations from peptide arrays and binding energy estimates (9, 10). However, our data show definitively that the backbone, while strongly contributing to the favorable energy of binding through hydrogen bonding network, can be accommodated in either an N to C or a C to N orientation with comparable binding energy. While it was known that peptide substrates could bind in a C to N orientation (12, 14, 15), the fact that the two possible orientations for the chaperone-bound substrate are energetically competitive is an important insight in our understanding of Hsp70–substrate interactions. In addition, we were fortunate to have expanded the atomic resolution data available for substrates bound to the DnaK SBD considerably, such that we can build detailed models for the mode of binding of sites within the full-length substrate, which is itself not amenable to crystallographic analysis.
Taken together, a picture emerges of the Hsp70 chaperone scanning accessible sequences of clients for the optimal fit of the substrate side chains to the SBD pockets, without regard for the backbone orientation (Fig. 7C). Moreover, our data show that multiple modes of binding for the same client sequences can coexist. Given the ability of the Hsp70 to bind and release in an adenosine triphosphate (ATP)-mediated fashion, the resulting image is that of a dynamic ensemble of diverse binding arrangements, the populations of which are governed by the energetics of side-chain interactions with the SBD pockets, which in turn, are dominated by sites 0, –1, and –2.
Importantly, the data presented show that binding of this unfolded protein substrate, proPhoAS4, to the DnaK SBD to a great extent recapitulates the binding of peptides corresponding to sites identified in peptide arrays with the exception of the strongly binding unknown site U and that binding to the unfolded protein substrate therefore occurs in large measure via a straightforward additivity of independent sites (Fig. 7D). These observations validate the use of peptides and peptide arrays to characterize Hsp70 chaperone–client interactions, with a few provisos. It is formally possible that the protein will behave identically to a collection of peptides of equal concentration presented all at once and consequently, that no new complexities will emerge as we consider how an unfolded protein is bound by this Hsp70. However, it is more likely that additional aspects of the interaction, such as effects of proximity of sites, possible influence of adjoining sequences, and perhaps unexpected binding sites not revealed in peptide array data, can be influential. Moreover, our observation that the modes of binding of peptides to the DnaK SBD include both N to C and C to N orientations, with comparable energies of binding, demands that peptide and peptide array data be interpreted as potentially reporting on either orientation. Whether these geometries are sampled with equal probability in cellulose-bound peptides in the array analyses is not at all clear.
While we would argue that some of our findings will have important general implications, such as the fact that the N to C and C to N binding orientations are very close in binding energy, we want to emphasize the risks of generalizing the conclusions reached based on binding of DnaK SBD to a single unfolded substrate. In particular, it is noteworthy that the potential DnaK binding sites on proPhoA identified first in peptide arrays (9) and now validated as true binding sites in the context of the protein substrate by our studies are well separated in sequence. Thus, this particular substrate does not provide any insight into issues that might arise when chaperones may interfere with one another on proximal sites. In addition, our data support the fact that proPhoAS4 retains very little residual structure (SI Appendix, SI Text and Fig. S1B). Most probably, this is the exception and not the rule for the unfolded substrates that may be scanned by Hsp70s in vivo. It will be important and potentially very informative to study selection of binding sites by Hsp70s when their accessibility is not the same because of residual structure.
A major part of the physiological Hsp70 chaperone system is missing in our work: partner cochaperones (2, 3). Of greatest concern is the absence of a J protein in these client binding studies. Many researchers have concluded that substrates are often first bound by J-protein cochaperones and subsequently delivered through a handoff to the corresponding Hsp70 (32). It is an essential follow-up to this work to assess any differences arising when the J-protein DnaJ is present and can hand off proPhoA to DnaK. We are currently exploring the impact of the presence of DnaJ on the binding behavior of DnaK.
From the current work, it is abundantly clear that any DnaK binding prediction strategy must consider the sequence information as if it were “read” by the chaperone in either an N to C or a C to N direction. Simply stated, this doubles the number of sequence possibilities for Hsp70 binding sites in the proteome. Our observation of a strong binding site neither predicted nor apparent in the peptide array data of Rüdiger et al. (9) points to the limits of our current understanding of DnaK binding site selection. We are intent on identifying this site and learning from it: for example, by “reading” the proPhoA sequence in the reverse direction for cryptic binding regions.
In addition to their importance for efforts to predict Hsp70 binding sites, our findings raise many issues of functional importance. One intriguing outgrowth is the question of whether the binding of an Hsp70 may in fact play a functional role in the folding mechanism of a given protein. Recent studies have demonstrated that Hsp70s may play active roles in facilitating productive folding (33). This role for Hsp70s may leverage selectivity-specific regions and in turn, evolutionary tuning of the relative affinity of the chaperone for the sites where chaperone binding minimizes specific misfolding pathways. As Hsp70s have coevolved with their cellular substrates in such a way that competitive advantages for the organism result, we should remain open minded about the range of possible specific chaperone–substrate binding interactions. Work is underway in our laboratory to follow up on the role of Hsp70 binding preferences in specific folding facilitation.
We are particularly struck by the observation that the chaperone may bind its substrates in either backbone orientation. The geometric consequences of this fundamental behavior are significant, particularly when the Hsp70 is involved in chaperone networks. For example, the handoff of a substrate from a J protein to an Hsp70 would require quite different interchaperone arrangements if the site of binding to the Hsp70 is complexed in an N to C or a C to N direction. Similarly, the handoff to a downstream chaperone, such as Hsp90, would have analogous geometric consequences depending on orientation of the substrate. Recent elegant cryoelectron microscopy studies have provided structural details for the complex of Hsc70s, the glucocorticoid receptor ligand binding domain (GR LBD), Hsp90, and the cochaperone Hop (34). Strikingly, binding of the GR LBD to the Hsc70 upstream of the Hsp90 is revealed to be C to N terminally oriented with a proline in the 0th pocket position of the Hsc70 SBD. This observation underlines the physiological relevance of our findings with proPhoA and peptides derived therefrom.
Also functionally provocative is the finding that DnaK binds strongly to the signal sequence in proPhoA. This result—while not surprising because of the overall hydrophobicity of this region and the high content of leucine and isoleucine, both strongly preferred in the central 0th pocket of the DnaK SBD—does present an intriguing cellular scenario. As the precursor to a secreted protein is synthesized, it is targeted to the secretory apparatus resident in the inner membrane by either SecA, in partnership with SecB, or Ffh, the bacterial homolog of the eukaryotic signal recognition particle (35, 36). These targeting species bind specifically to the signal sequence and are pictured as having the major role in recognizing and forming complexes with secretory proteins. However, under a variety of cellular conditions, their capacity may be limited. For example, if the secretory apparatus is not able to cope with a heavy secretory load, a buildup of precursor protein can ensue. In this case, the ability of DnaK to bind precursors via their signal sequences could provide a “backup” capacity to stockpile excess precursors and avoid their aggregation (21, 22). Whether this region binds DnaK in normal cellular function is unclear and intriguing, but nonetheless, our data show that peptide A, which encompasses the signal sequence, binds to DnaK in a purified preparation with the highest apparent affinity of all the peptides tested (Fig. 3B and SI Appendix, Fig. S4).
Materials and Methods
Wild-type E. coli DnaK and E. coli proPhoA were expressed and purified as described previously with some modifications (17, 37). SBD cysteine mutants and proPhoAS4 were obtained by site-directed mutagenesis using standard protocols. Peptides were synthetized by Biomatik, GenScript, or Ruifu Chemical, and their apparent affinity to the SBD was measured by fluorescence anisotropy using a competition assay. X-ray crystallography and NMR spectroscopy were carried out according to established protocols. Cross-linking experiments were specifically designed for this study. Detailed materials and methods are included in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sashrika Saini, Wenli Meng, and Anne Gershenson for thoughtful discussion and critical reading of the manuscript; Scott Garman and Meg Stratton for help with analysis of X-ray crystallographic data; and Jianhan Chen for discussion of binding site energetics in DnaK. We acknowledge the Mass Spectrometry Center and the Biophysical Characterization Core Facility of the Institute for Applied Life Sciences at the University of Massachusetts Amherst. C.Ö. was supported with start-up funds to Meg Stratton. Otherwise, this work was supported by NIH Grant R35 GM118161 (to L.M.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016962118/-/DCSupplemental.
Data Availability
The atomic coordinates for complexes of the DnaK SBD and peptides corresponding to binding sites on proPhoA (notated here as SBD–peptide abbreviation) have been deposited at the Protein Data Bank (PDB; ID codes SBD/A: 7N6J; SBD/Bsh: 7JMM; SBD/C: 7N6L; SBD/Dsh: 7JN8; and SBD/E: 7JN9).
References
- 1.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Rosenzweig R., Nillegoda N. B., Mayer M. P., Bukau B., The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Clerico E. M., et al., Hsp70 molecular chaperones: Multifunctional allosteric holding and unfolding machines. Biochem. J. 476, 1653–1677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer M. P., Intra-molecular pathways of allosteric control in Hsp70s. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer M. P., Gierasch L. M., Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 294, 2085–2097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn G. C., Chappell T. G., Rothman J. E., Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Blond-Elguindi S., Fourie A. M., Sambrook J. F., Gething M. J., Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J. Biol. Chem. 268, 12730–12735 (1993). [PubMed] [Google Scholar]
- 8.Fourie A. M., Sambrook J. F., Gething M. J., Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 269, 30470–30478 (1994). [PubMed] [Google Scholar]
- 9.Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B., Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Durme J., et al., Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput. Biol. 5, e1000475 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X., et al., Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahn M., et al., Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J. Mol. Biol. 425, 2463–2479 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Rosenzweig R., Sekhar A., Nagesh J., Kay L. E., Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. eLife 6, e28030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cupp-Vickery J. R., Peterson J. C., Ta D. T., Vickery L. E., Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J. Mol. Biol. 342, 1265–1278 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Tapley T. L., Cupp-Vickery J. R., Vickery L. E., Sequence-dependent peptide binding orientation by the molecular chaperone DnaK. Biochemistry 44, 12307–12315 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Peran I., et al., Unfolded states under folding conditions accommodate sequence-specific conformational preferences with random coil-like dimensions. Proc. Natl. Acad. Sci. U.S.A. 116, 12301–12310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saio T., Guan X., Rossi P., Economou A., Kalodimos C. G., Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344, 1250494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Rossi P., Saio T., Kalodimos C. G., Structural basis for the antifolding activity of a molecular chaperone. Nature 537, 202–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardis M. F., et al., Preprotein conformational dynamics drive bivalent translocase Docking and secretion. Structure 25, 1056–1067.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y., Rossi P., Kalodimos C. G., Structural basis for client recognition and activity of Hsp40 chaperones. Science 365, 1313–1319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wild J., Altman E., Yura T., Gross C. A., Dna K., DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 6, 1165–1172 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Wild J., Rossmeissl P., Walter W. A., Gross C. A., Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J. Bacteriol. 178, 3608–3613 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tugarinov V., Kay L. E., An isotope labeling strategy for methyl TROSY spectroscopy. J. Biomol. NMR 28, 165–172 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Sekhar A., Rosenzweig R., Bouvignies G., Kay L. E., Mapping the conformation of a client protein through the Hsp70 functional cycle. Proc. Natl. Acad. Sci. U.S.A. 112, 10395–10400 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekhar A., Nagesh J., Rosenzweig R., Kay L. E., Conformational heterogeneity in the Hsp70 chaperone-substrate ensemble identified from analysis of NMR-detected titration data. Protein Sci. 26, 2207–2220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis J. E., Voisine C., Craig E. A., Intragenic suppressors of Hsp70 mutants: Interplay between the ATPase- and peptide binding domains. Proc. Natl. Acad. Sci. U.S.A. 96, 9269–9276 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rousseau F., Serrano L., Schymkowitz J. W., How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 355, 1037–1047 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Clore G. M., Iwahara J., Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 109, 4108–4139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malamy M., Horecker B. L., The localization of alkaline phosphatase in E. coli K12. Biochem. Biophys. Res. Commun. 5, 104–108 (1961). [DOI] [PubMed] [Google Scholar]
- 30.Inouye H., Beckwith J., Synthesis and processing of an Escherichia coli alkaline phosphatase precursor in vitro. Proc. Natl. Acad. Sci. U.S.A. 74, 1440–1444 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inouye H., Barnes W., Beckwith J., Signal sequence of alkaline phosphatase of Escherichia coli. J. Bacteriol. 149, 434–439 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kampinga H. H., et al., Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 24, 7–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imamoglu R., Balchin D., Hayer-Hartl M., Hartl F. U., Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 11, 365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R. Y., et al., GR chaperone cycle mechanism revealed by cryo-EM: Inactivation of GR by GR:Hsp90:Hsp70:Hop client-loading complex. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.11.05.370247v1 (Accessed 5 November 2020).
- 35.Elvekrog M. M., Walter P., Dynamics of co-translational protein targeting. Curr. Opin. Chem. Biol. 29, 79–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsirigotaki A., De Geyter J., Šoštaric N., Economou A., Karamanou S., Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 15, 21–36 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Montgomery D. L., Morimoto R. I., Gierasch L. M., Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 286, 915–932 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates for complexes of the DnaK SBD and peptides corresponding to binding sites on proPhoA (notated here as SBD–peptide abbreviation) have been deposited at the Protein Data Bank (PDB; ID codes SBD/A: 7N6J; SBD/Bsh: 7JMM; SBD/C: 7N6L; SBD/Dsh: 7JN8; and SBD/E: 7JN9).