Significance
Genes often encode for proteins with specialized functions (e.g., lipase). However, the function of the protein, and hence the gene, may be critical for survival in diverse contexts (e.g., fasting and oxidative stress). Hence, how are common survival genes activated in multiple contexts? Based on genetics and mathematical modeling, we describe two modes of transcriptional activation: 1) convergent—a single transcriptional regulator activates the survival gene in multiple contexts—and 2) contextual—the activity/interaction of members of a network of transcriptional regulators is fine-tuned to activate the survival gene through molecular paths that are specific to a context. The results underscore the limitations of across-context molecular inferences and suggest an economic tactic to biological resilience.
Keywords: TFEB/HLH-30, FOXO/DAF-16, fat, fasting, oxidative stress
Abstract
Plasticity in multicellular organisms involves signaling pathways converting contexts—either natural environmental challenges or laboratory perturbations—into context-specific changes in gene expression. Congruently, the interactions between the signaling molecules and transcription factors (TF) regulating these responses are also context specific. However, when a target gene responds across contexts, the upstream TF identified in one context is often inferred to regulate it across contexts. Reconciling these stable TF–target gene pair inferences with the context-specific nature of homeostatic responses is therefore needed. The induction of the Caenorhabditis elegans genes lipl-3 and lipl-4 is observed in many genetic contexts and is essential to survival during fasting. We find DAF-16/FOXO mediating lipl-4 induction in all contexts tested; hence, lipl-4 regulation seems context independent and compatible with across-context inferences. In contrast, DAF-16–mediated regulation of lipl-3 is context specific. DAF-16 reduces the induction of lipl-3 during fasting, yet it promotes it during oxidative stress. Through discrete dynamic modeling and genetic epistasis, we define that DAF-16 represses HLH-30/TFEB—the main TF activating lipl-3 during fasting. Contrastingly, DAF-16 activates the stress-responsive TF HSF-1 during oxidative stress, which promotes C. elegans survival through induction of lipl-3. Furthermore, the TF MXL-3 contributes to the dominance of HSF-1 at the expense of HLH-30 during oxidative stress but not during fasting. This study shows how context-specific diverting of functional interactions within a molecular network allows cells to specifically respond to a large number of contexts with a limited number of molecular players, a mode of transcriptional regulation we name “contextualized transcription.”
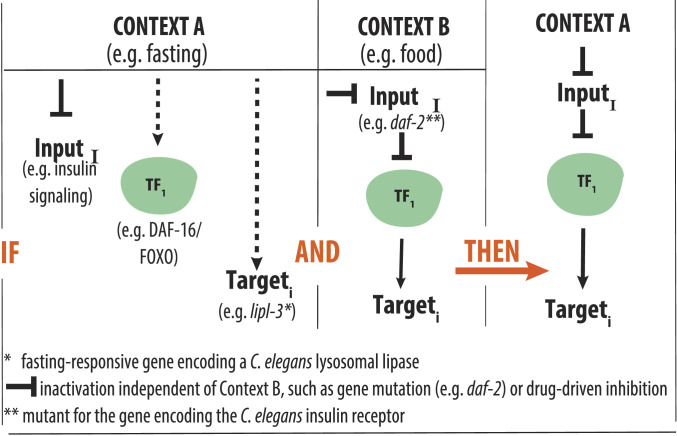
Organisms constantly face environmental challenges. Mounting effective adaptive responses to these challenges requires that the cells composing the organism receive information about the occurrence and intensity of the challenge. However, in multicellular organisms, most cells are not directly exposed to the challenge. Therefore, adaptive responses involve signaling pathways that sense and translate the stimulus or stressor (input) into genomic actions orchestrated by transcription factors (TFs) that promote homeostatic changes in gene expression (output). This sensing and communication theoretically work best if signals get transmitted into the cell without any loss of information. Therefore, signaling was traditionally thought of as linear and hence undistorted (1). Now, we know that signaling is far more complex, with different signaling pathways operating through independent and shared molecular components and biochemical mechanisms that ultimately repress or activate context-specific, as well as common, downstream genes. Nevertheless, we still mostly see signaling as pathways linearly linking an environmental condition or genetic perturbation (hereinafter referred to as context) to the activation of stable TF—target gene pairs that would, according to its stable nature, be activated across contexts. Under this paradigm, a common inference would look as follows (Fig. 1): If Context A is independently observed to 1) change the levels of InputI, 2) activate TF1, and 3) promote induction of Targeti; and in Context B, similar changes in InputI activate TF1, which in turn induces Targeti; then, we infer that Context A promotes induction of Targeti through activation of TF1. Although reasonable, the data used to build this kind of inferences are often obtained in contexts that can be, as the real example depicted in Fig. 1, as disparate as feeding and fasting. However, can we rely on input–output axes built upon data obtained in different contexts when we are aware that homeostatic responses are context specific?
Fig. 1.
Schematic representation of across-context inferences. Hypothetical inference described in the Introduction. In a real-life example, the literature shows that in fasted C. elegans (Context A), the following happens: 1) insulin signaling is reduced (12); 2) the TF DAF-16/FOXO is activated (2); and 3) the lysosomal lipase gene lipl-3 is induced (5). The literature also shows that in the presence of food (Context B), C. elegans carrying a hypomorphic mutation in the gene encoding the C. elegans insulin receptor DAF-2 show activation of DAF-16 (60) and DAF-16–mediated induction of lipl-3 (8). In this example, it would be reasonable to infer that induction of lipl-3 during fasting is mediated by DAF-16. However, as we show in this study, although all independent observations are true, the inference is false.
For example, several TFs have been experimentally validated to orchestrate adaptive responses to fasting and to genetic manipulations that mimic fasting in the nematode Caenorhabditis elegans. Among the most characterized fasting-activated TFs in C. elegans are DAF-16 (mammalian FOXO) (2), PHA-4 (mammalian FOXA) (3), NHR-49 (mammalian PPARɑ) (4), and HLH-30 (mammalian TFEB) (5). Several groups, including ours, have shown that these TFs mediate the activation of the C. elegans lysosomal lipases lipl-3 and lipl-4 in contexts that are related to but are not fasting. For instance, lipl-3 and lipl-4 are induced upon knockdown or mutation of the gene encoding the insulin receptor (daf-2), the Notch receptor (glp-1), and the intracellular nutrient sensor mTOR (6–10). In daf-2, glp-1, and mTOR-deficient animals, lipl-4 induction is mediated by DAF-16. As for lipl-3, Chen et al. suggested that DAF-16 would mediate its induction in daf-2 mutant animals (8). Therefore, lipl-3 and lipl-4 are downstream targets of several nutrient-sensing and nutrient-regulated growth pathways (SI Appendix, Fig. S1 A and B). Importantly, lipl-3 and lipl-4 encode for effectors of the C. elegans response to fasting, as LIPL-3 and LIPL-4 mobilize lipids in food-deprived worms, and the transcriptional program that leads to their induction is essential to C. elegans survival to fasting (5, 11). Based on the available information, several inferences could be made about the molecular pathways linking the expression of lipl-3 and lipl-4 to the feeding status of C. elegans. For instance, as illustrated in Fig. 1, the TF DAF-16 mediates the induction of lipl-3 downstream of reduced insulin signaling (daf-2 mutant) (8). Insulin signaling is reduced during fasting (12), and DAF-16 is activated during fasting (2). Hence, it is reasonable to infer that DAF-16 would promote the expression of lipl-3 in fasting C. elegans. If that were the case, how would the action of DAF-16 be coordinated with HLH-30, which is so far the only TF shown to be necessary for the induction of lipl-3 during fasting? On the other hand, if only DAF-16 or HLH-30 but not both would mediate the response of lipl-3 to fasting, how is the activity of the prevailing TF context specifically favored? Furthermore, if lipl-3 is activated in response to other stresses, would DAF-16, HLH-30, or other TFs execute the response in those contexts? Answering these questions would advance our understanding of how organisms mount adaptive responses to countless environmental and genetic perturbations using a limited number of molecular players.
In this study, we examined five signaling pathways that link nutritional status to growth in C. elegans. We refer to as inputs the contexts that inhibit or activate these signaling pathways (e.g., fasting, oxidative stress, or direct genetic inhibition/inactivation in the presence of food) and as output the level of transcription of the lysosomal lipase genes lipl-3 and lipl-4. We identified the TFs linking inputs to outputs, how the interactions between the TFs are diverted in different contexts to yield context-specific induction of the target gene, and mathematically expressed these roles via a discrete dynamic model. More broadly, the systematic investigation of the epistatic interactions between inputs, sensors, TFs, and the targets lipl-3 and lipl-4 demonstrates generalizable modes of transcriptional regulation that provide a plausible economic solution to the need to mount specialized adaptive responses while minimizing the number of dedicated molecular players.
Results
DAF-16 Is a Convergent Activator of lipl-4.
To identify the nutrient-sensing pathways and TFs that link food availability to the transcription of lipl-3 and lipl-4 required the use of thermosensitive mutants or RNA interference (RNAi) because mutation of several of the genes of interest leads to lethality (e.g., let-363) or sterility (e.g., glp-1). However, the Escherichia coli strains used to deliver double-stranded RNAs (dsRNA) in C. elegans can have phenotype-changing effects on nutrient sensing (13–16). We had previously characterized the lipl-3 and lipl-4 responses to fasting in animals fed E. coli OP50 (5); hence, we first show that fasting by withdrawal of an RNAi-competent derivative of OP50 (XU363) leads to induction of lipl-3 and lipl-4 (Fig. 2A). Thus, although some functional interactions may not be experimentally accessible when using RNAi (e.g., in neurons), RNAi-based epistasis studies of the lipl response to food availability can be performed using XU363-based RNAi.
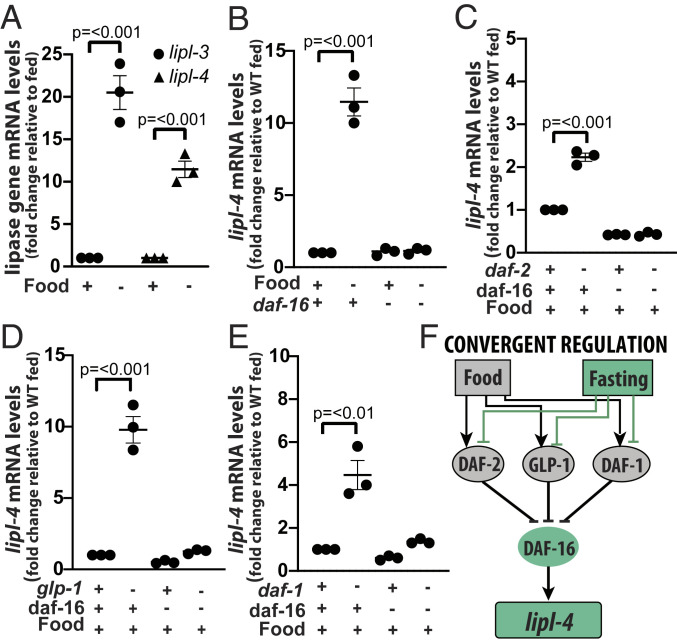
Fig. 2.
DAF-16 is a convergent activator of lipl-4. (A–E) qRT-PCR analysis of gene expression in young adult C. elegans normalized to housekeeping gene ama-1 or pmp-3 and relative to untreated as denoted in y-axis is depicted. Three or more independent biological replicates are presented in all panels. Error bars denote SEM. Statistical significance assessed via one-tailed unpaired parametric Student’s t test. Gene inactivations through mutation (gene name italicized) or RNAi (gene name in normal font) are represented with a minus sign in the treatment matrix. All RNAi treatments were started at the L1 stage. (A) WT animals grown on E. coli strain XU363 and then fasted for 6 h relative to fed (n = 3). (B) WT and daf-16(mu86) mutant animals fasted for 6 h relative to fed (n = 3). See results for other TFs in SI Appendix, Fig. S1C. (C) WT and daf-2(e1368) animals on empty vector control (EV) or RNAi against daf-16. (D) WT and glp-1(e2141) animals on EV or RNAi against daf-16. (E) WT and daf-1(m40) animals on EV or RNAi against daf-16. (F) Model of convergent regulation of lipl-4. Activating interactions are depicted as arrows and inhibitory interactions as blunt arrows.
We then screened TFs that mediate transcriptional responses to fasting, calorie restriction, starvation-triggered dauer formation, and control of lipid metabolism for their contribution to lipl-4 induction in fasting. Out of 11 TFs, only daf-16 was required for the induction of lipl-4 during fasting (Fig. 2B and SI Appendix, Fig. S1C). This result justified the use of daf-16–dependent induction as a probe to define which signaling pathways would link the feeding status of the worm to the expression of lipl-4 (see rationale in SI Appendix, Note S1).
Single inhibition of insulin signaling [daf-2(e1368)] and single inhibition of Notch signaling [glp-1(e2141)] was sufficient to promote induction of lipl-4 in feeding animals (Fig. 2 C and D); responses partially described by others and us for C. elegans feeding on E. coli OP50 (6, 8, 17). Importantly, these genetically triggered inductions were daf-16–dependent (Fig. 2 C and D). On the other hand, lipl-4 was reported to respond to mTOR (let-363) RNAi when the dsRNAs were delivered using E. coli HT115 (9, 10). However, we did not observe induction of lipl-4 when double-stranded RNAs against let-363 were delivered using E. coli XU363 (SI Appendix, Fig. S1D), even when reduced let-363 mRNA levels were confirmed (SI Appendix, Fig. S1E), and lipl-3 up-regulation was observed in the same samples (SI Appendix, Fig. S1F). Therefore, so far the data suggest that Notch and insulin signaling link lipl-4 expression to the feeding status of C. elegans and that DAF-16 would be a convergent transcriptional regulator of lipl-4.
To investigate whether the transcriptional regulation of lipl-4 converges onto DAF-16 in multiple contexts, we tested other nutrient sensors. C. elegans carrying a constitutively active form of the catalytic subunit of AMPK, AAK-2, did not show induction of lipl-4 (SI Appendix, Fig. S1G); therefore, we were not able to test the convergent model in this context. By contrast, reducing TGF-β signaling through loss-of-function mutation of the gene encoding the TGF-β receptor daf-1 did lead to lipl-4 induction (Fig. 2E). The classic outputs of the TGF-β pathway are DAF-3 (mammalian DPC4) and DAF-12 (mammalian LXR), and they both mediate responses to nutrients. However, neither of them mediated lipl-4 induction in daf-1 mutant C. elegans (SI Appendix, Fig. S1H). Instead, RNAi against daf-16 was negatively epistatic to daf-1 (Fig. 2E). The action of DAF-16 on the lipl-4 promoter is likely to be direct as DAF-16 binds 158 base pairs (bp) upstream of the lipl-4 transcription start site (SI Appendix, Table S3). Therefore, although our results do not rule out that other TFs may control lipl-4 in yet-to-be-tested contexts, the presented data enable us to propose that the transcriptional regulation of lipl-4 exemplifies the prevalent paradigm of transcriptional control, which assumes convergence of a transcriptional response onto a stable TF-target gene pair that acts across contexts (Fig. 2F). The convergent model supports wide-ranging inferences and meaningful extraction of general rules of transcriptional control from a relatively small number of experiments carried out in a limited number of experimental conditions.
Context Defines the TFs Regulating lipl-3.
We then investigated the transcriptional control of lipl-3. We previously reported that HLH-30 is necessary to induce the expression of lipl-3 in fasting C. elegans (5). Additionally, Chen et al. suggested that DAF-16 promotes lipl-3 induction in daf-2 mutant C. elegans fed ad libitum (8). Steinbaugh et al. proposed that SKN-1 promotes lipl-3 expression in glp-1 mutant C. elegans fed ad libitum (7). Furthermore, DAF-16 and SKN-1 were shown to generally promote the expression of lysosomal genes in daf-2 mutant worms (18). However, whether DAF-16, SKN-1, or other fasting-responsive TFs also contribute to the induction of lipl-3 in fasting C. elegans remains to be defined. Among the 11 TFs tested, only loss of hlh-30 function abrogated lipl-3 induction during fasting (Fig. 3A and SI Appendix, Fig. S2A). Unexpectedly and opposite to the most parsimonious inference, mutation of daf-16 led to further induction of lipl-3 in fasted worms (Fig. 3B), suggesting that DAF-16 is part of a regulatory axis that ultimately hinders lipl-3 induction.
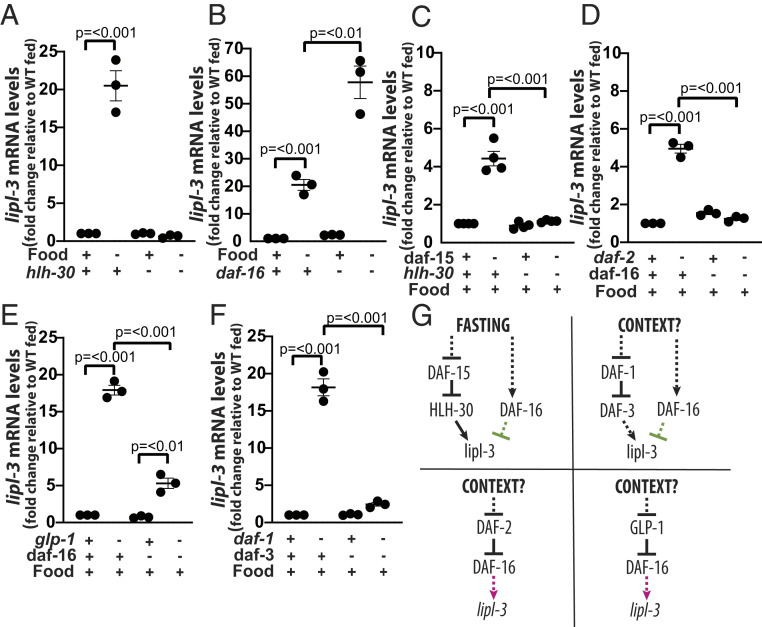
Fig. 3.
Context defines the TFs regulating lipl-3. (A–J) qRT-PCR analysis of gene expression in young adult C. elegans as described in Fig. 2. (A) WT and hlh-30(tm1978) mutant animals fasted for 6 h relative to fed (n = 3). (B) WT and daf-16(mu86) mutant animals fasted for 6 h relative to fed (n = 3). (C) WT and hlh-30(tm1978) mutant animals treated with empty vector control (EV) or RNAi against daf-15, the gene encoding the essential mTORC1component RAPTOR (n = 4). (D) WT and daf-2(e1368) animals treated with EV or RNAi against daf-16 (n = 3). (E) WT and glp-1(e2141) animals treated with EV or RNAi against daf-16 (n = 3). (F) WT and daf-1(m40) animals treated with EV or RNAi against daf-3 (n = 3). (K) Schematic representation of the contextualized transcriptional control of lipl-3 suggesting that the DAF-1-DAF-3, DAF-2-DAF-16, and GLP-1-DAF-16 axes would be important in yet-to-be-defined contexts but not during fasting.
As with lipl-4, we then used hlh-30 dependency as the probe to define which signaling input nodes link lipl-3 transcription to the feeding status of the worm (rationale in SI Appendix, Note S1). We did not observe lipl-3 induction in C. elegans carrying a constitutively active form of the catalytic subunit of AMPK, AAK-2 (SI Appendix, Fig. S2B). However, reduction of function of the intracellular nutrient sensor mTOR (let-363 RNAi) was sufficient to promote lipl-3 induction (SI Appendix, Fig. S1F). mTOR is present in cells in two complexes (mTORC1 and mTORC2), each having different functions (19); hence, we tested whether one or both mTOR complexes would be upstream of lipl-3. Inactivation of mTORC2 using RNAi against rict-1 (encoding C. elegans’s Rictor) did not alter the expression levels of lipl-3 (SI Appendix, Fig. S2C), while inactivation of mTORC1 using RNAi against daf-15 (encoding C. elegans’s Raptor) resulted in induction of lipl-3 (Fig. 3C). Similarly, impairing the function of the membrane receptor daf-1 was sufficient to promote lipl-3 induction (SI Appendix, Fig. S2D). In addition, similar to previous reports (7, 8), we found lipl-3 induced in daf-2 and glp-1 mutant worms, even when we fed animals E. coli XU363 (SI Appendix, Fig. S2 E and F). Therefore, mTORC1, insulin, Notch, and TGF-β signaling inhibit the expression of the lysosomal lipase lipl-3 in fed C. elegans.
Published studies of C. elegans showed the following: 1) fasting leads to inhibition of mTOR, glp-1, and daf-2 (12, 19, 20); 2) inhibition of mTOR, glp-1, and daf-2 leads to activation of HLH-30 (21); and 3) activated HLH-30 promotes the expression of autophagy and lysosomal genes (5, 21). Therefore, we hypothesized that HLH-30 would mediate the induction of lipl-3 in mTOR, glp-1, and daf-2–deficient C. elegans. Indeed, inhibition of mTORC1 (daf-15 RNAi) led to induction of lipl-3 in an hlh-30–dependent manner (Fig. 3C). Hence, the mTORC1-HLH-30 axis would link feeding status to the expression of lipl-3 in C. elegans. However, contrary to expectation, knockdown of hlh-30 did not affect the induction of lipl-3 in daf-1, daf-2, or glp-1 mutant animals (SI Appendix, Fig. S2 D–F), suggesting that a different TF or TFs would mediate lipl-3 induction in these genetic backgrounds and that these input sensors would mediate the activation of lipl-3 in other physiological contexts. Thus, at this point, we detoured to search for the TF/s promoting lipl-3 expression in the genetic contexts daf-2, glp-1, and daf-1.
We first tested DAF-16, SKN-1, and PHA-4 (7, 9, 22–24). Our qRT-PCR analyses showed, as suggested by Chen et al. (8), that loss of daf-16 function suppressed the induction of lipl-3 in daf-2 mutant animals (Fig. 3D). On the other hand, RNAi against skn-1 did not suppress lipl-3 induction in daf-2 mutant worms (SI Appendix, Fig. S3A). In the glp-1 mutant background, we observed that knockdown of pha-4 led to further induction of lipl-3 (SI Appendix, Fig. S3B) and that knockdown of skn-1 did not impair lipl-3 induction (SI Appendix, Fig. S3C). The latter was unexpected because Steinbaugh et al. previously proposed that skn-1 promotes the induction of lipl-3 in the glp-1(bn18ts) context (7). However, we here use a different glp-1 allele (e2141) and a different E. coli strain (E. coli XU363 in this study and E. coli HT115 in Steinbaugh’s study). We propose that the distinct E. coli strains may drive the discrepant results because different bacteria distinctively influence the nuclear localization of SKN-1::GFP (green fluorescent protein) (16). Thus, we then searched for a suppressor of lipl-3 induction in glp-1 mutant C. elegans fed E. coli XU363 and found that loss of daf-16 function suppressed most of the induction in this context (Fig. 3E). Thus, DAF-16 acts as an activator of lipl-3 expression upon inhibition of insulin and Notch signaling. Besides, we had found lipl-3 to be induced in daf-1 mutant animals (deficient in TGF-β signaling) (Fig. 3F and SI Appendix, Fig. S2D). Feeding daf-1 mutant animals RNAi against daf-3, daf-12, and daf-16 showed daf-3 to be negatively epistatic to daf-1 in the induction of lipl-3 (Fig. 3F), while daf-12 knockdown did not suppress and daf-16 knockdown further enhanced lipl-3 induction in this context (SI Appendix, Fig. S3D).
In summary (Fig. 3G), HLH-30 specifically acts as a transcriptional activator of lipl-3 downstream of the mTORC1 axis, and DAF-3 positively regulates lipl-3 downstream of the TGF-β axis. By contrast, DAF-16 has multiple, even opposing, roles on lipl-3 expression. DAF-16 positively regulates lipl-3 downstream of the insulin and Notch axes, while it negatively regulates lipl-3 during fasting, mTORC1 inhibition (SI Appendix, Fig. S3E), and downstream of the TGF-β axis. Therefore, at least so far, the transcriptional control of lipl-3 does not conform to a convergent model of transcriptional regulation.
The DAF-16-lipl-3–Activating Axis Promotes Resistance to Oxidative Stress.
DAF-16 activates lipl-3 downstream of inhibited insulin and Notch signaling (Fig. 3 D and E) and DAF-3 downstream of inhibited TGF-β signaling (Fig. 3F), whereas both DAF-16 and DAF-3 hinder the induction of lipl-3 during fasting (Fig. 3B and SI Appendix, Fig. S4A). We thus hypothesized that DAF-16 and DAF-3 would promote lipl-3 expression in a context or contexts other than fasting (Fig. 3G). We explored this hypothesis by testing whether lipl-3 would transcriptionally respond to other stresses. We found lipl-3 expression not increasing upon endoplasmic reticulum (ER), cold, heat, salt/osmotic stress, or anoxia (SI Appendix, Fig. S4B) but increasing in response to oxidative stress triggered by exposure to tert-butyl hydroperoxide (tBOOH) (Fig. 4A). Importantly, lipl-3 contributes to survival in animals exposed to tBOOH (Fig. 4B); hence, similar to its response to fasting, lipl-3 induction during oxidative stress would be a physiologically relevant response in C. elegans.
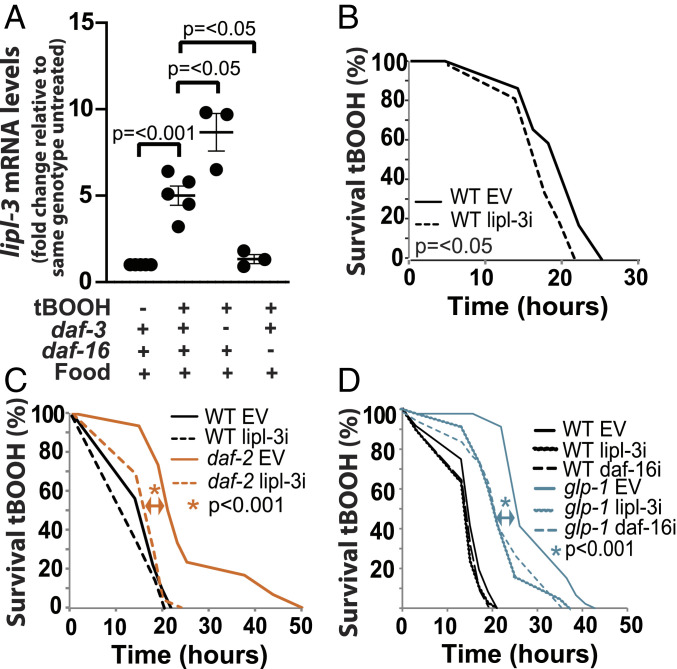
Fig. 4.
The DAF-16-lipl-3–activating axis promotes resistance to oxidative stress. qRT-PCR analysis of gene expression in young adult C. elegans as described in Fig. 2. (A) lipl-3 expression in WT, daf-3(mgDf90), and daf-16(mu86) mutant animals treated with tBOOH for 4 h relative to untreated (n = 3). (B–D) Survival to 5 mM tBOOH (n = 3) (SI Appendix, Table S7). (B) Survivorship of WT animals treated with EV or RNAi against lipl-3 (20 °C). (C) Survivorship of WT and daf-2(e1368) animals treated with EV or RNAi against lipl-3 (25 °C since L3s). (D) Survivorship of WT and glp-1(e2141) animals treated with EV or RNAi against daf-16 or lipl-3 (25 °C since L1s).
We then used the same rationale we used to investigate the feeding–fasting axis: if daf-3 and/or daf-16 were mediating the response to oxidative stress, then the lipl-3 response to oxidative stress should be daf-3 and/or daf-16 dependent. We found that loss of daf-3 function further enhanced the induction of lipl-3 in animals treated with tBOOH (Fig. 4A), suggesting that the daf-1-|daf-3→lipl-3 axis would operate in contexts other than fasting or oxidative stress. By contrast, loss of daf-16 function suppressed the induction of lipl-3 in animals treated with tBOOH (Fig. 4A), suggesting that the daf-2-|daf-16→lipl-3 and the glp-1-|daf-16→lipl-3 axes might be part of the response to changes in the redox status in C. elegans. We then tested this hypothesis. The contribution of daf-16 to resistance to oxidative stress in daf-2 animals has been extensively documented using several oxidative stress agents, including tBOOH. Nevertheless, given the context sensitivity of the system, we retested this observation in animals fed E. coli XU363. As expected, we found daf-16 suppressing daf-2–enhanced survival to tBOOH (SI Appendix, Fig. S4C). More importantly, we found lipl-3 contributing to daf-2 resistance to oxidative stress (Fig. 4C). Similarly, daf-16 and lipl-3 were negatively epistatic to glp-1–enhanced survival to tBOOH (Fig. 4D). Therefore, daf-2-|daf-16→lipl-3 and glp-1-|daf-16→lipl-3 compose input–output axes that maintain C. elegans’s redox status.
Literature-Based Network Predicts Multiple TF-TF Functional Interactions Regulating lipl-3.
We showed that DAF-16 positively regulates lipl-3 downstream of the insulin and Notch axes and during oxidative stress, while it negatively regulates lipl-3 downstream of the TGF-β axis and during fasting. The seemingly opposing roles that DAF-16 plays in the transcriptional regulation of lipl-3 is a noteworthy example of contextualized molecular function. Context-dependent roles for TFs and other molecules have been described before. For example, nuclear-hormone receptors (NHRs) promote different and even opposing transcriptional programs in different contexts (25). However, NHRs bind to alternative allosteric ligands and/or protein cofactors in different contexts, interactions that in turn define the subset of gene targets transcribed in each context. Even DAF-16 can promote growth in response to ultraviolet light stress (26) but arrest growth in response to starvation (27, 28). However, the consensus is that these distinct phenotypic outcomes are executed by at least partially distinctive downstream transcriptional programs (26, 28, 29). What is unique about the DAF-16-lipl-3 interaction is that daf-16 has opposite functional relationships with the same target gene. This is even more intriguing when considering that the different contexts investigated here coexist in fasting animals, as lack of food leads to reduced insulin and Notch signaling and activation of DAF-16 (12, 20).
We hypothesized that integrating all known interactions between sensors, TFs, and lipl-3 will allow us to understand the context-dependent interaction between DAF-16 and lipl-3. Hence, we reconstructed in silico the transcriptional network predicted to regulate lipl-3. We searched the literature for pairwise functional interactions connecting sensors, nutrient-responsive TFs, and lipl-3 (SI Appendix, Table S1). We built a lipl-3 regulatory network in which genes representing nutrient sensors, TFs, and lipl-3 are nodes, and their documented interactions are edges. We characterized each interaction in silico by its effect on the target and recorded whether it was mediated by one or more edges. Following the principle of parsimony, we evaluated every effect requiring more than one edge, and if single edges in the network already incorporated the published observation, then we did not expressly include the multiedge interaction in the network (SI Appendix, Supplemental Methods). The resulting network (which we will refer to as the inferred network) consists of the most parsimonious paths that include all the known interactions among the input, the sensors, the TFs, and lipl-3 (SI Appendix, Fig. S5 and Table S2).
The resulting regulatory network shows that signaling flux initiated by food and fasting can pass onto lipl-3 via an intricate network of TFs (SI Appendix, Fig. S5). Saliently, the network predicts that SKN-1 would independently regulate lipl-3 and that DAF-16, DAF-3, and HSF-1 would ultimately converge onto HLH-30 to promote lipl-3 expression. Since DAF-16, SKN-1, and DAF-3 do not mediate lipl-3 induction during fasting (Fig. 3B and SI Appendix, Figs. S2A and S4A), and HLH-30 and SKN-1 do not mediate the induction of lipl-3 in the glp-1 and daf-2 mutant contexts (Fig. 3 A and C and SI Appendix, Fig. S2 E and F), the inferred network seems to be inaccurate. The inaccuracies are most likely caused by the fact that we pooled together published interactions deduced from experiments done in different contexts. The inaccuracies of the inferred network illustrate the limitations of extrapolating epistatic interactions dissected in different contexts. Nevertheless, the inferred network includes potential functional interactions between DAF-16, DAF-3, SKN-1, HSF-1, MXL-3, and HLH-30 that may help us explain the context-dependent interactions between these TFs and lipl-3 and, more broadly, how context-dependent function can emerge from the same set of molecular players.
DAF-16 Represses lipl-3 Expression in Fed and Fasted C. elegans via HLH-30.
The generated literature-inferred network predicts that daf-16 functionally interacts with hlh-30 (SI Appendix, Fig. S5). Furthermore, work from the Riedel laboratory demonstrated that the DAF-16 and HLH-30 proteins directly interact and that daf-16 and hlh-30 can have opposing roles in the regulation of some target genes (29). However, this previous study did not focus on the roles that DAF-16 and HLH-30 play on specific target genes, and mutual regulation among daf-16 and hlh-30 has not been reported. Therefore, we first tested whether daf-16 and hlh-30 may be regulating each other. C. elegans overexpressing HLH-30 or carrying a loss of function mutation in hlh-30 show wild-type (WT) transcriptional levels of daf-16 (SI Appendix, Fig. S6A), as well as normal abundance and subcellular localization of DAF-16::GFP (SI Appendix, Fig. S6B), suggesting hlh-30 does not regulate daf-16 in our experimental setup.
By contrast, loss of daf-16 leads to a twofold increase in hlh-30 mRNA levels in fed animals and a twofold enhancement of hlh-30 induction during fasting when compared to fasted WT animals (Fig. 5A). Since we previously reported that the levels of induction of lipl-3 in fasting animals correlate with the levels of expression and activity of hlh-30 (5), and DAF-16 seems to repress hlh-30 expression, the results suggest that daf-16–mediated repression of hlh-30 could explain the seemingly repressing role of daf-16 in the expression of lipl-3. Indeed, we find that loss of function mutation of hlh-30 suppresses the induction of lipl-3 observed in daf-16–fed animals and the enhancement of induction observed in daf-16–fasted worms (Fig. 5B). These results place hlh-30 downstream of daf-16 in the regulation of lipl-3 in fed and fasting C. elegans. Furthermore, as supported by the presence of DAF-16 binding sites upstream of every hlh-30 isoform (SI Appendix, Table S3) and direct binding of DAF-16 in chromatin profiling by DNA adenine methyltransferase identification (DamID) surveys (30), hlh-30 is likely a direct target of DAF-16. Altogether, the results support a model in which DAF-16 is constitutively repressing hlh-30; hence, loss-of-function mutation of daf-16 leads to enhanced hlh-30 transcription and consequently lipl-3 induction independent of the feeding status of the worm.
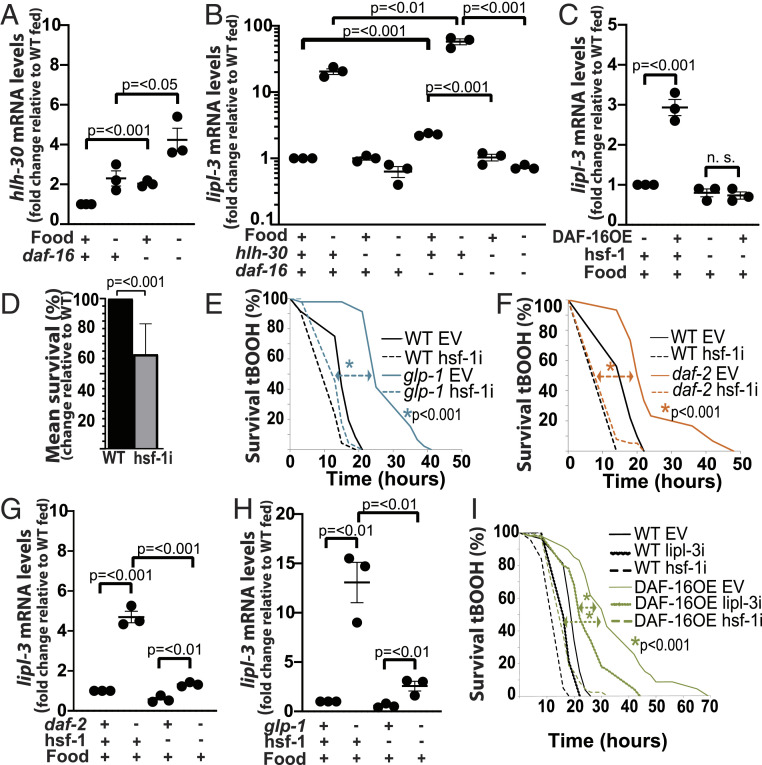
Fig. 5.
DAF-16 inhibits HLH-30 and promotes HSF-1 activity. (A–C, G and H) qRT-PCR analysis of gene expression in young adult C. elegans as described in Fig. 2. (E, F, and I) Survival to oxidative stress as described in SI Appendix. (A) hlh-30 expression in WT and daf-16(mu86) animals fasted for 6 h relative to fed (n = 3). (B) lipl-3 expression in WT, hlh-30(tm1978), daf-16(mu86), and hlh-30(tm1978);daf-16(mu86) double mutant animals fasted for 6 h relative to fed (n = 3). (C) lipl-3 expression in WT and DAF-16OE (GR1352) animals treated with empty vector control (EV) or RNAi against hsf-1 (n = 3). (D) Mean survival to 5 mM tBOOH of WT animals treated with EV or RNAi against hsf-1. Error bars denote SEM (n = 3). (E and F) Survival of WT and (E) glp-1(e2141) or (F) daf-2(e1368) animals treated with EV or RNAi against hsf-1 (n = 3). (G and H) lipl-3 expression in (G) daf-2(e1368) and (H) glp-1(e2141) animals treated with EV or RNAi against hsf-1 (n = 3). (I) DAF-16OE (GR1352) animals treated with EV or RNAi against hsf-1 or lipl-3 (n = 3).
DAF-16 Promotes lipl-3 Induction and Survival to Oxidative Stress via HSF-1.
DAF-16 seems to act as an activator of lipl-3 in glp-1 and daf-2 mutant C. elegans (Fig. 3 D and E) and in WT animals exposed to tBOOH (Fig. 4A). However, at least in the WT untreated condition reported by modEncode, there are no DAF-16 chromatin immunoprecipitation (ChIP) peaks up to 2 kb upstream of the lipl-3 coding sequence (SI Appendix, Table S3). Furthermore, using yeast 1-hybrid, we previously showed that DAF-16 does not bind to the lipl-3 promoter (5). Therefore, daf-16 may not be directly regulating lipl-3. Similarly, we find no predicted DAF-3 or SKN-1 binding sites in the lipl-3 promoter region (SI Appendix, Table S3). By contrast, MXL-3 and HLH-30 do directly bind to the lipl-3 promoter (5). However, MXL-3 is a repressor, and HLH-30 does not control lipl-3 in the daf-2 and glp-1 contexts (SI Appendix, Fig. S2 E and F); hence, alternative transcriptional regulators are likely to induce lipl-3 downstream of DAF-16 in the daf-2, glp-1, and oxidative stress contexts. From the most downstream transcriptional regulators predicted to modulate the expression of lipl-3 in our inferred network (SI Appendix, Fig. S5), HSF-1 could mediate DAF-16’s activating role on the lipl-3 promoter. Importantly, HSF-1 meets three criteria to play this role: 1) HSF is a stress-responsive TF; 2) there is a predicted HSF-1 binding site upstream of the lipl-3 start site (SI Appendix, Table S3); and 3) HSF-1 has been shown to orchestrate transcriptional responses downstream of glp-1 and daf-2 (31, 32). In support of DAF-16 regulating lipl-3 through hsf-1, we found that overexpression of DAF-16 was sufficient to promote induction of lipl-3 in fed C. elegans in an hsf-1–dependent manner (Fig. 5C).
Physiologically, we showed that daf-16 and lipl-3 contribute to survival to tBOOH in WT C. elegans (Fig. 4B and SI Appendix, Fig. S4C). Molecularly, we showed a daf-16–dependent induction of lipl-3 when C. elegans are treated with tBOOH (Fig. 4A). If hsf-1 links DAF-16 activation to the induction of lipl-3, as the model and data suggest, then hsf-1 should also contribute to survival to oxidative stress and to the induction of lipl-3 in glp-1, daf-2, and tBOOH-treated animals. In support of the first prediction, Servello et al. recently reported that hsf-1 contributes to C. elegans resistance to peroxides (33). We independently observed that hsf-1–deficient animals fed E. coli XU363 are more sensitive to tBOOH than WT worms (Fig. 5D). Additionally, we found that loss of hsf-1 function suppresses glp-1 and daf-2 resistance to tBOOH (Fig. 5 E and F), as well as the induction of lipl-3 observed in these mutants (Fig. 5 G and H). Finally, DAF-16OE animals are also resistant to tBOOH, and their resistance is hsf-1 and lipl-3 dependent (Fig. 5I). Therefore, the data place hsf-1 as a mediator of the adaptive transcriptional response to oxidative stress activated downstream of the insulin and Notch pathways and executed, at least in part, by lipl-3. Future studies may investigate whether DAF-16 directly or indirectly promotes HSF-1 activity.
Altogether, the data support a model in which HLH-30 and HSF-1 are regulating lipl-3 in the fasting and oxidative stress contexts, respectively. However, accurately defining context specificity requires one to test a regulatory axis downstream of the point of regulatory convergence. Therefore, we tested whether HLH-30 or HSF-1 would actually be the most downstream regulator in both contexts. Loss of hsf-1 function did not suppress the induction of lipl-3 in fasting C. elegans (SI Appendix, Fig. S6C); if anything, we observed a trend toward further induction of lipl-3 in fasted hsf-1 mutant animals. Similarly, although unable to promote lipl-3 induction during fasting, hlh-30–deficient worms were able to mount a robust lipl-3 response to tBOOH (SI Appendix, Fig. S6D). Furthermore, treating hsf-1 mutants with RNAi against hlh-30 did not further suppress or in any other way alter the suppressive effect of mutating hsf-1 on the induction of lipl-3 upon oxidative stress (SI Appendix, Fig. S6D). Similarly, loss of hsf-1 function did not alter the suppressive effect of knocking down hlh-30 on lipl-3’s induction during fasting (SI Appendix, Fig. S6C). Therefore, up to the extent that we have studied this axis, the transcription of lipl-3 conforms to a context-specific model of transcriptional regulation.
In extending the notion that one must consider where regulation converges to accurately classify a regulatory axis as convergent or contextualized, we tested the possibility that HSF-1 would be the ultimate convergent transcriptional regulator of lipl-4. However, the data showed that, in the tested conditions, HSF-1 is not part of the presented lipl-4 regulatory axes (SI Appendix, Note S2).
Our working model proposes that HSF-1 acts as a terminal regulator of lipl-3 in the oxidative stress axis, which in turn predicts the following: 1) HSF-1 promotes lipl-3 expression upon oxidative stress, 2) HSF-1 is activated by oxidative stress, 3) activation of HSF-1 is sufficient to promote lipl-3 induction, 4) HSF-1OE animals are resistant to oxidative stress, and 5) HSF-1OE resistance to oxidative stress is lipl-3 dependent. Support for these predictions follow 1) lipl-3 induction upon tBOOH treatment depends on hsf-1 (Fig. 6A); 2) HSF-1::GFP forms nuclear foci in animals treated with tBOOH (Fig. 6B), a phenotype previously associated with activation of HSF-1 (34); 3) Overexpression of HSF-1 is sufficient to promote lipl-3 induction (Fig. 6C); 4 and 5) HSF-1–overexpressing C. elegans are resistant to oxidative stress, and this resistance is lipl-3 dependent (Fig. 6D). Together, these data demonstrate that the TF-target pair composed of hsf-1 and lipl-3 is a relevant contributor to the C. elegans response to oxidative stress. Furthermore, the overlap between the players mediating the lipl-3 response to tBOOH and the ones composing the glp-1-|daf-16→hsf-1→lipl-3 and daf-2-|daf-16→hsf-1→lipl-3 epistatic axes, allows us to propose that these adaptive axes are tuned to protect the redox status of C. elegans.
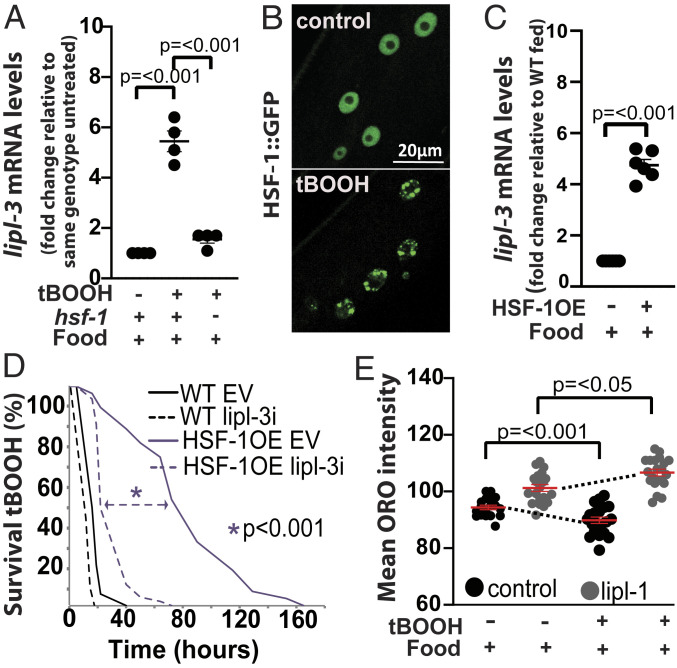
Fig. 6.
HSF-1 promotes lipl-3 induction and lipl-3–mediated survival to oxidative stress. (A–C) qRT-PCR analysis of gene expression in young adult C. elegans as described in Fig. 2. (A) lipl-3 expression in WT and hsf-1(sy441) mutant animals treated with 5 mM tBOOH for 4 h relative to untreated (n = 4). (B) Confocal images of adult worms expressing HSF-1::GFP (OG497) treated for 4 h with mock or 5 mM tBOOH. (C) lipl-3 expression in WT and HSF-1OE (AGD710) animals (n = 3). (D) Survivorship of WT and HSF-1OE (AGD710) animals treated with empty vector control (EV) or RNAi against lipl-3 since the L1 stage (n = 3). (E) Mean Oil red O (ORO) intensity in WT animals grown from L1 on EV or lipl-3 RNAi and exposed to 0.5 mM tBOOH for 4 h as day-1 adults. Statistical significance assessed via one-tailed unpaired parametric Student’s t test. Error bars denote SEM (n = 4).
lipl-3 Promotes Fat Mobilization upon Oxidative Stress.
Lipases contribute to survival of oxidative stress through two main mechanisms: 1) promoting sequestration of potentially toxic oxidized lipids and 2) providing energy and reducing power through fat mobilization (35, 36). Experimentally, in scenario 1 the content of lipid droplets increases (37), while in scenario 2, lipid droplet content is reduced. Therefore, we tested the response of C. elegans major fat stores to oxidative stress using the fat-specific dye Oil red O (38, 39). Supporting the notion that oxidative stress imposes an energetic demand provisioned by mobilization of fat stores, we observed a decline in Oil red O signal in animals treated with tBOOH (Fig. 6E). We then tested whether lipl-3 contributes to fat mobilization in C. elegans subject to oxidative stress. Indeed, knockdown of lipl-3 impaired fat mobilization during oxidative stress (Fig. 6E). Therefore, we propose that lipl-3 contributes to survival to oxidative stress through promoting lysosomal lipolysis, which would provide energy to mount an effective antioxidant response.
Multipoint Antagonism between the Oxidative Stress and Fasting Axes.
Our data suggest that fasting favors HLH-30–mediated regulation of lipl-3, and oxidative stress favors HSF-1–mediated regulation of lipl-3. Now, how does context tilt the balance in favor of one or the other TF? This question is particularly intriguing when considering that HLH-30 and HSF-1 share upstream regulators, including glp-1 (21, 32), daf-2 (21, 31), and mTOR (40, 41) and that all of these upstream regulators are simultaneously altered during fasting and oxidative stress. Hence, how does context determine the activity of intermediate players, when the upstream regulators are shared?
While food is abundant, HLH-30 is enriched in the cytoplasm, and lipl-3 is expressed at low levels (5). mTORC1 activity promotes cytoplasmic retention of the mammalian ortholog of HLH-30, TFEB (42–46), and knockdown of C. elegans’s mTOR (let-363 RNAi) promotes nuclear translocation of HLH-30 (21). Here, we more specifically show that inhibition of mTORC1 (via daf-15 RNAi) is sufficient to promote increased hlh-30 expression, nuclear translocation of HLH-30, and induction of lipl-3 (Fig. 3C and SI Appendix, Fig. S7 A and B). Therefore, one mechanism enabling HLH-30–mediated induction of lipl-3 during fasting is inhibition of mTORC1. However, for HLH-30 to promote lipl-3 expression, the activity of the lipl-3 repressor MXL-3 needs to be reduced, as these TFs compete for the same binding sites in the lipl-3 promoter (5). As mTORC1 coordinates the action of several transcriptional programs, we tested the hypothesis that mTORC1 may also modulate the activity of MXL-3. In support of this notion, RNAi against daf-15 leads to reduced MXL-3::GFP signal in fed animals (Fig. 7A), a response that mimics the MXL-3 physiological response to fasting (5). Therefore, as long as there is food and hence mTOR is active, HLH-30 action at the lipl-3 promoter would be disfavored through at least three paths: 1) food→mTORC1-|HLH-30 nuclear localization; 2) food→mTORC1→MXL-3–mediated outcompetition of HLH-30 at the lipl-3 promoter; and 3) unknown axis-|hlh-30 transcription. By contrast, during fasting, mTORC1 and MXL-3 activities would be reduced, and HLH-30 action on the lipl-3 promoter would be favored.
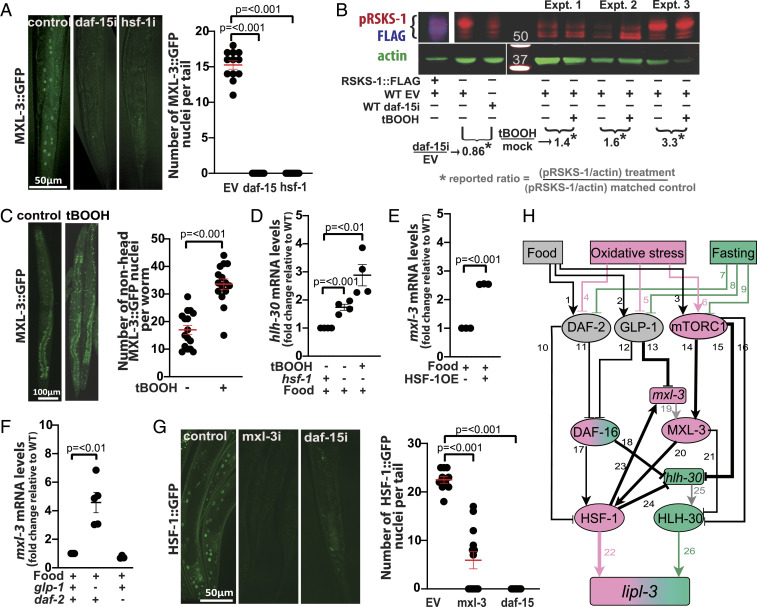
Fig. 7.
Multipoint antagonism between the oxidative stress and fasting axes. (A) Representative images of adult worms expressing MXL-3::GFP treated with empty vector control (EV) or RNAi against daf-15 or hsf-1 from the L1 stage. Mean number of GFP(+) nuclei per worm-tail ± SEM is depicted. (B) Western blot assessing phospho-RSKS-1 in worms treated with 5 mM tBOOH. RSKS-1 bands were ascertained based on the anti-FLAG signal observed in the lysates of RSKS-1::FLAG worms and from lysates of WT worms treated with RNAi against daf-15. (C) Representative images of adult worms expressing MXL-3::GFP treated with mock or 5 mM tBOOH for 4 h. Mean number of GFP(+) nuclei per worm (excluding head) ± SEM is depicted. (D–F) qRT-PCR analysis of gene expression in young adult C. elegans as detailed in Fig. 2. (D) hlh-30 expression in WT and hsf-1(sy441) mutant animals treated with tBOOH for 4 h relative to untreated (n = 4). (E) mxl-3 expression in WT and HSF-1OE (AGD710) animals (n = 3). (F) mxl-3 expression in WT, glp-1(e2141), and daf-2(e1368) animals (n = 3). (G) Representative images of adult worms expressing HSF-1::GFP (OG497) treated with EV or RNAi against mxl-3 or daf-15 from the L1 stage. Mean number of GFP(+) nuclei per worm-tail ± SEM is depicted. (H) Model of contextualized regulation of lipl-3. Shapes and arrowhead representations as in SI Appendix, Fig. S5. Colors of nodes represent activity in specific contexts as follows: pink = oxidative stress, green = fasting, and gray = inactive in both contexts. For more information see SI Appendix, Note S6 and Table S6.
In mammalian systems, mTORC1 is activated upon oxidative stress (47). We then tested the levels of kinase activity of mTORC1 using as readout the phosphorylation of its direct target RSKS-1 (mammalian S6K) (see Materials and Methods and SI Appendix, Note S3 for details). We observed increased levels of phosphorylated RSKS-1 (pRSKS-1) in C. elegans treated with 5 mM tBOOH for 4 h relative to mock treatment (Fig. 7B). Furthermore, based on the mTORC1-MXL-3 functional interaction demonstrated in Fig. 7A, we hypothesized that increased mTORC1 activity during oxidative stress would lead to increased activity of MXL-3. Supporting this hypothesis, we observed increased nuclear signal in MXL-3::GFP worms treated with tBOOH (Fig. 7C). Together, the data are in line with a model in which enhanced mTORC1-MXL-3 activity would disfavor HLH-30 activity on the lipl-3 promoter during oxidative stress.
We identified daf-2, glp-1, daf-16, hsf-1, and lipl-3 as nodes of an axis responsive to the redox status of C. elegans (denoted pink in Fig. 7H). Hence, we asked whether these nodes would also play a role in disfavoring HLH-30 action during oxidative stress. Bottom-up through the oxidative stress axis, the first player is HSF-1. Analysis of hlh-30 expression in an hsf-1 loss-of-function mutant shows induction of hlh-30 in the untreated and more so in tBOOH-treated animals (Fig. 7D). Furthermore, we found several potential HSF-1 binding sites up to −500 bp from the hlh-30 start site (SI Appendix, Table. S3), suggesting that HSF-1 may directly repress the expression of hlh-30. On the other hand, we found that hsf-1 deficiency leads to loss of MXL-3::GFP signal (Fig. 7A) and that HSF-1 overexpression is sufficient to promote mxl-3 induction (Fig. 7E). The presence of multiple HSF-1 binding sites up to −500 of the mxl-3 transcriptional initiation site, suggests that HSF-1 may directly activate mxl-3 transcription (SI Appendix, Table S3). Together, the results suggest that HSF-1 would curtail HLH-30 activity through at least two mechanisms: 1) repressing hlh-30 expression and 2) promoting mxl-3 expression, which in turn would promote MXL-3–mediated outcompetition of HLH-30 at the lipl-3 promoter.
As shown above, hsf-1 promotes lipl-3 transcription (Fig. 5 G and H), and MXL-3 and HSF-1 promote each other’s activities (Fig. 7 A and G). Therefore, we anticipated that loss of mxl-3 function would reduce HSF-1 activity and consequently lipl-3 expression and that mutation of hsf-1 in the mxl-3 background would further reduce lipl-3 expression. Thus, it was at first surprising to observe a synergistic induction of lipl-3 when both mxl-3 and hsf-1 were inactivated (SI Appendix, Fig. S7C). However, our model suggests an explanation for this observation. In the absence of MXL-3, HLH-30 can occupy its binding site at the lipl-3 promoter (5). In the absence of HSF-1, hlh-30 expression is elevated (Fig. 7D). Thus concomitant inactivation of mxl-3 and hsf-1 would compromise lipl-3 induction via the oxidative stress axis while augmenting it through the fasting axis. In line with this notion, RNAi against hlh-30 suppressed the synergistic induction of lipl-3 observed in the double mxl-3;hsf-1 mutant animals (SI Appendix, Fig. S7C). Also, surprisingly, despite the elevated levels of lipl-3 expression, mxl-3 animals showed similar sensitivity to tBOOH than WT worms (SI Appendix, Table S7), suggesting that lipl-3 is necessary but not sufficient to enhance oxidative stress resistance.
One step up in the oxidative stress axis, we find DAF-16 (Fig. 7H). We showed above that DAF-16 represses hlh-30 (Fig. 5A); hence, DAF-16 drives another inhibitory edge running from the oxidative stress to the fasting axis. On the other hand, loss-of-function mutation of daf-16 does not change the expression levels of mxl-3 (SI Appendix, Fig. S8A), and treatment with daf-16 RNAi does not change the abundance or localization of the MXL-3::GFP fusion protein (SI Appendix, Fig. S8B). Although these observations suggest lack of functional interaction between DAF-16 and MXL-3 in the regulation of lipl-3, future studies may focus on validating this preliminary conclusion, as requirement for a TF in a regulatory axis does not always manifest in regulation at the mRNA, protein, or localization levels of the functional interactors of interest.
Finally, we investigated whether the sensors would disfavor HLH-30 action during oxidative stress. We found that loss-of-function mutation of glp-1 promotes mxl-3 expression (Fig. 7F), which would indirectly promote outcompetition of HLH-30 at the lipl-3 promoter. By contrast, loss of daf-2 function did not increase mxl-3 expression (Fig. 7F). As for a more direct effect of the sensors on HLH-30, loss of daf-2 or glp-1 function did not increase HLH-30::GFP abundance (GFP intensity) or nuclear localization (SI Appendix, Fig. S8C), results that are in line with the lack of effect of tBOOH treatment on HLH-30::GFP expression (SI Appendix, Fig. S8D) but in contradiction with published results (29, 48). Hence, we repeated this experiment but now used a CRISPR knock-in of GFP downstream of HLH-30 (49). Confirming our original observation, we did not observe increased HLH-30::GFP intensity or nuclear localization in animals deficient in daf-2 or glp-1, or after treatment with tBOOH (SI Appendix, Fig. S8E). Nonetheless, meaningful biological differences may account for the discrepant results. Most notably, as described in SI Appendix, Note S4, in the experimental setup tested in this study, mTORC1 activity might be uncoupled from daf-2 and glp-1 or otherwise compensated.
Altogether, the data support the notion that the oxidative stress axis curtails the activity of HLH-30, at least, at four levels (Fig. 7H): 1) reduced GLP-1 activity (Notch signaling) promotes mxl-3 expression, which in turn would promote outcompetition of HLH-30 at the lipl-3 promoter; 2) reduced DAF-2 activity (insulin signaling) promotes DAF-16 activity, which in turn represses hlh-30 expression; 3) reduced DAF-2 activity promotes HSF-1 activity, which in turn represses hlh-30 expression; and 4) increased mTORC1 activity promotes 1) retention of HLH-30 in the cytoplasm and 2) increased activity of the HLH-30 competitor MXL-3.
We then investigated potential effects of the fasting axis on the oxidative stress axis. Loss of hlh-30 function does not change the levels of expression of hsf-1 (SI Appendix, Fig. S8F), or the abundance or localization of HSF-1::GFP (SI Appendix, Fig. S8G). We showed above that loss of hlh-30 function does not change the levels of expression of daf-16 or the abundance or localization of DAF-16::GFP (SI Appendix, Fig. S6 A and B). Similarly, loss of mxl-3 function does not change the levels of expression of daf-16 or the abundance or localization of DAF-16::GFP (SI Appendix, Fig. S8 H and I). Contrastingly, loss of mxl-3 function does lead to loss of HSF-1::GFP signal (Fig. 7G), suggesting a two-way activating edge between MXL-3 and HSF-1 (Fig. 7H). However, MXL-3 may not necessarily regulate hsf-1 directly, as no MXL-3 binding sites are found up to −5 kb of the hsf-1 transcription start site.
Finally, mTOR is the nutrient sensor linking feeding status to the activity of MXL-3 and HLH-30. Given that MXL-3 promotes HSF-1 activity (Fig. 7G) and mTOR activity positively correlates with MXL-3 abundance and nuclear localization (Fig. 7A), we hypothesized that mTOR may influence HSF-1 activity. In support of this notion, RNAi against daf-15 leads to reduced HSF-1::GFP signal (Fig. 7G). This observation is seemingly in contradiction with the report of Seo et al. (40), which shows hsf-1 suppressing the induction of hsp-16.1 and the longevity phenotype otherwise observed in animals treated with RNAi against mTOR or treated with the mTOR-inhibitor rapamycin (40). Testing in our experimental setup showed hsp-16.1 being down-regulated in animals treated with RNAi against mTOR and up-regulated in animals treated with tBOOH (SI Appendix, Fig. S9 A and B), results in line with mTOR being activated during oxidative stress and having a positive interaction with HSF-1 in our setting. Furthermore, a positive interaction between mTOR and HSF-1 has been reported across contexts and organisms. Most directly, rapamycin inactivates HSF1 to the point of erasing its genome-wide footprint in ChIP sequencing experiments, and of abrogating the expression of most of its targets across multiple cell lines (50, 51). Therefore, the precedent and our data on one side, and the documented role of HSF-1 in mTOR-driven longevity in C. elegans on the other, suggest, once again, a context-dependent interaction between two components of the C. elegans cytoprotective network.
In summary, the results are in line with a model in which (Fig. 7H) 1) food promotes mTOR activity, which in turn promotes both cytoplasmic retention of HLH-30 and MXL-3 activity. Together, these factors limit the activity of HLH-30 on the lipl-3 promoter in fed unstressed C. elegans; 2) Upon fasting, mTOR is inhibited, and consequently, HLH-30 can translocate into the nucleus and MXL-3 vacates the lipl-3 promoter, enabling HLH-30 to promote the transcription of lipl-3; and 3) Upon oxidative stress, mTOR activity is enhanced, likely more tightly restricting HLH-30 translocation into the nucleus. In parallel, a combination of glp-1 inhibition, mTOR activation, and activation of HSF-1 strengthens MXL-3 activity, which further limits HLH-30 activity at the lipl-3 promoter. Furthermore, MXL-3 promotes HSF-1 activity, building a self-reinforced hlh-30–restrictive loop. Simultaneously, the DAF-16-HSF-1 axis promotes repression of hlh-30 expression. The network-level promoted inhibition of HLH-30 and activation of HSF-1 makes HSF-1 a prevalent lipl-3 activator during oxidative stress. By contrast, during fasting, the edges that inhibit HLH-30 and activate HSF-1 are weakened, favoring HLH-30–mediated induction of lipl-3.
Finally, we asked how the experimental results we generated may fit or alter the literature-inferred network. We evaluated all the edges and nodes of the literature-inferred network and eliminated those that were contradicted by our results (SI Appendix, Note S5). This resulted in an improved network model of the transcriptional control of lipl-3 (Fig. 7H). We translated the network model of Fig. 7H into a discrete dynamic model in which inputs are binary (present/absent), and the activity of proteins and mRNAs is described by three levels calibrated to the basal activity corresponding to feeding (Materials and Methods and SI Appendix, Note S4). The model reproduces all of our observations in the different contexts studied here and expresses the contextualized transcriptional control of lipl-3 in a mathematical language (SI Appendix, Note S6). The model reduces context specificity to the opposing roles of MXL-3 on HSF-1 and HLH-30, as these provide the cell with an elegant mechanism for switching between the oxidant- and fasting-responsive state. In turn, the position of the switch would be determined by the signaling status of mTORC1 (Fig. 7H).
Discussion
Using systematic genetic epistasis and mathematical modeling, we show here that lipl-3 and lipl-4 represent two distinct modes of transcriptional control. We found that lipl-4 induction fits the paradigm in which a target gene is functionally linked to a specific terminal TF across several contexts, and we name this mode of transcriptional control “convergent.” It is worth noting that even if a transcriptional response is mediated by a stable TF-gene interaction across contexts, most likely the activation of this stable response will depend on a network of upstream factors that context specifically respond to each challenge and likely includes other TFs. Therefore, one would expect every gene’s response would ultimately result from the orchestrated action of several TFs, even if the other TFs are not directly promoting the transcription of the gene of interest (e.g., TF instead activates expression of an upstream node or the TF that in turns directly acts on the promoter of the gene of interest). This notion implies that classifying a regulatory axis as convergent may depend on whether the molecular factors under study are downstream or upstream of the point of convergence.
Unlike lipl-4, lipl-3 induction does not conform to the convergent paradigm. We name “contextualized” the mode of regulation represented by lipl-3, where the context defines the activity levels of two (or more) terminal TFs, one of which opposes the other, yet both have the same effect on the target gene in a particular context. Contextualized transcriptional regulation adds to the established strategies consisting of a TF activating distinctive transcriptional programs at different times (e.g., developmental stages), locations (e.g., tissues), or in different contexts as a result of interacting with alternative ligands or cofactors. In contextualized regulation, changing the flux of information among components of a regulatory axis defines the levels of activity of downstream TFs and in turn the level of transcriptional activation of downstream effectors. We propose that this mode of regulation contributes to plasticity in a wide range of contexts and that it likely represents a prevalent mode of regulation for stress-responsive downstream effectors.
In general, the response of each target gene to signaling activity may be unique; hence, the mechanisms that have evolved to limit the activation of targets to their proper contexts must operate at the level of individual promoters. In developmental biology, the distinct regulation of the transcriptional response of a particular target gene is defined by “zones of competence” (52). However, to the best of our knowledge, lipl-3 and lipl-4 are solely expressed in the intestinal cells of C. elegans (5, 11), and we are studying their transcriptional regulation at a single life stage in the ontogeny of the worm. Hence, space- and time-independent information is also capable of defining alternative axes of regulation for a single gene. Our data support the notion that plasma membrane and intracellular sensor-driven modulation of TF networks may allow cells to activate a single gene through alternative regulatory axes.
It is widely accepted that context affects the degree of activating or inhibitory action of a protein in a range from 0 to 1. However, we describe a scenario in which the environmental context can reverse the sign of the interaction. We define an intricate network of interactions between TFs (Fig. 7H) that provide an explanation for the seemingly opposing roles of DAF-16 in the expression of lipl-3. DAF-16 represses the expression of hlh-30, a gene whose protein product can effectively transcribe from the lipl-3 promoter in fasting conditions. On the other hand, DAF-16 promotes the activity of HSF-1, whose activity is promoted by inhibition of GLP-1 and DAF-2, activation of mTOR, and activation of MXL-3; all changes that are favored during oxidative stress. In a remarkable parallel to our work, the literature suggests a similar reversal for the DAF-16-sodh-1 interaction. sodh-1 had been identified as an effector positively regulated by DAF-16 in a daf-2 mutant background (53) and in response to oxidative stress (54). However, it was recently reported that in WT animals and during hypoxic stress, DAF-16 acts as a repressor of sodh-1 (55). Although no study has yet addressed this seemingly reversal of function of DAF-16 on the sodh-1 promoter, the published results suggest that diverting TF interactions may be an underappreciated strategy underlying plasticity. For example, the regulation of a critical longevity effector, fmo-2, is also likely to be contextualized. fmo-2 is induced by HLH-30 in response to fasting and hypoxia (56), and by NHR-49 in glp-1 mutants and in response to tBOOH (57) and to S. aureaus (58). Hence, again, contextualized transcriptional regulation may be common among genes affecting adaptive responses to multiple perturbations
Mechanistically, our work points to two interdependent strategies: 1) active repression of target promoter sequences in all inappropriate signaling contexts and 2) inability of signal-regulated TFs to activate transcription in any but the appropriate contexts. For mechanism 1, if food is available, mTOR promotes MXL-3 repressive activity on the lipl-3 promoter, an HLH-30–disfavoring action that is compounded by mTOR-mediated retention of HLH-30 in the cytoplasm. For mechanism 2, both HLH-30–inhibitory actions are relieved in the context of fasting. Significantly, HSF-1 is seemingly unaffected by MXL-3 presence at the lipl-3 promoter, whereas HLH-30 is outcompeted; observations that are in line with the HSF-1 binding site being 410 bp closer to the lipl-3 transcription start site than the MXL-3 binding site, whereas MXL-3 and HLH-30 bind to the exact same site in the lipl-3 promoter. Nevertheless, this mechanism would need to be directly tested as even though the binding sites of HLH-30 and MXL-3 were defined in the fed and the fasted contexts (5), none of the TFs at play has been biochemically investigated during oxidative stress.
We recognize that our study is limited to only several players and pathways and that the TF network we have created is not exhaustive, and there is further complexity that would need to be addressed in future studies. For instance, it is worth noting that HLH-30 may mediate the response to oxidative stress of targets other than lipl-3 (29), as well HSF-1 may regulate the expression of other genes during fasting (59). Such strict control over target gene expression may underlie two extraordinary features of cell signaling: 1) the capacity of a single pathway to elicit a large variety of gene expression patterns and hence to specify a large variety of cell responses using a reduced number of molecular players and 2) the capacity of a signaling pathway to control the specification of multiple distinct responses being directly dependent on its capacity to activate different, though perhaps overlapping, subsets of target genes in different contexts.
In summary, context is a critical variable in gene regulation; in particular for genes promiscuously activated or repressed across multiple physiological responses and stresses. All in all, we here demonstrate one more strategy organisms can use to mount adaptive responses to countless environmental and genetic perturbations using a limited number of molecular players.
Materials and Methods
All experimental procedures and statistical analyses are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Shawn Xu for kindly providing E. coli XU363. We thank K. Janes and M. Guertin for helpful reading of the manuscript. We are grateful to A. N. Chaudhry, M. Hilzendeger, A. Huckaby, A. Loperfito, M. Marraccini, G. Perry, T. Tabackman, J. Taylor, D. Martin, and R. Way for help with experiments and/or data analysis. We especially thank Nella Solodukhina for preparing reagents. We thank A. Bergland, G. Bloom, S. Siegrist, B. Winckler, and the E.J.O. laboratory members for the helpful discussions. We acknowledge the C. elegans strains provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and microscopy support from the University of Virginia Imaging Keck Center (NIH RR025616). This work was supported by NIH Grant DK087928, Pew Biomedical Scholar Award, Jeffress Trust Award, and the generous support of the W. M. Keck Foundation to E.J.O. The Jefferson Scholars Foundation Fellowship supported A.D-W., and NSF Grants MCB 1715826 and IIS 1814405 supported R.A.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104832118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Clark A. J., The mode of action of drugs on cells. Can. Med. Assoc. J. 29, 222 (1933). [Google Scholar]
- 2.Henderson S. T., Johnson T. E., daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975–1980 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Zhong M., et al., Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6, e1000848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R., A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U.S.A. 102, 13496–13501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Rourke E. J., Ruvkun G., MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M. C., O’Rourke E. J., Ruvkun G., Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinbaugh M. J., Narasimhan S. D., Robida-Stubbs S., Moronetti Mazzeo L. E., Dreyfuss J. M., Hourihan J. M., et al., Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. elife 4, e07836 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen A. T.-Y., et al., Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics 201, 613–629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapierre L. R., Gelino S., Meléndez A., Hansen M., Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 21, 1507–1514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seah N. E., et al., Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy 12, 261–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke E. J., Kuballa P., Xavier R., Ruvkun G., ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 27, 429–440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura K. D., Riddle D. L., Ruvkun G., The C. elegans DAF-2 insulin-like receptor is abundantly expressed in the nervous system and regulated by nutritional status. Cold Spring Harb. Symp. Quant. Biol. 76, 113–120 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G., Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23, 496–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizunuma M., Neumann-Haefelin E., Moroz N., Li Y., Blackwell T. K., mTORC2-SGK-1 acts in two environmentally responsive pathways with opposing effects on longevity. Aging Cell 13, 869–878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao R., et al., RNAi interrogation of dietary modulation of development, metabolism, behavior, and aging in C. elegans. Cell Rep. 11, 1123–1133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robida-Stubbs S., et al., TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amrit F. R. G., et al., DAF-16 and TCER-1 facilitate adaptation to germline loss by restoring lipid homeostasis and repressing reproductive physiology in C. elegans. PLoS Genet. 12, e1005788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y., Li M., Zhao D., Li X., Yang C., Wang X., Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. elife 9, e55745 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi C., et al., Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans. Genes Dev. 30, 307–320 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapierre L. R., et al., The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tissenbaum H. A., Ruvkun G., An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics 148, 703–717 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tullet J. M. A., et al., Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata S., Begun J., Troemel E. R., Ausubel F. M., DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics 178, 903–918 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antebi A., Nuclear receptor signal transduction in C. elegans. WormBook 1–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller M. M., et al., DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat. Cell Biol. 16, 1168–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baugh L. R., Sternberg P. W., DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16, 780–785 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kaplan R. E. W., et al., dbl-1/TGF-β and daf-12/NHR signaling mediate cell-nonautonomous effects of daf-16/FOXO on starvation-induced developmental arrest. PLoS Genet. 11, e1005731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X.-X., et al., DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat. Commun. 9, 4400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster E., et al., DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol. Syst. Biol. 6, 399 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu A.-L., Murphy C. T., Kenyon C., Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Shemesh N., Shai N., Ben-Zvi A., Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell 12, 814–822 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Servello F. A., Apfeld J., The heat shock transcription factor HSF-1 protects Caenorhabditis elegans from peroxide stress. Transl. Med. Aging 4, 88–92 (2020). [Google Scholar]
- 34.Morton E. A., Lamitina T., Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell 12, 112–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarc E., Petan T., Lipid droplets and the management of cellular stress. Yale J. Biol. Med. 92, 435–452 (2019). [PMC free article] [PubMed] [Google Scholar]
- 36.Bensaad K., et al., Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 9, 349–365 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Bailey A. P., et al., Antioxidant role for lipid droplets in a stem cell niche of drosophila. Cell 163, 340–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wählby C., et al., High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods 68, 492–499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ke W., Drangowska-Way A., Katz D., Siller K., O’Rourke E. J., The ancient genetic networks of obesity: Whole-animal automated screening for conserved fat regulators. Methods Mol. Biol. 1787, 129–146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo K., et al., Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell 12, 1073–1081 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Puertollano R., Ferguson S. M., Brugarolas J., Ballabio A., The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37, e98804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martina J. A., Chen Y., Gucek M., Puertollano R., MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Settembre C., et al., A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vega-Rubin-de-Celis S., Peña-Llopis S., Konda M., Brugarolas J., Multistep regulation of TFEB by MTORC1. Autophagy 13, 464–472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roczniak-Ferguson A., et al., The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nezich C. L., Wang C., Fogel A. I., Youle R. J., MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210, 435–450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarbassov D. D., Sabatini D. M., Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 280, 39505–39509 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Nakamura S., et al., Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 7, 10944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martina J. A., et al., A conserved cysteine-based redox mechanism sustains TFEB/HLH-30 activity under persistent stress. EMBO J. 40, e105793 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santagata S., et al., Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341, 1238303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lang B. J., et al., The functions and regulation of heat shock proteins: Key orchestrators of proteostasis and the heat shock response. Arch. Toxicol. 95, 1943–1970 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Barolo S., Posakony J. W., Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Zhang P., Judy M., Lee S.-J., Kenyon C., Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: Roles for GATA factors and lipid gene regulators. Cell Metab. 17, 85–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senchuk M. M., et al., Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 14, e1007268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heimbucher T., Hog J., Gupta P., Murphy C. T., PQM-1 controls hypoxic survival via regulation of lipid metabolism. Nat. Commun. 11, 4627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leiser S. F., et al., Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science 350, 1375–1378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goh G. Y. S., et al., NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell 17, e12743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wani K. A., Goswamy D., Taubert S., Ratnappan R., Ghazi A., Irazoqui J. E., NHR-49/PPAR-α and HLH-30/TFEB cooperate for C. elegans host defense via a flavin-containing monooxygenase. elife 10, e62775 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiao A., Jin X., Pang J., Moskophidis D., Mivechi N. F., The transcriptional regulator of the chaperone response HSF1 controls hepatic bioenergetics and protein homeostasis. J. Cell Biol. 216, 723–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogg S., et al., The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.