Significance
Our study provides detailed functional and spatial characteristics of immune cells in the LR-CHL microenvironment at single-cell resolution. We describe detailed T cell subset definitions and importantly identified a unique CD4+PD-1+CXCL13+CXCR5− TFH-like subset that surrounds HRS cells, appears in close proximity to CXCR5+ B cells, and is associated with poor clinical outcome. We also uncovered unique PD-1/PD-L1 axis biology in LR-CHL, namely a negative correlation between PD-L1 genetic alterations on HRS cells and PD-1 protein expression in the tumor microenvironment. Importantly, our findings contribute to a deeper understanding of cellular cross-talk in LR-CHL, which may aid in the development of novel biomarkers and targeted treatment strategies.
Keywords: Hodgkin lymphoma, single-cell analyses, CXCL13, PD-1, PD-L1
Abstract
Lymphocyte-rich classic Hodgkin lymphoma (LR-CHL) is a rare subtype of Hodgkin lymphoma. Recent technical advances have allowed for the characterization of specific cross-talk mechanisms between malignant Hodgkin Reed-Sternberg (HRS) cells and different normal immune cells in the tumor microenvironment (TME) of CHL. However, the TME of LR-CHL has not yet been characterized at single-cell resolution. Here, using single-cell RNA sequencing (scRNA-seq), we examined the immune cell profile of 8 cell suspension samples of LR-CHL in comparison to 20 samples of the mixed cellularity (MC, 9 cases) and nodular sclerosis (NS, 11 cases) subtypes of CHL, as well as 5 reactive lymph node controls. We also performed multicolor immunofluorescence (MC-IF) on tissue microarrays from the same patients and an independent validation cohort of 31 pretreatment LR-CHL samples. ScRNA-seq analysis identified a unique CD4+ helper T cell subset in LR-CHL characterized by high expression of Chemokine C-X-C motif ligand 13 (CXCL13) and PD-1. PD-1+CXCL13+ T cells were significantly enriched in LR-CHL compared to other CHL subtypes, and spatial analyses revealed that in 46% of the LR-CHL cases these cells formed rosettes surrounding HRS cells. MC-IF analysis revealed CXCR5+ normal B cells in close proximity to CXCL13+ T cells at significantly higher levels in LR-CHL. Moreover, the abundance of PD-1+CXCL13+ T cells in the TME was significantly associated with shorter progression-free survival in LR-CHL (P = 0.032). Taken together, our findings strongly suggest the pathogenic importance of the CXCL13/CXCR5 axis and PD-1+CXCL13+ T cells as a treatment target in LR-CHL.
Classic Hodgkin lymphoma (CHL) is a subtype of B cell lymphoma that is uniquely characterized by cross-talk of malignant cells with different types of noncancerous normal immune cells in the tumor microenvironment (TME). On the basis of the morphology and immunophenotype of the malignant Hodgkin and Reed-Sternberg (HRS) cells, infiltrating immune cells and fibroblastic elements, four histological subtypes of CHL are recognized: nodular sclerosis (NS), mixed cellularity (MC), lymphocyte rich (LR), and lymphocyte depleted (LD) (1). Lymphocyte-rich CHL (LR-CHL) is a rare subtype of Hodgkin lymphoma, which accounts for ∼5% of all CHL. The disease is more common in elderly males and exhibits less frequent mediastinal involvement and bulky disease when compared to other CHL subtypes (2–4). Histologically, LR-CHL is characterized by a predominant nodular pattern with few scattered HRS cells distributed in T cell–rich zones, with numerous small lymphocytes and an absence of eosinophils and neutrophils in the nodules (2). Clinically, patients often present with localized peripheral lymphadenopathy and it typically is associated with a favorable outcome (3).
In CHL, nonmalignant immune cell populations make up more than 99% of the tumor bulk and create a tumor-supportive milieu via cross-talk with the rare HRS cells (∼1%) (1, 5). The presence of specific immune cell types, including macrophages and T cells, as well as their spatial arrangement, plays a fundamental role in creating an immunosuppressive microenvironment in CHL. The presence of these immune cell types has been shown to have prognostic value and highlights the dependency of HRS cells on the TME for survival and immune evasion (6–11). CD4+ T cells are significantly enriched in CHL compared to reactive lymphadenopathies, which is consistent with previous literature that showed more frequent major histocompatibility class II (MHC-II) expression on HRS cells than MHC-I (12, 13). Of note, loss of MHC-II expression on HRS cells was found to be associated with inferior response to immune checkpoint inhibitors in CHL (12, 14). This indicates the importance of CD4+ T cells in CHL pathogenesis, and suggests MHC-II/CD4-dependent interactions between malignant cells and the TME.
Recent technical advances, including single-cell sequencing and spatial imaging analysis, revealed a high abundance of various types of immunosuppressive CD4+ T cells in the TME of CHL. These expressed coinhibitory receptors, including LAG3 and CTLA4 (15, 16). The interactions between these receptors and their ligands are believed to be the driving force behind the impaired immune response and unique microenvironment composition in CHL. Interestingly, despite the high efficacy of anti–PD-1 blockade in relapsed/refractory CHL (17–22), PD-1+ cells are not particularly abundant in Hodgkin lymphoma (HL) tissue except in LR-CHL (15, 23). PD-1+ T cells forming rosettes around HRS cells are reported to be present in approximately half of LR-CHL cases (2). However, the specific role of these PD-1+ T cells, their coexpression patterns with other coinhibitory receptors, and the overall TME composition, has not been well characterized in LR-CHL due to disease rarity.
Here, using single-cell RNA sequencing (scRNA-seq) and multicolor immunofluorescence (MC-IF), we identified LR-CHL–enriched immune cell subsets, including CXCL13+ T follicular helper (TFH)-like cells that were shown to be surrounding HRS cells in spatial analysis and were in close contact with CXCR5+ B cells. On the strength of an unprecedented number of single-cell transcriptomes in combination with multiplexed spatial assessment, we deciphered the unique immune cell architecture of the TME in LR-CHL with implications for previously uncharacterized treatment targets.
Results
The LR-CHL–Specific Immune Microenvironment at Single-Cell Resolution.
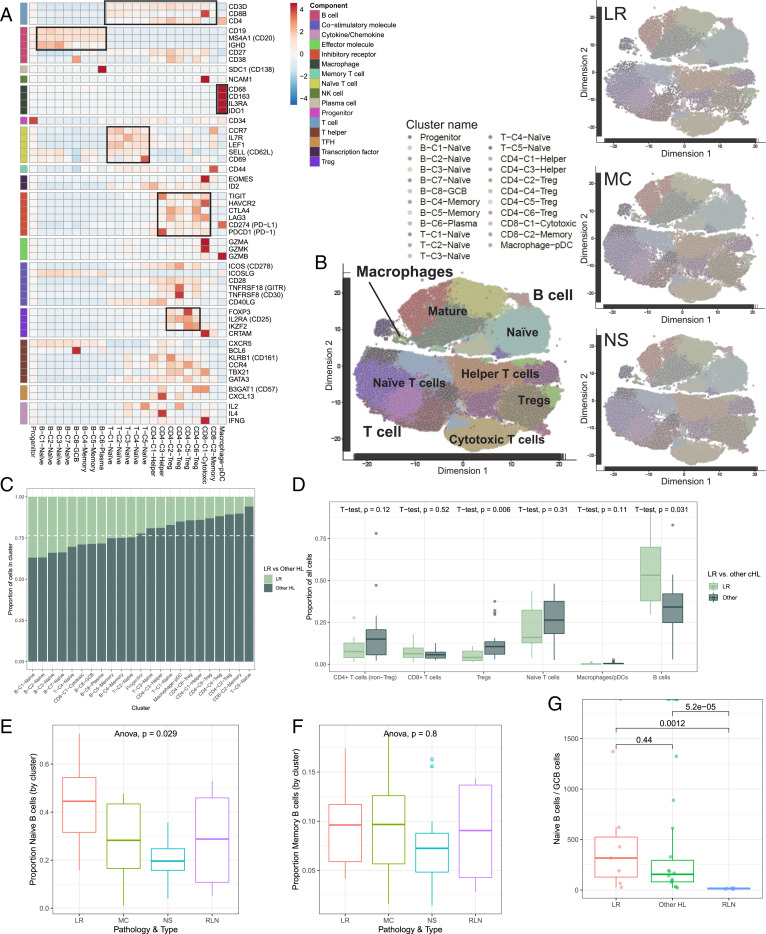
To investigate the specific immune cell profile of the LR-CHL TME, we utilized our previously published scRNA-seq cohort of CHL and sequenced an additional 7 LR-CHL cases for comparison (15). The resulting cohort contained data from 28 CHL patients, including 8 LR, 11 NS, and 9 MC, plus 5 reactive lymph nodes (RLNs) sequenced as normal comparators (SI Appendix, Tables S1 and S2). In total, transcriptomes were generated for 146,473 sorted live cells (SI Appendix, Table S3). After batch correction and normalization (Materials and Methods), unsupervised clustering of the single-cell expression profiles yielded a total of 23 clusters. We assigned each cluster to a cell type based on the expression of genes described in published transcriptome data of sorted immune cells (24) and known canonical markers (Fig. 1 A and B and SI Appendix, Fig. S1 and Dataset S1). This produced 13 T cell clusters, 8 B cell clusters, 1 macrophage/ plasmacytoid dendritic cell (pDC) cluster, and 1 progenitor cell cluster. Notably, we did not observe any clusters resembling HRS cells, likely due to size limitations in the microfluidics device or loss of HRS cells during the freezing and thawing process. While most immune cell phenotypes exhibited overlap among pathological subtypes, as demonstrated by clusters containing a mixture of cell types, we observed an enrichment of cells from LR-CHL in some specific cell clusters (Fig. 1 B and C). Of interest, regulatory T cells (Tregs), which we and others have observed as an enriched immune cell type in CHL (9, 15, 25, 26), were significantly decreased in LR-CHL compared to other CHL subtypes (P = 0.006, t test; Fig. 1D). All 4 Treg clusters, including those characterized by high LAG3 expression and those with high FOXP3 expression, had a low proportion of cells originating from LR samples, suggesting a relative paucity of Tregs as a unique feature of the LR-CHL TME (Fig. 1 C and D and SI Appendix, Fig. S2 A and B). Conversely, we found that B cell clusters were uniquely enriched in LR-CHL cells when compared to other CHL subtypes, and specifically all 4 naïve B cell clusters were dominated by cells derived from LR tumors (Fig. 1 C–E). While the proportion of cells assigned to naïve B cell clusters was significantly higher in LR-CHL samples compared to other CHL subtypes and RLNs, the proportion of memory B cells was comparable (Fig. 1F). We confirmed B cell enrichment in LR-CHL on the protein level by flow cytometry (SI Appendix, Fig. S2C). Intriguingly, the proportion of cells assigned to the germinal center B cell (GCB) cluster was significantly lower in LR-CHL compared to RLN samples (Fig. 1G and SI Appendix, Fig. S2D).
Fig. 1.
Immune cell atlas of the LR-CHL microenvironment at single-cell resolution. Cells from 28 CHL and 5 RLN cases were clustered using the PhenoGraph algorithm to identify groups of cells with similar expression patterns. (A) Heatmap summarizing mean expression (normalized and log transformed) of selected canonical markers in each cluster. Data have been scaled row-wise for visualization. The covariate bar on the Left side indicates the component associated with each gene, and black boxes highlight prominent expression of known subtype genes. (B) Single-cell expression of all cells from CHL and RLN in tSNE space (first two dimensions). Cells are colored according to PhenoGraph cluster. Subsets of cells from each CHL subtype are shown on the same coordinates. (C) Proportion of cells in each cluster originating from LR-CHL (light green) and other CHL (dark green) samples. The dashed white line represents the total proportion of cells from other CHL samples in the merged population. (D) The proportion of cells assigned to a given immune cell type (as determined by cluster annotation) was calculated for each sample. Boxplots summarize the distribution of the proportions for all samples, grouped by pathological subtype (LR-CHL or other CHL subtype). P values are shown Above and demonstrate a significant increase in the proportion of B cells present in LR-CHL compared to other CHL. (E and F) Boxplots summarizing the proportion of naïve (E) and memory (F) B cells relative to total cells in each sample, separated according to CHL subtype and RLNs. (G) Ratio of naïve B cells/germinal center B cells (by cluster assignment) according to pathological subtype. P values were calculated using t tests.
Single-Cell Expression Patterns of LR-CHL–Specific Immune Subsets.
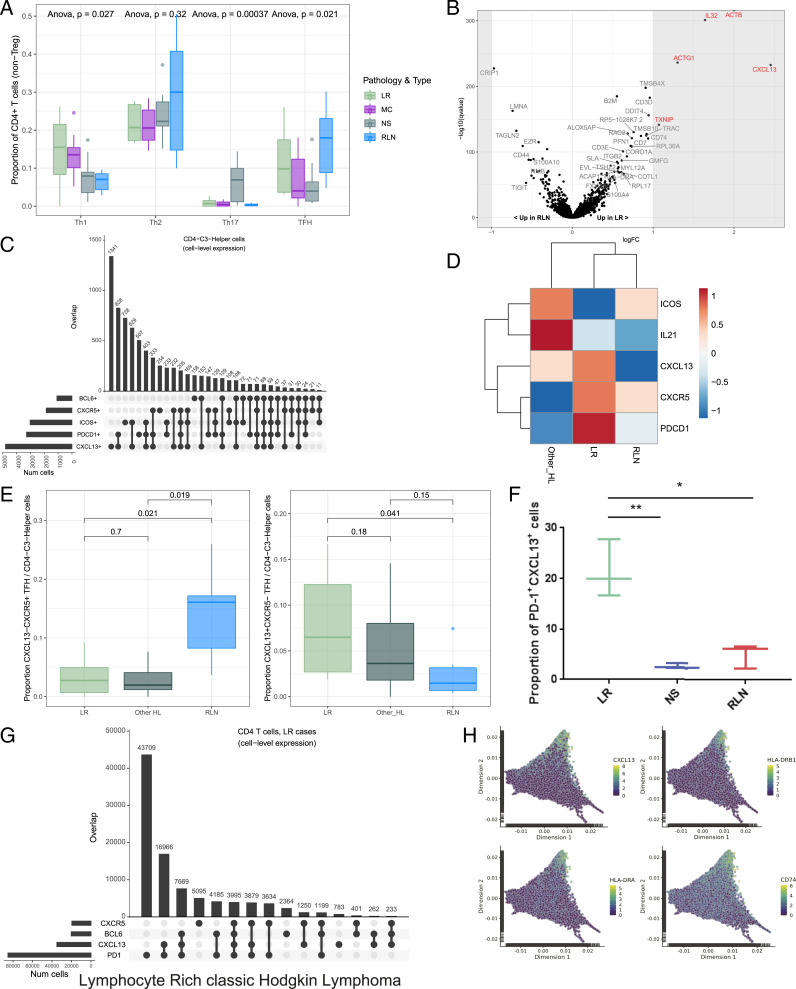
TFH cells play an important role in normal B cell development by supporting B cell differentiation and antibody production (27, 28). Our data demonstrated preferential enrichment of the TFH population in LR-CHL as compared to other CHLs (Fig. 2A). To investigate the characteristics of TFH cells in LR-CHL, we performed differential gene expression analysis between cells from LR-CHL and RLN samples in the cluster that most resembled a TFH profile (“CD4-C3-Helper”). Of note, CXCL13 was identified as the most up-regulated gene in LR-derived TFH cells compared to RLNs (Fig. 2B). CXCL13, which is the canonical ligand of CXCR5, is well known as a B cell attractant that works via the CXCL13/CXCR5 signaling axis and is highly expressed on follicular dendritic cells (FDCs) in germinal center lesions (29). Analyzing coexpression patterns on the single-cell level revealed that the majority of CXCL13+ T cells coexpressed PD-1 and ICOS, which are known as universal TFH markers, but coexpression with CXCR5, another common TFH marker, was rarely observed (Fig. 2C). Notably, the coexpression pattern of TFH markers was variable among disease subtypes, suggesting a potentially distinct role of TFH cells in each subtype (Fig. 2D). The proportion of TFH cells with a classical TFH profile coexpressing CXCR5 and PD-1 was high in RLNs, whereas TFH cells coexpressing PD-1 and CXCL13, but not CXCR5, were significantly more prevalent in LR-CHL (Fig. 2E and SI Appendix, Fig. S3). TFH coexpression patterns in LR-CHL were validated on the protein level by flow cytometry (FCM) using cell suspensions from primary CHL patients (n = 3) and RLN samples (n = 3) gated for CD4+ T cells (Fig. 2 F and G and SI Appendix, Fig. S4). We confirmed that PD-1+CXCL13+CD4+ T cells were significantly enriched in LR-CHL compared to both RLN and NS samples on the protein level (Fig. 2F). Intriguingly, differential expression between classical CXCR5+CXCL13− TFH cells and CXCL13+CXCR5− TFH-like cells revealed higher expression of MHC-II genes in the CXCL13+CXCR5− population (SI Appendix, Fig. S5). MHC-II expression is a known marker of T cell activation (30), indicating that the CXCL13+CXCR5− population exhibits an activated phenotype.
Fig. 2.
Detailed characterization and coexpression patterns of helper T cells in the tumor microenvironment of LR-CHL. (A) The proportion of helper T cells assigned to various cell subsets was calculated for each sample (see SI Appendix, Materials and Methods for assignment criteria). Boxplots summarize the distribution of the proportions for all samples, grouped by pathological subtype. P values, calculated using an Anova test, are shown Above. (B) Volcano plot showing differentially expressed genes between cells in the TFH cell cluster (CD4-C3-Helper) originating from LR-CHL vs. RLN samples. The y axis summarizes P values corrected for multiple testing using the Benjamini–Hochberg method (“q values”). Significant genes are labeled in red (q value <0.05 and absolute log2 fold change ≥1). (C) UpSet plot showing coexpression patterns of inhibitory receptors (CXCR5, PDCD1 [PD-1], CXCL13, ICOS, and BCL6) for individual cells in the TFH cluster. (D) Heatmap showing mean expression of TFH markers for cells in the CD4-C3-Helper cluster across all samples, grouped by pathological subtype. Expression values have been scaled row-wise for visualization. (E) Boxplots summarizing the proportion of classical TFH (Left) and CXCL13+ helper T cells (Right) in each sample, separated according to pathological subtype. P values, calculated with t tests, are shown Above. (F) Boxplot summarizing the proportion of PD-1+CXCL13+ cells from each cell suspension sample analyzed by flow cytometry, separated according to pathological subtype. Data are shown as the mean ± SEM (n = 3). *P < 0.05; **P < 0.01. (G) UpSet plot showing coexpression patterns on CD4+ T cells in LR-CHL by flow cytometry. (H) Cellular trajectories were inferred using diffusion map analysis of cells in CD4+ helper T cell clusters. Individual cells are shown in the first two resulting dimensions. Expression levels are shown for the four genes most positively correlated with dimension 2 score (SI Appendix, Materials and Methods).
To explore the functional role of CXCL13+ T cells, we next applied the diffusion map algorithm (31, 32) with the aim of characterizing differentiation states among helper T cells (Fig. 2H). CXCL13+ cells were enriched at the positive end of the second dimension, which was correlated with expression of genes representative of a terminal differentiation signature (SI Appendix, Fig. S6). The other most positively correlated genes tracking with dimension 2 were MHC-II genes, providing further evidence for an activated phenotype in the CXCL13+ T helper cells.
Spatial Assessment of CXCL13+ T Cells and HRS Cells.
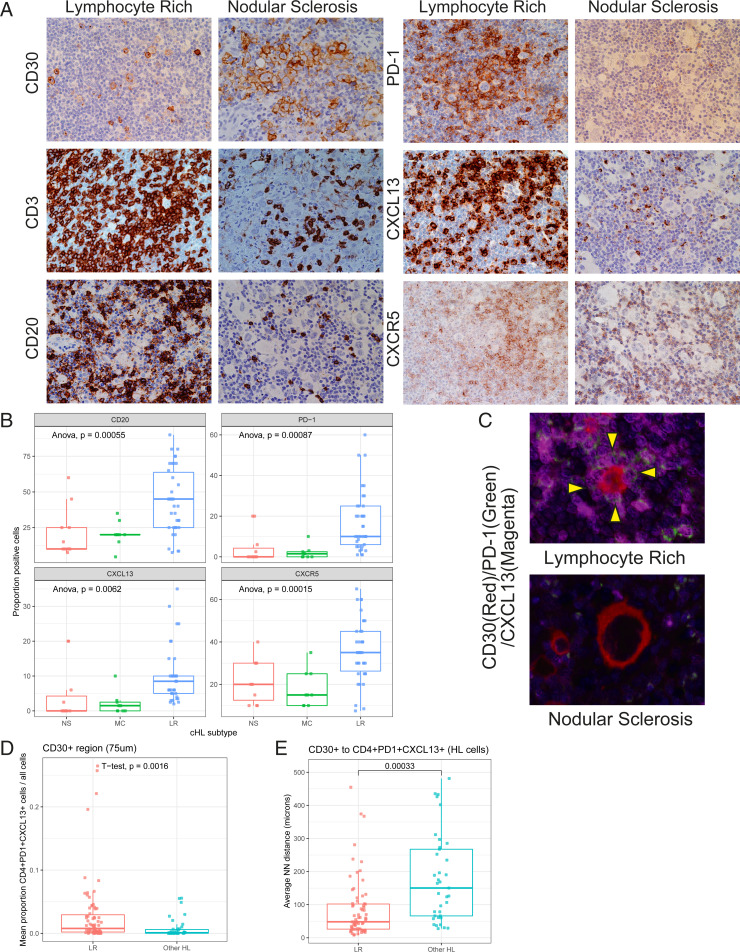
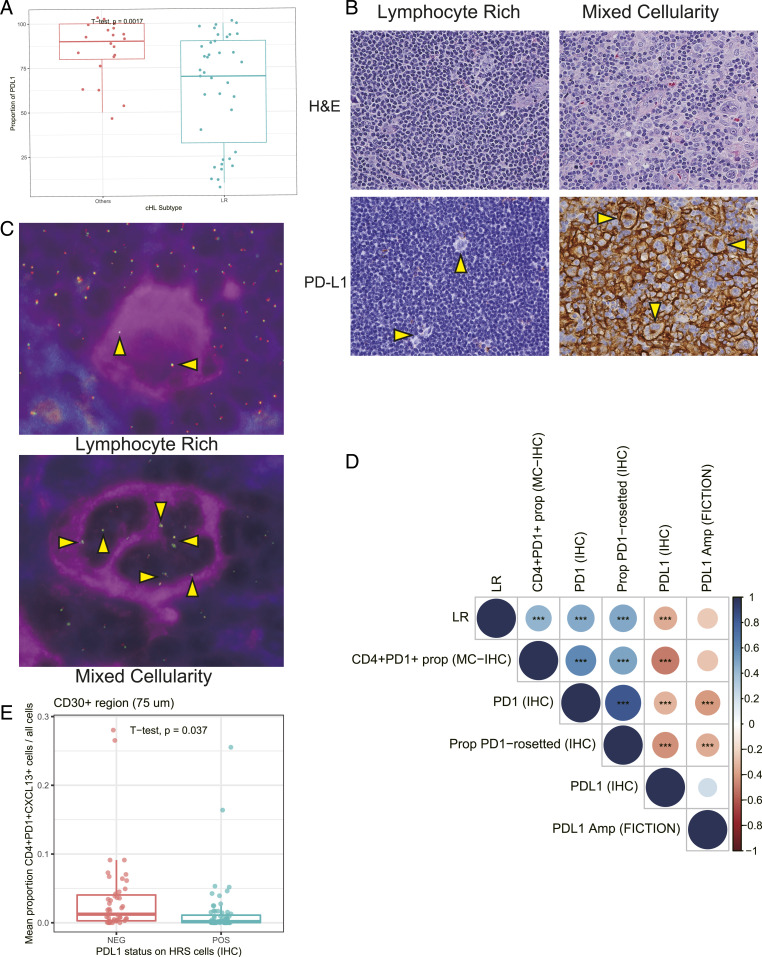
We next sought to validate our scRNA-seq findings in histologically intact tissue sections and understand the spatial relationship between CXCL13+ T cells and malignant HRS cells. We created a tissue microarray (TMA) from tumor tissue of 37 LR-CHL patients, which included 6 cases from our scRNA-seq cohort (SI Appendix, Table S4), and performed immunohistochemistry (IHC) on this TMA and the TMA from our previous scRNA-seq cohort (n = 26; 1 LR, 9 MC, 11 NS, 5 RLN) (15). IHC revealed that CXCL13+ T cells were significantly enriched in the LR-CHL TME compared to other subtypes (Fig. 3 A and B). Approximately half of the LR-CHL cases (46%) showed CXCL13+ T cells surrounding HRS cells (“rosettes”), whereas only 13% of patients with other CHL subtypes showed CXCL13+ T cell rosettes. Since PD-1+ T cell rosettes have been previously described as a specific feature of LR-CHL (2), we next evaluated PD-1 IHC on the CHL TMAs. Of note, all LR-CHL cases with CXCL13+ T cell rosettes also showed PD-1+ cell rosettes surrounding HRS cells, and PD-1+ cells were also significantly enriched in the LR-CHL TME compared to other CHLs. Consistent with scRNA-seq data, we also observed that CD20+ B cells were significantly enriched in LR-CHL (Fig. 3B). To validate coexpression patterns on the CXCL13+ T cells, we applied MC-IF on the same TMAs. We confirmed that most CD4+CXCL13+ T cells coexpressed PD-1, and the proportion of CD4+PD-1+CXCL13+ T cells in HRS-surrounding regions (i.e., within 75 μm of a CD30+ cell) was significantly increased in LR-CHL (Fig. 3 C and D). Similarly, the average distance between CD30+ cells (HRS cells) and their nearest CD4+PD-1+CXCL13+ T cell was significantly shorter in LR-CHL (Fig. 3E).
Fig. 3.
Spatial distribution of HRS cells and CXCL13+ T cells in LR-CHL. (A) IHC staining for major immune cell markers in representative cases with either LR-CHL (Left; LRCHL20) or nodular sclerosis CHL (Right; CHL03) (×400). (B) Boxplot showing proportions of positive cells by IHC for major immune cell markers according to disease subtype. P values were calculated using Anova tests. (C) Multicolor IF staining (CHL05 and LRCHL16) for CD30 (red), PD-1 (green), and CXCL13 (magenta) shows localization of CD4+PD-1+CXCL13+ T cells in rosettes around HRS cells in cases with LR-CHL. No rosettes are observed in cases of other CHL subtypes (e.g., nodular sclerosis shown here). (D) Boxplot showing the proportion of CD4+PD-1+CXCL13+ T cells in the region surrounding CD30+ cells (HRS) for each sample, separated by CHL subtype. The surrounding region was defined by a distance of 75 μm. (E) Average nearest neighbor (NN) distance from an HRS cell (defined by CD30+) to its closest CD4+PD-1+CXCL13+ cell was calculated per sample and plotted by pathological subtype. P values were calculated using t tests.
CXCL13/CXCR5 Interaction in LR-CHL.
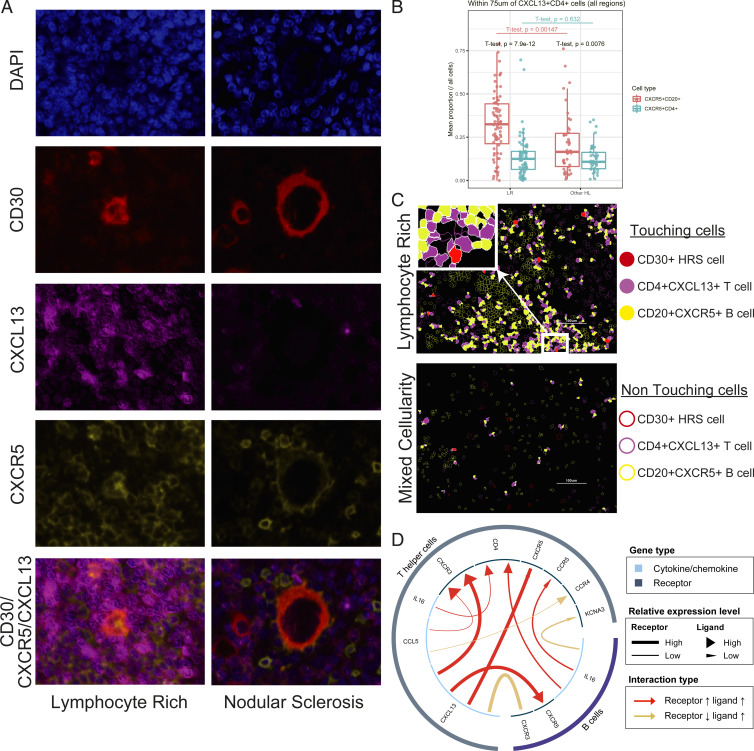
As CXCR5 is the primary receptor for CXCL13, we next investigated CXCR5+ cells in the TME of LR-CHL with the aim of characterizing their relationship with CD4+CXCL13+ T cells. MC-IF analysis revealed that the majority of CXCR5+ cells in the TME were B cells (CD20+) (SI Appendix, Fig. S7). In contrast to CD4+CXCR5+ T cells, CD20+CXCR5+ B cells were significantly enriched in regions surrounding CD4+CXCL13+ T cells (Fig. 4 A and B). Notably, CXCL13+ cells rarely coexpressed CXCR5, confirming a mostly mutually exclusive pattern between CXCR5 and CXCL13 in the TME of LR-CHL (Fig. 4A). Furthermore, the proportion of CD20+CXCR5+ cells in regions surrounding CD4+CXCL13+ T cells was significantly increased in LR-CHL when compared with other CHL subtypes, while the proportion of CD4+CXCR5+ cells was comparable between subtypes (Fig. 4 B and C). The iTALK tool (33) was used to predict receptor/ligand interactions enriched in LR-CHL compared to other CHLs and confirmed a significantly increased positive interaction between CXCL13 on helper T cells and CXCR5 on B cells (Fig. 4D), supporting the importance of the CXCR5/CXCL13 axis in the specific pathogenesis of LR-CHL. In contrast, TFH cells in a normal RLN germinal center showed a typical TFH cell phenotype (CXCR5+CXCL13−) (SI Appendix, Fig. S8).
Fig. 4.
CXCL13/CXCR5 interaction in LR-CHL. (A) Multicolor IF staining (CHL05 and LRCHL16) for CD30 (red), CXCL13 (magenta), and CXCR5 (yellow), shows localization of CXCR5+ cells near CXCL13+ cells in the region surrounding HRS cells in cases with LR-CHL. CXCL13+ cells (magenta) are rarely coexpressed with CXCR5 (yellow). (B) Boxplot showing the proportion of CD20+CXCR5+ B cells and CD4+CXCR5+ T cells in the region surrounding CD4+CXCL13+ T cells (within 75 μm) for each sample, separated by pathological subtype. t tests show comparisons both within the subtypes (LR or other HL) and across subtypes (LR vs. other HL). (C) Membrane map depicting CD4+CXCL13+ T cells (magenta), CD20+CXCR5+ B cells (yellow), and CD30+ HRS cells (red). Touching cells (CD30+ HRS cells/CD4+CXCL13+ T cells and CD4+CXCL13+ T cells/CD20+CXCR5+ B cells) are represented by filled shapes. (D) An enriched positive interaction between CXCL13 on T helper cells and CXCR5 on B cells in LR-CHL was predicted using the iTALK tool.
PD-1/PD-L1 Biology in LR-CHL Patients.
To investigate PD-1/PD-L1 biology in LR-CHL, we next investigated the expression of PD-L1 on HRS cells. HRS cells often exhibit overexpression of PD-L1 through copy number gains and amplifications of the 9p24.1 locus where its coding gene (CD274) resides (34–38). Surprisingly, regardless of the proportion of PD-1+ T cells, PD-L1 expression on HRS cells was significantly lower in LR-CHL when compared with other CHL subtypes (Fig. 5 A and B). Furthermore, we also performed copy number analysis of the CD274 and PDCD1LG2 (encoding PD-L2) genes in HRS cells using the FICTION technique (36), which enables quantitative assessment of the copy number of the CD274/PDCD1LG2 genes in CD30-labeled IHC sections on a TMA (Fig. 5C). Interestingly, LR-CHL cases showed fewer copy number amplifications of CD274/PDCD1LG2 (19%, 5/26 cases) compared to other CHL subtypes (43%, 9/21 cases), and CD274 copy number amplification status was positively correlated with PD-L1 protein expression on HRS cells (Fig. 5D). Of note, PD-L1 expression status of HRS cells was negatively correlated with the proportion of PD-1 rosettes in the TME, and CD4+PD-1+CXCL13+ T cells in the region surrounding HRS cells were significantly fewer in cases with PD-L1+ HRS cells (n = 34) (Fig. 5E). This might indicate a potential negative regulation and depletion of PD-1+ T cells if exposed to PD-L1+ HRS cells. Taken together these results suggest distinct PD-1/PD-L1–related biology in LR-CHL when compared to other CHL subtypes.
Fig. 5.
PD-L1 genomic alterations in HRS cells in LR-CHL. (A) Boxplot summarizing the proportion of PD-L1+ HRS cells by IHC in each sample, separated according to CHL subtype. (B) IHC staining for PD-L1 in representative CHL cases (×400; CHL20 and CHL25). (C) Combined immunofluorescence for CD30 (magenta) and fluorescence in situ hybridization (FISH) using bacterial artificial chromosome probes in the PD-L1 and PD-L2 region (green and red signals) shows PD-L1/L2 amplification in HRS cells in mixed cellularity CHL (Lower) (×400; CHL20) but not in lymphocyte-rich CHL (Upper) (×400; LRCHL01). Of 26 LR-CHL cases, 5 (19%) cases showed PD-L1/L2 amplification in HRS cells. (D) Dotplot showing correlation of PD-L1 alteration status in HRS cells with expression level of major immune cell markers (IHC). Dot size and color summarize Pearson correlation values, with positive correlations represented in blue and negative correlations represented in red. Asterisks represent associated P values (***P < 0.001). (E) Boxplot showing the proportion of CD4+PD-1+CXCL13+ T cells in the region surrounding CD30+ cells (HRS) for each sample, separated by PD-L1 expression status on HRS cells (IHC). Of the 58 CHL samples, 34 cases (59%) showed high PD-L1 expression on HRS cells. The surrounding region was defined by a distance of 75 μm.
TGF-β Induces CXCL13+ T Cells.
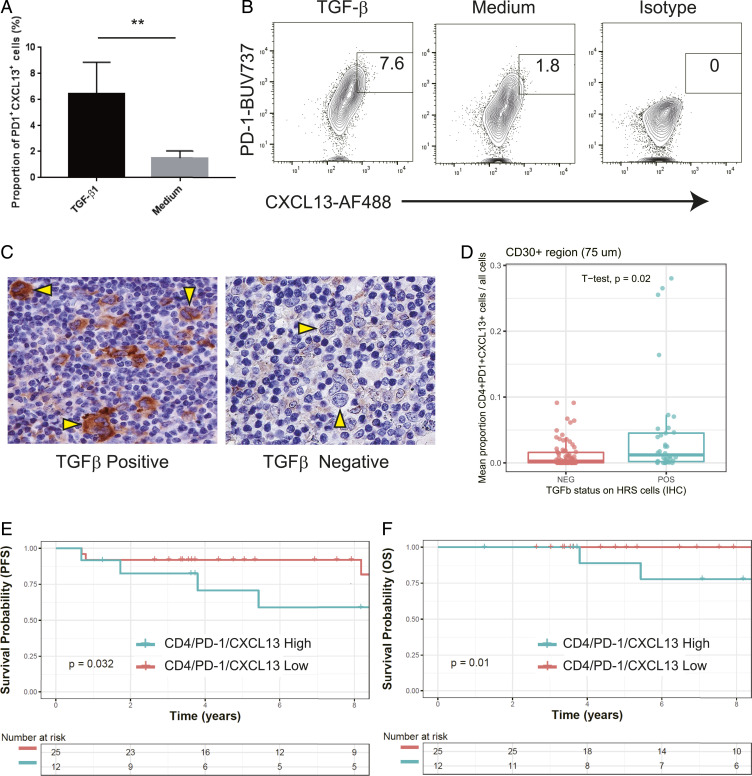
We hypothesized that cytokines or chemokines produced by HRS cells might influence the TME composition in LR-CHL. Consistent with previous literature (39), we confirmed that CD4+PD-1+CXCL13+ T cells were induced from naïve CD4 T cells by TGF-β in vitro (Fig. 6 A and B). In addition, IHC analysis revealed that in a subset of LR-CHL patients, HRS cells showed high expression of TGF-β (n = 12, 32%) (Fig. 6C). Notably, the proportion of CD4+PD-1+CXCL13+ T cells in the region surrounding HRS cells was significantly higher in cases with TGF-β+ HRS cells (P = 0.02, t test) (Fig. 6D). These results suggest that TGF-β may play a role in inducing the CD4+PD-1+CXCL13+ T cell population in the LR-CHL TME.
Fig. 6.
TGF-β induces a PD-1+CXCL13+ T cell population. (A) The proportion of PD-1+CXCL13+ cells among CD4+ T cells isolated from PBMCs after coculture with TGF-β or medium only. Data are shown as the mean ± SEM (n = 5) (**P < 0.01). (B) Representative flow cytometric analysis of PD-1 and CXCL13 expression on CD4+ T cells isolated from PBMCs cultured with TGF-β (Left), medium (Middle), or isotype control (Right). (C) IHC staining for TGF-β in representative cases with either positive (Left) or negative (Right) HRS cells (×400; CHL19 and LRCHL010). Of the 58 CHL cases, 18 cases (31%) showed high TGF-β expression on HRS cells. (D) Boxplot summarizing the proportion of CD4+PD-1+CXCL13+ cells from each cell suspension sample, separated according to TGF-β status on HRS cells (determined by IHC). (E and F) Patient outcomes based on proportion of CD4+PD-1+CXCL13+ T cells in LR-CHL patients. The Kaplan–Meier survival curves are shown for progression-free survival (E) and overall survival (F). P values were calculated using a log rank test.
Outcome Correlation of CD4+PD-1+CXCL13+ T Cells.
We finally investigated the prognostic value of the CD4+PD-1+CXCL13+ T cell population in LR-CHL (n = 37) patients uniformly treated with first-line ABVD (doxorubicin, bleomycin, vinblastine and dacarbazine)-like treatment. We observed significantly shortened progression-free survival (PFS) (5-y PFS 71% vs. 92%; P = 0.032) and overall survival (OS) (5-y OS 89% vs. 100%; P = 0.01) in patients with high levels of CD4+PD-1+CXCL13+ T cells in LR-CHL (Fig. 6 E and F and SI Appendix, Table S5). Importantly, an increased number of PD-1+ cells or CXCL13+ cells, measured as individual biomarkers, did not correlate with survival (SI Appendix, Fig. S9 and Table S6). This difference might reflect a distinct profile of CD4+PD-1+CXCL13+ T cells, as supported by our observations in the single-cell sequencing data, including unique TFH-like characteristics. The other clinical features such as age and advanced stage were not identified as prognostic factors for PFS in univariate analysis (SI Appendix, Table S6).
Discussion
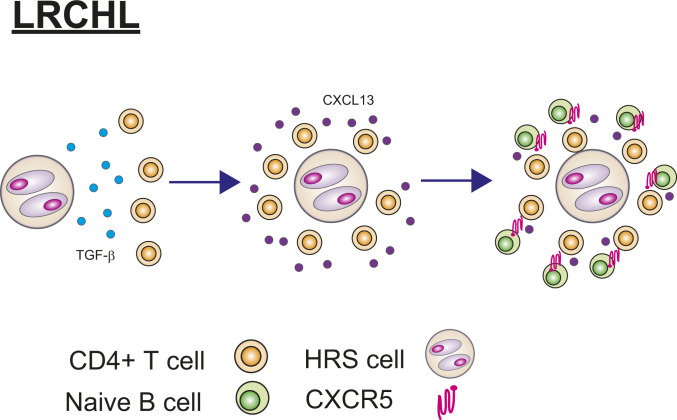
In this study, we comprehensively characterized immune cell populations in the TME of LR-CHL at both the RNA and protein levels. The relative rarity of LR-CHL has hampered its description in the past, and, to the best of our knowledge, this study utilizes one of the largest cohorts to date to investigate the TME of LR-CHL. We identified previously undescribed subpopulations specific to LR-CHL, including CD4+CXCL13+ T cells that are linked to unique pathological and clinical parameters. CD4+CXCL13+ T cells form rosettes surrounding HRS cells and coexpress PD-1. To date, PD-1–positive cells in the TME of LR-CHL were considered to be conventional TFH cells (1, 2, 40), but our data demonstrate a distinct phenotype of these CD4+PD-1+ T cells, implicating the CXCL13/CXCR5 axis. Collectively our results suggest a model in which the microenvironment of LR-CHL is highly organized and in part induced by CD4+CXCL13+ T cells, which in turn are induced by TGF-β secreted by HRS cells (Fig. 7).
Fig. 7.
Proposed model of CD4+PD-1+CXCL13+ T cell and HRS cell interactions in LR-CHL. HRS cells secrete TGF-β that induces a CXCL13+PD-1+ T cell population from CD4+ T cells, producing rosettes surrounding the HRS cells. CD4+PD-1+CXCL13+ T cells may in turn attract naïve CXCR5+ B cells.
Clinical trials have demonstrated high response rates of up to 87% by PD-1 blockade in relapsed and refractory CHL (17–22), indicating the importance of the PD-1/PD-L1 biology in the disease. Although high efficacy of PD-1 blockade might be associated with high frequency of 9p24.1 alterations in CHL (reported up to 90% response rates) (34–38), previous studies have demonstrated that the proportion of PD-1+ T cells is relatively low in CHL (15, 23). In this study, we demonstrated that LR-CHL shows a clearly distinct PD-1/PD-L1 profile when compared to other CHL subtypes, including more PD-1+ T cells and fewer PD-L1 genetic alterations in HRS cells. This is consistent with results from previous small case series in LR-CHL (41, 42). The response of PD-1 blockade by pathological subtype in CHL has not been reported, and further evaluation is warranted. Interestingly, we also observed that coexpression patterns on PD-1+CD4+ T cells are different among pathological subtypes, and CD4+PD-1+CXCL13+ T cells are specifically enriched in LR-CHL. We also observed a negative correlation between PD-L1 gene alterations on HRS cells and PD-1 protein expression in the TME of LR-CHL. This supports the notion that PD-L1 expression on HRS cells has a negative impact on PD-1+ T cells in LR-CHL, as suggested in a recent publication (43).
CXCL13, which is a ligand of CXCR5, is well known as a B cell attractant via the CXCL13/CXCR5 axis. Consistent with this known feature, CD4+ T cells in LR-CHL are located in close proximity to CXCR5+ B cells. Moreover, our scRNA-seq data demonstrated enrichment of naïve B cells, indicating that the naïve B cells attracted by CD4+CXCL13+ T cells might be prevented from entering the germinal center for antigen activation and maturation. The evidence of CD4+CXCL13+ T cells shaping TME composition may represent a LR-CHL–specific mechanism of immune dysfunction, suggesting that therapeutic targeting of these cells might reverse their immunosuppressive effects (44). Interestingly, therapeutic agents targeting CXCL13/CXCR5 are currently being explored in the context of autoimmune disease and non-Hodgkin lymphoma (40, 45). The characteristics of CD4+CXCL13+ T cells in LR-CHL are very similar to a CXCL13-producing TFH population that lacks CXCR5 expression identified in breast cancer (44). The scRNA-seq data also demonstrated that CD4+CXCL13+ T cells have an activated and terminally differentiated phenotype. Consistent with previous reports, CD4+PD-1+CXCL13+ T cells could be induced by TGF-β, a cytokine secreted by HRS cells.
Of clinical importance, our data demonstrated that the presence of CD4+PD-1+CXCL13+ T cells was associated with poor treatment outcome in LR-CHL, suggesting an important role of CD4+PD-1+CXCL13+ T cells in treatment response. In contrast, single IHC positivity of PD-1 and CXCL13 was not associated with outcome, suggesting the importance of identifying specific immune cell subsets using a multiple marker approach. However, it is still unclear whether CD4+CXCL13+ T cells are the main mediator for chemoresistance to standard chemotherapy, or whether this population is just an ancillary consequence of an HRS cell phenotype that drives poor outcome. In particular, a deeper understanding of receptor/ligand interactions linked to CD4+CXCL13+ T cells, including the CXCL13/CXCR5 and PD-1/PD-L1 axes, may be beneficial for future therapeutic and biomarker development.
In summary, our results reveal a unique TME composition in LR-CHL. Since the CXCL13/CXCR5 axis could affect multiple types of immune cells, including B cells, FDCs, and T cells, additional investigation into the biology of immune cell interactions will be crucial for future therapeutic development of alternative checkpoint inhibitors.
Materials and Methods
Additional detailed materials and methods are available in SI Appendix, Materials and Methods.
Tissue Samples.
For single-cell RNA sequencing, a total of 28 patients with histologically confirmed CHL (8 LR, 11 NS, 9 MC) and 5 patients with reactive lymphoid hyperplasia (but no evidence of malignant disease or systemic autoimmune disease) were included in this study. Patients were selected based on the availability of tissue that had been mechanically dissociated and cryopreserved as cell suspensions following diagnostic lymph node procedures at British Columbia (BC) Cancer.
The independent validation cohort of LR-CHL patients consisted of 31 newly diagnosed cases at BC Cancer between 2000 and 2018. The median follow-up time for living LR-CHL patients was 7 y (range: 1.2 to 17.4 y). Patient characteristics are summarized in SI Appendix, Tables S1, S2, S4, and S5.
This study was reviewed and approved by the University of British Columbia-BC Cancer Agency Research Ethics Board (H14-02304), in accordance with the Declaration of Helsinki. We obtained written informed consent from the patients or informed consent was waived for the samples used in this retrospective study.
Single-Cell RNA Sequencing and Library Preparation.
Samples were processed and libraries were prepared for scRNA-seq as previously described (15). In brief, sorted cells from cell suspensions were collected, and 8,700 cells per sample were loaded into a Chromium Single Cell 3′ Chip Kit v2 (PN-120236). Libraries were constructed using the Single Cell 3′ Library and Gel Bead Kit v2 (PN-120237) and Chromium i7 Multiplex Kit (PN-120262). For further details, see SI Appendix, Materials and Methods.
Normalization and Batch Correction.
Normalization and batch correction were performed as previously described (15). Briefly, CellRanger count data from all cells (n = 150,611) was read into R (v3.6.1) to create a single “SingleCellExperiment” object. To remove batch effects resulting from different library preparation chips, the fast mutual nearest neighbors (MNN) batch correction technique in the scran package (46) (v1.14.5) was utilized, grouping cells by their chip and using the expression of genes with positive biological components. For further details, see SI Appendix, Materials and Methods.
Clustering and Annotation.
Unsupervised clustering was performed with the PhenoGraph algorithm (47) as previously described (15). For visualization purposes, t-distributed stochastic neighbor embedding transformation was performed using the first 10 MNN-corrected components as input. For further details, see SI Appendix, Materials and Methods.
MC-IF on TMA, Scanning, and Image Analysis.
MC-IF was performed as previously described (15). In brief, TMA slides were deparaffinized and incubated with antibodies to each marker of interest (CXCR5, CXCL13, BCL6, CD20, PD-1, CD4, and CD30), followed by detection using Mach2 horseradish peroxidase and visualization using Opal fluorophores (SI Appendix, Table S7). To analyze the spectra for all fluorophores included, inForm image analysis software (v2.4.10; PerkinElmer) was used. For further details, see SI Appendix, Materials and Methods.
Cell Isolation and Purification of Human T Cells.
We purified CD4+ T lymphocytes from peripheral blood mononuclear cells (PBMCs) (see SI Appendix, Materials and Methods for details). Isolated naïve CD4+ cells were incubated in culture medium with or without TGF-β. At the end of day 5, we washed and analyzed the T cells using flow cytometry for characterization.
Flow Cytometry.
To characterize T cells in vitro, we stained cells with a panel of antibodies, including CD3, CD4, PD-1, and CXCL13 (see SI Appendix, Materials and Methods and Table S8 for details), and assessed them using flow cytometry (FACSymphony, BD). Flow cytometry data were analyzed using FlowJo software (v10.2; TreeStar).
Survival Analysis.
OS (death from any cause) and PFS (the time from initial diagnosis to the date of disease progression or relapse/death from any cause) were analyzed using the Kaplan–Meier method, and results were compared using a log rank test. Survival analyses were performed in the R Statistical Environment (v3.6.1). For further details, see SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This study is supported by Program Project grant funding from the Terry Fox Research Institute (Grant 1061), Large Scale Applied Research Project funding from Genome Canada (Grant 13124), Genome BC (Grant 271LYM), the Canadian Institutes of Health Research (CIHR) (Grant GP1-155873), the Canadian Cancer Society Research Institute (Grant 705288), a Foundation grant from CIHR (Grant 148393), the BC Cancer Foundation, and the Paul G. Allen Frontiers Group (Distinguished Investigator award to C.S., Grant 12829). T.A. was supported by a fellowship from CIHR and the Uehara Memorial Foundation. T.A. received research funding support from The Kanae Foundation for the Promotion of Medical Science. T.A. is the recipient of a Lymphoma Research Foundation Lymphoma Scientific Research Mentoring Program Scholar award. C.S. is the recipient of a Michael Smith Foundation for Health Research Career Investigator award. B.H.N. and K.M. were supported by the BC Cancer Foundation, Genome BC, and Canada’s Networks of Centres of Excellence (BioCanRx).
Footnotes
Competing interest statement: C.S. has performed consultancy for Seattle Genetics, Curis Inc., Roche, AbbVie, Juno Therapeutics, and Bayer and has received research funding from Bristol-Myers Squibb, Epizyme, and Trillium Therapeutics Inc. C.S. and D.W.S. are coinventors on a patent (“Method for determining lymphoma type”) using NanoString technology. D.W.S. has performed consultancy for Abbvie, AstraZeneca, Celgene, and Janssen and has received research funding from Janssen, NanoString, and Roche.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105822118/-/DCSupplemental.
Data Availability
Single cell RNA-seq counts (generated with CellRanger v2.1.0) and a merged ‘SingleCellExperiment’ R object is available in the European Genome-phenome Archive (EGA) (EGAS00001005541) via controlled access.
References
- 1.Swerdlow S. H., et al., The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127, 2375–2390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nam-Cha S. H., et al., Lymphocyte-rich classical Hodgkin’s lymphoma: Distinctive tumor and microenvironment markers. Mod. Pathol. 22, 1006–1015 (2009). [DOI] [PubMed] [Google Scholar]
- 3.de Jong D., et al., Lymphocyte-rich classical Hodgkin lymphoma (LRCHL): Clinico-pathological characteristics and outcome of a rare entity. Ann. Oncol. 17, 141–145 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Shimabukuro-Vornhagen A., et al., Lymphocyte-rich classical Hodgkin’s lymphoma: Clinical presentation and treatment outcome in 100 patients treated within German Hodgkin’s Study Group trials. J. Clin. Oncol. 23, 5739–5745 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Steidl C., Connors J. M., Gascoyne R. D., Molecular pathogenesis of Hodgkin’s lymphoma: Increasing evidence of the importance of the microenvironment. J. Clin. Oncol. 29, 1812–1826 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Nastoupil L. J., et al., Standard-of-care axicabtagene ciloleucel for relapsed or refractory large B-cell lymphoma: Results from the US lymphoma CAR T consortium. J. Clin. Oncol. 38, 3119–3128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan F. C., et al., Prognostic model to predict post-autologous stem-cell transplantation outcomes in classical Hodgkin lymphoma. J. Clin. Oncol. 35, 3722–3733 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Scott D. W., et al., Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J. Clin. Oncol. 31, 692–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves P., et al., Expression of FOXP3, CD68, and CD20 at diagnosis in the microenvironment of classical Hodgkin lymphoma is predictive of outcome. J. Clin. Oncol. 31, 256–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steidl C., et al., Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 362, 875–885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki T., Savage K. J., Steidl C., Biology in practice: Harnessing the curative potential of the immune system in lymphoid cancers. J. Clin. Oncol. 39, 346–360 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Diepstra A., et al., Association with HLA class I in Epstein-Barr-virus-positive and with HLA class III in Epstein-Barr-virus-negative Hodgkin’s lymphoma. Lancet 365, 2216–2224 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Roemer M. G., et al., Classical Hodgkin lymphoma with reduced β2M/MHC Class I expression is associated with inferior outcome independent of 9p24.1 status. Cancer Immunol. Res. 4, 910–916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roemer M. G. M., et al., Major histocompatibility complex Class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J. Clin. Oncol. 36, 942–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki T., et al., Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov. 10, 406–421 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Patel S. S., et al., The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative. Blood 134, 2059–2069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansell S. M., Nivolumab in the treatment of Hodgkin lymphoma. Clin. Cancer Res. 23, 1623–1626, 10.1158/1078-0432.CCR-16-1387 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Armand P., et al., Pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure: Long-term efficacy from the phase 1b keynote-013 study. Blood 128, 1108–1108 (2016). [Google Scholar]
- 19.Moskowitz C. H., et al., Pembrolizumab in relapsed/refractory classical hodgkin lymphoma: Primary end point analysis of the phase 2 keynote-087 study. Blood 128, 1107 (2016). [Google Scholar]
- 20.Timmerman J. M., et al., Checkmate 205 update with minimum 12-month follow up: A phase 2 study of nivolumab in patients with relapsed/refractory classical Hodgkin lymphoma. Blood 128, 1110–1110 (2016). [Google Scholar]
- 21.Younes A., et al., Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: A multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 17, 1283–1294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen R., et al., Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic Hodgkin lymphoma. J. Clin. Oncol. 35, 2125–2132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carey C. D., et al., Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 130, 2420–2430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novershtern N., et al., Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cader F. Z., et al., Mass cytometry of Hodgkin lymphoma reveals a CD4+ regulatory T-cell-rich and exhausted T-effector microenvironment. Blood 132, 825–836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaves P., et al., Defining characteristics of classical Hodgkin lymphoma microenvironment T-helper cells. Blood 122, 2856–2863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crotty S., T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boice M., et al., Loss of the HVEM tumor suppressor in lymphoma and restoration by modified CAR-T cells. Cell 167, 405–418.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazanietz M. G., Durando M., Cooke M., CXCL13 and its receptor CXCR5 in cancer: Inflammation, immune response, and beyond. Front. Endocrinol. (Lausanne) 10, 471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engleman E. G., Charron D. J., Benike C. J., Stewart G. J., Ia antigen on peripheral blood mononuclear leukocytes in man. I. Expression, biosynthesis, and function of HLA-DR antigen non-T cells. J. Exp. Med. 152, 99s–113s (1980). [PubMed] [Google Scholar]
- 31.Haghverdi L., Buettner F., Theis F. J., Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989–2998 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Coifman R. R., et al., Geometric diffusions as a tool for harmonic analysis and structure definition of data: Diffusion maps. Proc. Natl. Acad. Sci. U.S.A. 102, 7426–7431 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., et al., iTALK: An R package to characterize and illustrate intercellular communication. BioRxiv [Preprint] (2019). 10.1101/507871. Accessed 4 January 2019. [DOI]
- 34.Juszczynski P., et al., The AP1-dependent secretion of galectin-1 by Reed Sternberg cells fosters immune privilege in classical Hodgkin lymphoma. Proc. Natl. Acad. Sci. U.S.A. 104, 13134–13139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green M. R., et al., Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116, 3268–3277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steidl C., et al., MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 471, 377–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green MR, et al., Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: Implications for targeted therapy. Clin. Cancer Res. 18, 1611–1618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roemer M. G., et al., PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J. Clin. Oncol. 34, 2690–2697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi S., et al., TGF-β induces the differentiation of human CXCL13-producing CD4(+) T cells. Eur. J. Immunol. 46, 360–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klimatcheva E., et al., CXCL13 antibody for the treatment of autoimmune disorders. BMC Immunol. 16, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasse S., et al., Programmed cell death protein1 (PD1)-expression in the microenvironment of classical Hodgkin lymphoma at relapse after conventional chemotherapy and at relapse on anti-PD1 treatment. Haematologica 104, e45–e46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasse S., et al., Programmed cell death protein-1 (PD-1)-expression in the microenvironment of classical Hodgkin lymphoma at relapse during anti-PD-1-treatment. Haematologica 104, e21–e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng Q., et al., PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat. Commun. 11, 4835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu-Trantien C., et al., CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2, 91487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunse M., et al., CXCR5 CAR-T cells simultaneously target B cell non-Hodgkin’s lymphoma and tumor-supportive follicular T helper cells. Nat. Commun. 12, 240 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lun A. T., McCarthy D. J., Marioni J. C., A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000 Res. 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levine J. H., et al., Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA-seq counts (generated with CellRanger v2.1.0) and a merged ‘SingleCellExperiment’ R object is available in the European Genome-phenome Archive (EGA) (EGAS00001005541) via controlled access.