Significance
Epithelial cell–mediated chemokine production and subsequent neutrophil recruitment are important for pathogen clearance, which, however, are also closely related to severe inflammatory tissue damage during lung infection, especially influenza virus infection and the current pandemic coronavirus infection. Certain regulation and underlying mechanisms of chemokine expression in epithelial cells remain largely unknown. Here, by identifying the mouse long noncoding RNA lnc-Cxcl2 and human lnc-CXCL2-4-1 in virus-infected lung epithelial cells, we demonstrated a self-protecting mechanism in host lung epithelial cells for restraining viral infection–induced lung inflammation through feedback-suppressing chemokine expression. These findings provide better understandings of chemokine regulation and epithelial cell function during lung viral infection and will benefit the treatment of related lung infectious diseases.
Keywords: chemokine CXCL2, lung inflammation, neutrophil, epithelial cell
Abstract
Chemokine production by epithelial cells is important for neutrophil recruitment during viral infection, the appropriate regulation of which is critical for restraining inflammation and attenuating subsequent tissue damage. Epithelial cell expression of long noncoding RNAs (lncRNAs), RNA-binding proteins, and their functional interactions during viral infection and inflammation remain to be fully understood. Here, we identified an inducible lncRNA in the Cxcl2 gene locus, lnc-Cxcl2, which could selectively inhibit Cxcl2 expression in mouse lung epithelial cells but not in macrophages. lnc-Cxcl2–deficient mice exhibited increased Cxcl2 expression, enhanced neutrophils recruitment, and more severe inflammation in the lung after influenza virus infection. Mechanistically, nucleus-localized lnc-Cxcl2 bound to Cxcl2 promoter, recruited a ribonucleoprotein La, which inhibited the chromatin accessibility of chemokine promoters, and consequently inhibited Cxcl2 transcription in cis. However, unlike mouse lnc-Cxcl2, human lnc-CXCL2-4-1 inhibited multiple immune cytokine expressions including chemokines in human lung epithelial cells. Together, our results demonstrate a self-protecting mechanism within epithelial cells to restrain chemokine and neutrophil-mediated inflammation, providing clues for better understanding chemokine regulation and epithelial cell function in lung viral infection.
Chemokine production and chemokine-mediated neutrophil recruitment are critical for the elimination of invading pathogens and also determination of the infection outcome: homeostasis or inflammation (1, 2). High levels of chemokines and neutrophil recruitment have been found strongly correlated with the inflammatory tissue damage and fetal outcome of viral infection including influenza virus infection and the current pandemic coronavirus disease 2019 (COVID-19) (3–5). Besides, strategies that target the production of proinflammatory cytokines, chemokines, or downstream signaling have shown potential to suppress inflammation in COVID-19 (6, 7), reminding the urgent need for better understanding of the mechanism of chemokine expression and regulation in viral infectious diseases.
Chemokines that recruit neutrophils are mainly in the CXCL8 family, including CXCL1, CXCL2, and CXCL8 (human) (8). Epithelial cells, as the barrier between host and environment, play important roles in producing these chemokines (9, 10), especially in the respiratory tract where epithelial cells are the primary targets of many pathogens, including influenza virus and coronavirus (11–13). Moreover, a recently described human oral mucosa cell atlas has linked epithelial cells with inflammatory signatures to enhanced neutrophil recruitment during oral tissue inflammation (14). Thus, timely and appropriate regulation of chemokine expression in epithelial cells is critical for preventing inflammatory tissue damage while defending against the invading pathogens. However, the negative regulation of chemokines production in epithelial cells and its underlying mechanism remain to be elucidated in the interaction of host and pathogens, especially during lung viral infection.
Long noncoding RNAs (lncRNAs) can regulate gene expression at multiple levels in different physiological and pathological conditions (15). LncRNAs are classified into cis-acting and trans-acting according to the location of their transcription sites relative to their target positions. Cis-acting lncRNAs function as local effectors to regulate the expression of their neighboring genes, and many coexpressed neighboring messenger RNA (mRNA)–lncRNA pairs with positive correlation have been identified (16–18). However, whether cis-acting lncRNAs participate in the negative regulation of chemokine expression in epithelial cells and their potential functions in virus-induced inflammation remain to be investigated. Besides, interaction between RNA and RNA-binding proteins (RBPs) has been shown to play critical roles in the RNA-mediated regulation of transcription (19, 20). Thus, we wanted to identify cis-acting lncRNAs that transcribed near chemokine genes and their possible interactive RBPs in lung epithelial cells and tried to demonstrate their functions in virus infection–induced inflammation.
By analyzing lncRNA expression profile in virus-infected mouse lung epithelial cells, we identified an inducible lncRNA in the Cxcl2 gene locus, which we designated as lnc-Cxcl2, which could restrain neutrophil-mediated lung inflammation in a feedback manner during influenza virus infection. We demonstrated that the cis-acting lnc-Cxcl2 bound to the promoter of Cxcl2 and recruited the ribonucleoprotein La to inhibit the transcription of Cxcl2 in lung epithelial cells. However, we also found that unlike mouse lnc-Cxcl2, which acted in cis, human lnc-CXCL2-4-1 inhibited the expression of multiple cytokines including chemokines by binding La in human lung epithelial cells during influenza virus infection. Together, our findings uncover an epigenetic mechanism for inhibiting epithelial cell chemokine expression and attenuating lung inflammation during viral infection.
Results
lnc-Cxcl2 Expression Is Increased in the Nucleus of Mouse Lung Epithelial Cells in Response to Viral Infection.
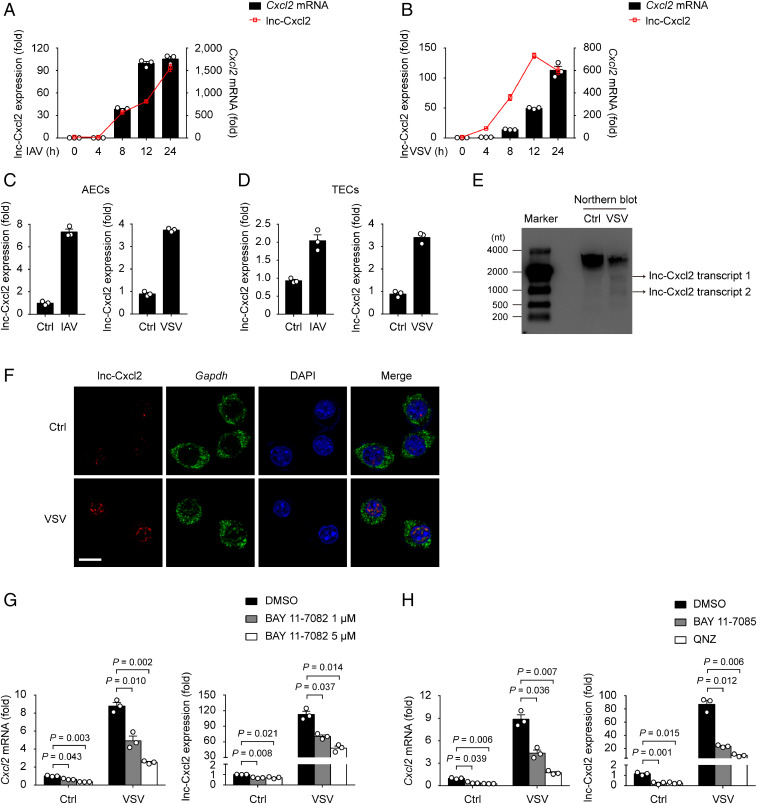
To investigate whether cis-acting lncRNAs participate in the regulation of Cxcl2 expression in epithelial cells during viral infection, we infected mouse lung epithelial cells (MLE-12 cells) with recombinant vesicular stomatitis virus (VSV) expressing GFP (GFP-VSV), collected GFP-positive cells 12 h later, and analyzed lncRNA expression profile by RNA sequencing. We focused on lncRNAs expressed near chemokine genes, which may function as local effectors, and found that the expression level of the Cxcl2 locus (Gene identity: ENSMUSG00000058427; chr5:90903870-90905938) in virus-infected lung epithelial cells was significantly higher than that in noninfected cells, indicating that there might be new transcripts within this region. Indeed, by using rapid amplification of complementary DNA (cDNA) ends (RACE) analysis, we found two previously uncharacterized transcripts in Cxcl2 gene locus, the longer transcript had 1,660 nucleotides (nt) whereas the shorter one had 977 nt (SI Appendix, Fig. S1 A and B and Table S1). These two transcripts had a same 5′ end started from the intron 2 region of Cxcl2, shared the same sequence with pre-mRNA of Cxcl2 containing the intron 3, and contained polyA signals in 3′ ends, which makes them different from the pre-mRNA of Cxcl2 (SI Appendix, Fig. S1 A and B). Moreover, analyses using Coding Potential Calculator (21) and Open Reading Frame Finder showed that both transcripts lacked coding potential (SI Appendix, Fig. S1 C and D). We thereafter designated the long and the short transcripts as lncRNA of Cxcl2 locus (lnc-Cxcl2).
To characterize lnc-Cxcl2, we first observed that the expression of lnc-Cxcl2 was increased in MLE-12 cells, primary mouse alveolar epithelial cells (AECs), and trachea epithelial cells (TECs) in response to both influenza A virus (IAV) and VSV infection (Fig. 1 A–D). By using Northern blot analysis, we found that both transcripts of lnc-Cxcl2 were induced in MLE-12 cells after VSV infection (Fig. 1E). We further calculated the absolute copy number of lnc-Cxcl2 and found that lnc-Cxcl2 has 15 ± 3 copies per uninfected MLE-12 cell and could reach up to 164 ± 8 copies after VSV infection (SI Appendix, Fig. S1E). In addition to epithelial cells, lnc-Cxcl2 expression was also increased in virus-infected mouse resident alveolar macrophages (AMs) and RAW264.7 cells (SI Appendix, Fig. S1 F and G).
Fig. 1.
lnc-Cxcl2 expression is increased in the nucleus of mouse lung epithelial cells in response to viral infection. (A and B) qPCR analysis of lnc-Cxcl2 and Cxcl2 mRNA expressions in MLE-12 cells infected with IAV (multiplicity of infection [MOI] = 1) (A) or VSV (MOI = 1) (B) for indicated hours. (C and D) qPCR analysis of lnc-Cxcl2 expression in AECs (C) and TECs (D) infected with IAV (MOI = 1) or VSV (MOI = 1) for 12 h. (E) Northern blot analysis of lnc-Cxcl2 in MLE-12 cells infected with VSV (MOI = 1) for 12 h. (F) RNA in situ hybridization analysis of lnc-Cxcl2 and Gapdh in MLE-12 cells infected with VSV (MOI = 1) for 12 h. (Scale bar, 10 μm.) (G) qPCR analysis of Cxcl2 mRNA and lnc-Cxcl2 expressions in MLE-12 cells treated with DMSO or BAY 11-7082 for 2 h followed by VSV infection (MOI = 1) for 12 h. (H) qPCR analysis of Cxcl2 mRNA and lnc-Cxcl2 expressions in MLE-12 cells treated with DMSO, BAY 11-7085 (5 μM), or QNZ (EVP4593) (10 nM) for 2 h followed by VSV infection (MOI = 1) for 12 h. Data are representative of three independent experiments (E and F) or three independent experiments with n = 3 biological replicates (A–D, G, and H; shown as mean ± SEM in A and B, shown as mean + SEM in C, D, G, and H), two-tailed unpaired Student’s t test. Ctrl, control.
To further investigate the cellular localization of lnc-Cxcl2, we separated different cell compartments and found that lnc-Cxcl2 was mainly located in the nucleus of MLE-12 cells (SI Appendix, Fig. S1H). Consistently, in situ hybridization analysis confirmed that lnc-Cxcl2 was located in the nucleus and its expression was increased after VSV infection (Fig. 1F). Then, we wondered how lnc-Cxcl2 was induced in response to viral infection. We found that addition of the NF-κB inhibitor reduced the expression of both Cxcl2 and lnc-Cxcl2 in response to viral infection (Fig. 1 G and H), indicating that lnc-Cxcl2, like Cxcl2, was induced via the NF-κB signaling pathway. Together, these results demonstrate that lnc-Cxcl2 is a nuclear lncRNA whose expression is increased in mouse lung epithelial cells in response to viral infection.
lnc-Cxcl2 Selectively Inhibits Cxcl2 Expression in Lung Epithelial Cells during Viral Infection.
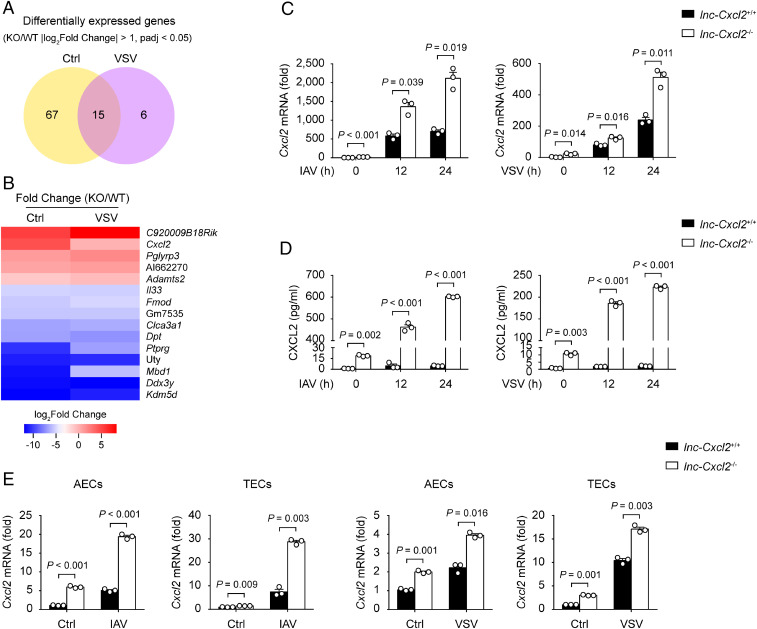
To investigate whether lnc-Cxcl2 regulated the expression of Cxcl2 during viral infection, we generated lnc-Cxcl2–deficient (lnc-Cxcl2−/−) MLE-12 cells using the CRISPR-Cas9 system (SI Appendix, Fig. S2A). lnc-Cxcl2−/− cells showed the same proliferation rate and similar proportion of cell death after VSV infection as wild-type (lnc-Cxcl2+/+) MLE-12 cells (SI Appendix, Fig. S2 B and C).
However, by performing transcriptome analysis, we found 21 genes dysregulated in lnc-Cxcl2−/− cells after viral infection (Fig. 2A and SI Appendix, Table S2), and Cxcl2 was among the up-regulated genes (Fig. 2B and SI Appendix, Table S2). Consistent with the transcriptome analysis data, we found that the Cxcl2 expression in lnc-Cxcl2−/− cells and the secreted CXCL2 level in the lnc-Cxcl2−/− cell supernatant were increased compared to those in lnc-Cxcl2+/+ cells in response to viral infection (Fig. 2 C and D). However, the expression of monocyte chemokine Ccl2, interferon Ifnb1, proinflammatory cytokine Tnf, and the other neutrophil chemokine Cxcl3 were not obviously changed (SI Appendix, Fig. S2 D and E).
Fig. 2.
lnc-Cxcl2 selectively inhibits Cxcl2 expression in lung epithelial cells during viral infection. (A) Venn diagram of differentially expressed genes between lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with VSV (multiplicity of infection [MOI] = 1) for 12 h. |log2Fold Change| > 1 and adjusted P value (padj) < 0.05. WT, wild type; KO, knockout. (B) Heatmap of differentially expressed genes between lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with VSV (MOI = 1) for 12 h. (C) qPCR analysis of Cxcl2 mRNA expression in lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with IAV (MOI = 1) or VSV (MOI = 1) for indicated hours. (D) Enzyme-linked immunosorbent assay of CXCL2 level in the supernatant of lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with IAV (MOI = 1) or VSV (MOI = 1) for indicated hours. (E) qPCR analysis of Cxcl2 mRNA expression in AECs and TECs from lnc-Cxcl2+/+ and lnc-Cxcl2−/− mice infected with IAV (MOI = 1) or VSV (MOI = 1) for 12 h. Data are representative of three independent experiments with n = 3 biological replicates (C and D; shown as mean + SEM) or two independent experiments with n = 3 biological replicates (E; shown as mean + SEM), two-tailed unpaired Student’s t test. Ctrl, control.
As tissue resident macrophage-derived chemokines also control the early recruitment of neutrophils in tissue inflammation (22), we detected whether lnc-Cxcl2 inhibited Cxcl2 expression in macrophages. By generating lnc-Cxcl2−/− RAW264.7 cells, we found that although lnc-Cxcl2 expression was increased after VSV infection, lnc-Cxcl2 deficiency did not change Cxcl2 expression in RAW264.7 cells (SI Appendix, Fig. S2F), indicating that lnc-Cxcl2 might selectively inhibit Cxcl2 expression in mouse lung epithelial cells but not in macrophages. To test this hypothesis, we generated lnc-Cxcl2−/− mice using CRISPR-Cas9 system (SI Appendix, Fig. S3A) and isolated primary AECs, TECs, and AMs from these mice. We found that lnc-Cxcl2 deficiency increased Cxcl2 expression in both IAV- and VSV-infected AECs and TECs but not in AMs (Fig. 2E and SI Appendix, Fig. S3B). Thus, these results demonstrate that lnc-Cxcl2 selectively inhibits Cxcl2 expression in mouse lung epithelial cells during viral infection.
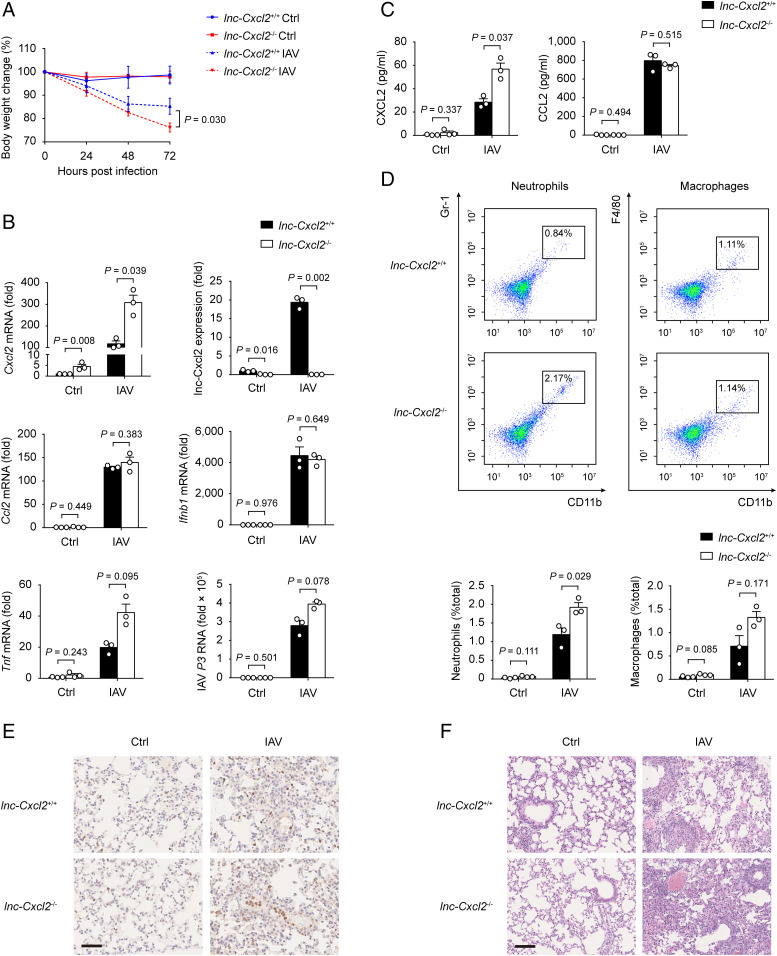
lnc-Cxcl2 Restrains Influenza Virus–Induced Lung Inflammation In Vivo.
We next investigated the in vivo function of lnc-Cxcl2. lnc-Cxcl2−/− mice were viable, fertile, and almost indistinguishable from lnc-Cxcl2+/+ littermates in appearance, body weight, and behavior. And the proportions of immune cells, including T cells, B cells, natural killer cells, neutrophils, macrophages, and dendritic cells in the spleen of lnc-Cxcl2+/+ and lnc-Cxcl2−/− mice were also similar (SI Appendix, Fig. S3C), suggesting that lnc-Cxcl2 deficiency did not affect immune cell development in mice. Then, we intranasally infected these mice with IAV, which targeted lung epithelial cells for infection and induced lung inflammation and tissue damage. We found that lnc-Cxcl2−/− mice showed more body weight loss compared to lnc-Cxcl2+/+ mice after IAV infection (Fig. 3A). And the expression of Cxcl2 but not other proinflammatory cytokines, like Ccl2, Ifnb1, or Tnf, in the lung of lnc-Cxcl2−/− mice was increased compared to lnc-Cxcl2+/+ mice after IAV infection, although viral load in the lungs was similar (Fig. 3B). Besides, the CXCL2 level and neutrophil numbers in the bronchoalveolar lavage fluid (BALF) of lnc-Cxcl2−/− mice were higher than that of lnc-Cxcl2+/+ mice (Fig. 3 C and D). Consistent with that, lnc-Cxcl2−/− mice had more neutrophil infiltration, more severe inflammation, and tissue damage in the lung compared to lnc-Cxcl2+/+ mice after IAV infection (Fig. 3 E and F). However, the expression of Cxcr2 in bone marrow neutrophils from lnc-Cxcl2+/+ and lnc-Cxcl2−/− mice was similar (SI Appendix, Fig. S3D), suggesting that more neutrophils recruited into the lung of lnc-Cxcl2−/− mice was due to the increased level of CXCL2.
Fig. 3.
lnc-Cxcl2 restrains influenza virus–induced lung inflammation in vivo. (A) Body weight change analysis of lnc-Cxcl2+/+ and lnc-Cxcl2−/− mice intranasally infected with IAV (3 × 105 PFU) for 72 h. (B) qPCR analysis of Cxcl2, lnc-Cxcl2, Ccl2, Ifnb1, and Tnf mRNA expressions and IAV load indicated by virus polymerase P3 expression in lungs from mice described in A. (C) Enzyme-linked immunosorbent assay of CXCL2 and CCL2 levels in the BALF from mice described in A. (D) Flow cytometric analysis of CD11b+Gr-1+ neutrophil and CD11b+F4/80+ macrophage propagations in BALF from mice described in A. (E) Immunohistochemistry staining of Gr-1+ neutrophils of lung sections from mice described in A. (Scale bar, 50 μm.) (F) Hematoxylin and eosin staining of lung sections from the mice described in A. (Scale bar, 100 μm.) Data are representative of three independent experiments with three mice per group (A–F; shown as mean ± SEM in A, shown as mean + SEM in B–D), two-way ANOVA analysis (A), two-tailed unpaired Student’s t test (B–D). Ctrl, control.
To further demonstrate the function of lnc-Cxcl2, we infected mice with lower doses of IAV and monitored body weight and inflammation for 2 wk. We found that 2 wk after IAV infection, lnc-Cxcl2−/− mice had delayed weight recovery and increased Cxcl2 expression in the lungs (SI Appendix, Fig. S3 E and F), which were accompanied by more neutrophil infiltration and unresolved inflammation compared to lnc-Cxcl2+/+ mice (SI Appendix, Fig. S3G). Together, these results demonstrate that lnc-Cxcl2 restrains influenza virus–induced lung inflammation through inhibiting Cxcl2 expression and neutrophil recruitment in vivo.
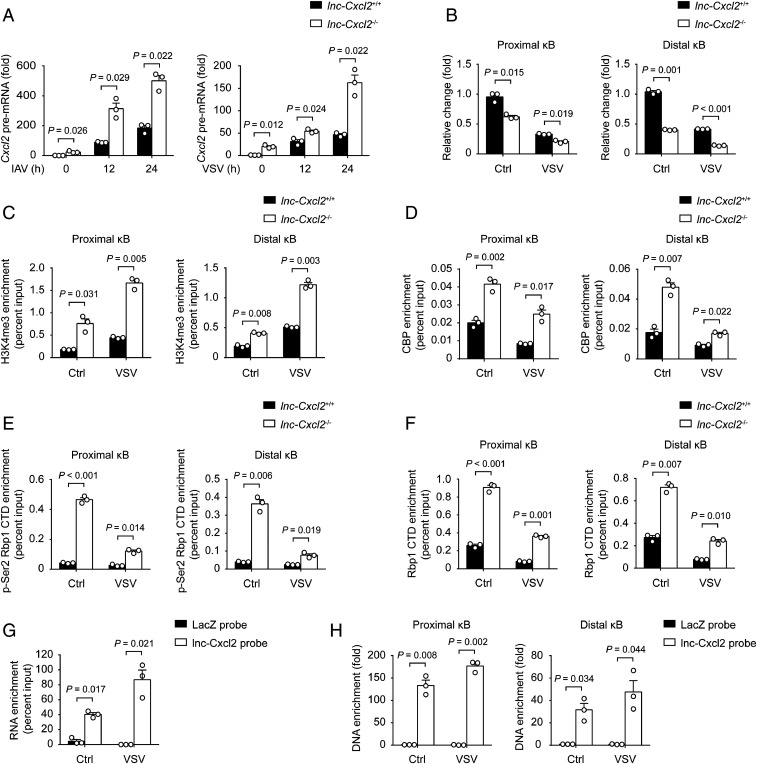
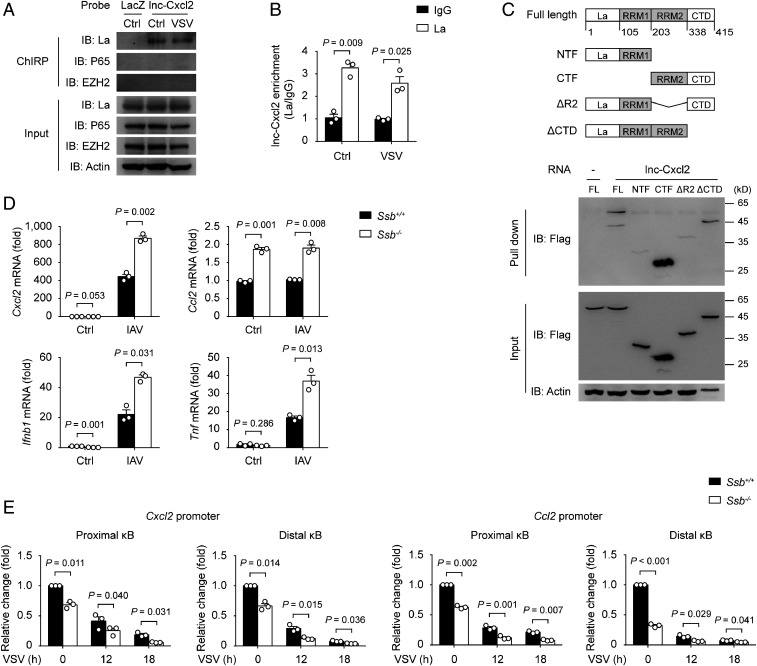
lnc-Cxcl2 Binds to and Maintains the Repressed Chromatin State of Cxcl2 Promoter in Cis.
We then investigated how lnc-Cxcl2 inhibited Cxcl2 expression during viral infection. We examined signalings that participate in the expression of Cxcl2 (23, 24) and found that lnc-Cxcl2 deficiency did not alter the nuclear localization of transcription factors like P65, P50, STAT1, STAT3, or c-JUN (SI Appendix, Fig. S4A). We wondered whether lnc-Cxcl2 regulated the transcription of Cxcl2 in cis. We found that lnc-Cxcl2 deficiency increased the Cxcl2 pre-mRNA level in response to viral infection (Fig. 4A), and the chromatin accessibility (indicated by DNase I sensitivity) of the Cxcl2 promoter (indicated by two κB sites, SI Appendix, Fig. S4B) was increased in lnc-Cxcl2−/− cells in response to VSV infection (Fig. 4B). Consistently, the H3K4me3 and CREB binding protein (CBP) levels in Cxcl2 promoter, which are two markers of active transcription, were increased in lnc-Cxcl2−/− cells in response to VSV infection (Fig. 4 C and D). The total and the phosphorylated RNA polymerase II levels in the Cxcl2 promoter were also increased in lnc-Cxcl2−/− cells (Fig. 4 E and F), suggesting that lnc-Cxcl2 inhibited the chromatin accessibility of Cxcl2 promoter and the transcription of Cxcl2. We further used small interfering RNA to silence the expression of lnc-Cxcl2 to further confirm the function of lnc-Cxcl2. As lnc-Cxcl2 shared the same sequence with pre-mRNA of Cxcl2, silencing the expression of lnc-Cxcl2 simultaneously down-regulated the expression of Cxcl2 (SI Appendix, Fig. S4C). Nevertheless, silencing the expression of lnc-Cxcl2 increased the chromatin accessibility of the Cxcl2 promoter compared to silencing the negative control (SI Appendix, Fig. S4D), indicating that lnc-Cxcl2 itself could inhibit the chromatin accessibility of the Cxcl2 promoter.
Fig. 4.
lnc-Cxcl2 binds to and maintains the repressed chromatin state of Cxcl2 promoter in cis. (A) qPCR analysis of Cxcl2 pre-mRNA expression in lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with IAV (multiplicity of infection [MOI] = 1) or VSV (MOI = 1) for indicated hours. (B) qPCR analysis of the DNase I sensitivity of the Cxcl2 promoter in lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with VSV (MOI = 1) for 12 h. (C–F) qPCR analysis of H3K4me3 (C), CBP (D), pSer2-RNA polymerase II carboxy-terminal domain (CTD) (E), and total RNA polymerase II CTD (F) enrichment levels in the Cxcl2 promoter in lnc-Cxcl2+/+ and lnc-Cxcl2−/− MLE-12 cells infected with VSV (MOI = 1) for 12 h. (G) qPCR analysis of lnc-Cxcl2 retrieved by antisense RNA probes in MLE-12 cells infected with VSV (MOI = 1) for 12 h. (H) qPCR analysis of enrichment level of Cxcl2 promoter that purified by lnc-Cxcl2 probes in MLE-12 cells infected with VSV (MOI = 1) for 12 h. Data are representative of three independent experiments with n = 3 biological replicates (A–F; shown as mean + SEM) or from three independent experiments (G and H; shown as mean + SEM), two-tailed unpaired Student’s t test. Ctrl, control.
To clarify how lnc-Cxcl2 regulated the chromatin accessibility of the Cxcl2 promoter, we examined whether lnc-Cxcl2 could directly bind to the Cxcl2 promoter. We isolated lnc-Cxcl2–binding chromatin by RNA purification using antisense probes that specifically target lnc-Cxcl2. We found that the probes successfully captured lnc-Cxcl2 especially after VSV infection (Fig. 4G), and the two regions containing κB sites of the Cxcl2 promoter were highly enriched in lnc-Cxcl2–binding DNA, whereas those of the Ccl2 promoter were not (Fig. 4H and SI Appendix, Fig. S4E), indicating that lnc-Cxcl2 selectively bound to the promoter of Cxcl2. Together, these results demonstrate that lnc-Cxcl2 binds to Cxcl2 promoter and maintains a repressed chromatin state of Cxcl2 promoter in cis during viral infection.
lnc-Cxcl2 Inhibits Cxcl2 Expression through Ribonucleoprotein La.
We next investigated whether lnc-Cxcl2 inhibited the chromatin accessibility through its interactive proteins. Using RNA pull-down assay followed by mass spectrometry (MS) analysis, we identified 13 lnc-Cxcl2–interacting nuclear proteins (SI Appendix, Table S3). We examined whether these proteins were involved in the regulation of Cxcl2 expression and found that only silencing the expression of Ssb, which encoded the ribonucleoprotein La, increased the expression of Cxcl2 (SI Appendix, Fig. S5A). To test the hypothesis that lnc-Cxcl2 inhibited Cxcl2 transcription through interacting with La, we first confirmed the interaction between lnc-Cxcl2 and La. We found that in vitro transcribed-lnc-Cxcl2 bound La but not transcription factor P65 or STAT3 (SI Appendix, Fig. S5B). Furthermore, by using antisense RNA probes to purify the endogenous lnc-Cxcl2 and its associated proteins, we found that lnc-Cxcl2 was associated with La in MLE-12 cells but not in RAW264.7 cells (Fig. 5A and SI Appendix, Fig. S5C). Conversely, immunoprecipitation analysis using antibody against La showed that endogenous La was associated with the endogenous lnc-Cxcl2 in MLE-12 cells (Fig. 5B).
Fig. 5.
lnc-Cxcl2 inhibits Cxcl2 expression through ribonucleoprotein La. (A) Immunoblot analysis of proteins purified by lnc-Cxcl2 probes in MLE-12 cells infected with VSV (multiplicity of infection [MOI] = 1) for 12 h. (B) qPCR analysis of lnc-Cxcl2 level purified by La antibody in MLE-12 cells infected with VSV (MOI = 1) for 12 h. (C) Schematic illustration of La truncations (Top), and the immunoblot analysis of lnc-Cxcl2 and La truncations interaction in HEK293T cells transfected with Flag-tagged La truncations (Bottom). FL, full length. (D) qPCR analysis of Cxcl2, Ccl2, Ifnb1, and Tnf mRNA expressions in Ssb+/+ and Ssb−/− MLE-12 cells infected with IAV (MOI = 1) for 12 h. (E) qPCR analysis of the DNase I sensitivity of Cxcl2 and Ccl2 promoters in Ssb+/+ and Ssb−/− MLE-12 cells infected with VSV (MOI = 1) for indicated hours. Data are representative of three independent experiments (A and C) or three independent experiments with n = 3 biological replicates (B, D, and E; shown as mean + SEM), two-tailed unpaired Student’s t test. Ctrl, control; IB, immunoblot.
La contains an N-terminal La motif, two RNA recognition motifs (RRM), and a C-terminal nuclear localization signal (25). The binding preference of the La motif and RRM1 domain in the N terminus correlates with the cellular localization of La: They recognize the UUU-3′OH of pre–transfer RNAs in the nucleus, whereas they recognize the poly(A) tail of mRNAs in the cytoplasm (26, 27). However, the function of the C-terminal part of La, especially the second RRM, has not been identified clearly. To detect which domain of La interacted with lnc-Cxcl2, we expressed different La truncations and found that only La fragments containing the second RRM bound to lnc-Cxcl2, suggesting that La interacted with lnc-Cxcl2 through RRM2 (Fig. 5C).
To investigate the function of La in regulating Cxcl2 expression, we generated La-deficient (Ssb−/−) MLE-12 cells (SI Appendix, Fig. S5D). To our surprise, La deficiency not only increased the expression of Cxcl2 but also other chemokines and proinflammatory cytokines, like Ccl2, Ifnb1, and Tnf in response to viral infection (Fig. 5D and SI Appendix, Fig. S5E). Then, we examined how La inhibited the expressions of these cytokines and found that La deficiency increased the chromatin accessibility of Cxcl2 and Ccl2 promoters (Fig. 5E). To clarify how La inhibited the chromatin accessibility, we tried to find La-interacting nuclear proteins using MS analysis. A total of 34 proteins were identified to interact with La in MLE-12 cells (SI Appendix, Table S4). Gene ontology analysis showed that the molecular functions of most of these proteins were linked to RNA binding and ribosome structure (SI Appendix, Fig. S5F), indicating that La might inhibit these cytokine transcriptions through forming an RBP complex and ribosomal protein complex in mouse epithelial cells. Together, although La inhibits the expressions of many cytokines, lnc-Cxcl2 can bind to the Cxcl2 promoter and inhibit Cxcl2 expression in cis through ribonucleoprotein La during viral infection.
lnc-CXCL2-4-1 Inhibits Immune Cytokine Expressions in Human Lung Epithelial Cells.
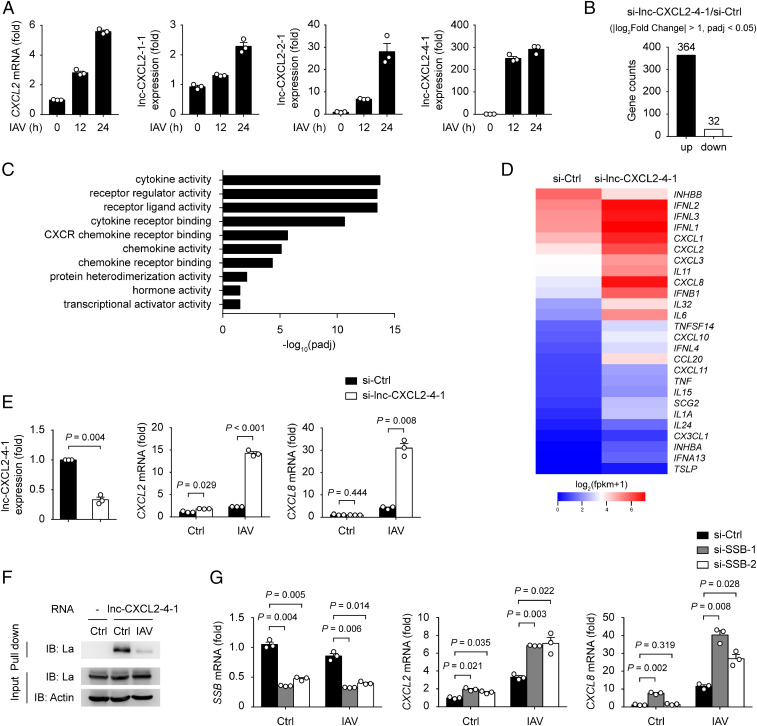
Although lncRNAs have been characterized as low conserved between different species, we asked whether a similar mechanism exists in human cells. By searching lncRNA databases, four transcripts related to CXCL2 were found in the downstream of human CXCL2, including lnc-CXCL2-1-1, lnc-CXCL2-2-1, lnc-CXCL2-3-1, and lnc-CXCL2-4-1 (28, 29). Among these, lnc-CXCL2-3-1 was predicted to have coding probability (29), thus only the other three transcripts were considered as noncoding RNAs; however, the detailed function of these three lncRNAs has not been clarified yet. Therefore, we first examined the expression of these lncRNAs in A549 human lung epithelial cells and found that the expression of lnc-CXCL2-4-1 was dramatically increased after IAV infection whereas the expressions of lnc-CXCL2-1-1 and lnc-CXCL2-2-1 were only slightly increased (Fig. 6A). We then tested whether lnc-CXCL2-4-1 participated in the regulation of chemokine expression. By performing transcriptome analysis, 364 genes were found to be up-regulated and 32 down-regulated in lnc-CXCL2-4-1–silenced A549 cells after IAV infection (Fig. 6B). Gene ontology analysis of these differentially expressed genes showed their molecular functions were enriched in cytokine activity, including chemokines, interferons, and other proinflammatory cytokines (Fig. 6 C and D). We further confirmed that silencing the expression of lnc-CXCL2-4-1 indeed increased the expression of CXCL2 and CXCL8 in A549 cells after IAV infection (Fig. 6E). We then wondered whether lnc-CXCL2-4-1 inhibited these cytokine expressions by binding La. We found that lnc-CXCL2-4-1 interacted with La, and silencing the expression of La increased both CXCL2 and CXCL8 expressions in A549 cells after IAV infection (Fig. 6 F and G), indicating that lnc-CXCL2-4-1 inhibited cytokine expressions through binding La in human lung epithelial cells.
Fig. 6.
lnc-CXCL2-4-1 inhibits cytokine expressions in human lung epithelial cells. (A) qPCR analysis of CXCL2 mRNA, lnc-CXCL2-1-1, lnc-CXCL2-2-1, and lnc-CXCL2-4-1 expressions in A549 cells infected with IAV (multiplicity of infection [MOI] = 1) for indicated hours. (B) Differentially expressed gene numbers between negative control (Ctrl) small interfering RNA (siRNA) and lnc-CXCL2-4-1 siRNA-transfected A549 cells infected with IAV (MOI = 1) for 12 h. |log2Fold Change| > 1 and adjusted P value (padj) < 0.05. (C) Gene ontology analysis of differentially expressed genes between negative ctrl siRNA and lnc-CXCL2-4-1 siRNA-transfected A549 cells infected with IAV (MOI = 1) for 12 h. Top 10 enriched molecular function gene ontology terms are shown (padj < 0.05). (D) Heatmap of differentially expressed cytokine genes between negative ctrl siRNA and lnc-CXCL2-4-1 siRNA-transfected A549 cells infected with IAV (MOI = 1) for 12 h. |log2Fold Change| > 1 and adjusted P value (padj) < 0.05. (E) qPCR analysis of lnc-CXCL2-4-1, CXCL2, and CXCL8 mRNA expressions in A549 cells transfected with negative ctrl siRNA or lnc-CXCL2-4-1 siRNA (100 nM) followed by IAV infection (MOI = 1) for 12 h. (F) Immunoblot analysis of lnc-CXCL2-4-1 and La interaction in A549 cells infected with IAV (MOI = 1) for 12 h. (G) qPCR analysis of SSB, CXCL2, and CXCL8 mRNA expressions in A549 cells transfected with negative ctrl siRNA or SSB siRNA (20 nM) followed by IAV infection (MOI = 1) for 12 h. Data are representative of three independent experiments (F) or three independent experiments with n = 3 biological replicates (A, E, and G; shown as mean + SEM), two-tailed unpaired Student’s t test. Ctrl, control; IB: immunoblot.
To further demonstrate the different regulatory function of mouse lnc-Cxcl2 and human lnc-CXCL2-4-1, we compared the secondary structure of these two lncRNAs. We found that although mouse lnc-Cxcl2 and human lnc-CXCL2-4-1 both have multiple stem loops in their secondary structure, human lnc-CXCL2-4-1 formed more stem-loop clusters, which might facilitate its regulation of multiple gene expression (SI Appendix, Fig. S6).
Taken together, these results demonstrate a host self-protecting mechanism in different species for restraining virus-induced lung inflammation through increasing the expression of an lncRNA, which can feedback inhibit chemokine production in lung epithelial cells.
Discussion
Epithelial cells act as the first line in defending against pathogens and maintaining homeostasis of regional tissue such as the lungs. The levels of epithelial cell–derived chemokines and subsequent neutrophil recruitment are closely related to the severity of many inflammatory diseases including infection, stress, asthma, and even cancer metastasis (30–32); thus, our findings of the inducible lncRNA, which can attenuate inflammatory damage through feedback inhibition of lung epithelial cell chemokine expression and neutrophil recruitment, will benefit not only the control of respiratory viral infections but also other related lung inflammatory diseases.
We demonstrate that lnc-Cxcl2 selectively inhibits Cxcl2 expression in epithelial cells but not in macrophages, indicating that a cell type–specific molecular mechanism may exist for lnc-Cxcl2, possibly relying on different intracellular and extracellular signals and metabolism-shaped chromatin architecture during viral infection. Thus, further investigations still need to be done to elucidate the selective role of lnc-Cxcl2 in different cells, especially by generating cell type–specific deficient mice. Besides, we also demonstrate that mouse lnc-Cxcl2 and human lnc-CXCL2-4-1 have different regulating profiles. Mouse lnc-Cxcl2 is located in the Cxcl2 gene locus and transcription factors, chromatin remodelers, and other nuclear proteins nearby may combine to facilitate it to regulate its neighboring gene expression in cis. However, human lnc-CXCL2-4-1 is located in the downstream of CXCL2 gene locus, which does not make it that easy to target to one specific chromatin site but rather to regulate multiple gene expression. These differences between cells and species indicate the complexity of lncRNA evolution and function, which needs to be considered in future studies.
We demonstrate that lnc-Cxcl2 inhibits Cxcl2 expression through binding La. La has been reported to associate with RNA polymerase III–transcribed genes and act as an initiation and termination factor for RNA polymerase III (33). However, whether and how La regulates RNA polymerase II–mediated transcription remains unclear. A previous study has found that conditional deletion of La in the mouse brain increases the expression of multiple immune genes, including chemokines (34). Consistent with this, we demonstrate that deletion of La in lung epithelial cells increases the expression of chemokines, interferons, and also some other proinflammatory cytokines during viral infection, confirming the regulating function of La in the RNA polymerase II–mediated transcription. Besides, our MS analysis suggests that La inhibits gene expression through forming two protein groups: the RBP group and ribosome structure protein group. As an RBP, it is possible for La to interact with other RBPs to assemble into RNP complexes (35). Cotranscriptional assembly of RNP complex is important for the maintenance of genome integrity by preventing the formation of R-loop during transcription (35, 36), which might simultaneously suppress the chromatin accessibility and shut down the transcription. On the other hand, noncanonical function of ribosome constituents in gene expression has also been found. Ribosomal protein S3 is an essential subunit of the NF-κB and can facilitate the selective control of gene expression by NF-κB (37). Suppressing the biogenesis of ribosome by interfering with ribosomal RNA accumulation can impair interferon expression without suppressing translation (38). Therefore, further study could focus on how La inhibits cytokine expression through binding these two protein complexes, and demonstrating this mechanism will greatly help understanding the function of La in inflammation and also autoimmune diseases more than just as an autoantigen.
Except for La, we also identify several other RBPs that interact with lnc-Cxcl2. Among them, the proteins which participated in RNA transcription and splicing are mostly enriched, like Ddx21, Ddx5, Ilf2, Tial1, Ybx1, and Eif4a3, suggesting that lnc-Cxcl2 may be processed through binding these proteins. Besides, some heterogeneous nuclear ribonucleoproteins (hnRNPs) that have been reported to participate in lncRNA-mediated gene regulation are also found, like hnRNPab, which can regulate Cxcl2 expression in bone marrow–derived macrophages (39). However, we did not detect the same effect of hnRNPab in MLE-12 cells, indicating that the function of hnRNPs may also be cell type specific. In addition, other proteins involved in gene regulation in different cells are also detected, like Eef1a1 which regulates gene expression in myelinating cells (40), while Pcbp1 promotes proinflammatory cytokine expression in T cells (41). Interaction with these proteins provides other functional possibilities for lnc-Cxcl2, and it will be worthy to investigate the role of lnc-Cxcl2 under different physiological and pathological conditions in the future.
Materials and Methods
Mice and Cell Lines.
C57BL/6 mice were from Beijing Vital River Laboratory (Beijing, China). lnc-Cxcl2–deficient mice were generated using the CRISPR-Cas9 system on a C57BL/6 background. Briefly, two guide RNAs respectively targeting the upstream of exon 3 (5′-TGGCCAGAGTTGCCCAGA-3) and downstream of exon 4 (5′-CTAACTGACCTGGAAAGG-3′) of Cxcl2 and a donor plasmid containing the sequence of exon 3 and exon 4 of Cxcl2 were used to delete the intron 3 of Cxcl2 and consequently delete lnc-Cxcl2. The genotypes of offspring mice were determined by PCR using the following primers: F: 5′-TTGCCTTGACCCTGAAGC-3′; R: 5′-GTACGATCCAGGCTTCCC-3′.
All mice were maintained in specific pathogen-free conditions. All mouse experiments were performed under the supervision of Institutional Animal Care and Use Committee, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences, Beijing, China (ACUC-A01-2017-004).
MLE-12 cells, RAW264.7 cells, human embryonic kidney cells (HEK293T cells), and A549 cells were from ATCC. lnc-Cxcl2–deficient MLE-12 cells and RAW264.7 cells were generated using guide RNAs targeting the intron 3 of Cxcl2: 5′-TCATTTGGAAAGGTTCAAGC-3′ and 5′-AGGCTAGAGATGGTTTCGTT-3′ for MLE-12 cells and 5′-ATCAAGGTTAACTTCAGTTC-3′ and 5′-AGGCTAGAGATGGTTTCGTT-3′ for RAW264.7 cells. La-deficient MLE-12 cells were generated using two guide RNAs targeting the exon 5 of Ssb: 5′-CATCAAGGGTGGCGTCAGTT-3′ and 5′-AAAGAATGGCTAGACGATAA-3′.
5′ and 3′ RACE.
A 5′ and 3′ RACE of lnc-Cxcl2 was performed using SMARTer RACE 5′/3′ Kit (Clontech Laboratories) following the manufacturer’s instructions. The cDNA template was from MLE-12 cells infected with VSV for 12 h. Primers used were as follows: 5′-R1:5′-GCTGGCGTTACAATTGTACCAGCTTGAACC-3′; 5′-R2: 5′-AGCGAGAGCCTACACCTGAATTTCAAGCCTC-3′; 3′-F1: 5′-CCTTCCTTAATGGATGGTCGCTGTGTGTCCC-3′; and 3′-F2: 5′-CCCCTGCCCTTTCCATGTCTGTGGGCTG-3′.
Northern Blot.
Total RNA was extracted from MLE-12 cells infected with VSV for 12 h; then, 20 μg RNA was used for electrophoresis in 1.5% denaturing agarose gel. After that, RNA was transferred to nylon membrane and crosslinked by ultraviolet light. The cross-linked membrane was prehybridized using PerfectHyb Plus hybridization buffer (Sigma-Aldrich) and further hybridized overnight by adding biotin-labeled RNA probes to the hybridization buffer. After wash, the membrane was exposed using Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) following the manufacturer’s instructions. Biotin-labeled RNA probes were synthesized by in vitro transcription using T7 RNA Polymerase and Biotin RNA Labeling Mix (Roche). The DNA template used to transcribe RNA probes was amplified according to lnc-Cxcl2 cDNA using the following primers: 5′-GGAAAGGTTCAAGCTGGTACAA-3′ and 5′-TAATACGACTCACTATAGGGACCTTGGTTTACTCTCCAAA-3′.
RNA In Situ Hybridization Assay.
MLE-12 cells were seeded into an eight-chambered dish and infected with VSV for 12 h. After infection, cells were fixed with 4% paraformaldehyde followed by permeabilization with 0.2% Triton X-100 in phosphate-buffered saline. Then, specific probe pools were used to hybridize lnc-Cxcl2, and Stellaris fluorescent in situ hybridization probes (Biosearch Technologies) were used to hybridize Gapdh mRNA in Stellaris hybridization buffer (Biosearch Technologies). Finally, nuclei were stained with DAPI. The fluorescence of RNA and nuclei were detected by Leica TCS SP8 gSTED 3X. The probes targeting lnc-Cxcl2 were as follows: 5′-ATTTTCAGAAATCGGGTGCC-3′, 5′-TGAAAGGAGCTAGTCGTACC-3′, 5′-TGACTTCGCGTACGTTGTAA-3′, 5′-CTGGGCGCTTTGAAACTAGA-3′, 5′-GGACTTTCCAGCAATAAACA-3′, 5′-AGTGGGTATCTTAAAGGCGG-3′, 5′-GGAAGGACTTTTACGTGTGT-3′, and 5′-AGATAACAGGCTTTACACCA-3′.
Influenza Virus Infection Model.
Six- to eight-week-old lnc-Cxcl2+/+ and lnc-Cxcl2−/− littermates with the same gender were used. For IAV infection, mice were first anesthetized using tribromoethanol intraperitoneally (0.25 mg/g body weight) and then intranasally inoculated with IAV (3 × 105 plaque-forming units [PFU] in 30 μL solution or 50 PFU in 20 μL solution). Mice in the control group were also anesthetized and intranasally administrated with solution instead. The body weight of all mice was measured every 24 h. After infection for indicated times, BALF was collected for cell infiltration and cytokine analysis; lung tissue was collected for RNA extraction and histopathological examination.
Chromatin Isolation by RNA Purification.
Chromatin isolation by RNA purification (ChIRP) assay was performed using Magna ChIRP RNA Interactome Kit (Millipore) following the manufacturer’s instructions with some modifications. In brief, cells were harvested and crosslinked with 1% formaldehyde. Then, the 100-mg cell pellet was resuspended in lysis buffer and sonicated to shear DNA. For lnc-Cxcl2–linked chromatin or protein isolation, 100 pmol biotin-labeled probe mix and streptavidin magnetic beads were added to hybridize and capture lnc-Cxcl2 separately. Obtained bead samples were divided into two parts, one for DNA or protein isolation and detection, the other for RNA isolation and quantification. The probes used to hybridize lnc-Cxcl2 were designed by ChIRP Probe Designer from LGC Biosearch Technologies, and the sequences were as follows: 5′-TTTCTTTAGGGTGAGCATGG-3′; 5′-TTCAGGGTCAAGGCAAACTT-3′; 5′-ATTTTCAGAAATCGGGTGCC-3′; 5′-TCGTACCCTTCTATCAATTC-3′; 5′-CGCTAGTTGGTTCTATCTAC-3′; 5′-AGTTAGACTTTACAGCCCAC-3′; 5′-CTGAATTTCAAGCCTCTCAC-3′; 5′-GACTTCGCGTACGTTGTAAA-3′; 5′-CTAGAATAGTAGAGCTGGCG-3′; 5′-AACGAAACCATCTCTAGCCT-3′; 5′-TTGCTGTTAGTGGGTATCTT-3′; and 5′-TCTTTGGTTCTTCCGTTGAG-3′.
Statistical Analysis.
Comparisons of data from two groups were performed using two-tailed unpaired Student’s t test. Comparisons of mouse body weight and cell proliferation data with different time points between two groups were performed using two-way ANOVA analysis. P values less than 0.05 were considered as statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81788101 and 81930041) and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2016-12M-1-003).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108276118/-/DCSupplemental.
Data Availability
The RNA sequencing data from this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession code GSE156949 and GSE179951 (42, 43). All other study data are included in the article and/or SI Appendix.
References
- 1.Griffith J. W., Sokol C. L., Luster A. D., Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Kolaczkowska E., Kubes P., Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Alon R., et al., Leukocyte trafficking to the lungs and beyond: Lessons from influenza for COVID-19. Nat. Rev. Immunol. 21, 49–64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laforge M., et al., Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 20, 515–516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang B. M., et al., Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat. Commun. 10, 3422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buszko M., et al., The dynamic changes in cytokine responses in COVID-19: A snapshot of the current state of knowledge. Nat. Immunol. 21, 1146–1151 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Cao X., COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 20, 269–270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokol C. L., Luster A. D., The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 7, a016303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki A., Foxman E. F., Molony R. D., Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 17, 7–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann R., et al., Differential regulation of the transcriptomic and secretomic landscape of sensor and effector functions of human airway epithelial cells. Mucosal Immunol. 11, 627–642 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Benam K. H., Denney L., Ho L. P., How the respiratory epithelium senses and reacts to influenza virus. Am. J. Respir. Cell Mol. Biol. 60, 259–268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polidoro R. B., Hagan R. S., de Santis Santiago R., Schmidt N. W., Overview: Systemic inflammatory response derived from lung injury caused by SARS-CoV-2 infection explains severe outcomes in COVID-19. Front. Immunol. 11, 1626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian Z., et al., Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 48, 742–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams D. W.et al.; NIDCD/NIDCR Genomics and Computational Biology Core , Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 184, 4090–4104.e15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Statello L., Guo C. J., Chen L. L., Huarte M., Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil N., Ulitsky I., Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Khyzha N., et al., Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 116, 16410–16419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarropoulos I., Marin R., Cardoso-Moreira M., Kaessmann H., Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571, 510–514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long Y., et al., RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 52, 931–938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao R., et al., Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong L., et al., CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345-9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Filippo K., et al., Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood 121, 4930–4937 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Burke S. J., et al., NF-κB and STAT1 control CXCL1 and CXCL2 gene transcription. Am. J. Physiol. Endocrinol. Metab. 306, E131–E149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaramillo M., Olivier M., Hydrogen peroxide induces murine macrophage chemokine gene transcription via extracellular signal-regulated kinase- and cyclic adenosine 5′-monophosphate (cAMP)-dependent pathways: Involvement of NF-kappa B, activator protein 1, and cAMP response element binding protein. J. Immunol. 169, 7026–7038 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Alfano C., et al., Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 11, 323–329 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Marrella S. A., et al., An interdomain bridge influences RNA binding of the human La protein. J. Biol. Chem. 294, 1529–1540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinayak J., et al., Human La binds mRNAs through contacts to the poly(A) tail. Nucleic Acids Res. 46, 4228–4240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L., et al., NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volders P. J., et al., LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 47, D135–D139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frey A., et al., More than just a barrier: The immune functions of the airway epithelium in asthma pathogenesis. Front. Immunol. 11, 761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernández-Santos N., et al., Lung epithelial cells coordinate innate lymphocytes and immunity against pulmonary fungal infection. Cell Host Microbe 23, 511–522.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., et al., Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30, 243–256 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Fairley J. A., et al., Human La is found at RNA polymerase III-transcribed genes in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 18350–18355 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blewett N. H., Iben J. R., Gaidamakov S., Maraia R. J., La deletion from mouse brain alters pre-tRNA metabolism and accumulation of Pre-5.8S rRNA, with neuron death and reactive astrocytosis. Mol. Cell. Biol. 37, e00588–e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montecucco A., Biamonti G., Pre-mRNA processing factors meet the DNA damage response. Front. Genet. 4, 102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida K., Kuwano Y., Nishikawa T., Masuda K., Rokutan K., RNA binding proteins and genome integrity. Int. J. Mol. Sci. 18, 1341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan F., et al., Ribosomal protein S3: A KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131, 927–939 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Bianco C., Mohr I., Ribosome biogenesis restricts innate immune responses to virus infection and DNA. eLife 8, e49551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carpenter S., et al., A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duman M., et al., EEF1A1 deacetylation enables transcriptional activation of remyelination. Nat. Commun. 11, 3420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., et al., Iron drives T helper cell pathogenicity by promoting RNA-binding protein PCBP1-mediated proinflammatory cytokine production. Immunity 49, 80–92.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Liu S., Liu J., Yang X., Jiang M., Cao X., lnc-Cxcl2 suppressed Cxcl2 expression in mouse lung epithelial cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156949. Deposited 27 August 2020.
- 43.Liu S., Liu J., Yang X., Cao X., Transcriptome analysis of genes that regulated by lnc-CXCL2-4-1. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179951. Deposited 12 July 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data from this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession code GSE156949 and GSE179951 (42, 43). All other study data are included in the article and/or SI Appendix.