Significance
Although germline heterozygous BRCA1 mutations predispose human carriers to cancer, heterozygous mouse BRCA1 mutations do not. We find that exposure to a source of upper gastrointestinal replication stress (RS) elicited a marked cancer incidence in Brca1+/−;Trp53+/− heterozygous mice but not in wild-type mice (Brca1+/+;Trp53+/−). Oral delivery of 4 nitroquinoline-1-oxide induced esophageal epithelial RS, an increased esophageal mutation rate, loss of esophageal Brca1 heterozygosity (LOH), and accelerated esophageal tumorigenesis. These data underscore the necessity of combining otherwise nontumorigenic BRCA1 heterozygosity with simultaneous induction of RS for the generation of not only BRCA1 mutant esophageal cancer, but also a dramatically accelerated form thereof. These results strongly imply that RS is both a major contributor to BRCA1 cancer development and a marked accelerant thereof.
Keywords: BRCA1, replication stress, haploinsufficiency, mouse model
Abstract
BRCA1 germline mutations are associated with an increased risk of breast and ovarian cancer. Recent findings of others suggest that BRCA1 mutation carriers also bear an increased risk of esophageal and gastric cancer. Here, we employ a Brca1/Trp53 mouse model to show that unresolved replication stress (RS) in BRCA1 heterozygous cells drives esophageal tumorigenesis in a model of the human equivalent. This model employs 4-nitroquinoline-1-oxide (4NQO) as an RS-inducing agent. Upon drinking 4NQO-containing water, Brca1 heterozygous mice formed squamous cell carcinomas of the distal esophagus and forestomach at a much higher frequency and speed (∼90 to 120 d) than did wild-type (WT) mice, which remained largely tumor free. Their esophageal tissue, but not that of WT control mice, revealed evidence of overt RS as reflected by intracellular CHK1 phosphorylation and 53BP1 staining. These Brca1 mutant tumors also revealed higher genome mutation rates than those of control animals; the mutational signature SBS4, which is associated with tobacco-induced tumorigenesis; and a loss of Brca1 heterozygosity (LOH). This uniquely accelerated Brca1 tumor model is also relevant to human esophageal squamous cell carcinoma, an often lethal tumor.
Germline BRCA1 mutations predispose humans to an elevated cancer risk and especially that of the breast and ovary (1, 2). In recent times the suggestion of an increased risk of esophageal cancer in BRCA1 mutation carriers has also been reported, e.g., in an individual with a germline BRCA1 mutation (3). Also, a complete clinical response to platinum treatment was observed in a patient with BRCA1 mutant esophageal cancer (4). Furthermore, the overall esophageal squamous cell carcinoma (ESCC) risk in BRCA1 carriers is significant (relative risk [RR] of 2.9 [95% CI 1.1 to 6.0]) (5, 6). This is in keeping with the observation that a relatively frequent loss of heterozygosity (LOH) is detected in the BRCA1-containing region of chromosome 17 in squamous cell carcinoma of the esophagus (7–9).
BRCA1 maintains genome integrity by engaging in multiple cellular processes, including the repair of DNA damage (10, 11), including double strand breaks (DSBs), stalled replication forks, and other abnormalities. Stalled forks, when not resolved, can lead to mutations or can collapse into DSBs (12–15). Both outcomes are components of what is commonly referred to as replication stress (RS), which, when chronic, can serve as a cancer driving force (16–18).
Loss of certain DNA damage repair functions in BRCA1 mutant tumor cells also renders these cells sensitive to platinum-based derivatives and PARP inhibitors (19, 20). Success of these agents in suppressing BRCA1 mutant tumor growth has made them therapeutic agents of choice for treating BRCA1 mutant cancer (21, 22). Loss of BRCA1 function either by germline deletion and/or promoter hypermethylation is now a predictive classifier of response to these agents (23).
Currently, multiple Brca1 mouse models facilitate the study of BRCA1 loss-associated tumorigenesis. Complete loss of BRCA1 is embryonically lethal (24); thus, successful, tumorigenic models either conditionally delete both alleles of Brca1 in a tissue of interest or express a hypomorphic mutant version of Brca1 (25–27). For example, conditional Brca1 loss can be driven by Cre-mediated deletion of two Brca1 floxed alleles in a tissue of choice. And mice bearing hypomorphic Brca1 mutant alleles, like delta 11, and those that express an incompletely functioning or a truncated version of the Brca1 protein instead of the full-length polypeptide also develop Brca1 tumors (26, 28, 29).
Often, BRCA1 knockout (KO) mice also incur a loss of p53 function, which results in accelerated tumor formation, and, in the case of hypomorphic Brca1 mutant mice, their survival as well (25, 27, 30). Mice in both models develop tumors, including mammary and ovarian tumors, which, on average, take ∼1 y to develop. Brca1 delta 11 (Brca1Δ11/Δ11) mice, which synthesize a markedly deleted but still partially functional Brca1 allele, also form esophageal tumors that can be accelerated by addition of the oxidative stress–inducing agent methyl-N-amylnitrosamine (MNAN) to their drinking water (31).
However, the role of BRCA1 haploinsufficiency has not been extensively evaluated in mouse models. One reason is that Brca1 heterozygous mice did not generate tumors more often or faster than their wild-type (WT) counterparts (27, 32). This is unlike the increased predisposition to cancer observed in human BRCA1 mutation carriers (who carry a germline loss-of-function mutation in a single BRCA1 allele). They manifest a significantly greater than normal breast and/or ovarian cancer incidence by age 70 (1, 2).
BRCA1 haploinsufficiency can be linked to increased genomic instability, in part, because of its defective participation in stalled fork repair and replication stress suppression (33) and, possibly, because of its role in regulating SIRT1 levels and affecting pRb pathway activation (34). Given the importance of RS development in tumorigenesis (16, 18, 35), this effect would be a logical contributor to BRCA1 mutant cancer development. Of note, BRCA1 heterozygous human cells are haploinsufficient for RS suppression (33), raising the possibility that this defect operates as a general contributor to the increased tumorigenicity observed in many germline BRCA1 heterozygous families.
To test this hypothesis, we have established a Brca1 mutant esophageal mouse cancer model that is capable of addressing the role of replication stress accumulation in BRCA1 mutant cancer. Here one allele of Brca1 and one of Trp53 were deleted through the action of Meox2Cre, which acts very early during embryogenesis (embryonic day 5 [E5]) (36) and results in the development of Brca1 and p53 heterozygosity in all tissues. Using this mouse model, we have found that BRCA1 deficiency in replication stress suppression is enhanced by exposure to 4-nitroquinoline-1-oxide (4NQO) in BRCA1 heterozygous tissue where it serves as an efficient and abnormally rapid driver of tumor formation.
Results
4NQO Induces Replication Stress in Mice Bearing Germline Conditional Brca1 and Trp53 Alleles.
Unlike the increased predisposition to cancer observed in human BRCA1 mutation carriers, heterozygous Brca1 mouse models do not manifest an increased cancer incidence (27). Given that 1) BRCA1 heterozygous cells (including Brca1 heterozygous mouse cells) are defective in stalled replication fork repair and RS suppression (33), and that 2) chronic RS is a general contributor to tumorigenesis in certain mammalian species (16, 18, 35), we reasoned that, if RS were a necessary driver of murine Brca1 mutant cancer, then increasing RS above the levels observed in Brca1 heterozygous mouse tissue might induce tumorigenesis in this tissue and not in the same tissue of wild-type control mice. Conceivably, in a typically short mouse life span (∼18 to 24 mo), insufficient, indigenous RS accumulates in Brca1 heterozygous animals to elicit tumorigenesis.
To investigate the role of RS in BRCA1 mutant tumorigenesis, we established a Brca1 heterozygous mouse model where RS was actively heightened, pharmacologically. Specifically, 4NQO, a known RS-inducing agent (37), was administered to mice by adding it to their drinking water. 4NQO is a known carcinogen, which primarily forms DNA adducts at guanine residues (38, 39).
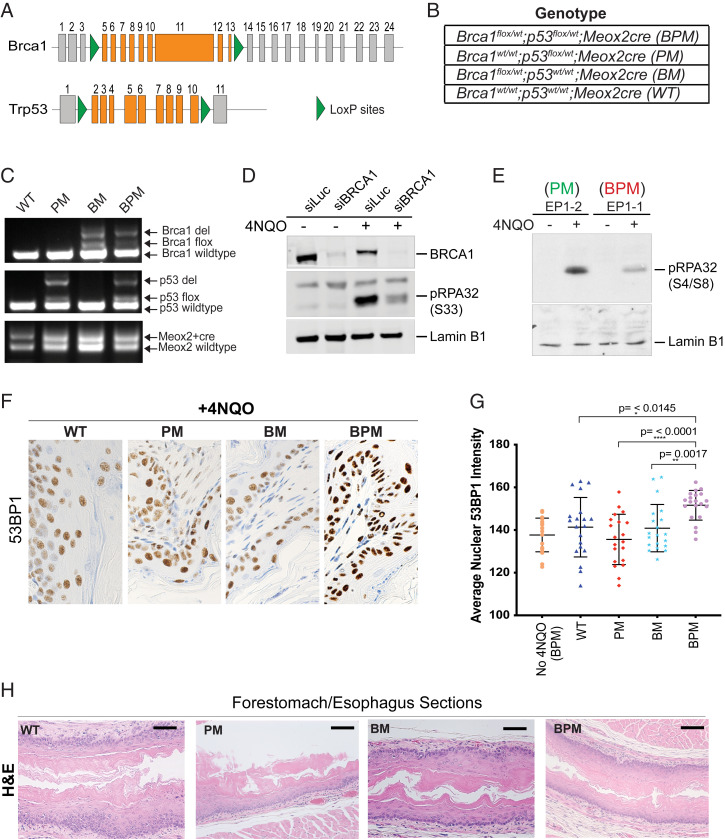
More specifically, conditional loss-of-function Brca1 and Trp53 mice were studied, wherein loxP sites flanked Brca1 exons 5 to 13 and Trp53 exons 2 to 10 (Fig. 1A) (27). We generated four different mouse cohorts to address the role of BRCA1 heterozygosity in tumor formation: Brca1flox/wt;Trp53flox/wt;Meox2Cre (hereafter referred to as BPM), Trp53flox/wt;Meox2Cre (PM), Brca1flox/wt;Meox2Cre (BM), and WT controls that also bore a Meox2Cre allele (Fig. 1B).
Fig. 1.
Use of 4NQO as a RS-inducing agent in mice carrying conditional Brca1 and Trp53 alleles. (A) Schematic representation of the location of loxP sites in the Brca1 and Trp53 loci in the mouse genome. (B) List of genotypes (BPM, PM, BM, and WT) used in this study. (C) PCR-based analysis to confirm germline deletion of the floxed segments of Brca1 and Trp53 by Meox2-driven Cre recombinase. Genomic DNA was extracted from mouse tail cuts and amplified using primers for the floxed and/or the deleted region of the gene. Details of the PCR primers and their locations are provided in Materials and Methods. A representative gel depicting the presence of WT, deleted (del), floxed (flox), and Meox2-Cre alleles is shown for all four genotypes (WT, PM, BM, and BPM). (D) Western blot analysis of the human esophageal squamous cell cancer line, KYSE410 nuclear extracts. The extracts were blotted for phosphorylated RPA (pRPA32, phosphorylated at S33), BRCA1, and Lamin B1 (loading control). pRPA32 accumulation was studied before and after 4NQO treatment in control (siLuc) and BRCA1-depleted (siBRCA1) cells. (E) Western blot analysis of phosphorylated, chromatin-associated RPA32 (pRPA32, phosphorylated at S4/S8) in extracts isolated from 4NQO-treated and -untreated MEFs. MEFs from Brca1flox/wt;Meox2Cre (Brca1+/−) and Brca1wt/wt;Meox2Cre (Brca1+/+) mice were cultured in the presence and absence of 4NQO for 3 h. In both D and E, cells were harvested, and equivalent amounts of lysate were electrophoresed, blotted, and the blot probed with an anti-pRPA32 antibody. A nonspecific band (D) and Lamin B1 (E) served as loading controls. (F and G) Effect of 4NQO treatment (4NQO delivered via the drinking water) on replication stress in BPM mice. Accumulation of 53BP1 in nuclei was used to assess induction of replication stress. Stomach sections were collected from mice who drank water with and without 4NQO for 2 d and analyzed for accumulation of 53BP1 by IHC. Forestomach/esophageal tissue sections from WT, PM, BM, and BPM mice (n = 2 mice for each genotype) were collected upon 4NQO exposure and stained for 53BP1 (IHC). Images were captured at 40X magnification and analyzed with ImageJ and MatLab_R2019b software. Mice heterozygous for Brca1 and Trp53 (BPM) showed enriched mean pixel intensity for 53BP1 compared with other genotypes. A two-tailed paired t test was used to determine statistical significance. (H) H&E staining of esophageal tissue. All four genotypes (WT, BM, PM, and BPM) show normal esophageal histology. (Scale bars in black, 200 mm.)
In these mice, Cre recombinase was expressed under the control of the endogenous Meox2 promoter, which has been shown to express as early as E5 (36). The early expression of Meox2Cre insured early deletion of the floxed Brca1 and/or Trp53 alleles from all mouse tissues. We confirmed the deletion of the floxed Brca1 and Trp53 alleles by genotyping tail cuts from these mice (Fig. 1C). All mice generated in this study were healthy and fertile.
To determine whether cells derived from mouse tissue were susceptible to 4NQO-induced replication stress, we studied mouse embryonic fibroblasts (MEFs). Upon replication stress development, stalled replication forks become coated with phosphorylated replication protein A (pRPA32), which is a prerequisite for efficient stalled replication fork repair (40). We showed earlier that BRCA1 heterozygous cells are defective in loading pRPA32 on chromatin following hydroxyurea (HU)-induced replication stress (33).
We first tested whether 4NQO is similarly capable of inducing replication stress. To address this question, we analyzed esophageal squamous cell carcinoma cell line KYSE410 and studied the loading of pRPA32 on chromatin as a manifestation of replication stress induction following 4NQO treatment. As shown in Fig. 1D, 4NQO induced pRPA32 (S33) loading in control cells (siLuc-treated cells), but not in siBRCA1- treated KYSE410 cells (Fig. 1D) confirming that, as shown before with other RS-inducing agents like hydroxyurea and ultraviolet (UV) (33, 41), pRPA32 loading following 4NQO treatment is BRCA1 dependent.
We next asked whether this is true for mouse cells as well. We found that MEFs derived from Brca1 heterozygous mice (Brca1+/−), but not those derived from Brca1 wild-type (Brca1+/+) MEFs, are defective in loading pRPA32 on chromatin following the induction of 4NQO-induced replication stress (Fig. 1E). These results confirmed that 4NQO is indeed a RS-inducing agent and that mouse Brca1 heterozygous mouse cells manifest a defective replication stress response, just like BRCA1 heterozygous human cells (33).
Increased Replication Stress in Forestomach Tissue of BPM Mice Exposed to Oral 4NQO.
Previous studies have employed two different modes of introducing 4NQO into mice to study carcinogen-induced tumor formation. 4NQO was either “painted” on to the tongues of mice or was added to their drinking water (42). We employed the latter approach. Having established that 4NQO induces replication stress in MEFs, we next asked whether 4NQO exposure elicits replication stress in vivo. Specifically, we analyzed mouse tissue and whether Brca1 heterozygous mouse tissue is especially susceptible to accumulating such DNA damage.
Given that we were introducing 4NQO into mice via their drinking water, we focused on the state of their esophageal/forestomach tissue. Two mice from each of the four, aforementioned mouse cohorts drank water containing 200 μg/mL of 4NQO for 2 d. The other animals served as controls. Mice were killed 24 h postoral 4NQO treatment, and their esophageal/forestomach tissue was harvested and analyzed for signs of replication stress. These sections were stained for the protein 53BP1 (SI Appendix, Fig. S1A), a marker of replication stress (43, 44), and a clear increase in 53BP1-positive nuclei was observed in tissue sections collected from the 4NQO-treated mice. Importantly, BPM mice revealed a higher incidence of strong 53BP1-positive nuclei compared with mice in the other cohorts (BM, PM, and WT) (Fig. 1 F and G). Stained sections were analyzed using ImageJ and Matlab, and 53BP1-positive pixel values for each image were determined by measuring levels that were above background (>150 pixels/μm2). These values were averaged for each sample (Fig. 1G).
Higher nuclear 53BP1 pixel intensities in BPM tissue compared with all the other genotypes, suggests that dual Brca1/p53 heterozygosity resulted in increased replication stress in the nuclei of BPM tissue compared with BM, PM, and/or WT tissue. Although there was an increased accumulation of 53BP1-positive nuclei in BPM mice, the hematoxylin and eosin (H&E) stains confirmed that such a short exposure to 4NQO (2 d) did not affect the normal histology of the esophageal tissue in mice from any of the four genotypes (Fig. 1H).
Brca1;Trp53 Double Heterozygous BPM Mice Are Prone to Esophageal and Forestomach Cancer when Faced with the Development of Replication Stress.
Having confirmed that there is increased replication stress in Brca1/p53 double heterozygous mouse esophagus/forestomach tissue upon 4NQO exposure, we then asked whether increased replication stress in the esophagus/stomach tissue of these mice is tumorigenic. Prior results of others have suggested that RS is potentially tumorigenic (45, 46). To study whether the codevelopment of Brca1 heterozygosity and upper gastrointestinal (GI)-focused replication stress is tumorigenic in the esophagus, we exposed mice to 4NQO-containing drinking water. The animals were administered 200 μg/mL 4NQO in drinking water for a period of 75 d and then followed for 45 d during which the animals received only normal, drug-free drinking water (Fig. 2A).
Fig. 2.
Brca1flox/wt;p53flox/wt;MeoX2cre double heterozygous mice (BPM) are prone to esophageal and/or forestomach cancer upon exposure to replication stress induced by 4NQO. (A) Schematic representation of the animal drug treatment protocol that was employed in this study. (B) The table lists the four genotypes (BPM, PM, BM, and WT) and the number of mice per cohort that were used in this study. (C) Representative macroscopic images that show formation of esophageal/forestomach tumors in BPM mice and lack of such tumors in WT mice after undergoing the same 4NQO treatment as detailed in A. (D) Macroscopic images of the forestomach in WT vs. BPM mice that underwent 4NQO treatment as detailed in A. WT mice show mostly tumor-free forestomach and BPM mice show large fused tumors in the forestomach. Detailed microscopic analysis of WT and BPM forestomach is provided in Fig. 3.
A common water stock was prepared and was equally distributed in drinking bottles. Bottles were replaced every 2 d. Mice from the various genotypes (BPM, PM, BM, and WT) (Fig. 2B) were housed in the same cage (four mice per cage) and all drank from the same water source. This ensured that all mice in our study were equivalently exposed to 4NQO, which was dissolved in a polyethylene glycol (PEG)/H2O solution. Control mice received PEG-containing water as a form of vehicle-only exposure. Equivalent exposure to 4NQO was confirmed by the observation that the incidence of benign tongue papillomatosis, a common occurrence in mice that drink 4NQO-containing water, was similar in mice of different genotypes (SI Appendix, Fig. S1B). All mice were killed at the end of a 45-d observation period, and their stomach, esophagus, and tongue tissue were harvested for analysis.
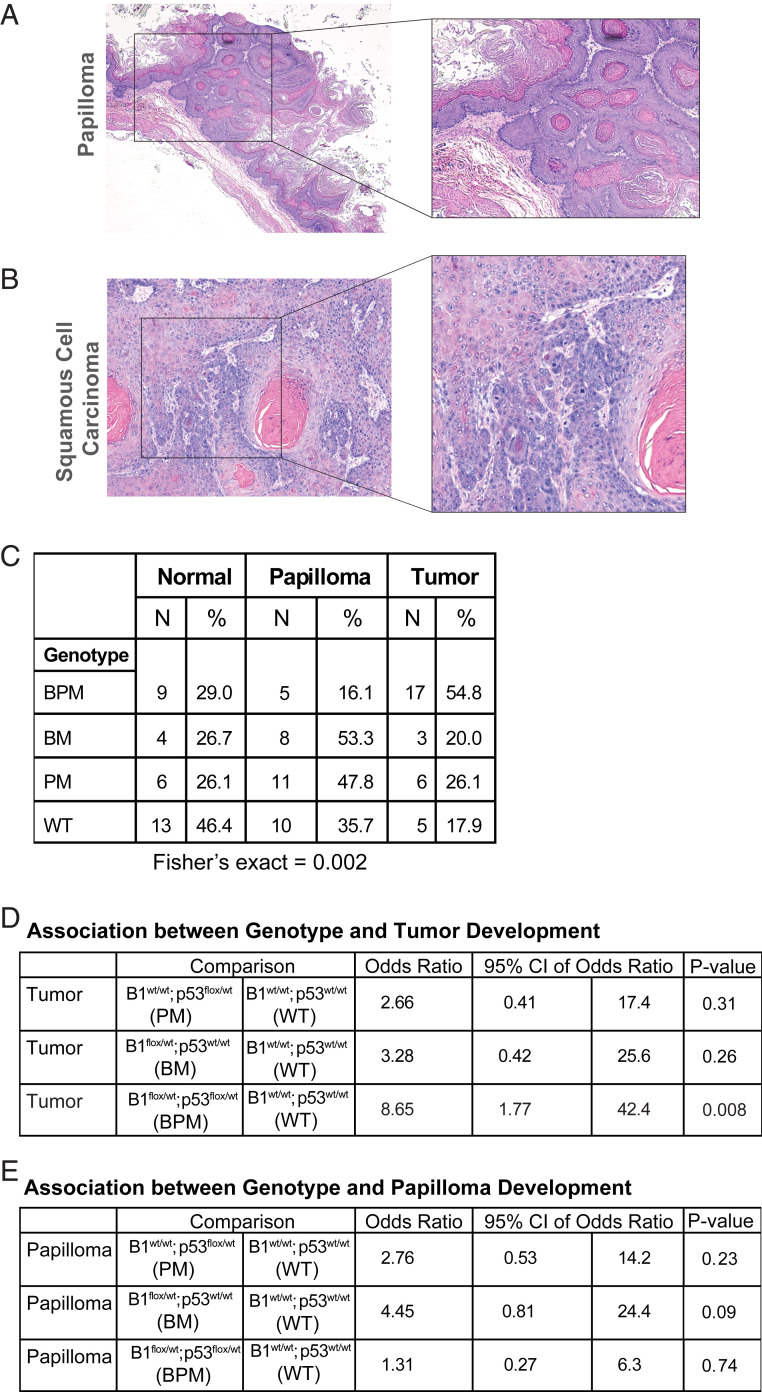
We worked with n = 107 mice divided among four genotypes, as detailed in Fig. 2B. They were scored for tumor formation at the time of death. We observed a significantly higher incidence of esophageal/forestomach tumorigenesis in BPM mice after 4NQO treatment compared with control mice (WT) (Fig. 2 C and D). Forestomach sections were further analyzed to determine the incidence of papilloma and/or tumor development (Fig. 3 A and B). After accounting for death arising from unknown causes during the course of the experiment, we analyzed n = 97 mice belonging to four different genotype cohorts: BPM (n = 31), BM (n = 15), PM (n = 23), and WT (n = 28) (Fig. 3C) in search of tumorigenesis in each cohort.
Fig. 3.
Brca1flox/wt;p53flox/wt;MeoX2cre double heterozygous mice (BPM) are prone to esophageal and/or forestomach carcinoma upon replication stress. (A and B) Microscopic analysis of lesions in the forestomach of mice following 4NQO treatment. Representative histological images (H&E stains) to document the existence of esophageal papilloma (A) and squamous cell carcinoma (B) observed in mice following 4NQO exposure. (A) Papilloma tissue growing upward into the lumen of the esophagus and (B) depicts small nests of squamous tumor cells invading the stroma. It also contains a tumor cyst filled with keratin. (C) Summary of genotype by outcome (normal, papilloma, and/or tumor). The lesions were classified as papilloma or squamous cell carcinoma based on similarity to representative images shown in A and B. (D) Association between genotype and tumor development. The table summarizes the results of a multinomial logistic regression model of tumor development. Compared with WT animals, mice with a Brca1flox/wt;Trp53flox/wt;Meox2Cre genotype (BPM) experienced a statistically significant likelihood of tumor development (OR: 8.7, 95% CI: 1.8 to 42.4, P = 0.008). (E) Association between genotype and papilloma development. Papilloma incidence in WT mice was compared with that in BM, PM, and BPM mice.
Given that both male and female mice were present in this study, we first tested whether males or females were affected differently. No tumor development gender bias was observed. Moreover, no mice in the PEG control arm (n = 16) formed tumors. By contrast, when different genotypes were compared for 4NQO-associated tumorigenesis, only Brca1;Trp53 double heterozygous (BPM) mice revealed a significantly increased incidence of tumorigenesis compared with WT mice (odds ratio [OR]: 8.7, 95% CI: 1.8 to 42.4, P = 0.008) (Fig. 3 D and E).
Although BPM mice revealed the highest frequency of tumorigenesis compared with other genotypes, they revealed low levels of oral papilloma development. In contrast to this observation, BM mice exhibited an increased incidence of oral papillomatosis relative to the other genotypes (Fig. 3 C and E). One possible explanation for the relatively low-level papilloma formation in BPM mice compared with the relatively high incidence in BM mice is that the loss of p53 in BPM mice allowed their precancerous cells to progress further along the path to tumorigenesis than did BM cells (which have lost Brca1 but expressed WT p53).
Esophageal Squamous Cell Carcinoma Reveals Krt14 and Phosphorylated Chk1 Expression.
The increased expression of keratins such as cytokeratin 14 (K14) is a known manifestation of esophageal carcinogenesis, especially in human squamous cell esophageal carcinoma (47, 48). Thus, we analyzed K14 expression in BPM esophageal tumors and confirmed that they were strongly K14 positive (Fig. 4A). PM tumors were similarly positive for K14 staining.
Fig. 4.
Cytokeratin 14 and phosphorylated CHK1 expression in esophageal squamous cell carcinoma (SCC) of mice. (A) Representative IHC sections to assess keratin 14 (Krt14) expression in forestomach tissue from nontumor and tumor tissue harvested from BPM mice. (B) Representative pictures of PM, BM, and BPM tumors stained with pCHK1 antibody. Upper panels show the H&E of the sections that were stained for pCHK1. Approximate region on the H&E slide that corresponds to the pCHK1-stained section is shown in the boxed region. (Scale bars in black, 200 mm and scale bars in white, 100 mm.) pCHK1 immunofluorescence analysis reveals increased pCHK1 expression in BPM esophageal/forestomach tumor tissue compared with PM or BM mice. (C) Percentage of pCHK1-positive cells was determined for tumor sections collected from mice of different genotypes. BPM tumor sections contained significantly more pChk1-positive cells per section than BM, PM, and WT cells (D).
We also tested whether phosphorylated CHK1, a reporter of DNA damage and of replication stress, was expressed in these tumors. Previous studies had shown that, in response to replication stress, ATR phosphorylates CHK1 kinase at S317 and S345 (49).
To determine whether tumors of any one genotype experienced higher levels of a given replication stress marker than others, we analyzed pChk1(S345) in esophageal tumors in mice of different genotypes (Fig. 4B). The overall percentage of cells with nuclear pChk1(S345) staining was significantly higher in BPM tumors compared with WT or BM or PM tumors (Fig. 4 B and C). This implies that higher levels of replication stress and, likely, DNA damage were present in Brca1/Trp53 double heterozygous (BPM) tumor cells than in tumor cells that appeared in any of the other cohorts. BPM tumors also stained positively for 53BP1, signaling signs of DNA damage in these tumors (Fig. 4D).
We also checked p63 status in BPM tumors, given that one of the traditional markers of esophageal squamous cell carcinoma is p63 (50). BPM tumor sections manifested strong p63 staining (Fig. 4D), while normal esophageal sections from BPM mice did not (SI Appendix, Fig. S1C).
Whole-Exome Sequencing Reveals an Increased C > A Mutation Frequency in 4NQO-Induced Tumors in BPM Mice.
To determine the mutational load and the abundance of relevant genomic alterations in tumors from BPM compared with PM mice, we carried out whole-exome sequencing (WES).
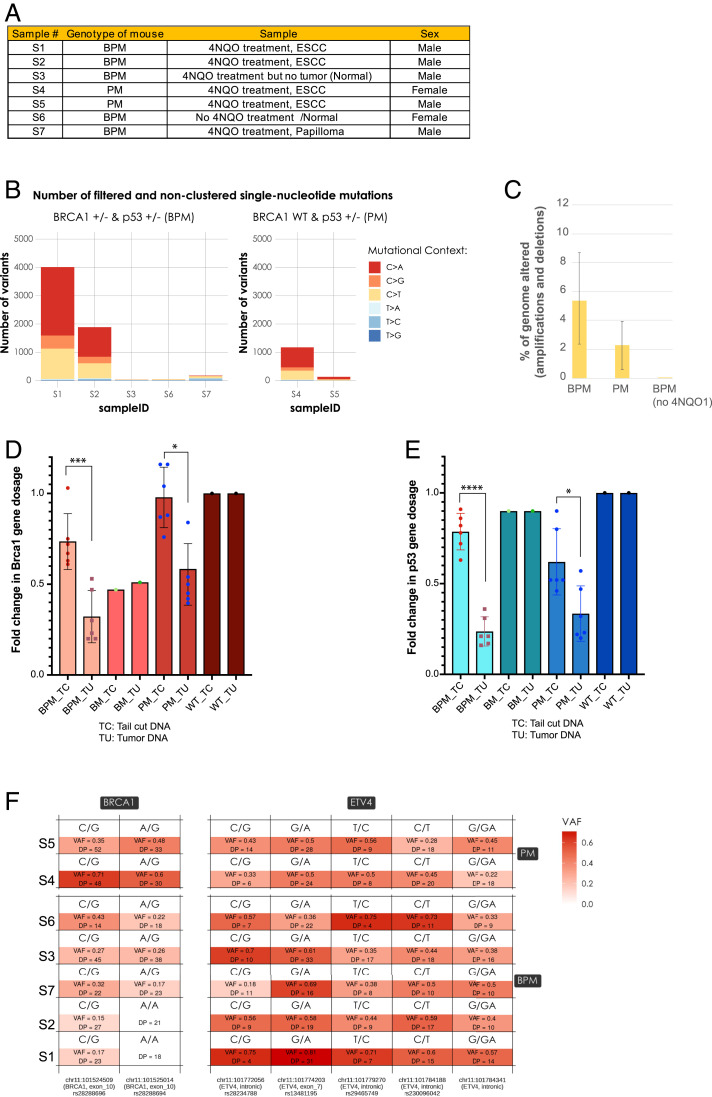
Given that mutational load closely correlates with tumor aggressiveness (51), we asked whether there were any differences in the nature and/or the extent of mutational signatures in tumors from one genotype compared with another. A sample list is shown in Fig. 5A. Genomic DNA used for WES was extracted from microdissected tumor sections of BPM mice (samples S1 and S2), PM mice (samples S4 and S5), of papilloma tissue from a BPM mouse (sample S7), and tissue from a normal forestomach/esophageal region of BPM mice (samples S3 and S6) (Fig. 5A).
Fig. 5.
DNA mutational signature and Brca1 and Trp53 LOH analysis in the esophageal tumors formed upon exposure to 4NQO. (A) List of mouse tissue samples that were used for WES. Genomic DNA was extracted from microdissected tumor sections (samples S1, S2, S4, and S5), papilloma sections (sample S7), and from the normal forestomach/esophageal region (samples S3 and S6). (B) Number of somatic mutations in the WES samples listed in A, after filtering on the quality, depth, and S-score parameters returned by isomut, and the removal of the clustered germline contaminants. Clusters were identified as a group of variants with a mean distance between the neighboring mutations less than 10% of the average distance between neighboring mutations across the entire exome. (C) Percentage genome altered (amplifications and deletions) was analyzed for samples S1 and S2 (BPM tumors), S4 and S5 (PM tumors), and S3 (BPM mouse, 4NQO exposure, no tumor) and S6 (BPM mouse no 4NQO exposure/no tumor). Each sample was compared with the same normal sample (sample S6). Copy number analysis was performed as described in Materials and Methods. (D and E) Forestomach/esophageal tumor tissue sections were microdissected and genomic DNA was extracted from them. Genomic DNA from TCs of the corresponding mice was also extracted. Tumor and the corresponding tail cut DNA were collected from n = 6 (BPM), n = 6 (PM), and n = 2 BM and WT mice. Actin primers were used to amplify an actin locus as an internal control. Primers for Brca1 and Trp53 were used to amplify the respective genes in both TCs and TU genomic DNA extracted from all relevant samples. The fold change in gene dosage (Brca1 and Trp53) was calculated by normalizing to WT tail cut genomic DNA. The Brca1 gene dosage is plotted in D and the Trp53 gene dosage is plotted in E. P value <0.05 was considered statistically significant (*P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001). (F) Variant allele frequency (VAF) analysis for single nucleotide polymorphisms (SNPs) in exon 10 (analogous to human exon 11) of the mouse Brca1 gene. S1 and S2 samples (BPM tumors) reveal a complete loss of the variant at position chr11:101524509 and a near complete loss of the variant at position chr11:101525014.
We analyzed the WES data in an effort to assess the roles of certain mutational signatures that exist in the relevant tumor material. Homologous recombination deficiency (HRD) signatures are enriched in cancers harboring biallelic inactivation of BRCA1 or BRCA2 (52). An HRD-related mutational signature, referred to as COSMIC signature 3, is characterized by a specific single nucleotide variation pattern and a distinct pattern of insertions and deletions. There was no accumulation of signature 3 in any of the tumor samples, including Brca1 mutant tumors (BPM). Previous studies with Brca1 mutant mouse tumors also have failed to detect the strong accumulation of an HRD signature unlike what was detected in human BRCA1 mutant tumors (53–55).
However, the WES analysis did reveal that C > A mutations were four- to fivefold more abundant in the 4NQO-induced tumors in BPM mice compared with PM mice (Fig. 5B). These mutations were observed only in tumor tissue as opposed to tissue of the same genotype that had been exposed to 4NQO but remained tumor free (e.g., in samples S3 and S7).
These findings are consistent with the clonal nature of the tumor cells and suggest that cells harboring C > A mutations were particularly prone to becoming tumorigenic of all of the cell types that were analyzed. Moreover, a four- to fivefold increase in C > A mutations in BPM tumors, compared with PM tumors strongly suggested that inefficient repair of 4NQO-induced damage in Brca1 heterozygous cells in BPM mice contributed to tumor formation. 4NQO and its metabolites have a strong preference to form adducts with guanine (38). Either 4NQO-guanine adducts, or its metabolite-driven 8-oxo-guanine (8-oxoG) and 8-hydroxydeoxyguanosine (8OHdG) DNA adducts (56, 57) could contribute to increased C > A mutations after 4NQO treatment. 8-oxoG and 8OHdG DNA adducts have been shown previously to accumulate in cells treated with 4NQO, and both are commonly associated with C > A transversions because of mispairing with adenine during DNA replication (58).
Furthermore, although there was a significant increase in C > A mutations in tumor cells, the contribution from other base substitutions was relatively low. This pattern is consistent with the presence of a single nucleotide variation-based mutational signature, SBS4 (59), that dominated the other mutational outcomes in the tumor. Of note, this signature accumulates in cancers that are strongly associated with tobacco exposure or appears in cell lines that have been exposed to benzo-o-pyrene, a tobacco smoke carcinogen (59). Both of these agents, like 4NQO, primarily form guanine adducts.
We also determined the frequency of indels (deletions and insertions) in these samples. Though we detected a strong correlation between 4NQO exposure and an increase in indel frequency, we did not detect a tissue genotype-based effect on indel frequency. This suggests that such events (insertions and deletions) do not necessarily depend on tissue genotype and are more dependent on exposure of the tissue to a relevant carcinogen.
Finally, in keeping with an increased C > A mutation signature in BPM- compared with PM-based tumor cells, we also found that the percentage of genome alterations (i.e., amplifications and deletions) was, on average, greater than twofold higher in BPM tumors than PM tumors (Fig. 5C).
Brca1 and Trp53 LOH in Esophageal Tumors Formed following 4NQO Exposure.
BRCA1 mutant cancers frequently reveal a loss of BRCA1 heterozygosity (LOH), largely arising from loss of the wild-type allele of BRCA1 and retention of the mutant allele (60–64). Recently, however, some reports have indicated that BRCA1 LOH might not be as common an event as previously suggested in Brca1 tumors (65). Studies where microdissected tumor cells were analyzed for BRCA1 LOH showed that BRCA1 LOH is a heterogeneous tumor-based outcome (66–68).
To address the extent of LOH of Brca1 and Trp53, we extracted genomic DNA from 4NQO-induced tumors and carried out qPCR. To enrich for tumor-associated DNA, tumor sections were microdissected in order to isolate tumor-enriched segments. Genomic DNA from tail cuts (TCs) of the corresponding mice was also extracted. DNA from the tumor (TU) and the corresponding TC was collected from n = 6 BPM, n = 6 PM, and n = 2 BM and WT mice. Actin primers were used to amplify the genomic actin locus DNA as an internal control.
Primers were designed such that they amplified regions within loxP site encompassed segments of Brca1 and Trp53. These primers were used to amplify a specific ∼100-bp region within the respective genes in both TCs and TU genomic DNA extracts from all samples. We chose this region of Brca1 and Trp53 because it allowed one to compare the gene dosage for Brca1 and Trp53 upon Cre-mediated deletion in the germline (i.e., tail cuts) with that in the tumors. The TCs served as positive controls to confirm loss of one allele of Brca1 and/or Trp53 upon action of Cre. The fold change in gene dosage (Brca1 and p53) was calculated by normalizing results to those obtained with WT tail cut genomic DNA.
As shown in Fig. 5D, qPCR-based analysis showed that BPM mice experienced a reduction in Brca1 gene dosage in the tumors compared with tail cuts, implying the loss of one or both Brca1 alleles in the tumors. No such additional loss of Brca1 or Trp53 was observed in the BM or the WT tumors compared with their tail cut genomic DNA. This could imply that the genomic instability in Brca1 heterozygous (BM) tumors is less than that observed in Brca1/Trp53 double het (BPM) mice, where there was additional loss (LOH) for both Brca1 and Trp53. This could also account for reduced tumor rate in BM mice compared with BPM.
Furthermore, we did detect a reduced Brca1 gene dosage in PM tumors compared with the corresponding PM tail cuts which, we believe, could be an indication of increased genomic instability in these tumors (Fig. 5D). Similarly, there was also p53 gene dosage loss in BPM compared with BM or WT tumors (Fig. 5E).
We also analyzed the WES sequencing data using GATK HaplotypeCaller (69) in genomic vcf mode from the Broad Institute and confirmed results generated through our qPCR analysis. There were two heterozygous single nucleotide polymorphisms (SNPs) (rs28288696 C/G and rs28288694 A/G) in these specimens that were located in Brca1, both of them in exon 10 (which is contained within the floxed region of Brca1) (Fig. 5F). Compared with their PM counterparts (S4 and S5), the BPM tumor samples (S1 and S2) show a strong reduction in coverage at both of these SNPs and the variant allele frequencies (VAFs) were near zero (Fig. 5F), indicating the presence of LOH in the region. In fact, in samples S1 and S2 the SNP rs28288694 completely disappeared, which strongly suggests the existence of LOH in this region. No such disappearance of SNPs or loss of allele frequency was observed in a neighboring gene, ETV4, where allele frequency for five SNPs was analyzed (Fig. 5F). This implies that the disappearance of the Brca1 region-specific SNP (rs28288694) in S1 and S2 cannot simply be attributed to a lack of sequencing depth. This is because the probability that the SNPs were present, with a contribution equal to that of the reference allele, but the sequencing did not capture them, was 7.6 × 10−6 and 9.5 × 10−7, respectively, assuming the diploidy of the two samples (Fig. 5F). These findings confirm that 1) in BPM tumors (S1 and S2 samples), only Brca1 heterozygous cells contributed to tumor formation and 2) that the loss of SNPs was specific to the Brca1 region.
Discussion
Germline heterozygous loss of BRCA1 is associated with an increased lifetime cancer risk and, especially, breast and ovarian cancer (1, 70). Increasing evidence suggests that BRCA1 loss is also associated with additional cancer types, including but not limited to pancreatic, prostate, colon, gastric, and esophageal cancer. Despite the well-established association between BRCA1 heterozygosity and cancer predisposition in humans, there are currently no such Brca1 heterozygous mouse models that faithfully recapitulate this high risk of tumor formation upon BRCA1 heterozygosity. Brca1 heterozygous mice are not tumor prone, and complete loss of Brca1 in mouse models by conditionally deleting it in the tissue of interest rarely triggers tumorigenesis, unless Trp53 is also deleted. This makes it difficult to use Brca1 mouse models to study the role of BRCA1 heterozygosity in tumor formation.
Here we report a heterozygous Brca1 mouse model that manifests a dramatic acceleration of disease and provides a potentially valuable tool to study the mechanism of BRCA1 mutant tumorigenesis. We find that BRCA1 heterozygosity in combination with the acute onset of chemically derived replication stress is a major BRCA1 tumor–driving process. We employed 4NQO, a well-established replication stress inducer, and found that exposure to this agent can elicit esophageal tumor formation in Brca1 heterozygous mice.
More specifically, oral delivery of 4NQO to doubly heterozygous Brca1/Trp53 (BPM) mice led to squamous cell carcinoma of the distal esophagus and forestomach, while no such 4NQO-promoted tumorigenesis developed in WT and PM mice. Thus, dual Brca1 and Trp53 heterozygosity combined with 4NQO replication stress represented an adequate tumorigenic force in the aforementioned animals (33). One interpretation of these results is that 4NQO provides an added source of replication stress that allows a subtumorigenic effect of this DNA damage-associated, BRCA1/p53 heterozygosity-driven process to 1) reach a tumor-inducing level and 2) to do so with remarkable speed.
The proposal of a link between achieving sufficient replication stress and Brca1 mutation–driven ESCC is supported by the high level expression of 53BP1 that was detected after persistent and direct exposure of esophageal/forestomach tissue to 4NQO. We also found that BPM-, but not PM-based tumors contained a high percentage of phosphorylated Chk1 (S345)-positive, RS-affected tumor cells, which is also in keeping with a role for chronic replication stress in BRCA1 mutant tumorigenesis and, possibly, in other cancers, as well (16, 18).
We also investigated the nature of mutational signatures in tumors formed in BPM and PM mice. Whole-exome sequencing data obtained from tumor and nontumor tissue genomic DNA showed that an SBS4 mutational signature was enriched in 4NQO- induced BPM tumors but not in PM tumors. This mutational signature is known to correlate with tobacco smoking and has also been associated with esophageal cancer (10). It is presumed to be an output of unresolved and unrepaired DNA adducts formed by tobacco exposure as well as in 4NQO-associated BPM tumors. Conceivably, if 4NQO leads to similar or related DNA adducts, when unrepaired, they could also give rise to a C > A mutational signature.
The fact that we observed a C > A mutational signature almost exclusively in BPM tumors and not in PM tumors strongly suggests that defective repair of 4NQO-induced lesions in Brca1/Trp53 double heterozygous cells contributed to tumor formation. However, why we did not detect a HRD-associated signature 3 in the BPM tumors is as yet unclear. This observation is similar to what has been reported previously for tumor lines derived from mice where both Brca1 alleles were conditionally deleted in the mammary gland. There too, only minor enrichment of signature 3 in the Brca1-deleted tumors was found (53–55).
A possible explanation for a lack of signature 3 in Brca1 mouse tumors is that the tumors grew fast enough through largely unhindered cell divisions, thereby preventing certain subclones from arising and surviving, and thereby contributing to the overall mutational signature of the tumor.
LOH analysis of the BPM tumor DNA did reveal a significant reduction in the Brca1 and Trp53 gene dosage compared with DNA from the tail cuts of the corresponding mice. Though significant, Brca1 LOH was not complete, implying the existence of intratumoral heterogeneity. This is in keeping with previous reports that have shown evidence that up to 25% of tumor nuclei retain positive BRCA1 staining in BRCA1 mutant human tumors (65, 68).
The BPM mouse model also serves as an excellent addition to existing esophageal squamous cell carcinoma mouse models. Distal esophageal and forestomach squamous cell cancers in mice share strong similarity with human ESCC. Previous attempts to model ESCC in mice have employed many different approaches, including genetically engineered mice [e.g., to inactivate p16 or p53 and/or to overexpress cyclin D1 (71)] or the use of carcinogens. It has been difficult to study ESCC in genetically engineered mice, partly because of its traditionally long latency, which ranges from 12 to 18 mo and normally appears to be necessary for tumor development. However, BPM tumors were detected between ∼90 and 120 d, representing a dramatic acceleration in tumorigenesis. Of note, this mouse model was generated in mice bearing a mixed genetic background (both C57BL/6 and FVB-N) (27) and the effect of genetic backgrounds on tumorigenesis could affect tumor latency in certain backgrounds.
The use of carcinogens, including 4NQO, has been helpful in replicating aspects of human ESCC in mice. For example, in the genetically engineered mice (Brca1/Trp53 double heterozygous mice) used in this study, ESCC was induced in a very short time (on average ∼90 to 120 d). By contrast, in 4NQO-free BPM mice, no esophageal tumors were detected within 18 mo, at which time observation was stopped. Moreover, the ∼90- to 120-d onset of tumor formation was directly dependent on both Brca1 heterozygosity and the induction of replication stress. Such a telescoped tumor model may offer an opportunity to gain a more rapid understanding of molecular events that lead to ESCC.
In conclusion, we have established a markedly accelerated mouse tumor model to study the effect of replication stress in BRCA1 heterozygous tissue and how it contributes to BRCA1 mutant tumorigenesis. Brca1 heterozygous cells are innately susceptible to accumulating exogenously delivered tumor-promoting replication stress (33), and we hypothesize that unrepaired replication stress effects result in an accumulation of genomic alterations that trigger the rapid onset of tumorigenesis in 4NQO-treated Brca1+/−;Trp53+/− mice. Replication stress in cases of esophageal cancer in BRCA1 mutation carriers can be generated by exogenous agents like tobacco and alcohol, both of which have been linked to esophageal cancer (72, 73). Although the role of endogenous replication stress in tumorigenesis is well established (74, 75), the nature of the endogenous replication stress-inducing agents that lead to BRCA1 mutant cancer is unclear, as is whether such agents contribute to the tissue specificity observed in human BRCA1 cancer (i.e., to the increased risk for breast and ovarian cancer). There is some indication that estrogen metabolites could be one such endogenous agent that causes increased replication stress in BRCA1 heterozygous cells (76). Clear identification of endogenous agents that contribute to increased replication stress in BRCA1 mutant cancer will be important in understanding the early steps that define BRCA1 mutant tumorigenesis.
Finally, although PM and BM mice also developed tumors, they did so at a much lower frequency than BPM mice. Moreover, only the BPM tumors revealed 1) an accumulation of 4NQO-induced C > A alterations and 2) evidence of increased Brca1 LOH. Both of these observations strongly suggest that the accelerated tumorigenesis observed in BPM mice is a direct consequence of 4NQO-induced replication stress coupled with Brca1 heterozygosity that, in turn, led to Brca1 LOH, an established Brca1 mutant cancer driver. Importantly, neither 4NQO-induced DNA damage nor Brca1 heterozygosity, on its own, was enough to drive tumorigenesis in this experimental setting.
That said, one might also consider the possibility that intraductal mammary 4NQO delivery could lead to Brca1 heterozygosity–driven mammary tumors. Such an accelerated tumor model system could prove to be invaluable in the detection of the earliest events in BRCA1 heterozygous mutation-driven breast cancer.
Materials and Methods
Cell Line and Culturing Condition.
The human esophageal squamous cell carcinoma KYSE410 cell line was provided by Adam J. Bass’ group at Columbia University, New York, NY. Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (GIBCO) and 1% penicillin/streptomycin (Thermo Fisher).
Mouse Models.
Brca1flox/wt (loxP sites flanking exons 5 to 13 of Brca1), and Trp53flox/wt (loxP sites flanking exons 1 to 10 of Trp53) mice were kindly provided by Jos Jonker’s group, Netherlands Cancer Institute, Amsterdam, The Netherlands. These mice were crossed with Meox2Cre deletor mice purchased from Jackson Labs (Stock No. 003755) to generate the four different genotypes (BPM, BM, PM, and WT). All experimental protocols were approved by the Dana-Farber Institutional Animal Care and Use Committee.
Experimental Conditions for 4NQO-Induced Tumorigenesis.
4NQO (Sigma-Aldrich, Cat. No. N8141) was dissolved in propylene glycol (Sigma-Aldrich, Cat. No. 398039-500ML) at 6 mg/mL and diluted in drinking water to a final concentration of 200 μg/mL. Mice were given access to the drinking water at all times during the treatment. Six- to eight-wk-old mice from different genotypes were cohoused such that same sex mice from different genotypes were in the same cage. The water was refilled with fresh 4NQO solution biweekly. Control groups received the same volume of propylene glycol (vehicle control) in their drinking water. After 75 d, 4NQO-containing water was replaced with regular drinking water, and all mice were monitored for 45 additional days and then killed for tissue/tumor collection.
Tissue Collection and Sectioning.
Whole stomach and distal esophagus from experimental mice, both tumor and normal, were harvested and fixed in 10% formalin. Macroscopic images of stomach and esophagus were recorded and photographed for each experimental mouse. The harvested tissue was embedded in paraffin wax, cut into 5-μm sections, and stained with H&E. Pathology processing and analyses were carried out at the Rodent Histopathology Core at Harvard Medical School.
Statistical Analyses.
The relationships between phenotype and outcome were assessed using Fisher’s exact test. More details are provided in SI Appendix.
Genomic DNA Extraction for LOH Analysis and WES.
Formalin-fixed paraffin-embedded tissue sections were deparaffinized with xylene, washed with ethanol, and rehydrated in deionized water. The tumor areas to be microdissected were identified on the unstained section by comparing the tissue with an adjacent section stained with H&E. Microdissection was performed under the microscope, using a sterile syringe needle and/or a scalpel blade. The isolated tissue was placed in a 0.5-mL PCR tube containing DNA extraction buffer. Genomic DNA from the microdissection sections was extracted using the QIAmp DNA FFPE tissue kit (Qiagen, 56404) according to the manufacturer’s instructions. Tail cut DNA was repurified with a DNeasy kit (Qiagen) and concentrated with a Microcon DNA Fast Flow Centrifugal Filter Unit with Ultracel membrane (Millipore, Cat. No. MRCF0R100).
Immunofluorescence and Immunohistochemistry.
Paraffin-embedded sections were deparaffinized in xylene and rehydrated by serial immersion in ethanol and ddH2O. Antigen retrieval was achieved by heating sections in a pressure cooker (Cuisinart CPC-600) for 12 min in pH 6 citrate buffer (Millipore-Sigma, Cat. No. C9999). Tissue sections were washed with phosphate buffered saline (PBS) twice for 10 min, and then blocked for 1 h in blocking buffer (PBS containing 1% bovine serum albumin [BSA], 0.3% Triton X-100, and 10% normal goat serum). Sections were incubated with primary antibody in antibody solution buffer (PBS containing 1% BSA, 0.3% Triton X-100, and 1% normal goat serum) overnight at 4 °C. For immunohistochemistry (IHC), a Novolink Polymer detection system (Leica) was used and counterstained with Gill’s Hematoxylin No. 3. For immunofluorescence (IF), tissue sections were subsequently incubated with Alexa Fluor 647 secondary antibody for 30 min at room temperature. Sections were mounted with VECTASHIELD antifade mounting medium containing DAPI (Vectorlabs H-1000). Primary antibodies used were: phospho-CHK1 Ser345 (Cell Signaling Technology, Cat. No. 2341T), 53BP1 (Bethyl Laboratories, Cat. No. A300-272A), and P63 (Santa Cruz, Cat. No. sc-25268).
Microscopy and Image Analysis.
Images were acquired with an Axio Imager.M2 (Carl Zeiss) equipped with an Axiocam 506 color camera, controlled by Zen software. Images were exported at 16-bit images for subsequent analysis. Image analysis was performed with FIJI and Matlab_9.7 R2019b. Pixel intensity analysis was determined by assessing the average pixel abundance per micrometer squared and averaged across images.
PCR Primers for Genotyping and LOH Analysis.
Details of PCR primers and the related methods are described in detail in SI Appendix.
Whole Exome Sequencing Data Analysis.
Alignment and postprocessing of the whole exomes, copy number analysis, somatic point mutation calling, somatic signature calling, indel analysis, coverage analysis, and variant allele frequencies in the direct vicinity of BRCA1 are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We express our sincerest thanks to Dr. Jos Jonkers of the Netherlands Cancer Institute for generously providing us with mice carrying floxed alleles of Brca1 and Trp53. This work was funded, in part, by grants from the NIH; The Gray Foundation; the Breast Cancer Research Foundation; the Susan G. Komen Foundation; the BRCA Foundation; Mr. Michael Robbins; Jeremy Maltby; Gia Lee; Bruce Rabb; and the Murray Winsten Foundation to D.M.L. It was also funded by University of Massachusetts Startup Funds and the Dana-Farber/Harvard Cancer Center (DF/HCC) Breast SPORE CEP to S.P.; the Breast Cancer Research Foundation (Grant ID: BCRF-20-159); Kræftens Bekæmpelse (Grant ID: R281-A16566); CDMRP Prostate Cancer Research Program (Grant W81XWH-18-2-00560); Det Frie Forskningsråd Sundhed og Sygdom (Grant ID: 7016-00345B); the Basser Foundation grant to Z.S.; and, in part, by the Dana-Farber/Harvard Cancer Center Kidney Cancer SPORE Grant P50-CA101942-12 (to S.S.).
Footnotes
Competing interest statement: D.M.L. is a scientific advisor to Constellation Pharma, a Science Partner of Nextech Ventures (Zurich, CH), a Science Advisory Board member of the Sidney Kimmel Cancer Center (Johns Hopkins School of Medicine), a Science Advisor to the Pezcoller Foundation (Trento, Italy), and a Special Advisor to the Director of Break Through Cancer, none of which poses a conflict of interest with the substance of this paper.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108421118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Antoniou A., et al., Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuchenbaecker K. B.et al.; BRCA1 and BRCA2 Cohort Consortium , Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Starr J., Ramnaraign B., Germline BRCA1 mutated esophageal squamous cell carcinoma. Rare Tumors 12, 2036361320972218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramnaraign B., Altshuler E., Complete clinical response of a patient with BRCA1-mutant cervical esophageal squamous cell carcinoma treated with oxaliplatin-based chemotherapy highlights the importance of performing genomic profiling in cancer treatment Curr. Prob. Cancer Case Rep. 3, 100069 (2021). [Google Scholar]
- 5.Mersch J., et al., Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 121, 269–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran A., et al., Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam. Cancer 11, 235–242 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Petty E. M., Kalikin L. M., Orringer M. B., Beer D. G., Distal chromosome 17q loss in Barrett’s esophageal and gastric cardia adenocarcinomas: Implications for tumorigenesis. Mol. Carcinog. 22, 222–228 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Dunn J., et al., Multiple target sites of allelic imbalance on chromosome 17 in Barrett’s oesophageal cancer. Oncogene 18, 987–993 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Mori T., et al., Frequent loss of heterozygosity in the region including BRCA1 on chromosome 17q in squamous cell carcinomas of the esophagus. Cancer Res. 54, 1638–1640 (1994). [PubMed] [Google Scholar]
- 10.Deng C.-X., Wang R.-H., Roles of BRCA1 in DNA damage repair: A link between development and cancer. Hum. Mol. Genet. 12, R113–R123 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Roy R., Chun J., Powell S. N., BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branzei D., Foiani M., The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17, 568–575 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Branzei D., Foiani M., The checkpoint response to replication stress. DNA Repair (Amst.) 8, 1038–1046 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Jones R. M., Petermann E., Replication fork dynamics and the DNA damage response. Biochem. J. 443, 13–26 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Osborn A. J., Elledge S. J., Zou L., Checking on the fork: The DNA-replication stress-response pathway. Trends Cell Biol. 12, 509–516 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Gorgoulis V. G., et al., Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Halazonetis T. D., Gorgoulis V. G., Bartek J., An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Negrini S., Gorgoulis V. G., Halazonetis T. D., Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Tan D. S. P., Kaye S. B., Chemotherapy for patients with BRCA1 and BRCA2-mutated ovarian cancer: Same or different? American Society of Clinical Oncology educational book. American Society of Clinical Oncology. Annual Meeting 10.14694/EdBook_AM.2015.35.114, 114–121 (2015). [DOI] [PubMed]
- 20.Yap T. A., Plummer R., Azad N. S., Helleday T., The DNA damaging revolution: PARP inhibitors and beyond. American Society of Clinical Oncology educational book. American Society of Clinical Oncology. Annual Meeting 39, 185–195 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Bryant H. E., et al., Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Farmer H., et al., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Kondrashova O.et al.; Australian Ovarian Cancer Study (AOCS) , Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 9, 3970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gowen L. C., Johnson B. L., Latour A. M., Sulik K. K., Koller B. H., Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 12, 191–194 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Dine J., Deng C.-X., Mouse models of BRCA1 and their application to breast cancer research. Cancer Metastasis Rev. 32, 25–37 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Kim S. S., et al., Hyperplasia and spontaneous tumor development in the gynecologic system in mice lacking the BRCA1-Delta11 isoform. Mol. Cell. Biol. 26, 6983–6992 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X., et al., Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12111–12116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig T., Fisher P., Ganesan S., Efstratiadis A., Tumorigenesis in mice carrying a truncating Brca1 mutation. Genes Dev. 15, 1188–1193 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachelier R., et al., Normal lymphocyte development and thymic lymphoma formation in Brca1 exon-11-deficient mice. Oncogene 22, 528–537 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Perets R., et al., Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell 24, 751–765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao L., et al., Absence of full-length Brca1 sensitizes mice to oxidative stress and carcinogen-induced tumorigenesis in the esophagus and forestomach. Carcinogenesis 28, 1401–1407 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Michalak E. M., Jonkers J., Studying therapy response and resistance in mouse models for BRCA1-deficient breast cancer. J. Mammary Gland Biol. Neoplasia 16, 41–50 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Pathania S., et al., BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat. Commun. 5, 5496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedic M., et al., Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat. Commun. 6, 7505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartkova J., et al., DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Tallquist M. D., Soriano P., Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26, 113–115 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Duan H., et al., E3 ligase RFWD3 is a novel modulator of stalled fork stability in BRCA2-deficient cells. J. Cell Biol. 219, e201908192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Downes D. J., et al., Characterization of the mutagenic spectrum of 4-nitroquinoline 1-oxide (4-NQO) in Aspergillus nidulans by whole genome sequencing. G3 (Bethesda) 4, 2483–2492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikenaga M., Ichikawa-Ryo H., Kondo S., The major cause of inactivation and mutation by 4-nitroquinoline 1-oxide in Escherichia coli: Excisable 4NQO-purine adducts. J. Mol. Biol. 92, 341–356 (1975). [DOI] [PubMed] [Google Scholar]
- 40.Maréchal A., Zou L., RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pathania S., et al., BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell 44, 235–251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang X. H., Knudsen B., Bemis D., Tickoo S., Gudas L. J., Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin. Cancer Res. 10, 301–313 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Sotiriou S. K., et al., Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol. Cell 64, 1127–1134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Her J., Ray C., Altshuler J., Zheng H., Bunting S. F., 53BP1 mediates ATR-Chk1 signaling and protects replication forks under conditions of replication stress. Mol. Cell. Biol. 38, e00472-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaillard H., García-Muse T., Aguilera A., Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Macheret M., Halazonetis T. D., DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 10, 425–448 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Schoop R. A. L., Noteborn M. H. M., Baatenburg de Jong R. J., A mouse model for oral squamous cell carcinoma. J. Mol. Histol. 40, 177–181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao X. L., et al., Cytokeratin-14 contributes to collective invasion of salivary adenoid cystic carcinoma. PLoS One 12, e0171341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinhardt H. C., Yaffe M. B., Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 21, 245–255 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DiMaio M. A., Kwok S., Montgomery K. D., Lowe A. W., Pai R. K., Immunohistochemical panel for distinguishing esophageal adenocarcinoma from squamous cell carcinoma: A combination of p63, cytokeratin 5/6, MUC5AC, and anterior gradient homolog 2 allows optimal subtyping. Hum. Pathol. 43, 1799–1807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barroso-Sousa R., et al., Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann. Oncol. 31, 387–394 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Davies H., et al., HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huo Y., et al., Genetic interactions among Brca1, Brca2, Palb2, and Trp53 in mammary tumor development. NPJ Breast Cancer 7, 45 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hámori L., et al., Establishment and characterization of a Brca1-/-, p53-/- mouse mammary tumor cell line. Int. J. Mol. Sci. 21, 1185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Póti Á., et al., Correlation of homologous recombination deficiency induced mutational signatures with sensitivity to PARP inhibitors and cytotoxic agents. Genome Biol. 20, 240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arima Y., et al., 4-Nitroquinoline 1-oxide forms 8-hydroxydeoxyguanosine in human fibroblasts through reactive oxygen species. Toxicol. Sci. 91, 382–392 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Nunoshiba T., Demple B., Potent intracellular oxidative stress exerted by the carcinogen 4-nitroquinoline-N-oxide. Cancer Res. 53, 3250–3252 (1993). [PubMed] [Google Scholar]
- 58.Viel A., et al., A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine 20, 39–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexandrov L. B.et al.; PCAWG Mutational Signatures Working Group; PCAWG Consortium , The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schildkraut J. M., et al., Loss of heterozygosity on chromosome 17q11-21 in cancers of women who have both breast and ovarian cancer. Am. J. Obstet. Gynecol. 172, 908–913 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Merajver S. D., et al., Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin. Cancer Res. 1, 539–544 (1995). [PubMed] [Google Scholar]
- 62.Cornelis R. S.et al.; The Breast Cancer Linkage Consortium , High allele loss rates at 17q12-q21 in breast and ovarian tumors from BRCAl-linked families. Genes Chromosomes Cancer 13, 203–210 (1995). [DOI] [PubMed] [Google Scholar]
- 63.Merajver S. D., et al., Somatic mutations in the BRCA1 gene in sporadic ovarian tumours. Nat. Genet. 9, 439–443 (1995). [DOI] [PubMed] [Google Scholar]
- 64.Neuhausen S. L., Marshall C. J., Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 54, 6069–6072 (1994). [PubMed] [Google Scholar]
- 65.Maxwell K. N., et al., BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 8, 319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.King T. A., et al., Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann. Surg. Oncol. 14, 2510–2518 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Clarke C. L., et al., Mapping loss of heterozygosity in normal human breast cells from BRCA1/2 carriers. Br. J. Cancer 95, 515–519 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martins F. C., et al., Evolutionary pathways in BRCA1-associated breast tumors. Cancer Discov. 2, 503–511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKenna A., et al., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mann G. J.et al.; Kathleen Cuningham Consortium for Research in Familial Breast Cancer , Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 8, R12 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishida K., et al., Current mouse models of oral squamous cell carcinoma: Genetic and chemically induced models. Oral Oncol. 73, 16–20 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Freedman N. D., et al., A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. Am. J. Epidemiol. 165, 1424–1433 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Uhlenhopp D. J., Then E. O., Sunkara T., Gaduputi V., Epidemiology of esophageal cancer: Update in global trends, etiology and risk factors. Clin. J. Gastroenterol. 13, 1010–1021 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Tubbs A., Nussenzweig A., Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeman M. K., Cimprich K. A., Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savage K. I., et al., BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 74, 2773–2784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.