Abstract
Objective
To investigate the potential correlation between the Arg188His (rs3218536) polymorphism of X-ray repair cross-complementing 2 (XRCC2) and colorectal cancer (CRC) risk, as the association remains unclear.
Methods
The CNKI, PubMed, EMBASE and Cochrane library databases were systematically searched for relevant studies published up to July 2021. Data were extracted from included studies, and analysed for pooled or subgroup odds ratios (ORs) with 95% confidence intervals (CIs) using STATA 12.0 software.
Results
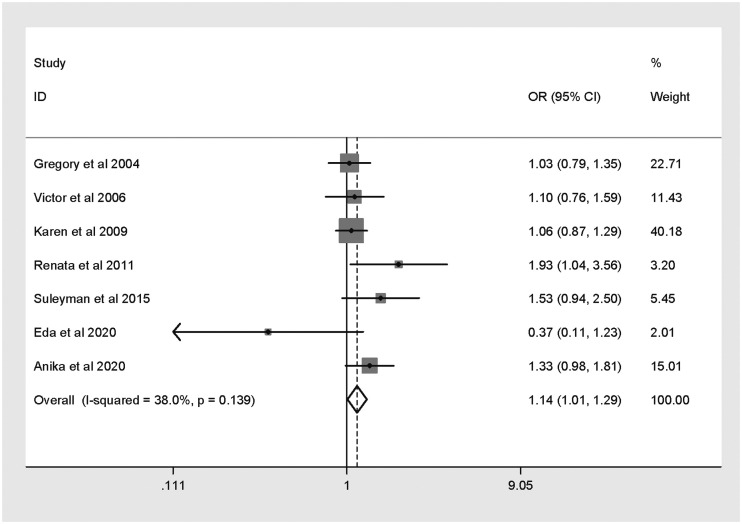
Seven published studies were included. Pooled analysis revealed that the XRCC2 Arg188His polymorphism was associated with increased CRC risk (His versus Arg: OR 1.14, 95% CI 1.01, 1.29). Trial Sequential Analysis to test the power of the results showed that they were unreliable and the meta-analysis required additional studies.
Conclusion
The current meta-analysis suggests that the XRCC2 Arg188His polymorphism may be a risk factor for CRC.
Keywords: XRCC2, cancer, meta-analysis, cancer risk, rs3218536, polymorphism
Introduction
Colorectal cancer (CRC) ranks third in prevalence among malignancies and fifth among frequently seen fatal cancers globally. 1 CRC is generally detected at a late stage, which, together with the high incidence, leads to high mortality rates. Although alcoholism, excess red meat uptake and smoking are recognized to be possible CRC risk factors, the precise aetiology of CRC remains unknown.2,3 Many patients with CRC are not characterised with the above carcinogenic risk factors, suggesting a critical involvement of genetic susceptibility in the CRC pathogenic mechanism. 4
Multiple mechanisms are involved in the repair of DNA double-strand breaks, but typically, homologous recombination repair (HRR) accounts for a critical pathway that exerts its function at S phase in the somatic mammalian cell cycle. 5 A sister chromatid is supplied as a template in HRR, with the alignment of homologous DNA sequence, and defects in HRR are associated with human malignant tumors. 6 Many critical molecules are found to be involved in the HRR process, 7 including various DNA repair protein RAD51 paralogs (X-ray repair cross-complementing [XRCC] protein 2 [XRCC2], XRCC3, RAD51 homolog 2 [RAD51B], RAD51 homolog 3 [RAD51C], and RAD51 homolog 4 [RAD51D]) that are thought to be pivotal proteins in HRR. 8
Located at chromosome 7q36.1, X-ray repair cross-complementing 2 (XRCC2) represents a potential functional gene related to tumour formation. 9 The XRCC2/3 proteins can interact with RAD51 and also stabilize the latter; in addition, they participate in the HRR of DNA double-strand breaks, as well as cross-link repair within mammalian cells. 10 Polymorphisms in the XRCC2 gene are found to relate to numerous malignant tumours, including thyroid carcinoma, and breast, gastric, and prostate cancer.11,12 An XRCC2 G-to-A polymorphism, localized within exon 3, results in histidine (His) being replaced by arginine (Arg) (Arg188His, single nucleotide polymorphism database [dbSNP] ID rs3218536). 11 This polymorphism has been widely investigated to explore its potential impact on cancer susceptibility. 13
Numerous epidemiological articles have evaluated the relationship between the XRCC2 Arg188His polymorphism and incidence of CRC, however, results are inconsistent between the studies. Meta-analysis is an efficient approach for detecting relationships that may not be revealed by small studies, and in particular, studies that evaluate polymorphisms with less common allele frequencies. Therefore, the present meta-analysis aimed to examine the relationship between the XRCC2 Arg188His polymorphism and CRC risk among relevant articles published to date.
Materials and methods
Study retrieval strategy
The meta-analysis was registered with PROSPERO (database ID: 266483). The CNKI, PubMed, EMBASE and Cochrane library databases were systemically searched for relevant articles published up to July 2021 (last search update, 11 July 2021) using the following keywords: ‘colorectal cancer’ or ‘cancer, neoplasm or tumor’ ‘X-ray repair cross-complementing group 2 or XRCC2’ and ‘polymorphism or variant’. The search was limited to English language papers. In addition, studies were identified by a manual search of references from the original studies. Articles were screened and assessed by two independent researchers (ZW and YW) on the basis of a standard protocol, and any discrepancies were resolved by discussion until a consensus was reached.
Study inclusion and exclusion criteria
The study inclusion criteria were as follows: (1) case–control studies including patients with colorectal cancer and normal subjects; (2) studies focussed on the relationship between Arg188His polymorphism and CRC risk; and (3) adequate available genotyping information. Study exclusion criteria comprised: (1) non-case–control studies evaluating the relationship between the Arg188His polymorphism and CRC incidence; (2) letters, case reports, editorial articles or reviews; and (3) articles with insufficient raw data or no available data. Articles published by identical research groups were examined for any overlap of study participants between publications. For studies with partial overlap, the latest publication was adopted.
Assessment of study quality
The methodological quality of each included article was assessed using the Newcastle-Ottawa quality assessment scale. 14 An ultimate score of 6 stars or more was regarded as a high-quality study.
Data extraction
Data were extracted from each included article by two independent reviewers (LA and CX). Any disagreement was resolved by mutual negotiation. Extracted data included first author, publication date, country, genotype frequency in cases and controls, case and control numbers, Hardy–Weinberg Equilibrium (HWE) evidence among controls and quality score.
Statistical analyses
Statistical analyses were performed with STATA 12 software (Stata statistical software release 12; StataCorp LP, College Station, TX, USA), and P <0.05 was considered to indicate statistical significance. The relationship between the XRCC2 Arg188His polymorphism and CRC incidence was estimated according to the pooled odds ratios (ORs) with 95% confidence intervals (CIs) for His versus Arg, the additive model (Arg/His versus Arg/Arg, His/His versus Arg/Arg), the recessive model (His/His versus Arg/His + Arg/Arg) and the dominant model (His/His + Arg/His versus Arg/Arg), respectively. In addition, the I2 statistic was used to investigate and measure heterogeneity, with I2 >50% indicating obvious heterogeneity. The random effects model was utilized for calculating pooled OR in the presence of heterogeneity; otherwise, the fixed effects model was used. Furthermore, result stability was examined through one-way sensitivity analysis, where studies were eliminated one at a time to examine the impact of each study on the combined OR. 15 To evaluate publication bias, funnel plots were analysed using Begg’s test and Egger’s test.
Trial sequential analysis
Trial sequential analysis (TSA) was performed using TSA software, version 0.9.5.10 (https://ctu.dk/tsa/), with the following parameters: type I error (5%), 95% CI, relative risk reduction (20%), and type II error (20%). Evidence was confirmed as reliable if the cumulative Z-curve surpassed the required information size (RIS) line or passed through the sequential monitoring boundary of the trial; otherwise, more articles were deemed to be required. 16
Results
Study characteristics
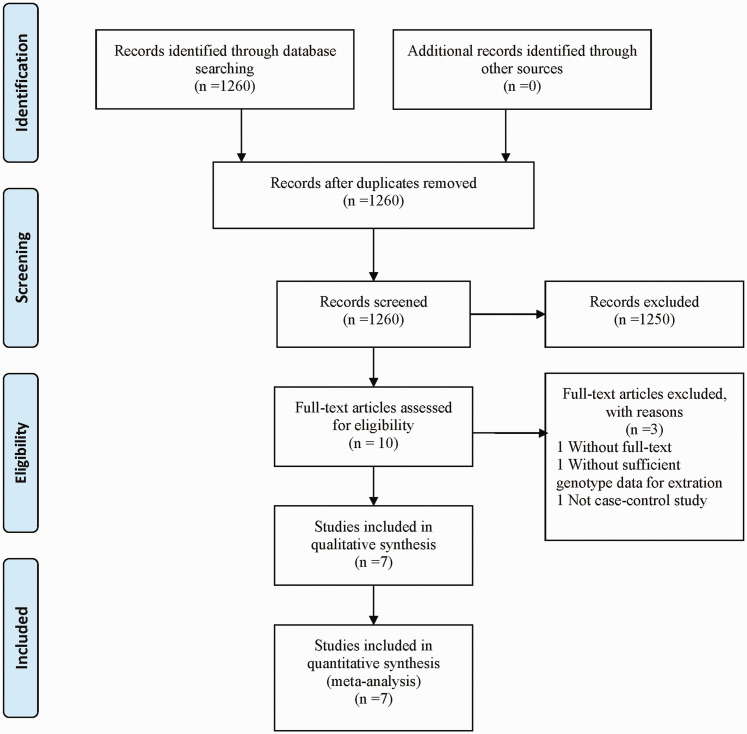
A total of 1260 articles reporting the association between the XRCC2 Arg188His polymorphism and CRC risk were identified through database searches (Figure 1). After removal of duplicates and assessment of articles for eligibility criteria, a total of seven case–control studies involving 2331 patients and 2830 normal subjects met the study criteria and were enrolled into the final analyses (summarised in Table 1).17–23 Controls were mostly derived from healthy people. Of the included articles, three studies comprised a sample size of more than 500 participants, while four studies comprised a sample size of less than 500 participants. Four of seven studies involved Caucasian study populations and three studies involved Asian participants. In addition, HWE tests to examine control genotype distribution revealed consistent HWEs, except for the studies by Moreno et al., 18 and Cetinkunar et al. 21 According to the Newcastle-Ottawa quality assessment scale for case–control studies, overall scores of the included studies ranged from six to eight stars, thus, all studies were defined as high-quality.
Figure 1.
Flowchart summarising selection of studies included in the meta-analysis.
Table 1.
Summary of study and population characteristics for studies included in the meta-analysis.
| Author, year | Country | Ethnicity | Cases | Controls | Case genotype |
Control genotype |
Case genotype |
Control genotype |
HWE P | NOS Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arg | His | Arg | His | Arg/Arg | Arg/His | His/His | Arg/Arg | Arg/His | His/His | |||||||
| Tranah, 2004 17 | USA | Caucasian | 376 | 725 | 656 (87) | 96 (13) | 1270 (88) | 180 (12) | 302 | 52 | 22 | 582 | 106 | 37 | 0.12 | 6 |
| Moreno, 2006 18 | Italy | Caucasian | 350 | 316 | 631 (90) | 69 (10) | 575 (91) | 57 (9) | 287 | 57 | 6 | 265 | 45 | 6 | 0.02 | 6 |
| Curtin, 2009 19 | USA | Caucasian | 1209 | 1380 | 2213 (92) | 205 (8) | 2538 (92) | 222 (8) | 1014 | 185 | 10 | 1167 | 204 | 9 | 0.98 | 7 |
| Krupa, 2011 20 | Poland | Caucasian | 100 | 100 | 168 (84) | 32 (16) | 182 (91) | 18 (9) | 75 | 18 | 7 | 84 | 14 | 2 | 0.14 | 8 |
| Cetinkunar, 2015 21 | Turkey | Asian | 71 | 86 | 94 (66) | 48 (34) | 129 (75) | 43 (25) | 32 | 30 | 9 | 54 | 21 | 11 | 0.00 | 6 |
| Balkan, 2020 22 | Turkey | Asian | 40 | 40 | 76 (95) | 4 (5) | 70 (88) | 10 (12) | 36 | 4 | 0 | 30 | 10 | 0 | 0.36 | 6 |
| Hridy, 2020 23 | Bangladesh | Asian | 185 | 183 | 233 (63) | 137 (37) | 254 (69) | 112 (31) | 63 | 107 | 15 | 83 | 88 | 12 | 0.07 | 7 |
Data presented as n or n (%) prevalence.
Arg, arginine; His, histidine; HWE, Hardy–Weinberg equilibrium; NOS, Newcastle-Ottawa quality assessment scale.
Pooled meta-analysis and subgroup analysis
Results of pooled and subgroup analyses are presented in Table 2. In pooled analysis (Figure 2), the XRCC2 Arg188His polymorphism was found to be significantly associated with CRC risk (His versus Arg: OR 1.14, 95% CI 1.01, 1.29). As confounders potentially affected the pooled analysis, subgroup analyses were also conducted. Subgroup analysis based on ethnicity showed no relationship between the XRCC2 Arg188His polymorphism and CRC risk among Asian or Caucasian populations (Table 2). In addition, the polymorphism was not shown to be related to CRC risk in studies stratified by HWE. Sensitivity analysis showed that no study impacted the combined OR, indicating that the results were stable (data not shown).
Table 2.
Pooled and subgroup analyses of the association between the XRCC2 Arg188His polymorphism and colorectal cancer risk.
| Study n | His versus Arg | His/His versus Arg/Arg | Arg/His versus Arg/Arg | Dominant model | Recessive model | |
|---|---|---|---|---|---|---|
| OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | ||
| Total | 7 | 1.14 (1.01, 1.29) | 1.34 (0.95, 1.89) | 1.19 (0.92, 1.54) | 1.15 (0.99, 1.32) | 1.22 (0.87, 1.71) |
| Ethnicity | ||||||
| Asian | 3 | 1.19 (0.73, 1.93) | 1.53 (0.81, 2.88) | 1.30 (0.58, 2.94) | 1.27 (0.62, 2.60) | 1.14 (0.62, 2.08) |
| Caucasian | 4 | 1.09 (0.95, 1.26) | 1.26 (0.84, 1.91) | 1.06 (0.89, 1.25) | 1.08 (0.92, 1.26) | 1.26 (0.83, 1.89) |
| HWE | ||||||
| Yes | 5 | 1.14 (0.91, 1.44) | 1.39 (0.94, 2.05) | 1.08 (0.92, 1.27) | 1.15 (0.86, 1.52) | 1.31 (0.89, 1.92) |
| No | 2 | 1.13 (0.90, 1.42) | 1.16 (0.55, 2.45) | 1.58 (0.79, 3.19) | 1.43 (0.83, 2.44) | 0.95 (0.46, 1.97) |
His, histidine; Arg, arginine; OR, odds ratio; CI, confidence interval; HWE, Hardy–Weinberg equilibrium.
Figure 2.
Forest plot showing meta-analysis of the association between the X-ray repair cross-complementing 2 (XRCC2) gene Arg188His polymorphism and colorectal cancer risk for histidine versus arginine.
Publication bias
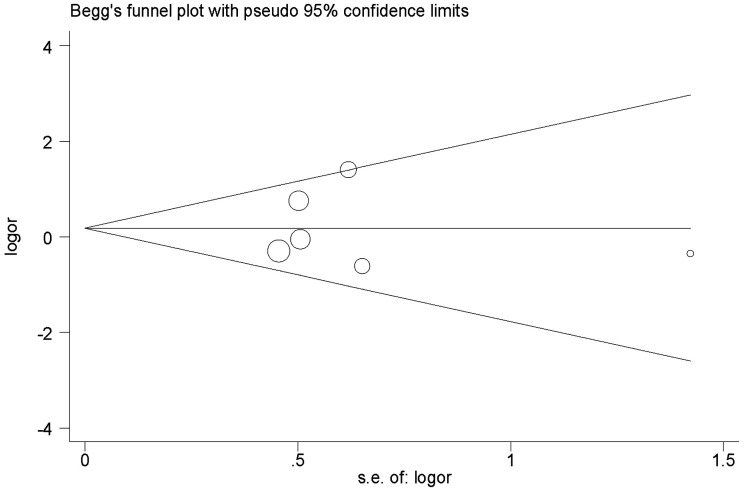
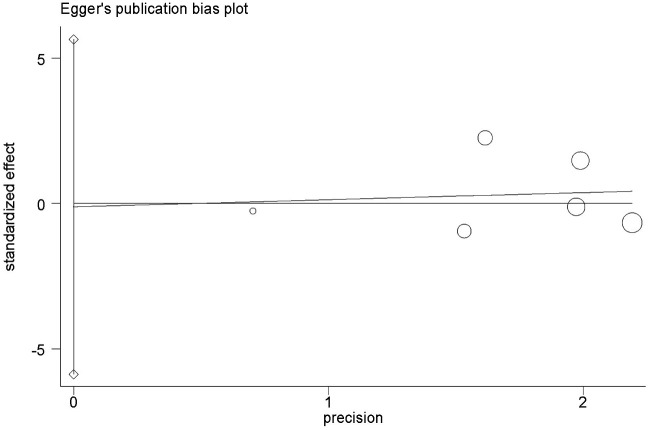
No obvious evidence of publication bias was revealed using Begg's test (P = 1.0; Figure 3) or Egger’s test (P = 0.96; Figure 4).
Figure 3.
Begg’s funnel plot analysis of bias in publications reporting the X-ray repair cross-complementing 2 (XRCC2) gene Arg188His polymorphism for histidine versus arginine. OR, odds ratio; SE, standard error.
Figure 4.
Egger’s funnel plot analysis of bias in publications reporting the X-ray repair cross-complementing 2 (XRCC2) gene Arg188His polymorphism for histidine versus arginine.
TSA results
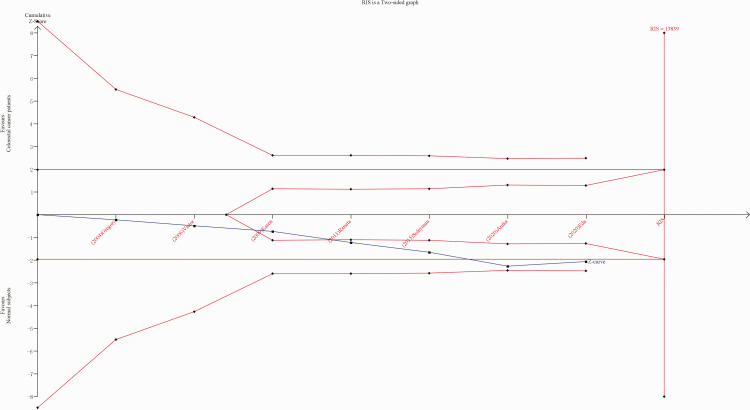
Trial sequential analysis, conducted to diminish the random errors and fortify the robustness of the present results, showed that the cumulative z-curve did not surpass RIS, and TSA and RIS thresholds were not crossed (Figure 5). This indicated unreliable results, and the need for more studies to be included.
Figure 5.
Trial sequential analysis (TSA) of associations between the X-ray repair cross-complementing 2 (XRCC2) gene Arg188His polymorphism and the risk of colorectal cancer for histidine versus arginine. Red broken line, TSA boundary value; blue broken line, cumulative Z curve; traditional boundary value line parallel to the horizontal axis, Z = 1.96; and the rightmost vertical line, RIS line. RIS, required information size.
Discussion
The most recent previously published meta-analysis on the association between the XRCC2 gene Arg188His polymorphism and CRC risk was by Zhang et al., 2014. 24 Their meta-analysis included two studies comprising a total of 1309 cases and 1480 controls, for Arg188His, and their pooled OR suggested a null association between Arg188His and CRC (OR for Arg versus His: 1.33, 95% CI 0.75, 2.34). The present study, with pooled analysis of a larger sample size, found a significantly increased CRC risk associated with the Arg188His polymorphism, which was not consistent with the previous meta-analysis. Similar to other disorders, CRC is induced by the complex associations between certain genes, and between genes and the environment. 23 Such associations should be further investigated in future studies.
The present pooled meta-analysis combining seven published studies found that the XRCC2 gene Arg188His polymorphism may result in an increased risk of CRC susceptibility. Considering the possible effects of potential confounders, subgroup analyses were also performed. In studies grouped according to HWE, the XRCC2 gene Arg188His polymorphism was not found to be related to CRC risk. In addition, in studies grouped according to population ethnicity, no correlation between the polymorphism and CRC risk was found in either Caucasian or Asian populations. TSA was used to check the reliability of conclusions in the present study. The cumulative Z-curve of the Arg188His polymorphism failed to reach the trial sequential monitoring boundary and RIS line, suggesting that additional large-sample, multi-ethnic studies are required to verify any associations.
Certain limitations should be noted in the present meta-analysis. First, no predefined uniform standard was used to select control subjects. Control cases were mainly derived from normal subjects, but some were patients. Secondly, only studies published in English were included, so relevant articles published in other languages were possibly omitted, and this may have resulted in the relatively small sample size. Thirdly, the present meta-analysis enrolled published articles only, while some related articles may remain unpublished, possibly resulting in publication bias. Lastly, this study did not investigate the influences of interactions between genes, and between genes and the environment.
In summary, through analysis of pooled data, the XRCC2 gene Arg188His polymorphism was found to be associated with CRC incidence. As few articles and restricted evidence are available, additional large studies with high methodological quality should be included in future meta-analyses to validate the association between the XRCC2 gene Arg188His polymorphism and CRC risk.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD: Shenyong Su https://orcid.org/0000-0001-7985-8998
References
- 1.Liang PS Chen TY andGiovannucci E.. Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis. Int J Cancer 2009; 124: 2406–2415. [DOI] [PubMed] [Google Scholar]
- 2.Chan DS, Lau R, Aune D, et al. Red and processed meat and colorectal cancer incidence: meta-analysis of prospective studies. PLoS One 2011; 6: e20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner H Kloor M andPox CP.. Colorectal cancer. Lancet 2014; 383: 1490–1502. [DOI] [PubMed] [Google Scholar]
- 4.Thacker J andZdzienicka MZ.. The XRCC genes: expanding roles in DNA double-strand break repair. DNA Repair (Amst) 2004; 3: 1081–1090. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C Kinzler KW andVogelstein B.. Genetic instabilities in human cancers. Nature 1998; 396: 643–649. [DOI] [PubMed] [Google Scholar]
- 6.Pires E Sung P andWiese C.. Role of RAD51AP1 in homologous recombination DNA repair and carcinogenesis. DNA Repair (Amst) 2017; 59: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerbinskaite A, Mukhopadhyay A, Plummer ER, et al. Defective homologous recombination in human cancers. Cancer Treat Rev 2012; 38: 89–100. [DOI] [PubMed] [Google Scholar]
- 8.Suwaki N Klare K andTarsounas M.. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol 2011; 22: 898–905. [DOI] [PubMed] [Google Scholar]
- 9.Zhan P, Wang Q, Qian Q, et al. XRCC3 Thr241Met gene polymorphisms and lung cancer risk: a meta-analysis. J Exp Clin Cancer Res 2013; 32: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan L, Li Q, Li X, et al. Association studies between XRCC1, XRCC2, XRCC3 polymorphisms and differentiated thyroid carcinoma. Cell Physiol Biochem 2016; 38: 1075–1084. [DOI] [PubMed] [Google Scholar]
- 11.Yu KD, Chen AX, Qiu LX, et al. XRCC2 Arg188His polymorphism is not directly associated with breast cancer risk: evidence from 37,369 subjects. Breast Cancer Res Treat 2010; 123: 219–225. [DOI] [PubMed] [Google Scholar]
- 12.Lin WY, Camp NJ, Cannon-Albright LA, et al. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J Med Genet 2011; 48: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romanowicz-Makowska H, Smolarz B, Zadrozny M, et al. The association between polymorphisms of the RAD51-G135C, XRCC2-Arg188His and XRCC3-Thr241Met genes and clinico-pathologic features in breast cancer in Poland. Eur J Gynaecol Oncol 2012; 33: 145–150. [PubMed] [Google Scholar]
- 14.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010; 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Hu Y, Zhang M, et al. Polymorphisms in ERCC2 and ERCC5 and risk of prostate cancer: a meta-analysis and systematic review. J Cancer 2018; 9: 2786–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wetterslev J Jakobsen JC andGluud C.. Trial sequential analysis in systematic reviews with meta-analysis. BMC Med Res Methodol 2017; 17: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tranah GJ, Giovannucci E, Ma J, et al. XRCC2 and XRCC3 polymorphisms are not associated with risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev 2004; 13: 1090–1091. [PubMed] [Google Scholar]
- 18.Moreno V, Gemignani F, Landi S, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 2006; 12: 2101–2108. [DOI] [PubMed] [Google Scholar]
- 19.Curtin K, Lin WY, George R, et al. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol Biomarkers Prev 2009; 18: 2476–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krupa R, Sliwinski T, Wisniewska-Jarosinska M, et al. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer-a case control study. Mol Biol Rep 2011; 38: 2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cetinkunar S, Gok I, Celep RB, et al. The effect of polymorphism in DNA repair genes RAD51 and XRCC2 in colorectal cancer in Turkish population. Int J Clin Exp Med 2015; 8: 2649–2655. [PMC free article] [PubMed] [Google Scholar]
- 22.Balkan E, Bilici M, Gundogdu B, et al. ERCC2 Lys751Gln rs13181 and XRCC2 Arg188His rs3218536 gene polymorphisms contribute to subsceptibility of colon, gastric, HCC, lung and prostate cancer. J BUON 2020; 25: 574–581. [PubMed] [Google Scholar]
- 23.Hridy AU, Shabnaz S, Asaduzzaman MD, et al. Genetic variations of RAD51 and XRCC2 genes increase the risk of colorectal cancer in Bangladeshi population. Asian Pac J Cancer Prev 2020; 21: 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang H, Peng Y, et al. The Arg188His polymorphism in the XRCC2 gene and the risk of cancer. Tumour Biol 2014; 35: 3541–3549. [DOI] [PubMed] [Google Scholar]