Abstract
Circular RNAs with covalently linked ends are generated from many eukaryotic protein-coding genes when the pre-mRNA splicing machinery backsplices. These mature transcripts are resistant to digestion by exonucleases and typically have much longer half-lives than their associated linear mRNAs. Circular RNAs thus have great promise as sensitive biomarkers, including for detection of transcriptional activity. Here, we show that circular RNAs can serve as markers of readthrough transcription events in Drosophila and human cells, thereby revealing mechanistic insights into RNA polymerase II transcription termination as well as pre-mRNA 3′ end processing. We describe methods that take advantage of plasmids that generate a circular RNA when an upstream polyadenylation signal fails to be used and/or RNA polymerase II fails to terminate. As a proof-of-principle, we show that RNAi-mediated depletion of well-established transcription termination factors, including the RNA endonuclease Cpsf73, results in increased circular RNA output from these plasmids in Drosophila and human cells. This method is generalizable as a circular RNA can be easily encoded downstream of any genomic region of interest. Circular RNA biomarkers, therefore, have great promise for identifying novel cellular factors and conditions that impact transcription termination processes.
Keywords: backsplicing, circRNA, Cpsf73, pre-mRNA 3′ end processing, RNAi screening, transcription termination
1. Introduction
In recent years, it has become increasingly clear that pre-mRNAs from many eukaryotic genes can be alternatively spliced to yield circular RNAs [reviewed in 1, 2–4]. Such transcripts have covalently linked ends and are produced when the pre-mRNA splicing machinery “backsplices” and joins a splice donor (5′ splice site) to an upstream splice acceptor (3′ splice site). Most endogenous circular RNAs are rarely generated and are expressed at low levels [5]. Nonetheless, these transcripts are naturally resistant to digestion by exonucleases and typically have longer half-lives (e.g. >10-fold longer) than their associated linear mRNAs [6]. This enables some circular RNAs to accumulate to high levels [7–12] and have physiological functions, including modulating microRNA activity or innate immunity [9, 13–16]). The unusual stability of circular RNAs further endows this class of transcripts with promising biotechnological characteristics, including as sensitive biomarkers. Here, we build upon recent work [17] and describe detailed methods for how circular RNAs can be engineered to serve as markers of readthrough transcription events in Drosophila and human cells. In particular, we show that many factors that control RNA polymerase II termination/pre-mRNA 3′ end processing can be identified when RNAi screening is combined with plasmids that produce a circular RNA when an upstream polyadenylation signal fails to be used.
Transcription termination by RNA polymerase II is tightly linked with pre-mRNA 3′ end processing in eukaryotes [reviewed in 18, 19]. To generate the mature 3′ end of a typical eukaryotic mRNA, only two enzymatic activities, cleavage and polyadenylation, are required. These activities are carried out by an RNA endonuclease (Cpsf73 in Drosophila, CPSF3 in humans) and a poly(A) polymerase (hrg in Drosophila, PAPOLA in humans), respectively. A large complex is nonetheless assembled on the pre-mRNA, which includes several multisubunit sub-complexes: cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor Im (CF Im), cleavage factor IIm (CF IIm), as well as many (>50) other factors that may mediate crosstalk with other cellular processes [20]. This complexity enables tight control over polyadenylation signal usage. In fact, alternative polyadenylation events are widespread across eukaryotes and known to be globally regulated by cell proliferation, differentiation, stress, and extracellular cues [reviewed in 21, 22]. For example, proximal polyadenylation signals tend to be used in cancer cells, resulting in production of mRNAs with short 3′ untranslated regions (UTRs) that escape microRNA-mediated repression and promote oncogenic transformation [23, 24]. In contrast, there is progressive lengthening of 3′ UTRs during embryonic development due to the use of more distal polyadenylation signals [25].
Upon reaching a polyadenylation signal, RNA polymerase II sometimes precisely terminates because traversing this sequence (typically composed of the hexanucleotide motif AAUAAA along with a downstream G/U-rich motif) can slow or pause transcription elongation [26, 27]. However, RNA polymerase II often continues transcribing. Termination of transcription thus must occur further downstream, and this can sometimes be 10 kb or more downstream of the polyadenylation signal [28, 29]. Some of the underlying mechanistic details of the termination process still await discovery, but it is thought that cleavage of the nascent pre-mRNA by the Cpsf73 endonuclease provides an entry site for a 5′−3′ exonuclease (Rat1 in Drosophila, XRN2 in humans) [30–32]. This exonuclease then digests the downstream RNA in order to catch up to the elongation complex and cause it to terminate.
Recent work has revealed that osmotic stress [33, 34] and some viral infections [35] inhibit the usage of many polyadenylation signals, resulting in widespread transcriptional readthrough and production of transcripts with very long 3′ UTRs (often >45 kb). To better understand how termination mechanisms are regulated and impacted by cellular conditions, there is a pressing need for novel methods that can easily and accurately quantify the relative amounts of properly terminated vs. readthrough transcripts produced from a given gene. This has most commonly been done by quantifying transcripts downstream of the polyadenylation signal [33, 34], but this can be difficult using standard approaches that measure steady-state RNA levels (e.g. RT-qPCR, Northern blots, or RNA-seq) as these downstream RNAs are typically of variable sizes and transient in nature. Another option has thus been to use gain-of-function reporter genes in which a fluorescent protein (e.g. GFP) [36, 37] or a selectable marker [38] is expressed when an upstream polyadenylation signal fails to be used. To further improve this gain-of-function approach, we reasoned that the unusual stability of circular RNAs could be harnessed to allow these transcripts to serve as biomarkers of specific readthrough transcription events, especially considering that some endogenous circular RNAs (e.g. from the human PAIP2 gene) are naturally generated from readthrough transcripts [17]. Here, we describe methods and proof-of-principle experiments that demonstrate circular RNAs derived from the Drosophila laccase2 (straw) gene can indeed be encoded downstream of a polyadenylation signal and used in RNAi screening efforts to identify factors that control transcription termination/pre-mRNA 3′ end processing.
2. Materials and methods
2.1. Reporter plasmids
For experiments in Drosophila cells, the previously described Hy_pMT Laccase2 Exons 1–3 plasmid (Addgene 91799) was used [17] (Fig. 1). For experiments in human cells, the pCRII-TOPO CMV-cGFP-SV40 pA + Laccase2 Exon 2 plasmid (Addgene 91802) was generated by inserting Drosophila laccase2 exon 2 along with its flanking intronic sequences downstream of the SV40 polyadenylation signal (between BlpI and AflII) of the previously described pCRII-TOPO CMV-cGFP-SV40 Poly(A) Sense expression plasmid (Addgene 46836) [39] (Fig. 3).
Fig. 1. A circular RNA-based reporter plasmid for studying readthrough transcription.
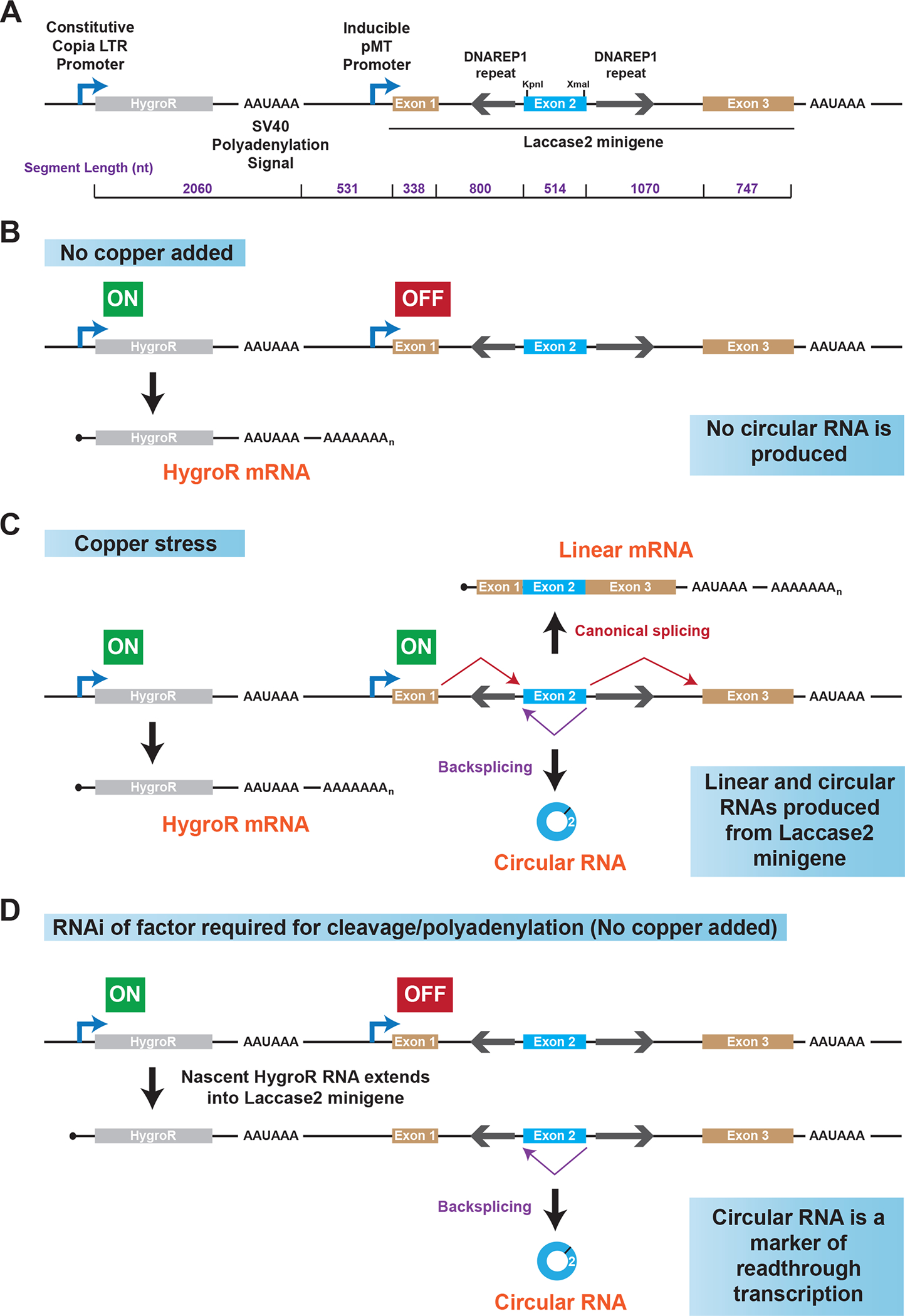
(A) Schematic of the Hy_pMT Laccase2 Exons 1–3 plasmid. The length of each indicated region is denoted in purple. (B) When Drosophila cells are grown under standard conditions (no exogenous copper added to the cell culture medium), the Copia LTR promoter is active, resulting in production of mature (capped and polyadenylated) HygroR mRNAs. The pMT promoter is off and no transcripts are produced from the laccase2 minigene. (C) When exogenous copper is added, the inducible pMT promoter becomes activated and the transcribed pre-mRNAs can be alternatively spliced to yield a three-exon linear mRNA or a circular RNA from exon 2 from the laccase2 minigene. (D) If the SV40 polyadenylation signal located downstream of the HygroR open reading frame fails to be used, there is readthrough transcription into the downstream laccase2 minigene. This can result in production of the circular RNA even though no exogenous copper has been added to cells.
Fig. 3. Identification of factors required for transcription termination in human cells.
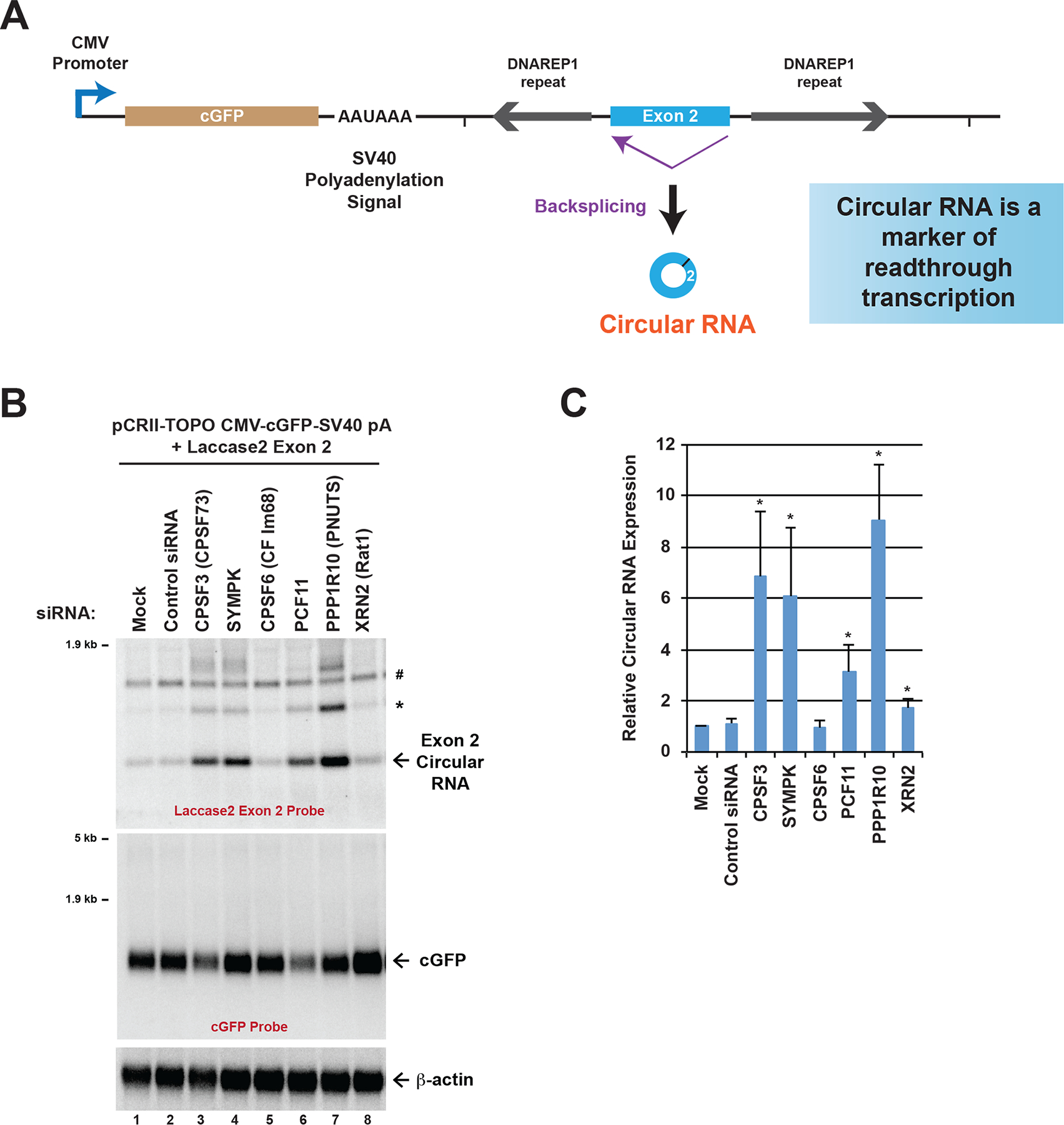
(A) Schematic of the pCRII-TOPO CMV-cGFP-SV40 pA + Laccase2 Exon 2 expression plasmid. Readthrough of the SV40 poly(A) signal can result in the production of the laccase2 circular RNA. (B) HeLa cells were transfected with the indicated siRNAs followed by plasmid transfection and total RNA isolation (48 h after siRNA transfection, 24 h after plasmid transfection). Northern blots were used to examine expression of the laccase2 circular RNA and the mature cGFP mRNA. *Concatenated and/or intertwined circular RNA. #Cross-reactive band that was observed when no plasmid was transfected. (C) Laccase2 circular RNA levels were quantified using ImageQuant from four independent Northern blot experiments and were normalized to the Mock samples. Data are shown as mean ± SD. * p<0.05.
2.2. Drosophila cell culture
Drosophila DL1 cells were grown at 25°C in Schneider’s Drosophila medium (Thermo Fisher Scientific 21720024) supplemented with 1% penicillin-streptomycin (Thermo Fisher Scientific 15140122), L-glutamine (Thermo Fisher Scientific 35050061), and 10% fetal bovine serum (Cytiva SH30396.03). To generate the DL1 cell line stably maintaining the Hy_pMT Laccase2 Exons 1–3 plasmid, 2 × 106 cells were plated in complete media in 6-well dishes and 2 μg of plasmid was transfected using Effectene (Qiagen 301427). Beginning the following day, cells were selected and maintained in complete medium containing 150 μg/mL hygromycin B.
2.3. Drosophila RNAi
Double-stranded RNAs (dsRNAs) from the DRSC (Drosophila RNAi Screening Center) were generated by in vitro transcription (MEGAscript kit, Thermo Fisher Scientific AMB13345) of PCR templates containing the T7 promoter sequence on both ends [40]. Primer sequences are provided in Table S1. Transcription reactions were incubated at 37°C for 8 h followed by digestion with 1 μL DNase I at 37°C for 1 h. dsRNAs were purified using the RNeasy Mini Kit (Qiagen 74106) following the manufacturer’s RNA cleanup protocol and quantified using a spectrophotometer (1 OD260 Unit = 45 μg/mL).
DL1 cells stably maintaining the Hy_pMT Laccase2 Exons 1–3 plasmid were grown in complete medium until confluent. Cells were dislodged from the flask and counted using a hemocytometer. Knockdown experiments were then set up in 12-well dishes by adding 2 μg of dsRNA to 500,000 cells in 0.5 mL Schneider’s Drosophila medium without penicillin-streptomycin, L-glutamine, or fetal bovine serum [see 40 for a detailed protocol]. After incubation at 25°C for 45–60 min, 1 mL of complete Drosophila medium was added. Cells were then incubated for 3 days and, where indicated, a final concentration of 500 μM copper sulfate was added for the final 14 h to induce transcription from the Metallothionein A promoter (pMT). Total RNA was isolated using TRIzol (Thermo Fisher Scientific 15596018) as per the manufacturer’s instructions.
2.4. Mammalian cell culture and transfections
HeLa cells were grown at 37°C, 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing high glucose (Thermo Fisher Scientific 11995065), supplemented with penicillin-streptomycin (Thermo Fisher Scientific 15140122) and 10% fetal bovine serum (Cytiva SH30396.03). Lipofectamine RNAiMAX (Thermo Fisher Scientific 13778150) and Lipofectamine 2000 (Thermo Fisher Scientific 11668019) were used to transfect siRNAs and plasmids, respectively, as per the manufacturer’s instructions. HeLa cells were plated in 6-well dishes and first transfected with siRNAs (30 pmol/well). siRNA sequences are provided in Table S2. On the following day, cells were transfected with the indicated expression plasmid (1 μg/well). 48 h after siRNA transfection (24 h after plasmid transfection), total RNA was isolated using TRIzol as per the manufacturer’s instructions.
2.5. Northern blotting
Northern blots using oligonucleotide probes and NorthernMax reagents (Thermo Fisher Scientific) were performed as previously described in detail [41]. Probes used for Drosophila cell experiments: Laccase2 Exon 2 XmaI (5′-GCTGAGCTCCCCGGG-3′), Drosophila β-actin (5′-AGCACAGTGTTGGCGTACAG-3′), and HygroR (5′-GACATATCCACGCCCTCCTA-3′). Probes used for human cell experiments: Laccase2 Exon 2 (5′-GCTAGGATTGAGGATGGAGCTCC-3′), cGFP (5′-TCCATGCCGTGGGTGATGCC-3′), and human β-actin (5′-AGCACTGTGTTGGCGTACAG-3′). All blots were viewed with a Typhoon 9500 scanner (GE Healthcare) and quantified using ImageQuant.
3. Results and Discussion
3.1. Design of a laccase2 expression plasmid that can be used to identify regulators of readthrough transcription in Drosophila cells
The Drosophila laccase2 (straw) gene generates a 490-nt circular RNA from exon 2 that accumulates to high levels in vivo in the nervous system as well as in commonly used Drosophila cell culture lines, including S2 and DL1 [7, 42, 43]. To identify cis-acting regulatory sequences and trans-acting factors that control laccase2 circular RNA levels, we previously generated a plasmid (Hy_pMT Laccase2 Exons 1–3) consisting of a three-exon laccase2 minigene as well as a hygromycin resistance (HygroR) expression cassette (Fig. 1) [17]. The HygroR gene is constitutively transcribed, while the laccase2 minigene is under control of the copper-inducible Metallothionein A promoter (pMT) (Fig. 1A) [44]. Thus, when Drosophila DL1 cells stably maintaining the plasmid (due to selection with hygromycin) are grown under basal conditions, little transcription of the laccase2 minigene is observed (Fig. 1B; Fig. 2A, Lane 1). Upon addition of exogenous copper to the cell culture medium, the pMT promoter is induced, allowing the laccase2 minigene to produce a three-exon linear mRNA as well as a circular RNA from exon 2 (Fig. 1C), and both of these transcripts are easily detectable by Northern blotting (Fig. 2A, Lane 2). It should be noted that KpnI and XmaI restriction sites were inserted into exon 2 (Fig. 1A) so that minigene-derived transcripts could be distinguished from endogenous laccase2 transcripts. Using mutagenesis of the plasmid, we previously showed that the complementary DNAREP1_DM transposable elements in the flanking introns help drive laccase2 backsplicing (Fig. 1A) [17, 43]. We further demonstrated that the efficiency of the backsplicing reaction is tuned by a number of RNA binding proteins, including the SR protein SF2, as well as the levels of core spliceosome components.
Fig. 2. Use of a circular RNA-based reporter to identify factors required for transcription termination in Drosophila cells.

(A) A Drosophila DL1 cell line stably maintaining the Hy_pMT Laccase2 Exons 1–3 plasmid was treated with the indicated dsRNAs for 3 d and, where noted, CuSO4 was added for the last 14 h. Components of the CPSF, CstF, CF Im, and CF IIm complexes are indicated. Total RNA was isolated and Northern blots were used to examine expression of transcripts derived from the laccase2 minigene. β-Actin was used as a loading control. Representative blots are shown. (B) Circular RNA levels were quantified using ImageQuant from three independent experiments and were normalized to the “β-gal, No copper” samples. Data are shown as mean ± SD. ** p<0.01, * p<0.05. (C) Northern blots were used to examine expression of HygroR transcripts when no CuSO4 was added. (D) Summary of factors required for transcription termination of the HygroR mRNA [Model based on 52]. Depletion of shaded factors caused downstream circular RNA levels to increase, whereas depletion of unshaded factors had no or minimal effect.
Besides being a useful tool for defining how splicing decisions are controlled at the Drosophila laccasse2 locus, this plasmid can be used to study transcription termination mechanisms when cells are grown in the absence of exogenous copper [17]. This is because the HygroR expression cassette is encoded upstream of the laccase2 minigene (Fig. 1). Transcription from the constitutive Copia LTR promoter (Fig. 1A) typically does not significantly extend into the laccase2 minigene due to the presence of the SV40 polyadenylation signal. However, if this polyadenylation signal fails to be used and/or RNA polymerase II fails to efficiently terminate, the nascent HygroR transcript extends further downstream (Fig. 1D). A nascent HygroR-laccase2 fusion transcript is produced that can be subjected to backsplicing to generate the laccase2 circular RNA. Circular RNA biogenesis has thus become independent of the pMT promoter, thereby allowing the mature circular RNA to be observed even when exogenous copper (which activates the pMT promoter) has not been added to the cell culture medium. Importantly, readthrough transcription from the HygroR cassette should not induce the expression of the ~2-kb canonically spliced (exon 1-2-3) laccase2 reporter linear mRNA, as these exons will instead be part of longer HygroR-laccase2 fusion transcripts (Fig. 1D; transcripts labeled HygroR-Laccase2 Fusion in Fig. 2).
By quantifying the expression of transcripts derived from the HygroR and laccase2 minigenes, one thus can easily distinguish factors that regulate HygroR transcription termination from factors that affect pMT transcription, backsplicing, or otherwise non-specifically alter the local chromatin environment. In short, depletion of a gene required for HygroR transcription termination should cause all of the following expression changes when cells are grown in the absence of exogenous copper: (i) extension of the HygroR mRNA into the downstream laccase2 minigene, (ii) production of the laccase2 circular RNA, and (iii) no change in expression of the ~2-kb canonically spliced (exon 1-2-3) laccase2 reporter mRNA.
3.2. RNAi screening in Drosophila cells to identify genes that control transcription termination
To test if the Hy_pMT Laccase2 Exons 1–3 plasmid can indeed be used to identify regulators of transcription termination, we first took advantage of the HygroR cassette to generate a Drosophila DL1 cell line that stably maintains the plasmid [17]. These cells were then soaked with in vitro transcribed double-stranded RNAs (dsRNAs) for 3 days to individually deplete proteins with well-established roles in transcription termination/pre-mRNA 3′ end processing (Fig. 2). As a negative control for all comparisons, cells were soaked with dsRNAs against E. coli β-galactosidase (β-gal). After 3 days of dsRNA treatment (with or without exogenous copper added for the last 14 h), total RNA was isolated and subjected to Northern blotting using a probe complementary to exon 2 of the laccase2 minigene (Fig. 2A) or to the HygroR mRNA (Fig. 2C). As expected, in the absence of exogenous copper, HygroR mRNA was robustly expressed (Fig. 2C, Lane 1) but there was minimal expression of the laccase2 circular RNA or the ~2-kb canonically spliced (exon 1-2-3) laccase2 reporter (Fig. 2A, Lane 1). This indicates that the inducible pMT promoter is off under these conditions, thereby allowing gain of the laccase2 circular RNA to serve as a marker of readthrough transcription.
We predicted that depletion of the RNA endonuclease Cpsf73 should inhibit 3′ end cleavage of the HygroR nascent RNA, leading to readthrough transcription into the downstream laccase2 minigene (Fig. 1D) [17]. Indeed, Cpsf73 depletion resulted in a >20-fold increase in the level of the laccase2 circular RNA observed in the absence of exogenous copper (Fig. 2A, Lane 3; Fig. 2B) [17]. There was no change in the expression of the canonically spliced (exon 1-2-3) laccase2 reporter mRNA, and this ~2-kb linear transcript could only be observed when the pMT promoter was activated by addition of exogenous copper to the cell culture medium (Fig. 2A, even lanes). As expected, Cpsf73 depletion also led to reduced expression of the canonical HygroR mRNA (Fig. 2C, Lane 2) and a larger (>4-kb) HygroR-laccase2 fusion transcript was instead detected (Fig. 2A, C). Readthrough transcripts are generally predicted to be of variable sizes, but the observation of a discrete band suggests that some HygroR-laccase2 fusion transcripts terminate at the polyadenylation signal downstream of the laccase2 minigene. It should further be noted that the laccase2 circular RNA transcript was much more abundant than the observed HygroR-laccase2 fusion transcript, likely reflecting the long half-life of the circular RNA (Fig. 2A). Collectively, these data confirm that readthrough transcription from the HygroR expression cassette leads to backsplicing of the laccase2 minigene and that the mature circular RNA can serve as a marker of readthrough transcription events.
We then examined the effect of depleting other well-characterized components of the pre-mRNA 3′ end processing machinery [reviewed in 18, 19] (Fig. 2D). Like depletion of Cpsf73, depletion of the scaffolding protein Symplekin (Symp) as well as the entire CPSF (Cpsf100, Cpsf160, Cpsf30, Wdr33, and Fip1) and CF IIm (ClpI and Pcf11) complexes led to >10-fold increases in the level of the circular RNA observed in the absence of exogenous copper (Fig. 2A, B). Only two of the three components of the CstF complex (CstF64 and CstF77, but not CstF50) affected circular RNA expression (Fig. 2A, Lanes 21–26) and no effect on the circular RNA was observed when the CF Im complex was down-regulated (Fig. 2A, Lanes 27–30). No HygroR mRNA was detected when CF Im25 or CF Im68 were depleted (Fig. 2C, Lanes 8–9), indicating that the lack of change in circular RNA levels may be due to CF Im25 or CF Im68 being required for transcription from the Copia LTR promoter.
Consistent with the poly(A) polymerase (hrg) acting only to polyadenylate the mature mRNA after Cpsf73 cleavage, depletion of hrg caused the mature HygroR mRNA to be shorter (Fig. 2C, Lane 4) but had only a minor effect on downstream circular RNA levels (Fig. 2A, Lanes 17–18). This result highlights the specificity of the laccase2 minigene reporter and its ability to distinguish factors that act at distinct steps in the process of transcription termination/pre-mRNA 3′ end processing. As an additional example of specificity, we previously showed that depletion of the 5′−3′ exonuclease Rat1 resulted in increased circular RNA expression, but no change in HygroR mRNA levels and no accumulation of HygroR-laccase2 fusion transcripts [17]. These phenotypes exactly fit the torpedo model of transcription termination [30–32]. We thus conclude that circular RNA-based plasmids can be used to efficiently and accurately study transcription termination mechanisms and readthrough transcription in Drosophila cells.
3.3. Use of circular RNAs to define transcription termination mechanisms in human cells
We next addressed whether circular RNAs could likewise be used as markers of readthrough transcription events in human cells. A coral GFP (cGFP) reporter plasmid (pCRII-TOPO CMV-cGFP-SV40 pA + Laccase2 Exon 2) was generated in which laccase2 exon 2 and its flanking intronic repeat sequences were inserted downstream of a SV40 polyadenylation signal (Fig. 3A). This design is simpler than the Drosophila reporter (Fig. 1A), but is conceptually similar in that the CMV promoter should only produce a polyadenylated cGFP mRNA (and not the circular RNA) under normal conditions. However, if the SV40 polyadenylation signal fails to be used and/or RNA polymerase II fails to efficiently terminate, then the laccase2 circular RNA can be generated via backsplicing of the readthrough transcript.
HeLa cells were transfected with siRNAs to individual transcription termination factors as well as with the pCRII-TOPO CMV-cGFP-SV40 pA + Laccase2 Exon 2 plasmid, followed by total RNA isolation and Northern blotting (Fig. 3B). As in Drosophila, depletion of CPSF3 (CPSF73), Symplekin (SYMPK), and PCF11 resulted in significant readthrough past the SV40 poly(A) signal and generation of the downstream circular RNA (Fig. 3B, C). This confirms that this plasmid-based system can identify well-established pre-mRNA 3′ end processing factors in human cells. We further found that depletion of PPP1R10 (PNUTS), which is known to modulate protein phosphatase-1 (PP1) activity in the nucleus [45], resulted in increased circular RNA expression (Fig. 3B, C). In yeast [46] and humans [20], PP1 (Glc7 in yeast) was originally proposed to be dispensable for pre-mRNA 3′ cleavage, but required for polyadenylation (and thus PPP1R10 depletion was predicted to have no effect on circular RNA expression in our assay). Instead, our data are consistent with a transcriptional profiling study performed in mouse cells [47] and suggest that PNUTS/PP1 is involved in additional steps in termination (e.g. pre-mRNA 3′ cleavage) [48].
It should be noted that some siRNAs resulted in reduced expression of the mature cGFP mRNA (as expected), but not all siRNAs that resulted in a termination defect did so (Fig. 3B). In addition, no stable cGFP-laccase2 fusion transcript could be observed by Northern blotting. Gain of circular RNA expression was thus overall a more robust and reproducible readout of transcription termination defects. This highlights the inherent power and specificity of using circular RNAs as markers of readthrough transcription.
4. Conclusion
We have outlined methodologies for how circular RNAs can be encoded downstream of polyadenylation signals and used to study transcription termination mechanisms in Drosophila and human cells. This approach takes advantage of the long half-lives and discrete sizes of circular RNAs, which make these transcripts very sensitive biomarkers of transcriptional activity. Nonetheless, because readthrough transcripts must be backspliced in order to be detected, a potential limitation of this approach is that factors required for backsplicing (or that modulate backsplicing efficiency) may be missed if one only quantifies circular RNA levels. This underscores the importance of also directly examining whether linear readthrough transcripts can be detected, e.g. using Northern blotting (Fig. 2C) or RT-qPCR, from the reporter plasmids.
Our circular RNA-based, gain-of-function plasmid methods are generalizable and modular in design, making them applicable for studying likely any polyadenylation signal or RNA polymerase II transcription termination mechanism. One can, for example, easily replace the SV40 polyadenylation signal that was used here (Fig. 1A, 3A) with a sequence of the reader’s interest. Likewise, the laccase2 circular RNA sequence can be swapped with other sequences that can backsplice, including an exon encoding a fluorescent protein (e.g. GFP) [43] or a fluorogenic RNA aptamer (e.g. Broccoli) [49] to enable a more high-throughput approach. We prefer to use Northern blots as they enable visualization of all the transcripts (including their sizes), but RT-qPCR using primers that amplify across the backsplicing junction [50] or catalytically dead CRISPR-Cas13 proteins [51] can alternatively be used to detect/quantify circular RNAs generated from readthrough transcription. Together, these approaches open the opportunity for further genetic (e.g. RNAi or CRISPR/Cas9) or chemical-based screens to identify factors or cellular conditions that modulate transcription termination events.
Supplementary Material
Highlights.
Readthrough transcription can yield circular RNAs in Drosophila and human cells
Plasmid-based methods to quantify readthrough transcription events
The long half-lives of circular RNAs simplifies detection of readthrough transcripts
Proof-of-principle experiments identify many known transcription termination factors
Acknowledgements
We thank all members of the Wilusz laboratory for helpful discussions and suggestions. This work was supported by NIH grants R35-GM119735 and K99-GM131028.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Xiao MS, Ai Y, Wilusz JE, Biogenesis and Functions of Circular RNAs Come into Focus, Trends Cell Biol 30(3) (2020) 226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J, The biogenesis, biology and characterization of circular RNAs, Nat Rev Genet 20(11) (2019) 675–691. [DOI] [PubMed] [Google Scholar]
- [3].Chen LL, The expanding regulatory mechanisms and cellular functions of circular RNAs, Nat Rev Mol Cell Biol 21(8) (2020) 475–490. [DOI] [PubMed] [Google Scholar]
- [4].Wilusz JE, A 360 degrees view of circular RNAs: From biogenesis to functions, Wiley Interdiscip Rev RNA (2018) e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL, The Biogenesis of Nascent Circular RNAs, Cell Rep 15(3) (2016) 611–24. [DOI] [PubMed] [Google Scholar]
- [6].Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y, Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor, Nucleic Acids Res 44(3) (2016) 1370–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC, Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation, Cell Rep 9(5) (2014) 1966–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, Rajewsky N, A map of human circular RNAs in clinically relevant tissues, J Mol Med (Berl) 95(11) (2017) 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N, Circular RNAs are a large class of animal RNAs with regulatory potency, Nature 495(7441) (2013) 333–8. [DOI] [PubMed] [Google Scholar]
- [10].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE, Circular RNAs are abundant, conserved, and associated with ALU repeats, RNA 19(2) (2013) 141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO, Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types, PLoS One 7(2) (2012) e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N, Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed, Mol Cell 58(5) (2015) 870–85. [DOI] [PubMed] [Google Scholar]
- [13].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, Natural RNA circles function as efficient microRNA sponges, Nature 495(7441) (2013) 384–8. [DOI] [PubMed] [Google Scholar]
- [14].Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, Qu B, Zhou Z, Shen N, Yang L, Chen LL, Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity, Cell 177(4) (2019) 865–880 e21. [DOI] [PubMed] [Google Scholar]
- [15].Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N, Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function, Science 357(6357) (2017). [DOI] [PubMed] [Google Scholar]
- [16].Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z, A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion, Immunity 48(4) (2018) 688–701 e7. [DOI] [PubMed] [Google Scholar]
- [17].Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, Cherry S, Wilusz JE, The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting, Mol Cell 68(5) (2017) 940–954 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Proudfoot NJ, Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut, Science 352(6291) (2016) aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shi Y, Manley JL, The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site, Genes Dev 29(9) (2015) 889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL, Molecular architecture of the human pre-mRNA 3’ processing complex, Mol Cell 33(3) (2009) 365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tian B, Manley JL, Alternative polyadenylation of mRNA precursors, Nat Rev Mol Cell Biol 18(1) (2017) 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Venters CC, Oh JM, Di C, So BR, Dreyfuss G, U1 snRNP Telescripting: Suppression of Premature Transcription Termination in Introns as a New Layer of Gene Regulation, Cold Spring Harb Perspect Biol 11(2) (2019) a032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mayr C, Bartel DP, Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells, Cell 138(4) (2009) 673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB, Proliferating cells express mRNAs with shortened 3 ‘ untranslated regions and fewer microRNA target sites, Science 320(5883) (2008) 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ji Z, Lee JY, Pan Z, Jiang B, Tian B, Progressive lengthening of 3’ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development, Proc Natl Acad Sci U S A 106(17) (2009) 7028–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang H, Rigo F, Martinson HG, Poly(A) Signal-Dependent Transcription Termination Occurs through a Conformational Change Mechanism that Does Not Require Cleavage at the Poly(A) Site, Mol Cell 59(3) (2015) 437–48. [DOI] [PubMed] [Google Scholar]
- [27].Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ, Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing, Cell 161(3) (2015) 526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, Cramer P, TT-seq maps the human transient transcriptome, Science 352(6290) (2016) 1225–8. [DOI] [PubMed] [Google Scholar]
- [29].Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL, Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition, Mol Cell 60(2) (2015) 256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Connelly S, Manley JL, A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II, Genes Dev 2(4) (1988) 440–52. [DOI] [PubMed] [Google Scholar]
- [31].West S, Gromak N, Proudfoot NJ, Human 5’ --> 3’ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites, Nature 432(7016) (2004) 522–5. [DOI] [PubMed] [Google Scholar]
- [32].Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S, The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II, Nature 432(7016) (2004) 517–22. [DOI] [PubMed] [Google Scholar]
- [33].Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA, Widespread Inducible Transcription Downstream of Human Genes, Mol Cell 59(3) (2015) 449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rosa-Mercado NA, Zimmer JT, Apostolidi M, Rinehart J, Simon MD, Steitz JA, Hyperosmotic stress alters the RNA polymerase II interactome and induces readthrough transcription despite widespread transcriptional repression, Mol Cell 81(3) (2021) 502–513.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rutkowski AJ, Erhard F, L’Hernault A, Bonfert T, Schilhabel M, Crump C, Rosenstiel P, Efstathiou S, Zimmer R, Friedel CC, Dolken L, Widespread disruption of host transcription termination in HSV-1 infection, Nature communications 6 (2015) 7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z, FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3’ end processing of histone pre-mRNAs, Mol Cell 36(2) (2009) 267–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Peart N, Wagner EJ, Gain-of-function reporters for analysis of mRNA 3’-end formation: Design and optimization, BioTechniques 60(3) (2016) 137–40. [DOI] [PubMed] [Google Scholar]
- [38].Steinmetz EJ, Conrad NK, Brow DA, Corden JL, RNA-binding protein Nrd1 directs poly(A)-independent 3’-end formation of RNA polymerase II transcripts, Nature 413(6853) (2001) 327–31. [DOI] [PubMed] [Google Scholar]
- [39].Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA, A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails, Genes Dev 26(21) (2012) 2392–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tatomer DC, Liang D, Wilusz JE, RNAi Screening to Identify Factors That Control Circular RNA Localization, Methods Mol Biol 2209 (2021) 321–332. [DOI] [PubMed] [Google Scholar]
- [41].Tatomer DC, Liang D, Wilusz JE, Inducible Expression of Eukaryotic Circular RNAs from Plasmids, Methods Mol Biol 1648 (2017) 143–154. [DOI] [PubMed] [Google Scholar]
- [42].Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S, circRNA biogenesis competes with pre-mRNA splicing, Mol Cell 56(1) (2014) 55–66. [DOI] [PubMed] [Google Scholar]
- [43].Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE, Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins, Genes Dev 29(20) (2015) 2168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bunch TA, Grinblat Y, Goldstein LS, Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells, Nucleic Acids Res 16(3) (1988) 1043–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Allen PB, Kwon YG, Nairn AC, Greengard P, Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit, J Biol Chem 273(7) (1998) 4089–95. [DOI] [PubMed] [Google Scholar]
- [46].He X, Moore C, Regulation of yeast mRNA 3’ end processing by phosphorylation, Mol Cell 19(5) (2005) 619–29. [DOI] [PubMed] [Google Scholar]
- [47].Austenaa LM, Barozzi I, Simonatto M, Masella S, Della Chiara G, Ghisletti S, Curina A, de Wit E, Bouwman BA, de Pretis S, Piccolo V, Termanini A, Prosperini E, Pelizzola M, de Laat W, Natoli G, Transcription of Mammalian cis-Regulatory Elements Is Restrained by Actively Enforced Early Termination, Mol Cell 60(3) (2015) 460–74. [DOI] [PubMed] [Google Scholar]
- [48].Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover-Cutter K, Brannan K, Bentley DL, Control of RNA Pol II Speed by PNUTS-PP1 and Spt5 Dephosphorylation Facilitates Termination by a “Sitting Duck Torpedo” Mechanism, Mol Cell 76(6) (2019) 896–908.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Litke JL, Jaffrey SR, Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts, Nat Biotechnol 37(6) (2019) 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dodbele S, Mutlu N, Wilusz JE, Best practices to ensure robust investigation of circular RNAs: pitfalls and tips, EMBO Rep 22(3) (2021) e52072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang LZ, Wang Y, Li SQ, Yao RW, Luan PF, Wu H, Carmichael GG, Chen LL, Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems, Mol Cell 76(6) (2019) 981–997.e7. [DOI] [PubMed] [Google Scholar]
- [52].Mandel CR, Bai Y, Tong L, Protein factors in pre-mRNA 3’-end processing, Cell Mol Life Sci 65(7–8) (2008) 1099–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


