Abstract
Liver cancer is the second most lethal malignancy worldwide. Cell lines and murine models are the most common tools for modeling human liver carcinogenesis. Most recently, organoids with a three-dimensional structure derived from primary tissues or cells have been applied to liver cancer research. Organoids can be generated from induced pluripotent stem cells, embryonic or adult, healthy or diseased tissues. In particular, liver organoids have been widely employed in mechanistic studies aimed at delineating the molecular pathways responsible for hepatocarcinogenesis. The introduction of clustered regularly interspaced palindromic repeats (CRISPR)-associated protein 9 (Cas9) and microengineered miniorganoid technologies into liver organoids for cancer study has significantly accelerated these investigations. Translational advances have been made by utilizing liver tumor organoids for anticancer drug screening, biobanking, omics profiling, and biomarker discovery. This review summarizes the latest advances and the remaining challenges in the use of organoid models for the study of liver cancer.
Keywords: organoids, primary liver cancer, basic research, translational science
Liver cancer is the sixth most common cancer and the second most lethal malignancy worldwide.1 Primary liver cancer (PLC) includes a heterogeneous group of tumors with distinct histological features and clinical outcomes, with hepatocellular carcinoma (HCC) representing 80% of all PLCs, and intrahepatic cholangiocarcinoma (iCCA) covering most of the remaining cases.2 A combined hepatocellular–cholangiocarcinoma (HCC/CC) subtype has also been identified among PLCs.3 PLCs are characterized by a complex and diverse genetic landscape of alterations, including high degree of aneuploidy, DNA copy number variations, somatic mutations, and epigenetic changes, all contributing to hepatocarcinogenesis at various degrees.4 This molecular heterogeneity implies the need for patient-tailored treatments, an approach known as precision medicine.5 Frequent genetic alterations in PLCs include, among many, TP53, cell cycle-related genes, such as CCND1 and CDKN2A, and chromatin-remodeling genes ARID1A and ARID2.4,6 HCC is further characterized by mutations leading to the activation of WNT/β-Catenin pathway and TERT, whereas mutations of IDH1/2 and KRAS genes, as well as FGFR2 fusions frequently occur in iCCA.7–9
Both in vitro and in vivo models have been employed to study liver cancer pathogenesis. In vitro techniques include cell lines and organoids, whereas in vivo techniques include genetic modified animals and patient-derived xenograft (PDX) models. Among them, the organoid system has increasingly been used to investigate the molecular mechanisms associated with liver carcinogenesis and examine the therapeutic efficacy of anti-tumor therapies.10,11
Liver Organoids as a Model for Liver Disease Investigation
An organoid is defined as a three-dimensional (3D) structure derived from primary tissues or cells. It consists of organ-specific cell types, thus mimicking in vivo tissue structures.12–14 Organoids can be propagated for a relatively long time while maintaining their cellular complexity and 3D structure. They can be amplified from a small tissue sample and adapted to high-throughput research. In the last few years, a wide variety of human and murine organoids have been established from different organs to study numerous physiologic and pathologic conditions.14
Embryonic stem cells (ESC), induced pluripotent stem cells (iPSC), and primary tissues are the main sources for the generation of tissue-specific organoids.15 The culturing and expansion of normal liver organoids was first achieved by Huch et al with mouse Lgr5+ liver stem cells.16 Subsequently, Takebe et al generated liver organoids from human iPSCs in combination with endothelial and mesenchymal cells in a Matrigel matrix.17 Two years later, the Clevers’ group reported a novel in vitro system to generate long-term organoid cultures derived from human bipotent progenitor liver cells.18 The authors optimized the culturing system and provided protocol for culturing and establishing self-renewing human and mouse adult liver organoids.18 Liver cells can grow in a gel-based extracellular matrix (ECM) when supplemented with essential growth factors, including hepatocyte growth factor (HGF), epidermal growth factor (EGF), fibroblast growth factor 10 (FGF10), and R-spondin-1 (Rspo1).19 However, these liver organoids are primarily generated from biliary epithelial cells (BEC). Aiming at establishing a long-term expansion system for hepatocyte originated liver organoids, Hu et al established a protocol for generating hepatic organoids from Axin2+ hepatocytes with a modified culture medium. In brief, Wnt agonists such as Rspo1 and CHIR99021; EGF, FGF7, FGF10, and transforming growth factor-α (TGF-α); ROCK inhibitor Y27632; HGF and TGF-β inhibitor A83–01 were supplemented to the medium. The study by Hu et al emphasized the critical role of Wnt/Rspo1 and HGF signaling pathways in regulating primary hepatocyte expansion in both mouse and human organoid models.20 In parallel, Peng et al revealed that tumor necrosis factor-α (TNFα) enabled the in vitro expansion of primary mouse hepatocytes in 3D culture in the absence of Rspo-1. Single-cell RNA sequencing (RNAseq) of these 3D culture hepatocytes demonstrated the expression of numerous hepatocellular markers but not of other cell types.21 Since then, several protocols for normal liver organoids have been established by modifying the culturing system or the stem cells oforigin.22–24 For instance, Sun et al transfected fibroblasts with FOXA3, HNF1A, and HNF4A and expanded them by SV40 large T antigen (SV40LT) to generate human-induced hepatocytes (hiHeps). These cells were able to self-organize into liver organoids with hepatic architecture and functions in a hepatocyte-maintaining medium.25
Thus, in summary, liver organoids were developed using several approaches and cells of origin (►Fig. 1) and are widely applied for functional studies.26
Fig. 1.
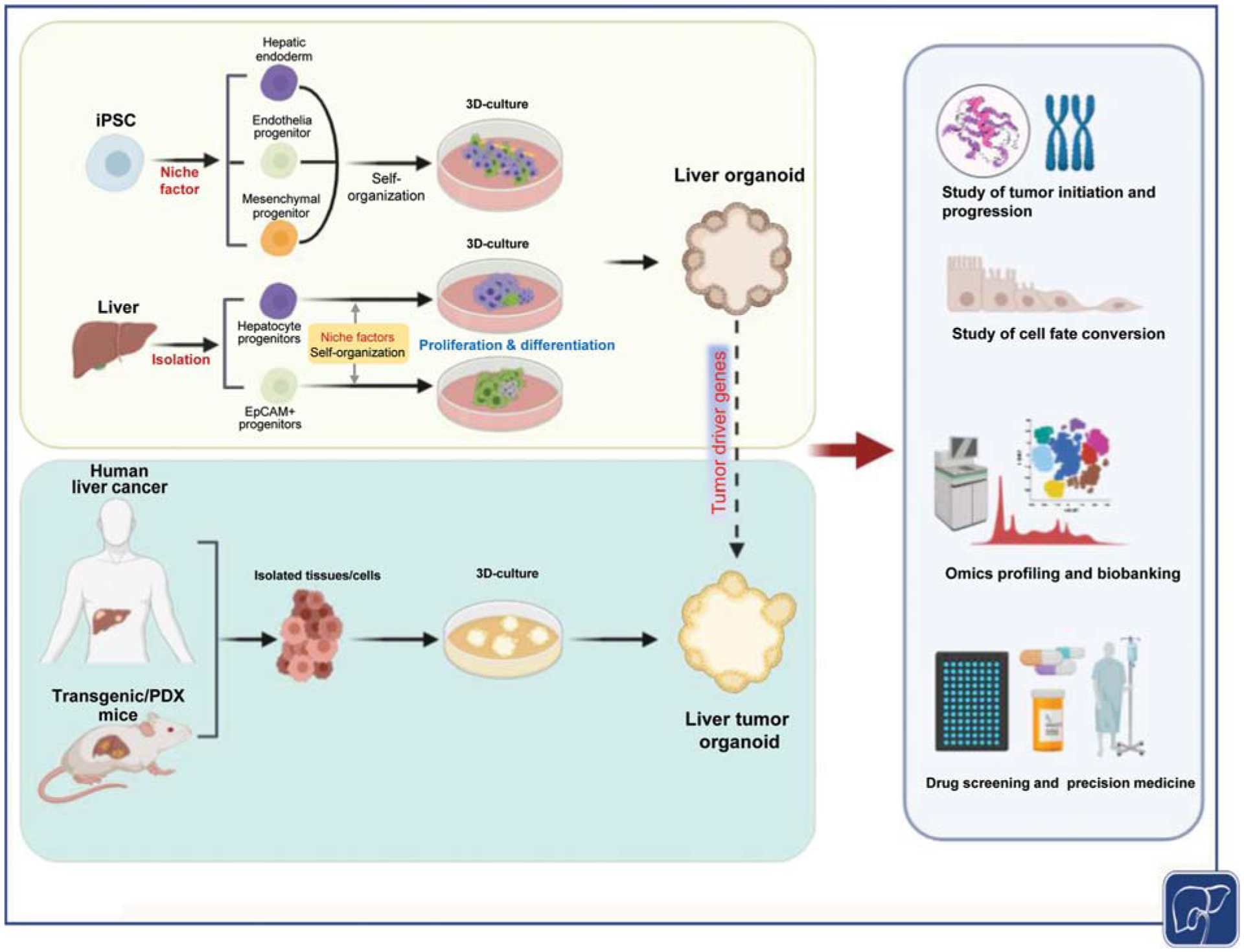
Establishment and applications of liver tumor organoids. Liver organoids can be generated from induced pluripotent stem cells (iPSC) or liver tissue originated progenitor cells. Liver tumor organoids can be induced in normal liver organoids upon transfection with tumor driver genes or from isolated human or murine liver tumor cells. Liver tumor organoids can be used for mechanistic study of tumor development, cell fate conversion, omics profiling, biobanking, drug screening, and precision medicine.
Generation of Liver Tumor Organoids for the Study of Human Liver Cancer
Early studies on liver organoids have been focusing on testing and validating the accuracy and genetic features of the system. Recently, human and mouse liver tumor organoids (also termed as “liver tumoroids”) have been established through various methods (►Fig. 1 and ►Table 1).
Table 1.
Summary of HCC and iCCA organoids for the study of liver cancer
| Cancer type | Cell/tissue of origin | Key factors for derivation | Applications |
|---|---|---|---|
| HCC | Human HCC specimens27,28 | Matrigel-based liver organoid culturing system; dexamethasone and Y27632; removal of R-spondin-1, Noggin and Wnt3a |
|
| Primary human hepatocytes25 | FOXA3, HNF1A, and HNF4A; SV40LT that inactivates p53 and RB; c-MYC overexpression |
|
|
| HCC-PDX cells37 | 3D bioengineered sponge system |
|
|
| Human HCC cell lines41 | Matrigel based; HCC cells and nonparenchymal cells; assembly culture medium |
|
|
| DEN-induced murine liver tumor tissues38 | Matrigel-based culturing system |
|
|
| CCA | Human iCCA specimens27,28 | Matrigel-based culturing system |
|
| Primary human hepatocytes25 | FOXA3, HNF1A, and HNF4A; SV40LT that inactivates p53 and RB; RAS overexpression |
|
|
| KrasG12D /Tp53−/− murine iCCA tumor40 | Matrigel-based culturing system |
|
Abbreviations: HCC, hepatocellular carcinoma; iCCA, intrahepatic cholangiocarcinoma; FOXA3, forkhead box A3; HNF1A, HNF1 homeobox A; HNF4A, HNF4 homeobox A; SV40LT, SV40 large T antigen; PDX, patient derived xenograft.
Patient-Derived Liver Cancer Organoids
In 2017, Broutier et al reported the development of long-term organoid cultures derived from patients with PLC.27 Specifically, patient-derived organoid (PDO) cultures were established from eight individuals with PLC, belonging to three tumor histotypes (HCC, iCCA, and HCC/iCCA). Using histological, immunohistochemical, and genomic analyses, the authors showed that PDOs retained the organ site-specific histologic and genetic traits. In addition, PDOs exhibited a similar biological behavior when transplanted into immunocompromised mice.27 In a similar study, Nuciforo et al reported long-term organoid cultures from needle biopsies of HCC patients with various etiologies and tumor stages. Similar to that described by Broutier et al, HCC organoids retained the morphology and the expression pattern of HCC tumor markers and preserved the genetic heterogeneity of the originating tumors. Interestingly, the success rate for the generation of HCC organoids was 26% based on the number of cultured tumor biopsies (10 of 38 HCC biopsies),28 a percentage that is significantly lower when compared with the reported success rates for pancreatic cancer (75–83%)29 and colorectal cancer (90%).30 The lower success rate using HCC specimens may be due to the fact that hepatocytes, the cell of origin of HCCs, lack the features of epithelial stem cells needed for their propagation in the organoid culture system.31 Of note, the study by Nuciforo et al adopted the system developed by Broutier et al.27 Although the authors attempted to optimize the system to improve the success rate by removing compounds with potential negative effects on HCC proliferation such as Forskolin, N-acetyl-L-cysteine, nicotinamide, and HGF from the medium, and adding FGF19, a factor with potential growth-promoting effects for HCCs, these efforts had limited effects on the establishment of additional HCC organoid lines.28 These protocols have been subsequently applied in several studies.32–36
In addition to the traditional Matrigel organoid system, Fong et al attempted to generate HCC patient-derived xenograft originated organoids (HCC-3DPDX) by applying a 3D sponge system. The system consisted of a cellulose-based hydrogel with interconnected macropores fabricated by leveraging the ability of hydroxypropyl cellulose (HPC) derivatives to undergo thermal-induced phase separation and photo-crosslinking. Methacrylate (MA) was used in the photo-crosslinkable group and dichloromethane was used for the reaction to synthesize MA-HPC in the system. In total, 14 HCC-3DPDX lines were generated from the authors’ previously built HCC-PDX biobank. Organoids rapidly formed by Day 2 and remained viable for 1 week, and a few lines maintained viability for almost 3 weeks in culture.37 HCC-3DPDX lines feasibly maintain various cellular compartments of HCC samples and retain genomic profiles and intratumoral heterogeneity of their in vivo counterpart. Therefore, they might represent a promising tool to determine individualized treatments. However, technical complexities of this bioengineering approach may restrict its widespread application.
Murine Liver Tumor-Derived Organoids
Cao et al reported the establishment of organoids from mouse primary liver tumors induced by the hepatocarcinogen diethylnitrosamine (DEN).38 The author succeeded in culturing 91 lines from 129 liver tissue/tumors. These organoids could be grown in long-term cultures in vitro and ~20% formed tumors in immunodeficient mice upon serial transplantation, confirming their tumorigenic and self-renewal properties. Interestingly, single cells from the tumor organoids had high efficiency of organoid initiation, and a single organoid derived from a cancer cell was able to initiate a tumor in mice, indicating the enrichment of tumor-initiating cells in the tumor organoids. Furthermore, these organoids recapitulated, to some extent, the heterogeneity of liver cancer in patients, with respect to phenotype, cancer cell composition, and treatment response.38 Recently, Saborowski et al were able to establish a iCCA organoid from the mouse iCCA model consisting of KrasG12D mutation and loss of Tp53.39 The organoid was cultured using the protocol from Broutier et al and used to probe candidate cancer in genes in vivo by applying RNA interference or CRISPR/Cas9 technology in liver organoids. In a proof-of-principle study, they showed that loss of function mutation of phosphatase and tensin homolog (Pten) either by shRNA against Pten or CRISPR/Cas9 mediated knockout of Pten was able to promote iCCA organoid growth.40
Primary Human Hepatocytes (PHH)-Modified Liver Tumor Organoids
As described previously, Sun et al generated hiHeps via reprogramming fibroblasts.25 These hiHeps were subsequently used to investigate the genes that may promote liver tumor imitation. Specifically, the authors found that HCC could be formed via c-MYC overexpression, while iCCA could be induced by oncogenic RAS-driven lineage conversion.25 Studies on liver tumor initiation using hiHeps organoids are further discussed in the section “Study of liver tumor initiation and progression mechanisms.”
Cancer Cells Self-Assembled Liver Tumor Organoids
Recently, Wang et al sought to demonstrate that liver nonparenchymal cells, including endothelial cells and fibroblasts, support HCC tumor organoid formation in vitro. For this purpose, HCC and nonparenchymal cells were coseeded in a thick layer of Matrigel. Strikingly, HCC and nonparenchymal cells self-organized into 3D cell clusters, exhibiting a remarkable intrinsic organizing capacity. At the molecular level, HCC cells cultured together with nonparenchymal cells were characterized by higher expression of neoangiogenesis inducers (vascular endothelial growth factor 2 [VEGFR2], VEGF, hypoxia inducible factor-α), tumor-related inflammatory cytokines (CXCR4, CXCL12, TNF-α), and epithelial-mesenchymal transition markers (TGF-β, Vimentin, MMP9) when compared with organoids containing only HCC cells,41 thus more closely recapitulating the alterations of the human disease. Although further validation is required, the described findings underline the usefulness of this system to study the interactions between cancer cells and nonparenchymal cells in hepatocarcinogenesis as well as for preclinical testing of innovative therapies.
Application of Liver Tumor Organoids for the Study of Liver Cancer
Liver tumor organoids have been used to investigate the mechanisms responsible for tumor initiation and progression. Extensive studies have also utilized liver tumor organoids to improve therapy response prediction in patients. Other applications of organoids for liver tumor include, but are not limited to, omics profiling, biomarker discovery, and biobanking (►Fig. 1 and ►Table 1).
Study of Liver Tumor Initiation and Progression Mechanisms
Cancer cell lines and organoids derived from PLCs roughly recapitulate the mutations and expression profiles of liver tumors but cannot be used to study cancer initiation. In contrast, organoids derived from normal liver tissues can be used to model liver cancer pathogenesis by sequentially introducing cancer driving mutations.42 The advantage of using such models is that they enable the investigation of the effects of driver mutations in an isogenic genetic background. For instance, Sun et al transfected hiHeps originated liver organoids with c-MYC oncogene. Of note, forced transfection of c-MYC led to HCC formation in the system. Mechanistically, alterations of mitochondrial-associated endoplasmic reticulum (ER) membranes were observed, suggesting that excessive mitochondrial-ER coupling may be the key mechanism underlying c-MYC dependent hepatocarcinogenesis. In addition, a mutant form of RAS (RASG12V) was able to transform hiHeps into iCCA organoids. Subsequently, these iCCA organoids were able to form tumors in mice after orthotopic transplantation. Further analysis suggested that iCCAs developed via hepatocyte-based lineage conversion. At the molecular level, the NOTCH and JAK/STAT cascades were found to be responsible for the observed hepatocyte-iCCA conversion.25 The results provide evidence that at least some human iCCAs may derive from mature hepatocytes. BRCA1 associated protein-1 (BAP1) is a deubiquitinating enzyme that is implicated in iCCA development.43 In a recent study, using CRISPR-Cas9 based gene-editing, Artegiani et al deleted BAP1 in normal human cholangiocyte organoids.44 It was found that deletion of BAP1 alone led to loss of several epithelial characteristics of the organoids. When loss of BAP1 was combined with other common iCCA mutations, including TP53, PTEN, SMAD4, and NF1, it resulted in iCCA development when the organoids were transplanted into mice.44 Excitingly, Artegiani et al later reported a CRISPR-Cas9-mediated homology-independent organoid transgenesis (CRISPR-HOT) methodology for fast and efficient generation of knock-in human organoids.45 Combining tubulin tagging with TP53 knockout revealed that TP53 is involved in controlling hepatocyte ploidy and mitotic spindle fidelity, providing evidence that TP53 mutation-induced aneuploidy may promote liver cancer.45 Thus, organoid technology in combination with CRISPR/Cas9 strategy is a powerful tool to investigate cancer gene function in a human context.46
PDO liver tumoroids are also widely used to characterize the molecular mechanisms leading to tumor progression. For instance, Chan et al demonstrated that silencing of protein methyltransferase 6 (PRMT6) induced cancer stemness in HCC-derived PDOs. Further analysis suggested that PRMT6 functions via CRAF-ERK signaling.32 In a follow-up study, it was revealed that PRMT6 regulates ERK-dependent PKM2 expression and Warburg effect in HCC PDOs. The results provide a mechanistic link among tumorigenicity, sorafenib resistance, and glucose metabolism.33 Similarly, using PDO iCCA organoids, Li et al reported that silencing of mitochondrial fusion regulatory genes, including optic atrophy 1 (OPA1) and mitofusin 1 (MFN1), inhibited the fusion process in iCCA tumor organoids. This led to the inhibition of cell growth in vitro and tumor formation in vivo after tumor cell engraftment in mice. Mechanistically, it was discovered that the increased mitochondrial fusion in liver cancer cells altered metabolism and fueled tumor cell growth.34 Furthermore, Li et al silenced kinesin family member 15 (KIF15) in HCC organoids and revealed a significant reduction in sphere formation as well as the decreased expression of stemness-related genes. The results indicated that KIF15 might promote the CSC phenotype and malignancy in HCC.47
Hepatic and Biliary Lineage Commitment Studies
Both hepatocytes and cholangiocytes exhibit elevated plasticity, and cell fate conversion from hepatocytes to cholangiocytes has been reported under pathological conditions.48 To investigate whether iCCA cells can be converted to functional hepatocytes, Saito et al established organoids derived from human iCCAs and cultured them under conditions suitable for hepatocyte differentiation. Noticeably, the organoids acquired functions of mature hepatocytes, such as albumin secretion, bile acid production, and increased CYP3A4 activity. Moreover, the malignant potential of iCCA cells was markedly reduced after hepatocytic differentiation. Removal of Rspo1, the ligand of LGR5, induced differentiation of iCCA cells into functional hepatocytes, suggesting that inhibition of the Wnt signaling pathway may induce hepatocyte differentiation.18,49 Consistently, Rimland et al generated intrahepatic bile duct (IHBD) organoids using culture conditions promoting Wnt signaling. The authors found that IHBD organoids expressed the stem cell/progenitor markers LGR5 and PROM1 as well as the ductal markers KRT19 and KRT7. Interestingly, IHBDs were found to express low levels of hepatocyte markers under differentiation conditions, suggesting the critical role of Wnt signaling in promoting hepatocyte differentiation.50
Ochiai et al isolated mouse liver cells and cultured them with a modified protocol to generate cholangiocyte lineage committed organoids. The combination of Kras overexpression with suppression of one of the tumor suppressor genes commonly inactivated in iCCA (Cdkn2a, Pten, Apc, and Tp53) resulted in iCCA formation in these organoids, supporting the oncogenic potential of these genetic events for iCCA development. The authors also demonstrated that mutant Pik3ca and FGFR2-AHCYL1 fusion possess limited oncogenic potential in these liver-derived organoids, probably requiring additional mutations or a proper hepatic niche to induce iCCA formation.51 Li et al generated a murine HCC model consisting of codeletion of Pten and Tp53 together with knocking-in prominin-1 (Prom1). HCC tumor organoids were subsequently derived from these mouse HCCs. Subsequently, the authors investigated whether these HCC organoids may acquire iCCA-like traits following their expansion and orthotopic transplantation in mice. Invasive and metastatic tumors developed following this procedure and exhibited iCCA-related pathologic and molecular features, which were absent in the primary HCC tumors. This model partly recapitulates the molecular features of human HCC/iCCA mixed tumors and suggest that this peculiar tumor subtype is the consequence of the evolution of HCC into another cancer entity with iCCA features.52
Drug Screening and Precision Medicine
The available options for PLC management have been elegantly reviewed elsewhere.53,54 As for molecular therapies, sorafenib55 and lenvatinib56 multikinase inhibitors are first-line therapies, while regorafenib, cabozantinib, ramucirumab, and nivolumab are second-line therapies for HCC.57 Excitingly, a recent phase III clinical trial (IMbrave150) showed that atezolizumab (an anti-PDL1 antibody) combined with bevacizumab (an anti-VEGF-A antibody) resulted in better overall and progression-free survival outcomes than sorafenib in unresectable HCC,58 holding the promises for immunotherapy as a crucial therapy for HCC treatment. Pemigatinib,59 a FGFR1–3 inhibitor, has been recently approved as the first ever target therapy for iCCA patients with FGFR2 fusion mutations.
The tumor organoid culture preserves intratumor heterogeneity60 and the tumor microenvironment.61 Consequently, organoids may be better suited for drug screening than cancer cell lines; in addition, organoids are significantly cheaper and more efficient than murine tumor models. Organoids have also been applied as the platform to evaluate drug efficacy, opening the doors for precision oncology. Since the liver is the major organ responsible for drug metabolism, liver organoids have been also used to dissect the potential side effects of candidate therapies.
Broutier et al performed a proof-of-principle drug screening using 29 drugs and identified liver PDOs sensitivity to ERK inhibition, which was validated in xenograft models.40 Chen et al showed that immunosuppressant mycophenolic acid (MPA) effectively inhibited the initiation and growth of mouse liver tumoroids.62 It has been hypothesized that intratumor genetic heterogeneity may contribute to the high failure rate of chemotherapy. Recently, Li et al evaluated the functional heterogeneity in a cohort of primary human liver cancer organoids and how it might contribute to drug responsiveness. For this purpose, a total of 27 liver cancer organoids were established and tested with 129 cancer drugs. The results suggest that the intratumor drug response heterogeneity does not correlate with molecular profiles variations in these liver cancer organoids.63 Additional studies identified nine pan-effective drugs from five classes of antineoplastic agents, including histone deacetylase inhibitors, proteasome inhibitors, DNA topoisomerase II inhibitors, protein translation inhibitors, and RNA synthesis inhibitors, as potential drugs for liver cancer treatment.63 To better understand the limited efficacy of the multikinase inhibitor sorafenib on liver cancer, Lee et al established sorafenib-resistant HCC-PDO lines. They discovered that the Src homology 2 domain-containing phosphatase (SHP2) blocker SHP099 is able to annul sorafenib resistance by impeding RTK-induced reactivation of the MEK/ERK and AKT signaling pathways. These encouraging results highlighted the potential application of liver tumor organoids for the study of chemoresistance and the development of alternative therapeutic approaches.64 Another mechanism of resistance to sorafenib treatment by HCC was discovered by Wang et al.35 The authors demonstrated that CD44-positive HCC PDOs are resistant to sorafenib, and sorafenib increases CD44 levels. A subsequent drug screening conducted by the same group showed that GANT61, a Hedgehog signaling inhibitor, potently suppresses HCC PDO viability. Noticeably, a highly synergistic, antigrowth effect in vitro and in vivo was observed when sorafenib and GANT61 were concomitantly administered to CD44-positive HCC PDOs and cell lines.35 Together with studies on chemoresistance, investigation has been conducted using liver tumor organoids also to identify molecular markers of chemosensitivity. For instance, Wang et al generated four HCC PDO lines with differential long-chain acyl-CoA dehydrogenase (ACADL) expression and demonstrated that organoids with low expression of ACADL were sensitive to the YAP inhibitor Verteporfin.36 Saito et al conducted a drug screening among 3 iCCA PDO lines among 339 clinical used medicines and identified 22 compounds able to suppress iCCA organoid growth. Among these compounds, anticancer agents such as antimicrotubule agents, tyrosine kinase inhibitors, antimetabolite agents, mammalian target of rapamycin (mTOR) inhibitors, and proteasome inhibitors were found. Intriguingly, they also proved two antifungal compounds (amorolfine and fenticonazole) as promising drugs against iCCA.65 More recently, Antonia et al applied a high-throughput drug screening platform with AI-enhanced robotics to iCCA PDOs, reporting responsiveness to the mTOR inhibitor Sapanisertib (INK128) by all iCCA PDO lines. Intriguingly, INK128 was synergistic with gemcitabine in PDOs derived from one patient.66
Another prospective direction of applying organoids to the study of liver cancer is using omics profiling data gathered from organoid to indirectly guide drug repurpose. For instance, Guan et al performed single-cell RNA-sequencing and metabolomic analysis on developing organoid cultures at the iPSC, hepatoblast, and mature organoid stage. Data analysis indicated that early hepatoblasts are selectively sensitive to the cytotoxic effect of the antihistamine drug meclizine. Importantly, administration of meclizine in combination with the glycolysis inhibitor PFK158 suppressed liver carcinoma cell line growth in vitro and in a xenotransplantation model without major side effects, supporting the clinical efficacy of meclizine as a repurposed anticancer drug when used in combination therapy.67
Omics Profiling and Biomarker Discovery
Biomarkers have a pivotal function in assisting diagnosis and providing guidance in precision medicine. Indeed, the identification of reliable biomarkers able to accurately predict drug responsiveness is the key for effective cancer treatments. By using molecular profiling approaches, including transcriptomics, proteomics, and metabolomics, several studies revealed the promising application of liver PDOs as a biomarker discovery tool. Using liver PDOs in combination with genome-wide RNAseq analysis, Broutier et al reported C19ORF48, UBE2S, DTYMK (for HCC), and C1QBP and STMN1 (for iCCA) as biomarkers associated with poor prognosis for PLC.27 Moreover, Li et al demonstrated that liver tumor organoids generated from high-grade tumors exhibited significantly increased extracellular vesicle secretion, suggesting that the identification of tumor-specific extracellular vesicle proteins in plasma might be a promising tool for liver cancer detection.52
Discussion and Future Directions
Similar to that described in other organs and tumor types, organoid technologies are emerging as a powerful tool to model liver diseases, elucidate molecular mechanisms underlying hepatocarcinogenesis, and screen for drug sensitivity for liver cancer treatment. Despite the encouraging results reached using this technology to date, however, important limitations are still present and should be circumvented (►Table 2). In the setting of translational research, large sample sizes are required to ensure feasible resemblance of distinct genetic characteristics among tumors, to provide reliable drug screening results. Several large collections of pancreatic68 and colorectal cancer69 PDOs have been established for omics profiling, allowing to link genetic and transcriptional information to drug responsiveness. However, current high-throughput screening results for liver PDOs are still limited by the small sample sizes.38 Therefore, the generation of liver tumor organoid biobanks is highly necessary. Another concern for applying organoid based drug screening results to the clinical setting is whether the in vitro outcomes truly represent patient responsiveness. Thus, clinical trials comparing the predictive value of the organoid treatment response to the clinical outcome of the corresponding liver cancer patients should be conducted.
Table 2.
Summary of advantages and current limitations of liver organoids for the study of liver cancer
| Advantages | Limitations |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
Liver organoids are able to perform most hepatic functions, including albumin secretion, glycogen storage, and drug metabolism.70 However, the capacity of liver organoids in terms of hepatic function is not comparable with that of the mature adult liver. Indeed, transcriptomic analysis showed that liver organoids are molecularly similar to fetal liver or hepatoblasts rather than to the adult liver.16 Thus, the current in vitro technologies require further improvements to promote a complete maturation and performance of liver organoids comparable to those of the adult liver tissues. One important reason responsible for the different functionality between normal liver and organoids might be that liver organoids fail to recapitulate the complex network of the normal liver. Emerging studies have implied the possibility to integrate the microengineered miniorganoid systems (also called as “organoid-on-a chip”) for liver organoid studies.71,72 These novel technologies may provide new tools for improved engineered liver organoids. Furthermore, development of more comprehensive liver organoids including mesenchymal, vascular, and nervous components will presumably allow a better understanding of the hepatocyte-stroma crosstalk and the effects of their interplay. This approach could be highly relevant also to dissect out the functional interaction between liver cancer cells and the tumor microenvironment.
Recent advances in immunotherapy suggest that this treatment approach might become the first-line therapy for multiple tumor types, including HCC and iCCA.73,74 One of the major limitations of liver tumor organoids is the lack of tumor infiltrating immune cells and, therefore, they may have limited value in studying immunotherapy or provide guidance in terms of immunotherapy effectiveness. In a recent study, it has been shown that tumor organoids can be reconstituted with native embedded infiltration of immune cells, including T cells, B cells, NK cells, and macrophages.75 Importantly, these reconstituted tumor organoids were able to accurately model the therapeutic features and effects of anti-PD1 and/or anti-PD-L1 antibodies.75 Although additional studies supported the use of immune-enhanced tumor organoids for immunotherapy testing,76,77 further investigation is needed on this topic.
Overall, liver organoids are an invaluable system to investigate and provide critical insights into liver cancer molecular pathogenesis, tumor diagnosis and prognosis, as well as preclinical drug testing.
Main Concepts and Learning Points.
Liver organoids can be generated from induced pluripotent stem cells or liver tissue originated progenitor cells.
HCC and iCCA organoids can be induced in normal liver organoids upon transfection with oncogenes or be isolated and cultured from human or murine liver tumor tissues.
Liver organoids as well as liver tumor organoids are valuable systems to elucidate the molecular mechanisms of hepatic carcinogenesis and investigate the therapeutic strategies against liver cancers.
Funding
This study is supported by grants from the National Institute of Health (P30DK026743, R01CA190606, R01CA239251) and West China Hospital, Sichuan University Post-Doctoral Research Project (2020HXBH006).
Footnotes
Conflict of Interest None declared.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(06):394–424 [DOI] [PubMed] [Google Scholar]
- 2.Hirohashi S, Ishak K, Kojiro M, et al. World Health Organization Classification of Tumours. In: Stanley R, Hamilton SR, Aaltonen LA, eds. Pathology and Genetics of Tumours of the Digestive System. Lyon, France: IARC Press; 2000:159–172 [Google Scholar]
- 3.Gu Q, Yu X, Chen H, Chen G. Clinicopathological features of combined hepatocellular-cholangiocarcinoma with sarcomatous change: case report and literature review. Medicine (Baltimore) 2018;97(03):e9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller M, Bird TG, Nault J-C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol 2020;72(05): 990–1002 [DOI] [PubMed] [Google Scholar]
- 5.Dhanasekaran R, Nault J-C, Roberts LR, Zucman-Rossi J. Genomic medicine and implications for hepatocellular carcinoma prevention and therapy. Gastroenterology 2019;156(02):492–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marquardt JU, Andersen JB. Liver cancer oncogenomics: opportunities and dilemmas for clinical applications. Hepat Oncol 2015; 2(01):79–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ally A, Balasundaram M, Carlsen R, et al. ; Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017; 169(07):1327–1341.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farshidfar F, Zheng S, Gingras M-C, et al. ; Cancer Genome Atlas Network. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep 2017;18(11):2780–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labib PL, Goodchild G, Pereira SP. Molecular pathogenesis of cholangiocarcinoma. BMC Cancer 2019;19(01):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho BX, Pek NMQ, Soh B-S. Disease modeling using 3D organoids derived from human induced pluripotent stem cells. Int J Mol Sci 2018;19(04):936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau HCH, Kranenburg O, Xiao H, Yu J. Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastroenterol Hepatol 2020;17(04):203–222 [DOI] [PubMed] [Google Scholar]
- 12.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345(6194):1247125. [DOI] [PubMed] [Google Scholar]
- 13.Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 2014;15(10):647–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clevers H Modeling development and disease with organoids. Cell 2016;165(07):1586–1597 [DOI] [PubMed] [Google Scholar]
- 15.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol 2016;18(03):246–254 [DOI] [PubMed] [Google Scholar]
- 16.Huch M, Dorrell C, Boj SF, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013;494(7436):247–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499(7459):481–484 [DOI] [PubMed] [Google Scholar]
- 18.Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160(1–2):299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science 2008;322(5907):1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu H, Gehart H, Artegiani B, et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 2018;175(06):1591–1606.e19 [DOI] [PubMed] [Google Scholar]
- 21.Peng WC, Logan CY, Fish M, et al. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell 2018;175(06):1607–1619.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbadawy M, Yamanaka M, Goto Y, et al. Efficacy of primary liver organoid culture from different stages of non-alcoholic steatohepatitis (NASH) mouse model. Biomaterials 2020;237:119823. [DOI] [PubMed] [Google Scholar]
- 23.Ayabe H, Anada T, Kamoya T, et al. Optimal hypoxia regulates human iPSC-derived liver bud differentiation through intercellular TGFB signaling. Stem Cell Reports 2018;11(02):306–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang RR, Zheng YW, Li B, et al. Hepatic stem cells with self-renewal and liver repopulation potential are harbored in CDCP1-positive subpopulations of human fetal liver cells. Stem Cell Res Ther 2018;9(01):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Wang Y, Cen J, et al. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat Cell Biol 2019;21(08):1015–1026 [DOI] [PubMed] [Google Scholar]
- 26.Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut 2019;68(12):2228–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broutier L, Mastrogiovanni G, Verstegen MMA, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 2017;23(12):1424–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nuciforo S, Fofana I, Matter MS, et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep 2018; 24(05):1363–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boj SF, Hwang C-I, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160(1–2):324–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161(04):933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science 2019;364(6444):952–955 [DOI] [PubMed] [Google Scholar]
- 32.Chan LH, Zhou L, Ng KY, et al. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep 2018;25(03):690–701.e8 [DOI] [PubMed] [Google Scholar]
- 33.Wong TL, Ng KY, Tan KV, et al. CRAF methylation by PRMT6 regulates aerobic glycolysis driven hepatocarcinogenesis via ERK-dependent PKM2 nuclear relocalization and activation. Hepatology 2020;71(04):1279–1296 [DOI] [PubMed] [Google Scholar]
- 34.Li M, Wang L, Wang Y, et al. Mitochondrial fusion via OPA1 and MFN1 supports liver tumor cell metabolism and growth. Cells 2020;9(01):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Wang Y, Xun X, et al. Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient-derived organoids. J Exp Clin Cancer Res 2020;39(01):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Qin W, Jiang Y, et al. ACADL plays a tumor-suppressor role by targeting Hippo/YAP signaling in hepatocellular carcinoma. NPJ Precis Oncol 2020;4(01):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong ELS, Toh TB, Lin QXX, et al. Generation of matched patient-derived xenograft in vitro-in vivo models using 3D macroporous hydrogels for the study of liver cancer. Biomaterials 2018;159:229–240 [DOI] [PubMed] [Google Scholar]
- 38.Cao W, Liu J, Wang L, et al. Modeling liver cancer and therapy responsiveness using organoids derived from primary mouse liver tumors. Carcinogenesis 2019;40(01):145–154 [DOI] [PubMed] [Google Scholar]
- 39.Saborowski A, Wolff K, Spielberg S, et al. Murine liver organoids as a genetically flexible system to study liver cancer InVivo and In Vitro. Hepatol Commun 2019;3(03):423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broutier L, Andersson-Rolf A, Hindley CJ, et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc 2016;11(09):1724–1743 [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Takeishi K, Li Z, et al. Microenvironment of a tumor-organoid system enhances hepatocellular carcinoma malignancy-related hallmarks. Organogenesis 2017;13(03):83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii M, Clevers H, SatoT. Modeling human digestive diseases with CRISPR-Cas9-modified organoids. Gastroenterology 2019;156 (03):562–576 [DOI] [PubMed] [Google Scholar]
- 43.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45(12): 1470–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artegiani B, van Voorthuijsen L, Lindeboom RGH, et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 2019;24(06):927–943.e6 [DOI] [PubMed] [Google Scholar]
- 45.Artegiani B, Hendriks D, Beumer J, et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol 2020;22(03):321–331 [DOI] [PubMed] [Google Scholar]
- 46.Yang Q, Oost KC, Liberali P. Engineering human knock-in organoids. Nat Cell Biol 2020;22(03):261–263 [DOI] [PubMed] [Google Scholar]
- 47.Li Q, Qiu J, Yang H, et al. Kinesin family member 15 promotes cancer stem cell phenotype and malignancy via reactive oxygen species imbalance in hepatocellular carcinoma. Cancer Lett 2020; 482(482):112–125 [DOI] [PubMed] [Google Scholar]
- 48.Suzuki A Evidence of cell-fate conversion from hepatocytes to cholangiocytes in the injured liver: in-vivo genetic lineage-tracing approaches. Curr Opin Gastroenterol 2015;31(03):247–251 [DOI] [PubMed] [Google Scholar]
- 49.Saito Y, Nakaoka T, Muramatsu T, et al. Induction of differentiation of intrahepatic cholangiocarcinoma cells to functional hepatocytes using an organoid culture system. Sci Rep 2018;8(01):2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rimland CA, Tilson SG, Morell CM, et al. Regional differences in human biliary tissues and corresponding in vitro derived organoids. Hepatology 2020. Doi: 10.1002/hep.31252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochiai M, Yoshihara Y, Maru Y, et al. Kras-driven heterotopic tumor development from hepatobiliary organoids. Carcinogenesis 2019;40(09):1142–1152 [DOI] [PubMed] [Google Scholar]
- 52.Li L, Qian M, Chen IH, et al. Acquisition of cholangiocarcinoma traits during advanced hepatocellular carcinoma development in mice. Am J Pathol 2018;188(03):656–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villanueva A Hepatocellular carcinoma. N Engl J Med 2019;380 (15):1450–1462 [DOI] [PubMed] [Google Scholar]
- 54.Nault JC, Cheng AL, Sangro B, Llovet JM. Milestones in the pathogenesis and management of primary liver cancer. J Hepatol 2020;72(02):209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llovet JM, Ricci S, Mazzaferro V, et al. ; SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359(04):378–390 [DOI] [PubMed] [Google Scholar]
- 56.Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 2018; 391(10126):1163–1173 [DOI] [PubMed] [Google Scholar]
- 57.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15(10):599–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RS, Qin S, Ikeda M, et al. ; IMbrave150 Investigators. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382(20):1894–1905 [DOI] [PubMed] [Google Scholar]
- 59.Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21(05):671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bu L, Baba H, Yoshida N, et al. Biological heterogeneity and versatility of cancer-associated fibroblasts in the tumor microenvironment. Oncogene 2019;38(25):4887–4901 [DOI] [PubMed] [Google Scholar]
- 61.Yuan Y, Jiang Y-C, Sun C-K, Chen Q-M. Role of the tumor microenvironment in tumor progression and the clinical applications (Review). Oncol Rep 2016;35(05):2499–2515 [DOI] [PubMed] [Google Scholar]
- 62.Chen K, Sheng J, Ma B, et al. Suppression of hepatocellular carcinoma by mycophenolic acid in experimental models and in patients. Transplantation 2019;103(05):929–937 [DOI] [PubMed] [Google Scholar]
- 63.Li L, Knutsdottir H, Hui K, et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 2019;4(02):e121490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leung CON, Tong M, Chung KPS, et al. Overriding adaptive resistance to sorafenib through combination therapy with Src homology 2 domain-containing phosphatase 2 blockade in hepatocellular carcinoma. Hepatology 2019. Doi: 10.1002/hep.30989 [DOI] [PubMed] [Google Scholar]
- 65.Saito Y, Muramatsu T, Kanai Y, et al. Establishment of patient-derived organoids and drug screening for biliary tract carcinoma. Cell Rep 2019;27(04):1265–1276.e4 [DOI] [PubMed] [Google Scholar]
- 66.Antonia RJ, Toriguchi K, Karelehto E, et al. Patient-derived organoids for personalized drug screening in intrahepatic cholangiocarcinoma. J Clin Oncol 2020;38(4_suppl):581–581 [Google Scholar]
- 67.Guan Y, Chen X, Wu M, et al. The phosphatidylethanolamine biosynthesis pathway provides a new target for cancer chemotherapy. J Hepatol 2020;72(04):746–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 2018;8(09):1112–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kondo J, Ekawa T, Endo H, et al. High-throughput screening in colorectal cancer tissue-originated spheroids. Cancer Sci 2019; 110(01):345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakabe K, Takebe T, Asai A. Organoid medicine in hepatology. Clin Liver Dis (Hoboken) 2020;15(01):3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science 2019; 364(6444):960–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skardal A, Murphy SV, Devarasetty M, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep 2017;7(01):8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hilmi M, Vienot A, Rousseau B, Neuzillet C. Immune therapy for liver cancers. Cancers (Basel) 2019;12(01):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hou J, Zhang H, Sun B, Karin M. The immunobiology of hepatocellular carcinoma in humans and mice: basic concepts and therapeutic implications. J Hepatol 2020;72(01):167–182 [DOI] [PubMed] [Google Scholar]
- 75.Neal JT, Li X, Zhu J, et al. Organoid modeling of the tumor immune microenvironment. Cell 2018;175(07):1972–1988.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Votanopoulos KI, Forsythe S, Sivakumar H, et al. Model of patient-specific immune-enhanced organoids for immunotherapy screening: feasibility study. Ann Surg Oncol 2020;27(06):1956–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hai J, Zhang H, Zhou J, et al. Generation of genetically engineered mouse lung organoid models for squamous cell lung cancers allows for the study of combinatorial immunotherapy. Clin Cancer Res 2020. Doi: 10.1158/1078-0432.CCR-19-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]


