Significance
With geological sea-level fluctuations driven by climate change, the distribution of mangrove forests has expanded and contracted through time. We studied an inland, isolated mangrove forest located 170 km away from the nearest coastline in the interior of the rainforests of the Yucatan Peninsula (Mexico). Combining multiple lines of evidence, we demonstrate that this extant forest is a relict from a past, warmer world when relative sea levels were 6 to 9 m higher than at the present. Our finding highlights the extensive landscape impacts of past climate change on the world’s coastline and opens opportunities to better understand future scenarios of relative sea level rise.
Keywords: mangrove, freshwater, interglacial, Pleistocene
Abstract
Climatic oscillations during the Pleistocene played a major role in shaping the spatial distribution and demographic dynamics of Earth's biota, including our own species. The Last Interglacial (LIG) or Eemian Period (ca. 130 to 115 thousand years B.P.) was particularly influential because this period of peak warmth led to the retreat of all ice sheets with concomitant changes in global sea level. The impact of these strong environmental changes on the spatial distribution of marine and terrestrial ecosystems was severe as revealed by fossil data and paleogeographic modeling. Here, we report the occurrence of an extant, inland mangrove ecosystem and demonstrate that it is a relict of the LIG. This ecosystem is currently confined to the banks of the freshwater San Pedro Mártir River in the interior of the Mexico–Guatemala El Petén rainforests, 170 km away from the nearest ocean coast but showing the plant composition and physiognomy typical of a coastal lagoon ecosystem. Integrating genomic, geologic, and floristic data with sea level modeling, we present evidence that this inland ecosystem reached its current location during the LIG and has persisted there in isolation ever since the oceans receded during the Wisconsin glaciation. Our study provides a snapshot of the Pleistocene peak warmth and reveals biotic evidence that sea levels substantially influenced landscapes and species ranges in the tropics during this period.
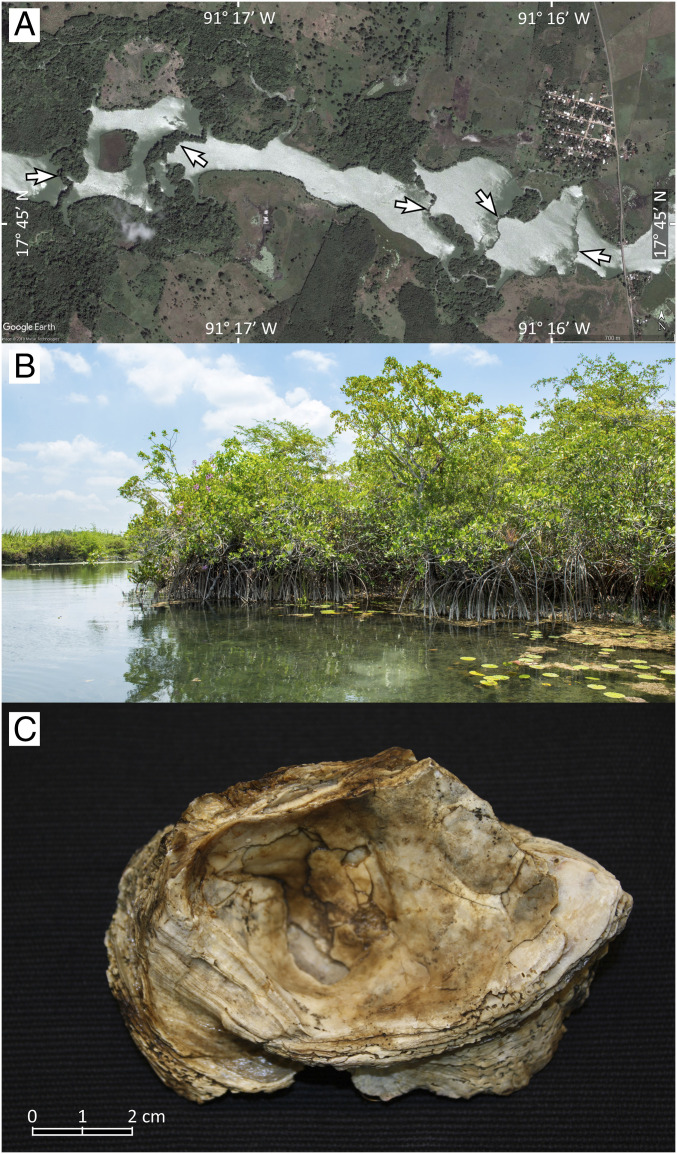
Most of the San Pedro Mártir River (hereafter referred to as the San Pedro River) flows within the Yucatan Platform, a massive karst (limestone) block that forms the Yucatan Peninsula and is composed in this region mostly of calcite of Paleocene and Eocene origin (33 to 66 Ma) (1). The San Pedro River runs in a northwest direction from its sources in the El Petén rainforests in Guatemala to the Reforma Waterfalls in Tabasco, Mexico (17°45'20” N, 91°18'02” W). At that point, the river descends in elevation abruptly, from ca. 17 m above current sea level to ca. 5 m downstream as it leaves the Yucatan Platform and descends into the sedimentary Tabasco Lowlands. From Reforma upstream, the river is crossed by tufa dams, formed by deposits of calcium carbonate precipitated from calcium-saturated waters of the river (SI Appendix, Table S1) (2). The tufa dams interrupt the water flow and form a chain of terraced and slow-flowing lakes (Fig. 1). Below the Reforma Waterfalls, the river drains first into a series of alluvial terraces of pebble conglomerates and sand, mostly of Pleistocene origin (3, 4), and, further downstream, into the Tabasco Lowlands, formed by Holocene sediments of alluvial, fluvial, or palustrine origin (5).
Fig. 1.
Geomorphology and geology of the San Pedro River. (A) Aerial image of the Lower San Pedro River 1 km upstream from the Reforma Waterfalls showing (in white arrows) the tufa dams that traverse its course (image © Google Earth). (B) A stand of Rhizophora mangle on the banks of the Upper San Pedro River with the Mayan water lily (Nymphaea ampla, a freshwater plant) growing with the mangroves. (C) Fossil specimen of a Crassostrea oyster found on clayey sediments of an ancient lagoon bottom, around 10 m above current sea level.
Red mangroves (Rhizophora mangle) occur along the karstic banks and lakes of the San Pedro River between the Reforma Waterfalls and Guatemala’s El Petén Department, where they enter some 8 km past the Tabasco–Guatemala border on to the community of San Andrés. Downstream of the Reforma Waterfalls, they do not occur again for 170 km until the Usumacinta estuary, where the regional watershed drains into the Gulf of Mexico. The constrained distribution of mangroves in the karst part of the San Pedro River is also known in other calcium-rich substrates (6). In addition to their ability to grow on seawater, red mangroves can establish and grow well in calcic waters without any need for seawater (7). Their capacity to cope with calcic substrates is possibly related to R. mangle’s ability to exclude ions at the root level, a mechanism that allows them to cope with high salinity water but also with excess calcium (8). Phosphorus immobilization, and not substrate salinity, is the most important factor limiting red mangrove growth in calcium-rich substrates (9, 10). Red mangroves, in short, can live and reproduce in nonoceanic karstic environments if they can colonize them, but the question remains how these obligate ocean-dispersed plants got so deep inland initially.
We hypothesized that if red mangroves reached the San Pedro River above the Reforma Waterfalls at some point in the past and then from there dispersed upstream along the river’s karstic system of lakes and dams, sea ingression during past Pleistocene interglacial events must have left an imprint in the plants’ genomes and in the local geology. In order to test this hypothesis, we used information from different analyses, including a) a population genetic analysis using single-nucleotide polymorphisms (SNPs) of the San Pedro River mangrove population and of other populations along the coasts of the Yucatan Peninsula to infer the demographic history and divergence time of the San Pedro River mangrove population; b) a study of the sediments in the Pleistocene coastal terraces that surround the lower part of the San Pedro River to assess for geological evidence of past sea incursions; c) a floristic survey of the plants that grow on the banks of the San Pedro River, looking for other plant species with coastal habitat affinities; and d) a Geographic Information System (GIS) analysis of the level of flooding of the Yucatan Peninsula under different sea level rise scenarios.
Results
Population Genetic Analysis.
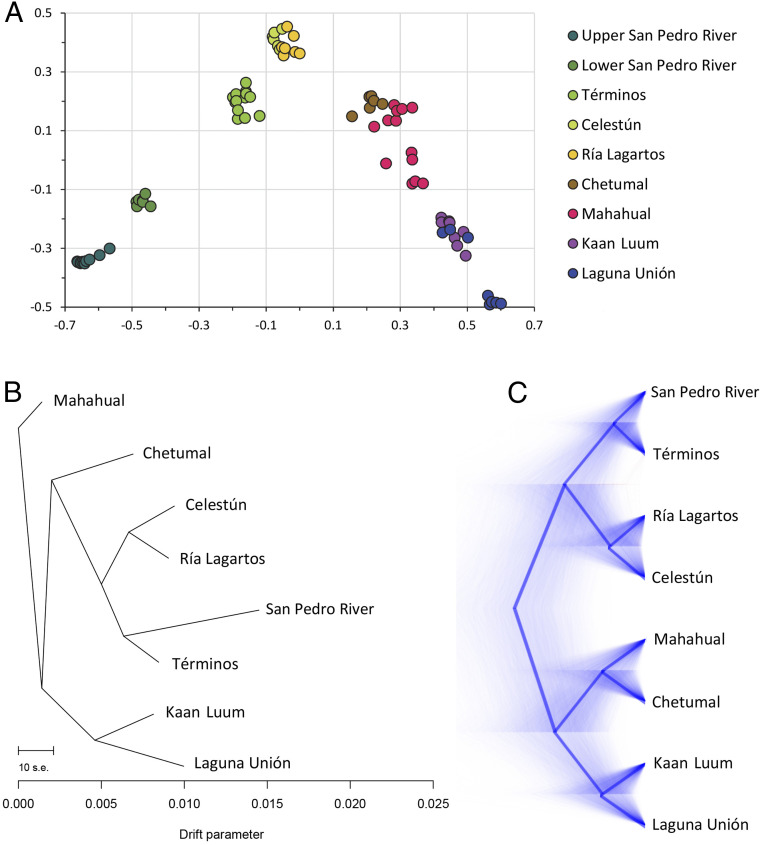
Using principal component analysis (PCA, Fig. 2A), we visualized the genome-wide variation of 79 trees in 11 sites around the Yucatan Peninsula (SI Appendix, Table S2 and Fig. S1). The analysis yielded two eigenvectors capturing more variance than predicted by chance alone according to the broken-stick test. Eigenvector 1 (20.2% of total variation) separated mangrove populations along a longitudinal axis (west to east, Gulf of Mexico sites to the left, and Caribbean mangroves to the right) and was significantly correlated with longitude (r = 0.88, P < 0.0001). Similarly, eigenvector 2 (10.7% of total variation) roughly separated individuals along a latitudinal axis (north to south) and was significantly correlated with latitude (r = 0.42, P = 0.0001). All individuals from the San Pedro River clustered together, distinctly separated from coastal populations but nearest to Términos Lagoon in the Gulf of Mexico. Similarly, mangroves from the two inland lagoons derived from large karstic sinkholes, Laguna Unión and Kaan Luum, also clustered together, genetically distant from their nearest coastal locality, Mahahual. Mangroves in the San Pedro River (and those in isolated inland lagoons in the Yucatan Platform) are genetically distinct and have differentiated in isolation from the nearest red mangrove populations along the coasts. No significant differences among the eigenvectors of datasets subject to different filtering criteria were found in the sensitivity analysis, suggesting that the observed results are robust (SI Appendix, Tables S6 and S7).
Fig. 2.
Population genetic analysis. (A) Statistical summary of genetic data from 77 individuals based on principal component axis one (eigenvector 1) and axis two (eigenvector 2). Individuals are colored according to sample locality. (B) Pattern of genetic ancestry among R. mangle populations in the Yucatan Peninsula inferred using TreeMix: maximum likelihood population tree displaying relationships between populations assuming no migration events. (C) Pattern of genetic ancestry among R. mangle populations in the Yucatan Peninsula inferred using SNAPP: claudogram of all loci trees is displayed in the background and a consensus tree in thick lines in the foreground.
A population tree using a TreeMix analysis confirmed that the San Pedro River population is more closely related to the mangroves in Términos Lagoon (Fig. 2B). This result indicates that the population from Rio San Pedro descend from a common ancestor with the population from Términos Lagoon. These two sites, in turn, are related to the two nearest sampling sites, Celestún and Ría Lagartos, in the northern part of the Peninsula. In this analysis, the mangroves in the two inland lagoons (Kaan Luum and Laguna Unión) near the Caribbean coast showed as distinctly different from all other populations. This tree explains 91% of the variance in relatedness between populations. We examined the residuals of this model to identify aspects of ancestry not captured by the bifurcating tree (SI Appendix, Fig. S2A). Some likely admixed populations stand out in this analysis, in particular Ría Lagartos in the boundary between the Gulf of Mexico and the Caribbean, where local mangroves have genetic traits of both large marine ecosystems. We sequentially added migration edges to the tree and found that four migration edges are enough to explain the remaining variance (χ2 = 5.834, degrees of freedom = 1, P = 0.01; SI Appendix, Fig. S2B). All migration edges are statistically significant (P values < 1 × 10−5), and the admixed tree explained 98.5% of the variance in relatedness between populations. In ecological terms, the presence of significant migration edges means that several populations show signs of genetic admixture. Given that our sampling across the Yucatan coastline is not exhaustive, we hesitate to overinterpret this result, except to note that the relationship among Gulf Coast, Caribbean, and inland lagoon populations of R. mangle in the region likely involves complex patterns of gene flow that merit further investigation.
The coalescent model (using the SNAPP package) to reconstruct the genealogical tree of population splits among all sampled localities gave results consistent with both the PCA and the TreeMix admixture graph in which the mangroves from the San Pedro River appear as a sister population to those from the Términos Lagoon. Like the PCA, this analysis distinctly separated the Gulf of Mexico from the Caribbean populations and maintained the two inland lagoons as a separate, isolated population within the Caribbean coast (Fig. 2C). Together, these results indicate that gene flow in red mangroves is strongly spatially structured across the Yucatan Peninsula.
Consistent with the tree-based analyses, the nontree-based clustering analysis using Rmaverick also showed that individuals from the San Pedro River share genetic ancestry with individuals from the Gulf of Mexico rather than the Caribbean (SI Appendix, Fig. S3). At a clustering level of five groups (K = 5), most individuals from the San Pedro River belong to group 3, with some individuals sharing ancestry with individuals from Términos Lagoon in group 2. This analysis suggests that the eight sampling localities correspond to five “genetic populations” in which individuals from a given population may show admixture with geographically close sampling localities. The overall pattern of the groups shows marked geographic structure, with one set of populations in the Gulf of Mexico (groups 2 and 3) and the other set in the Caribbean (groups 1, 4, and 5).
Using a mean generation time of 100 y (Methods and SI Appendix, Table S3) and a median mutation rate of 3.16 × 10−9, we found an estimated divergence time (i.e., the time since the two populations shared a common ancestor) of 98 ka with a confidence range of 5 to 273 ka between the San Pedro mangroves and their sister lineage in Términos Lagoon. Despite the uncertainty surrounding estimates of absolute divergence time from genomic data as well as the ongoing sorting of ancestral alleles in these populations, our results are consistent with the hypothesis that the population of the San Pedro River is a relict from the Last Interglacial (ca. 120 K years). Variations in the assumptions of generation times and mutation rates still produced time ranges that included the Last Interglacial within the range (SI Appendix, Table S4). Furthermore, the coalescence model indicates that the San Pedro River population underwent a strong reduction in effective population size after becoming isolated from other populations (SI Appendix, Fig. S4).
Geologic Exploration.
Five of our eight sampling sites in the Pleistocene terraces (SI Appendix, Fig. S1) that surround the lower part of the San Pedro River in Balancán, Tabasco, showed loose conglomerates of polymictic rounded gravel. The uniform size of the pebble clasts, their rounded shape, and the horizontal regularity of the sediment profile suggest a marine rather than an alluvial origin (11). The presence of a water-eroded marine gastropod shell in site 5 strongly supports an oceanic origin for these gravelly terraces (although too eroded for identification, the conical shape and grooved external structure suggest a species within the clade Neogastropoda). These pebble deposits are currently being quarried for construction gravel and are clearly visible on aerial images. They cover a band 25-km long and around 2-km wide (i.e., ca. 5,000 ha in area) between the Yucatan Platform and the lowland sediments.
Two sites, found northwards in a flat, rolling plain currently dedicated to pasturelands because of the low fertility of the soil, had a substrate of quartzite sand where the lack of distinguishable soil layering suggests aeolian deposition in a system of coastal paleo-dunes. Finally, the presence of a clayey sediment rich in shell fragments and large oyster shells unequivocally confirms the presence of an ancient coastal lagoon in site 7 near the Reforma Waterfalls (Fig. 1C). The shells are identical to those of modern Crassostrea virginica, the Atlantic oyster common in estuaries around the Gulf of Mexico. This site lies 14 m above current sea level and, taking into consideration that the oyster layer was found 4 m below the ground surface in a dugout well, it shows that a paleo-coastal lagoon bed exists here around 10 m above current sea level, confirming that ocean intrusion reached the Lower San Pedro River (Fig. 1C, Fig. 3B, and SI Appendix, Fig. S5). Taken together, our geological findings point strongly to a past marine ingression in the lower basin of the San Pedro River adjacent to the relictual mangrove population.
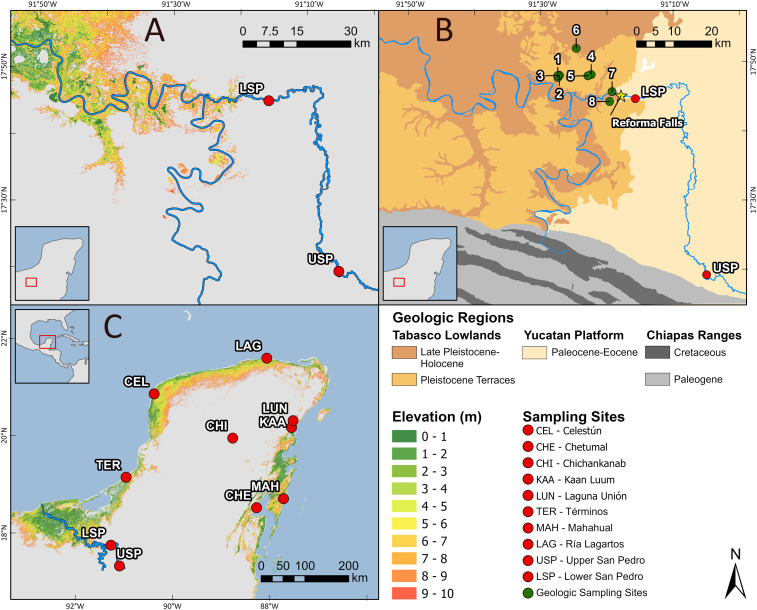
Fig. 3.
Sampling sites, geology, and sea level models. (A) Sea level rise model of the San Pedro River region showing marine ingression under different scenarios of sea level flooding height. (B) Geologic map of the San Pedro River region showing the sedimentary late Pleistocene terraces and the Paleocene–Eocene limestone karst of the Yucatan Platform, where the mangrove relicts survive. The numbered dots indicate sampling sites for geologic analysis, described in SI Appendix, Table S5. (C) Sea level rise model for the whole Yucatan Peninsula showing marine ingression under different flooding scenarios. The labeled dots indicate sampling sites for molecular analysis, and the numbered dots indicate geologic sampling sites (SI Appendix, Fig. S1 and Tables S2 and S5).
Floristic Survey.
The total riparian flora of the San Pedro River is composed of 359 species grouped in 87 families that include ferns, gymnosperms, and angiosperms. Of these, a subgroup of 112 species in 51 families (31% of the total flora) belong to coastal taxa, commonly distributed in environments such as coastal lagoons or beachfronts. The most species-rich families in this group are the orchids (Orchidaceae) and the legumes (Fabaceae) with 10 coastal species each as well as the sedges (Cyperaceae) with six species and the bromeliads (Bromeliaceae) with five species of coastal distribution. Within this group of species that are preferentially distributed along tropical ocean coasts, the river harbors some species that are obligate or quasi-obligate coastal dwellers, such as the golden leather fern (Acrostichum aureum), the mangrove orchid (Myrmecophila tibicinis), the Barbados sea grape (Coccoloba barbadensis), the Everglades palm (Acoelorrhaphe wrightii), the coastal rosewood (Dalbergia brownei), and the buttonwood mangrove (Conocarpus erectus). Hence, the San Pedro River contains, apart from the red mangrove itself, a large number of coastal species that form a relict ecosystem of past ocean intrusions.
Sea Level Modeling.
The ocean ingression simulation on the digital model of the terrain showed that a flooding of ca. 9 m above current sea level would reach the Pleistocene terraces around the Reforma Waterfalls, where the marine fossils were found, inundating most of the Tabasco Lowlands (Fig. 3).
Discussion
Two studies (12, 13) have described red mangrove ecosystems along the Mexican Caribbean coast separated from the sea by sandbars as “fossil lagoons,” implying that they are thought to have been connected to the sea in the past. Other authors (14) have described an inland red mangrove ecosystem in Barbuda some 15 km from the coast, around 6 to 9 m above the current level of the sea, associated with inland lithified beach ridges of Pleistocene origin, and have advanced the hypothesis that they might represent a vegetation relict of the period of formation of the ridges. A similar study (15) on the inland mangroves of Inagua (Bahamas) noted that inland mangroves in Caribbean islands are always found in karstic, calcium-rich environments. Woodroffe described the occurrence of a unique stand of oriental mangrove Bruguiera gymnorrhiza (Rhizophoraceae) on a freshwater spring 24 to 37 m above current sea level on a terrace that has been dated as formed during the Last Interglacial at Christmas Island, Indian Ocean and discussed the hypothesis that this stand is a coastal lagoon relict that has persisted at this freshwater site since then (6). Finally, a recent study on community structure of swamp forests throughout the larger Usumacinta River basin in Mexico (16) described the presence of an “isolated, relictual population” of Rhizophora mangle along the limestone banks of the San Pedro River, underscoring the urgent need to preserve these mangrove stands and their rich associated flora.
Our findings at the San Pedro River confirm these hypotheses and bring detail to the time of establishment of this inland mangrove forest. The population genetic analysis confirms that San Pedro River mangroves are a relict of a coastal ecosystem that colonized the river’s tufa lakes, possibly during the Last Interglacial, and stayed behind along the riverbanks after the oceans receded during the Wisconsin glaciation. The genetic data indicates the presence of two quite distinct lineages of R. mangle: a Gulf of Mexico population and a Caribbean population with the two lagoons in the north of the Yucatan Peninsula, Celestún and Ría Lagartos, showing transitional traits. The San Pedro River population is clearly derived from mangroves in the Gulf of Mexico, but it is interesting to note that the two inland lagoons we sampled in the Caribbean coast (Kaan Luum and Laguna Unión) were also markedly distinct from the truly coastal populations, suggesting that these lagoons might also be Eemian relicts and that the descriptor of “fossil lagoons” used by other scientists is possibly correct. More research remains to be done to understand the formation and timing of these sinkhole lagoons.
Studying fossilized coastal coral reefs in the Yucatan Peninsula, Blanchon et al. (17) found evidence for a rapid sea level rise event during the Last Interglacial, implying an episode of rapid ice loss with sea level rise rates higher than 36 mm ⋅ yr−1. At such rates, the Tabasco Lowlands could have become flooded under ocean water on the scale of centuries, not millennia. The mangrove ecosystem must have colonized the shifting coasts in only a few generations. The Last Interglacial flooding hypothesis is also in agreement with the fact that Chichankanab, another inland karst lagoon similar to Kaan Luum and Laguna Unión but separated from the Caribbean coast by a 40-m-high ridge, does not harbor red mangroves.
The “bathtub” flooding model used in our study does not include dynamic processes such as tidally forced direct ocean flooding, seasonal wave inundation, and coastal erosion. These factors could have increased substantially the transgression of the ocean along the San Pedro River during the LIG (18), facilitating mangrove dispersal upriver. Although the Yucatan Platform has experienced little tectonic uplift (19), seismicity (17), or neotectonic activity (20) during the Pleistocene, the Gulf of Mexico region is located in the forebulge of the North American ice sheets and, as a result, may have experienced substantial glacial isostatic adjustments during the Eemian (21). Furthermore, the Balancán terraces are on the transition between the limestone of the Yucatan Platform and the sedimentary basin of the Tabasco Lowlands that formed during the Holocene. The possibility exists that the massive accumulation of sediments could have produced some elevational changes on the basin’s edges, where the Balancán terraces lie. Despite these potential sources of variation, the 10-m elevation of the fossil lagoon sediments we found coincides closely with the 6- to 9-m relative sea level rise reported by other studies for the Eemian highstand (21, 22).
The karstic plains around the San Pedro River were severely deforested during the 1970s as part of a governmental plant to develop tropical cattle husbandry. Because of their swampy nature, the banks of the river were spared and still maintain large tracts of their original vegetation. Our findings reveal the importance of this ecosystem to understand the migrations of coastal plants during the Pleistocene and the extensive impacts of past interglacials on the line of the world’s coasts.
Methods
Field Methodology for Mangrove DNA Analysis.
At each site (SI Appendix, Table S2), single leaf samples were collected from ca. 10 individual red mangrove, Rhizophora mangle, trees. These samples were later bagged with silica gel for preservation.
Coastal sites.
Five sites, Términos Lagoon, Chetumal, Mahahual, Celestún, and Ría Lagartos, were chosen to represent coastal red mangrove populations across the Yucatan Peninsula. This way, we could have a genetic representation of the entire region to assess general genetic diversity among different coastal mangrove populations and be able to compare them with the river populations.
Additionally, samples were collected and analyzed in two inland lagoons created by giant cenotes (karstic sinkholes) near the Caribbean coast of the state of Quintana Roo: Laguna Unión and Kaan Luum. These lagoons contain large, isolated stands of red mangrove in freshwater, like the San Pedro River, but much closer to the coastline and with a freshwater level only a few centimeters higher than the mean level of the sea. These samples serve to provide comparison to a third category of mangroves of the Yucatan Peninsula, those found in isolated inland waters with a relatively close proximity (<20 km) to the ocean.
Chichankanab exploration.
We visited the inland Chichankanab Lagoon, a Ramsar site in the middle of the Yucatan Peninsula formed by a long and narrow freshwater lake that runs along a fault in the karstic platform some 140 km from the Caribbean coast. This lake has been cited as harboring red mangroves (23), but no herbarium specimens or photo document exist to validate this claim. The goal of our visits to the lagoon was to verify the presence of R. mangle and, if found, to collect herbarium specimens and leaf samples.
San Pedro River exploration.
Similar to our exploration of the Chichankanab Lagoon, we visited the Upper and the Lower San Pedro River to validate reports on the presence of red mangroves along the river, some 140 km from the nearest coast at the Gulf of Mexico.
Red Mangrove Generation Time.
We selected two sites in the coastal banks of Laguna del Cacahuate near the Mexico–Guatemala border, one of the places along the San Pedro River with the most pristine mangrove stands (17°17'59” N, 91°05'08” W). Each site was ∼4,000 m2 in size, and in each one, we selected the three largest (and presumably oldest) trees. In each tree, we searched for the primary stem (because of the stilt root growth of red mangroves, a tree can have multiple secondary stems) and, using a Haglöf increment borer (Haglöf) with a 350-mm bit and 4.3-mm core diameter, we drilled the trunk at ca. 1 m aboveground until we reached the pith at the center. We then extracted the full core and separated a 3-mm section of the innermost core containing the pith. The pith samples were oven dried at 60 °C in preparation for radiocarbon analysis.
Radiocarbon dating was performed at the Keck Carbon Cycle Accelerator Mass Spectrometry Facility in the Earth System Science Department at the University of California, Irvine. Wood shavings of the pith samples were pretreated using sonication (ultrasound bath) and then with a standard acid–base–acid protocol (24) in order to remove mobile wood fractions. Finally, they were processed in the mass accelerator following standard radiocarbon protocols. Results from the Keck laboratory were delivered as a fraction of the Modern Standard D14C and conventional radiocarbon age following Stuiver and Polach (25). We then calibrated the results using the Calibomb online application (http://calib.org/CALIBomb/) to transform radiocarbon dates into calendar ages. All dates were calculated in Gregorian, or calendar, years and then were converted to tree ages (years B.P.) using 2019 (the year of the analysis) as the reference point. Because the samples were taken 1 m aboveground, 10 y were added to the estimated age under the assumption that it takes around a decade for mangrove seedlings to establish and start forming a woody stem at that height (SI Appendix, Table S3).
Generation time was estimated as the mean age at which members of a given cohort are expected to reproduce. If lx is the survivorship function (probability that an individual survives to age x) and mx is the age-specific fecundity, then the cohort’s age-specific reproductive function (kx) is defined as kx = lx × mx, and generation (T) is calculated as (26)
Because the survivorship function and the age-specific fecundities for R. mangle are not known, we made the following assumptions: a) the probability of survival from 1 y to the next was constant (q) for all ages in reproductive plants, b) fecundity (estimated as the number of propagules a plant produces every year) is proportional to the size of reproductive plants’ canopy, and c) based on our measures of diameter and age, we estimate that red mangroves start reproducing at around 30 y of age, and fecundities will increase linearly with age as the canopy grows until the plants reach 60 y. From that age onwards, we assumed that fecundities are constant until the trees die.
If annual survivorship probabilities are constant for all ages (q), then the survivorship function simplifies to lx = qx, and the reproductive function becomes kx = qx × mx. To get an estimate of annual survivorship, we used our radiocarbon longevity estimates in which some of the older adult plants reached ages close to 300 y. Based on this fact, we numerically tried different values of q in order to get a survivorship curve with ca. 1 to 2% of any given cohort reaching 200 y and a much lower probability (ca. 0.1%) of plants in any cohort reaching 300 y or more. The age-specific fecundity, mx, was calculated as a linearly increasing function between 30 and 60 y of age, stabilizing to a constant value after that point. Different values of q gave, in all cases, estimates of generation times very close to 100 y, so we took this value as our working estimate of generation time.
Population Genomics Analyses.
To investigate the demographic history of the red mangroves in the Yucatan Peninsula, we sequenced SNPs across the genome from 79 mature trees in four main study regions: two sites at San Pedro River, three sites in the Gulf of Mexico, three sites in the Caribbean coast of the Yucatan Peninsula, and two inland lagoons 15 km inshore of the Quintana Roo coastal area (Fig. 2 and SI Appendix, Table S2).
Genomic DNA was extracted from ground leaf tissue using a modified CTAB (cetlytrimethylammonium bromide buffer) protocol (27), which included up to five prewashes until the supernatant was clear. The final concentration of DNA was measured by using a Qubit fluorometer v.3.0 (Invitrogen by Thermo Fisher Scientific). The quality of DNA was quantified using an Agilent 2200 TapeStation (Agilent Technologies). We used genotype by sequencing (GBS) to characterize genetic variation in SNPs across all samples. To prepare the libraries, we used the ApeK1 enzyme (New England Biolabs) to digest the DNA and followed the protocol in Elshire et al. for subsequent steps (28). Fragments were sequenced as 100 bases paired-end reads on an Illumina HiSeq 4000 at the University of California, Los Angeles, High-Throughput Sequencing Broad Stem Cell Research Center.
The raw GBS reads were processed using iPyrad (29) v0.7.28 using the MiniConda environment containing MUSCLE (30) v3.8.31 and VSEARCH v2.7.0 (31). To reduce the chance of false homology, we assembled the reads de novo using different clustering thresholds and maximum coverage values above which clusters were excluded. Results did not differ across runs, so we chose a conservative approach and applied 0.90 and 8,000 for the clustering threshold and maximum coverage, respectively, to generate the assembly that we used for downstream analyses. Full parameters can be found in the parameter file included in a public repository at https://github.com/zapata-lab/ms_rhizophora. The de-multiplexed fastq files were submitted to the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA692272 (https://www.ncbi.nlm.nih.gov/bioproject/692272).
We applied multiple filters to limit the amount of missing data and ensure the high quality of SNPs in downstream analyses. Processed data from iPyrad were filtered using VCFtools v0.1.14 (32) following the approach outlined below. Scripts to reproduce these filtering analyses can be found in a public repository at https://github.com/zapata-lab/ms_rhizophora. This repository includes our own scripts and modified scripts from Pina-Martins et al. (33).
From the full matrix assembled in iPyrad, we required that each SNP had to be represented in at least 80% of the individuals, and each individual had to be represented in at least 75% of the SNPs. We further required that the minimum allele frequency (MAF) of each SNP had to be at least 0.03 to be retained. To minimize the effects of linkage disequilibrium, we used only one SNP per locus by discarding all but the SNP closest to the center of the sequence in each locus. Additionally, we retained only biallelic loci. Finally, we did a sensitivity analysis on the filtering thresholds used by repeating the multivariate analysis with different filtering thresholds (60, 70, 80, and 90% for SNPs, 25, 50, and 75% for individual plants, and 0.01, 0.03, and 0.05 for MAF). A detailed description of the sensitivity analysis is provided in SI Appendix, Tables S6 and S7.
Demographic and Population Genomic Analyses.
To assess patterns of genetic variation among sampling localities, we used PCA (34) based on the genetic covariance matrix from genotypes using SNPRelate (35). We used eigenvectors 1 and 2 to produce a visual summary of the SNP data in fewer dimensions.
To evaluate the hypothesis that the R. mangle population from San Pedro River is more closely related to populations from the Gulf of Mexico coast rather than to the Caribbean coast of the Yucatan Peninsula, we reconstructed population genealogies using two approaches specifically designed for SNP data. First, we used TreeMix v1.12 to infer maximum likelihood admixture trees based on the observed variance–covariance matrix of allele frequencies (36). This approach is especially powerful at the population level because it allows population splits in the presence of postdivergence admixture/migration. The resulting population tree has branch lengths proportional to the amount of genetic drift per branch. With this approach, inference is based on “shared genetic drift” between sets of populations under the premise that shared drift implies a shared evolutionary history. We added stepwise migration edges to each tree and used a likelihood ratio test to assess model fit. Additionally, we performed a round of global rearrangements to the population tree and calculated the SE of migration edges.
Second, we inferred a population tree using a finite-sites mutation model per locus, integrating information over all possible loci/gene trees. For this analysis, we used the Bayesian Markov Chain Monte Carlo (MCMC) sampler SNAPP (37), which we ran through the BEAST2 package (38). SNAPP uses a Yule prior distribution for the population tree and its parameters. This prior has a single parameter (λ) representing the branching rate. We used a Γ distribution as hyper prior to accommodate uncertainty in λ. To set the parameters α and β describing the Γ distribution, we used the pylue Python script (https://github.com/joaks1/pyule) to calculate the expected height of the population tree. Based on this analysis, we used α = 1 and β = 3,000. We ran six independent MCMC chains for 1M generations, sampling every 200 samples each, which were combined for final parameter estimation. Based on convergence analysis of the likelihood trace over generations, we used a burn-in of 20%.
In addition to tree-based approaches, we assessed whether the R. mangle individuals from the San Pedro River shared a higher proportion of genetic ancestry with the Gulf of Mexico rather than Caribbean individuals using Rmaverick (39). This program is an implementation of a model-based clustering algorithm to assign individuals to populations on the basis of their genotypes while simultaneously estimating population allele frequencies. Rmaverick is more accurate than alternative approaches like STRUCTURE (40) to estimate the number of subpopulations (K) because it uses thermodynamic integration rather than cross entropy that has larger SEs (38). We ran the Rmaverick MCMC sampler exploring values of K from 1 to 8, using 30,000 sampling iterations with 20% burn-in and 20 rungs.
To investigate whether the divergence time between the R. mangle population from the San Pedro River and its closest relative at Términos Lagoon overlapped with the estimated time of the Last Interglacial, or Eemian period (ca. 120 K years), we sampled divergence times for nodes in the population tree during the SNAPP tree estimation. The nodes in this tree correspond to the most recent common ancestor of descendant populations. SNAPP also samples population size estimates for ancestral and extant populations. Under the hypothesis that the San Pedro River is a relict of the Eemian, we predict a reduction in the population size since this population diverged from its sister population. From the SNAPP model, we obtained the divergence time parameter in coalescent units (τ, the mean number of substitutions per individual) as well as the population size parameter (Θ, the total number of substitutions in the population). Scaling these parameters by mutation rate (µ) and generation time (g), divergence times (T) and effective population sizes (Ne) can be obtained using the formulae τ = T.µ/g and θ = 4Ne.µ. Generation times were estimated as described previously. Average mutation rates for SNPs (41) vary between µ = 10−8 and µ = 10−9 mutations per nucleotide per generation. These values are consistent with average estimates of mutation rates across organisms (42). Because mutation rates are probabilities, we used the geometric median as our central estimate (µ = 3.16 × 10−9) and then tried different values along the range to test whether the CIs for divergence time included the time of the Last Interglacial (SI Appendix, Table S4).
The scripts and files to reproduce all demographic and population genomic analyses can be found in a public repository at https://github.com/zapata-lab/ms_rhizophora. This repository includes our own scripts and a script from Daren Card (2015, https://github.com/darencard/RADpipe/blob/master/treemixVarianceExplained.R) to estimate the variance explained by each TreeMix model.
Geology of the San Pedro River.
On October 17 and 18, 2018, we made observations and took samples in eight sites (SI Appendix, Table S5 and Fig. S1) within the Balancán terraces. In all sites, with the exception of site 7, samples were collected and bagged from the upper 2 to 3 m of sediments. In site 7, located in the transitional escarpment between the limestone of the Yucatan Platform and the lower terraces, samples were collected from a dugout well at 4 to 5 m depth in a clayey layer rich in shell fragments buried under an upper surficial layer of loose limestone rubble. Because of the abundant presence of rounded gravel in a large region where it is elsewhere absent, the terraces around the San Pedro River are strewn by open quarries from where gravel is extracted. Five sites (1, 2, 3, 5, and 8) were sampled in open quarries. Site 4 was a 2-m deep water hole for cattle. Finally, in site 6, a sandy, gently undulating plain, soil observations were done by digging a soil observation pit and sampling the profile to 1.5 m deep. Samples were collected for biological identification of fossil marine shells and for macro- and microscopic observations in the laboratory. The elevation of the sampling sites was determined using NASA’s ICESat-2 data, consulted through the map interface “OpenAltimetry” (https://openaltimetry.org/) developed by Scripps Institution of Oceanography (ICESat-2 elevations are measured relative to the WGS84 ellipsoid datum). The exact elevation of the satellite measurements nearest to the sampling point were downloaded and used for interpolation.
Additionally, two water samples were collected from the Lower San Pedro River near the Provincia Bridge (17°44'52.25” N, 91°15'46.04” W), 4 km east of the Reforma Waterfalls. The samples were analyzed in the Environmental Sciences Research Laboratory of the University of California, Riverside for main cations (Ca2+, Mg2+, Na+, K+) and anions (Cl−, SO42−). Bicarbonate (HCO3−) was estimated indirectly as the difference between cations and anions.
Geographic Analyses and Maps.
We downloaded the INEGI (Instituto Nacional de Estadística, Geografía e Informática) digital geologic map (1) of the region and simplified merging areas of similar geologic age and origin to highlight the presence of a) the Paleocene–Eocene limestones of the Yucatan Platform where the San Pedro River runs, b) the Pleistocene terraces where the marine fossils were found, and c) the geologically recent Holocene sediments that are part of the Tabasco Lowlands.
To explore the consequences of relative sea level rise across the Yucatan Peninsula during past interglacials, a digital elevation model (DEM) for the Yucatan Peninsula, based on Shuttle Radar Topography Mission (SRTM) data, was downloaded from the Consultative Group on International Agricultural Research (CGIAR) online database (43). SRTM along-track elevations are also measured above the WGS84 ellipsoid datum (ITRF2014 reference frame). The average accuracy of SRTM altimetry estimates compared to ICESat measurements is ±1.03 m (44). SRTM error, however, increases with the slope of the ground surface, so for the area of our study with a very flat topography, the error of our DEM is less than 1 m. Using the CGIAR DEM data and a GIS, we simulated flooding levels of the region, assuming different values of relative sea level rise up to 10 m in elevation, in 1-m intervals. The results were presented in the form of a map.
List of Vascular Plant Species from San Pedro Mártir River, Tabasco, Mexico.
The main course of the San Pedro River was surveyed from the Mexico–Guatemala border in the municipality of Tenosique to the confluence with the Usumacinta River in the municipality of Balancán by water. Reconnaissance on foot was also conducted in sites where the conditions allowed, and lagoons linked to the river were also explored.
Plants growing on the shore of the river and lagoons up to 50 m away from the water were collected, including rooted or floating hydrophytes. Nonflowering individuals (e.g., orchids, bromeliads, cacti, and arums) were collected live and cultivated at the Botanical Gardens of the University of Tabasco (UJAT) until flowering, at which time they were identified and pressed into herbarium specimens. Specimen identification was conducted at the UJAT herbarium. Difficult or doubtful identifications were checked with specialists, primarily in the families Bromeliaceae, Cucurbitaceae, Cicadaceae, Fabaceae, Lauraceae, Malpighiaceae, Orchidaceae, and Sapindaceae. The specimens collected were deposited at the UJAT herbarium with duplicates distributed in the main herbaria in Yucatan (CICY), Veracruz (XAL), and Mexico City (MEXU). We completed the species list with specimens deposited by other researchers in UJAT and MEXU as well as with records from online databases in the United States (F, MO, and K herbaria) and with the species listed by Lundell (45) deposited in the Wisconsin State Herbarium (WIS). Nomenclature follows The Plant List (46). Mickel y Smith’s classification (2004) was followed for pteridophytes and the Angiosperm Phylogeny Group’s III (2009) for flowering plants (47, 48). Plants were classified according to their predominant habitat into a) coastal (i.e., plants chiefly found in coastal ecosystems but rarely seen inland) and b) inland (i.e., plants predominantly found in the regional rainforests of Tabasco and Guatemala). Additionally, species were classified according to their life form into a) aquatic, b) herb, c) shrub, d) tree, e) epiphyte, f) vine, g) liana, and h) palm.
Supplementary Material
Acknowledgments
We thank John Southon, Keck Carbon Cycle Accelerator Mass Spectrometry facility at University of California (UC) Irvine, for the radiocarbon analyses of mangrove piths and David Lyons, Environmental Sciences Research Laboratory UC Riverside, for the analysis of water samples. We also thank Ana and Pedro Ezcurra for help and advice during the first field trip; Ben Fiscella Meissner for his assistance and direction collecting graphic material; M. González-Aguilar, M. Campos-Díaz, N. Morales-Rodríguez, and the staff at Herbario UJAT for their help collecting samples and field support; L. Parra-Gómez, L. Godínez-García, and the staff from Universidad Politécnica Mesoamericana for their help at Laguna del Cacahuate; Pedro and Freddy for logistical support; and the Gulf of California Marine Program, El Centro para la Biodiversidad Marina y la Conservación A.C., and UC Institute for Mexico and the United States for providing administrative and organizational support for our study. O.A.-O. was supported by a Pew Marine fellowship, and some of this research is part of his research work. Funding was provided by the David and Lucile Packard Foundation, the Baum Foundation, National Geographic Society, and Gina Rogers personal donation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024518118/-/DCSupplemental.
Data Availability
Molecular sequencing data have been deposited in the publicly accessible repository GitHub, https://github.com/zapata-lab/ms_rhizophora (49). The floristic list data for the San Pedro River Region is included in this paper as Dataset S1. All other study data are included in the article and/or supporting information.
References
- 1.SGM , Digital map from “Carta geológico-minera E15-9, Tenosique.” Servicio Geológico Mexicano (2006). https://mapserver.sgm.gob.mx/Cartas_Online/geologia/113_E15-9_GM.pdf. Accessed 22 July 2019.
- 2.Ford T. D., Tufa: A freshwater limestone. Geol. Today 5, 60–63 (1989). [Google Scholar]
- 3.Solís-Castillo B., et al., Holocene sequences in the Mayan Lowlands—A provenance study using heavy mineral distributions. E&G Quaternary Sci. J. 62, 84–97 (2013). [Google Scholar]
- 4.Solís-Castillo B., Ortiz-Pérez M., Solleiro-Rebolledo E., Unidades geomorfológico-ambientales de las Tierras Bajas Mayas de Tabasco-Chiapas en el río Usumacinta: Un registro de los procesos aluviales y pedológicos durante el Cuaternario. Bol. Soc. Geol. Mex. 66, 279–290 (2014). [Google Scholar]
- 5.Thom B. G., Mangrove ecology and deltaic geomorphology: Tabasco, Mexico. J. Ecol. 55, 301 (1967). [Google Scholar]
- 6.Woodroffe C. D., Relict mangrove stand on Last Interglacial terrace, Christmas Island, Indian Ocean. J. Trop. Ecol. 4, 1–17 (1988). [Google Scholar]
- 7.Koch M. S., Snedaker S. C., Factors influencing Rhizophora mangle L. seedling development in Everglades carbonate soils. Aquat. Bot. 59, 87–98 (1997). [Google Scholar]
- 8.Hanagata N., Takemura T., Karube I., Dubinsky Z., Salt/water relationships in mangroves. Isr. J. Plant Sci. 47, 63–76 (1999). [Google Scholar]
- 9.White P. J., “Ion uptake mechanisms of individual cells and roots” in Marschner’s Mineral Nutrition of Higher Plants, Marschner H., Ed. (Elsevier, ed. 3, 2011), pp. 7–47. [Google Scholar]
- 10.Feller I. C., McKee K. L., Whigham D. F., O’Neill J. P., Nitrogen vs. phosphorus limitation across an ecotonal gradient in a mangrove forest. Biogeochemistry 62, 145–175 (2003). [Google Scholar]
- 11.Nemec W., Steel R., “Alluvial and coastal conglomerates: Their significant features and some comments on gravelly mass-flow deposits” inSedimentology of Gravels and Conglomerates, Koster E. H., Steel R. J., Eds. (Canadian Society of Petroleum Geologists, Memoir 10, 1984), pp. 1–31. [Google Scholar]
- 12.Trejo-Torres J. C., Durán R., Olmsted I., “Manglares de la Península de Yucatán” in Biodiversidad Marina y Costera de México, Salazar-Vallejo S. I., González N. E., Eds. (CONABIO and CIQRO, Mexico, 1993), pp. 660–672. [Google Scholar]
- 13.Lara-Domínguez A. L., et al., Structure of a unique inland mangrove forest assemblage in fossil lagoons on the Caribbean Coast of Mexico. Wetlands Ecol. Manage. 13, 111–122 (2005). [Google Scholar]
- 14.Stoddart D. R., Bryan G. W., Gibbs P. E., Inland mangroves and water chemistry, Barbuda, West Indies. J. Nat. Hist. 7, 33–46 (1973). [Google Scholar]
- 15.Lugo A. E., The inland mangroves of Inagua. J. Nat. Hist. 15, 845–852 (1981). [Google Scholar]
- 16.Martínez-Camilo R., et al., An assessment of the spatial variability of tropical swamp forest along a 300 km long transect in the Usumacinta River Basin, Mexico. Forests 11, 1238 (2020). [Google Scholar]
- 17.Blanchon P., Eisenhauer A., Fietzke J., Liebetrau V., Rapid sea-level rise and reef back-stepping at the close of the last interglacial highstand. Nature 458, 881–884 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Anderson T. R., et al., Modeling multiple sea level rise stresses reveals up to twice the land at risk compared to strictly passive flooding methods. Sci. Rep. 8, 14484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephenson S. N., Roberts G. G., Hoggard M. J., Whittaker A. C., A Cenozoic uplift history of Mexico and its surroundings from longitudinal river profiles. Geochem. Geophys. Geosyst. 15, 4734–4758 (2014). [Google Scholar]
- 20.Szabo B. J., Ward W. C., Weidie A. E., Brady M. J., Age and magnitude of the late Pleistocene sea-level rise on the eastern Yucatan Peninsula. Geology 6, 713–715 (1978). [Google Scholar]
- 21.Simms A. R., Last interglacial sea levels within the Gulf of Mexico and northwestern Caribbean Sea. Earth Syst. Sci. Data 13, 1419–1439 (2021). [Google Scholar]
- 22.Dyer B., et al., Sea-level trends across The Bahamas constrain peak last interglacial ice melt. Proc. Natl. Acad. Sci. U.S.A. 118, e2026839118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabrera-Cano E. F., Sánchez-Vázquez A. M., Rhizophoraceae de la Península de Yucatán (Universidad Autónoma de Yucatán: Sostenibilidad Maya, 2000). [Google Scholar]
- 24.Southon J., Magana A., A comparison of cellulose extraction and ABA pretreatment methods for AMS 14C dating of ancient wood. Radiocarbon 52, 1371–1379 (2010). [Google Scholar]
- 25.Stuiver M., Polach H. A., Reporting of 14C data. Radiocarbon 10, 355–363 (1977). [Google Scholar]
- 26.Charlesworth B., Evolution in Age-Structured Populations (Cambridge University Press, ed. 2, 1994). [Google Scholar]
- 27.Li J. T., Yang J., Chen D. C., Zhang X. L., Tang Z. S., An optimized mini-preparation method to obtain high-quality genomic DNA from mature leaves of sunflower. Genet. Mol. Res. 6, 1064–1071 (2007). [PubMed] [Google Scholar]
- 28.Elshire R. J., et al., A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eaton D. A. R., Overcast I., ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592–2594 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rognes T., Flouri T., Nichols B., Quince C., Mahé F., VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danecek P., et al., The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pina-Martins F., Baptista J., Pappas G. Jr, Paulo O. S., New insights into adaptation and population structure of cork oak using genotyping by sequencing. Glob. Change Biol. 25, 337–350 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng X., et al., A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryant D., Bouckaert R., Felsenstein J., Rosenberg N. A., RoyChoudhury A., Inferring species trees directly from biallelic genetic markers: Bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 29, 1917–1932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouckaert R., et al., BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verity R., Nichols R. A., Estimating the number of subpopulations (K) in structured populations. Genetics 203, 1827–1839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard J. K., Stephens M., Donnelly P., Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brumfield R. T., Beerli P., Nickerson D. A., Edwards S. V., The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol. Evol. 18, 249–256 (2003). [Google Scholar]
- 42.Lynch M., Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarvis A., Reuter H. I., Nelson A., Guevara E., Data from “Hole-filled seamless SRTM data V4.” International Centre for Tropical Agriculture (2008). https://srtm.csi.cgiar.org. Accessed 18 May 2020.
- 44.Huang X., Xie H., Liang T., Yi D., Estimating vertical error of SRTM and map-based DEMs using ICESat altimetry data in the eastern Tibetan Plateau. Int. J. Remote Sens. 32, 5177–5196 (2011). [Google Scholar]
- 45.Lundell C. L., Flora of eastern Tabasco and adjacent Mexican areas. Contrib. Univ. Mich. Herb. 8, 5–74 (1942). [Google Scholar]
- 46.Alkin B., et al., The Plant List, Version 1.1 (2013). http://www.theplantlist.org/. Accessed 11 March 2020.
- 47.Mickel J. T., Smith A. R., The Pteridophytes of Mexico (New York Botanical Garden Press, 2004). [Google Scholar]
- 48.The Angiosperm Phylogeny Group , An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121 (2009). [Google Scholar]
- 49.Zapata F., Rhizophora mangle from Yucatan. GitHub. https://github.com/zapata-lab/ms_rhizophora. Deposited 9 June 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Molecular sequencing data have been deposited in the publicly accessible repository GitHub, https://github.com/zapata-lab/ms_rhizophora (49). The floristic list data for the San Pedro River Region is included in this paper as Dataset S1. All other study data are included in the article and/or supporting information.