Significance
Lysosomes are organelles that also act as cell-signaling hubs. They regulate functions ranging from antigen presentation to autophagy. Spherical lysosomes can spontaneously elongate into tubules in starving or inflamed immune cells. We describe a DNA-based reagent, denoted Tudor, that tubulates lysosomes in macrophages without triggering either an immune response or autophagy. Chemical imaging revealed that tubular lysosomes differ from vesicular ones in terms of their pH, calcium, and proteolytic activity. Tudor revealed a role for tubular lysosomes in that they enhance MMP9 secretion and phagocytosis in resting macrophages. The ability to tubulate lysosomes in resting immune cells without starving or inflaming them may help reveal new insights into how tubular lysosomes function.
Keywords: DNA nanotechnology, lysosomes, macrophages, phagocytosis, MMP9
Abstract
Lysosomes adopt dynamic, tubular states that regulate antigen presentation, phagosome resolution, and autophagy. Tubular lysosomes are studied either by inducing autophagy or by activating immune cells, both of which lead to cell states where lysosomal gene expression differs from the resting state. Therefore, it has been challenging to pinpoint the biochemical properties lysosomes acquire upon tubulation that could drive their functionality. Here we describe a DNA-based assembly that tubulates lysosomes in macrophages without activating them. Proteolytic activity maps at single-lysosome resolution revealed that tubular lysosomes were less degradative and showed proximal to distal luminal pH and Ca2+ gradients. Such gradients had been predicted but never previously observed. We identify a role for tubular lysosomes in promoting phagocytosis and activating MMP9. The ability to tubulate lysosomes without starving or activating immune cells may help reveal new roles for tubular lysosomes.
The multiple functions of lysosomes have been linked to differences in their size, shape, location, abundance, and composition (1, 2). Although lysosomes are generally vesicular, they can form tubules several microns long (3). New roles are emerging for such tubular lysosomes (TLs) (4–6). They facilitate antigen presentation, promote the efflux of bacterial peptides to activate NOD-like receptors, and undergo scission to produce proto-lysosomes (7). TLs are found in Drosophila melanogaster, Caenorhabditis elegans, and Leishmania mexicana, suggesting that lysosome tubulation is conserved across phyla (6, 8). TLs are generally observed when the cell undergoes either autophagy or immune activation, and their luminal biochemistry in different contexts varies remarkably (2, 4, 8). Thus, if we could tubulate lysosomes without inducing either autophagy or the immune response we would be able to better connect the nature and functionality of TLs independent of cell state. This is especially because autophagy and the immune response activate different transcription programs, up-regulating distinct sets of lysosomal genes. Here we describe a serendipitously discovered DNA nanodevice that tubulates lysosomes without activating macrophages. This nanodevice, Tudor (Tubular lysosome DNA reporter) binds Ku70/80 heterodimers at the plasma membrane and tubulates lysosomes via a pathway that is distinct from that triggered by lipopolysaccharide (LPS) that invariably activates macrophages (9).
Tudor enables the comparison of the proteolytic properties of TLs and vesicular lysosomes (VLs) in activated and resting macrophages. Since lysosomal ionic content regulates proteolysis, we also analyzed luminal pH and Ca2+ levels at single-lysosome resolution (10–14). Our observations revealed that there are different classes of TLs. TLs play important roles in the late stages of phagocytosis (15). They enhance antigen presentation and facilitate phagolysosome resolution (16). Using Tudor, we found that TLs poise macrophages for phagocytosis by aiding phagosome formation and phagosome–lysosome fusion. Our studies identify an additional molecular player, matrix metalloproteinase 9 (MMP9), as being pivotal to the tubulation cascade. Tudor treatment activates MMP9 and triggers lysosome tubulation. Tubulation in turn promotes extracellular MMP9 activity, and the resultant positive feedback leads to sustained MMP9 secretion. MMP9 secretion by macrophages drives pathophysiologies such as cancer, atherosclerosis, and renal fibrosis (17–19). Our studies reveal TLs aid the early stages of phagocytosis and promote extracellular MMP9 activity.
Results and Discussion
A DNA Nanodevice, Tudor, Tubulates Lysosomes.
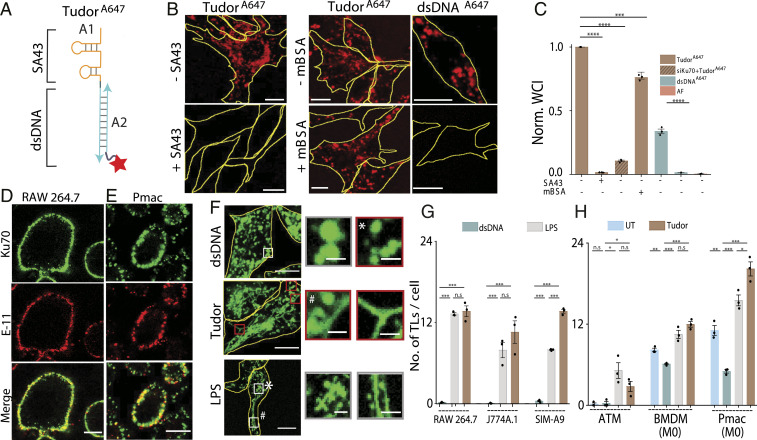
We describe the design, endocytic uptake pathway and activity of a DNA nanodevice that triggers lysosomes tubulation in macrophages without activating them. This DNA nanodevice, Tudor, is designed so that complementary DNA strands A1 and A2 display an actuator domain and a fluorescent reporter, respectively (Fig. 1A and SI Appendix, Table S1). The actuator domain in A1 is a 43-base-long DNA aptamer called SA43 that binds the Ku70/80 dimer at the plasma membrane (20). Strand A1 is hybridized to A2, which displays an Alexa 647 fluorophore, to form a 24-base pair duplex (double-stranded DNA [dsDNA]) (21). The dye on Tudor enables one to evaluate tubulation, its uptake efficiency, and its subcellular distribution (Fig. 1A). The formation, integrity, and purity of Tudor were confirmed by gel electrophoresis (SI Appendix, Fig. S1).
Fig. 1.
A DNA nanodevice, Tudor, tubulates lysosomes. (A) Schematic of Tudor containing modules A1 bearing SA43 and A2 bearing Alexa 647N (red star). (B) TudorA647 and dsDNAA647 uptake in RAW 264.7 cells in the presence or absence of competitors (3 µM SA43 or 60 equiv mBSA). (C) Normalized whole-cell intensities (WCI) from B (N = 100 cells), AF: autofluorescence. (D and E) Images of Ku70 (green) and Pan Cadherin (E-11, red) in (D) RAW 264.7 cells and (E) Pmacs. (F) TMR dextran-labeled RAW 264.7 cells in the presence of dsDNA, Tudor, and LPS. Boxed regions are magnified (Right), VLs (*) and TLs (#). (G and H) Number of TLs per cell when treated with dsDNA, Tudor, or LPS (N = 20 cells); ****P < 0.00005; ***P < 0.0005; **P < 0.005; *P < 0.05 (unless otherwise mentioned these are one-way ANOVA with Tukey post hoc test). Unless otherwise mentioned, n.s: nonsignificant, all error bars represent SEM from three independent experiments. (Scale bars, 10 μm; Inset scale bars, 2 μm.)
Tudor is internalized by receptor-mediated endocytosis in RAW 264.7 cells. Tudor uptake is not via scavenger receptors (22) but rather via Ku70/80. Unlike dsDNA, Tudor uptake could not be competed out by excess maleylated bovine serum albumin (mBSA), a ligand for scavenger receptors (23–25) (Fig. 1 B and C). However, depleting Ku70 or addition of excess, unlabeled SA43 completely abolished the uptake, revealing that Tudor internalization was mediated by engaging Ku70/80 at the cell surface (Fig. 1 B and C and SI Appendix, Fig. S2). In addition to the nucleus, the Ku70/80 heterodimer is also seen on the surface of many cell types including macrophages (26). Immunostaining without permeabilizing the plasma membrane revealed cell-surface Ku70 in diverse macrophages such as RAW 264.7, J774A.1, and SIM-A9.1 (SI Appendix, Fig. S3) as well as primary macrophages such as naïve murine bone marrow-derived macrophages (BMDMs), peritoneal macrophages (Pmacs), and their polarized states including the M1 phenotype (LPS/interferon γ, proinflammatory) or the M2 phenotype (interleukin 4, antiinflammatory) (Fig. 1 D and E and SI Appendix, Fig. S3).
Upon treating macrophages with 100 nM Tudor for 4 h, we found that it labeled vesicular as well as tubular organelles. When lysosomes in macrophages were prelabeled with TMR dextran they colocalized with internalized Tudor, revealing that the vesicular and tubular Tudor-containing compartments were lysosomes (Fig. 1F and SI Appendix, Fig. S4). TLs formed by Tudor treatment phenocopied those induced by LPS treatment (Fig. 1 F and G). LPS tubulates lysosomes by a well-defined pathway but it also activates macrophages via TLR4 (3, 27). Tubulation was not observed when RAW 264.7 cells were either treated with dsDNA or an aptamer against a different cell surface protein, MUC-1 (28) (Fig. 1 F and G and SI Appendix, Table S2). Treating RAW 264.7 cells with CpG DNA, which activates macrophages via TLR9 (21), also failed to induce tubulation, suggesting that Tudor-induced tubulation was specifically triggered by SA43 and was not due to generic immunogenicity of DNA (SI Appendix, Fig. S5). We found that Tudor-induced TL formation was not linked to DNA damage. DNA damage induced by etoposide failed to tubulate lysosomes even after 8 h (SI Appendix, Fig. S6) (29). Tudor tubulated lysosomes in macrophages such as J774A.1 and SIM-A9 as well as primary macrophages such as BMDMs, Pmacs, and adipose tissue macrophages (Fig. 1G and SI Appendix, Figs. S7 and S8).
Tudor Tubulates Lysosomes in Macrophages without Immune Activation.
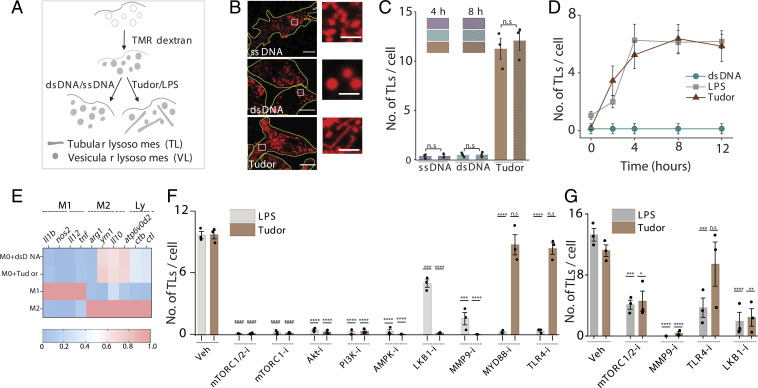
We then tested whether Tudor treatment activated macrophages by quantifying the number of TLs per cell (Methods, Fig. 2 A–C, and SI Appendix, Figs. S9 and S10). The extent and kinetics of Tudor-induced tubulation phenocopied those induced by LPS, suggesting that both agents could potentially act via a similar pathway (Fig. 2D). Messenger RNA (mRNA) expression data for Tudor-treated resting BMDMs or Pmacs (M0) revealed no significant up-regulation of M1 or M2 markers, indicating that Tudor did not polarize them toward either state (Fig. 2E and SI Appendix, Fig. S11). Additionally, Tudor treatment did not activate autophagy, ruling out the autophagy-induced lysosome tubulation cascade (SI Appendix, Fig. S12).
Fig. 2.
Tudor nonimmunogenically tubulates lysosomes in macrophages. (A) Schematic of tubulation assay. (B) TMR-dextran–labeled lysosomes in RAW 264.7 upon treatment with single-stranded DNA (ssDNA), dsDNA, and Tudor. (Inset) Magnified images of VLs and TLs. (C) Number of TLs per cell, from B, at 4 and 8 h posttreatment (N = 25 cells). (D) Number of TLs per cell versus time of treatment with Tudor, LPS, or dsDNA in RAW 264.7 cells (N = 20 cells). (E) Heat maps of fold change of mRNA levels of M1, M2, and lysosomal markers (Ly) in BMDM (M0) upon treatment with dsDNA and Tudor. Fold changes of M1 and M2 markers are normalized to M1 and M2 BMDMs, respectively; fold changes of lysosomal markers are normalized to M0+dsDNA. (F and G) Number of TLs per cell upon Tudor or LPS treatment with pharmacological inhibitors for the indicated protein (protein-i) in RAW 264.7 cells (N = 20 cells) (F) and in M0 BMDMs (n = 20 cells) (G). ****P < 0.00005; ***P < 0.0005; **P < 0.005; *P < 0.05; n.s: nonsignificant. (Scale bars, 10 µm; Inset scale bars, 4 µm.)
To identify the players responsible, we quantified TLs in RAW 264.7 cells treated with different pharmacological inhibitors (SI Appendix, Table S3). Unlike LPS, Tudor-induced tubulation was independent of TLR4 as well as MYD88 (Fig. 2F and SI Appendix, Fig. S13A). Pharmacological inhibition of TLR5, TLR3, TLR2/TLR6, and TLR1/TLR2 prior to Tudor treatment revealed that these TLRs were not on the pathway (SI Appendix, Fig. S13 B and D). Treatment with Dynasore and Tudor still led to efficient tubulation, suggesting that Tudor acts primarily at the plasma membrane (SI Appendix, Fig. S14) (26). Ku80 in the Ku heterodimer activates MMP9 at the plasma membrane by interacting with its hemopexin domain (30). Indeed, we found that that Tudor treatment activated MMP9 (SI Appendix, Fig. S13E) and treating cells with MMP9 inhibitor-I (31) abolished both Tudor and LPS-induced lysosome tubulation (Fig. 2 F and G and SI Appendix, Fig. S13A). These findings were recapitulated in primary BMDMs (Fig. 2G and SI Appendix, Fig. S13E).
To test whether LPS and Tudor worked via common players late in the tubulation pathway, we depleted mTOR, Akt, and PI3K by small interfering RNA (siRNA) as well as pharmacologically inhibited them with Torin-1 (32), Akt-I (3), or ZSTK474 (33), respectively (SI Appendix, Figs. S15 and S16). Indeed, both LPS and Tudor-mediated tubulation activated the PI3K–Akt–mTOR cascade, which was also confirmed by the increased phosphorylation levels of these proteins (Fig. 2F and SI Appendix, Fig. S17).
PI3K can be activated via cSrc, c-Cbl, RAC1, or JAK1/2 (26, 34, 35). Treatment with inhibitors for cSrc (Dasatinib), RAC1 (RAC1i), and JAK1/2 (barcitinib) revealed that neither Tudor nor LPS acted via these proinflammatory signaling pathways (SI Appendix, Fig. S13 C and D). We therefore targeted antiinflammatory pathways involving PI3K and Akt (36). In our hands, AMPK was significantly phosphorylated in both LPS- and Tudor-treated RAW 264.7 macrophages (SI Appendix, Fig. S17) and inhibiting AMPK with dorsomorphin (compound C) (37) impeded tubulation, despite prior work showing that AMPK activation prevents LPS-mediated tubulation (Fig. 2F) (3). SiRNA knockdown of AMPK reaffirmed its involvement in Tudor-induced lysosomal tubulation (SI Appendix, Fig. S15). AMPK activity is regulated by LKB1 or TGFβ activating kinase 1 (TAK1) (38). We found that Tudor-induced tubulation required LKB1 activity but not TAK1 (Fig. 2F and SI Appendix, Figs. S13 A, C, and D, S15B, and S17E). Our studies reveal two additional players, LKB1 and AMPK, that negatively regulate mTOR, in the tubulation cascade.
TLs Are Less Degradative.
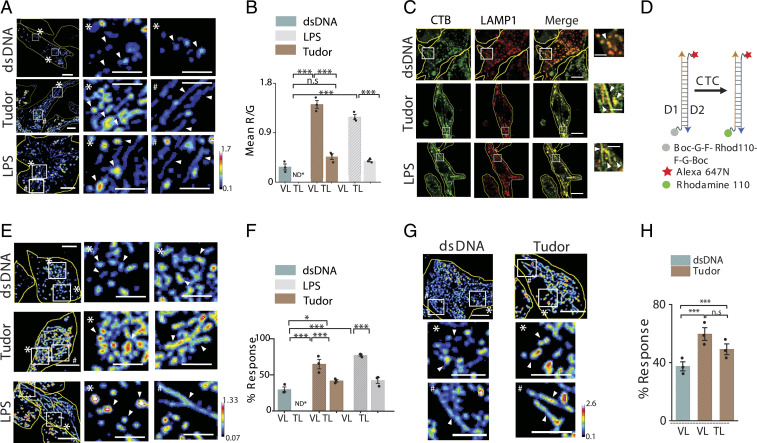
TLs facilitate functions ranging from antigen presentation to autophagy, yet it is still unclear whether tubulation changes the luminal biochemistry. LPS stimulation induces TL formation but also up-regulates many lysosomal enzymes (39). We therefore quantitatively mapped enzymatic activity at the resolution of single lysosomes in live RAW 264.7 macrophages treated with either LPS or Tudor. We allowed cells with Alexa 488-dextran–labeled lysosomes to endocytose DQ BSA and compared the proteolytic activity of TLs and VLs (Fig. 3 A and B and SI Appendix, Fig. S18). Regardless of how they were tubulated, increase in proteolysis was confined to VLs while that in TLs was unaffected (Fig. 3B).
Fig. 3.
Enzymatic cleavage maps show low degradation in TLs. (A) Pseudocolor R/G images of Alexa 488 dextran- (G) and DQ Red BSA- (R) labeled lysosomes in dsDNA, Tudor, or LPS-treated RAW 264.7 cells (Left). White boxes are magnified (Right). (B) Mean R/G ratios of single lysosomes in A (N = 50 cells; n = 200 lysosomes). (C) RAW 264.7 cells immunostained for CTB (green) and LAMP1 (red) upon treatment with dsDNA, Tudor, or LPS. White boxes are magnified (Right). (D) A DNA-based CTC activity reporter consisting of DNA duplex with sensing module (caged Rhodamine 110, gray), normalizing module (Alexa 647N, red), and cleaved module (Rhodamine 110, green). (E) Pseudocolored G/R images of CTC activity reporter in RAW 264.7 cells pretreated with dsDNA, Tudor, or LPS (Left). White boxes are magnified (Right). (F) Percent response of CTC reporter in VLs and TLs in dsDNA-, Tudor-, and LPS-treated cells (N = 50 cells, n = 500 lysosomes). (G) Pseudocolored G/R images of CTC activity reporter in labeled BMDMs (M0) pretreated with dsDNA or Tudor (Top). White boxes are magnified below. (H) Percent response in VLs and TLs of dsDNA- and Tudor-treated cells (N = 50, n = 500 lysosomes), *P < 0.05; ***P < 0.0005. ND*: not defined; n.s: nonsignificant. White arrowheads show VLs (*) and TLs (#). (Scale bars, 10 μm; zoomed scale bars, 2 μm.)
DNA-based enzyme activity reporters can selectively address the contribution of specific lysosomal proteases (40). Cathepsin C (CTC) plays key roles in inflammation, interleukin 1β production, tumor necrosis factor α production, and macrophage reprogramming (41, 42). We therefore used a previously published ratiometric DNA-based CTC reporter to probe CTC activity in VLs and TLs in RAW 264.7 cells and BMDMs (Fig. 3 D–H and SI Appendix, Figs. S19 and S20) (40). The CTC activity reporter is a DNA duplex bearing a reference dye and a Rhodamine 110 dye caged by phenylglycyl (FG) groups that are cleaved by CTC activity in lysosomes (SI Appendix, Table S4). In Tudor- or LPS-treated cells, VLs show much higher proteolysis (CTC activity) compared to TLs in the same cells. In fact, the level of proteolysis in TLs upon LPS/Tudor treatment is comparable to the basal level of hydrolysis observed in VLs of dsDNA-treated (i.e., resting) cells (Fig. 3 E and F and SI Appendix, Fig. S19 A–C). These trends were recapitulated in BMDMs (Fig. 3 G and H and SI Appendix, Fig. S19 D–K).
Importantly, the differences in lysosomal activity between TLs and VLs was not due to the relative abundance of hydrolases (4), since immunofluorescence revealed that cathepsin B (CTB) content was similar across lysosomes regardless of whether cells were treated with dsDNA, LPS, or Tudor (Fig. 3C and SI Appendix, Fig. S21 A–C). We quantified the total numbers of VLs and TLs per cell, Feret length, percentage of TLs per cell, and average number of VLs and TLs per cell (SI Appendix, Fig. S22). We found no perceptible differences in number of VLs in Tudor-treated or dsDNA-treated cells. However, the fraction of TLs consistently increases upon treatment with either Tudor or LPS.
Luminal pH and Ca2+ Maps Reveal Two Major Kinds of TLs.
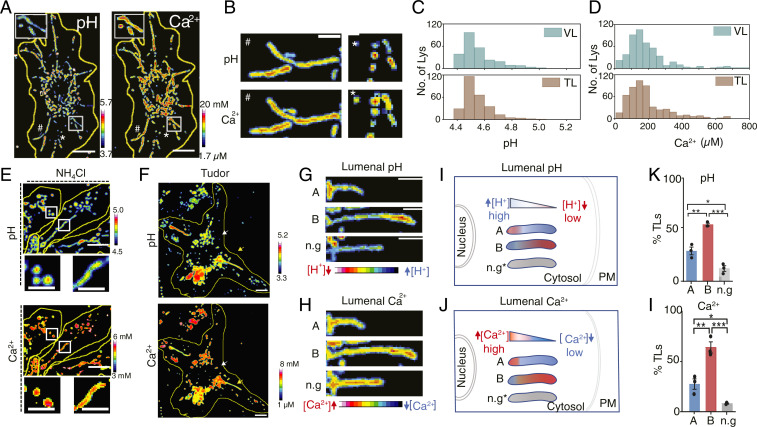
The differential proteolytic activity despite their comparable cathepsin content led us to map the luminal pH of TLs and VLs (43). We also mapped Ca2+, since TLs undergo active fission and fusion that implicate the lysosomal Ca2+ channels P2X4, TPC2, and TRPML1 (44–46). Moreover, these Ca2+ channels strongly depend on mTOR activity, which is part of the tubulation cascade (47). We used a DNA-based, pH-correctable Ca2+ sensor, CalipHluor2.0 (SI Appendix, Fig. S23A), which carries 1) a pH-sensing dye, DCF (SI Appendix, Fig. S24), 2) a Ca2+-sensing dye, Rhod-5F, and 3) a reference dye, Atto647N, for quantitative ratiometry (SI Appendix, Tables S1 and S4). We measured the stability of CalipHluor2.0 and found that it was ∼95% intact at time t = 2 h within lysosomes of RAW 264.7 cells (SI Appendix, Fig. S25). We mapped luminal Ca2+ and pH by treatment with Tudor then labeling lysosomes with CalipHluor2.0 and imaging cells in three channels, G, O, and R (Fig. 4A and SI Appendix, Figs. S23 and S26 A–C). According to the lysosomal pH and Ca2+ maps, the overall luminal pH and Ca2+ levels in TLs were comparable to those of VLs (Fig. 4 C and D).
Fig. 4.
Heterogeneity of ionic gradient within TLs. (A) pH and Ca2+ images of CalipHluor2.0-labeled RAW 264.7 cells treated with Tudor. (B) pH and Ca2+ maps of TLs and VLs. (C and D) Lysosomal (C) pH and (D) Ca2+ distributions in VLs of dsDNA-treated RAW 264.7 and TLs of Tudor-treated cells (∼20 cells, ∼300 VLs, ∼100 TLs). (E) pH and Ca2+ images of Tudor-treated RAW 264.7 cells in the presence of 10 mM NH4Cl. (F) pH and Ca2+ images of Tudor-treated RAW 264.7 cells showing both VLs and TLs. (G and H) pH and Ca2+ maps of TLs. (I and J) Schematic of the different TL populations according to their luminal gradients and orientation in the cell. n.g.: no gradient. (K and L) Percentage of TL populations (N = 20 cells, n = 100 TLs). ***P < 0.0005; **P < 0.005; *P < 0.05. (Scale bars, 10 μm; zoomed scale bars, 2 μm.)
Interestingly, the pH and the Ca2+ maps of TLs revealed a striking gradient of each ion along the long axis of the tubule, unlike VLs where they were homogenous (Fig. 4 A–H). Areas of high acidity in TLs corresponded to low Ca2+, and vice versa. NH4Cl neutralizes lysosomal pH and releases luminal Ca2+ from lysosomes (48). While NH4Cl treatment greatly reduced the number of TLs, pH and Ca2+ gradients were dissipated in the few TLs that did form (Fig. 4E and SI Appendix, Fig. S26B), indicating that the gradients were not an imaging artifact. Moreover, the ion gradients dynamically changed as TLs underwent growth or deformation. However, regions of high Ca2+ always correlated with regions of low acidity (Fig. 4 G and H). Such Ca2+ gradients are consistent with the hypothesis that tubulation requires the stringent control of TRPML1 activity since either overexpressing or inhibiting TRPML1 disintegrates TLs (49). We posit that the tight regulation of TRPML1 observed by others could potentially function to sculpt the Ca2+ gradient.
Further analysis revealed at least three kinds of TL populations. Since TLs are stretched along microtubules, ∼95% of TLs radiated from the nucleus to the plasma membrane. We therefore classified radially oriented TLs based on whether their high Ca2+ termini were nearer the nucleus (population A) or the plasma membrane (population B) or those with no ion gradient (n.g., Fig. 4 I and J). Nearly 53% of TLs fell under population B, ∼29% had the reverse orientation (population A), and ∼13% showed no gradients (Fig. 4 K and L). Our findings were recapitulated with LPS-induced TLs (SI Appendix, Fig. S26 C–G).
These different orientations suggest either that there are different tubulating mechanisms or different kinds of TLs, or both. Tubulation requires opposing pulls generated by Arl8b-SKIP-kinesin along the plus end of microtubules and Rab7-RILP-dynein complexes along the minus end (27, 50). In fact, Arl8b-SKIP and Rab7-RILP complexes regulate the spatial positions of VLs in cells (51). Further, peripheral VLs regulate plasma membrane repair while perinuclear VLs fuse with autophagosomes for autophagy (49, 52). Thus, these two populations of TLs also could function differently.
TLs Promote Phagocytosis and Phagosome–Lysosome Fusion.
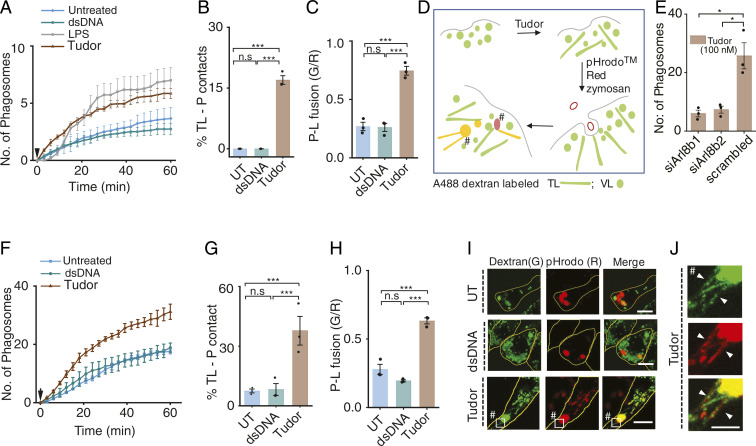
Phagocytosis drives key functions including antigen presentation and pathogen killing in innate immune cells (53). When macrophages are treated with LPS, inflammation is initiated and phagocytosis is enhanced (3, 54, 55). However, since LPS also tubulates lysosomes, we do not know whether increased phagocytosis is due to inflammation or lysosome tubulation, or both. To test this, we treated RAW 264.7 cells and Pmacs with either dsDNA, LPS, or Tudor and quantified phagocytosis of pHrodo Red–conjugated zymosan (Fig. 5A and SI Appendix, Figs. S27 and S28). We found that lysosome tubulation alone was sufficient to enhance phagocytosis (Fig. 5 A and E and SI Appendix, Figs. S27 and S28). We then probed the effect of lysosome tubulation on phagosome maturation by quantifying TL–phagosome contacts and content mixing, where lysosomes and phagosomes were labeled with Alexa 488 dextran and pHrodo-Red zymosan, respectively (SI Appendix, Materials and Methods and Fig. 5D). In Pmacs, ∼30% of TLs contacted phagosomes, while in RAW 264.7 cells it was 15 to 20% (Fig. 5 B–F, H, and I and SI Appendix, Fig. S29). More contacts also correlated with more content exchange characteristic of more productive fusion events (Fig. 5 C and G). Tudor treatment did not alter fluid-phase endocytosis (SI Appendix, Figs. S27D and S29 G–I). Overall, our results show that TLs promote phagocytosis and aid phagosome–lysosome fusion.
Fig. 5.
Tubulation promotes phagocytosis and phagosome–lysosome fusion in macrophages. (A) Average number of phagosomes versus time in RAW 264.7 cells treated with dsDNA, LPS, or Tudor. Arrow shows zymosan addition (N = 30 cells). (B) Percentage TLs contacting phagosomes (% TL-P contact) (N = 50 cells,). (C) Phagosome–lysosome fusion (P-L fusion) given by mean G/R value in RAW 264.7 cells (n = 60 cells). (D) Schematic of fusion assay in Tudor-treated cells where lysosomes and phagosomes are labeled with Alexa 488 dextran (G) and pHrodo Red zymosan (R), respectively. (E) Number of phagosomes in Tudor-treated RAW 264.7 cells depleted of Arl8b (N = 100 cells). (F) Average number of phagosomes versus time in Pmac (M0) treated with dsDNA, Tudor, or LPS. Arrow shows zymosan addition (N = ∼30 cells). (G) Percentage TLs contacting phagosomes in M0 Pmacs (n = 30 cells). (H) Extent of phagosome lysosome fusion (P-L fusion) (N = 30 cells). (I) Lysosomes (G) and phagocytosed zymosan particles (R) with or without dsDNA or Tudor treatment in RAW 264.7 cells (J) Inset of TLs-P contacts indicated by #. ***P < 0.0005; *P < 0.05 (one-way ANOVA with Tukey post hoc test). n.s: nonsignificant; n = 100 phagosomes wherever mentioned.
To explicitly test the role of TLs in aiding phagocytosis, we knocked down Arl8b, an adaptor that connects lysosomes to kinesin, which lies downstream of mTOR activation and upstream of lysosome tubulation. Knocking down Arl8b suppressed lysosome tubulation and phagocytosis in Tudor-treated cells, demonstrating that tubulation promotes phagocytosis (Fig. 5E and SI Appendix, Figs. S30 and S31).
MMP9 Activation Drives Tudor-Mediated Tubulation.
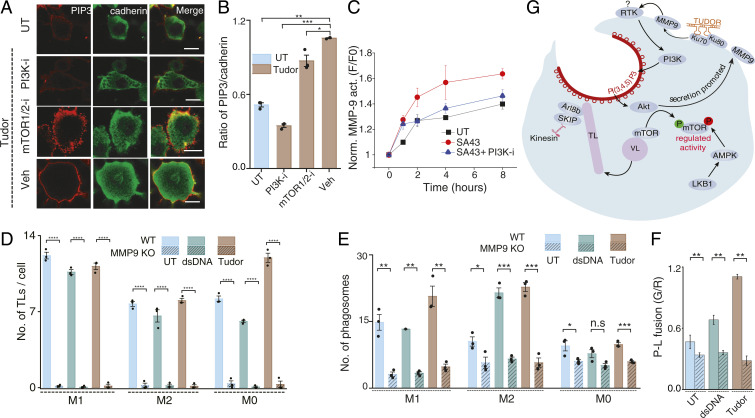
To examine how TL formation aided phagocytosis, we pharmacologically inhibited mTOR and PI3K in Tudor-treated cells and observed a drastic decrease in phagocytosis in both cases (SI Appendix, Fig. S31). PI3K promotes phagocytosis by converting PI(4,5)P2 on the inner leaflet of the plasma membrane to PI(3,4,5)P3, which facilitates phagosome cup formation (56). Tudor treatment increased PI(3,4,5)P3 on the plasma membrane, consistent with PI3K activity. Importantly, inhibiting mTOR did not affect PI(3,4,5)P3 levels yet prevented lysosomal tubulation and impeded phagocytosis (Fig. 6 A and B and SI Appendix, Fig. S31).
Fig. 6.
Role of the Tudor-mediated pathway in enhancing phagocytosis. (A) Untreated (UT) or Tudor-treated RAW 264.7 cells, immunostained for PI(3,4,5)P3 (red) and Cadherin (green) in the presence of indicated inhibitors. (B) Ratio of mean WCI of PI(3,4,5)P3 to Cadherin in A (N = 50 cells). (C) Kinetics of extracellular MMP9 activity in Tudor-treated RAW 264 cells with and without a PI3K inhibitor (PI3K-i); UT (untreated). (D) Number of TLs per cell in UT, dsDNA, or Tudor-treated wild-type (WT) and MMP9 KO BMDMs (M1, M2, and M0 states) (N = 45 cells). (E) Number of phagosomes in UT or dsDNA- or Tudor-treated WT and MMP9 KO BMDMs (M1, M2, M0 states) (N = 100 cells). (F) Extent of phagosome lysosome fusion (P-L fusion) (N = 30 cells) in UT or dsDNA- or Tudor-treated WT and MMP9 KO M2 BMDMs. (G) Proposed model of Tudor triggered lysosome tubulation pathway promoting phagocytosis. ****P < 0.00005; ***P < 0.0005; **P < 0.005; *P < 0.05; n.s: nonsignificant. (Scale bars, 10 μm.).
PI3K and Akt activation enhances MMP9 secretion (57). To test whether Tudor activated any secreted MMP9 which could feed back and further tubulate lysosomes, we probed the kinetics of extracellular MMP9 activity in SA43-treated macrophages with and without a PI3K inhibitor (Methods). Inducing lysosome tubulation elevated extracellular MMP9 activity (Fig. 6C). Note that Tudor fully activates the tubulation cascade at t ∼4 h posttreatment (Fig. 2D). Interestingly, extracellular MMP9 activity also reached a maximum at t = 4 h, coinciding with maximal lysosome tubulation (Fig. 6C). The enhancement is lost when PI3K is inhibited, which also abolishes tubulation. This demonstrates that lysosome tubulation and MMP9 secretion synergistically increase extracellular MMP9 activity, sustaining tubulation.
Since MMP9 is transcriptionally up-regulated when either nuclear factor κB is activated during inflammation or Nrf2 is activated during starvation, we tested whether Tudor treatment increased MMP9 expression (58, 59). This was not the case, which is consistent with Tudor not triggering autophagy-mediated lysosome tubulation or inflammation (SI Appendix, Fig. S32). This suggested the importance of extracellular MMP9 in lysosome tubulation and its downstream effects on macrophage function. Accordingly, we tested the effect of Tudor on primary M1, M0, and M2 macrophages derived from MMP9 knockout mice. Indeed, in all three types of macrophages Tudor treatment failed to tubulate lysosomes (Fig. 6D and SI Appendix, Fig. S33), and phagocytosis (Fig. 6E and SI Appendix, Fig. S34) and phagosome–lysosome fusion were diminished (Fig. 6F and SI Appendix, Fig. S35).
A Model for How TLs Promote Phagocytosis.
We propose a model where Tudor acts as an extracellular ligand for plasma membrane-resident Ku70/80 (Fig. 6G). When Tudor binds the Ku heterodimer, Ku80 activates MMP9. Activated MMP9 interacts with and activates diverse receptor tyrosine kinases (RTKs) at the plasma membrane. We posit that MMP9 could activate PI3K via an unidentified RTK, possibly PAR-1 (60). PI3K activation, in turn, promotes phagocytosis through multiple mechanisms. First, it enriches PI(3,4,5)P3 abundance (61). Second, PI(3,4,5)P3 activates Akt, secreting more MMP9. This sets up positive feedback whereby Tudor at the cell surface activates newly secreted MMP9. Third, PI3K activity leads to mTOR activation tubulating lysosomes (3, 60). Suppressing any of these steps impedes phagosome formation and phagosome–lysosome fusion.
Our results also revealed that, like lysosomal Ca2+ channels, mTOR activity is also stringently regulated and underpins TL formation. LKB1 and AMPK activity contributed to lysosome tubulation. AMPK negatively regulates mTOR by phosphorylation at S722 and S792 (62). This likely prevents hyperphosphorylation and runaway mTOR activation, which, in turn, likely toggles lysosomal Ca2+ channel activity for effective tubulation (Fig. 6G) (63). Lysosomal Ca2+ channels induce local Ca2+ surges to facilitate phagosome formation by activating dynamin or providing the extra membrane needed for phagocytosis (64). With their Ca2+-rich termini localized near the cell periphery, TLs could facilitate both processes.
Summary
Thus far, TLs have been studied either by inducing autophagy or immune activation, both of which lead to transcriptional changes in the cell. Tudor acutely tubulates lysosomes in macrophages without activating either inflammation or autophagy, allowing one to study metabolic adaptation of lysosomes more precisely. Metabolic adaptation impacts cellular metabolism, which in turn shapes innate immune cell memory. Cellular memory is responsible for the exacerbated response of an innate immune cell to repeated encounters of the same pathogen through epigenetic, transcriptional, and functional reprogramming (65), which is accompanied by major changes in cell metabolism (66) and metabolic adaptation of their lysosomes. Immune activation also generates transcriptional changes and complicates the study of innate immune memory pathways that lack gene rearrangements. Tudor provides a route to study such pathways.
The overall proteolytic activity of TLs is lower than in VLs. While lysosomal proteolysis is known to increase upon LPS stimulation (67), our studies reveal that this elevation is confined to VLs. The hypoactive proteolysis in TLs and hyperactive proteolysis within VLs suggest that both forms likely perform distinct functions in phagocytosis. Quantitative pH and Ca2+ maps of TLs revealed spatial gradients along the tubule. Such spatial gradients of Ca2+ have been previously observed in primary cilia, a subcellular structure similar in shape and size to TLs (68). This active Ca2+ gradient along the long axis of the cilium enables mechanosensation (68). Specifically in TLs, Saffi and Botelho have predicted that both pH and Ca2+ gradients should arise from continuously toggling the activity of P2X4 and TRPML1 (69). Our studies now provide experimental proof of this model (69). What the model did not predict, but our observations now reveal, is that there is more than one class of TLs.
We found that TLs play a role in the early stages of phagosome formation whereby they poise resting macrophages for phagocytosis. Tudor activates PI3K, enriching PI(3,4,5)P3 on the cell membrane, promoting phagocytosis (56). Our studies reveal that MMP9 activation is a crucial player in the lysosome tubulation cascade. MMP9 activation proceeds via positive feedback, leading to the sustained tubulation needed to support the extensive cell membrane ruffling and remodeling needed for phagocytosis. The ruffled border in osteoclasts is actually formed by secretory lysosomes where SNX10 is implicated in transporting secretory lysosomes to the plasma membrane (70). In fact, SNX10 promotes MMP9 secretion in osteoclasts (71). We therefore suggest a model where the movement of the much larger TLs on force-generating microtubules could similarly push against the fluid, PI(3,4,5)P3-rich cell membrane, thereby causing the large-scale remodeling needed to engulf phagocytic cargo.
Accordingly, our studies mechanistically connect lysosome tubulation with MMP9 activation in macrophages. MMP9 is a key player in macrophage biology as its secretion by macrophages plays an important role in numerous diseases. In cancer, MMP9 secretion by macrophages promotes the epithelial–mesenchymal transition in pancreatic, gastric, colon, and bone cancers (3, 19, 49, 72). MMP9 secreted by macrophages in tumors activates the PI3K–Akt pathway via protease activated receptor-1 (PAR-1) (72). In atherosclerosis, MMP9 secretion by macrophages enhances elastin degradation and induces significant plaque disruption (18). In renal fibrosis, MMP9 contributes to pathogenesis through osteopontin cleavage (17). Given the different types of TL populations, it is possible their roles in cell function may be more widespread than previously anticipated. The ability to switch on or switch off lysosome tubulation using Tudor or MMP9 inhibition will help uncover new roles of TLs and potentially modulate macrophage function.
Methods
Competition assays, tubulation assays, inhibitor assays, TL quantification, immunofluorescence, gene expression analysis, MMP9 activity, characterization of DNA nanodevices, quantitative maps of enzymatic activity, pH and calcium in TLs, and phagocytosis assays are described in SI Appendix, Materials and Methods. Details of cell lines and primary cells, reagents, and all chemicals with supplier information are detailed in SI Appendix, Materials and Methods and SI Appendix tables. SI Appendix also contains supplementary notes and figures with figure legends as well as the legend for Movie S1.
Supplementary Material
Acknowledgments
We thank Prof. J. W. Szostak, J. Zou, and C. Labno for valuable discussions, and the Integrated Light Microscopy and Biophysics Cores at The University of Chicago. This work was supported by the University of Chicago Women’s Board (Y.K.); FA9550-19-0003 from the Air Force Office of Scientific Research (Y.K.); NIH grants R21NS114428 (Y.K.), 1R01NS112139-01A1 (Y.K.), R01DK102960 (L.B.), and R01HL137998 (L.B.); and the Ono Pharma Foundation Breakthrough Science Award (Y.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113174118/-/DCSupplemental.
Data Availability
All data related to the study are included in the article and supporting information. The raw data supporting Figs. 1–6, respectively, are available for public access at Figshare (80–85): https://figshare.com/articles/dataset/Figure_1/16571561/1, https://figshare.com/articles/dataset/Figure_2/16571570/2, https://figshare.com/articles/dataset/Figure_3/16571579/2, https://figshare.com/articles/dataset/Figure_4/16571582/2, https://figshare.com/articles/dataset/Figure_5/16571585/2, and https://figshare.com/articles/dataset/Figure_6/16574231/2.
References
- 1.Perera R. M., Zoncu R., The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 32, 223–253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hipolito V. E. B., Ospina-Escobar E., Botelho R. J., Lysosome remodelling and adaptation during phagocyte activation. Cell. Microbiol. 20, e12824 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Saric A., et al., mTOR controls lysosome tubulation and antigen presentation in macrophages and dendritic cells. Mol. Biol. Cell 27, 321–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L., et al., Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonet-Ponce L., et al., LRRK2 mediates tubulation and vesicle sorting from lysosomes. Sci. Adv. 6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson A. E., Shu H., Hauswirth A. G., Tong A., Davis G. W., VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. elife 4, e07366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura N., et al., Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509, 240–244 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Miao R., Li M., Zhang Q., Yang C., Wang X., An ECM-to-nucleus signaling pathway activates lysosomes for C. elegans larval development. Dev. Cell 52, 21–37.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Meng F., Lowell C. A., Lipopolysaccharide (LPS)-induced macrophage activation and signal transduction in the absence of Src-family kinases Hck, Fgr, and Lyn. J. Exp. Med. 185, 1661–1670 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayanaswamy N., et al., A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat. Methods 16, 95–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung K., Chakraborty K., Saminathan A., Krishnan Y., A DNA nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 14, 176–183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnan Y., Zou J., Jani M. S., Quantitative imaging of biochemistry in situ and at the nanoscale. ACS Cent. Sci. 6, 1938–1954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modi S., Nizak C., Surana S., Halder S., Krishnan Y., Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 8, 459–467 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty K., et al., Tissue-specific targeting of DNA nanodevices in a multicellular living organism. elife 10, e67830 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantegazza A. R., et al., TLR-dependent phagosome tubulation in dendritic cells promotes phagosome cross-talk to optimize MHC-II antigen presentation. Proc. Natl. Acad. Sci. U.S.A. 111, 15508–15513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin-Konigsberg R., et al., Phagolysosome resolution requires contacts with the endoplasmic reticulum and phosphatidylinositol-4-phosphate signalling. Nat. Cell Biol. 21, 1234–1247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan T. K., et al., Matrix metalloproteinase-9 of tubular and macrophage origin contributes to the pathogenesis of renal fibrosis via macrophage recruitment through osteopontin cleavage. Lab. Invest. 93, 434–449 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Gough P. J., Gomez I. G., Wille P. T., Raines E. W., Macrophage expression of active MMP-9 induces acute plaque disruption in apoE-deficient mice. J. Clin. Invest. 116, 59–69 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L., Ye Y., Zhu X., MMP-9 secreted by tumor associated macrophages promoted gastric cancer metastasis through a PI3K/AKT/Snail pathway. Biomed. Pharmacother. 117, 109096 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Aptekar S., et al., Selective targeting to glioma with nucleic acid aptamers. PLoS One 10, e0134957 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veetil A. T., et al., DNA-based fluorescent probes of NOS2 activity in live brains. Proc. Natl. Acad. Sci. U.S.A. 117, 14694–14702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukasawa M., et al., SRB1, a class B scavenger receptor, recognizes both negatively charged liposomes and apoptotic cells. Exp. Cell Res. 222, 246–250 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Saminathan A., et al., A DNA-based voltmeter for organelles. Nat. Nanotechnol. 16, 96–103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha S., Prakash V., Halder S., Chakraborty K., Krishnan Y., A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 10, 645–651 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Modi S., et al., A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 4, 325–330 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Chan Y. G. Y., Cardwell M. M., Hermanas T. M., Uchiyama T., Martinez J. J., Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 11, 629–644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mrakovic A., Kay J. G., Furuya W., Brumell J. H., Botelho R. J., Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic 13, 1667–1679 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Jani M. S., Zou J., Veetil A. T., Krishnan Y., A DNA-based fluorescent probe maps NOS3 activity with subcellular spatial resolution. Nat. Chem. Biol. 16, 660–666 (2020). [DOI] [PubMed] [Google Scholar]
- 29.van Maanen J. M., Retèl J., de Vries J., Pinedo H. M., Mechanism of action of antitumor drug etoposide: A review. J. Natl. Cancer Inst. 80, 1526–1533 (1988). [DOI] [PubMed] [Google Scholar]
- 30.Monferran S., Paupert J., Dauvillier S., Salles B., Muller C., The membrane form of the DNA repair protein Ku interacts at the cell surface with metalloproteinase 9. EMBO J. 23, 3758–3768 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee E.-J., Kim H.-S., Inhibitory mechanism of MMP-9 gene expression by ethyl pyruvate in lipopolysaccharide-stimulated BV2 microglial cells. Neurosci. Lett. 493, 38–43 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., et al., Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513, 440–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong D. X., Yamori T., ZSTK474, a novel phosphatidylinositol 3-kinase inhibitor identified using the JFCR39 drug discovery system. Acta Pharmacol. Sin. 31, 1189–1197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbibe L., et al., Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Yamada O., Ozaki K., Akiyama M., Kawauchi K., JAK-STAT and JAK-PI3K-mTORC1 pathways regulate telomerase transcriptionally and posttranslationally in ATL cells. Mol. Cancer Ther. 11, 1112–1121 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y. P., Brown J. R., Sag D., Zhang L., Suttles J., Adenosine 5′-monophosphate-activated protein kinase regulates IL-10-mediated anti-inflammatory signaling pathways in macrophages. J. Immunol. 194, 584–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi C.-O., et al., Resveratrol activates AMPK and suppresses LPS-induced NF-κB-dependent COX-2 activation in RAW 264.7 macrophage cells. Anat. Cell Biol. 44, 194–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shackelford D. B., Shaw R. J., The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jakoš T., Pišlar A., Jewett A., Kos J., Cysteine cathepsins in tumor-associated immune cells. Front. Immunol. 10, 2037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dan K., Veetil A. T., Chakraborty K., Krishnan Y., DNA nanodevices map enzymatic activity in organelles. Nat. Nanotechnol. 14, 252–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korkmaz B., et al., Therapeutic targeting of cathepsin C: From pathophysiology to treatment. Pharmacol. Ther. 190, 202–236 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Liu Q., et al., Cathepsin C promotes microglia M1 polarization and aggravates neuroinflammation via activation of Ca2+-dependent PKC/p38MAPK/NF-κB pathway. J. Neuroinflammation 16, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty K., Leung K., Krishnan Y., High lumenal chloride in the lysosome is critical for lysosome function. elife 6, e28862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Q., et al., Calcium release through P2X4 activates calmodulin to promote endolysosomal membrane fusion. J. Cell Biol. 209, 879–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., Zhao Z., Gu M., Feng X., Xu H., Release and uptake mechanisms of vesicular Ca2+ stores. Protein Cell 10, 8–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., et al., TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P., Gu M., Xu H., Lysosomal ion channels as decoders of cellular signals. Trends Biochem. Sci. 44, 110–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen K. A., Myers J. T., Swanson J. A., pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 115, 599–607 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Li X., et al., A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaniuk N. A., et al., Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell. Microbiol. 13, 1812–1823 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Cabukusta B., Neefjes J., Mechanisms of lysosomal positioning and movement. Traffic 19, 761–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson D. E., Ostrowski P., Jaumouillé V., Grinstein S., The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212, 677–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guilliams M., et al., Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hipolito V. E. B., et al., Enhanced translation expands the endo-lysosome size and promotes antigen presentation during phagocyte activation. PLoS Biol. 17, e3000535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thekkan S., et al., A DNA-based fluorescent reporter maps HOCl production in the maturing phagosome. Nat. Chem. Biol. 15, 1165–1172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levin R., Grinstein S., Canton J., The life cycle of phagosomes: Formation, maturation, and resolution. Immunol. Rev. 273, 156–179 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Cheng C.-Y., Hsieh H.-L., Hsiao L.-D., Yang C.-M., PI3-K/Akt/JNK/NF-κB is essential for MMP-9 expression and outgrowth in human limbal epithelial cells on intact amniotic membrane. Stem Cell Res. (Amst.) 9, 9–23 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Shihab P. K., et al., TLR2 and AP-1/NF-kappaB are involved in the regulation of MMP-9 elicited by heat killed Listeria monocytogenes in human monocytic THP-1 cells. J. Inflamm. 12, 32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y., et al., Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. elife 9, e55745 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castellano E., Downward J., RAS interaction with PI3K: More than just another effector pathway. Genes Cancer 2, 261–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carricaburu V., et al., The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc. Natl. Acad. Sci. U.S.A. 100, 9867–9872 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inoki K., Kim J., Guan K.-L., AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 52, 381–400 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Munson M. J., et al., mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 34, 2272–2290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis L. C., Morgan A. J., Galione A., NAADP-regulated two-pore channels drive phagocytosis through endo-lysosomal Ca2+ nanodomains, calcineurin and dynamin. EMBO J. 39, e104058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Netea M. G., et al., Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Viola A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A., The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Creasy B. M., McCoy K. L., Cytokines regulate cysteine cathepsins during TLR responses. Cell. Immunol. 267, 56–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su S., et al., Genetically encoded calcium indicator illuminates calcium dynamics in primary cilia. Nat. Methods 10, 1105–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saffi G. T., Botelho R. J., Lysosome fission: Planning for an exit. Trends Cell Biol. 29, 635–646 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Lacombe J., Karsenty G., Ferron M., Regulation of lysosome biogenesis and functions in osteoclasts. Cell Cycle 12, 2744–2752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou C., et al., SNX10 plays a critical role in MMP9 secretion via JNK-p38-ERK signaling pathway. J. Cell. Biochem. 118, 4664–4671 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Tekin C., et al., Macrophage-secreted MMP9 induces mesenchymal transition in pancreatic cancer cells via PAR1 activation. Cell Oncol. (Dordr.) 43, 1161–1174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saminathan A., et al., Figure 1. Dataset. Figshare. 10.6084/m9.figshare.16571561.v1. Deposited 9 June 2021. [DOI]
- 81.Saminathan A., et al., Figure 2. Dataset. Figshare. 10.6084/m9.figshare.16571570.v2. Deposited 9 June 2021. [DOI]
- 82.Saminathan A., et al., Figure 3. Dataset. Figshare. 10.6084/m9.figshare.16571579.v2. Deposited 9 June 2021. [DOI]
- 83.Saminathan A., et al., Figure 4. Dataset. Figshare. 10.6084/m9.figshare.16571582.v2. Deposited 9 June 2021. [DOI]
- 84.Saminathan A., et al., Figure 5. Dataset. Figshare. 10.6084/m9.figshare.16571585.v2. Deposited 9 June 2021. [DOI]
- 85.Saminathan A., et al., Figure 6. Dataset. Figshare. 10.6084/m9.figshare.16574231.v2. Deposited 9 June 2021. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data related to the study are included in the article and supporting information. The raw data supporting Figs. 1–6, respectively, are available for public access at Figshare (80–85): https://figshare.com/articles/dataset/Figure_1/16571561/1, https://figshare.com/articles/dataset/Figure_2/16571570/2, https://figshare.com/articles/dataset/Figure_3/16571579/2, https://figshare.com/articles/dataset/Figure_4/16571582/2, https://figshare.com/articles/dataset/Figure_5/16571585/2, and https://figshare.com/articles/dataset/Figure_6/16574231/2.