Abstract
Exonic splicing enhancers (ESEs) are important cis elements required for exon inclusion. Using an in vitro functional selection and amplification procedure, we have identified a novel ESE motif recognized by the human SR protein SC35 under splicing conditions. The selected sequences are functional and specific: they promote splicing in nuclear extract or in S100 extract complemented by SC35 but not by SF2/ASF. They can also function in a different exonic context from the one used for the selection procedure. The selected sequences share one or two close matches to a short and highly degenerate octamer consensus, GRYYcSYR. A score matrix was generated from the selected sequences according to the nucleotide frequency at each position of their best match to the consensus motif. The SC35 score matrix, along with our previously reported SF2/ASF score matrix, was used to search the sequences of two well-characterized splicing substrates derived from the mouse immunoglobulin M (IgM) and human immunodeficiency virus tat genes. Multiple SC35 high-score motifs, but only two widely separated SF2/ASF motifs, were found in the IgM C4 exon, which can be spliced in S100 extract complemented by SC35. In contrast, multiple high-score motifs for both SF2/ASF and SC35 were found in a variant of the Tat T3 exon (lacking an SC35-specific silencer) whose splicing can be complemented by either SF2/ASF or SC35. The motif score matrix can help locate SC35-specific enhancers in natural exon sequences.
Accurate removal of introns from pre-mRNA requires multiple cis elements, including the splice sites, polypyrimidine tract, branch site, and other intronic and exonic sequences that have positive or negative effects on splicing (10, 31, 44); reviewed in references (1 and 2). Positive-acting sequences, termed exonic splicing enhancers (ESEs) (37, 39, 42), have been identified primarily in exons associated with regulated splicing. These exons are typically adjacent to introns with weak intronic splicing signals and require ESEs for their inclusion. Deletion of an ESE often causes exon skipping or, in the case of terminal exons, suppresses removal of the last intron. One of the first characterized ESEs is located in the M2 3′-terminal exon of the mouse immunoglobulin M (IgM) gene (39). This 73-nucleotide (nt) ESE, which is highly purine rich, is required for inclusion of the alternatively spliced M2 exon. However, deletion of just the purine-rich sequences within this ESE does not abolish splicing completely. The M2 ESE also functions in a heterologous context to enhance splicing of a Drosophila melanogaster doublesex intron (39).
A SELEX procedure has been used to identify sequences that can function as ESEs (36). A 20-nt sequence of the internal duplicated exon of a model pre-mRNA was replaced by 20 nt of random sequence. The randomized pre-mRNAs were incubated under splicing conditions in nuclear extract, and functional enhancer elements that promoted splicing were selected. A large number of sequences, both purine rich and non-purine rich were obtained, and the two types of sequences stimulated exon inclusion to similar extents. A similar approach was used in an in vivo system involving transfection of a troponin minigene with random sequences in place of a natural ESE (9). Purine-rich sequences and a novel class of AC-rich ESE sequences were identified. The AC-rich sequences are efficient splicing enhancers and can also function in a heterologous gene context.
Considerable evidence suggests that ESEs interact specifically with a family of RNA-binding proteins called SR proteins, which are characterized by one or two RNA recognition motifs (RRMs) and a C-terminal Arg-Ser-rich domain (12, 18, 27, 34, 37, 38). SR proteins are essential splicing factors required for both constitutive and alternative splicing (11, 17, 43). SR proteins can determine alternative splice site selection by antagonizing the activity of hnRNP A/B proteins. High concentrations of SR proteins usually favor the use of proximal splice sites and exon inclusion, whereas high concentrations of hnRNP A/B proteins tend to favor distal splice sites and exon skipping (5, 22, 25). SR proteins also specifically recognize ESEs, and the resulting complex may then stimulate U2AF binding to the weak polypyrimidine tract of the upstream 3′ splice site. The ESE–SR protein–U2AF interaction is thought to be important during the early stages of spliceosome assembly (8, 16, 41, 45), although recent evidence suggests that, in at least some cases, including the IgM M2 exon, ESEs act in part by neutralizing exonic silencer elements (7, 15). SF2/ASF and SC35 are two of the best characterized among the nine human SR proteins identified to date. Both proteins have been implicated in many aspects of constitutive and regulated splicing. Both are found in the prespliceosomal E complex and can interact with U1-70K and U2AF by RS domain-mediated protein-protein interactions. The RRMs of these two proteins are responsible for their unique substrate specificities (6, 26).
A better understanding of the functional interactions between ESEs and SR proteins depends on knowledge of the sequence specificity of all SR proteins. To this end, we recently performed an iterative selection under splicing conditions to identify exon sequences that can enhance splicing in the presence of each of three SR proteins. We identified three novel classes of functional ESE motifs recognized specifically by SF2/ASF, SRp40, and SRp55. The consensus motifs indicated that individual SR proteins recognize distinct and highly degenerate sequences (20). The three SR proteins we studied previously are closely related, i.e., they all have two tandem RRMs. To extend this analysis, we have now determined the sequence specificity of an additional, extensively studied SR protein, SC35, which has a single N-terminal RRM.
MATERIALS AND METHODS
Preparation of HeLa cell extract and recombinant SR proteins.
HeLa nuclear and S100 extracts were prepared as described (23). Recombinant SC35 expressed in baculovirus was generously provided by K. Lynch and T. Maniatis and by R.-M. Xu.
Selection and amplification procedure.
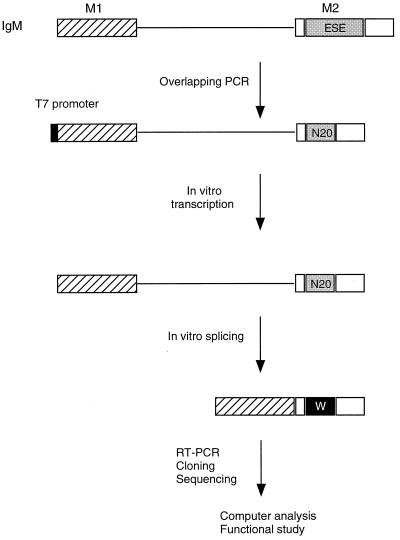
The amplification and selection procedure was carried out as described (20). Briefly, the natural ESE of the IgM pre-mRNA was replaced by 20 nt of random sequence by overlap-extension PCR with plasmid μMΔ DNA (39) as a template. The resulting PCR product was used for in vitro transcription to generate a 32P-labeled random pre-mRNA pool. Twenty femtomoles of the pre-mRNA pool was incubated under in vitro splicing conditions in S100 extract plus recombinant SC35 in a 25-μl reaction mixture. The RNA was separated by denaturing polyacrylamide gel electrophoresis, and the spliced mRNAs were excised and eluted from the gel in 0.5 M ammonium acetate plus 0.1% sodium dodecyl sulfate and reamplified by reverse transcription-PCR (RT-PCR). Reverse transcription was carried out by using Superscript II as described by the manufacturer (Life Technologies). PCR was performed by using high-fidelity Pfu polymerase as specified by the manufacturer (Stratagene). The PCR product was subcloned into the vector PCR-Blunt (Stratagene) and sequenced by using a Dye Terminator Cycle Sequencing kit (Perkin-Elmer) and an automated ABI 377 sequencer. Selected winner sequences were rebuilt into DNA templates for transcription of pre-mRNAs by overlap-extension PCR, as done initially for the random sequences (20).
Sequence analysis and construction of score matrices.
The selected sequences of each SR protein winner pool plus a portion of the flanking nucleotides were aligned by using Gibbs sampler (19). The identified consensus motif was then used to generate a score matrix. The compositional bias of the initial RNA pool was taken into account. For details of the sequence analysis, see reference (20).
In vitro splicing.
PCR products carrying an SP6 or T7 promoter were used for in vitro transcription. 5′-capped transcripts were incubated in 25-μl splicing reaction mixtures as previously described (24). Each reaction mixture had 4 μl of nuclear extract or 7 μl of S100 extract. For S100 complementation assays, 20 pmol of specific SR protein was used. Splicing reactions were carried out at 30°C for 4 h. The RNA was then extracted, loaded on 6 or 12% polyacrylamide gels, and visualized by autoradiography (20). DNA templates for IgM M1-M2 pre-mRNAs with a D2 variant containing an SC35 consensus match or with the 6-24 winner sequence (29) were made by overlap-extension PCR with primers M2-D2HXL (GTGAAATGACTCTCAGCATggggacatactcggcccctgCTAGTAAACTTATTCTTACGT) and M2-SCH24 (GTGAAATGACTCTCAGCATtttgcggtctccggcctccCTAGTAAACTTATTCTTACGT), respectively (shared flanking sequences are in uppercase letters). DNA templates for pre-mRNAs in an IgM C3-C4 context were made by PCR on pμC3-C4 plasmid DNA (40) with an SP6 promoter primer and the following antisense primers: Ca (TGGCAGCAGGTACACAGC), CaCb (gtggctgactccctcagg), D2 (ctgcggccgagtatgtccccTGGCAGCAGGTACACAGC) D2C (caggggccgagtatgtccccTGGCAGCAGGTACACA GC) and 6-24 (ggaggccggagaccgcaaaTGGCAGCAGGTACACAGC). RNAs were made as described above.
RESULTS
Identification of ESE motifs recognized by SC35 under splicing conditions.
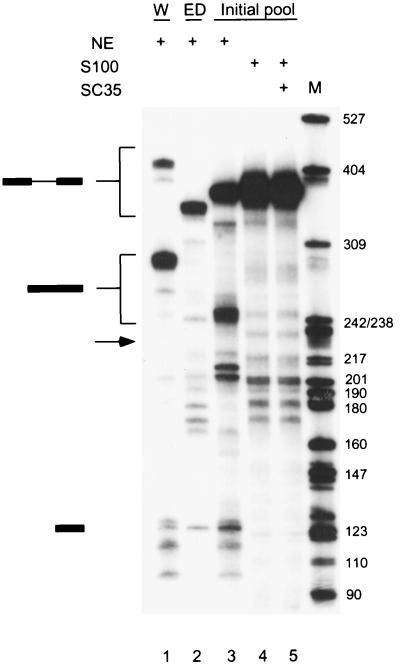
To study the sequence specificity of ESE recognition by SC35 under splicing conditions, a functional SELEX procedure (20) was used (Fig. 1). Functional ESEs were selected in the context of a well-characterized mouse immunoglobulin μ heavy chain minigene transcript, comprising the last intron flanked by the M1 and M2 exons (39). The natural ESE in the M2 exon was replaced by 20 nt of random sequence by overlap-extension PCR. The random RNA pool, a library of pre-mRNAs representing 1.2 × 1010 different molecules, was spliced in nuclear extract or in S100 extract complemented by SC35. As previously reported, the wild-type IgM pre-mRNA spliced very efficiently in nuclear extract, with the mature mRNA representing greater than 90% of the RNA after a 4-h incubation (Fig. 2, lane 1). In contrast, the mutant with a deletion of ESE (ED) did not splice at all under the same conditions (Fig. 2, lane 2), confirming that the natural ESE of the IgM pre-mRNA is essential for splicing (39). The initial RNA pool was spliced in nuclear extract with an apparent efficiency of about 20% (Fig. 2, lane 3), whereas no splicing was detected in the S100 extract alone (lane 4). When the S100 extract was complemented by SC35, splicing of the initial RNA pool remained undetectable by autoradiography (lane 5). However, we assumed that a very small fraction of the RNA pool was correctly spliced, and we excised a gel slice corresponding to the position of spliced mRNA, using the product in lane 3 as a marker. RNA was eluted from the gel slice and amplified by RT-PCR. The amplified products were cloned, and 30 clones were sequenced. The resulting sequences were analyzed by using the program GIBBS sampler to determine a consensus sequence (19, 20). A score matrix was generated according to the frequency of each nucleotide at each position of the consensus motif, adjusted for the compositional bias of the initial random pool. This score matrix was used to identify high-score motifs within each winner sequence, taking into account the randomized sequence region and a small portion of the flanking sequences.
FIG. 1.
Experimental procedure for functional SELEX. The structure of the IgM M1-M2 minigene pre-mRNA is shown. The characterized natural ESE (73 nt) was replaced by 20 nt of randomized sequence by overlap-extension PCR (20). A T7 promoter (black box) was built into the PCR product. In vitro-transcribed RNA was then incubated under splicing conditions. Any spliced mRNA molecules must contain a functional ESE or winner sequence, designated by the white W in a black box. The spliced mRNA molecules were purified from a denaturing polyacrylamide gel, reamplified by RT-PCR, and cloned. Individual clones were sequenced and analyzed by a sampling algorithm to define a common motif, and a subset was rebuilt into minigene templates, transcribed, and assayed for splicing in vitro.
FIG. 2.
Splicing of the initial RNA pool. Twenty femtomoles of wild-type IgM minigene pre-mRNA (W; lane 1), pre-mRNA with a deletion of the ESE (ED; lane 2), or a pre-mRNA pool representing 1.2 × 1010 different molecules with a randomized 20-nt segment within exon M2 were spliced in 25-μl reaction mixtures in nuclear extract (lane 3), S100 extract alone (lane 4), or S100 extract complemented by 20 pmol of recombinant SC35 (lane 5). The structures of the precursor, intermediates, and products are indicated next to the autoradiogram. The expected size of the spliced mRNA product of the ESE deletion mutant is indicated by an arrow.
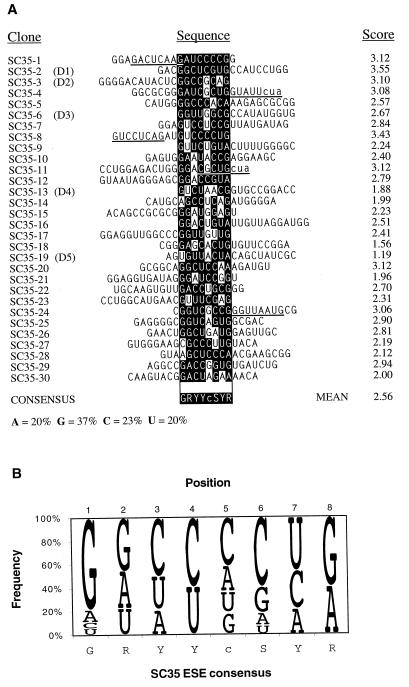
The SC35 winner sequences after a single round of selection yielded the short degenerate octamer consensus motif GRYYcSYR (Fig. 3). The C-residue content within the randomized 20-nt segment increased from 19% in the initial pool to 23% after a single round of selection. This change in C composition occurred at the expense of slight reductions in the content of G, A, and U residues. As reported previously for our similar analysis of other SR proteins, the SC35 consensus sequence is highly degenerate. Several of the winner sequences have more than one high-score motif (Fig. 3A). The scores of the 30 SC35 winner sequences range from 1.19 to 3.55, with a mean score of 2.56 ± 0.56. Thirty individual sequences cloned and randomly selected from the initial pool (20) gave a range of scores from 0.64 to 3.23, with a mean score of 1.62 ± 0.72, when searched by the same score matrix. Only 3 sequences in the control pool had scores higher than the mean of the winner pool, whereas 16 sequences in the winner pool had scores higher than this mean, and 28 had scores higher than the mean of the control pool. The difference in the means of the scores between the two sequence pools is highly significant (P < 10−7, t test with df = 58).
FIG. 3.
Analysis of the SC35-selected sequences. (A) Sequence alignment and identification of a consensus motif. The consensus motif and score matrix were derived as described previously (20). The sequences were aligned on the basis of the highest score motif for each sequence. Nucleotides matching the consensus are shown as white on a black background; mismatched nucleotides are not shaded. The scores of the aligned motifs are indicated on the right. Additional motifs present in some of the sequences with a score greater than 1.62 (the mean score of the random pool) are underlined. In two cases these include a trinucleotide contributed by the 3′-flanking sequence, which is indicated with lowercase letters (cua). The consensus shown is only an approximation that indicates the most frequent nucleotide(s) at each position. The lowercase c at position 5 denotes a slight preference for this nucleotide over the other three nucleotides, which occur at similar frequencies. Y, pyrimidine; S, G or C; R, purine. The nucleotide composition of the selected pool is shown at the bottom. The nucleotide composition of the initial RNA pool was as follows: A, 21%; G, 39%; C, 19%, and U, 21%. (B) Representation of the SC35 ESE score matrix and consensus motif. The diagram shows the frequency of each nucleotide at each position of the octamer consensus, adjusted for the compositional bias of the initial pool (20). The height of each letter is proportional to its frequency, and the nucleotides are shown from top to bottom in decreasing order of frequency. This method of displaying nucleotide frequencies is based on references (3) and (13).
The highest possible score for a single octamer is 3.95, corresponding to the sequence GGCCCCUG (Fig. 3B). This precise sequence does not occur in any of the 30 winner sequences analyzed. The absence of a perfect motif in the selected sequences may reflect the small sample size or the fact that a linear consensus sequence or nucleotide frequency matrix assumes an independent contribution at each position, an assumption that may or may not fit the actual recognition mechanism (33).
The SC35-selected sequences are functional and specific ESEs.
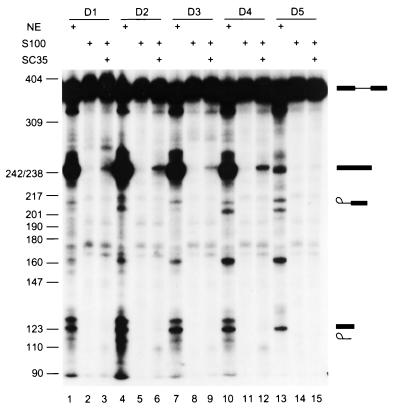
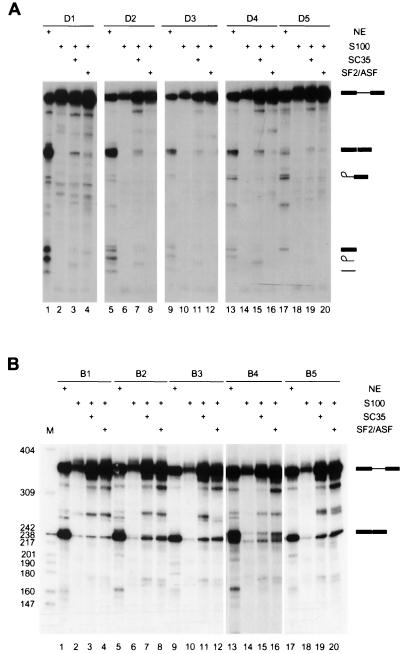
We next tested whether the individual sequences selected in the presence of SC35 could function as true splicing enhancers. Five sequences with a range of scores were arbitrarily chosen from the 30 analyzed sequences and individually rebuilt into IgM M1-M2 pre-mRNAs with the same structure as those in Fig. 1 and 2, using overlap-extension PCR and in vitro transcription (20). Each pre-mRNA was then incubated under splicing conditions in nuclear extract or in S100 extract complemented by SC35 (Fig. 4). Four of the five SC35 winner sequences activated IgM pre-mRNA splicing very efficiently in nuclear extract (Fig. 4, lanes 1, 4, 7, and 10). They also promoted IgM pre-mRNA splicing in S100 extract complemented by SC35, albeit less efficiently (Fig. 4, lanes 3, 6, 9, and 12), but not in S100 extract alone (Fig. 4, lanes 2, 5, 8, and 11). One winner sequence from the SC35 winner pool, D5, enhanced splicing less efficiently in nuclear extract (Fig. 4, lane 13) and gave only trace activity in the complementation assay (Fig. 4, lane 15). In general, the splicing efficiency correlated with the motif scores shown in Fig. 3. D1 and D2 have the highest scores; D3 and D4 have intermediate scores; and D5 has the lowest score among the 30 sequences analyzed (Fig. 3). However, the correlation between splicing efficiency and motif scores is not linear, presumably reflecting sequence context effects. Also, D3 has a higher score than D4, and although they spliced with similar efficiency in nuclear extract, D4 spliced more efficiently in the complementation assay. Sixteen sequences from the random RNA pool were also analyzed for enhancer activity (20). All of them spliced in nuclear extract poorly or not at all. In most cases the pre-mRNAs showed partial degradation, suggesting that spliceosomal complexes did not assemble on these RNAs (H.-X. Liu and A. R. Krainer, unpublished data).
FIG. 4.
Activity of the selected ESE motifs. SC35 SELEX winners were rebuilt into the IgM M1-M2 minigene by overlap-extension PCR (as described in the legend for Fig. 1), and transcripts corresponding to individual winners were spliced in HeLa nuclear extract (lanes 1, 4, 7, 10, and 13), in S100 extract alone (lanes 2, 5, 8, 11, and 14), or in S100 extract complemented by recombinant SC35 (lanes 3, 6, 9, 12, and 15).
Next, we determined the SR protein specificity of the SC35-selected ESEs. Pre-mRNAs with the different winner sequences were separately incubated under splicing conditions in S100 extract complemented by SC35, SF2/ASF, SRp40, or SRp55. All of the tested SC35 winners promoted splicing with higher efficiency in S100 extract when the extract was complemented by SC35 (Fig. 5A, lanes 3, 7, 11, 15, and 19), SRp40, or SRp55 (Liu and Krainer, unpublished). When the extract was complemented by SF2/ASF, the splicing efficiencies were much lower (Fig. 5A, lanes 4, 8, 12, 16, and 20). In contrast, five SF2/ASF-selected winners promoted splicing in S100 extract complemented by either SF2/ASF (Fig. 5B, lanes 4, 8, 12, 16, and 20) (20) or SC35 (Fig. 5B, lanes 3, 7, 11, 15, and 19) with comparable efficiencies. These SF2/ASF winners promoted splicing very poorly or not at all in the presence of SRp40 or SRp55 (20).
FIG. 5.
Specificity of the selected ESE motifs. (A) Splicing of SC35-selected ESEs was analyzed in nuclear extract (lanes 1, 5, 9, 13, and 17), S100 extract alone (lanes 2, 6, 10, 14, and 17), or S100 extract plus recombinant SC35 (lanes 3, 7, 11, 15, and 19) or recombinant SF2/ASF (lanes 4, 8, 12, 16, and 20). (B) Splicing of SF2/ASF-selected ESEs was examined in nuclear extract (lanes 1, 5, 9, 13, and 17), S100 extract alone (lanes 2, 6, 10, 14, and 18), or in S100 extract plus SC35 (lanes 3, 7, 11, 15, and 19) or SF2/ASF (lanes 4, 8, 12, 16, and 20).
Comparison of SC35 ESE motifs and activity in different exonic contexts.
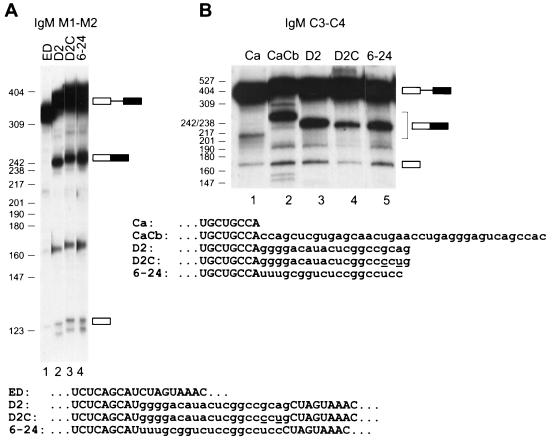
To test whether an octamer with the highest possible SC35 ESE score has enhancer activity and to compare this consensus with a previously identified one, we analyzed the D2 winner containing the motif GGCCGCAG, a variant of D2 with two transversions that create the maximum score consensus GGCCCCUG, and one of the 19mer winners (6-24) selected by Schaal and Maniatis (29). These three sequences were first tested in the context of the IgM M2 exon (Fig. 6A). All three sequences strongly promoted splicing of exons M1 and M2 in nuclear extract (Fig. 6A, lanes 3 to 5), in contrast to the lack of detectable splicing with the parent pre-mRNA in which the natural ESE was deleted (Fig. 6A, lane 1).
FIG. 6.
Comparison of SC35 winner sequences and SC35 ESE motif in two different exonic contexts. (A) The 20-nt D2 winner sequence (see Fig. 3), a variant of D2 with two nucleotide changes to introduce a maximum-score consensus (D2C), and a 19-nt SC35 SELEX winner sequence (6-24) described in a previous study (29) were inserted into the IgM M1-M2 minigene in place of the natural ESE in exon M2, and the corresponding transcripts were spliced in nuclear extract (lanes 2 to 4). The control pre-mRNA lacking an ESE (ED) is shown in lane 1. (B) The same D2, D2C, and 6-24 sequences were tested in the context of an IgM C3-C4 minigene (26). The Ca pre-mRNA includes the first 38 nt of the C4 exon (lane 1). In the remaining pre-mRNAs, this segment of the C4 exon is followed by the next 38 nt of the C4 exon, which comprise a natural SC35-dependent ESE (CaCb; lane 2) or it is followed by the D2, D2C, or 6-24 sequence (lanes 3 to 5). The sequences of the relevant portions of the 3′ exons are shown below each panel. The two nucleotide changes in D2C, compared to D2, are underlined. The mobilities of the pre-mRNAs, mRNAs, and 5′ exon intermediates are indicated next to each autoradiogram.
Next we tested the same three ESEs in a different exonic context, namely the C4 exon derived from a different region of the IgM pre-mRNA. When this exon is divided into three segments, Ca, Cb, and Cc, the Cc segment is dispensable, whereas the Cb segment behaves as an SC35-specific ESE (26). Indeed, a shortened 3′ exon consisting of the Ca and Cb segments of C4 spliced to exon C3 much more efficiently in nuclear extract than one consisting of Ca alone (Fig. 6B, lanes 1 and 2). When the Cb segment was replaced by each of the above three ESEs, all of them promoted splicing above the background of Ca alone (Fig. 6B, lanes 4 to 6). However, the D2 winner ESE was as strong as the natural Cb ESE, the 6-24 ESE was slightly less efficient, and the perfect consensus was the least active. These results show that both our SC35 motif and a winner sequence identified in a previous study (29) can function in different exonic contexts, although the precise context can influence the extent of enhancement.
Distribution of SC35 ESE motifs in natural genes.
To determine whether the selected ESE motifs are relevant to splicing of natural pre-mRNA substrates, we conducted a search of SC35 high-score motifs in natural genes. Only scores higher than the lowest score of the SC35 winner pool are shown (Fig. 7, green vertical bars). For comparison, we also show the high-score SF2/ASF motifs in the same genes (Fig. 7, blue vertical bars) (20). The first natural sequence we examined was the M2 exon of the IgM gene. The search result indicated that there are many SC35 ESE motifs within the segment comprising the previously characterized natural ESE (Fig. 7A, magenta horizontal bar). The distribution of high-score SC35 motifs differs from that of SF2/ASF motifs. SF2/ASF-specific motifs are present at a higher density within the natural ESE than in the flanking regions. In contrast, high-score SC35 motifs have a relatively even distribution across the M2 exon. Both SR proteins can promote splicing of this pre-mRNA in S100 extract (Liu and Krainer, unpublished). The presence of ESE motifs in regions lacking enhancer activity shows that although the motifs may be necessary, they are not sufficient for ESE function (see Discussion).
FIG. 7.
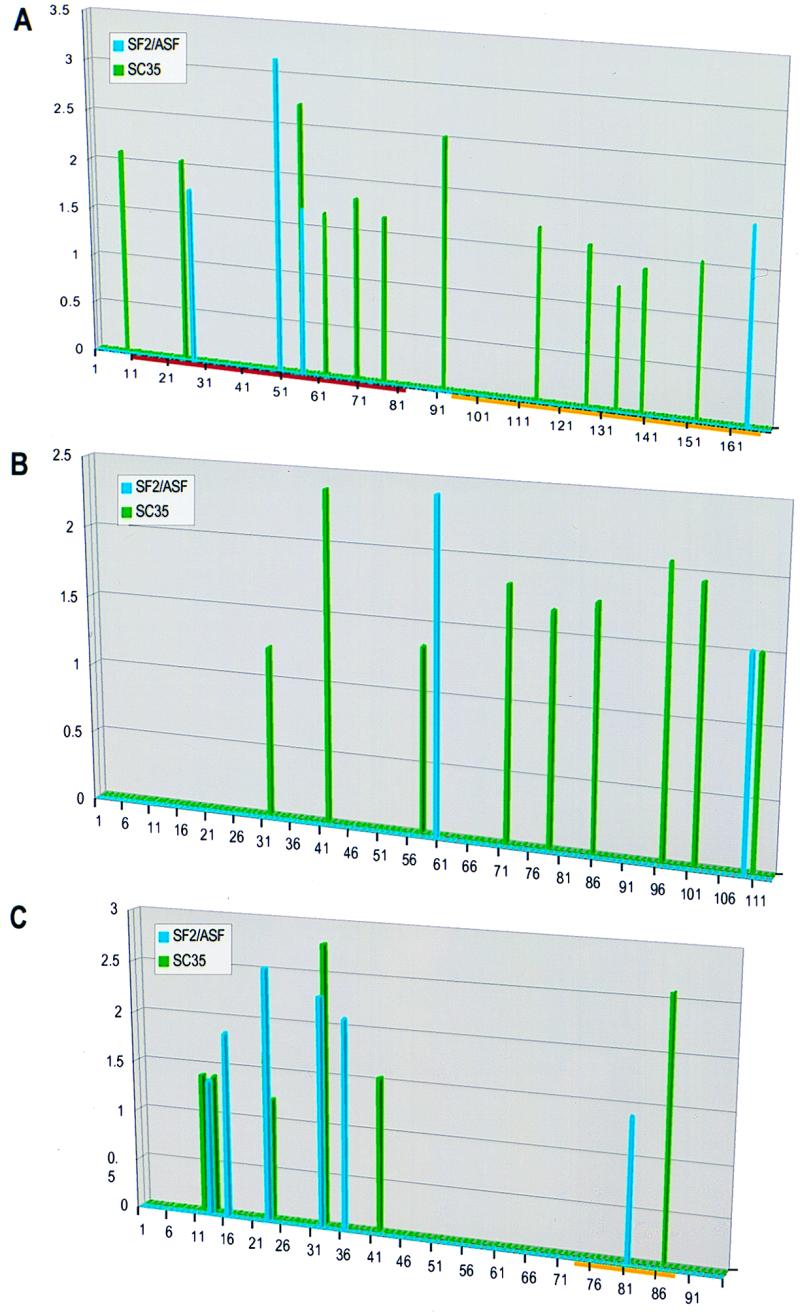
Correlation between predicted ESE motifs in natural genes and SR protein specificity of the pre-mRNAs. Score matrices derived for SC35 and SF2/ASF were used to search the sequences of natural genes. The resulting scores (y axis) were plotted against the nucleotide position along each exon (x axis). The vertical bars indicate the first nucleotide of each motif. SC35 high-score motifs are shown in green, and SF2/ASF ones are shown in blue. Since different score matrices were used for each protein, the numerical scores of the two different proteins cannot be compared. (A) High-score motifs in the IgM gene M2 exon. The characterized ESE is indicated by the horizontal magenta bar, and the yellow bar indicates the region comprising a recently described silencer (15, 39). (B) High-score motifs in the IgM gene C4 exon. (C) High-score motifs in the tat gene T3 exon. The horizontal yellow bar indicates the position of the SC35-specific silencer.
To address the issue of whether the identified ESE motifs are specific to SC35, we searched two additional pre-mRNA substrates that are known to have different SR protein specificities. Splicing of the IgM C3-C4 pre-mRNA is activated in S100 extract when complemented by SC35 but not by SF2/ASF (26). In contrast, splicing of the human immunodeficiency virus Tat T2-T3 pre-mRNA is activated by SF2/ASF but not by SC35 in S100 extract (6, 26). When an SC35-specific splicing silencer in the 3′ region of the T3 exon is deleted, both SF2/ASF and SC35 can activate T2-T3 splicing in S100 extract. Detailed analysis of the splicing of these two pre-mRNAs indicated that the C4 and T3 exons determine the SR protein specificity (26). Our search result matches the experimental data (Fig. 7B and C). Many high-score motifs matching the consensus of SC35 were found in the C4 exon, but only two well-separated SF2/ASF motifs were found in this exon (Fig. 7B). Interestingly, in a deletion mutant missing the first 38 nt of the C4 exon, splicing of C3-C4 was activated by both SF2/ASF and SC35 (26). Consistent with this result, the SF2/ASF motif near position 61 is closer to the 3′ splice site in the deletion mutant. High-score motifs for both SF2/ASF and SC35 were found in the T3 exon of the tat gene (Fig. 7C). Curiously, a single SC35 high-score motif is present within the SC35-specific silencer region.
Finally, we studied the distribution of SC35 high-score motifs in human exons versus introns. A total of 570 genes, representing 2,626 exons (426 kb) and 2,079 introns (1,295 kb), were extracted from the ALLSEQ database (4) and analyzed. Scores equal to or higher than the mean score of the winner pool were taken into account. High-score motifs appeared more frequently in exons than in introns. An average of nine SC35 high-score motifs were found per kilobase of exon compared to only 5.9 per kilobase of intron. This comparison was statistically significant because of the large database size (P < 10−10).
DISCUSSION
We have identified a novel ESE motif recognized by the human SR protein SC35. Several lines of evidence point to the biological relevance of the selected ESE motifs. First, they are functional ESEs. All of the SELEX winners we have tested promote splicing in nuclear extract and in S100 extract plus the cognate SR protein. In nuclear extract, the SELEX winners function as potent ESEs. Second, the SC35 motifs are present within exon segments containing natural ESEs and are more frequently found in exons than in introns, suggesting that they may contribute to exon definition by the spliceosome. Third, the SC35 motifs are specific, i.e., they are not recognized by all SR proteins. The SC35-selected ESEs were recognized by SC35, SRp40, or SRp55, but not by SF2/ASF under splicing conditions. In addition, the distribution of high-score motifs of SF2/ASF and SC35 in the IgM C4 and Tat T3 exons correlated with the observed SR protein specificity of the corresponding substrates (26). This result also suggests that the score matrices we have generated have some predictive value. We have previously analyzed the predictive value of the SR-specific score matrices derived for other SR proteins (20). Statistically, high-score motifs of SR proteins are present at a higher density in natural ESEs than in the flanking regions. Experimentally, SR proteins specifically recognize their cognate ESE motifs when these are placed in the context of the IgM M2 exon, replacing the natural ESE. The present study confirms and extends our previous work to two natural ESEs in IgM and human immunodeficiency virus Tat exons. In addition, we have now shown that SC35 winner sequences and a maximum-score SC35 motif can promote splicing in different exonic contexts.
The specific interaction between SR proteins and ESEs has also been described in other systems. During assembly of enhancer complexes in vitro (Enh complex, which resembles the E complex), the enhancer sequences determine the specific pattern of SR proteins that can be UV cross-linked to the RNA (32). Female-specific alternative splicing of the Drosophila doublesex pre-mRNA requires six 13-nt repeat elements and a purine-rich element. UV cross-linking analysis showed that SR proteins, along with Tra and Tra-2, assemble on the ESEs in a stepwise and sequence-specific manner (21). The fact that SR proteins are expressed in a tissue-specific manner (14, 43), together with the specific recognition of ESEs by individual SR proteins may contribute quantitatively to the regulation of gene expression.
The SC35 SELEX winners have the consensus GRYYcSYR, which is a highly degenerate sequence. Even though SC35 has a single RRM, a SELEX protocol based on RNA binding yielded two different nonamer consensus sequences, AGSAGAGTA and GTTCGAGTA, which share the last five nucleotides (35). These two motifs differ significantly from the more degenerate consensus identified by functional SELEX. Although the second motif has a partial fit to the above consensus, neither motif has a good score, consistent with the observation that the high-affinity binding sequences fail to enhance splicing of RNA substrates in nuclear extract or in S100 extract plus SC35, even when present in several copies (35). Therefore, it appears that high-affinity SC35-binding sites are not optimal for function. Perhaps RNA-binding selection does not achieve an interaction geometry compatible with SC35 enhancement function, or it is essential to coselect sequences that in addition to binding SC35 can also accommodate putative coactivators or fail to bind silencing factors.
Nevertheless, our data argue that SC35 has limited but defined sequence specificity in recognizing functional sequences. Despite the fact that this protein has a single RRM, the functional recognition motif is degenerate, as was the case for the two-RRM SR proteins SF2/ASF, SRp40, and SRp55 (20). Therefore, the degeneracy of the ESE motifs recognized by those proteins is probably not attributable to the recognition of distinct motifs by each of their RRMs. The sequence degeneracy of the ESEs is consistent with the fact that they must coexist with a very wide variety of unrelated open reading frames and must be recognized by a discrete set of SR proteins (20, 28, 29).
Schaal and Maniatis recently used a similar functional SELEX approach to select ESEs that could function in the context of the Drosophila doublesex pre-mRNA in HeLa nuclear extract (29). The selected 18-nt winner sequences were then individually analyzed by S100 complementation assays to define their SR protein specificity. Two round 6 winner sequences were the most active in the presence of SC35. By comparing these two sequences to each other and to an SC35-dependent ESE present in human β-globin exon 2, the authors proposed the SC35 heptamer consensus UGCNGYY, which is also a highly degenerate sequence. Although this heptamer motif is substantially different from our consensus octamer motif, some versions of the degenerate heptamer consensus have high scores, as defined in the present study. We therefore searched the two published winner sequences (29) by using our SC35 score matrix. Both sequences had multiple high-score motifs, some of which were nonoverlapping, consistent with the fact that they had undergone six rounds of selection for splicing. In the case of the 6-24 sequence, the highest score (3.13) corresponded to the octamer GGUCUCCG, which has a 4-nt overlap with the UGCGGUC sequence that fits the heptamer consensus. In the case of the 6-38 sequence, the second highest score (1.56) corresponds to the octamer UGCCGCC, of which the first 7 nt fit the heptamer consensus; the highest score (2.44) was for the nonoverlapping octamer GGACCGGA. Similarly, within the 18-nt β-globin fragment in which Schaal and Maniatis characterized an SC35-dependent ESE that comprises the heptamer UGCUGUU (28), the highest score (1.36) corresponds to the octamer UGAUGCUG, which includes the first 5 nt of the heptamer.
We conclude that despite the very different pre-mRNA contexts, types of extract used for the selection, and number of selection rounds, the SC35 ESEs identified by the two approaches are remarkably consistent. We believe, however, that our octamer motif has greater predictive value because it was derived from a much larger number of winner sequences. Also, the use of a nucleotide frequency matrix derived from 30 sequences allows identification of putative SC35 ESEs that do not precisely match the consensus at every position. Thus, our SC35 score matrix finds high-score motifs in both of the winner sequences and the β-globin segment characterized by Schaal and Maniatis (28, 29), whereas of our 30 SC35 winner sequences (Fig. 3), only no. 14 has a precise match to the heptamer consensus they defined.
The IgM M2 exon has a higher density of SF2/ASF and SRp40 high-score motifs within the natural ESE segment than in the flanking sequences. In contrast, the SRp55 high-score motifs do not correlate with the location of the ESE (20). In the case of SC35, the high-score motifs also have a relatively even distribution across the exon. The different motif distributions may reflect different mechanisms of SR protein-ESE recognition. Although for some pre-mRNAs any SR protein can complement splicing in the S100 extract (30, 43), each SR protein may function by slightly different mechanisms. Some SR proteins may require multiple binding sites to function, and the optimal distance from the 3′ splice site to the SR protein-binding site may also be protein specific. The fact that ESE motifs are not found exclusively in natural exonic segments required for ESE activity indicates that the motifs are not sufficient for ESE function. It appears that sequence context, structure, or position effects are also very important.
Examples of sequence context effects that can influence ESE activity are provided by exonic splicing silencers. These inhibitory elements probably coexist with splicing enhancers in many exons, and they may also be SR protein dependent and function in a cell-type specific manner. For example, an SC35-dependent silencer sequence has been mapped in the tat gene T3 exon (26). This silencer element includes within it an SC35-specific ESE motif (Fig. 7C). We speculate that binding of SC35 to this region prevents the function of other splicing factors, although it is presently unclear how this element acts at a distance and suppresses the effect of SC35-dependent ESEs but not of SF2/ASF-dependent ESEs. Recently, the 3′ portion of the IgM M2 exon was also shown to comprise a silencer element that binds U2 snRNP and antagonizes the upstream ESE (15). The silencer element, so far mapped to a fragment between nt 94 and 167 (Fig. 7A) in the M2 exon, overlaps with several SC35 high-score motifs and with one SF2/ASF high-score motif.
The similar arrangement of adjacent ESE and exonic splicing silencer elements seen in the IgM M2 and Tat T3 exons may turn out to be a common feature of many vertebrate cellular and viral exons. To improve the predictive value of the SR protein-specific ESE motifs, it will be necessary to gain a better understanding of the influence of sequence context and position, as well as of the mechanistic basis for the function of splicing enhancers and silencers.
ACKNOWLEDGMENTS
We thank Y. Shimura for the gift of IgM plasmids, K. Lynch, T. Maniatis, R.-M. Xu, and A. Mayeda for recombinant SR proteins, J. Yin for DNA sequencing, A. Mayeda and members of our laboratory for valuable ideas, and M. Hastings for helpful comments on the manuscript.
This work was supported by NIH grants to A.R.K. (GM42699) and M.Q.Z. (HG01696), by an Advanced Fellowship from The Wellcome Trust to S.L.C. (045401), by a fellowship from the Human Frontiers Science Program to L.C. (LT0066/1997-M), and by a fellowship from the U.S. Army Medical Research and Matériel Command under DAMD 17-96-1-6172 to H.-X.L.
REFERENCES
- 1.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 2.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 3.Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–559. [Google Scholar]
- 4.Burset M, Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- 5.Cáceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 6.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X-D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew S L, Liu H-L, Mayeda A, Krainer A R. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc Natl Acad Sci USA. 1999;96:10655–10660. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiara M D, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- 9.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. . (Erratum, 17:3468.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freyer G A, O'Brien J P, Hurwitz J. Alterations in the polypyrimidine sequence affect the in vitro splicing reactions catalyzed by HeLa cell-free preparations. J Biol Chem. 1989;264:14631–14637. [PubMed] [Google Scholar]
- 11.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 12.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorodkin J, Heyer L J, Brunak S, Stormo G D. Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- 14.Hanamura A, Cáceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 15.Kan J L, Green M R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 17.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 18.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 20.Liu H-X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 22.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. . (Erratum, 13:4458.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayeda A, Krainer A R. Preparation of Hela cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- 24.Mayeda A, Krainer A R. Mammalian in vitro splicing assays. Methods Mol Biol. 1999;118:315–322. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 25.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 26.Mayeda A, Screaton G R, Chandler S D, Fu X-D, Krainer A R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaal T D, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaal T D, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Screaton G R, Cáceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seif I, Khoury G, Dhar R. BKV splice sequences based on analysis of preferred donor and acceptor sites. Nucleic Acids Res. 1979;6:3387–3398. doi: 10.1093/nar/6.10.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stormo G D. Consensus patterns in DNA. Methods Enzymol. 1990;183:211–237. doi: 10.1016/0076-6879(90)83015-2. [DOI] [PubMed] [Google Scholar]
- 34.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 35.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 38.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 39.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 40.Watakabe A, Inoue K, Sakamoto H, Shimura Y. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin μ chain pre-mRNAs. Nucleic Acids Res. 1989;17:8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 42.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 44.Zhuang Y A, Goldstein A M, Weiner A M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci USA. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]