Abstract
The ubiquitin-related SUMO-1 modifier can be covalently attached to a variety of proteins. To date, four substrates have been characterized in mammalian cells: RanGAP1, IκBα, and the two nuclear body-associated PML and Sp100 proteins. SUMO-1 modification has been shown to be involved in protein localization and/or stabilization and to require the activity of specialized E1-activating and E2 Ubc9-conjugating enzymes. SUMO-1 homologues have been identified in various species and belong to the so-called Smt3 family of proteins. Here we have characterized the Drosophila homologues of mammalian SUMO-1 and Ubc9 (termed dSmt3 and dUbc9, respectively). We show that dUbc9 is the conjugating enzyme for dSmt3 and that dSmt3 can covalently modify a number of proteins in Drosophila cells in addition to the human PML substrate. The dSmt3 transcript and protein are maternally deposited in embryos, where the protein accumulates predominantly in nuclei. Similar to its human counterpart, dSmt3 protein is observed in a punctate nuclear pattern. We demonstrate that Tramtrack 69 (Ttk69), a repressor of neuronal differentiation, is a bona fide in vivo substrate for dSmt3 conjugation. Finally, we show that both the modified and unmodified forms of Ttk69 can bind to a Ttk69 binding site in vitro. Moreover, dSmt3 and Ttk69 proteins colocalize on polytene chromosomes, indicating that the dSmt3-conjugated Ttk69 species can bind at sites of Ttk69 action in vivo. Altogether, these data indicate a high conservation of the Smt3 conjugation pathway and further suggest that this mechanism may play a role in the transcriptional regulation of cell differentiation in Drosophila flies.
Ubiquitination is a well-known process of posttranslational modification of proteins. The covalent conjugation of the small protein ubiquitin can regulate the function and the stability of its target proteins (for a review, see reference 7). The reaction involves the formation of an isopeptide bond between the carboxyl-terminal glycine residue of ubiquitin and the ɛ-amino group of a lysine residue of an acceptor protein. This covalent attachment is carried out by a multistep pathway. Initially, ubiquitin is activated by the ATP-dependent formation of a high-energy thioester intermediate between the ubiquitin-activating enzyme (E1) and the C terminus of ubiquitin. Next, ubiquitin is transferred from the E1 to a cysteine residue of a ubiquitin-conjugating enzyme (E2) through transacetylation. Finally, ubiquitin is transferred from E2 to its target protein. This last transfer step may require the participation of an E3 ligase. All known functions of ubiquitin, including its role in selective protein degradation, are thought to be mediated by this pathway.
A number of proteins with homology to ubiquitin have been discovered in recent years. These ubiquitin-like proteins (Ubls) are thought to have some properties of ubiquitin, including the ability to be conjugated to other proteins. The reactions involving these variants appear to have much in common with those of ubiquitin, but the Ubls have novel regulatory functions not necessarily linked to proteolysis. One of these Ubls, SUMO-1 (also named Smt3C, UBL1, PIC1, GMP1, and sentrin), has been discovered in a number of independent studies (3, 28, 30, 32, 36, 43), and recently several groups have shown that SUMO-1 can be covalently conjugated to a variety of proteins in a manner analogous to that for ubiquitin.
The exact function of SUMO-1 conjugation is unknown. However, SUMO-1-modified proteins display altered subcellular targeting and/or stability (for reviews, see references 25 and 38). The IκBα inhibitor was recently reported to be modified by SUMO-1 at the same residue as the one used for ubiquitination, thus rendering the protein resistant to proteasomal degradation (8). SUMO-1 has been found to be covalently linked to RanGAP1, the activating protein of the RanGTPase involved in the regulation of nucleocytoplasmic trafficking. Conjugation of SUMO-1 to RanGAP1 targets the protein from its otherwise cytosolic localization to the nuclear pore complex (30, 32). In addition, SUMO-1 has been found to be attached to PML and Sp100, two proteins that localize to the so-called PML nuclear bodies (NBs) (also referred to as ND10 or PODs) (34, 44). The SUMO-1 modification of PML was shown to target the protein from the nucleoplasm to the NBs (34). A number of observations suggest that the NBs perform critical cellular functions. In particular, these nuclear structures are disrupted in a retinoic acid-reversible manner in the hematopoietic malignancy acute promyelocytic leukemia (10, 27, 47). Moreover, NBs are highly responsive to environmental stimuli such as heat shock and interferons and are the specific subnuclear targets for DNA tumor viral early gene products (reviewed in reference 41).
Analysis of the Saccharomyces cerevisiae SUMO-1 homologue, ScSmt3, indicates that SUMO-1 modification may play a role in meiosis and/or mitosis control. ScSmt3 was first isolated as a high-copy-number suppressor of a temperature-sensitive allele of MIF2, a gene encoding a centromere-binding protein (33). Strains in which ScSmt3 is deleted are lethal but can be rescued by SUMO-1, demonstrating the conservation of the pathway. In addition, like SUMO-1, ScSmt3 modifies multiple proteins in yeast. Analysis of temperature-sensitive mutants indicated that under nonpermissive conditions, cells arrest at the G2-M transition of the cell cycle (25). Accordingly, mammalian SUMO-1 has been shown to localize to the mitotic spindle apparatus in dividing cells (32). Several groups have identified the SUMO-1/ScSmt3 E2-specific enzyme as Ubc9 (9, 16, 23, 39). It was shown that Ubc9 could form a thioester bond with SUMO-1/ScSmt3 but not with ubiquitin, confirming the specificity of the pathway. In yeast, repression of Ubc9 synthesis prevents cell cycle progression at the G2 or early M phase, causing the accumulation of enlarged budded cells with a single nucleus, a short spindle, and replicated DNA (42).
To clarify further the role of Smt3 conjugation, we identified the Drosophila Smt3 and Ubc9 homologues (dSmt3 and dUbc9) and showed that dUbc9 is the functional analogue of E2 in the dSmt3 pathway. The dSmt3 protein, which can be conjugated to a number of cellular substrates, is in part localized in subnuclear foci, suggesting a conservation of NB-type structures in invertebrates. Finally we demonstrate that the zinc finger transcriptional repressor Tramtrack 69 (Ttk69) is a substrate for dSmt3 modification. Ttk69 and dSmt3 proteins colocalize at polytene chromosome sites in vivo, and the conjugated form of Ttk69 can bind Ttk69 DNA sites in vitro. These findings not only identify Ttk69 as the first Drosophila protein found to undergo this type of posttranslational modification but also provide a possible link between this process and the modulation of transcriptional regulation.
MATERIALS AND METHODS
cDNA isolation and characterization.
A Drosophila cDNA library (larva, third instar; American Type Culture Collection) was used as template DNA for PCR amplification with primers based on the sequences of the expressed sequence tags (ESTs) provided by the Berkeley Drosophila Genome Project (BDGP) (LD07716 for dSmt3; CK01148 and LD12093 for dUbc9; BDGP/HHMI EST Project, unpublished data). PCR products were subcloned and sequenced by standard procedures. For each PCR product, five independent clones were sequenced. The amino acid alignment was carried out with the PILEUP program.
For transfection studies in Drosophila SL2 cells and human HeLa cells, the indicated hemagglutinin epitope (HA)-tagged, His-tagged, or untagged cDNAs were cloned in pPACPL vector (a gift from N. Dostatni) or in pSG5 (Stratagene). Ttk69 cDNA was subcloned in pRmHa-3 vector (5). For bacterial expression, the cDNAs were cloned into pGEX-2TK (Pharmacia). Details concerning each construction are available upon request.
Northern analysis.
Drosophila flies were maintained at 25°C. Total RNA was isolated from material at 4-h intervals during embryogenesis and at 24-h stages of larval development and from pupae and adult flies, using Trizol as instructed by the manufacturer (Gibco BRL). Poly(A)+ RNAs were purified by using the PolyATract mRNA isolation system (Promega). Poly(A)+ RNA (5 μg) was separated on formaldehyde-agarose gel and transferred to a Hybond-N membrane. The blot was hybridized overnight with 32P-labeled dSmt3 or dUbc9 probes and washed at room temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) before autoradiography. The ribosomal protein gene 49 transcript was used as an RNA-loading control by reprobing the blot with the corresponding probe (35).
Antibodies.
The polyclonal anti-PML antibody (47) and the anti-PML monoclonal antibody (MAb) 5E10 (45) were described previously. MAb 12CA5 (Boehringer Mannheim) was used against the HA tag. Two rabbit polyclonal anti-dSmt3 antisera (78 and 80) were obtained following immunization of two female rabbits with purified glutathione S-transferase (GST)–dSmt3 emulsified with Freund's complete adjuvant. Two and four weeks later, animals were given a booster injection with the same amount of protein in Freund's incomplete adjuvant. Antisera were obtained 1 week after the last booster injection. Serum 78 was used for all experiments. Ttk88- and Ttk69-specific antibodies were raised against a GST-Ttk88 fusion containing the zinc fingers and C-terminal domain of Ttk88 and the R113 derivative of Ttk69 containing amino acids 286 to 641, respectively. Antibodies were raised in rabbits and rats as described previously (15). Animals were subjected to an initial inoculation with protein in Freund's complete adjuvant, followed, at 2-week intervals, by four inoculations with protein suspended in Freund's incomplete adjuvant. Two weeks after the last boost, animals were sacrificed and sera were collected. Either sera were stored at −20°C or sodium azide was added to 1 mM and sera were stored at 4°C. Prior to use, rabbit sera were further purified over protein A-Sepharose.
In situ hybridization and immunolocalization on embryos and pupae.
Control wild-type flies were w[1118] flies. The A101 line carries a P[lacZ, ry+] enhancer-trap allele of neuralised that specifically expresses nuclear β-galactosidase in pI and its progeny cells. In situ hybridization on w[1118] embryos was performed as previously described (40), using a digoxigenin labeled RNA probe synthesized from a full-length cDNA plasmid. Embryos and dissected nota from 24 h after puparium formation (APF) pupae were processed as described previously (14). Primary antibodies were rabbit anti-dSmt3 (1/300), rat anti-Ttk69 (1/100); mouse anti-cut (2B10; 1/500; Developmental Studies Hybridoma Bank [DSHB]) and rabbit-anti-β-galactosidase (1/500; Cappel). All secondary conjugated antibodies were from Jackson Laboratories (1/200). Images were obtained on a Leica TCS 4D confocal microscope or on a Leica DMLB microscope using a Micromax camera (Princeton Instruments). All images were processed with NIH Image and Photoshop programs.
Thioester assays.
GST fusion proteins (dSmt3GG and dUbc9) were expressed in Escherichia coli BL21 and affinity purified on glutathione-Sepharose (Pharmacia). dSmt3GG was radioactively labeled while bound to glutathione-Sepharose by using protein kinase A (Sigma) in the presence of [γ-32P]ATP. The radiolabeled fusion proteins and the GST-dUbc9 fusion protein were cleaved by thrombin to yield free dSmt3GG and dUbc9. Upon cleavage, thrombin was inactivated by incubation at 75°C for 15 min. SL2 cell extracts were prepared in 1% NP-40, 20 mM Tris-HCl (pH 8), 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.01% phenylmethylsulfonyl fluoride, and aprotinin (1 μg/ml). Reaction mixtures contained 10 μg of SL2 cell extracts, 300 ng of 32P-labeled dSmt3GG, and 300 ng of dUbc9 in 20 mM Tris-HCl (pH 7.6)–50 mM NaCl–4 mM ATP–10 mM MgCl2–0.2 mM DTT. After 5 min at 25°C, reactions were terminated by incubating the mixtures for 15 min at 30°C in 50 mM Tris-HCl (pH 7.6)–4 M urea–2% SDS–10% glycerol or by boiling the mixtures in the buffer above containing 100 mM DTT instead of urea. Reaction mixtures were separated by polyacrylamide gel electrophoresis (PAGE) on SDS–14% polyacrylamide gels, and radioactively labeled bands were visualized by autoradiography.
Preparation of cell extracts, immunoprecipitation, and Western blotting.
For direct Western blots, cells were washed twice in cold phosphate-buffered (PBS), scraped in SDS sample buffer, and then boiled for 10 min. For immunoprecipitations, cells were washed twice in cold PBS, lysed in SDS 1%–100 mM Tris HCl (pH 7.5), and boiled for 5 min. The lysates were sonicated briefly, diluted 1:10 in PBS, and cleared by centrifugation. Supernatants were precleared with 40 μl of protein A-Sepharose (Pharmacia) for 2 h at 4°C, centrifuged, and incubated for 3 h with the appropriate antibodies and 30 μl of protein A-Sepharose. The protein A-Sepharose beads were sedimented by a brief centrifugation and washed four times with ice-cold radioimmunoprecipitation assay buffer, and proteins were recovered by boiling in SDS sample buffer. Proteins from whole-cell extracts or immunoprecipitates were separated by SDS-PAGE and transferred to Hybond-C extra (Amersham) membranes. Membranes were blocked in 5% nonfat dry milk in PBS–0.05% Tween (PBS-Tw) and incubated for 2 h with the various antibodies diluted in PBS-Tw. MAb 12CA5 was used at a dilution of 1/2,500, anti-dSmt3 was used at 1/1,000, rabbit polyclonal anti-PML was used at 1/2,000, and rat polyclonal anti-Ttk69 was used at 1/2,000. After incubation with the primary antibody, blots were extensively washed in PBS-Tw and incubated for 1 h with the appropriate peroxidase-coupled secondary antibodies (Amersham). Enhanced chemiluminescence reagents (Amersham) were used for detection.
Nickel precipitation of PML and Ttk69.
PML or Ttk69 was coexpressed with untagged dSmt3, HA-tagged dSmt3 (dSmt3HA), or His-tagged dSmt3 (dSmt3His) in SL2 cells. Cell lysates from the transfected cells were prepared in the lysis buffer (6 M guanidinium HCl, 100 mM NaH2PO4, 10 mM Tris-HCl [pH 7.8]). After sonication, the lysates were incubated with nickel-charged agarose resin beads (Qiagen) overnight at room temperature. The beads were washed twice with washing buffer (pH 7.8) containing 8 M urea and then with a buffer (pH 6.3) containing 8 M urea. Finally, the beads were washed twice with PBS and treated with SDS sample buffer for SDS-PAGE. The proteins were analyzed by Western blotting using the rabbit anti-PML or rat anti-Ttk69 polyclonal antibody.
Cell cultures, transfections, and immunolocalization studies.
HeLa cells were grown at 37°C in 5% CO2 in Dulbecco's modified minimal medium (Gibco BRL), supplemented with antibiotics, glutamate, and 10% fetal calf serum. Drosophila SL2 cells were cultured at 23°C in Schneider medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. Transfections were performed by the calcium phosphate precipitation method using 5 μg of expression vector DNA; 24 h after transfection, the medium was replaced by fresh culture medium, and the cells were further incubated for 24 to 48 h. For immunofluorescence studies, cells were grown on round coverslips in six-well plates. Cells were fixed in 3.7% paraformaldehyde in PBS for 10 min at room temperature and then permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. After fixation and permeabilization, cells were rinsed twice in PBS and once in PBS-Tw, incubated with primary antibodies for 1 h, washed in PBS and PBS-Tw, and further incubated with the appropriate secondary antibodies conjugated with either fluorescein isothiocyanate (FITC; Sigma) or Texas red (Amersham). Primary and secondary antibodies were used at a dilution of 1:200 except for the anti-PML MAb 5E10 (1:2). After three washed in PBS, the samples were mounted in VectaShield (Vector Laboratories, Burlington, Calif.). Confocal laser scanning microscopy was performed with a Leica SM microscope, using excitation wavelengths of 488 nm (for FITC) and 543 nm (for Texas red). The two channels were recorded independently, and pseudocolor images were generated and superimposed. The acquired digital images were processed with Adobe Photoshop version 5.1 software.
Antibody staining of polytene chromosomes.
Protein distributions on polytene chromosomes were analyzed by the protocol of Andrew and Scott (1). Briefly, salivary glands were dissected from wandering third-instar larvae in 0.7% saline, fixed for 30 s in PBT (PBS–0.1% Tween 20) containing 3.7% formaldehyde, and then transferred to 45% acetic acid containing 3.7% formaldehyde and 0.1% Tween 20 for 3.5 min prior to spreading. Slides were blocked in three changes of blocking solution (PBT–10% fetal calf serum) followed by primary antibody incubation for 2 h at room temperature. Rabbit anti-dSmt3 and rat anti-Ttk69 antibodies were used at 1/200 dilution in blocking solution. Slides were washed three times in blocking solution prior to secondary antibody incubation for 30 min at room temperature. FITC-conjugated anti-rabbit and Cy3-conjugated anti-rat antibodies (Jackson Immunoresearch) were used at 1/200 dilution in blocking solution. Slides were washed in three changes of PBT and DNA counterstained by brief incubation in Hoechst 33258 (10 μg/ml; Sigma) prior to mounting in 90% glycerol containing phenylenediamine (1 mg/ml; Sigma). Preparations were viewed in a Zeiss Axiophot microscope, and images were captured with a cooled charge-coupled device camera (Photometrics). Images were analyzed with the IPlab program.
DNA affinity precipitation assays.
In vitro modification of Ttk69 was performed as described previously (8) except that reticulocyte lysates were used for in vitro translation of Ttk69. DNA affinity precipitation assays using biotinylated oligonucleotides were performed essentially as described by Franza et al. (13). Briefly, binding reactions were assembled using [35S]methionine-labeled Ttk69 protein modified in vitro, poly(dI-dC) competitor DNA at 40-fold excess (by weight) over the specific oligonucleotide, and an appropriate amount of buffer B (50 mM Tris-HCl [pH 8.0], 0.01% NP-40, 20% glycerol, 1.5 mM MgCl2, 50 mM KCl, 1 mM DTT 1 mM phenylmethylsulfonyl fluoride) to form a 60-μl volume. After 15 min at 37°C, 20 pmol of biotinylated oligonucleotide was added, and incubation was continued for 20 min. Streptavidin magnetic beads (Promega) were added, and the mixture was incubated for a further 20 min at room temperature. Reaction mixes were clarified by using magnetic stands, and the beads were washed three times in buffer B. Proteins were eluted from the beads by boiling in Laemmli sample buffer and analyzed by SDS-PAGE and autoradiography. The oligonucleotides used were FTZ (5′-[biotin]AACAGAAGCCAAGGACACAGGCGACGCGTG3′ and 5′-CACGCGTCGCCTGTGTCCTTGGCTTCTGTT-3′) and control (5′-[biotin]TCGACGTGACTCAGCGCGCATCGTGACTCAGCGCGC-3′ and 5′-TCGAGCGCGCGCTGAGTCACGATGCGCGCTGAGTCACG-3′). The FTZ sequence is a Ttk69 binding site located in the ftz proximal enhancer (BS6) (19), and the control sequence is a c-Jun binding site (twice-iterated sequence of the AP-1 binding site).
RESULTS
dSmt3 and dUbc9 are coexpressed during Drosophila development.
To gain a better understanding of the function of SUMO-1 modification, we wished to characterize the Smt3/Ubc9 pathway in Drosophila. Homology search of the Drosophila genome database revealed two ESTs encoding proteins with similarity to ScSmt3 and Ubc9. The corresponding cDNAs were isolated by PCR from a Drosophila cDNA library and sequenced. The Drosophila cDNA sequence homologous to SUMO-1/ScSmt3 (dSmt3) encodes a 90-amino-acid polypeptide that is 47 and 56% identical to human Smt3c/SUMO-1 and yeast ScSmt3, respectively. Figure 1A shows the amino acid alignment of dSmt3 and other members of the Smt3 family. The Drosophila homologue of Ubc9, dUbc9, is 85 and 53% identical to the mammalian and the yeast Ubc9 proteins, respectively (not shown), and was independently isolated in a yeast two-hybrid screen using the heat shock protein hsp23 as a bait protein (22).
FIG. 1.
Conservation of dSmt3 with Smt3 proteins from other organisms and expression analysis of dSmt3 and dUbc9. (A) Sequence alignment of dSmt3 and other members of the Smt3-related protein family. Residues identical in two or more of the aligned proteins are boxed in the upper panel. CelSmt3, Caenorhabditis elegans Smt3; hsSUMO1, hsSmt3A, and hsSmt3B, human SUMO-1, -2, and -3 (also named HsSmt3C, -A, and -B, respectively) (25). (B) Developmental Northern blot analysis of dSmt3 and dUbc9. Poly(A)+ RNAs (5 μg) prepared from various developmental stages were hybridized sequentially with 32P-labeled dSmt3 and dUbc9 probes. Numbers during embryogenesis refer to hours of development after fertilization: L1, L2, and L3, first-, second-, and third-instar larvae; Ad., adult. The lower panel shows the same Northern blot hybridized with a ribosomal protein gene 49 sequence.
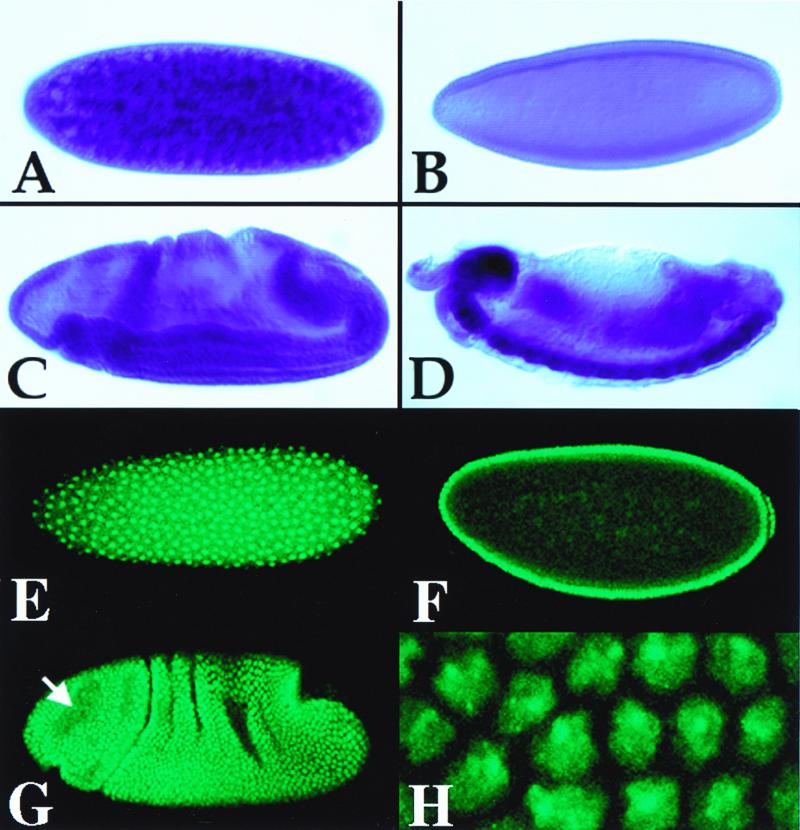
Northern blot analysis of dUbc9 and dSmt3 revealed that the mRNAs encoding the two proteins are expressed in similar patterns during Drosophila development (Fig. 1B). dSmt3 and dUbc9 transcripts were abundant in early embryos, after which levels declined throughout development. In adult females, however, dSmt3 and dUbc9 transcripts were again expressed, indicating a strong maternal contribution to the embryo. The developmental expression of dSmt3 was further examined both by in situ hybridization on whole-mount embryos and by indirect immunofluorescence using a polyclonal anti-dSmt3 antiserum (the specificity of this antibody is demonstrated in Fig. 4). Both dSmt3 mRNA and protein were detected in preblastoderm and blastoderm-stage embryos (Fig. 2A and E). No such labeling was observed after control incubations with the preimmune serum (data not shown). Prior to embryonic stage 11, a uniform distribution of dSmt3 mRNA and protein was seen (Fig. 2B, C, F, and G). Subsequently, a significantly higher level of dSmt3 transcripts was observed in the central nervous system (CNS) (Fig. 2D). In contrast, levels of dSmt3 protein uniformly declined throughout embryogenesis, and we found no evidence of CNS-specific accumulation of dSmt3 (not shown). The dSmt3 protein was predominantly nuclear, where it localized in dots (Fig. 2H; see also below). During mitosis, dSmt3 redistributed throughout the cell volume and did not appear to colocalize with mitotic spindles (Fig. 2G), unlike the reported behavior of SUMO-1 (32).
FIG. 4.
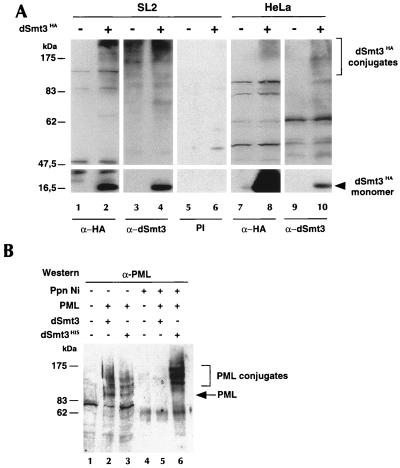
dSmt3-protein conjugates in Drosophila and human cells. (A) dSmt3 is conjugated to multiple proteins in SL2 and HeLa cells. dSmt3HA was transfected in SL2 or HeLa cells. Protein extracts from untransfected (−) and transfected (+) cells were subjected to Western blot analysis using anti-HA (α-HA) MAb, anti-dSmt3 (α-dSmt3) antiserum, or the corresponding preimmune serum (PI). Positions of the free dSmt3HA and its conjugates are indicated. (B) Modification of PML by dSmt3 in SL2 cells. dSmt3His or untagged dSmt3 was coexpressed with PML in SL2 cells. Extracts were precipitated by nickel-agarose beads (Ppn Ni). Crude extracts (1/10; lanes 1 to 3) and Ni-agarose precipitates (lanes 4 to 6) were analyzed by Western blotting using anti-PML antibodies. Positions of PML and its dSmt3-conjugated forms are indicated.
FIG. 2.
Localization of dSmt3 transcripts and protein in embryos. Lateral views of wild-type embryos hybridized with an anti-dSmt3 digoxigenin-labeled RNA probe (A to D) or incubated with anti-dSmt3 antibodies (E to H). A high level of maternal transcript and protein was seen in preblastoderm embryos (stage 2; A and E). The gross distribution of both the transcript and protein was uniform throughout the embryo both at the cellular blastoderm stage (stage 4; B and F) and in gastrulating embryos (stage 7; C and G). Analysis of the protein subcellular distribution revealed that dSmt3, which initially accumulated in the cytoplasm (not shown), rapidly partitioned to nuclei (E). Within the nucleus, dSmt3 localized to a small number of apically localized dots, clearly visible on a surface view of a stage 5 embryo (H). At the onset of mitosis, the dSmt3 protein redistributed throughout the cytoplasm and did not colocalize with mitotic spindles (the arrow in panel F indicates mitotic domain B). Preferential accumulation of dSmt3 transcripts in the CNS is shown in a late stage 15 embryo (D). Anterior pole is at left; dorsal is up. Stages were determined according to Campos-Ortega and Hartenstein (6).
dUbc9 is the conjugating enzyme for dSmt3.
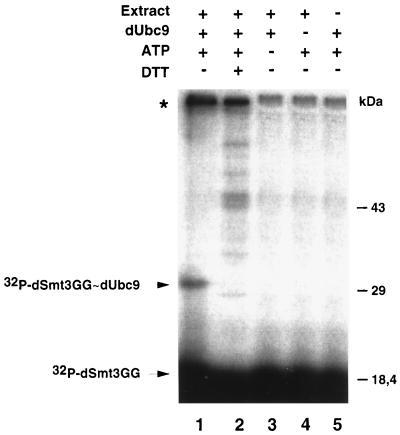
Ubc9 homologues in yeast and mammals were found to conjugate ScSmt3/SUMO-1 but not ubiquitin. This process is mediated through the formation of a thioester bond between the two proteins (9, 23, 39). To test whether dUbc9 could function as a dSmt3-conjugating enzyme, we performed a thioester formation assay. dUbc9 and an activated form of dSmt3 that lacks two amino acids from the C terminus (dSmt3GG) (24, 26, 31) were expressed as GST fusions in bacteria and purified. Incubation of dUbc9 with 32P-dSmt3GG, ATP, and a Drosophila SL2 extract promoted formation of a ∼30-kDa band (Fig. 3, lane 1). Generation of this product was ATP dependent and required Drosophila nuclear extracts, which presumably supply an E1 activity (Fig. 3, lanes 3 and 5). Moreover, this product was destroyed by incubation with DTT (Fig. 3, lane 2). These data are consistent with the ∼30-kDa product being the 32P-dSmt3GG-dUbc9 thioester complex. An additional high-molecular-weight band was also obtained in the presence of nuclear extracts in an ATP-dependent fashion (asterisk in lanes 1 and 2). This band was insensitive to DTT, suggesting that it corresponds to a protein covalently modified by 32P-dSmt3GG.
FIG. 3.
Thioester complex between dSmt3 and dUbc9. Thioester reactions contained 32P-labeled dSmt3GG, dUbc9, protein extracts from Drosophila SL2 cells, and ATP. After 5 min at 25°C, reactions were stopped in the absence or presence of a reducing agent and the products were subjected to SDS-PAGE followed by autoradiography. Positions of the free 32P-dSmt3GG and of the 32P-dSmt3GG-dUbc9 thioester complex are indicated. The band indicated by an asterisk probably represents a 32P-dSmt3GG-conjugated form of Drosophila protein present in the cell extract.
Formation of dSmt3 conjugates in Drosophila and human cells.
To further characterize dSmt3, a rabbit polyclonal antiserum was raised against recombinant, bacterially expressed dSmt3 protein. Western blotting confirmed the specificity of this antiserum. Lysates prepared from SL2 cells transfected with dSmt3HA were probed with either anti-HA antibodies, anti-dSmt3 antibodies, or the corresponding preimmune serum (Fig. 4A). A ∼15-kDa band corresponding to the dSmt3HA monomer was equally recognized by both the anti-HA and anti-dSmt3 antibodies in transfected cells (lanes 2 and 4). This band was not detected with the preimmune serum (lane 6). In addition to the ∼15-kDa band, several high-molecular-weight bands were detected by the anti-HA antibody in transfected extracts (compare lane 1 with lane 2). These bands also were revealed by the anti-dSmt3 antibody in both transfected and untransfected cells (lanes 3 and 4) and are likely conjugates between dSmt3 and cellular proteins. Interestingly, free dSmt3 was not detected in untransfected cells, suggesting that most endogenous dSmt3 is present as protein conjugates.
To determine whether dSmt3 could also conjugate proteins in human cells, dSmt3HA was expressed in HeLa cells, and cell extracts were analyzed by Western blotting using anti-HA or anti-dSmt3 antibodies. In addition to free dSmt3HA, a number of high-molecular-weight dSmt3HA conjugates could be detected with either of the two antibodies (Fig. 4A, lanes 8 and 10). A similar conjugation pattern was observed in SUMO-1HA-transfected HeLa cells (data not shown). These results strongly suggest that dSmt3 and SUMO-1 are able to modify common target proteins in human cells.
Reciprocally, we wished to examine whether the Drosophila dSmt3 conjugation system could recognize and modify human SUMO-1 substrates in Drosophila cells. To this end, the human PML protein was coexpressed in SL2 cells with untagged dSmt3 or dSmt3His. Cell extracts were incubated with nickel-charged agarose beads to recover the putative PML-dSmt3His conjugates. Total cell extracts and the pellet fractions were then compared by Western blotting using an anti-PML polyclonal antibody (Fig. 4B). Untransfected cells served as a negative control (lane 1). In crude extracts from cells cotransfected with PML and dSmt3His, several PML-reactive bands were detected (lane 3). The three upper ones were retained on Ni-agarose beads (lane 6), demonstrating that these bands correspond to dSmt3His-PML conjugates. As anticipated, the unmodified 100-kDa PML form was not recovered on the nickel-agarose beads. When dSmt3His was replaced by dSmt3, PML conjugates were still formed (lane 2) but, as expected, could not be retained on the beads (lane 5), thus confirming the specificity of the binding of dSmt3His. Collectively, the results shown in Fig. 4 indicate that dSmt3 can be processed and conjugated in human cells and that PML can be modified by dSmt3 in Drosophila cells. They suggest the existence of an evolutionarily conserved pathway of protein modification.
Subcellular localization of dSmt3 and dUbc9.
Recently it was shown that in mammalian cells, SUMO-1 in part localizes to NBs and that conjugation to PML is involved in targeting this protein to NBs (34). This prompted us to examine the subcellular distribution of dSmt3 and dUbc9 in Drosophila cells. A dUbc9HA expression vector was transfected into SL2 cells, and immunofluorescence studies were performed with anti-HA and anti-dSmt3 antibodies. The ectopically expressed dUbc9HA protein was found to be predominantly localized in the nucleus. While the dUbc9HA intranuclear signal was largely homogeneous, in a significant fraction of cells, the protein was also concentrated in one to five nuclear foci (Fig. 5A). In some cells, a rim staining of the nuclear envelope occasionally could be observed (data not shown). When stained for endogenous dSmt3, most cells exhibited a diffuse nuclear signal over which was overlaid a punctate nuclear labeling (Fig. 5B). As a control, staining of the SL2 cells with the corresponding preimmune serum did not reveal any staining. Superimposition of the dUbc9HA and dSmt3 signals demonstrated a partial colocalization of the two proteins particularly in the nuclear foci (Fig. 5C). A similar speckled nuclear distribution pattern was observed in embryos. Indeed, at the cellular blastoderm stage, dSmt3-containing foci were found at the apical nuclear pole (Fig. 2H).
FIG. 5.
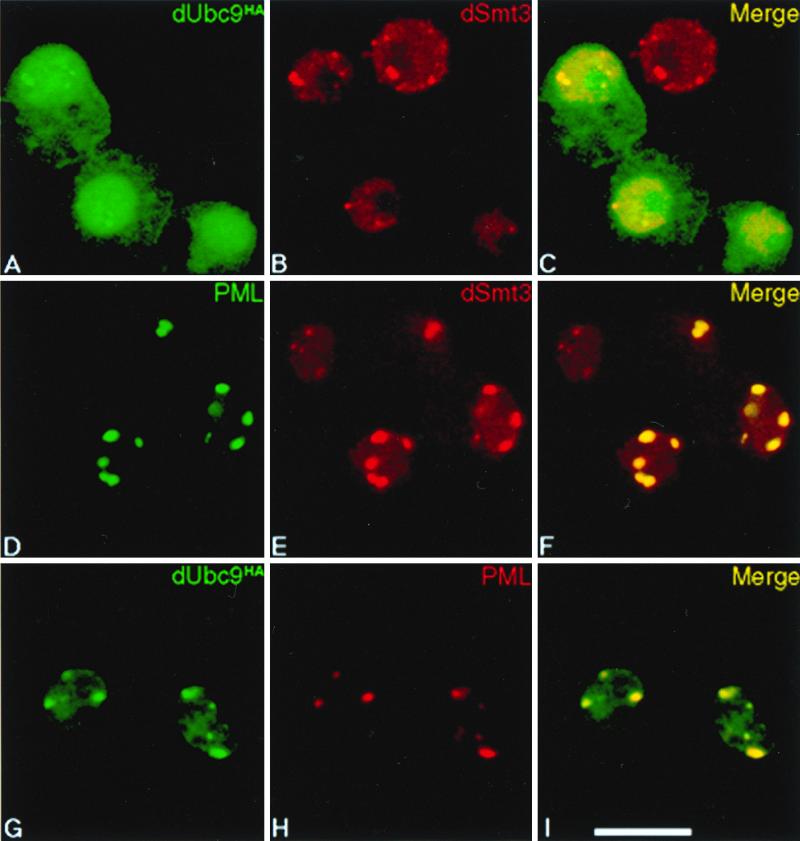
dSmt3 concentrates in nuclear speckles. SL2 cells were transfected with plasmid constructs expressing dUbc9HA (A to C), PML (D to F), or PML together with dUbc9HA (G to I). Cells overexpressing dUbc9HA were revealed with an anti-HA MAb (A and G). PML was revealed either with the anti-PML MAb 5E10 (D) or a rabbit anti-PML antiserum (H). Endogenous dSmt3 was visualized with a rabbit polyclonal anti-dSmt3 antiserum (B and E). The secondary antibodies used were conjugated to FITC (left, green) or to Texas red (center, red). Confocal overlay of red and green panels yields yellow in the right panels (C, F, and I). (Scale bar = 10 μm).
When the human PML protein was expressed in SL2 cells, it adopted a punctate nuclear distribution pattern (Fig. 5D) indistinguishable from that observed in human cells (not shown). Concomitantly, the diffuse nuclear dSmt3 signal became much weaker and an intense punctate pattern became apparent in most cells (Fig. 5E). A clear overlap between PML and dSmt3 was noted in these enlarged foci (Fig. 5F). Similarly, cotransfection of dUbc9HA and PML in SL2 cells led to almost complete recruitment of dUbc9HA to the PML-containing foci (Fig. 5G to I).
Ttk69 is modified by conjugation to dSmt3.
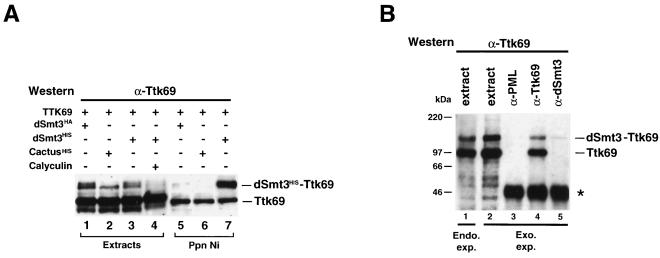
The dUbc9 protein has been recently shown to interact in a yeast two-hybrid screen with Seven in Abstentia (Sina), a RING finger protein which selects proteins for degradation through ubiquitination (20, 21). In addition, Sina can form a complex with at least one of the isoforms, Ttk88, of the transcriptional repressor Ttk, thereby promoting Ttk degradation (29, 46). We therefore hypothesized that Sina could target dUbc9 to Ttk, leading to the covalent modification of the latter by dSmt3. To test this possibility, SL2 cells were cotransfected with a vector expressing either the Ttk88 or the Ttk69 isoform of Ttk together with a vector expressing the dSmt3His protein. Extracts from cells transfected either with an unrelated His-tagged protein (CactusHis) or with dSmt3HA served as negative controls. Both the Ni-agarose precipitates and the unprecipitated extracts were analyzed by Western blotting using an appropriate anti-Ttk polyclonal antibody. We could not detect any modification of Ttk88 (data not shown). In contrast, the anti-Ttk69 antibody revealed two Ttk69-positive bands in crude extracts: the major Ttk69 isoform migrating with an apparent molecular mass of ∼100 kDa and an additional, larger species with an apparent mass of ∼120 kDa (Fig. 6A, lanes 1 to 3). (In cells cotransfected with dSmt3HA or dSmt3His, this larger form appeared as a doublet [lanes 1 and 3].) The 120-kDa species was retained on Ni-agarose beads in extracts from Ttk69- and dSmt3His-cotransfected cells (lane 7), demonstrating that this band corresponds to a Ttk69 protein covalently attached to dSmt3His. When dSmt3HA (lane 5) or cactusHis (lane 6) was substituted for dSmt3His, although a systematic residual binding of the unmodified 100-kDa Ttk69 species was visible, the 120-kDa conjugated form was not retained on the beads, thus confirming the specificity of the Ttk69-dSmt3His conjugate. Treatment of the cells with calyculin A (a potent inhibitor of serine/threonine phosphatases) abrogated the formation of the 120-kDa conjugate (lane 4), indicating that as had been described for the modification of PML and IκBα (8, 34), hyperphosphorylation prevents the attachment of dSmt3 to Ttk69. Interestingly, in the presence of calyculin A, the non-dSmt3-modified Ttk69 species migrated more slowly, suggesting that it had become hyperphosphorylated.
FIG. 6.
The Ttk69 transcriptional repressor is covalently modified by dSmt3. (A) Extracts from SL2 cells cotransfected with a vector expressing the Ttk69 protein with a vector expressing dSmt3HA (lanes 1 and 5), CactusHis (lanes 2 and 6), or dSmt3His (lanes 3, 4, and 7) were subjected to precipitation with Ni-agarose beads, and the precipitates were analyzed by Western blotting with the rat anti-Ttk69 polyclonal antibody. Aliquots of the corresponding unprecipitated extracts (1/10) were loaded in lanes 1 to 4. In lane 4, the cells had been incubated with 1.25 μM calyculin A prior to protein extraction. The 120-kDa doublet observed in crude extracts (lanes 1 and 3) corresponds to the Ttk69 protein conjugated either to the endogenous dSmt3 protein (lower band of the doublet) or to the transfected HA- or His-tagged dSmt3 product (upper band of the doublet). (B) SDS lysates from untransfected SL2 cells (lane 1) or from SL2 cells cotransfected with dSmt3 and Ttk69 were immunoprecipitated with the rabbit polyclonal anti-PML antibody (lane 3), the rabbit anti-Ttk69 antibody (lane 4), or the rabbit anti-dSmt3 antibody (lane 5). An aliquot of the transfected cell extract (1/200 of the material used for immunoprecipitation) was loaded in lane 2. Immunoprecipitates and cell extracts were fractionated by electrophoresis and analyzed by Western blotting with a rat anti-Ttk69 antibody. Immunoglobulins are marked by an asterisk. The 100-kDa unmodified and 120-kDa dSmt3-modified forms of Ttk69 are indicated.
Coimmunoprecipitation studies confirmed the conjugation of dSmt3 to Ttk69. Extracts from SL2 cells cotransfected with Ttk69 and dSmt3 were immunoprecipitated with rabbit polyclonal antibodies directed against the Ttk69, dSmt3, or PML protein and analyzed by Western blotting using a rat anti-Ttk69 polyclonal antiserum (Fig. 6B). In anti-dSmt3 precipitates, we observed a Ttk69-immunoreactive band of 120 kDa (lane 5) which comigrates with the modified form of Ttk69 present in both the crude extracts (lane 2) and the anti-Ttk69 precipitates (lane 4). In contrast, the major, unmodified, 100-kDa Ttk69 form was not precipitated by the dSmt3 antiserum (compare lane 5 with lanes 2 and 4). Under identical conditions, no Ttk69-reactive species were immunoprecipitated with the unrelated anti-PML antibodies (lane 3). Substantial amounts of dSmt3-Ttk69 conjugates were also detected in nontransfected SL2 cell extracts after direct SDS lysis (lane 1), confirming that both endogenous and ectopically expressed Ttk69 protein are modified by dSmt3.
Expression of Ttk69 and dSmt3 in sense organ cells.
Ttk has been shown to act as a repressor of neuronal fate determination in the Drosophila peripheral nervous system (PNS) (17, 18). In addition to antagonizing neurogenesis, it has been suggested that Ttk69 may also facilitate nonneuronal cell type specification (37). We thus were interested in comparing the expression profiles of Ttk69 and dSmt3 in neuronal and nonneuronal cells. To this aim, we performed immunofluorescence analysis of cells of the external sensory bristles as they represent an amenable model for analyzing the mechanisms of cell fate determination in the PNS (reviewed in reference 2). At 24 hs APF, i.e., soon after precursor cell divisions, a rat-specific anti-Ttk69 antiserum detected a high level of Ttk69 expression in all three nonneuronal cells (sheath, shaft, and socket cells) of the bristle but not in the differentiating neuron (Fig. 7A to C; see also reference 37). Immunofluorescence analysis using the rabbit polyclonal anti-dSmt3 antiserum revealed that dSmt3 was also expressed at a higher level in cells of the sensory organ lineage than in the surrounding epidermal cells. In contrast to Ttk69, however, dSmt3 was observed in both the sensory neuron and the three associated support cells (Fig. 7D to F). As had been shown in embryos (Fig. 2) and SL2 cells (Fig. 5), dSmt3 was found predominantly in nuclear dots in the surrounding epidermal cells (Fig. 7E). The specific accumulation of dSmt3 in sense organ cells is consistent with a role of dSmt3 in cell fate determination in the PNS. The expression of dSmt3 in the Ttk69 negative neuronal cells suggests that dSmt3 probably not only modifies Ttk69 but also may target other protein substrates presumably involved in neuronal differentiation.
FIG. 7.
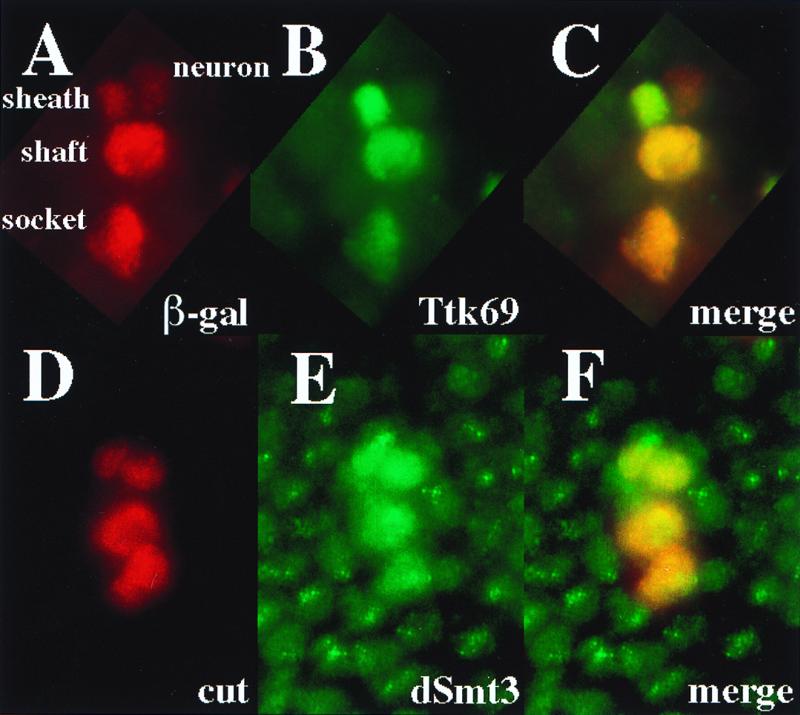
Distribution of dSmt3 and Ttk69 expression in sensory bristles. (A to C) Specific accumulation of Ttk69 (green; B) in sense organ cells in A101 pupae at 24 h APF. Sense organ cells were identified based on lacZ expression in the enhancer-trap line A101 (red; A). (D to F) Distribution of dSmt3 (green; E) in sense organ cells in pupal nota at 24 h APF. Sense organ cells were identified based on cut expression (red; D).
Colocalization of Ttk69 and dSmt3 at chromosomal sites.
In an effort to determine in vivo consequences of dSmt3 modification of Ttk69, the distribution of Ttk69 and dSmt3 was determined by double-label immunofluorescence microscopy on third-instar polytene chromosomes. Ttk69 protein had been previously shown to be highly expressed in third-instar salivary glands (4). Antibody staining of polytene chromosomes revealed that Ttk69 bound to a large number of euchromatic sites (Fig. 8A), an observation consistent with the pleiotropic nature of Ttk overexpression and loss-of-function phenotypes as well as with the developmentally complex Ttk expression profile (2, 4, 15, 18). Similarly, many euchromatic sites were stained with antibody against dSmt3 (Fig. 8C). No such staining was obtained with the corresponding preimmune serum (not shown). A merge of the Ttk69 and dSmt3 staining patterns revealed partial overlap between the Ttk69 and dSmt3 distributions (Fig. 8B). Most sites of Ttk69 accumulation also reacted with dSmt3 antibodies (Fig. 8, arrows), suggesting that Ttk69 bound at these sites was modified by dSmt3. However, a number of Ttk69-reactive sites did not show appreciable accumulation of dSmt3 (Fig. 8A and B, white arrowheads), indicating that unmodified Ttk69 was bound. Reciprocally, dSmt3 was found to be associated with a number of sites in the genome, including the chromocenter, which show no concentration of Ttk69 (Fig. 8B and C, black arrowheads), suggesting the existence of additional DNA-bound protein substrates for dSmt3 in Drosophila. The effects of the modification by dSmt3 on centromere structure and components remain to be clarified.
FIG. 8.
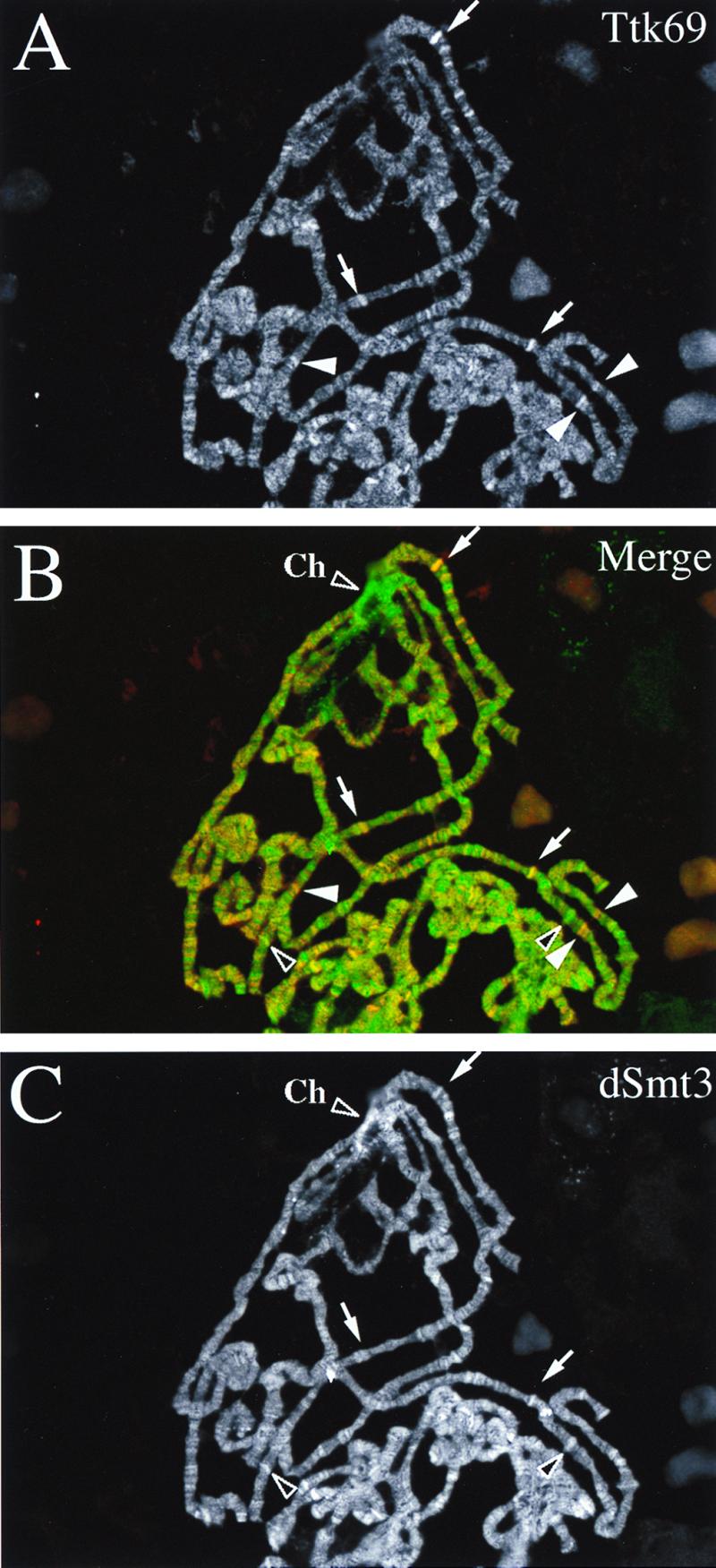
Distribution of Ttk69 and dSmt3 on polytene chromosomes. Salivary gland polytene chromosomes from wild-type third-instar larvae were simultaneously stained with antibodies against Ttk69 visualized with cyanin (A) and dSmt3 visualized with fluorescein (C). Both antibodies reacted against a large number of euchromatic sites on polytene chromosomes. (B) The merged image reveals that while most sites of Ttk69 accumulation (red staining) also were stained with the dSmt3 antibodies (green staining) (arrows), a number of Ttk69 sites did not show appreciable accumulation of dSmt3 (white arrowheads), and a number of dSmt3 sites (including the chromocenter [Ch]) were not stained with the Ttk69 antibodies (black arrowheads).
Binding of both unmodified and modified forms of Ttk69 on DNA sites in vitro.
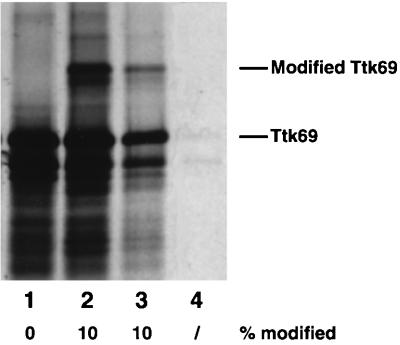
After having established that dSmt3 and Ttk69 colocalize on polytene chromosomes, we wished to see whether Ttk69-conjugated proteins could bind to Ttk69 DNA sites in vitro. We first used a system described by Desterro et al. (8) to reconstitute the covalent modification of Ttk69. [35S]Met-labeled Ttk69 generated by in vitro translation (Fig. 9, lane 1) was incubated with an assay mix containing recombinant Ubc9, SUMO-1, and a fraction of HeLa cells containing E1 activity, leading to the appearance of an upper band corresponding to the Ttk69-conjugated protein (Fig. 9, lane 2). To investigate the DNA-binding properties of the modified Ttk69 protein, we performed a DNA affinity precipitation assay (13) using either an oligonucleotide containing a Ttk69 binding site present in the ftz proximal enhancer (19) or an unrelated oligonucleotide containing a c-Jun binding site. The mixture of conjugated and unconjugated Ttk69 proteins was incubated with each oligonucleotide in the presence of nonspecific competitor poly(dI-dC). Specific DNA-protein complexes were recovered with streptavidin magnetic beads and analyzed by SDS-PAGE. Conjugated and unconjugated Ttk69 proteins were equally recovered with the oligonucleotide containing the Ttk69 binding site (lane 3). In contrast, the unrelated oligonucleotide did not precipitate the Ttk69 proteins (lane 4). Thus, it seems unlikely that posttranslational modification by dSmt3 influences the DNA-binding capacity of Ttk69.
FIG. 9.
Both modified and unmodified Ttk69 proteins bind to DNA in vitro. [35S]Met-labeled Ttk69 generated by in vitro translation (lane 1) was incubated with a reaction mix leading to the covalent modification of Ttk69 (lane 2). This reaction product was incubated with a biotinylated oligonucleotide containing a Ttk69 binding site (lane 3) or with an unrelated oligonucleotide (lane 4). The DNA-protein complexes were recovered by using streptavidin magnetic beads, separated on an SDS-polyacrylamide gel, and revealed by autoradiography. The percentage of modified versus unmodified Ttk69 is shown at the bottom.
DISCUSSION
The two principal conclusions from this study are as follows. First, the SUMO-1/Ubc9 protein modification pathway, previously characterized in humans and yeast, is functionally conserved in Drosophila. This result shows that this novel pathway is conserved from lower to higher eukaryotes. Second, we show that the transcriptional repressor Ttk69 is covalently modified by the Drosophila SUMO-1 homologue. These data suggest that the modification by dSmt3 may interfere with transcriptional mechanisms involved in sense organ development.
The SUMO-1/Ubc9 pathway is conserved in Drosophila.
In the present work, we have identified and characterized the Drosophila homologues, dSmt3 and dUbc9, of the mammalian SUMO-1 modifier and its conjugating enzyme Ubc9, respectively. Both the structure and the function of these proteins appear to be highly conserved relative to their counterparts in yeast and humans. We observe that in a manner analogous to that for the ScSmt3 protein in yeast cells and to the SUMO-1 protein in human cells, dSmt3 in Drosophila cells is conjugated to multiple proteins. In addition, we show by immunofluorescence studies that dSmt3 localizes in SL2 cells in a pattern similar to that of SUMO-1 in human cells. In particular, in SL2 cells, dSmt3 and dUbc9 are localized in punctate nuclear foci as well as in the diffuse nuclear fraction of the nucleoplasm. The former pattern is very similar to the localization of SUMO-1-modified PML and Sp100 in NBs in mammalian cells (3, 34, 44). Moreover, when expressed in mammalian cells, dSmt3 is targeted to the NBs (data not shown) and conversely human PML expressed in SL2 cells concentrates in subnuclear foci. Although we cannot exclude that PML, by self-aggregating, may form nuclear structures without participation of Drosophila proteins, one could also hypothesize that NB-like structures exist in Drosophila cells and that these specialized assemblies perform an evolutionarily conserved function.
We have also characterized the dSmt3-conjugating enzyme as dUbc9, a protein that shows high homology to the yeast and human Ubc9. The Drosophila protein appears functionally equivalent to its mammalian and yeast counterparts (9, 23, 39). Indeed, it was found to form a thioester bond-containing covalent intermediate with dSmt3 (Fig. 3) but not with ubiquitin (not shown), indicating that dUbc9 is the functional analog of E2s in the dSmt3 pathway.
Protein targets of the dSmt3/dUbc9 pathway.
Rather targeting proteins for degradation, the dSmt3/SUMO-1 modifications target some of their protein conjugates to nuclear macromolecular complexes including NBs for PML (34) and the nuclear pore complex for RanGAP1 (30, 32). Support for a targeting role in Drosophila is provided by the phenotype of semushi, a lethal mutant of dUbc9. In this mutant, the nuclear import of bicoid is blocked during early embryogenesis, resulting in a misregulation of the segmentation genes that are bicoid targets (11). A second lethal mutation, lesswright, is a dominant suppressor of the female meiotic mutation noddtw (S. Apionishev and R. S. Rasooly, submitted for publication), but in this case, the molecular basis for the phenotype has not been established. In other organisms, Ubc9 has been shown to interact with numerous proteins in two-hybrid assays (38), suggesting that a significant number of proteins can be modified by this pathway. Our results in Drosophila cells that show a large number of dSmt3-reactive bands in a Western blot support this view.
Interestingly, we were unable to detect the presence of any dSmt3 monomers in extracts of untransfected cells by using anti-dSmt3 antibodies, suggesting either that dSmt3 itself is limiting in Drosophila or that its level and conjugation are tightly regulated. Consistent with this view, transfection of dSmt3 does not lead to an augmentation of the number and the quantity of its modified products but leads to an accumulation of dSmt3 monomers. However, exogenously expressed dSmt3 protein does increase the proportion of modified Ttk69 protein present. Taken together, these data suggest that the dSmt3/dUbc9 modification pathway is tightly regulated in Drosophila. It is likely that the final levels of dSmt3 modification reflect a balance between substrate availability and associated targeting/regulatory cofactors. Undoubtedly, one component of such a complex is the RING finger protein Sina. Sina has been shown to interact with dUbc9 via N-terminal sequences. In addition, Sina can bind to itself and a number of target proteins through a C-terminal domain (20, 21). In this way, Sina could recruit dUbc9 to dSmt3 substrates. However, Sina has also been implicated in ubiquitin-dependent proteolysis (21, 29, 46). Consistent with this, Sina interacts with the ubiquitin-conjugating enzyme UbcD1 in a yeast two-hybrid assay (21). It is possible that the interactions of Ubc9 and UbcD1 with Sina are competitive. Alternatively, one may hypothesize that the interaction of Sina with dUbc9 targets dUbc9 for proteolytic degradation. Understanding how these different interactions are regulated could provide insight into the differential targeting of proteins either for proteolysis or to specialized nuclear structures. In this regard, our analysis of the posttranslational modification of isoforms of the transcriptional repressor Ttk could be informative.
The pattern of dSmt3 mRNA and protein expression during oogenesis and early embryonic development and the restriction of mRNA to the CNS in the later stages are reminiscent of housekeeping proteins whose expression correlates with the embryonic mitotic cycles. While the relatively high levels of expression in the external sensory lineage would also be consistent with this view, they could also be indicative of a particular requirement for the protein during sense organ differentiation. Indeed, the modification of Ttk69, a known repressor of neuronal differentiation, shows that the dSmt3/dUbc9 pathway could be directly involved in the determination or stabilization of a differentiated state.
The transcriptional repressor Ttk69 is a target for dSmt3 modification.
The identification of the transcriptional repressor Ttk69 as a substrate of the dSmt3 conjugation pathway suggests that this mode of posttranslational modification may play a direct role in the modulation of transcriptional regulation. Supporting this possibility, the localization of dSmt3 at particular chromosomal sites shows that the dSmt3 modification can be chromosome associated. Its partial colocalization with Ttk69 and the ability of the dSmt3-modified Ttk69 protein to bind Ttk69 sites are also consistent with the binding of modified Ttk69 to a subset of Ttk69 recognition elements. Although Ttk69 is the first transcription factor shown to be modified by the SUMO-1/Smt3 homologues, it seems likely that SUMO-1 also modifies several transcription factors in mammalian cells, as suggested by the observed interaction in a two-hybrid assay of Ubc9 with E1A, IκBα, WT1, Jun, p53, ATF2, ETS-1, the glucocorticoid receptor, and other nuclear proteins (reference 38 and references therein) and thus may perform a more general role in transcriptional regulation. Our data also indicate that the pattern of covalent modification of Ttk69 may be more complex. In particular, we note that Ttk69 can be phosphorylated as well as conjugated with dSmt3. Notably, general inhibition of serine/threonine phosphorylation prevents dSmt3 conjugation (8, 34), although it is uncertain whether this is a consequence of a reduction in substrate availability or conjugating activity.
The biological role and consequences of the conjugation of dSmt3 to Ttk69 are unclear. Among several possibilities would be effects on the targeting of the repressor to specific chromosomal sites or on its interaction with specific protein partners. Another attractive hypothesis is that dSmt3 modification might antagonize the degradation of Ttk69 by a proteasome-dependent pathway. Indeed, it has recently been suggested that in human cells, SUMO-1 modification of IκBα might serve to block signal-induced ubiquitination and thus degradation of IκBα (8). In this context it is intriguing that Sina interacts directly with and destabilizes the other isoform of Ttk, Ttk88 (29, 46), but that no comparable interaction of Sina and Ttk69 is observed in a two-hybrid assay (46). Nevertheless, Ttk69 levels are stabilized in SL2 cells by MG132, an inhibitor of proteasome-mediated proteolysis (F. Lehembre and A. Dejean, unpublished observations). We therefore suggest that dSmt3 modification might provide a mechanism for the differential stabilization of splicing isoforms, such as Ttk69 and Ttk88, that are transcribed from the same promoter. Genetic analysis of dSmt3 mutants in Drosophila should hopefully lead to a better understanding of the role of dSmt3 modification in the transcriptional regulation of sense organ development.
ACKNOWLEDGMENTS
We greatly acknowledge Rebekah Rasooly for helpful discussions. We are indebted to Amy Tang, Nathalie Dostatni, Ruth Steward, and Roel van Driel for the generous gift of antibodies and expression vectors used in these experiments. We are grateful to Veronique Brodu for providing the SL2 cell line. We thank Emmanuelle Perret for excellent help with confocal microscopy. We thank Pierre Tiollais for support and all members of our groups for stimulating discussions and for providing reagents.
This work was supported by grants from the CNRS (ATIPE), the Association pour la Recherche contre le Cancer, and the European Economic Community (Biomed 2). F.L. was supported by a fellowship from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche. S.M. was supported by a fellowship from the Association for International Cancer Research. P.B. acknowledges the support of the Emanual Bradlow Foundation.
REFERENCES
- 1.Andrew D J, Scott M P. Immunological methods for mapping protein distributions on polytene chromosomes. Methods Cell Biol. 1994;44:353–370. doi: 10.1016/s0091-679x(08)60923-1. [DOI] [PubMed] [Google Scholar]
- 2.Badenhorst P, Harrison S, Travers A. End of the line? Tramtrack and cell fate determination in Drosophila. Genes Cells. 1996;1:707–716. doi: 10.1111/j.1365-2443.1996.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 3.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 4.Brown J L, Wu C. Repression of Drosophila pair-rule segmentation genes by ectopic expression of tramtrack. Development. 1993;117:45–58. doi: 10.1242/dev.117.1.45. [DOI] [PubMed] [Google Scholar]
- 5.Bunch T A, Grinblat Y, Goldstein L S B. Characterization of an endogenous metallothionein gene in cultured Drosophila melanogaster cells and the potentials of using its inducible promoter. Nucleic Acids Res. 1988;16:1043–1059. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos-Ortega J A, Hartenstein V. The embryonic development of Drosophila melanogaster. Berlin, Germany: Springer-Verlag; 1985. [Google Scholar]
- 7.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 8.Desterro J M, Rodriguez M S, Hay R T. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 9.Desterro J M, Thomson J, Hay R T. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 10.Dyck J A, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 11.Epps J L, Tanda S. The drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr Biol. 1998;8:1277–1280. doi: 10.1016/s0960-9822(07)00538-6. [DOI] [PubMed] [Google Scholar]
- 12.Fairall L, Harrison S D, Travers A A, Rhodes D. Sequence-specific DNA binding by a two zinc-finger peptide from the Drosophila melanogaster Tramtrack protein. J Mol Biol. 1992;226:349–366. doi: 10.1016/0022-2836(92)90952-g. [DOI] [PubMed] [Google Scholar]
- 13.Franza B R, Josephs S F, Gilman M Z, Ryan W, Clarkson B. Characterization of cellular proteins recognizing the HIV enhancer using a microscale DNA-affinity precipitation assay. Nature. 1987;330:391–395. doi: 10.1038/330391a0. [DOI] [PubMed] [Google Scholar]
- 14.Gho M, Lecourtois M, Geraud G, Posakony J W, Schweisguth F. Subcellular localisation of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development. 1996;122:1673–1682. doi: 10.1242/dev.122.6.1673. [DOI] [PubMed] [Google Scholar]
- 15.Giesen K, Hummel T, Stollewerk A, Harrison S, Travers A, Klambt C. Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronaldifferentiation genes. Development. 1997;124:2307–2316. doi: 10.1242/dev.124.12.2307. [DOI] [PubMed] [Google Scholar]
- 16.Gong L, Kamitani T, Fujise K, Caskey L S, Yeh E T. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. FEBS Lett. 1997;417:297–300. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 17.Guo M, Bier E, Jan L Y, Jan Y N. tramtrack acts downstream of numb to specify distinct daughter cell. Neuron. 1995;14:913–925. doi: 10.1016/0896-6273(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 18.Guo M, Jan L Y, Jan Y N. Control of daughter cell fates during asymmetric division: interaction. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 19.Han W, Yu Y, Altan N, Pick L. Multiple proteins interact with the fushi tarazu proximal enhancer. Mol Cell Biol. 1993;13:5549–5559. doi: 10.1128/mcb.13.9.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu G, Fearon E R. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu G, Zhang S, Vidal M, Baer J L, Xu T, Fearon E R. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joanisse D R, Inaguma Y, Tanguay R M. Cloning and developmental expression of a nuclear ubiquitin-conjugating enzyme (DmUbc9) that interacts with small heat shock proteins in Drosophila melanogaster. Biochem Biophys Res Commun. 1998;244:102–9. doi: 10.1006/bbrc.1998.8214. [DOI] [PubMed] [Google Scholar]
- 23.Johnson E S, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 24.Johnson E S, Schwienhorst I, Dohmen R J, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson P R, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 26.Kamitani T, Nguyen H P, Yeh E T. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- 27.Koken M H M, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, De Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, De Thé H. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapenta V, Chiurazzi P, van der Spek P, Pizzuti A, Hanaoka F, Brahe C. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40:362–366. doi: 10.1006/geno.1996.4556. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Li Y, Carthew R W, Lai Z C. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 30.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 31.Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meluh P B, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell P, Rosbash M. Sequence, structure, and codon preference of the drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei C F, Chang H M, Yeh E T. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 37.Ramaekers G, Usui K, Usui-Ishihara A, Ramaekers A, Ledent V, Ghysen A, Dambly-Chaudiere C. Lineage and fate in Drosophila: role of the gene tramtrack in sense organ development. Dev Genes Evol. 1997;207:97–106. doi: 10.1007/s004270050096. [DOI] [PubMed] [Google Scholar]
- 38.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 39.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proteins Suppl. 1997;1:43–49. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweisguth F, Posakony J W. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 41.Seeler J S, Dejean A. The PML nuclear bodies: actors or extras? Curr Opin Gen Dev. 1999;9:362–367. doi: 10.1016/s0959-437x(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 42.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 43.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 44.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuurman N, De Graaf A, Floore A, Josso A, Humbel B, De Jong L, Van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 46.Tang A H, Neufeld T P, Kwan E, Rubin G M. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 47.Weis K, Rambaud S, Lavau C, Jansen J, Carvahlo T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]